Abstract
Analyzing genetic differences between closely related populations can be a powerful way to detect recent adaptation. The very large sample size of the UK Biobank is ideal for using population differentiation to detect selection and enables an analysis of the UK population structure at fine resolution. In this study, analyses of 113,851 UK Biobank samples showed that population structure in the UK is dominated by five principal components (PCs) spanning six clusters: Northern Ireland, Scotland, northern England, southern England, and two Welsh clusters. Analyses of ancient Eurasians revealed that populations in the northern UK have higher levels of Steppe ancestry and that UK population structure cannot be explained as a simple mixture of Celts and Saxons. A scan for unusual population differentiation along the top PCs identified a genome-wide-significant signal of selection at the coding variant rs601338 in FUT2 (p = 9.16 × 10−9). In addition, by combining evidence of unusual differentiation within the UK with evidence from ancient Eurasians, we identified genome-wide-significant (p = 5 × 10−8) signals of recent selection at two additional loci: CYP1A2-CSK and F12. We detected strong associations between diastolic blood pressure in the UK Biobank and both the variants with selection signals at CYP1A2-CSK (p = 1.10 × 10−19) and the variants with ancient Eurasian selection signals at the ATXN2-SH2B3 locus (p = 8.00 × 10−33), implicating recent adaptation related to blood pressure.
Introduction
Detecting signals of selection can provide biological insights into adaptations that have shaped human history.1, 2, 3, 4 Searching for genetic variants that are unusually differentiated between populations is a powerful way to detect recent selection on standing variation;5 this approach has been applied for detecting signals of selection linked to lactase persistance6, 7 (MIM: 223100), fatty-acid decomposition8 (MIM: 612795), hypoxia response9, 10, 11 (MIM: 609070), malaria resistance12, 13, 14 (MIM: 61162), and other traits and diseases.15, 16, 17, 18
Leveraging population differentiation to detect selection is particularly powerful in analyses of closely related subpopulations with large sample sizes.19 Here, we analyzed 113,851 samples of UK ancestry from the UK Biobank (see Web Resources) in conjunction with recently published datasets on People of the British Isles (PoBI)20 and ancient DNA21, 22, 23, 24 to draw inferences about population structure and recent selection. We employed a recently developed selection statistic that detects unusual population differentiation along continuous principal components (PCs) instead of between discrete subpopulations25 and combined our results with independent results from ancient Eurasians.23 We detected three interesting signals of selection and were able to show that genetic variants at these and previously reported23 signals of selection are strongly associated with diastolic blood pressure (DBP [MIM: 145500]) in UK Biobank samples.
Material and Methods
UK Biobank Dataset
The UK Biobank phase 1 data release contains 847,131 SNPs and 152,729 samples. We removed SNPs that were multi-allelic, had a genotyping rate less than 99%, or had a minor allele frequency (MAF) less than 1%. We also removed samples with non-British ancestry (including admixed samples) and samples with a genotyping rate less than 98%. This left 510,665 SNPs and 118,650 samples, a dataset that we call “QC∗.” Using PLINK226 (see Web Resources), we removed SNPs not in Hardy-Weinberg equilibrium (p < 10−6), and we pruned SNPs to have a linkage disequilibrium (LD) of r2 < 0.2. We then generated a genetic relationship matrix (GRM) and removed one of each pair of samples with relatedness greater than 0.05. This dataset, which we call “LD,” contained 210,113 SNPs and 113,851 samples. We combined the full set of SNPs from the QC∗ dataset and the set of unrelated samples from the LD dataset to produce the final “QC” dataset.
PoBI and POPRES Datasets
The UK PoBI dataset consisted of 2,039 samples from the 4,371 samples collected as part of the PoBI project.20 These 2,039 samples were a subset of 2,886 samples that had been genotyped on the Illumina Human 1.2M Duo genotyping chip, and after 2,510 passed quality-control procedures, 2,039 showed that all four grandparents had been born within 80 km of each other. This dataset allows us to examine the population genetics of the UK prior to the migrations of the late 19th and early 20th centuries. We also examined 2,988 European POPRES samples from the LOLIPOP and CoLaus collections.27 These samples were genotyped on the Affymetrix GeneChip 500K Array. The POPRES dataset allowed us to compare the UK Biobank population structure with that of continental Europe.
Ancient-DNA Datasets
Ancient DNA was gathered from several regions. Nine Steppe samples were collected from the Yamna oblast in Russia,22 seven western European hunter-gatherers were collected from Loschbour,21 26 Neolithic farmer samples were collected from the Anatolian region,22 and ten Saxon samples were collected from three sites in the UK.24 DNA was extracted from bone tissue, PCR amplified, and then purified by a hybrid capture approach.22, 23, 24 The resulting DNA was sequenced on an Illumina MiSeq, HiSeq, or NextSeq platform. Sequenced reads were aligned to the human genome (GRCh37) with the Burrows-Wheeler Aligner, and called SNPs were intersected with the SNPs found on the Human Origins Array.28
PC Analysis
We ran PC analysis (PCA) on the UK Biobank LD dataset by using the FastPCA software in EIGENSOFT25 (see Web Resources). We identified several artifactual PCs dominated by regions of long-range LD (Figure S1). Removing loci with significant or suggestive selection signals (Table S1) along with their flanking 1 Mb regions from the LD dataset and rerunning PCA eliminated these artifactual PCs (Figure S2). We refer to the resulting dataset with 202,486 SNPs and 113,851 samples as the “PC” dataset. This dataset allowed us to better capture axes of variation that correspond to population structure rather than artifacts due to LD.
PC Projection
We projected PoBI20 (642,288 SNPs and 2,039 samples from 30 populations), POPRES27 (453,442 SNPs and 4,079 samples from 60 populations), and ancient-DNA22, 23 (159,588 SNPs and 52 samples from 4 populations) samples onto the UK Biobank PCs via PC projection.29 The SNPs in the UK Biobank QC dataset were intersected with those in the projected dataset, and A/T and C/G SNPs were removed because of strand ambiguity (75,254, 37,593, and 24,467 SNPs for PoBI, POPRES, and ancient DNA, respectively). The intersected set of SNPs was stringently LD pruned for r2 < 0.05 with PLINK226 (see Web Resources), leaving 27,769, 20,914, and 15,722 SNPs for PoBI, POPRES, and ancient DNA, respectively. SNP weights were computed for the intersected set of SNPs, and these weights were then used for projecting the new samples onto the UK Biobank PCs.29
Analysis of Population Clusters
After running PCA, we clustered individuals by k-means clustering into six clusters on five PCs. We labeled clusters by comparing the centroids of each cluster with the centroids of the projected PoBI populations, as well as by visual inspection. We then analyzed these clusters by running TreeMix30, 31 with default settings (see Web Resources) in order to assess the hierarchical population structure between the clusters.
Pairwise Discrete Subpopulation-Based Selection Statistic
Although we focused primarily on the PCA-based selection statistic from Galinsky et al.25 (see below), we also applied the discrete subpopulation-based selection statistic from Bhatia et al.,19 which we briefly review here. Suppose two populations with genetic distance FST are descended from a single ancestral population. The allele frequencies at a particular SNP in these two populations (p1 and p2) follow a normal distribution such that , where is the ancestral allele frequency of this SNP. As a result, the difference in allele frequency also follows a normal distribution with mean 0 and variance . Thus, the sample allele-frequency difference , where and are the number of observed haplotypes from each population. By estimating and , we can assess the statistical significance of unusually large values of . We applied this statistic to pairs of population clusters.
PCA-Based Selection Statistic
We applied the PCA-based selection statistic from Galinsky et al.,25 which we briefly review here. PCA is equivalent to the singular-value decomposition , where is the normalized genomic matrix, is the matrix of left singular vectors, is the matrix of right singular vectors, and is a diagonal matrix of singular values. The singular values are related to the eigenvalues of the GRM by the relationship , where is the number of SNPs used for computing the GRM . The matrix has the properties and . By the central limit theorem, the elements of follow a normal distribution, and after rescaling by , they follow a chi-square (1 degree of freedom [df]) distribution. In other words, the statistic for the ith SNP at the kth PC follows a chi-square (1 df) distribution.25 One benefit of this statistic is that the PCs can be generated on one set of SNPs (here, we used the PC dataset described earlier in order to capture axes of variation related to true population structure), and the selection statistic can be calculated on another set of SNPs (we used the QC dataset in order to maximize the set of SNPs evaluated for signals of selection).
We clustered signals of selection by considering all SNPs for which the p value with respect to at least one PC was less than an initial threshold (which we set at 10−6) and by clustering together SNPs within 1 Mb of each other. SNPs with signals on different PCs but in close proximity were clustered together because loci often have signals of selection on multiple PCs. We defined genome-wide-significant loci on the basis of clusters that contained at least one SNP with a p value smaller than the genome-wide significance threshold. Because we analyzed 5 PCs and 510,665 SNPs, the genome-wide significance threshold was 0.05/(5 × 510,665) = 1.96 × 10−8. We defined suggestive loci on the basis of clusters with at least two SNPs crossing the initial threshold (but none crossing the genome-wide significance threshold).
Combined Selection Statistic
We intersected the chi-square (4 df) ancient Eurasian selection statistics for 1,004,613 SNPs from Mathieson et al.23 with the PC-based chi-square (1 df) UK Biobank selection statistics for 510,665 QC SNPs, producing a list of 115,066 SNPs. For each SNP and each PC, we added the ancient Eurasian selection statistics to the UK Biobank selection statistics for that PC, producing chi-square (5 df) statistics, which we corrected by using genomic control.
Association Tests
Association analyses were performed with PLINK226 with the top five PCs as covariates and the “--linear” or “--logistic” flags.
Results
Population Structure in the UK Biobank
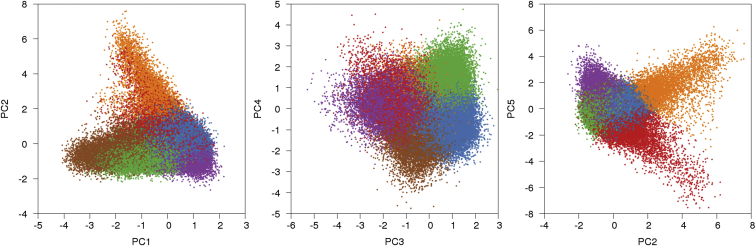
We restricted our analyses of population structure to 113,851 UK Biobank samples of UK ancestry and 202,486 SNPs after quality-control filtering and LD pruning (see Material and Methods). We ran PCA on this data by using our FastPCA implementation25 (see Web Resources). By visually examining plots of the top ten PCs (Figure S2) and observing that the eigenvalues for the top five PCs were above background levels, we determined that the top five PCs represent geographic population structure (Figure 1). PC1–PC4 were also strongly correlated with birth coordinate (Table S2). The eigenvalue for PC1 was 20.99, which corresponds to the eigenvalue that would be expected at this sample size for two discrete subpopulations of equal size with an FST of 1.76 × 10−4 (Table S2).
Figure 1.
Results of PCA with k-Means Clustering
The top five PCs in UK Biobank data are displayed. Samples were clustered with these PCs into six clusters by k-means clustering (see Table 1). PC5 is plotted against PC2 because PC5 primarily separated the orange and red clusters, which were separated from the other clusters by PC2.
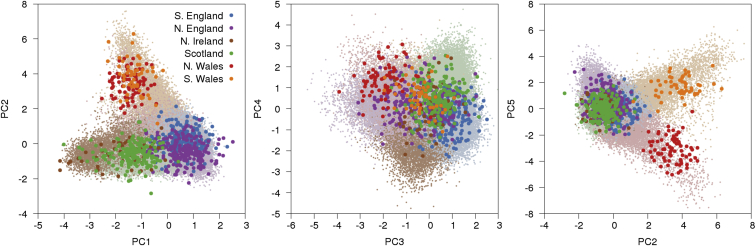
We ran k-means clustering on these five PCs to partition the samples into six clusters, given that PCs can differentiate populations (Figure 1, Table 1, and Figure S3). To identify the populations underlying the six clusters, we projected the PoBI dataset,20 comprising 2,039 samples from 30 regions of the UK, onto the UK Biobank PCs (Figure 2 and Figure S4). The individuals in the PoBI study were from rural areas of the UK and had all four grandparents born within 80 km of each other, allowing a glimpse into the genetics of the UK before the increase in mobility in the 20th century. We selected representative PoBI sample regions that best aligned with the six UK Biobank clusters by comparing centroids of each projected population region with those from the UK Biobank clusters via visual inspection (see Material and Methods and Table 1). The largest cluster represented southern and eastern England, three clusters represented different regions in the northern UK (northern England, Northern Ireland, and Scotland), and two clusters represented North and South Wales. The PCs separated the six UK clusters along two general geographical axes: a north-south axis and a Welsh-specific axis. PC1 and PC3 both separated individuals on north-south axes of variation, with southern England on one end and one of the northern UK clusters on the other. PC2 separated the Welsh clusters from the rest of the UK. PC4 separated the Scotland cluster from the Northern Ireland cluster. PC5 separated the North Wales and South Wales (also known as Pembrokeshire) clusters from each other. To confirm these clusterings, we ran TreeMix30, 31 on our UK Biobank clusters (Figure S5), as well as on the UK Biobank clusters and PoBI populations (Figure S6), and found that the Celtic subpopulations were grouped separately from the Saxon-related subpopulations; surprisingly, the North and South Wales clusters were separated by TreeMix, possibly because the North Wales cluster potentially contains Saxon-related samples (we note the low values between North Wales and southern England; see values in Table S6). Overall, our results were generally similar to those from the PoBI study;20 for example, both analyses identified a Welsh axis of differentiation (PC2) and split North and South Wales (PC5). We also observed some differences due to the different sampling schemes; in particular, the UK Biobank dataset contained many more Irish and Scottish samples (driving variation along PC1) and fewer Orkney samples, which affected the clustering of PoBI samples.
Table 1.
Correspondence between UK Biobank Clusters and PoBI Populations
| Color | Count | Cluster Name | PoBI Populations |
|---|---|---|---|
| Purple | 19,452 | northern England | Yorkshire, Lancashire |
| Blue | 41,494 | southern England | Hampshire, Devon, Norfolk |
| Brown | 12,895 | Northern Ireland | Northern Ireland |
| Green | 21,215 | Scotland | Argyll and Bute, Banff and Buchan, Orkney |
| Red | 14,190 | North Wales | North Wales |
| Orange | 4,605 | South Wales (Pembrokeshire) | northern and southern Pembrokeshire |
We report the PoBI population that most closely corresponds to each UK Biobank cluster (see main text).
Figure 2.
Results of PCA with Projection of PoBI Samples
The top five PCs in UK Biobank data are displayed with PoBI samples projected onto them. PoBI populations that visually best matched the clusters from k-means clustering were used for assigning names to the six clusters (Table 1).
We next analyzed UK Biobank population structure in conjunction with ancient DNA samples. Modern European populations are currently thought to have descended from three ancestral populations: Steppe, Mesolithic Europeans, and Neolithic farmers.21, 22 We projected ancient samples from these three populations as well as ancient Saxon samples24 onto the UK Biobank PCs (see Figure 3, Figure S7, and Material and Methods). These populations were primarily differentiated along PC1 and PC3, indicating higher levels of Steppe ancestry in northern UK populations. Additionally, the lack of any correlation between ancient samples and PC2 suggests that Welsh populations are not differentially admixed with any ancient population in our dataset and most likely underwent Welsh-specific genetic drift. We confirmed these findings by projecting pan-European POPRES27 samples onto the UK Biobank PCs (see Material and Methods and Figure S8). We note that the Irish and Scottish POPRES populations were projected on top of their corresponding UK Biobank population clusters. Of the continental European populations, Russians (who have the most Steppe ancestry) were projected farther in the Steppe direction along PC1 and PC3 than Spanish and Italians (who have the least Steppe ancestry22). Additionally, none of the continental European populations projected onto the same regions as the Welsh on PC2 and PC5. We ran TreeMix on the UK Biobank clusters and ancient populations (Figure S9) as well as the UK Biobank clusters and POPRES populations (Figure S10); although using the ancient populations to infer population structure was challenging, the UK Biobank samples were most closely grouped with the Scottish and Irish POPRES samples.
Figure 3.
Results of PCA with Projection of Ancient Samples
The top five PCs in UK Biobank data are displayed with ancient samples projected onto them.
In addition to the impact of ancient Eurasian populations migrating to the UK, we know that the genetics of the UK has been strongly affected by Anglo-Saxon migrations since the Iron Age,24 such that the Angles arrived in eastern England and the Saxons arrived in southern England. The Anglo-Saxons interbred with the native Celts, which explains much of the genetic landscape in the UK. We analyzed a variety of samples from predominantly Celtic (Scotland and Wales) and Anglo-Saxon (southern and eastern England) populations from modern Britain in conjunction with the PoBI samples20 and ten ancient Saxon samples from eastern England24 in order to assess the relative amounts of Steppe ancestry. We computed statistics28 of the form
where the Steppe and Neolithic farmer populations are from Lazaridis et al.21 and Haak et al.,22 population 1 (Pop1) is either a modern Celtic (Scotland or Wales) or ancient Saxon population, and population 2 (Pop2) is a modern Anglo-Saxon (southern and eastern England) population (Table 2 and Table S3). This statistic is sensitive to Steppe ancestry, such that positive values indicate more Steppe ancestry in Pop1 than in Pop2. We consistently obtained significantly positive statistics, implying that both the modern Celtic samples and the ancient Saxon samples have more Steppe ancestry than the modern Anglo-Saxon samples from southern and eastern England. This indicates that southern and eastern England are not exclusively a genetic mix of Celts and Saxons, which each have more Steppe ancestry. There are a variety of possible explanations, but one is that the present genetic structure of Britain, although subtle, is quite old and that southern England in Roman times already had less Steppe ancestry than Wales and Scotland.
Table 2.
Results of f4 Statistics in Ancient and Modern British Samples
| Grouping | Pop1 |
Pop2 |
||
|---|---|---|---|---|
| Hampshire | Devon | Norfolk | ||
| Ancient | Saxon | 2.543 | 3.732 | 5.118 |
| Scotland | Argyll and Bute | 3.323 | 6.223 | 9.560 |
| North Wales | North Wales | 1.918 | 5.239 | 8.490 |
| South Wales | northern Pembrokeshire | 1.759 | 4.430 | 7.124 |
We report f4 statistics of the form f4 (Steppe, Neolithic farmer; Pop1, Pop2), representing a Z score whose positive values indicate more Steppe ancestry in Pop1 than in Pop2. Samples for Pop1 were either modern Celtic (Scotland and Wales) or ancient Saxon. Samples for Pop2 were modern Anglo-Saxon (southern and eastern England).
Signals of Natural Selection
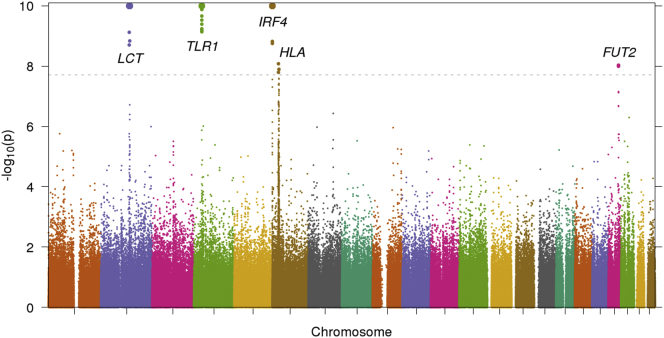
We searched for signals of selection by using a recently developed selection statistic that detects unusual population differentiation along continuous PCs.25 Notably, this statistic is able to detect selection signals at genome-wide significance. We analyzed the top five UK Biobank PCs (which were computed with LD-pruned SNPs) and computed selection statistics at 510,665 SNPs, reflecting the set of SNPs after quality control but before LD pruning (see Material and Methods). The Manhattan plot for PC1 is reported in Figure 4, and additional plots are provided in Figure S11. We detected genome-wide-significant signals of selection at FUT2 (MIM: 182100) and at several loci with widely known signals of selection (Table 3). Loci with suggestive signals of selection (p < 10−6) are reported in Table S4. FUT2 has also previously been reported as a target of natural selection;36, 37 those results focused on frequency differences between highly diverged continental populations, whereas our results implicate much more recent selection because UK Biobank populations diverged much more recently than continental populations. FUT2 encodes fucosyltransferase 2, an enzyme that affects the Lewis blood group. The SNP with the most significant p value, rs601338, is a coding variant where the variant rs601338∗G encodes the secretor allele and the rs601338∗A variant encodes the nonsecretor allele, which protects against the Norwalk norovirus.38, 39 This SNP also affects the progression of HIV infection40 (MIM: 609423) and is associated with vitamin B12 levels,41 Crohn disease42 (MIM: 266600), celiac disease (MIM: 212750), and inflammatory bowel disease,43 possibly because of changes in energy metabolism in the gut microbiome.44 rs601338∗A is more common in northern UK samples (Table S5). The GERA45 and PoBI20 datasets do not include rs601338 but exhibited similar allele-frequency patterns at rs492602 and rs676388 (Table S5), two linked FUT2 SNPs whose allele frequencies vary on a north-south axis in UK Biobank data. All three SNPs had genome-wide-significant signals of selection in the UK Biobank, and rs601338 and rs492602 were also genome-wide significant when we analyzed the six UK Biobank clusters described above with a selection test based on unusual differentiation between pairs of discrete subpopulations (Table S6). On the other hand, rs492602 and rs676388 were only suggestively significant (p < 1.00 × 10−6) in selection tests using the GERA dataset (Table S7), emphasizing the advantage of analyzing more closely related subpopulations in very large sample sizes in the UK Biobank dataset.
Figure 4.
Selection Statistics for UK Biobank along PC1
A Manhattan plot with −log10(p) values is displayed. Values above the significance threshold (dotted line, p = 1.96 × 10−8, α = 0.05 after correction for 5 PCs and 510,665 SNPs) are displayed as larger points and are labeled with the locus they correspond to (see Table 3). −log10(p) values larger than 10 are truncated at 10 for easier visualization and are displayed as even larger points.
Table 3.
Top Signals of Selection for UK Biobank along PC1–PC5
| Locus | Chromosome | Position (Mb) | PC | Top SNP | p Value |
|---|---|---|---|---|---|
| LCT6 | 2 | 134.9–137.2 | 1 | rs7570971 | 3.96 × 10−15 |
| TLR132 | 4 | 38.8–38.9 | 1 | rs4833095 | 7.96 × 10−15 |
| 2 | 1.27 × 10−8 | ||||
| 3 | 7.89 × 10−9 | ||||
| 4 | 1.54 × 10−11 | ||||
| IRF433, 34 | 6 | 0.4–0.5 | 1 | rs62389423 | 2.31 × 10−43 |
| HLA35 | 6 | 31.1–32.9 | 1 | rs9366778 | 8.45 × 10−9 |
| FUT2 | 19 | 49.2–49.2 | 1 | rs601338 | 9.16 × 10−9 |
We report the top signal of natural selection for each locus reaching genome-wide significance (1.96 × 10−8) along any of the top five PCs. Neighboring SNPs < 1 Mb apart with genome-wide-significant signals were grouped together into a single locus.
To detect additional signals of selection, we combined our PC-based selection statistics from the UK Biobank data with a previously described selection statistic that detects unusual allele-frequency differences after the admixture of ancient Eurasian populations by identifying SNPs whose allele frequencies are inconsistent with admixture proportions inferred from genome-wide data.23 For each of PC1–PC5 in the UK Biobank, we summed our chi-square (1 df) selection statistics for that PC with the chi-square (4 df) selection statistics from Mathieson et al.23 to produce chi-square (5 df) statistics combining these independent signals (see Material and Methods). We confirmed the independence of the two selection statistics by examining the correlations between the two selection statistics and checking that the combined statistics were not substantially inflated, obtaining λGC values of 1.04–1.06 (Table S8; see Figure S12 for a probability-probability plot). In order to produce maximally conservative statistics, we corrected our combined statistics with these λGC values. We looked for signals that were genome-wide significant in the combined selection statistic but not in either of the constituent UK Biobank or ancient Eurasian selection statistics. Results are reported in Table 4.
Table 4.
Top Signals of Selection for Combined Selection Statistics
| Locus | Chromosome | Position (Mb) | PC | Top SNP | Combined p Value | UK Biobank p Value | Ancient Eurasian p Value |
|---|---|---|---|---|---|---|---|
| F12 | 5 | 33.9–34.0 | 4 | rs2545801 | 2.05 × 10−10 | 3.99 × 10−5 | 5.35 × 10−8 |
| CYP1A2-CSK | 15 | 75.0–75.1 | 2 | rs1378942 | 4.65 × 10−8 | 1.05 × 10−2 | 1.08 × 10−7 |
Restricting to loci that were not genome-wide significant in either the UK Biobank or the ancient Eurasian selection statistics, we report the top selection statistic for each locus reaching genome-wide significance. Neighboring SNPs < 1 Mb apart with genome-wide significant signals were grouped together into a single locus.
We detected genome-wide-significant signals of selection at the F12 (MIM: 610619) and CYP1A2 (MIM: 124060)-CSK (MIM: 124095) loci. We are not currently aware of previous evidence of selection at F12. F12 codes for coagulation factor XII, a protein involved in blood clotting.46 The SNP at the F12 locus, rs2545801, was suggestively significant in the ancient Eurasian analysis (p = 5.35 × 10−8), and combining it with the UK Biobank selection statistic on PC2 produced a genome-wide-significant signal. This SNP has been associated with activated partial thromboplastin time, a measure of blood-clotting speed where shorter time is a risk factor for strokes.47 An additional significant SNP at F12, rs2731672, affects expression of F12 in the liver48 and is associated with plasma levels of factor XII.49 The CYP1A2-CSK locus has previously been reported as a target of natural selection in comparisons of inter-continental allele and haplotype frequencies,50, 51 but our results implicate much more recent selection. The two detected SNPs at this locus are in strong LD (r2 = 0.858). The top SNP, rs1378942, is in an intron in CSK. This SNP has a greatly varying allele frequency across continents51 and is associated with blood pressure52, 53 and systemic sclerosis54 (an autoimmune disease affecting connective tissue; MIM: 181750). The second SNP, rs2472304 in CYP1A2, is associated with esophageal cancer55 (MIM: 133239) and caffeine consumption56 and might mediate the protective effect of caffeine on Parkinson disease57 (MIM: 168600).
Using the top five PCs as covariates, we tested SNPs with genome-wide-significant signals of selection in the constituent UK Biobank or ancient Eurasian scans or the combined scan for association with 15 phenotypes in the UK Biobank dataset (see Table S9 and Material and Methods). The top SNP at F12 (rs2545801) was associated with height (p = 4.8 × 10−11), and the top SNP at CYP1A2-CSK (rs1378942) was associated with DBP (p = 3.6 × 10−19) and hypertension (p = 4.8 × 10−9), consistent with previous findings.58 We detected additional associations with DBP (p = 8.0 × 10−33) and hypertension (p = 1.3 × 10−9) at the ATXN2 (MIM: 601517)-SH2B3 (MIM: 605093) locus, which was reported as under selection in the ancient Eurasian scan. The top SNP in ATXN2-SH2B3, rs3184504, is known to be associated with blood pressure.59 We note that PC1 and PC3 were strongly associated with height in the UK Biobank dataset, and PC3 and PC4 were associated with DBP (Table S10). GRK460 (MIM: 137026), AGT60 (MIM: 106150), and ATP1A114 (MIM: 182310) have also been reported to be under selection and to be associated with DBP or hypertension. None of the SNPs in GRK4 or ATP1A1 were found to be under selection or associated with DBP or hypertension in our analyses. The AGT SNP rs699 was associated with DBP (p = 7.2 × 10−10) and nominally associated with hypertension (p = 4.8 × 10−4), although it did not produce a significant signal of selection in our analyses.
Discussion
In this study, we used PCA to analyze the population structure of a large UK cohort (N = 113,851). We detected five PCs representing geographic population structure that partitioned this cohort into six subpopulation clusters. Projecting ancient samples onto these PCs revealed greater Steppe ancestry in northern UK samples. No ancient samples were found to vary along the Welsh-specific axis, suggesting that the Welsh populations differ from the rest of the UK as a result of drift rather than different levels of admixture. We also determined that UK population structure cannot be explained as a simple mixture of Celts and Saxons.
We leveraged the subtle population structure and large sample size of the UK Biobank dataset to detect signals of natural selection. We determined that the rs601338∗A allele of FUT2 is more common in northern UK samples, suggesting that pathogens might have exerted selective pressure in those populations. Combining a selection statistic that detects selection via population differentiation within the UK with a separate statistic that detects selection since ancient population admixture in Europe, we were able to detect selection at two additional loci, F12 and CYP1A2-CSK. We additionally found associations with DBP at CYP1A2-CSK and at the ATXN2-SH2B3 locus, implicated in a previous selection scan.
We conclude by noting three limitations in our work. First, we employed PCA, a widely used method for analyzing population structure,25, 29, 61 but haplotype-based methods such as fineSTRUCTURE could be more powerful;20, 62, 63 recent advances in computationally efficient phasing64, 65 increase the prospects for applying such methods to biobank-scale data. Second, we employed methods designed to detect selection at individual loci but did not employ methods to detect polygenic selection;66, 67, 68, 69, 70 our observation that top PCs were correlated with height and DBP in the UK Biobank dataset, which could potentially be consistent with the action of polygenic selection on these traits, motivates further analyses of possible polygenic selection. Finally, the PC-based test for selection that we employed assumes that allele frequencies vary linearly along a PC. The spatial ancestry analysis (SPA) method71, 72, 73 allows for a logistic relationship between allele frequency and ancestry and is not constrained by this limitation. However, the advantage of the PC-based test for selection over SPA is that it provides an assessment of statistical significance (p values), allowing for the detection of genome-wide-significant signals, a key consideration in genome scans for selection.
Acknowledgments
We thank Iain Mathieson and David Reich for helpful discussions and Stephan Schiffels for technical assistance with Saxon samples. This research was conducted with the UK Biobank Resource (application numbers 10438 and 14292) and was funded by NIH grant R01 HG006399.
Published: October 20, 2016
Footnotes
Supplemental Data include 12 figures and 10 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.09.014.
Contributor Information
Kevin J. Galinsky, Email: galinsky@fas.harvard.edu.
Alkes L. Price, Email: aprice@hsph.harvard.edu.
Web Resources
EIGENSOFT v.6.1.3, http://www.hsph.harvard.edu/alkes-price/software
Genome Reference Consortium Human Build 37 (GRCh37), https://www.ncbi.nlm.nih.gov/assembly/2758/
OMIM, http://www.omim.org
TreeMix, https://bitbucket.org/nygcresearch/treemix/wiki/Home
UK Biobank, http://www.ukbiobank.ac.uk
Supplemental Data
References
- 1.Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z.P., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen R., Hellmann I., Hubisz M., Bustamante C., Clark A.G. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novembre J., Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat. Rev. Genet. 2009;10:745–755. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheinfeldt L.B., Tishkoff S.A. Recent human adaptation: genomic approaches, interpretation and insights. Nat. Rev. Genet. 2013;14:692–702. doi: 10.1038/nrg3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shriver M.D., Kennedy G.C., Parra E.J., Lawson H.A., Sonpar V., Huang J., Akey J.M., Jones K.W. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum. Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bersaglieri T., Sabeti P.C., Patterson N., Vanderploeg T., Schaffner S.F., Drake J.A., Rhodes M., Reich D.E., Hirschhorn J.N. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tishkoff S.A., Reed F.A., Ranciaro A., Voight B.F., Babbitt C.C., Silverman J.S., Powell K., Mortensen H.M., Hirbo J.B., Osman M. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fumagalli M., Moltke I., Grarup N., Racimo F., Bjerregaard P., Jørgensen M.E., Korneliussen T.S., Gerbault P., Skotte L., Linneberg A. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 9.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X.P., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bigham A., Bauchet M., Pinto D., Mao X., Akey J.M., Mei R., Scherer S.W., Julian C.G., Wilson M.J., López Herráez D. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorenzo F.R., Huff C., Myllymäki M., Olenchock B., Swierczek S., Tashi T., Gordeuk V., Wuren T., Ri-Li G., McClain D.A. A genetic mechanism for Tibetan high-altitude adaptation. Nat. Genet. 2014;46:951–956. doi: 10.1038/ng.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamblin M.T., Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 2000;66:1669–1679. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayodo G., Price A.L., Keinan A., Ajwang A., Otieno M.F., Orago A.S.S., Patterson N., Reich D. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am. J. Hum. Genet. 2007;81:234–242. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurdasani D., Carstensen T., Tekola-Ayele F., Pagani L., Tachmazidou I., Hatzikotoulas K., Karthikeyan S., Iles L., Pollard M.O., Choudhury A. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamason R.L., Mohideen M.-A.P.K., Mest J.R., Wong A.C., Norton H.L., Aros M.C., Jurynec M.J., Mao X., Humphreville V.R., Humbert J.E. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 16.Perry G.H., Dominy N.J., Claw K.G., Lee A.S., Fiegler H., Redon R., Werner J., Villanea F.A., Mountain J.L., Misra R. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock A.M., Witonsky D.B., Alkorta-Aranburu G., Beall C.M., Gebremedhin A., Sukernik R., Utermann G., Pritchard J.K., Coop G., Di Rienzo A. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 2011;7:e1001375. doi: 10.1371/journal.pgen.1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko W.-Y., Rajan P., Gomez F., Scheinfeldt L., An P., Winkler C.A., Froment A., Nyambo T.B., Omar S.A., Wambebe C. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am. J. Hum. Genet. 2013;93:54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatia G., Patterson N., Pasaniuc B., Zaitlen N., Genovese G., Pollack S., Mallick S., Myers S., Tandon A., Spencer C. Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection. Am. J. Hum. Genet. 2011;89:368–381. doi: 10.1016/j.ajhg.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leslie S., Winney B., Hellenthal G., Davison D., Boumertit A., Day T., Hutnik K., Royrvik E.C., Cunliffe B., Lawson D.J., Wellcome Trust Case Control Consortium 2. International Multiple Sclerosis Genetics Consortium The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P.H., Schraiber J.G., Castellano S., Lipson M. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiffels S., Haak W., Paajanen P., Llamas B., Popescu E., Loe L., Clarke R., Lyons A., Mortimer R., Sayer D. Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun. 2016;7:10408. doi: 10.1038/ncomms10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galinsky K.J., Bhatia G., Loh P.-R., Georgiev S., Mukherjee S., Patterson N.J., Price A.L. Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 2016;98:456–472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson M.R., Bryc K., King K.S., Indap A., Boyko A.R., Novembre J., Briley L.P., Maruyama Y., Waterworth D.M., Waeber G. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. 2008;83:347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickrell J.K., Pritchard J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickrell J.K., Patterson N., Barbieri C., Berthold F., Gerlach L., Güldemann T., Kure B., Mpoloka S.W., Nakagawa H., Naumann C. The genetic prehistory of southern Africa. Nat. Commun. 2012;3:1143. doi: 10.1038/ncomms2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heffelfinger C., Pakstis A.J., Speed W.C., Clark A.P., Haigh E., Fang R., Furtado M.R., Kidd K.K., Snyder M.P. Haplotype structure and positive selection at TLR1. Eur. J. Hum. Genet. 2014;22:551–557. doi: 10.1038/ejhg.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton P.R., Clayton D.G., Cardon L.R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D.P., McCarthy M.I., Ouwehand W.H., Samani N.J., Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickrell J.K., Coop G., Novembre J., Kudaravalli S., Li J.Z., Absher D., Srinivasan B.S., Barsh G.S., Myers R.M., Feldman M.W., Pritchard J.K. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Bakker P.I.W., McVean G., Sabeti P.C., Miretti M.M., Green T., Marchini J., Ke X., Monsuur A.J., Whittaker P., Delgado M. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrer-Admetlla A., Sikora M., Laayouni H., Esteve A., Roubinet F., Blancher A., Calafell F., Bertranpetit J., Casals F. A natural history of FUT2 polymorphism in humans. Mol. Biol. Evol. 2009;26:1993–2003. doi: 10.1093/molbev/msp108. [DOI] [PubMed] [Google Scholar]
- 37.Fumagalli M., Cagliani R., Pozzoli U., Riva S., Comi G.P., Menozzi G., Bresolin N., Sironi M. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome Res. 2009;19:199–212. doi: 10.1101/gr.082768.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorven M., Grahn A., Hedlund K.-O., Johansson H., Wahlfrid C., Larson G., Svensson L. A homozygous nonsense mutation (428G-->A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J. Virol. 2005;79:15351–15355. doi: 10.1128/JVI.79.24.15351-15355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlsson B., Kindberg E., Buesa J., Rydell G.E., Lidón M.F., Montava R., Abu Mallouh R., Grahn A., Rodríguez-Díaz J., Bellido J. The G428A nonsense mutation in FUT2 provides strong but not absolute protection against symptomatic GII.4 Norovirus infection. PLoS ONE. 2009;4:e5593. doi: 10.1371/journal.pone.0005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kindberg E., Hejdeman B., Bratt G., Wahren B., Lindblom B., Hinkula J., Svensson L. A nonsense mutation (428G-->A) in the fucosyltransferase FUT2 gene affects the progression of HIV-1 infection. AIDS. 2006;20:685–689. doi: 10.1097/01.aids.0000216368.23325.bc. [DOI] [PubMed] [Google Scholar]
- 41.Hazra A., Kraft P., Selhub J., Giovannucci E.L., Thomas G., Hoover R.N., Chanock S.J., Hunter D.J. Common variants of FUT2 are associated with plasma vitamin B12 levels. Nat. Genet. 2008;40:1160–1162. doi: 10.1038/ng.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGovern D.P.B., Jones M.R., Taylor K.D., Marciante K., Yan X., Dubinsky M., Ippoliti A., Vasiliauskas E., Berel D., Derkowski C., International IBD Genetics Consortium Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum. Mol. Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parmar A.S., Alakulppi N., Paavola-Sakki P., Kurppa K., Halme L., Färkkilä M., Turunen U., Lappalainen M., Kontula K., Kaukinen K. Association study of FUT2 (rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue Antigens. 2012;80:488–493. doi: 10.1111/tan.12016. [DOI] [PubMed] [Google Scholar]
- 44.Tong M., McHardy I., Ruegger P., Goudarzi M., Kashyap P.C., Haritunians T., Li X., Graeber T.G., Schwager E., Huttenhower C. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn’s disease risk polymorphism. ISME J. 2014;8:2193–2206. doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banda Y., Kvale M.N., Hoffmann T.J., Hesselson S.E., Ranatunga D., Tang H., Sabatti C., Croen L.A., Dispensa B.P., Henderson M. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1285–1295. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renné T., Schmaier A.H., Nickel K.F., Blombäck M., Maas C. In vivo roles of factor XII. Blood. 2012;120:4296–4303. doi: 10.1182/blood-2012-07-292094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang W., Schwienbacher C., Lopez L.M., Ben-Shlomo Y., Oudot-Mellakh T., Johnson A.D., Samani N.J., Basu S., Gögele M., Davies G. Genetic associations for activated partial thromboplastin time and prothrombin time, their gene expression profiles, and risk of coronary artery disease. Am. J. Hum. Genet. 2012;91:152–162. doi: 10.1016/j.ajhg.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Innocenti F., Cooper G.M., Stanaway I.B., Gamazon E.R., Smith J.D., Mirkov S., Ramirez J., Liu W., Lin Y.S., Moloney C. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerrero J.A., Rivera J., Quiroga T., Martinez-Perez A., Antón A.I., Martínez C., Panes O., Vicente V., Mezzano D., Soria J.-M., Corral J. Novel loci involved in platelet function and platelet count identified by a genome-wide study performed in children. Haematologica. 2011;96:1335–1343. doi: 10.3324/haematol.2011.042077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wooding S.P., Watkins W.S., Bamshad M.J., Dunn D.M., Weiss R.B., Jorde L.B. DNA sequence variation in a 3.7-kb noncoding sequence 5′ of the CYP1A2 gene: implications for human population history and natural selection. Am. J. Hum. Genet. 2002;71:528–542. doi: 10.1086/342260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding K., Kullo I.J. Geographic differences in allele frequencies of susceptibility SNPs for cardiovascular disease. BMC Med. Genet. 2011;12:55. doi: 10.1186/1471-2350-12-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S., Wellcome Trust Case Control Consortium Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabara Y., Kohara K., Kita Y., Hirawa N., Katsuya T., Ohkubo T., Hiura Y., Tajima A., Morisaki T., Miyata T., Global Blood Pressure Genetics Consortium Common variants in the ATP2B1 gene are associated with susceptibility to hypertension: the Japanese Millennium Genome Project. Hypertension. 2010;56:973–980. doi: 10.1161/HYPERTENSIONAHA.110.153429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin J.-E., Broen J.C., Carmona F.D., Teruel M., Simeon C.P., Vonk M.C., van ’t Slot R., Rodriguez-Rodriguez L., Vicente E., Fonollosa V., Spanish Scleroderma Group Identification of CSK as a systemic sclerosis genetic risk factor through Genome Wide Association Study follow-up. Hum. Mol. Genet. 2012;21:2825–2835. doi: 10.1093/hmg/dds099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Q., Ratnasinghe L.D., Hong H., Perkins R., Tang Z.-Z., Hu N., Taylor P.R., Tong W. Decision forest analysis of 61 single nucleotide polymorphisms in a case-control study of esophageal cancer; a novel method. BMC Bioinformatics. 2005;6(Suppl 2):S4. doi: 10.1186/1471-2105-6-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cornelis M.C., Monda K.L., Yu K., Paynter N., Azzato E.M., Bennett S.N., Berndt S.I., Boerwinkle E., Chanock S., Chatterjee N. Genome-wide meta-analysis identifies regions on 7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet. 2011;7:e1002033. doi: 10.1371/journal.pgen.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popat R.A., Van Den Eeden S.K., Tanner C.M., Kamel F., Umbach D.M., Marder K., Mayeux R., Ritz B., Ross G.W., Petrovitch H. Coffee, ADORA2A, and CYP1A2: the caffeine connection in Parkinson’s disease. Eur. J. Neurol. 2011;18:756–765. doi: 10.1111/j.1468-1331.2011.03353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong K.-W., Jin H.-S., Lim J.-E., Kim S., Go M.J., Oh B. Recapitulation of two genomewide association studies on blood pressure and essential hypertension in the Korean population. J. Hum. Genet. 2010;55:336–341. doi: 10.1038/jhg.2010.31. [DOI] [PubMed] [Google Scholar]
- 59.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., International Consortium for Blood Pressure Genome-Wide Association Studies. CARDIoGRAM consortium. CKDGen Consortium. KidneyGen Consortium. EchoGen consortium. CHARGE-HF consortium Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 61.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lawson D.J., Hellenthal G., Myers S., Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M., Lawson D., UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loh P.R., Palamara P.F., Price A.L. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 2016;48:811–816. doi: 10.1038/ng.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Connell J., Sharp K., Shrine N., Wain L., Hall I., Tobin M., Zagury J.F., Delaneau O., Marchini J. Haplotype estimation for biobank-scale data sets. Nat. Genet. 2016;48:817–820. doi: 10.1038/ng.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pritchard J.K., Pickrell J.K., Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pritchard J.K., Di Rienzo A. Adaptation - not by sweeps alone. Nat. Rev. Genet. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turchin M.C., Chiang C.W., Palmer C.D., Sankararaman S., Reich D., Hirschhorn J.N., Genetic Investigation of ANthropometric Traits (GIANT) Consortium Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 2012;44:1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berg J.J., Coop G. A population genetic signal of polygenic adaptation. PLoS Genet. 2014;10:e1004412. doi: 10.1371/journal.pgen.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson M.R., Hemani G., Medina-Gomez C., Mezzavilla M., Esko T., Shakhbazov K., Powell J.E., Vinkhuyzen A., Berndt S.I., Gustafsson S. Population genetic differentiation of height and body mass index across Europe. Nat. Genet. 2015;47:1357–1362. doi: 10.1038/ng.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang W.-Y., Novembre J., Eskin E., Halperin E. A model-based approach for analysis of spatial structure in genetic data. Nat. Genet. 2012;44:725–731. doi: 10.1038/ng.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baran Y., Quintela I., Carracedo A., Pasaniuc B., Halperin E. Enhanced localization of genetic samples through linkage-disequilibrium correction. Am. J. Hum. Genet. 2013;92:882–894. doi: 10.1016/j.ajhg.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baran Y., Halperin E. A Note on the Relations Between Spatio-Genetic Models. J. Comput. Biol. 2015;22:905–917. doi: 10.1089/cmb.2015.0080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.