Abstract
DNA methylation is globally reprogrammed after fertilization, and as a result, the parental genomes have similar DNA-methylation profiles after implantation except at the germline differentially methylated regions (gDMRs). We and others have previously shown that human blastocysts might contain thousands of transient maternally methylated gDMRs (transient mDMRs), whose maternal methylation is lost in embryonic tissues after implantation. In this study, we performed genome-wide allelic DNA methylation analyses of purified trophoblast cells from human placentas and, surprisingly, found that more than one-quarter of the transient-in-embryo mDMRs maintained their maternally biased DNA methylation. RNA-sequencing-based allelic expression analyses revealed that some of the placenta-specific mDMRs were associated with expression of imprinted genes (e.g., TIGAR, SLC4A7, PROSER2-AS1, and KLHDC10), and three imprinted gene clusters were identified. This approach also identified some X-linked gDMRs. Comparisons of the data with those from other mammals revealed that genomic imprinting in the placenta is highly variable. These findings highlight the incomplete erasure of germline DNA methylation in the human placenta; understanding this erasure is important for understanding normal placental development and the pathogenesis of developmental disorders with imprinting effects.
Keywords: DNA methylation, germline differentially methylated region, human placenta, genomic imprinting, X-chromosome inactivation, whole-genome bisulfite sequencing, RNA sequencing
Introduction
Mammalian gametes have unique DNA-methylation profiles, different from those of somatic cells. The DNA-methylation patterns acquired during gametogenesis are predominantly erased by demethylation after fertilization, followed by de novo methylation at implantation.1, 2, 3 Consequently, the parental genomes have similar DNA-methylation profiles after implantation except at the germline differentially methylated regions (gDMRs). gDMRs are characterized by parent-of-origin-dependent, allele-specific DNA methylation initiated in the gametes and maintained after fertilization. Canonical gDMRs regulate the strict parent-of-origin-dependent, allele-specific expression of imprinted genes in cis.4, 5 Currently, about 80 and 25 gDMRs have been identified in the human and mouse, respectively.6, 7, 8, 9
Recently, we and others reported genome-wide DNA-methylation analyses of human gametes and early embryos.10, 11, 12 These data show that regions hypermethylated in oocytes and hypomethylated in sperm (oocyte-specific methylated regions) maintained levels of methylation very similar to those of the known gDMRs in human blastocysts. Most of these oocyte-specific methylated regions become hyper- or hypo-methylated in the embryonic lineage after implantation. These data demonstrated that many oocyte-specific methylated regions were transiently maintained as gDMRs during human preimplantation development but were lost in the embryonic lineage.
Regulation of DNA methylation in the extraembryonic lineage differs from that of the embryonic lineages. For example, the placenta has lower methylation levels than embryonic tissues,13 and partially methylated domains cover ∼40% of the genome in the human placenta.14, 15 In addition, more than half of the known human gDMRs are maintained in a placenta-specific manner.7, 8, 9 Aberrant DNA methylation, including abnormal imprinting, in the human placenta is associated with various developmental disorders, including miscarriage, preeclampsia, intrauterine growth restriction (IUGR), and imprinting disorders such as hydatidiform mole, Beckwith-Wiedemann syndrome (MIM: 130650), and Silver-Russell syndrome (MIM: 180860).16, 17, 18, 19, 20 In spite of the potential clinical importance of these epigenetic alterations, there is insufficient information on how germline DNA methylation is reprogramed and how the unique methylation profile is established and maintained in the human placenta.
In this study, we performed genome-wide allelic DNA methylation analyses and transcriptome-wide allelic expression analyses. We identified hundreds of gDMRs in the human placenta, and some of these were confirmed to be associated with imprinted gene expression.
Material and Methods
Sample Collection
Human placentas and peripheral blood were obtained from healthy women who had provided signed informed consent, and approval of the Ethics Committee of Tohoku University School of Medicine was obtained (research licenses 2012-1-538 and 2015-1-219). First- (5–11 weeks’ gestation, n = 23) and second (21 weeks’ gestation, n = 1)-trimester placentas were obtained from elective termination of pregnancies with live fetuses. Term placentas (n = 2) were obtained after elective caesarean section.
Isolation of Cytotrophoblast cells
Cytotrophoblast (CT) cells were isolated from fresh placental tissues as described previously21 but with some modifications. In brief, for first-trimester placentas, whole placental villi were cut into small pieces, washed several times in 0.9% NaCl, and enzymatically digested in a solution containing equal amounts of TrypLE (Life Technologies) and Accumax (Innovative Cell Tech). Single-cell suspensions were prepared with a 70 μm mesh filter, and CT cells were immunomagnetically purified with the EasySep PE selection kit (StemCell Technologies) and PE-conjugated anti-CD49f antibody (clone GoH3, Miltenyi Biotec). CD49f is a specific surface marker of CT cells,22 and >90% of the purified CT cells were positive for both CD49f and CK7 (clone SP52, Abnova Corporation), a pan-trophoblast marker. The cell purity was assessed by flow cytometry (FACSAria II, BD Bioscience). Thy1 (CD90)-positive stromal cells were also isolated from first-trimester placentas in a similar way to CT cells except for the use of anti-Thy1 antibody (clone DG3, Miltenyi Biotec) instead of anti-CD49f antibody.
For second-trimester and term placentas, about 50 g placental villous tissue was cut into small pieces and washed with 0.9% NaCl until the supernatant was clear. The washed tissue was enzymatically digested as described above. Single-cell suspensions were prepared by filtration, and red blood cells and cell debris were removed via Percoll density gradient (10%–70%; GE Healthcare Life Sciences) centrifugation. After removal of CD45-positive leukocytes with the EasySep Human CD45 Depletion Kit (StemCell Technologies), CD49f-positive CT cells were immunomagnetically purified as described above.
Whole-Genome Bisulfite Sequencing
Whole-genome bisulfite sequencing (WGBS) was performed via the paired-end post-bisulfite adaptor-tagging (PBAT) method.23, 24 In brief, genomic DNA (100–300 ng) was purified with phenol and chloroform extraction and ethanol precipitation. Genomic DNA spiked with 0.5% (w/w) unmethylated lambda phage DNA (Promega) was used for library preparation according to the PBAT protocol. Concentrations of the PBAT products were quantified with the KAPA Library Quantification Kit for Illumina platforms (Kapa Biosystems). PBAT libraries were sequenced on the Illumina HiSeq 1500 platform (HCS v2.0.5, RTA v1.17.20; Illumina, CA, USA) with 101 bp paired-end reads. After the first four bases and the last base were trimmed, the paired-end reads were aligned to the reference genome (UCSC hg19) with Bismark (v.0.9.0) under default parameters.25 The Y chromosome was excluded from the reference genome. After the paired-end alignment, unmapped reads were further aligned as single-end reads. The methylation level of each cytosine was calculated with the Bismark methylation extractor (the option “no overlap” was used for paired-end reads). For each cytosine-phosphate-guanine (CpG) site, combining reads from both strands allowed calculation of the methylation level. Methylation levels of CpGs covered with ≥ 5 reads were analyzed.
Targeted Bisulfite Sequencing
Genomic DNA (∼1 μg) was sheared into 400 bp fragments with Covaris M220 (Covaris, Woburn, MA). The fragments were end repaired and A-tailed by the NEBNext Ultra End Repair/dA-Tailing Module (NEB, Ipswich, MA) and ligated to custom sequencing adapters using the NEBNext Ultra Ligation Module (NEB). The sequencing adapters were a mixture of two cytosine-methylated adapters (Adaptor-T and Adaptor-C) in equal amounts. Adaptor-T with a 3′-T overhang was prepared by annealing (5′-CTA CAC GAC GCT CTT CCG ATC TT-3′) and (5′-AGA TCG GAA GAG CAC ACG TCT GAA-3′; 5′ phosphorylated)). Adaptor-C with a 3′-C overhang was prepared by annealing (5′- CTA CAC GAC GCT CTT CCG ATC TC-3′) and (5′-AGA TCG GAA GAG CAC ACG TCT GAA-3′; 5′ phosphorylated). Adaptor-C was added to increase the ligation efficiency because both A- and G-tailing take place by the dA-tailing. Target regions (candidate mDMRs (n = 1,594), candidate pDMRs (n = 187) and known gDMRs (n = 76)) were enriched using the SureSelect Custom Target Enrichment kit (Agilent Technologies, Santa Clara, CA). The enriched fragments were treated with sodium bisulfite using the EZ DNA Methylation-Lightning Kit (Zymo Research, Orange, CA) and amplified for 10 cycles using the KAPA HiFi HotStart Uracil+ ReadyMix PCR Kit (KAPA Biosystems, Boston, MA). The libraries were sequenced on the Illumina HiSeq 2500 platform (Illumina) with 101-bp single-end or paired-end reads. For the single-end reads, the first 2 and last 1 bases were trimmed. For the paired end reads, the first 2 and last 1 bases were trimmed from the forward reads and the first 5 and last 1 bases were trimmed from the reverse reads. The trimmed reads were aligned to the reference genome with Bismark using default parameters. For female samples, the Y chromosome was excluded from the reference genome. The mean percentage of off-target sequences was 82.6% (standard deviation (SD): 1.4). The percentages of off-target sequences were relatively high because repeat sequences were not excluded from the targeted regions. The mean percentage of the targeted regions covered with at least 10 reads was 73.5% (SD: 2.4). The methylation level of each cytosine was calculated using the Bismark methylation extractor. For each CpG site, reads from both strands were combined to calculate the methylation level. Methylation levels of CpGs covered with ≥ 5 reads were analyzed.
WGS and SNP Calling
To identify SNPs distinguishing parental alleles, WGS was performed with CT samples and the mother’s peripheral blood cells. Genomic DNA was purified with phenol and chloroform extraction and ethanol precipitation, and WGS libraries were constructed with the TruSeq Nano DNA LT Sample Prep Kit (Illumina) according to the manufacturer’s instructions. Libraries were sequenced on the Illumina HiSeq 2500 platform (Illumina) with 101 bp paired-end reads. The reads were aligned to the reference genome with Bowtie2 (v.2.1.0) under default parameters.26 The Y chromosome was excluded from the reference genome.
Only uniquely mapped reads were kept, and identical reads were treated as a single read to remove PCR duplicates. Overlapping paired-end reads were clipped by clipOverlap from BamUtil (v.1.0.12). SNPs were called with UnifiedGenotyper from GenomeAnalysisTK (v.3.3-0).27 For each SNP covered with ≥ 20 reads, the numbers of reads containing the reference allele ([read_ref]) and reads containing the major variant allele ([read_var]) were counted. The proportion of reads containing the reference allele ([proportion_ref]) was then calculated as follows: [proportion_ref] = [read_ref]/([read_ref] + [read_var]). We used only SNPs with 0.65 > [proportion_ref] > 0.35 in a CT sample and with [proportion_ref] > 0.99 or < 0.01 in the mother’s peripheral blood cells to distinguish parental alleles. To estimate maternal cell contamination, we used SNPs with [proportion_ref] > 0.95 or < 0.05 in a CT sample and with 0.65 > [proportion_ref] > 0.35 in the mother’s peripheral blood cells.
Exome Sequencing and SNP Calling
To identify SNPs distinguishing parental alleles, we performed exome sequencing with CT samples and the mother’s peripheral blood cells. Exome-sequencing libraries were constructed with the SureSelect Human All Exon V5+UTRs Kit (Agilent Technologies) according to the manufacturer’s instructions. For samples analyzed by targeted bisulfite sequencing, the SureSelect Custom Target Enrichment library described above was mixed with the SureSelect Human All Exon V5+UTRs library at a 1:4 ratio. Libraries were sequenced on the Illumina HiSeq 2500 platform (Illumina) with 101 bp paired-end reads. The reads were aligned to the reference genome, and SNPs were called as described above.
Allelic DNA-Methylation Analysis
Single-end and paired-end reads containing SNPs were extracted from the uniquely mapped reads obtained by the WGBS and targeted bisulfite sequencing analyses. C>T SNPs were not considered for reads aligned to the top strand, and G>A SNPs were not considered for reads aligned to the bottom strand. The reads were classified into maternal and paternal reads according to the SNP alleles (reads containing both maternal and paternal alleles were discarded). The methylation level of each cytosine was calculated from the maternal and paternal reads separately. For each CpG site, we combined reads from both strands to calculate the methylation level. For allelic methylation analyses, we used only methylation levels of CpGs that were covered with ≥ 5 reads for both maternal and paternal alleles and that were not overlapping SNPs. Only regions or windows containing ≥ 5 informative CpGs were analyzed. In cases where there were two or more SNPs in a region or window, we considered all the maternal and paternal reads overlapping these SNPs when we calculated the allelic methylation levels.
For windows, the statistical significance of the methylation differences between parental alleles was assessed with a Student’s t test with BH’s False Discovery Rate (FDR) correction. For candidate gDMRs, we combined the data from first-trimester CT (1st-CT) or We also combined data from second-trimester and term CT (2nd/term-CT) samples and calculated the mean maternal and paternal methylation levels for each CpG site. Then, the BH-corrected p value was determined for each candidate gDMR as described above. BH-corrected p < 0.05 was considered statistically significant.
RNA Sequencing and Allelic Expression Analysis
Total RNA was extracted with the RNeasy Mini Kit and RNase-free DNase (QIAGEN, CA, USA) and used for library construction with the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) according to the manufacturer’s protocol. RNA integrity was assessed via TapeStation 2200 (Agilent Technologies), and all samples had a RNA Integrity Number Equivalent (RINe) value of >8.0. The libraries were sequenced on the Illumina HiSeq 2500 platform (Illumina) with 101 bp paired-end reads. The reads were aligned to the reference genome with TopHat (v.2.0.13)28 and the Refseq gene annotation. For female samples, the Y chromosome was excluded from the reference genome. Expression levels (FPKM) of Refseq genes were calculated with Cufflinks (v. 2.2.1).28 For allelic expression analyses, only uniquely mapped reads were kept, and overlapping paired-end reads were clipped by clipOverlap. We analyzed only SNPs covered with ≥20 reads, and the numbers of paternal and maternal reads were counted for each gene. To identify candidate imprinted genes, we combined the data from 1st- CT or 2nd/term-CT samples and summed the numbers of paternal and maternal reads for each gene. The allelic expression differences were then analyzed via the binomial test with BH correction. BH-corrected p < 0.05 was considered statistically significant. We defined genes showing <35% mean [M-expression] ratios and statistically significant allelic expression differences as paternally expressed and those showing >65% as maternally expressed according to the criteria applied in a previous study.29
Conventional Bisulfite Sequencing
We treated DNA samples with sodium bisulfite by using the EZ DNA Methylation-Gold Kit (Zymo Research) and amplified them by performing PCR with the TaKaRa EpiTaq HS (Takara Bio). The PCR products were cloned into the pGEM-T Easy vector (Promega), and individual clones were sequenced. The following primers were used: the LOC389906-ds DMR (5′-TTA ATG GGG TAA AGG GG TTA GA-3′ and 5′-ACC AAA TAA ACC CCA CCC AAA C-3′) and the NUDT10 (MIM: 300527) DMR (5′-TTT TGT AAG GTG GGA ATT TGT TGA-3′ and 5′-CTC CTA AAA CCA AAA ACC TCC T-3′).
Annotations of Genomic Regions
Annotation of Refseq genes was downloaded from the UCSC Genome Browser. Promoters were defined as regions 1 kb upstream and downstream of transcription start sites of Refseq transcripts. The gene bodies were defined as transcribed regions of Refseq transcripts except for promoters. When several Refseq transcripts were assigned to a Refseq gene, the transcribed regions were merged into a single gene body. Promoters and gene bodies of Refseq genes shorter than 300 bp (these genes encoded microRNAs or small nucleolar RNAs in most cases) were excluded from our analyses. Regions and names of the 80 known gDMRs were defined as previously reported.7, 8, 9, 10 Among 101 mDMRs identified by Hanna et al.,9 only mDMRs whose maternal methylation was confirmed in normal placental samples are included in the known gDMRs. The list of known imprinted genes was obtained from the Catalogue of Parental Origin Effects database and a previous study.8 GO analyses were performed with the Database for Annotation, Visualization, and Integrated Discovery (DAVID).30
External Data
The WGBS data of human oocytes, sperm, blastocysts, and cord blood cells were from our previous study.10 We also included available WGBS data of human H9 ES cells (GEO: GSM706059),31 a term placenta analyzed without a cell purification step (GSM1134682),7 mouse oocytes (DDBJ Sequence Read Archive (DRA) accession number: DRA000570),32 sperm, blastocysts (DRA: DRA000484)33 and placenta (GEO: GSM1051161).13
Graphical Presentation
Methylation levels of CpGs were visualized with Integrative Genomics Viewer (IGV) software.34 Violin plots were generated with the vioplot package. The bee swarm plots were generated with the beeswarm package in R.
Results
Global DNA-Methylation Patterns of Parental Genomes
The chorionic villi of the first-trimester placenta are mainly composed of cytotrophoblast (CT) and syncytiotrophoblast (ST) cells but also contain non-trophoblastic cell types, including stromal cells and maternal immune cells.35 ST cells are formed by fusion of many CT cells and are difficult to isolate. In this study, CT cells were immunomagnetically enriched and used for the analysis.
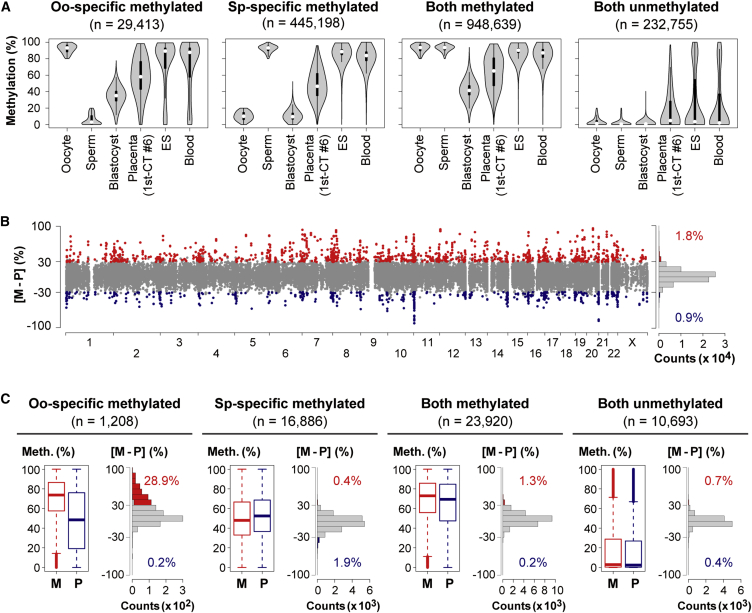
We performed whole-genome bisulfite sequencing (WGBS) of CT cells isolated from one first-trimester placenta (sample ID: 1st-CT #6 (♀); see Table S1) at high coverage (an average of 41 reads per CpG site). We compared the methylation profile of the CT cells with those of human gametes, blastocysts, cord blood cells,10 and ES cells.31 We used a system of sliding windows of 20 CpGs with a step size change of 10 CpGs to characterize global DNA-methylation changes as described previously.10 The mean length of the 20 CpG windows was 2.0 kb, and windows of more than 10 kb were excluded from the analysis. We focused on windows hypermethylated (≥80%) or hypomethylated (≤20%) in one or both gametes (Figure 1A). Windows hypermethylated in oocytes and hypomethylated in sperm (oocyte-specific methylated windows) maintained intermediate methylation levels in blastocysts (median = 35.1%). An intermediate pattern was also apparent in CT cells (median = 58.0%) (Figure 1A). Therefore, we inferred that some oocyte-specific methylated regions might maintain maternal allele-specific DNA methylation in CT cells. In contrast, sperm-specific methylated windows were nearly completely demethylated in blastocysts (median = 9.5%) (Figure 1A), suggesting the loss of paternal allele-specific DNA methylation after fertilization.
Figure 1.
Genome-wide Profiling of Allelic DNA Methylation
(A) Violin plots of methylation levels of windows hypermethylated (≥80%) or hypomethylated (≤20%) in one or both gametes (window size = 20 CpGs, step size = 10 CpGs). We compared the methylation profile of the CT cells (1st-CT #6 [♀]) with those of human gametes, blastocysts, cord blood cells (DRA003802),10 and ES cells (GSM706059).31 Oo-specific (Sp-specific) methylated windows are defined as windows hypermethylated in oocytes (sperm) and hypomethylated in sperm (oocytes). Thin and thick lines are boxplots, and white dots indicate the median.
(B) Chromosomal distribution of maternally and paternally methylated windows in 1st-CT #6. The x and y axes show chromosome numbers and the maternal methylation level minus the paternal methylation level ([M − P] level), respectively. Windows showing ≥30% [M − P] levels and statistically significant allelic methylation differences (BH-corrected p < 0.05) are shown in red, and those with ≤ −30% [M − P] levels are in blue. The other windows are in gray. A histogram of the distribution of the [M − P] levels and the proportions of maternally and paternally methylated windows are also shown.
(C) Boxplots of methylation levels of the maternal (M) and paternal (P) alleles of windows in 1st-CT #6. Boxes represent lower and upper quartiles, and horizontal lines indicate the median. Whiskers extend to the most extreme data points within 1.5 times the interquartile range from the boxes. The open circles indicate the data points outside the whiskers. Histograms of the distribution of the [M − P] levels are also shown (colored as defined in [B]).
To identify single-nucleotide polymorphisms (SNPs) that would allow the analysis of DNA methylation on the two parental alleles, we performed whole-genome sequencing (WGS) of the CT cells (1st-CT #6) and the mother’s peripheral blood cells. We successfully analyzed the allelic DNA-methylation patterns of 75,391 windows (∼3% of all windows) (Figure 1B). The mean length of these windows was 1.2 kb, and the mean number of informative CpGs per window was 7.7. Consistent with the global methylation changes, the maternal alleles of oocyte-specific methylated regions had higher methylation levels than the paternal alleles (Figure 1C). We defined maternally or paternally methylated windows as those showing ≥30% or ≤−30% [M − P] level (the maternal methylation level minus the paternal methylation level) and statistically significant allelic methylation differences (Benjamini-Hochberg (BH)-corrected p < 0.05, Student’s t test). We found that 28.9% of oocyte-specific methylated windows showed maternal methylation, whereas only 0.2% of them showed paternal methylation (Figure 1C). For the other types of windows, the parental alleles had similar methylation levels (Figure 1C). To confirm these findings, we further performed WGBS of an additional CT sample (sample ID: 1st-CT #9 (♀)) at relatively low coverage (an average of 20 reads per CpG site). In this sample, 27.5% of oocyte-specific methylated windows showed maternal methylation, and the allelic methylation levels were strongly correlated between 1st-CT #6 and 1st-CT #9 (Pearson’s r > 0.9) (Figure S1). Therefore, more than one-quarter of oocyte-specific methylated windows might maintain parent-of-origin-dependent maternal DNA methylation in CT cells.
Identification of gDMRs
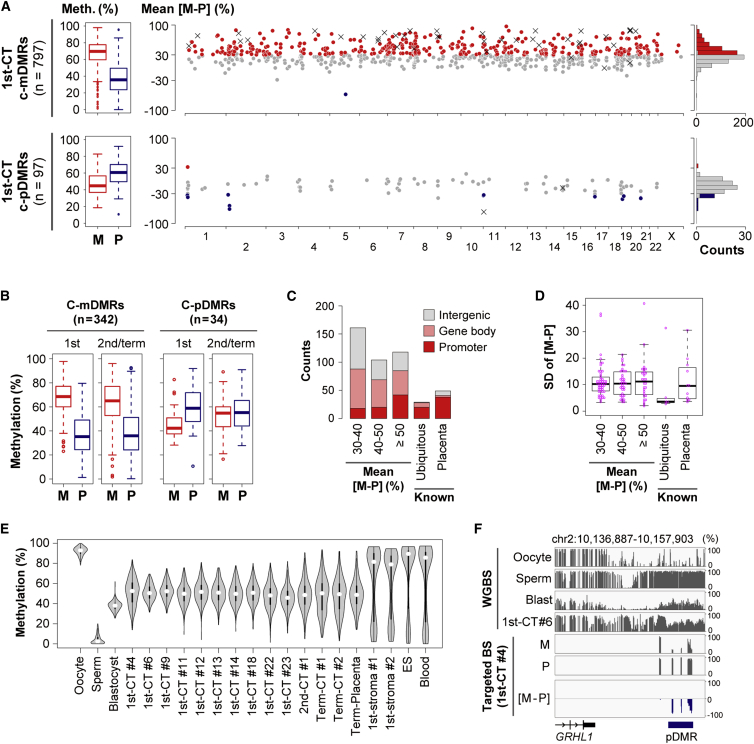
To analyze allelic DNA methylation in more placental samples, we focused on oocyte- and sperm-specific methylated windows showing methylation patterns similar to those of known gDMRs (25%–65% methylation in blastocysts and 30%–70% methylation in CT cells). After merging these windows, we obtained 3,676 candidate mDMRs and 1,229 candidate pDMRs except for known gDMRs (Table S2). We selected 1,594 candidate mDMRs and 187 candidate pDMRs containing at least two windows for further analyses (Figure S2A). These candidate gDMRs were enriched, treated with bisulfite, and analyzed by high-throughput sequencing (targeted bisulfite sequencing). Among 80 known gDMRs identified in previous reports,7, 8, 9, 10 76 gDMRs also met the criteria described above. These 76 gDMRs were also included in the targeted bisulfite sequencing analyses (Table S2).
We analyzed allelic DNA methylation of candidate gDMRs in CT samples obtained from eight 1st-CT, one 2nd-CT, and two term-CT placentas (Figure S3). In all, 797 and 449 candidate mDMRs were successfully analyzed for 1st-CT and 2nd/term-CT samples, respectively (Tables S3 and S4). Allelic DNA-methylation patterns of 42 and 32 known gDMRs were also obtained for 1st-CT and 2nd/term-CT samples, respectively (Tables S5 and S6). Most known gDMRs were confirmed to show ≥30% or ≤−30% [M − P] levels. We defined candidate mDMRs or pDMRs showing ≥30% or ≤−30% mean [M − P] levels and statistically significant allelic methylation differences (BH-corrected p < 0.05, Student’s t test) as confirmed gDMRs. These cutoff criteria for confirmed gDMRs are more stringent than those applied in a previous study9 (Figure S2B). In total, 48.1% (383/797) and 33.2% (149/449) of the candidate mDMRs were confirmed to be mDMRs in 1st-CT and 2nd/term-CT samples, respectively (Figure 2A and Figure S4A). In all, the allelic methylation was successfully analyzed for 904 regions, of which 440 were confirmed to be mDMRs in 1st-CT and/or 2nd/term-CT samples. Among the 440 confirmed mDMRs, 439 mDMRs contained two or more CpGs with [M − P] ≥ 30%. Although one mDMR had only one CpG with [M − P] ≥ 30%, the other CpGs within this mDMR consistently showed >20% [M − P] levels. We found that the maternally biased DNA methylation was predominantly maintained throughout gestation (Figure 2B). Whereas known mDMRs are predominantly located at promoter regions, most of the confirmed mDMRs were in gene bodies or intergenic regions (Figure 2C). However, when compared to the distribution of all 20 CpG windows (i.e., the background distribution), the distribution of confirmed mDMRs was significantly enriched at promoter regions (p = 1.0 × 10−15: Chi-square test) (Figure S4B). We searched sequence motifs in the confirmed mDMRs by using MEME-ChIP36 and identified only one motif (Figure S5). The motif was similar to the binding sites of several transcription factors, such as EGR1 and SP2, suggesting that some of the confirmed mDMRs might be transcriptional regulatory elements.
Figure 2.
Identification of gDMRs
(A) Allelic DNA methylation patterns of candidate DMRs (c-DMRs) obtained from ten 1st-CT samples. Boxplots of methylation levels of the maternal (M) and paternal (P) alleles, chromosomal distribution of mean [M − P] values, and histograms of the distribution of the [M − P] values are shown. In the chromosome maps, red circles indicate c-DMRs showing ≥30% [M − P] levels and statistically significant allelic methylation differences (BH-corrected p < 0.05). Similarly, blue circles indicate those with ≤ −30% [M − P] values. The other c-DMRs are shown as gray circles. Each x indicates a known gDMR.
(B) Box plots of allelic DNA methylation levels of candidate gDMRs covered by both 1st-CT and 2nd/term-CT samples.
(C) Classification of confirmed mDMRs according to their mean [M − P] values and location. The data of the 1st-CT samples were used. Known ubiquitous and placenta-specific mDMRs are also shown for comparison.
(D) Variations in allelic methylation between samples. We calculated the SD value for each confirmed mDMR by using the data of the 1st-CT samples. Only mDMRs for which there were data from three or more samples were analyzed. Box plots with individual data points are shown. Known ubiquitous and placenta-specific mDMRs are also included for comparison.
(E) Violin plots of methylation levels of confirmed mDMRs. 1st-CT #6 and #9 were analyzed by WGBS. Additional CT samples and stromal cells obtained from two first-trimester placentas were analyzed via targeted bisulfite sequencing. External WGBS data were used for human gametes, blastocysts, cord blood cells (DRA003802),10 a term placenta analyzed without a cell purification step (GSM1134682),7 and ES cells (GSM706059).31 Only confirmed mDMRs covered by all the samples were analyzed (n = 404).
(F) DNA methylation patterns of a confirmed pDMR located downstream of GRHL1 (MIM: 609786). Each vertical bar represents a CpG site. The pDMR was hypomethylated in oocytes, hypermethylated in sperm, and paternally methylated in CT cells.
To test the possibility that the allele-specific methylation of some mDMRs was not parent-of-origin dependent but haplotype dependent, we examined haplotype-dependent allele-specific methylation (hap-ASM). In our dataset, 306 SNPs overlapping 149 candidate mDMRs were available for the hap-ASM analysis. No region was found to show hap-ASM (Figure S6). Although not all candidate mDMRs were subjected to the hap-ASM analysis as a result of the lack of informative SNPs, this result supports the conclusion that few regions with hap-ASM were included in the candidate mDMRs.
To examine the variation in allelic methylation levels between samples, we focused on confirmed mDMRs where there were data for three or more 1st-CT samples and calculated the SD values of [M − P] levels (Figure 2D). The [M − P] levels of these mDMRs are shown in Figure S7. The SD values of most of the confirmed mDMRs were 5%–15%, regardless of their mean [M − P] levels. Known placenta-specific mDMRs had SD values similar to those of the confirmed mDMRs. The SD values of known ubiquitous mDMRs (mDMRs maintained in both embryos and placentas) were lower than those of the confirmed mDMRs and known placenta-specific mDMRs. Some of the confirmed mDMRs with >15% SD values showed polymorphic imprinting (Figure S7), consistent with a previous study revealing that some placenta-specific mDMRs show polymorphic imprinting.9 On the other hand, the allelic methylation levels of mDMRs with <10% SD values were relatively consistent across samples (Figure S7).
To obtain an insight into the regulation of the confirmed mDMRs in embryonic lineages, we analyzed stromal cells obtained from first-trimester placentas, which are derived from the inner cell mass (ICM) of the blastocyst. We found that about 70% of the confirmed mDMRs were hyper- or hypomethylated in stromal cells (Figure 2E). Similarly, about 80% of the confirmed mDMRs were hyper- or hypomethylated in ES and cord blood cells (Figure 2E). These data suggest that the allelic imbalance of most of the confirmed mDMRs might not be maintained in embryonic lineages.
Allelic methylation patterns of 97 and 43 candidate pDMRs were also obtained for 1st-CT and 2nd/term-CT samples, respectively. The paternal alleles (median = 60.7%) had higher methylation levels than the maternal alleles (median = 44.8%) in 1st-CT samples (Figure 2A), and several confirmed pDMRs were identified (Figure 2F and Figure S4). However, the allelic imbalance was predominantly lost, and very few regions maintained paternally biased DNA methylation in 2nd/term-CT samples (Figure 2B).
In the analyses described above, we used only CpGs covered with ≥ 5 reads (5×) for both maternal and paternal alleles. To see whether this coverage was sufficient to provide reliable allelic methylation data, we also examined the coverages at 8× and 10×. We found that the 8× and 10× data were highly correlated with the 5× data (r > 0.95), suggesting that coverage at 5× was sufficient to accurately represent the allelic methylation status (Figure S8).
Transcriptome-wide Identification of Imprinted Genes
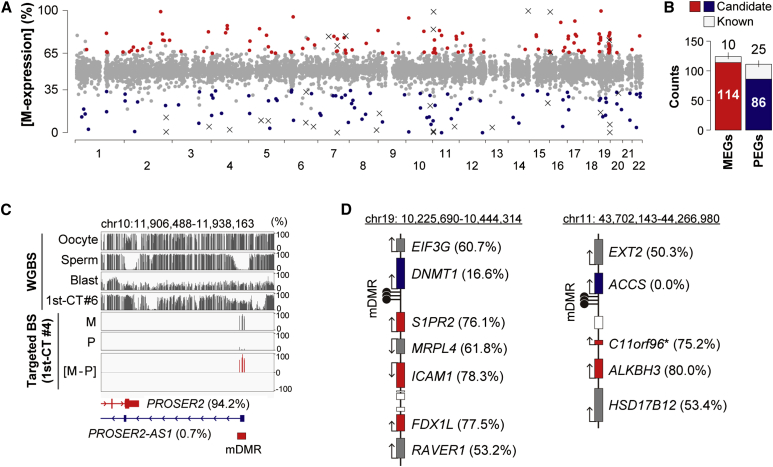
We were next interested in whether the identified gDMRs were associated with expression of imprinted genes. RNA-sequencing-based allelic expression analyses were performed with 1st -CT samples obtained from 18 female and three male placentas. For ∼75% of genes with ≥1 FPKM (fragments per kilobase of exon per million fragments mapped), allelic expression was successfully analyzed in at least one sample (Figure S9A). For each gene, the proportion of reads derived from the maternal allele to total reads ([M-expression] ratio) was calculated (Figure 3A and Table S7).
Figure 3.
Transcriptome-wide Screening of Imprinted Genes
(A) Chromosomal distribution of candidate imprinted genes. The x axis and y axis show chromosome numbers and the maternal expression ratio ([M-expression] ratio), respectively. Genes showing >65% [M-expression] ratios and statistically significant allelic expression differences (BH-corrected p < 0.05) are shown in red, and those with <35% [M-expression] ratios are in blue. Each x indicates a known imprinted gene with an [M-expression] ratio > 65% or < 35%. The other genes are in gray.
(B) Counts of candidate and known imprinted genes.
(C) DNA methylation patterns of the PROSER2-AS1 DMR. [M-expression] ratios of associated genes are shown in parentheses.
(D) The DNMT1 and ACCS imprinted gene clusters. Genes with [M-expression] ratios > 65% and < 35% are shown in red and blue, respectively. Genes with 35%–65% [M-expression] ratios are in gray. Genes without available allelic expression ratios are shown as white boxes without gene symbols. An asterisk indicates that it was unclear whether C11orf96 was really maternally expressed because the [contamination] rate of C11orf96 was not available.
We defined genes showing <35% mean [M-expression] ratios and statistically significant allelic expression differences (BH-corrected p < 0.05, binomial test) as candidate paternally expressed genes (PEGs) and identified 111 candidate PEGs, of which 25 were known PEGs (Figure 3B). It is possible that biased allelic expression of some genes is regulated by mechanisms other than genomic imprinting (e.g., polymorphisms in cis-regulatory elements). Therefore, we further selected candidate PEGs by using the following criteria: (1) location of confirmed or candidate mDMRs at promoter regions and (2) paternally biased expression consistently observed in at least three samples. We defined genes meeting (1) and/or (2) as strong candidate PEGs (n = 30) (Table S8). Most of the strong candidate PEGs were associated with confirmed or candidate mDMRs (26/30), suggesting maternal allele-fspecific silencing by DNA methylation.
We found 445 genes with maternally biased expression (>65% [M-expression] ratios). This result should be carefully interpreted because it was difficult to exclude the maternal cell contamination completely. Genes very highly expressed in contaminating maternal cells could be mistakenly identified as maternally expressed genes (MEGs).37 To avoid misidentification, we utilized SNPs that were homozygous in CT samples and heterozygous in mothers, and we estimated the expression rate from the contaminating maternal cells (contamination rate) for each gene (see Figures S9B–S9D for details). Genes with >10% mean [contamination] rates tended to show maternally biased expression, and immune-related gene ontology (GO) terms were significantly enriched in these genes. Therefore, we defined candidate MEGs as genes showing > 65% mean [M-expression] ratios, significant allelic expression differences and ≤10% mean [contamination] rates and identified 124 candidate MEGs. The candidate MEGs included ten known MEGs (Figure 3B). Although few candidate MEGs had confirmed or candidate gDMRs at their promoter or gene body regions, several candidate MEGs were found to be linked to PEGs with mDMRs (Figures 3C and 3D). One of these clusters contained DNMT1 (MIM: 126375), and the DMR and imprinted expression of DNMT1 have already been reported.38, 39 Our data revealed that DNMT1 was located within an imprinted gene cluster with three candidate MEGs. In consideration of these data, candidate MEGs were further selected according to the following criteria: (1) maternally biased expression consistently observed in at least three samples and (2) location in the imprinted gene clusters. We defined genes meeting criteria (1) and/or (2) as strong candidate MEGs (n = 14) (Table S8). Strong candidate imprinted genes associated with confirmed mDMRs are highlighted in Table 1.
Table 1.
Confirmed mDMRs Associated with Strong-Candidate Imprinted Genes
|
Confirmed mDMRs |
Strong-Candidate Imprinted Genes |
|||||
|---|---|---|---|---|---|---|
| Chr | Start | End | [M − P] | Gene (MIM Number) | [M-expression] | Cluster |
| 12 | 4,433,379 | 4,433,928 | 96.5% | TIGAR (610775) | 23.6% | - |
| 3 | 27,504,162 | 27,506,728 | 86.3% | SLC4A7 (603353) | 22.9% | - |
| 10 | 11,934,845 | 11,934,845 | 83.9% | PROSER2-AS1 (NA) | 0.7% | PROSER2-AS1 |
| - | - | - | - | PROSER2 (NA) | 94.2% | PROSER2-AS1 |
| 7 | 129,746,868 | 129,749,725 | 82.2% | KLHDC10 (615152) | 78.0% | - |
| 10 | 115,999,021 | 115,999,895 | 73.3% | VWA2 (NA) | 13.0% | - |
| 8 | 8,559,607 | 8,560,147 | 70.9% | CLDN23 (609203) | 23.9% | - |
| 11 | 93,582,929 | 93,583,380 | 61.5% | VSTM5 (NA) | 11.5% | - |
| 14 | 24,562,609 | 24,563,883 | 56.4% | PCK2 (614095) | 12.7% | - |
| 7 | 151,327,824 | 151,330,524 | 56.0% | PRKAG2 (602743) | 8.4% | - |
| 15 | 93,198,061 | 93,201,008 | 47.7% | FAM174B (NA) | 31.6% | - |
| 11 | 44,087,195 | 44,088,588 | 46.5% | ACCS (608405) | 0.0% | ACCS |
| - | - | - | - | ALKBH3 (610603) | 80.0% | ACCS |
Confirmed mDMRs are arranged according to their mean [M − P] levels. See Table S8 for the full list of strong-candidate imprinted genes. NA: not available.
Skewed X-Chromosome Inactivation (XCI) and Identification of X-Linked mDMRs
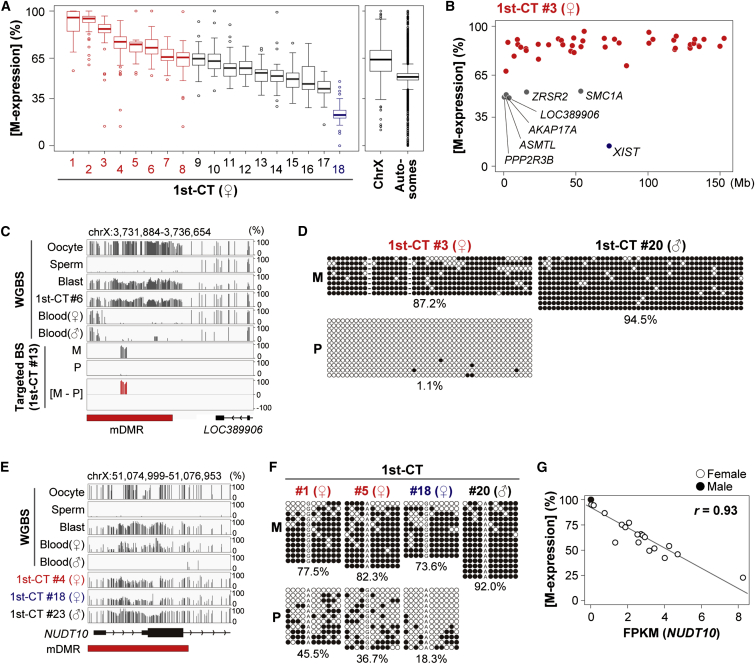
Allelic expression profiles of X-linked genes were analyzed in 1st-CT samples obtained from 18 female placentas (Figure 4A and Table S9). Most X-linked genes had similar [M-expression] ratios in each CT sample; exceptions were XIST [MIM: 314670] and escape genes (Figures 4A and 4B). In most of the 1st-CT samples (15/18), X-linked genes had >50% median [M-expression] ratios (Figure 4A). Therefore, the paternal X chromosomes were preferentially inactivated in the majority of the CT samples, although three CT samples showed <50% median [M-expression] ratios (1st-CT numbers 16–18 in Figure 4A).
Figure 4.
Allelic Regulation of X-Linked Genes and gDMRs
(A) Boxplots of maternal expression ratios ([M-expression] ratios) of X-linked genes. (Left) 18 1st-CT samples were analyzed, and samples with >65% and <35% median [M-expression] ratios are shown in red and blue, respectively. (Right) Summary of allelic expression of X-linked and autosomal genes from the 18 1st-CT samples. X-linked genes had higher [M-expression] ratios than autosomal genes (p < 2.2 × 10−16, Mann Whitney U-test).
(B) A chromosome map of [M-expression] ratios of X-linked genes (1st-CT #3). Genes with >65% [M-expression] ratios are shown in red, and those with 35%–65% [M-expression] ratios are in gray with gene symbols. The [M-expression] ratio of XIST was 14.4% (shown in blue).
(C) DNA methylation patterns of the LOC389906-ds DMR. LOC389906 is an escape gene as shown in (B).
(D) Bisulfite sequencing analysis of the LOC389906-ds DMR. Black and white circles indicate methylated and unmethylated residues, respectively. The percentages of methylated CpG sites are indicated.
(E) DNA methylation patterns of the NUDT10 DMR.
(F) Bisulfite sequencing analysis of the NUDT10 DMR.
(G) A high correlation between NUDT10 expression levels and the median [M-expression] ratios of X-linked genes. 18 female and three male 1st-CT samples were analyzed. Pearson’s r was 0.93.
Our DNA-methylation analyses identified several candidate mDMRs on the X chromosome (Table S2). By using conventional bisulfite sequencing, we confirmed maternal-allele-specific DNA methylation of one confirmed mDMR downstream of LOC389906; this mDMR was designated the LOC389906-ds DMR (Figures 4C and 4D). LOC389906 was biallelically expressed in CT cells (Figure 4B). The allelic DNA methylation patterns of the LOC389906-ds DMR were not affected by the XCI status or gender (Figures 4C-D). The LOC389906-ds DMR was hypomethylated in blood cells (Figure 4C) and thus might be placenta specific. It was not found to be closely associated with allele-specific expression of any RefSeq genes.
We also analyzed a candidate mDMR at the promoter region of NUDT10 in detail (this mDMR was designated the NUDT10 DMR) (Figure 4E). Similar to the LOC389906-ds DMR, the NUDT10 DMR had a highly methylated maternal allele in CT cells regardless of XCI status or gender (Figure 4F). In contrast, the paternal allele of the NUDT10 DMR showed variable methylation levels across the samples (Figure 4F). Interestingly, the expression levels of NUDT10 were well correlated with the XCI status in female CT samples. NUDT10 had very low expression levels or was undetectable in female CT samples with predominant paternal XCI and also in male CT samples (Figure 4G). Although allelic expression of NUDT10 was unavailable in our CT samples because of the lack of SNPs in the exonic regions, these data suggested that the maternal allele of NUDT10 was repressed and that the paternal allele was expressed in an XCI status-dependent manner. The expression level of NUDT10 might be a good marker for analysis of the XCI status in CT cells. Similar to the LOC389906-ds DMR, the NUDT10 DMR was unmethylated in male blood cells (Figure 4E).
Evolutionary Variability of mDMRs and Imprinted Genes in Mammalian Placentas
Using previously reported mouse WGBS data,13, 32, 33 we analyzed DNA methylation levels of mouse regions orthologous to the human candidate mDMRs identified in our study. In all, 723 mouse regions were successfully analyzed, but only five had methylation patterns similar to those of known mDMRs (hypermethylated in oocytes, hypomethylated in sperm, 25%–65% methylation in blastocysts, and 30%–70% methylation in the placenta) (Figure S10A).
We also identified 44 strong candidate imprinted genes (30 PEGs and 14 MEGs) in the human placental CT cells. None of these are reported to be imprinted in the mouse placenta. In addition, among the known mouse placenta-specific imprinted genes, only one (Ppp1r9a) was confirmed to be imprinted in human CT cells (Figure S10B). Placental imprinting has not been explored in detail in mammalian species other than humans and mice, but a recent study of RNA-sequencing-based screening of placental imprinted genes in hybrids of the horse and donkey identified 78 candidate imprinted genes.29 Of these candidates, only MUC1 (MIM: 158340) and FKBP5 (MIM: 602623) showed imprinted expression patterns in the human placenta (Figure S10C). Neither gene was associated with candidate gDMRs in human CT cells. Further studies are required to determine whether MUC1 and FKBP5 are conserved imprinted genes. Taken together, these data suggest that the placental imprinting of genes is also poorly conserved across mammalian species.
Discussion
Mechanisms of Maintenance of Oocyte-Specific Methylation in the Human Placenta
In this study, we identified 3,676 candidate mDMRs. We successfully analyzed allelic methylation for 904 regions and confirmed 440 regions as mDMRs. These data imply that there are ∼1,800 mDMRs present in the human placenta. Although the allelic methylation biases of most confirmed mDMRs were relatively weaker than those of known mDMRs, the biases were maintained in the placenta throughout gestation. Most of the confirmed mDMRs identified in this study were probably placenta-specific given that these regions were predominantly hypermethylated in embryonic cells. Determination of which gDMRs identified in this study are maintained during the preimplantation stage and identification of the stage at which the gDMRs are lost in the embryonic lineage will require analysis of allelic DNA methylation in the human trophectoderm and ICM.
Recently, Hanna et al. analyzed triploid placentas by using the Illumina Infinium HumanMethylation450 array (450K array) and, in combination with reduced representation bisulfite sequencing (RRBS) data of human gametes and blastocysts, identified 101 putative mDMRs (11 of them were shown to be maternally methylated in normal placental samples).9 For 31 of them, the allelic methylation patterns were available in our dataset. We confirmed maternal methylation of 25 of the 31 putative mDMRs. The analysis by Hanna et al. was restricted to CpG islands covered by both the 450K array and RRBS data. Our WGBS-based analysis is consequently more comprehensive than that of Hanna et al. It should also be noted that our cutoff criteria for mDMRs were more stringent than those used by Hanna et al. (Figure S2B). Our data strongly suggest that there are ∼18 times more mDMRs in the human placenta than expected by Hanna et al.
The placenta-specific maintenance of mDMRs can be mainly explained by two mechanisms: (1) incomplete demethylation of the maternal alleles during preimplantation development and (2) incomplete de novo methylation and/or inefficient methylation maintenance of the paternal alleles after implantation (Figure 5). In human CT cells, the DNA methyltransferases and UHRF1 (MIM: 607990), which targets DNMT1 to hemimethylated sites,40 were all highly expressed (FPKM was >100 for DNMT1, >10 for DNMT3A [MIM: 602769] and DNMT3B [MIM: 602900], and ∼10 for UHRF1). Therefore, site-specific exclusion of methyltransferases and/or selective recruitment of factors involved in demethylation might be important for the maintenance of mDMRs in the human placenta. Lack of transcription might be one of the mechanisms because gene body methylation positively correlates with gene expression levels in the human placenta.14 Consistent with this hypothesis, more than one-third of the confirmed mDMRs identified in this study were located in intergenic regions. Additionally, the candidate mDMRs identified in this study were significantly enriched (1,917/3,676, p < 2.2 × 10−16: chi-square test) in the partially methylated domains where genes are preferentially repressed.14 In contrast to the oocyte-specific methylated regions, most sperm-specific methylated regions did not maintain paternally biased methylation in the human placenta, which can be explained by the global and nearly complete demethylation of the paternal genome after fertilization (Figure 5).
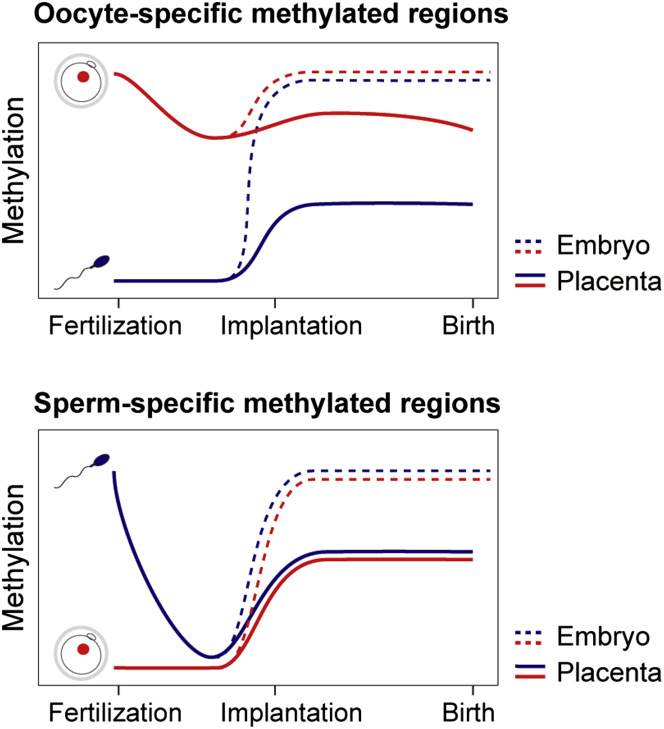
Figure 5.
Schematic Illustration of Allelic DNA Methylation Levels of Oocyte- and Sperm-Specific Methylated Regions during Human Development
For oocyte-specific methylated regions, the maternal alleles maintain high methylation levels throughout human development. The paternal alleles are predominantly hypermethylated in the embryonic lineage after implantation, but the de novo methylation and/or methylation maintenance of the paternal alleles are incomplete in the placenta. Consequently, many oocyte-specific methylated regions maintain maternally biased DNA methylation in the placenta. In contrast, sperm-specific methylated regions are demethylated during preimplantation development, and very few regions maintain paternally biased DNA methylation after implantation.
Functions of Placenta-Specific mDMRs and Imprinted Genes
We identified 44 strong candidate imprinted genes (30 PEGs and 14 MEGs), and seven of them were located in three imprinted gene clusters. The strong candidate imprinted genes included several genes that might play important roles in placental development. For example, CUL7 (MIM: 609577) was paternally expressed in the human placenta and encodes an E3 ubiquitin ligase scaffold protein. Cul7-deficient mouse embryos exhibit IUGR, and their placentas show defects in the differentiation of the trophoblast lineage and an abnormal vascular structure.41 CUL7 is reported to show abnormal expression in human IUGR placentas.42 Another example is CYP2J2 (MIM: 601258), which encodes an arachidonic acid epoxygenase. CYP2J2 and its metabolites are elevated in preeclamptic human placentas, and preeclamptic features are improved by a CYP epoxygenase inhibitor in rat models of preeclampsia.43 These data suggest that abnormal expression of some of the candidate imprinted genes could increase the risk of pregnancy complications such as IUGR and preeclampsia.
Recently, Sanchez-Delgado et al. reported ten novel imprinted genes.8 For five of them (CMTM3 [MIM: 607886], RHOBTB3 [MIM: 607353], RASGRF1 [MIM: 606600], SCIN [MIM: 613416], and ZFP90 [MIM: 609451]), the allelic expression patterns were available in our dataset. Three (CMTM3, RHOBTB3, and ZFP90) were confirmed to be imprinted. SCIN was not classified as imprinted in our study, although there was paternally biased expression ([M-expression] = 36.1%). Although RASGRF1 was found to be biallelically expressed ([M-expression] = 50.0%) in one informative sample, the imprinting of RASGRF1 is already known to be polymorphic.8 Overall, our data are consistent with those of Sanchez-Delgado et al.
It should be noted that there were some limitations in our RNA sequencing analyses. First, splicing variant- or transcription-start-site-specific imprinting was difficult to analyze. Second, developmental-stage- and cell-type-specific imprinted genes were difficult to identify. Finally, small RNAs, RNAs without poly-A tails, and other RNAs unannotated in the Refseq database were not analyzed in this study. For these reasons, we speculate that there could be more imprinted genes and transcripts in the human placenta. The gDMRs identified in this study should provide a useful platform with which to identify these additional imprinted genes and transcripts in future studies.
Many of the confirmed mDMRs identified in this study, especially those located in gene bodies or intergenic regions, were not found to be closely associated with allele-specific expression. Likewise, only some of the known placenta-specific mDMRs were confirmed to be associated with allele-specific expression.7, 8 Although the functional roles of such mDMRs are currently unclear, there are several possibilities. Our approach might not have captured all the imprinted genes. Consequently, some of the mDMRs might in fact regulate allele-specific expression of unidentified genes or transcripts. Furthermore, gDMRs can act over several megabases, 44 and thus it is also possible that some mDMRs are linked to imprinted genes located at a distance. Additionally, some mDMRs could act as enhancers given that a motif similar to the binding sites of transcription factors such as EGR1 and SP2 was enriched in mDMRs. In such cases, their effects on allelic expression biases might not be strong enough to be detected in this study. Finally, some mDMRs could be by-products of incomplete reprogramming of germline methylation and have no functional importance in the regulation of gene expression.
Genomic Imprinting on the X Chromosome
There have been many reports investigating XCI patterns in the human placenta.45 Some studies suggest random XCI, whereas others support preferential paternal XCI. Our RNA sequencing analyses have several advantages over previous studies because (1) the contamination of non-trophoblast cells was minimized, (2) purifying CT cells from whole placentas avoided sampling biases (the XCI patterns might be mosaic), and (3) chromosome-wide allelic expression data were available. We observed variable degrees of preferential paternal XCI in most of our samples. Although studies with a larger sample size and different ethnic groups are needed to support a firm conclusion, our data lend support to weakly skewed paternal XCI in the human placenta.
Neither X-linked imprinted genes nor gDMRs have been identified in the human genome, but it has been proposed that there might be imprinted genes on the human X chromosome. For example, in Turner’s syndrome, the degree of the deficit in social cognition is dependent on the parent of origin of the missing X chromosome.46 In this study, we identified some X-linked mDMRs and also a putative paternally expressed imprinted gene, NUDT10. This gene encodes a diphosphoinositol polyphosphate phosphohydrolase, and its role in placental development is currently unknown. Further studies are required to explain the functional importance of X-linked mDMRs and imprinted genes and to clarify the underlying mechanisms of functional differences of the parental X chromosomes.
Evolutionary Variability of Genomic Imprinting in Mammalian Placentas
This and previous studies suggest that placenta-specific gDMRs and imprinted genes are poorly conserved between humans and mice.7, 37, 47 We identified many mDMRs in the human placenta, whereas few placenta-specific gDMRs have been reported in the mouse. In mouse blastocysts, both the maternal and paternal genomes are significantly demethylated,48, 49 whereas the maternal genome is demethylated to a much lesser extent than the paternal genome in human blastocysts.10 The differential regulation of the demethylation of the maternal genome between humans and mice may, in part, explain the increased number of mDMRs and imprinted genes in the human placenta. To further understand the evolutionary variation of placental imprinting, it will be important to compare allelic DNA methylation and expression patterns in placentas from a number of mammalian species.
Conclusions
In conclusion, we have revealed the unique patterns of allelic DNA methylation in the human placenta. The findings are fundamental to our understanding of normal placental development as well as developmental disorders with imprinting effects. Our study also highlights the variability of genomic imprinting in mammalian placentas. The critical role of the placenta in transport and endocrine production is conserved among placental mammals, but the placental anatomy and trophoblastic subtypes are highly variable.50 Genomic imprinting might have continually coevolved within the placenta in a species-specific manner, contributing to the diversification of mammalian placentas.
Acknowledgments
We thank all the individuals and their families who participated in this study. We also thank Professor K. Nakayama, Dr. R. Funayama, Ms. N. Miyauchi, Ms. A. Kitamura, Ms. M. Tsuda, Ms. M. Kikuchi, Ms. M. Nakagawa, and Mr. K. Kuroda for technical assistance and Professor Rosalind M. John for support and valuable suggestions. We are also grateful to the Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support. This work was supported by Grants-in-Aid for Scientific Research (KAKENHI) (15K15592), the Japan Agency for Medical Research and Development (AMED) (15ek0109132, 15ek0109101h0001), Banyu Life Science Foundation International, Smoking Research Foundation, and the Takeda Science Foundation to TA and KAKENHI (26112502, 15K10657) to HO. This work was also supported by the Core Research for Evolutional Science and Technology (CREST) from AMED (HS and TA).
Published: October 27, 2016
Footnotes
Supplemental Data include ten figures, nine tables, and Supplemental Materials and Methods and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.08.021.
Contributor Information
Hiroaki Okae, Email: okaehiro@m.tohoku.ac.jp.
Takahiro Arima, Email: tarima@med.tohoku.ac.jp.
Accession Numbers
All sequencing data reported in this paper have been deposited in the DDBJ Japanese Genotype-phenotype Archive under accession number JGAS00000000038.
Web Resources
Catalogue of Parental Origin Effects database, http://igc.otago.ac.nz/home.html
OMIM, http://www.omim.org
UCSC Genome Browser, https://genome.ucsc.edu
Vioplot, http://wsopuppenkiste.wiso.uni-goettingen.de/∼dadler
Supplemental Data
The number of uniquely mapped reads and mean coverage per CpG site are shown.
Regions selected for the targeted bisulfite sequencing are indicated. Names of the known gDMRs are also indicated.
Ten 1st-CT samples were analyzed. For each gDMR, the methylation levels of the maternal (M) and paternal (P) alleles and the [M − P] level are shown. The candidate mDMRs and pDMRs were ranked according to their [M − P] levels. The statistical significance (BH-corrected p value) of the methylation differences between parental alleles is shown. Associated genes are also indicated. All informative candidate gDMRs, including those that were not confirmed to be imprinted, are indicated. NA: not available.
Three 2nd- or term-CT samples were analyzed. For each gDMR, the methylation levels of the maternal (M) and paternal (P) alleles and the [M − P] value are shown. The candidate mDMRs and pDMRs were respectively ranked according to their [M − P] levels. The statistical significance (BH-corrected p value) of the methylation differences between parental alleles is shown. Associated genes are also indicated. All informative candidate gDMRs, including those that were not confirmed to be imprinted, are indicated. NA: not available.
Known gDMRs are classified as described in the Materials and Methods. For each gDMR, the methylation levels of the maternal (M) and paternal (P) alleles and the [M − P] level are shown. The statistical significance (BH-corrected p value) of the methylation differences between parental alleles is also indicated. NA: not available.
Known gDMRs are classified as described in the Materials and Methods. For each gDMR, the methylation levels of the maternal (M) and paternal (P) alleles and the [M − P] value are shown. The statistical significance (BH-corrected p value) of the methylation differences between parental alleles is also indicated. NA: not available.
For each gene, the proportion of reads derived from the maternal allele to total reads ([M-expression] ratio), [contamination] rate and FPKM are shown. The statistical significance (BH-corrected p value) of the allelic expression differences is also shown. NA: not available.
For each gene, the mean [M-expression] ratio, BH-corrected p value, and expression allele are shown. The numbers of samples with maternal (<35% [M-expression] ratios), paternal (>65% [M-expression] ratios) and biallelic (35%–65% [M-expression] ratios) expression are also indicated. Allelic DNA methylation patterns of associated gDMRs, which were obtained from the 1st-CT samples, are also included. Maternal methylation of the CYP2J2 DMR was previously reported.6 NA: Not available; M: maternal; P: paternal.
For each gene, the [M-expression] ratio, [Contamination] rate, and FPKM are shown. Genes in the pseudoautosomal regions are also indicated. NA: not available.
References
- 1.Saitou M., Kagiwada S., Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 2.Messerschmidt D.M., Knowles B.B., Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasaki H., Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson-Smith A.C. Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 5.John R.M., Surani M.A. Imprinted genes and regulation of gene expression by epigenetic inheritance. Curr. Opin. Cell Biol. 1996;8:348–353. doi: 10.1016/s0955-0674(96)80008-1. [DOI] [PubMed] [Google Scholar]
- 6.Proudhon C., Duffié R., Ajjan S., Cowley M., Iranzo J., Carbajosa G., Saadeh H., Holland M.L., Oakey R.J., Rakyan V.K. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol. Cell. 2012;47:909–920. doi: 10.1016/j.molcel.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Court F., Tayama C., Romanelli V., Martin-Trujillo A., Iglesias-Platas I., Okamura K., Sugahara N., Simón C., Moore H., Harness J.V. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Delgado M., Martin-Trujillo A., Tayama C., Vidal E., Esteller M., Iglesias-Platas I., Deo N., Barney O., Maclean K., Hata K. Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with NLRP7 Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting. PLoS Genet. 2015;11:e1005644. doi: 10.1371/journal.pgen.1005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanna C.W., Peñaherrera M.S., Saadeh H., Andrews S., McFadden D.E., Kelsey G., Robinson W.P. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 2016;26:756–767. doi: 10.1101/gr.196139.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okae H., Chiba H., Hiura H., Hamada H., Sato A., Utsunomiya T., Kikuchi H., Yoshida H., Tanaka A., Suyama M., Arima T. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 2014;10:e1004868. doi: 10.1371/journal.pgen.1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., Yan J., Ren X., Lin S., Li J. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 12.Smith Z.D., Chan M.M., Humm K.C., Karnik R., Mekhoubad S., Regev A., Eggan K., Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hon G.C., Rajagopal N., Shen Y., McCleary D.F., Yue F., Dang M.D., Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder D.I., Blair J.D., Lott P., Yu H.O., Hong D., Crary F., Ashwood P., Walker C., Korf I., Robinson W.P., LaSalle J.M. The human placenta methylome. Proc. Natl. Acad. Sci. USA. 2013;110:6037–6042. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroeder D.I., Jayashankar K., Douglas K.C., Thirkill T.L., York D., Dickinson P.J., Williams L.E., Samollow P.B., Ross P.J., Bannasch D.L. Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas. PLoS Genet. 2015;11:e1005442. doi: 10.1371/journal.pgen.1005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson W.P., Price E.M. The human placental methylome. Cold Spring Harb. Perspect. Med. 2015;5:a023044. doi: 10.1101/cshperspect.a023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob K.J., Robinson W.P., Lefebvre L. Beckwith-Wiedemann and Silver-Russell syndromes: opposite developmental imbalances in imprinted regulators of placental function and embryonic growth. Clin. Genet. 2013;84:326–334. doi: 10.1111/cge.12143. [DOI] [PubMed] [Google Scholar]
- 18.Frost J.M., Moore G.E. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher R.A., Hodges M.D. Genomic imprinting in gestational trophoblastic disease--a review. Placenta. 2003;24(Suppl A):S111–S118. doi: 10.1053/plac.2002.0939. [DOI] [PubMed] [Google Scholar]
- 20.Tomizawa S., Sasaki H. Genomic imprinting and its relevance to congenital disease, infertility, molar pregnancy and induced pluripotent stem cell. J. Hum. Genet. 2012;57:84–91. doi: 10.1038/jhg.2011.151. [DOI] [PubMed] [Google Scholar]
- 21.Kliman H.J., Nestler J.E., Sermasi E., Sanger J.M., Strauss J.F., 3rd Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology. 1986;118:1567–1582. doi: 10.1210/endo-118-4-1567. [DOI] [PubMed] [Google Scholar]
- 22.Bischof P., Irminger-Finger I. The human cytotrophoblastic cell, a mononuclear chameleon. Int. J. Biochem. Cell Biol. 2005;37:1–16. doi: 10.1016/j.biocel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Miura F., Enomoto Y., Dairiki R., Ito T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012;40:e136. doi: 10.1093/nar/gks454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura F., Ito T. Highly sensitive targeted methylome sequencing by post-bisulfite adaptor tagging. DNA Res. 2015;22:13–18. doi: 10.1093/dnares/dsu034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Miller D.C., Harman R., Antczak D.F., Clark A.G. Paternally expressed genes predominate in the placenta. Proc. Natl. Acad. Sci. USA. 2013;110:10705–10710. doi: 10.1073/pnas.1308998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang D.W., Sherman B.T., Tan Q., Collins J.R., Alvord W.G., Roayaei J., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O’Malley R., Castanon R., Klugman S. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirane K., Toh H., Kobayashi H., Miura F., Chiba H., Ito T., Kono T., Sasaki H. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013;9:e1003439. doi: 10.1371/journal.pgen.1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi H., Sakurai T., Imai M., Takahashi N., Fukuda A., Yayoi O., Sato S., Nakabayashi K., Hata K., Sotomaru Y. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgiades P., Ferguson-Smith A.C., Burton G.J. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- 36.Machanick P., Bailey T.L. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okae H., Hiura H., Nishida Y., Funayama R., Tanaka S., Chiba H., Yaegashi N., Nakayama K., Sasaki H., Arima T. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum. Mol. Genet. 2012;21:548–558. doi: 10.1093/hmg/ddr488. [DOI] [PubMed] [Google Scholar]
- 38.Das R., Lee Y.K., Strogantsev R., Jin S., Lim Y.C., Ng P.Y., Lin X.M., Chng K., Yeo G.Sh., Ferguson-Smith A.C., Ding C. DNMT1 and AIM1 Imprinting in human placenta revealed through a genome-wide screen for allele-specific DNA methylation. BMC Genomics. 2013;14:685. doi: 10.1186/1471-2164-14-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuen R.K., Jiang R., Peñaherrera M.S., McFadden D.E., Robinson W.P. Genome-wide mapping of imprinted differentially methylated regions by DNA methylation profiling of human placentas from triploidies. Epigenetics Chromatin. 2011;4:10. doi: 10.1186/1756-8935-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 41.Arai T., Kasper J.S., Skaar J.R., Ali S.H., Takahashi C., DeCaprio J.A. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:9855–9860. doi: 10.1073/pnas.1733908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gascoin-Lachambre G., Buffat C., Rebourcet R., Chelbi S.T., Rigourd V., Mondon F., Mignot T.M., Legras E., Simeoni U., Vaiman D., Barbaux S. Cullins in human intra-uterine growth restriction: expressional and epigenetic alterations. Placenta. 2010;31:151–157. doi: 10.1016/j.placenta.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 43.Herse F., Lamarca B., Hubel C.A., Kaartokallio T., Lokki A.I., Ekholm E., Laivuori H., Gauster M., Huppertz B., Sugulle M. Cytochrome P450 subfamily 2J polypeptide 2 expression and circulating epoxyeicosatrienoic metabolites in preeclampsia. Circulation. 2012;126:2990–2999. doi: 10.1161/CIRCULATIONAHA.112.127340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartolomei M.S., Ferguson-Smith A.C. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011;3:3. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreira de Mello J.C., de Araújo E.S., Stabellini R., Fraga A.M., de Souza J.E., Sumita D.R., Camargo A.A., Pereira L.V. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS ONE. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knickmeyer R.C. Turner syndrome: advances in understanding altered cognition, brain structure and function. Curr. Opin. Neurol. 2012;25:144–149. doi: 10.1097/WCO.0b013e3283515e9e. [DOI] [PubMed] [Google Scholar]
- 47.Monk D., Arnaud P., Apostolidou S., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. USA. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L., Zhang J., Duan J., Gao X., Zhu W., Lu X., Yang L., Zhang J., Li G., Ci W. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furukawa S., Kuroda Y., Sugiyama A. A comparison of the histological structure of the placenta in experimental animals. J. Toxicol. Pathol. 2014;27:11–18. doi: 10.1293/tox.2013-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The number of uniquely mapped reads and mean coverage per CpG site are shown.
Regions selected for the targeted bisulfite sequencing are indicated. Names of the known gDMRs are also indicated.
Ten 1st-CT samples were analyzed. For each gDMR, the methylation levels of the maternal (M) and paternal (P) alleles and the [M − P] level are shown. The candidate mDMRs and pDMRs were ranked according to their [M − P] levels. The statistical significance (BH-corrected p value) of the methylation differences between parental alleles is shown. Associated genes are also indicated. All informative candidate gDMRs, including those that were not confirmed to be imprinted, are indicated. NA: not available.
Three 2nd- or term-CT samples were analyzed. For each gDMR, the methylation levels of the maternal (M) and paternal (P) alleles and the [M − P] value are shown. The candidate mDMRs and pDMRs were respectively ranked according to their [M − P] levels. The statistical significance (BH-corrected p value) of the methylation differences between parental alleles is shown. Associated genes are also indicated. All informative candidate gDMRs, including those that were not confirmed to be imprinted, are indicated. NA: not available.
Known gDMRs are classified as described in the Materials and Methods. For each gDMR, the methylation levels of the maternal (M) and paternal (P) alleles and the [M − P] level are shown. The statistical significance (BH-corrected p value) of the methylation differences between parental alleles is also indicated. NA: not available.
Known gDMRs are classified as described in the Materials and Methods. For each gDMR, the methylation levels of the maternal (M) and paternal (P) alleles and the [M − P] value are shown. The statistical significance (BH-corrected p value) of the methylation differences between parental alleles is also indicated. NA: not available.
For each gene, the proportion of reads derived from the maternal allele to total reads ([M-expression] ratio), [contamination] rate and FPKM are shown. The statistical significance (BH-corrected p value) of the allelic expression differences is also shown. NA: not available.
For each gene, the mean [M-expression] ratio, BH-corrected p value, and expression allele are shown. The numbers of samples with maternal (<35% [M-expression] ratios), paternal (>65% [M-expression] ratios) and biallelic (35%–65% [M-expression] ratios) expression are also indicated. Allelic DNA methylation patterns of associated gDMRs, which were obtained from the 1st-CT samples, are also included. Maternal methylation of the CYP2J2 DMR was previously reported.6 NA: Not available; M: maternal; P: paternal.
For each gene, the [M-expression] ratio, [Contamination] rate, and FPKM are shown. Genes in the pseudoautosomal regions are also indicated. NA: not available.