Abstract
Intracranial aneurysms (IAs) are the result of focal weakness in the artery wall and have a complex genetic makeup. To date, genome-wide association and sequencing studies have had limited success in identifying IA risk factors. Distinct populations, such as the French-Canadian (FC) population, have increased IA prevalence. In our study, we used exome sequencing to prioritize risk variants in a discovery cohort of six FC families affected by IA, and the analysis revealed an increased variation burden for ring finger protein 213 (RNF213). We resequenced RNF213 in a larger FC validation cohort, and association tests on further identified variants supported our findings (SKAT-O, p = 0.006). RNF213 belongs to the AAA+ protein family, and two variants (p.Arg2438Cys and p.Ala2826Thr) unique to affected FC individuals were found to have increased ATPase activity, which could lead to increased risk of IA by elevating angiogenic activities. Common SNPs in RNF213 were also extracted from the NeuroX SNP-chip genotype data, comprising 257 FC IA-affected and 1,988 control individuals. We discovered that the non-ancestral allele of rs6565666 was significantly associated with the affected individuals (p = 0.03), and it appeared as though the frequency of the risk allele had changed through genetic drift. Although RNF213 is a risk factor for moyamoya disease in East Asians, we demonstrated that it might also be a risk factor for IA in the FC population. It therefore appears that the function of RNF213 can be differently altered to predispose distinct populations to dissimilar neurovascular conditions, highlighting the importance of a population’s background in genetic studies of heterogeneous disease.
Introduction
Intracranial aneurysms (IAs [MIM: 105800]) are vascular abnormalities characterized by dilations or ballooning of intracranial arteries. Their pathology is not clear, but lesions are usually characterized by very thin (or absent) tunica media and internal elastic lamina within the arterial walls at IA sites.1 The worldwide prevalence of IAs is 1%–3%.2, 3 Moreover, this condition entails severe consequences, particularly when subarachnoid hemorrhages (SAHs) occur (incidence rate = 0.5%–1%4) because 30%–45% of SAHs are fatal within 30 days.5 The annual incidence of SAHs is 4–9 per 100,000 worldwide, and 80% of all spontaneous SAHs are caused by rupture of the aneurysm.6 The formation and rupture of IAs are related to complex risk factors, including smoking and alcohol consumption, hypertention,7 and other vascular diseases,8 a very small percentage of which are related to infections9 and trauma.10 Many studies have provided evidence suggesting that a large fraction of IA and SAH cases involve underlying genetic risk factors. First-degree relatives of individuals with aneurysmal SAHs have a 4- to 7-fold higher risk of being affected than the general population.11 Several genome-wide association studies (GWASs) have examined sporadic IAs, and these have identified several candidate loci (e.g., 2q33.1, 8q11.23, and 9p23.1;12, 13 18q11.2, 13q13.1, and 10q24.32;14 7p21.1;15 regions near rs1930095, rs7781293, rs7550260, and rs9864101;16 and 4q31.2217). Unfortunately, follow-up replication studies suggest that these loci are unlikely to explain a large fraction of IAs. It has been reported that the French-Canadian (FC) population has higher IA and SAH incidence rates, and affected individuals usually aggregate in families, especially large pedigrees.18 In a study conducted in the Saguenay-Lac-Saint-Jean region of Québec, it was observed that 30% of IA-affected individuals had a family history.19 It is believed that IAs are genetically heterogeneous, and therefore population-specific variants might play a big part in their pathogenesis, especially in familial aggregates. In order to explore those hypotheses, we conducted whole-exome sequencing (WES) and target sequencing experiments in a group of familial IA subjects of FC descent. We focused our attention on the identification of rare, or FC-specific, variations as genetic factors that might explain the high prevalence and familial aggregation of IAs in this population.
Material and Methods
Discovery Cohort
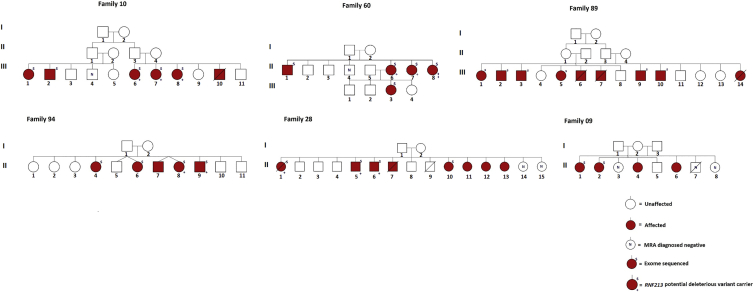
The discovery cohort of this study included 26 FC individuals (from six families) who had received a positive IA diagnosis (Figure 1); diagnostics were confirmed either by magnetic resonance angiography (1.5T) or by surgical confirmation (clipped or coiled). Subjects were excluded if they had received a personal diagnosis or had a history of polycystic kidney disease (MIM: 173900), Ehlers-Danlos syndrome type IV (MIM: 130050), neurofibromatosis type 1 (MIM: 162200), Marfan syndrome (MIM: 154700), or other intracranial vascular malformations, including moyamoya disease (MMD [MIM: 607151]).
Figure 1.
Six Pedigrees with 26 IA-Affected Individuals from the Initial Cohort
Written consent was obtained for all the participants, and the study was approved by the Comité d’Éthique de la Recherche du Centre Hospitalier de l’Université de Montréal (Notre Dame Hospital 04.101, Québec).
Control Population Cohort
Exome control populations for this study were selected from our in-house exome database, which includes 189 healthy FC individuals. Additionally, SNP-chip genotype data for 1,988 unrelated FC control individuals with no obvious cerebrovascular disease were also included in this study.
WES and Variant Prioritization
Genomic DNA for each individual was extracted from peripheral-blood lymphocytes with the Gentra Puregene Blood Kit (QIAGEN). A 50 μL DNA sample at a concentration of 100 ng/μL from each sample was captured by the Agilent SureSelect V4 Capture Kit. The library was subsequently paired-end sequenced with 100 bp reads on an Illumina HiSeq 2000; three samples per lane were used to ensure an average coverage depth of 100×. All high-throughput and Sanger sequencing was performed at the McGill University and Génome Québec Innovation Centre.
Raw fastq files were aligned to the human reference sequence (NCBI Genome build GRCh37) with the Burrows-Wheeler Aligner (BWA),20 and all PCR duplicates were removed from the alignments. The aligned reads were converted to binary format for further analysis with SAMtools.21 Single-nucleotide variants and indels were called with the UnifiedGenotyper from the Genome Analysis Toolkit (GATK)22 v.2.7. Variant annotation was performed with ANNOVAR23 with references to GRCh37, dbSNP version 132, the 1000 Genomes Project (1KGP, 2012 data release),24 69 Complete Genomics (2012 update), and the NHLBI Exome Sequencing Project Exome Variant Server (EVS), including ∼6,500 exomes (2013 update). Finally, variant segregation analysis was performed with an in-house segregation program, and more than 1,000 exomes of different ethnicities from our lab were used as controls.
The sequencing quality was determined with GATK’s DepthOfCoverage Walker. Quality control (QC) excluded variants with a sequencing depth < 10 or a genotype quality < 90 and variants present in a selection of frequently mutated genes, pseudo-genes, and genes in repetitive regions. After QC, preliminary analysis focused on variants that potentially affect protein function (missense and splice variants and small indels). Then, variants from all aforementioned databases were removed so the analysis could focus on FC-specific variants. PolyPhen-225 and GERP++26 were used for predicting the deleterious level of the functional variants.
Subsequently, FC-specific, potentially deleterious variants shared by two or more affected individuals from each family were prioritized for further analysis, and genes harboring two or more of those variants were further selected as genes of interest.
Variants from these genes of interest in 106 FC control exomes were selected according to the same criteria, and a gene-based burden test using the variable-threshold (VT) method27 implemented in Variant Association Tools (VAT)28 was performed on these genes.
Validation Cohort and Target Resequencing
After variant prioritization and selection of the most promising candidate gene, the whole coding regions of two isoforms of RNF213 (ring finger protein 213 [MIM: 613768; GenBank: NM_020954 and NM_001256071]) were sequenced in a validation cohort of 223 IA-affected individuals and 88 IA-negative control individuals. 74 of the affected subjects had sporadic IAs, and the rest had a family history. The mean age of onset for familial IAs was 53.5 ± 12 years, whereas that for sporadic IAs was 55.4 ± 9 years. Of the individuals with detailed clinical information, 37.4% with a family history and 29% without a family history were indicated to have multiple aneurysms. On the other hand, 40.8% of subjects with familial IAs were indicated to have hypertension, whereas 47.7% of subjects with sporadic IAs were shown to have high blood pressure. Other environmental factors, such as drinking (22.4% familial versus 40% sporadic) and smoking (60.5% familial versus 76.9% sporadic), also seemed to play a larger part in sporadic IAs (Table S1). Targeted resequencing was performed with the Fluidigm Access Array System. 122 pairs of primers, each under 300 bp in length, were designed to cover a total of 58 exons of RNF213 (Table S2). 50 ng of DNA for each sample was used for multiplex PCR, and products were paired-end sequenced on an Illumina MiSeq platform.
Bioinformatic analysis of the Fluidigm sequencing data was performed to first remove bad QC reads and adapter sequences. Subsequently, the reads were aligned to GRCh37 with the BWA v.0.7.5,29 and alignments of each sample were combined. The GATK UnifiedGenotyper22 was used for variant calling in the coding intervals of RNF213. The annotation and segregation steps were followed in a manner similar to that for WES. All variants were validated by the Integrative Genomics Viewer (IGV),30 and rare (minor allele frequency [MAF] ≤ 0.01) missense variants were further validated by Sanger sequencing.
Genotyping RNF213 Common SNPs
SNP genotyping data from the NeuroX SNP-chip (Illumina Human OmniExpress bead chip as backbone), which contains 719,885 SNPs, was obtained for 257 IA-affected and 1,988 control individuals. Principle-component analysis (PCA) was performed with smartPCA software (implemented in EIGENSOFT)31 to confirm the FC ethnicity of the samples used, and SNPs within the RNF213 region were extracted from the genome-wide data. Additionally, RNF213 common SNPs from 33 different populations were also obtained from our in-house database, the Gene Expression Omnibus (GEO),32, 33, 34, 35 and 1KGP phase 3 data24 (Table S3).
Statistical and Population Analysis
For further selection of the genes that are most likely to be risk candidates for IA, a gene-based burden test using the VT method27 implemented in VAT28 was performed for genes prioritized in the initial cohort.
The Sequence Kernel Association Test (SKAT)36 was used to evaluate the effect of RNF213 variants with weighted scores; this unified statistical test allows both rare and common variants to contribute to the overall statistic by calculating the logistic weights for each variant and applying them to the analysis according to the following formula:
The SKAT optimal test (SKAT-O) was then used to evaluate the statistical significance of RNF213 functional variants between affected and control individuals.
A Cochran-Armitage trend test was performed with Plink 1.937 to compare the allele distributions of all RNF213 common SNPs in IA-affected and control individuals; an adjustment was made for multiple testing. Pairwise FST was calculated with Arlequin 3.538 between different populations to measure the population differentiation according to two different sets of their RNF213 variations: (1) RNF213 common SNPs from FC IA subjects, control individuals, and 33 other populations and (2) RNF213 functional variants from FC IA subjects, control individuals, and 25 populations from 1KGP phase 3 data. Variation burden of RNF213 between FC and 1KGP phase 3 populations was also calculated by the VT method27 implemented in VAT.28
Expression-Vector Design
The RNF213 full-length cDNA cloned into pcDNA3.1+ was obtained from a previous study.39 Two AAA+ module fragments (amino acids 2,370–2,632 and 2,717–3,004) were individually amplified with Gibson-assembly-adapted primers; the first primer pair was 5′-GGT TCC GCG TGG ATC CCC GGA ATT Cgt gcc ctt caa tgt cga ctt tga taa ac-3′ and 5′-GTC AGT CAC GAT GCG GCC GCT CGA Ggc gag tcc cgt ttt cat cta gg-3′, and the second primer pair was 5′-GGT TCC GCG TGG ATC CCC GGA ATT Cag cag gct gct tct gga tg-3′ and 5′-GTC AGT CAC GAT GCG GCC GCT CGA Gtc tat ttg aag cct ttg ctg cag caa aga cc-3′. The two AAA+ amplified fragments were assembled and cloned into the EcoRI and XhoI sites of the pGEX-4T1 vector (GE Healthcare) with the Gibson assembly method.40 All clones were sequence validated.
Mutagenesis
PCR site-directed mutagenesis was performed to introduce two variants, p.Arg2438Cys and p.Ala2826Thr, into the first and second AAA+ plasmids, respectively. Four additional previously described41 variants (p.Lys2426Ala, p.Glu2488Ala, p.Lys2775Ala, and p. Glu2845Ala), which are known to cause loss of function (LOF) of the AAA+ ATPase, were also introduced as controls.
Protein Purification
The wild-types, variants, and LOF controls of the two AAA+ fragments and empty pGEX-4T1 plasmid were transformed into BL21-CodonPlus cells, the protein was inducted at 16°C overnight, and the N-terminal glutathione S-transferase (GST)-tagged protein was extracted with Glutathione Sepharose 4B beads (GE Healthcare) in PBS buffer containing PMSF protease inhibitor. PBS was carefully removed after protein extraction, and GST was eluted with buffer containing 50 mM Tris-HCl and 10 mM reduced glutathione. A Bradford protein assay was used for protein quantification.
ATPase Assay
An ATPase assay was performed with BIOMOL Green (Enzo Life Sciences) for the detection of free phosphate. The GST-tagged wild-types, variants, and LOF controls were incubated in a 50 μL buffer containing 300 mM KCl, 10 mM MgCl2, 50 mM HEPES-KOH (PH 7.5), and 5 mM ATP at 37°C for 20–40 min. The reaction was terminated by the addition of 100 μL BIOMOL Green reagent and incubated at room temperature for 10 min. OD630nm was measured for the wild-types, variants, and LOF controls, and a standard curve was made with phosphate standard according to the manufacturer’s protocol. ATPase activity (Vmax) was measured by PO4 quantity (nmol) per mg of protein per minute of reaction.
Results
Variant-Segregation Analysis and RNF213 Recurrent Mutations
The average base depth of coverage of the 26 IA samples from the discovery cohort was 101×, and 89.5% of the total target region was covered at 20× (Table S4). After QC and variant prioritization, 79, 109, 103, 15, 99, and 67 FC-specific deleterious variants segregated across more than two affected individuals in each of the six families (Table 1, row G). 35 of these variants defined 19 genes with variants in more than one family, and 11 of these 19 contained 25 distinct variants (Table 2). Interestingly, one of the 11 genes, RNF213, revealed five distinct variants in four families (Table 2 and Figure 1). These RNF213 variants (GenBank: NM_001256071.2) included a nonsense mutation (c.11413del [p.Glu3806Argfs∗27]), an exonic splicing mutation (c.2017C>T [p.Arg673Trp]), and three missense mutations (c.13577T>C [p.Ile4526Thr], c.6980A>G [p.Asn2327Ser], and c.3134C>T [p.Ser1045Leu] [GenBank: NM_020954.3 and NP_066005.2]). Only the c.6980A>G and c.3134C>T variants were observed once each in 189 FC control individuals. Variation burden of the 11 genes is displayed in Table 3. After Bonferroni correction for multiple testing, only RNF213 still differed significantly between affected and control individuals (p = 0.013).
Table 1.
Exome-Variant Filtration Steps and Results of Initial Cohort
| Family 60 | Family 89 | Family 10 | Family 9 | Family 28 | Family 94 | |
|---|---|---|---|---|---|---|
| QC Filtration Steps | ||||||
| A: variants in at least one affected family | 33,061 | 38,609 | 37,706 | 31,975 | 33,090 | 32,727 |
| B: functional variants | 18,455 | 21,857 | 21,255 | 17,602 | 18,417 | 18,118 |
| C: FC-specific variants | 1,489 | 2,041 | 1,797 | 825 | 1,089 | 882 |
| D: variants after exclusion of most variable exonic genes | 999 | 1,388 | 1,157 | 595 | 670 | 627 |
| E: variants in fewer than five samples from in-house database | 403 | 581 | 509 | 309 | 360 | 294 |
| F: variants in at least two affected families | 178 | 258 | 226 | 50 | 236 | 201 |
| G: deleterious variantsa (genes) | 79 (78) | 109 (108) | 103 (102) | 15 (15) | 99 (90) | 67 (64) |
| H: genes with cross-family recurrent variants (variants) | 6 (6) | 7 (7) | 6 (6) | 2 (2) | 8 (9) | 11 (13) |
| Results of Initial Cohort for All Six Families | ||||||
| Genes with cross-family recurrent variants (variants) | 19 (35) | |||||
| Genes with different variants | 11 | |||||
| Genes with at least two different variants | 1 (RNF213)b | |||||
PP2 > 0.8 or GERP > 2 and frameshift.
p = 0.01 (VT test) after Bonferroni correction (5,000 permutations).
Table 2.
Prioritized Genes with FC-Specific Deleterious Variants
| Gene (MIM) | Position |
Variant (GenBank) |
Family | PolyPhen-2 Score | GERP++ Score | |
|---|---|---|---|---|---|---|
| cDNA | Amino Acid | |||||
| ABCA10 (612508) | chr17: 67,215,903 | c.313A>G (NM_080282) | p.Asn105Asp (NP_525021) | 60 | 0.995 | 1.83 |
| chr17: 67,221,498 | c.2T>C (NM_080282) | p.Met1Thr (NP_525021) | 94 | 0.998 | 3.6 | |
| AIM1 (601797) | chr6: 107,008,779 | c.4733G>T (NM_001624) | p.Gly1578Val (NP_001615) | 89 | 1 | 3.4 |
| chr6: 106,968,391 | c.2084C>A (NM_001624) | p.Ser695Tyr (NP_001615) | 94 | 0.966 | 5.62 | |
| CDAN1 (607465) | chr15: 43,028,521 | c.548C>T (NM_138477) | p.Ser183Leu (NP_612486) | 89 | 0.127 | 3.91 |
| chr15: 43,023,250 | c.1880C>T (NM_138477) | p.Ala627Val (NP_612486) | 28 | 0.007 | 5.49 | |
| GPATCH8 (614396) | chr17: 42,477,055 | c.2390G>A (NM_001002909) | p.Arg797Gln (NP_001002909) | 28 | 0.658 | 2.67 |
| chr17: 42,475,271 | c.4174G>A (NM_001002909) | p.Gly1392Ser (NP_001002909) | 10 | 0.98 | 4.57 | |
| HELZ2 (611265) | chr20: 62,196,325 | c.2143C>T (NM_033405) | p.Arg715Trp (NP_208384) | 28 | 0.858 | 4.11 |
| chr20: 62,195,122 | c.3346C>T (NM_033405) | p.Arg1116Trp (NP_208384) | 60 | 1 | −0.842 | |
| RNF213 (613768) | chr17: 78,272,125 | c.2017C>T (NM_001256071) | p.Arg673Trp (NP_001243000) | 28 | 0.997 | 0.06 |
| chr17: 78,319,115 | c.6980A>G (NM_001256071) | p.Asn2327Ser (NP_001243000) | 60 | 0.006 | 2.19 | |
| chr17: 78,293,222 | c.3134C>T (NM_020954.3) | p.Ser1045Leu (NP_066005.2) | 10 | 0.496 | 2.628 | |
| chr17: 78,353,451 | c.13577T>C (NM_001256071) | p.Ile4526Thr (NP_001243000) | 94 | 0.291 | 2.81 | |
| chr17: 78,336,958 | c.11413del (NM_001256071) | p.Glu3806Argfs∗27 (NP_001243000) | 60 | – | – | |
| OR11H1 | chr22: 16,449,405 | c.400G>A (NM_001005239) | p.Ala134Thr (NP_001005239) | 89 | 0.852 | 0.664 |
| chr22: 16,449,399 | c.406G>A (NM_001005239) | p.Asp136Asn (NP_001005239) | 10 | 0.988 | 1.84 | |
| PLEC (601282) | chr8: 144,998,555 | c.5542C>T (NM_201384) | p.Leu1848Phe (NP_958786) | 60 | 0.726 | 4.06 |
| chr8: 144,996,050 | c.7939G>T (NM_201384) | p.Asp2647Tyr (NP_958786) | 89 | 0.992 | 0.966 | |
| RTTN (610436) | chr18: 67,806,837 | c.2786G>A (NM_173630) | p.Arg929Lys (NP_775901) | 94 | 0.985 | 4.96 |
| chr18: 67,742,698 | c.4454A>G (NM_173630) | p.His1485Arg (NP_775901) | 89 | 0.043 | 2.92 | |
| SF3A2 (600796) | chr19: 2,248,232 | c.1082C>G (NM_007165) | p.Ala361Gly (NP_009096) | 28 | 0.474 | 2.57 |
| chr19: 2,245,459 | c.260C>T (NM_007165) | p.Ala87Val (NP_009096) | 94 | 0.024 | 4.54 | |
| ZNF335 (610827) | chr20: 44,577,719 | c.3902C>T (NM_022095) | p.Ala1301Val (NP_071378) | 60 | 0.201 | 4.91 |
| chr20: 44,596,594 | c.593A>G (NM_022095) | p.Asp198Gly (NP_071378) | 10 | 0.977 | 3.92 | |
Table 3.
Mutation Burden of 11 Prioritized Genes between FC IA-Affected and Control Individuals
| Gene |
Risk/Total Allelesa |
VT Statistic | p Value (Unadjusted) | SE | Permutations | |
|---|---|---|---|---|---|---|
| Affected Individuals | Control Individuals | |||||
| ABCA10 | 2/36 | 2/212 | 0.709677 | 0.0775845 | 0.347336 | 5,000 |
| AIM1 | 2/36 | 3/212 | 0.569837 | 0.126873 | 0.339132 | 1,000 |
| CDAN1 | 2/36 | 2/212 | 0.709677 | 0.0771846 | 0.346911 | 5,000 |
| GPATCH8 | 2/36 | 1/212 | 0.903274 | 0.0259948 | 0.345287 | 5,000 |
| HELZ2 | 2/36 | 4/212 | 0.569837 | 0.135864 | 0.340307 | 1,000 |
| OR11H1 | 2/36 | 3/212 | 1.20892 | 0.0195961 | 0.398189 | 5,000 |
| PLEC | 2/36 | 15/212 | 0.121577 | 0.532468 | 0.376399 | 1,000 |
| RNF213 | 5/36 | 4/212 | 1.50576 | 0.00119976b | 0.354706 | 5,000 |
| RTTN | 2/36 | 5/212 | 0.371868 | 0.260739 | 0.344665 | 1,000 |
| SF3A2 | 2/36 | 2/212 | 0.903274 | 0.0313937 | 0.325226 | 5,000 |
| ZNF335 | 2/36 | 1/212 | 0.903274 | 0.0269946 | 0.378224 | 5,000 |
Risk alleles are those harboring FC-specific deleterious variants; total alleles include the founder alleles of six IA-affected families.
Corrected p = 0.013.
Target Resequencing
After Fluidigm capture and sequencing, the average coverage for the RNF213 targeted region of each individual ranged between 6,828× and 227,160×. Only 4% of individuals had ≤70% of the total target region covered at less than 100×.
272 exonic variants were first observed across the validation cohort of 311 samples (223 affected and 88 control individuals), and after IGV examination, 142 variants were used for the study (the remaining variants were deemed to be false positives because they were either located in adapters or introduced by PCR duplicates). Non-coding and synonymous variants were further removed from the 142 variants, leaving 60 variants for the final analysis. Additionally, 44 functional RNF213 variants were also found in 189 FC population control individuals.
SKAT Analysis and RNF213 Rare Variants in FC Individuals
In total, 72 functional (missense, indel, and splice) RNF213 variants were observed in FC individuals, including 233 IA subjects (10 from nuclear families of the discovery cohort and 223 from the validation cohort) and 277 control individuals (88 from the validation cohort and 189 from exome control populations) (Table S5). Hardy-Weinberg equilibrium (HWE) and a missingness test were performed with PLINK 1.9, but no variant was removed.
The SKAT-O test using logistic weights to compare RNF213 functional variants showed significantly different allelic distributions between the 233 FC IA-affected and 277 control individuals (p = 0.008), and the difference was even more significant when only rare functional variants (p = 0.006) were compared. When FC affected individuals were compared to only 88 IA-negative controls, the difference remained significant (p = 0.018); adding sex as a covariate slightly increased the difference (p = 0.013).
It is also noteworthy that among the 34 rare functional RNF213 variants that were observed in 233 IA subjects, 18 were absent from all control individuals (Table S5), and 13 of these 18 were predicted to be probably deleterious (PolyPhen-2 score > 0.8 or GERP++ score > 2) (Table 4). On the other hand, 30 rare functional variants were observed in 277 control individuals, and 14 (only 4 of which were predicted to be deleterious) were absent from affected individuals (Table S5 and Table 4). Hence, there is a significant difference between the number of deleterious variants observed in affected and control individuals (p = 0.00006, binomial test). 25 FC IA individuals from 249 discovery and validation cohorts carry 14 rare deleterious RNF213 variants (13 plus c.11413del [p.Glu3806Argfs∗27]) that are absent in FC control individuals. Of these 25 individuals, 9 had multiple IAs, 8 had SAHs, 8 had hypertension, and 3 had hypercholesterolemia; 2 of these last 3 also had myocardial infarctions. Only 6 individuals had no other recorded clinical histories besides smoking.
Table 4.
RNF213 Deleterious Variants Found Exclusively in Affected or Control Individuals
| Position | Variant | Affected Individuals (n = 233) | Control Individuals (n = 277) | PolyPhen-2 Score | GERP++ Score | Frequency in EVS |
|---|---|---|---|---|---|---|
| 78,264,463 | c.1208_1210del (p.403_404del) | 1 | 0 | 1 | 0 | 0.001917 |
| 78,268,746 | c.1699A>G (p.Met567Val) | 1 | 0 | 0.999 | 2.72 | 0 |
| 78,272,125 | c.2017C>T (p.Arg673Trp) | 3 | 0 | 0.997 | 0.06 | 0 |
| 78,305,962 | c.3674A>G (p.Asp1225Gly) | 1 | 0 | 0.919 | 0 | 0 |
| 78,319,447 | c.7312C>T (p.Arg2438Cys) | 1 | 0 | 0.655 | 2.35 | 0.000077 |
| 78,320,611 | c.8476G>A (p.Ala2826Thr) | 1 | 0 | 1 | 5.59 | 0 |
| 78,321,697 | c.9562G>A (p.Val3188Met) | 1 | 0 | 0.96 | 5.36 | 0 |
| 78,321,844 | c.9709C>A (p.Gln3237Lys) | 3 | 0 | 0.264 | 2.61 | 0 |
| 78,345,744 | c.12496G>A (p.Asp4166Asn) | 1 | 0 | 1 | 5.02 | 0.000923 |
| 78,346,877 | c.12854G>A (p.Ser4285Asn) | 1 | 0 | 0.024 | 2.7 | 0.000077 |
| 78,348,389 | c.13074G>A (p.Lys4358Lys) | 1 | 0 | 1 | 0 | 0 |
| 78,353,451 | c.13577T>C (p.Ile4526Thr) | 3 | 0 | 0.290848 | 2.81 | 0 |
| 78,363,707 | c.15275G>A (p.Arg5092Gln) | 1 | 0 | 0.998 | 3.14 | 0.000077 |
| 78,321,941 | c.9806G>A (p.Arg3269Gln) | 0 | 2 | 1 | 3.02 | 0.000077 |
| 78,328,364 | c.10850C>T (p.Ala3617Val) | 0 | 1 | 0.555 | 4.79 | 0 |
| 78,291,060 | c.2884G>A (p.Glu962Lys) | 0 | 1 | 0.068 | 3.66 | 0 |
| 78,321,560 | c.9425T>C (p.Val3142Ala) | 0 | 1 | 0.998 | 5.09 | 0 |
RNF213 Common Variants in a Population-Based Study
Among 257 IA-affected and 1,988 control individuals with SNP-chip genotyping data, PCA analysis showed that 3 IA-affected and 92 control individuals presented with an admixture of East Asian and African ancestry, and these samples were excluded from subsequent analysis (Figure S1). We performed a gene-wide association test on 38 polymorphic SNPs across the RNF213 loci in 254 affected and 1,896 control individuals, and the SNP rs6565666 appeared to be significantly associated with IA subjects (Cochran-Armitage trend test: p = 0.03 after Bonferroni correction; Table S6).
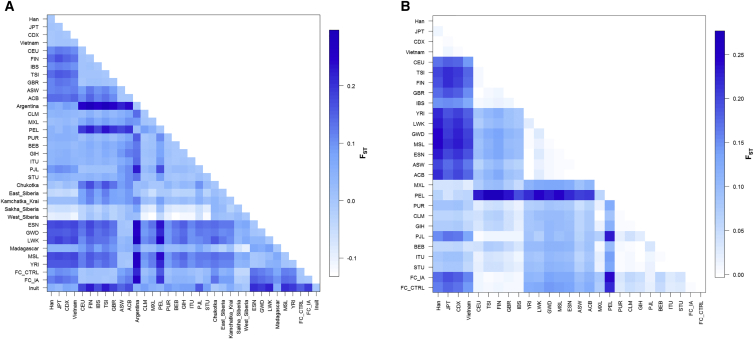
The pairwise FST comparisons between 1KGP phase 3 populations and FC individuals suggest that the common SNPs of RNF213 vary between populations. FC individuals have a higher number of distinct variations than do South Americans, followed by Africans and East Asians. However, East Asians share fewer RNF213 functional variations with the FC population than they do with the African and Caribbean populations (Figure 2). Overall, eight RNF213 SNPs appear to be outliers in the frequency changes across the worldwide populations, and four of these SNPs are located in the 3′ region. It also appeared that more functional variants of RNF213 were possibly affected by genetic drift (Figure S2).
Figure 2.
Matrix of Pairwise FST of RNF213 across Worldwide Populations
FST was calculated for RNF213 common variants from the Omni chip (A) and functional variants from exome sequencing and (B) between each two populations. FC_CTRL and FC_IA are the FC control cohort and the IA cohort, respectively; other populations are indicated by the three-letter codes used by 1KGP. The allele distributions of RNF213 common SNPs vary among populations, suggesting the existence of genetic drift. Among others, there is a big difference in the allelic distributions of RNF213 functional variants between Asians and FC populations.
A burden test comparing the FC population to other populations showed a similar trend—East Asians (followed by Peruvians, Indians, and Finnish) appeared to have the most significant difference of derived RNF213 allele frequencies in comparison to the overall FC population. As expected, FC IA individuals also had a significantly higher burden of RNF213 than control individuals did (Table 5).
Table 5.
RNF213 Variable Thresholds between FC and 1KGP Populations
| Population | Sample | No. of Variants | Total MAC | Statistic | p Value | SE | Permutations |
|---|---|---|---|---|---|---|---|
| CHB | 380 | 55 | 2,333 | 3.95287 | 0.00019996 | 0.364289 | 5,000 |
| JPT | 381 | 54 | 2,453 | 1.79685 | 0.00119976 | 0.346035 | 5,000 |
| CDX | 370 | 54 | 2,306 | 3.21125 | 0.00019996 | 0.343601 | 5,000 |
| KHV | 376 | 51 | 2,322 | 3.61053 | 0.00019996 | 0.355326 | 5,000 |
| CEU | 376 | 54 | 2,187 | 0.955313 | 0.113629 | 0.346662 | 3,000 |
| TSI | 384 | 54 | 2,151 | 1.23624 | 0.0319936 | 0.364394 | 5,000 |
| GBR | 368 | 52 | 2,108 | 1.0242 | 0.0657868 | 0.349268 | 5,000 |
| FIN | 376 | 51 | 2,104 | 1.48321 | 0.00679864 | 0.358871 | 5,000 |
| IBS | 384 | 57 | 2,250 | 1.01387 | 0.124875 | 0.388177 | 1,000 |
| YRI | 385 | 83 | 2,666 | 0.741073 | 0.361638 | 0.421002 | 1,000 |
| LWK | 376 | 80 | 2,545 | −0.0442278 | 0.936064 | 0.415337 | 1,000 |
| GWD | 390 | 81 | 2,763 | −0.0641889 | 0.924076 | 0.433297 | 1,000 |
| MSL | 362 | 82 | 2440 | −0.177864 | 0.973027 | 0.401114 | 1,000 |
| ESN | 376 | 84 | 2,598 | −0.6186 | 0.998002 | 0.436067 | 1,000 |
| ASW | 338 | 82 | 2,173 | −1.80752 | 1 | 0.39357 | 1,000 |
| ACB | 373 | 82 | 2,552 | −0.38227 | 0.99001 | 0.43374 | 1,000 |
| MXL | 341 | 55 | 2,060 | 0.660489 | 0.275724 | 0.302976 | 1,000 |
| PUR | 381 | 66 | 2,335 | −0.359017 | 0.995005 | 0.379015 | 1,000 |
| CLM | 371 | 69 | 2,293 | −0.673233 | 1 | 0.420994 | 1,000 |
| PEL | 362 | 58 | 2,313 | 1.48289 | 0.0039992 | 0.350589 | 5,000 |
| GIH | 380 | 53 | 2,312 | 1.47606 | 0.00559888 | 0.355099 | 5,000 |
| PJL | 373 | 56 | 2,169 | 1.07995 | 0.0631874 | 0.351286 | 5,000 |
| BEB | 363 | 58 | 2,212 | 0.293378 | 0.743257 | 0.33014 | 1,000 |
| STU | 379 | 56 | 2,278 | 0.959633 | 0.135864 | 0.362751 | 1,000 |
| ITU | 379 | 57 | 2,271 | 0.434712 | 0.601399 | 0.349177 | 1,000 |
| FC_IA | 513 | 72 | 2,938 | 1.69234 | 0.0125975 | 0.454633 | 5,000 |
Abbreviations are as follows: MAC, minor allele count; CHB, Han Chinese in Beijing, China; JPT, Japanese in Tokyo, Japan; CDX, Chinese Dai in Xishuangbanna, China; KHV, Kinh in Ho Chi Minh City, Vietnam; CEU, Utah residents with northern and western European ancestry from the CEPH collection; TSI, Toscani in Italia; GBR, British in England and Scotland; FIN, Finnish in Finland; IBS, Iberian population in Spain; YRI, Yoruba in Ibadan, Nigeria; LWK, Luhya in Webuye, Kenya; GWD, Gambian in western divisions in the Gambia; MSL, Mende in Sierra Leon; ESN, Esan in Nigeria; ASW, Americans of African ancestry in southwest USA; ACB, African Caribbeans in Barbados; MXL, Mexican ancestry from Los Angeles, USA; PUR, Puerto Ricans from Puerto Rico; CLM, Colombians from Medellin, Colombia; PEL, Peruvians from Lima, Peru; GIH, Gujarati Indian from Houston, Texas; PJL, Punjabi from Lahore, Pakistan; BEB, Bengali from Bangladesh; STU, Sri Lankan Tamil from the UK; ITU, Indian Telugu from the UK; and FC_IA, French-Canadian individuals with intracranial aneurysms.
Functional Characterization of Selected RNF213 Variants
Among the 14 RNF213 variants predicted to be deleterious and specific to IA individuals, only two variants, p.Arg2438Cys (c.7312C>T) and p.Ala2826Thr (c.8476G>A), were located in the two distinct AAA+ modules of the protein. These two variants were therefore the most suitable for a functional test measuring the ATPase activity of RNF213. As previously reported,41 this activity can be detected with a recombinant fragment containing amino acids 2,370–2,632 and 2,717–3,004.
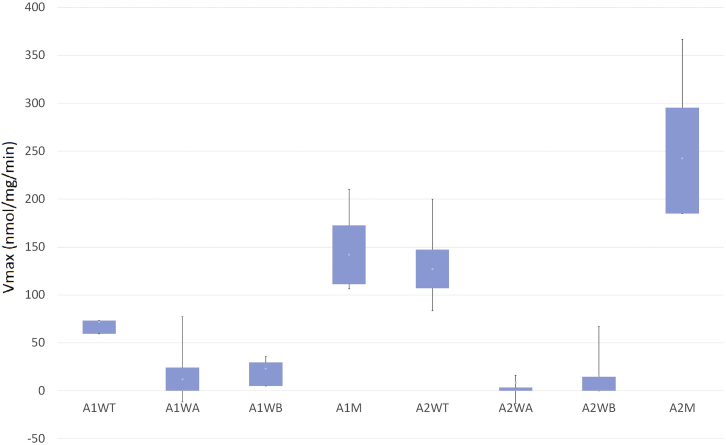
The baseline of ATP hydrolysis was determined by GST. As a result, the average ATPase activity of the first AAA+ wild-type protein (A1WT) and the p.Arg2438Cys variant (A1M) was estimated to be 72.0 and 142.1 nmol/mg/min, respectively, whereas that of the LOF variants in the walker A domain (A1WA) and walker B domain (A1WB) was estimated to be 12.2 and 23.1 nmol/mg/min, respectively. The average ATPase activity of the second AAA+ wild-type protein (A2WT) and the p.Ala2826Thr variant (A2M) was estimated to be 127.1 and 242.6 nmol/mg/min, respectively, whereas that of the A2WA and A2WB variants was as low as 8.8 and 15.5 nmol/mg/min, respectively. Compared to their respective wild-type proteins, both variants showed significant increases in ATPase activity (Mann-Whitney-Wilcoxon Test: pA1M-A1WT = 0.015, pA2M-A2WT = 0.0003); the differences were also significant in comparison to activity of the LOF variants (Mann-Whitney-Wilcoxon test: pA1M-A1WA = 0.002, pA1M-A1WB = 0.002, pA2M-A2WA = 0.0007, pA2M-A2WB = 0.0009) (Figure 3).
Figure 3.
ATPase Activity of the Wild-Types, Variants, and LOF Controls of the Two AAA+ Domains of RNF213
Box-and-whisker plot showing ATPase activity measured as nmol of free-phosphate release per mg protein per minute. A1WT and A2WT are the wild-types of the first and second AAA+ modules, respectively; A1M and A2M are the p.Arg2438Cys and p.Ala2826Thr variants, respectively. A1WA, A1WB, A2WA, and A2WB are four LOF controls (p. Lys2426Ala, p. Glu2488Ala, p. Lys2775Ala, and p. Glu2845Ala, respectively) located in the walker A and B domains of the first and second AAA+ modules. The results were generated by eight ATPase assays from two independent cell cultures; each sample is measured in duplicate. Boxes show the interquartile range, and whiskers show maximum and minimum measurements.
Discussion
A major weakness of using a GWAS approach for IAs is that the effects of rare variants and population-specific variants are not well addressed. Recently, two studies42, 43 using exome sequencing on multiple IA-affected families attempted to address the issue. Although no recurrent mutations were found, using different hypotheses and different populations, they reported 68 genes42 and 78 genes.43 Only one gene, ITGB6 (MIM: 147558), was found to be common across the two lists established by these two studies; nonetheless, neither study deemed this to be an interesting IA candidate gene. We believe that genetic heterogeneity is most likely at play in the development of IAs in distinct populations. Therefore, we conducted this research exclusively in FC individuals and focused on finding genes that harbor founder mutations or haplotypes in FC individuals as candidates for predisposition to IAs. Although our exome sequencing results for the first six FC families strongly suggest that rare, deleterious RNF213 variants predispose to IAs in FC individuals, some other interesting variants are worth noting, such as ABCA10 (MIM: 612508), which is expressed predominately in intracranial vascular endothelia cells.44
Functional Characterization of RNF213 and Variants
RNF213 has been identified as an important candidate for MMD in the Japanese population.39, 45 MMD is a rare cerebrovascular disorder characterized by stenosis of the internal carotid arteries and abnormal angiogenesis. It is a population-specific condition with an extremely low prevalence in Western populations. A founder mutation, c.14429G>A (p.Arg4810Lys) (rs112735431), previously annotated as c.14576G>A (p.Arg4859Lys) (GenBank: XM_005257545.3 and XP_005257602.2),45 was found to be significantly and specifically associated with the disease in the East Asian population.39 However, in a study looking at French MMD subjects, there was no evidence linking RNF213 to MMD,46 and another recent study further reported that ZXDC and OBSCN had higher variation burden in affected individuals of European descent, suggesting the complexities of the MMD genetic etiology.47
RNF213 encodes an AAA-type ATPase with E3 ubiquitin ligase activity, and studies have suggested a role in vascular-wall construction.39 A functional study focused on RNF213 p.Arg4810Lys suggested that the founder mutation is a MMD risk factor through reduced angiogenic activity48, 49 and induced mitotic abnormalities.50 When exploring the functions of RNF213, studies have found the gene to be linked to a variety of artery-wall developments. Several studies conducted on Rnf213-knockout mice have shown thinning of the intima and media layer after ligation of the common carotid artery,51 thinning in vascular walls and increased Mmp9 expression,52 or enhanced post-ischemic angiogenesis;53 a recent report also showed that RNF213 was associated with inflammatory responses and angiogenesis.49, 54 All of these results suggest that RNF213 is involved in vascular remodeling processes. A recent study even showed that RNF213 is associated with aneurysm formation after anastomotic surgery.55
In addition to being associated with MMD, RNF213 has also been associated with other vascular disorders,56 such as fibromuscular dysplasia (MIM: 135580),57 high blood pressure,58 intracranial major artery stenosis,59, 60 and heterogeneous intracerebral vasculopathy.61 The p.Arg4810Lys variant in RNF213 is suggested to be a risk factor for MMD by causing stenosis;60, 62 other variants, however, might have different effects on the artery. Some studies have suggested that other RNF213 variants are associated with different types of MMD, i.e., rare variant p.Ala4399Thr, which is intermediately frequent in East Asians, is associated with hemorrhage-type MMD,63 and p.Arg4810Lys is associated with the ischemia type.
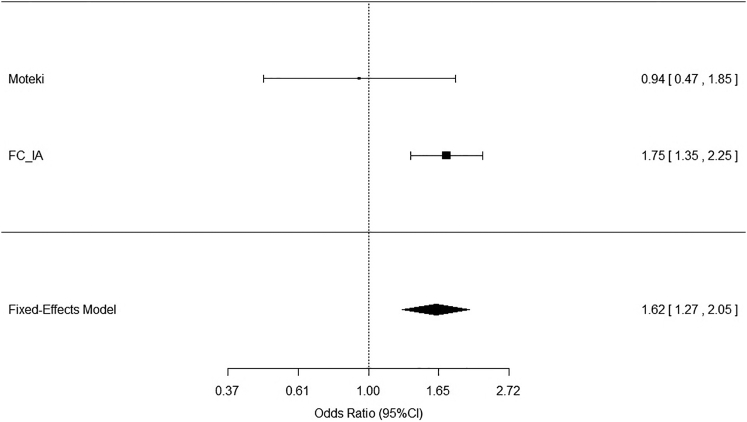
Rare variants in RNF213 have been found in MMD-affected individuals without p.Arg4810Lys and who are of Japanese,64 Taiwanese,65 and non-Asian56 ethnicities. Of the 30 rare functional variants found in 649 MMD-affected and control individuals in Japan, 16 were observed only in affected individuals; additionally, 3 novel variants were found in 31 Taiwanese MMD subjects, and 7 rare variants were found in 24 individuals with diverse ethnicities. Compared to those of the Japanese case-control study,64 our results show a significantly larger proportion of RNF213 mutations in affected individuals than in controls individuals (Cochran-Mantel-Haenszel test: p < 0.0001) (Figure 4).
Figure 4.
Meta-analysis of Two RNF213 Studies in Different Populations
Forest plot of Moteki et al.64 and the current study. Result show significantly more RNF213 mutations in FC IA-affected individuals (Cochran-Mantel-Haenszel test: p < 0.0001). Error bars represent confidence intervals, shown on the right.
AAA+ ATP Domain Variants and the Risk of IAs
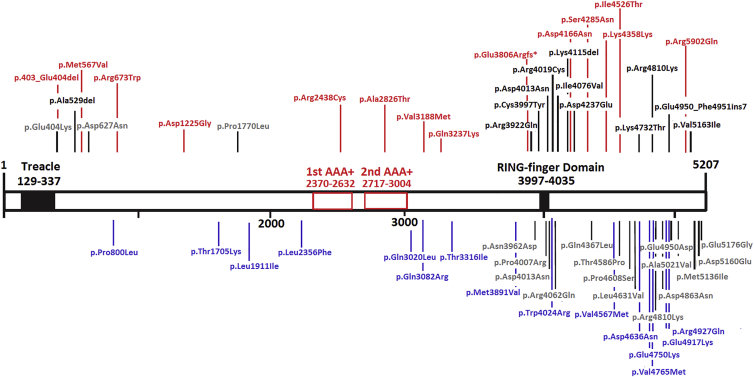
One of the very important functions of RNF213 is most likely mediated by the two AAA+ regions, which are encoded by exon 29 and exhibit ATPase activity.41 In our IA study, we found several deleterious variants in the AAA+ domains, whereas other studies focused on MMD-affected individuals did not (Figure 5).56, 66 We can assume that variants in different locations of RNF213 act as risk factors for different cerebrovascular disorders by affecting gene function differently. Whereas Cecchi et al.56 suggested that the C-terminal domain of RNF213 is the main risk region for MMD, we believe that exon 29, which encodes AAA+ domains, might be the risk region for IAs.
Figure 5.
RNF213 Rare Functional Variants Exclusively Found in IA- and MMD-Affected Individuals
Adapted from Cecchi et al.56 and Koizumi et al.66 14 variants found in the current study are marked in red, variants reported in Moteki et al.64 are marked in blue, MMD variants reported in Cecchi et al. are marked in black, and MMD variants found in other studies are marked in gray. The results show that FC IA-affected individuals harbor more deleterious mutations in the AAA+ domains than other populations do.
In our study, we found that a gain of ATPase activity could lead to increased risk of IAs. In contrast, a recent study suggested that p.Glu2488Gln in the first AAA+ domain and the p.Arg4810Lys variant reduced protein ATPase activity in the MMD model49 and thus inhibited angiogenesis. The discordance could be due to the pathological differences between MMD and IAs; although both have disrupted vascular walls, the main characteristic of MMD is progressive stenosis accompanied by aberrant angiogenesis of collateral vasculature,67 whereas IA lesions are characterized by vascular-wall weakness and dilatation accompanied by vascular remodeling and inflammatory response. Actually, the increased angiogenic activity might be a characteristic of the formation of aneurysm lesions,68 and some studies have suggested high expression of angiogenic factors in IA tissues.69, 70 Another paper also stated that the ATP concentration is lower in aneurysm walls than in normal artery tissue.71 We hypothesize that increased ATPase activity of RNF213 could lead to an increased or unbalanced level of angiogenesis and therefore be associated with the formation of aneurysmal lesions. Although Rnf213-knockout models inflict different angiogenesis alternations under different circumstances,49, 53 we imply that RNF213 is critical in vascular-wall remodeling, which can explain the pathogeneses of both MMD and IAs. Additionally, the increased ATP hydrolysis could also affect the oxidative phosphorylation process,72 which plays an important role in hypertension73 and heart failure. In our study, both IA individuals carrying variants in the AAA+ domains showed signs of hypertension; one was recorded to also suffer from multiple cardiovascular problems, including hypercholesterolemia, myocardial infarction, and arrhythmia.
RNF213 Variants in General Populations
We found that RNF213 SNP rs6565666 was associated with IAs in FC individuals. Interestingly, compared to other populations, FC IA individuals showed a gradual decrease in the frequency of the non-ancestral allele (A), which matches the out-of-Africa migration pattern (Figure S3) and could be the result of genetic drift, given that this non-ancestral allele might also be a risk allele for IAs (corrected p = 0.007 for the dominant model of allele A). The Finnish have a relatively high frequency of allele A (MAF = 0.26) in comparison to other European populations (CEU MAF = 0.18), which correlates with their high incidence of IA.74, 75
When further exploring the distributions of RNF213 variants across populations, we noted that groups of Native American populations (Inuit, Argentinians, and Peruvians) diverged the most from other populations, which fits the migration patterns. However, when we focused on potential functional variants of RNF213, East Asians (Chinese, Japanese, and Vietnamese) seemed to have the highest diversity (Figure 2). These results seem to agree with the fact that RNF213 East-Asian-specific variants are risk factors for MMD, which also has a high prevalence in East Asians. The significant difference in RNF213 burden between the FC and East Asian populations further suggests that the variations in this gene lead to different levels of risk in these two populations. The potential genetic drift of RNF213 variations could explain the difference between MMD prevalence and RNF213 founder variants in East Asians, but not in other populations. Exploring differences in RNF213 variants between populations also provided additional evidence that the risk gene associated with IAs could be ethnically different.75 Additionally, because some studies have suggested that the Inuit have a high prevalence of IAs, it would be interesting to see whether other Native Americans also have this predisposition. Nevertheless, given our observations, we can also assume that RNF213 might not be a risk factor for IAs among those populations.
Because of the limited number of subjects, we could not determine any founder mutation that was both highly frequent in FC individuals and significantly associated with IAs. The control populations used in this study were mostly unscreened for IAs, which could also have led to an underlying bias in our results. Only 10% of the affected individuals carry rare, deleterious variants in RNF213, suggesting that this gene is only a risk factor for some of the affected individuals and that IA pathogenesis is very complex and heterogeneous. Additionally, because RNF213 mutations have not been reported in IA subjects of other ethnicities and the risk genes for MMD also differ across populations, RNF213 variations might be associated with different vascular disorders in different populations. We further assume that different genes are likely to contribute to IAs in other populations.
In conclusion, our study supports a role for RNF213 as a risk factor for IAs, possibly via variants that affect different portions of the protein and lead to increased ATPase activity.
Acknowledgments
P.X. and Z.G.O. are recipients of a fellowship from the Canadian Institutes of Health Research. G.A.R. funded this study from the Canadian Hearth and Stroke Foundation. G.A.R. holds the Canada’s Research Chair in Neurogenetics and the Wilder Penfield Chair in Neuroscience. The NeuroX SNP genotyping was partially funded by a grant from the Michael J. Fox Foundation to Z.G.O.
Published: October 13, 2016
Footnotes
Supplemental Data include three figures and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.09.001.
Web Resources
NCBI Genome build GRCh37, ftp://gsapubftp-anonymous@ftp.broadinstitute.org/bundle/2.8/b37/human_g1k_v37.fasta.gz
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Stehbens W.E. The Pathology of Intracranial Arterial Aneurysms and Their Complications. In: Fox J.L., editor. Intracranial Aneurysms. Springer-Verlag New York; 1983. pp. 272–357. [Google Scholar]
- 2.Rinkel G.J. Natural history, epidemiology and screening of unruptured intracranial aneurysms. J. Neuroradiol. 2008;35:99–103. doi: 10.1016/j.neurad.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Vernooij M.W., Ikram M.A., Tanghe H.L., Vincent A.J., Hofman A., Krestin G.P., Niessen W.J., Breteler M.M., van der Lugt A. Incidental findings on brain MRI in the general population. N. Engl. J. Med. 2007;357:1821–1828. doi: 10.1056/NEJMoa070972. [DOI] [PubMed] [Google Scholar]
- 4.Rinkel G.J., Djibuti M., Algra A., van Gijn J. Prevalence and risk of rupture of intracranial aneurysms: a systematic review. Stroke. 1998;29:251–256. doi: 10.1161/01.str.29.1.251. [DOI] [PubMed] [Google Scholar]
- 5.Qureshi A.I., Suri M.F., Nasar A., Kirmani J.F., Divani A.A., He W., Hopkins L.N. Trends in hospitalization and mortality for subarachnoid hemorrhage and unruptured aneurysms in the United States. Neurosurgery. 2005;57:1–8. doi: 10.1227/01.neu.0000163081.55025.cd. discussion 1–8. [DOI] [PubMed] [Google Scholar]
- 6.Feigin V.L., Lawes C.M., Bennett D.A., Barker-Collo S.L., Parag V. Worldwide stroke incidence and early case fatality reported in 56 population-based studies: a systematic review. Lancet Neurol. 2009;8:355–369. doi: 10.1016/S1474-4422(09)70025-0. [DOI] [PubMed] [Google Scholar]
- 7.Teunissen L.L., Rinkel G.J., Algra A., van Gijn J. Risk factors for subarachnoid hemorrhage: a systematic review. Stroke. 1996;27:544–549. doi: 10.1161/01.str.27.3.544. [DOI] [PubMed] [Google Scholar]
- 8.Brown R.D., Jr., Wiebers D.O., Forbes G.S. Unruptured intracranial aneurysms and arteriovenous malformations: frequency of intracranial hemorrhage and relationship of lesions. J. Neurosurg. 1990;73:859–863. doi: 10.3171/jns.1990.73.6.0859. [DOI] [PubMed] [Google Scholar]
- 9.Clare C.E., Barrow D.L. Infectious intracranial aneurysms. Neurosurg. Clin. N. Am. 1992;3:551–566. [PubMed] [Google Scholar]
- 10.Benoit B.G., Wortzman G. Traumatic cerebral aneurysms. Clinical features and natural history. J. Neurol. Neurosurg. Psychiatry. 1973;36:127–138. doi: 10.1136/jnnp.36.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krischek B., Inoue I. The genetics of intracranial aneurysms. J. Hum. Genet. 2006;51:587–594. doi: 10.1007/s10038-006-0407-4. [DOI] [PubMed] [Google Scholar]
- 12.Bilguvar K., Yasuno K., Niemelä M., Ruigrok Y.M., von Und Zu Fraunberg M., van Duijn C.M., van den Berg L.H., Mane S., Mason C.E., Choi M. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat. Genet. 2008;40:1472–1477. doi: 10.1038/ng.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foroud T., Koller D.L., Lai D., Sauerbeck L., Anderson C., Ko N., Deka R., Mosley T.H., Fornage M., Woo D., FIA Study Investigators Genome-wide association study of intracranial aneurysms confirms role of Anril and SOX17 in disease risk. Stroke. 2012;43:2846–2852. doi: 10.1161/STROKEAHA.112.656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasuno K., Bilguvar K., Bijlenga P., Low S.K., Krischek B., Auburger G., Simon M., Krex D., Arlier Z., Nayak N. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat. Genet. 2010;42:420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foroud T., Lai D., Koller D., Van’t Hof F., Kurki M.I., Anderson C.S., Brown R.D., Jr., Connolly E.S., Eriksson J.G., Flaherty M., Familial Intracranial Aneurysm Study Investigators Genome-wide association study of intracranial aneurysm identifies a new association on chromosome 7. Stroke. 2014;45:3194–3199. doi: 10.1161/STROKEAHA.114.006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akiyama K., Narita A., Nakaoka H., Cui T., Takahashi T., Yasuno K., Tajima A., Krischek B., Yamamoto K., Kasuya H. Genome-wide association study to identify genetic variants present in Japanese patients harboring intracranial aneurysms. J. Hum. Genet. 2010;55:656–661. doi: 10.1038/jhg.2010.82. [DOI] [PubMed] [Google Scholar]
- 17.Low S.K., Takahashi A., Cha P.C., Zembutsu H., Kamatani N., Kubo M., Nakamura Y. Genome-wide association study for intracranial aneurysm in the Japanese population identifies three candidate susceptible loci and a functional genetic variant at EDNRA. Hum. Mol. Genet. 2012;21:2102–2110. doi: 10.1093/hmg/dds020. [DOI] [PubMed] [Google Scholar]
- 18.Halal F., Mohr G., Toussi T., Napoléon Martinez S. Intracranial aneurysms: a report of a large pedigree. Am. J. Med. Genet. 1983;15:89–95. doi: 10.1002/ajmg.1320150112. [DOI] [PubMed] [Google Scholar]
- 19.Mathieu J., Pérusse L., Allard P., Prévost C., Cantin L., Bouchard J.M., DeBraekeleer M. Epidemiological study of reptured intracranial aneurysms in the Saguenay-Lac-Saint-Jean region (Quebec, Canada) Can. J. Neurol. Sci. 1996;23:184–188. doi: 10.1017/s0317167100038488. [DOI] [PubMed] [Google Scholar]
- 20.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.J., Sunyaev S.R. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G.T., Peng B., Leal S.M. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am. J. Hum. Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 32.Eichstaedt C.A., Antão T., Pagani L., Cardona A., Kivisild T., Mormina M. The Andean adaptive toolkit to counteract high altitude maladaptation: genome-wide and phenotypic analysis of the Collas. PLoS ONE. 2014;9:e93314. doi: 10.1371/journal.pone.0093314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierron D., Razafindrazaka H., Pagani L., Ricaut F.X., Antao T., Capredon M., Sambo C., Radimilahy C., Rakotoarisoa J.A., Blench R.M. Genome-wide evidence of Austronesian-Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc. Natl. Acad. Sci. USA. 2014;111:936–941. doi: 10.1073/pnas.1321860111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardona A., Pagani L., Antao T., Lawson D.J., Eichstaedt C.A., Yngvadottir B., Shwe M.T., Wee J., Romero I.G., Raj S. Genome-wide analysis of cold adaptation in indigenous Siberian populations. PLoS ONE. 2014;9:e98076. doi: 10.1371/journal.pone.0098076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen M., Li Y., Lindgreen S., Pedersen J.S., Albrechtsen A., Moltke I., Metspalu M., Metspalu E., Kivisild T., Gupta R. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ionita-Laza I., Lee S., Makarov V., Buxbaum J.D., Lin X. Sequence kernel association tests for the combined effect of rare and common variants. Am. J. Hum. Genet. 2013;92:841–853. doi: 10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Excoffier L., Lischer H.E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 39.Liu W., Morito D., Takashima S., Mineharu Y., Kobayashi H., Hitomi T., Hashikata H., Matsuura N., Yamazaki S., Toyoda A. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE. 2011;6:e22542. doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 41.Morito D., Nishikawa K., Hoseki J., Kitamura A., Kotani Y., Kiso K., Kinjo M., Fujiyoshi Y., Nagata K. Moyamoya disease-associated protein mysterin/RNF213 is a novel AAA+ ATPase, which dynamically changes its oligomeric state. Sci. Rep. 2014;4:4442. doi: 10.1038/srep04442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farlow J.L., Lin H., Sauerbeck L., Lai D., Koller D.L., Pugh E., Hetrick K., Ling H., Kleinloog R., van der Vlies P., FIA Study Investigators Lessons learned from whole exome sequencing in multiplex families affected by a complex genetic disorder, intracranial aneurysm. PLoS ONE. 2015;10:e0121104. doi: 10.1371/journal.pone.0121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan J., Hitomi T., Takenaka K., Kato M., Kobayashi H., Okuda H., Harada K.H., Koizumi A. Genetic study of intracranial aneurysms. Stroke. 2015;46:620–626. doi: 10.1161/STROKEAHA.114.007286. [DOI] [PubMed] [Google Scholar]
- 44.Lindner C., Sigrüner A., Walther F., Bogdahn U., Couraud P.O., Schmitz G., Schlachetzki F. ATP-binding cassette transporters in immortalised human brain microvascular endothelial cells in normal and hypoxic conditions. Exp. Transl. Stroke Med. 2012;4:9. doi: 10.1186/2040-7378-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamada F., Aoki Y., Narisawa A., Abe Y., Komatsuzaki S., Kikuchi A., Kanno J., Niihori T., Ono M., Ishii N. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J. Hum. Genet. 2011;56:34–40. doi: 10.1038/jhg.2010.132. [DOI] [PubMed] [Google Scholar]
- 46.Hervé D., Philippi A., Belbouab R., Zerah M., Chabrier S., Collardeau-Frachon S., Bergametti F., Essongue A., Berrou E., Krivosic V. Loss of α1β1 soluble guanylate cyclase, the major nitric oxide receptor, leads to moyamoya and achalasia. Am. J. Hum. Genet. 2014;94:385–394. doi: 10.1016/j.ajhg.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shoemaker L.D., Clark M.J., Patwardhan A., Chandratillake G., Garcia S., Chen R., Morgan A.A., Leng N., Kirk S., Chen R. Disease Variant Landscape of a Large Multiethnic Population of Moyamoya Patients by Exome Sequencing. G3 (Bethesda) 2015;6:41–49. doi: 10.1534/g3.115.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hitomi T., Habu T., Kobayashi H., Okuda H., Harada K.H., Osafune K., Taura D., Sone M., Asaka I., Ameku T. Downregulation of Securin by the variant RNF213 R4810K (rs112735431, G>A) reduces angiogenic activity of induced pluripotent stem cell-derived vascular endothelial cells from moyamoya patients. Biochem. Biophys. Res. Commun. 2013;438:13–19. doi: 10.1016/j.bbrc.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi H., Matsuda Y., Hitomi T., Okuda H., Shioi H., Matsuda T., Imai H., Sone M., Taura D., Harada K.H. Biochemical and Functional Characterization of RNF213 (Mysterin) R4810K, a Susceptibility Mutation of Moyamoya Disease, in Angiogenesis In Vitro and In Vivo. J. Am. Heart Assoc. 2015;4:4. doi: 10.1161/JAHA.115.002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hitomi T., Habu T., Kobayashi H., Okuda H., Harada K.H., Osafune K., Taura D., Sone M., Asaka I., Ameku T. The moyamoya disease susceptibility variant RNF213 R4810K (rs112735431) induces genomic instability by mitotic abnormality. Biochem. Biophys. Res. Commun. 2013;439:419–426. doi: 10.1016/j.bbrc.2013.08.067. [DOI] [PubMed] [Google Scholar]
- 51.Sonobe S., Fujimura M., Niizuma K., Nishijima Y., Ito A., Shimizu H., Kikuchi A., Arai-Ichinoi N., Kure S., Tominaga T. Temporal profile of the vascular anatomy evaluated by 9.4-T magnetic resonance angiography and histopathological analysis in mice lacking RNF213: a susceptibility gene for moyamoya disease. Brain Res. 2014;1552:64–71. doi: 10.1016/j.brainres.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Sonobe S., Fujimura M., Niizuma K., Fujimura T., Furudate S., Nishijima Y., Kure S., Tominaga T. Increased vascular MMP-9 in mice lacking RNF213: moyamoya disease susceptibility gene. Neuroreport. 2014;25:1442–1446. doi: 10.1097/WNR.0000000000000289. [DOI] [PubMed] [Google Scholar]
- 53.Ito A., Fujimura M., Niizuma K., Kanoke A., Sakata H., Morita-Fujimura Y., Kikuchi A., Kure S., Tominaga T. Enhanced post-ischemic angiogenesis in mice lacking RNF213; a susceptibility gene for moyamoya disease. Brain Res. 2015;1594:310–320. doi: 10.1016/j.brainres.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 54.Ohkubo K., Sakai Y., Inoue H., Akamine S., Ishizaki Y., Matsushita Y., Sanefuji M., Torisu H., Ihara K., Sardiello M., Hara T. Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells. Sci. Rep. 2015;5:13191. doi: 10.1038/srep13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukushima Y., Miyawaki S., Inoue T., Shimizu S., Yoshikawa G., Imai H., Saito N., Tsutsumi K. Repeated de novo aneurysm formation after anastomotic surgery: Potential risk of genetic variant RNF213 c.14576G>A. Surg. Neurol. Int. 2015;6:41. doi: 10.4103/2152-7806.153709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cecchi A.C., Guo D., Ren Z., Flynn K., Santos-Cortez R.L., Leal S.M., Wang G.T., Regalado E.S., Steinberg G.K., Shendure J., University of Washington Center for Mendelian Genomics RNF213 rare variants in an ethnically diverse population with Moyamoya disease. Stroke. 2014;45:3200–3207. doi: 10.1161/STROKEAHA.114.006244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiando S.R., Barlassina C., Cusi D., Galan P., Lathrop M., Plouin P.F., Jeunemaitre X., Bouatia-Naji N. Exome sequencing in seven families and gene-based association studies indicate genetic heterogeneity and suggest possible candidates for fibromuscular dysplasia. J. Hypertens. 2015;33:1802–1810. doi: 10.1097/HJH.0000000000000625. discussion 1810. [DOI] [PubMed] [Google Scholar]
- 58.Koizumi A., Kobayashi H., Liu W., Fujii Y., Senevirathna S.T., Nanayakkara S., Okuda H., Hitomi T., Harada K.H., Takenaka K. P.R4810K, a polymorphism of RNF213, the susceptibility gene for moyamoya disease, is associated with blood pressure. Environ. Health Prev. Med. 2013;18:121–129. doi: 10.1007/s12199-012-0299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyawaki S., Imai H., Shimizu M., Yagi S., Ono H., Mukasa A., Nakatomi H., Shimizu T., Saito N. Genetic variant RNF213 c.14576G>A in various phenotypes of intracranial major artery stenosis/occlusion. Stroke. 2013;44:2894–2897. doi: 10.1161/STROKEAHA.113.002477. [DOI] [PubMed] [Google Scholar]
- 60.Miyawaki S., Imai H., Takayanagi S., Mukasa A., Nakatomi H., Saito N. Identification of a genetic variant common to moyamoya disease and intracranial major artery stenosis/occlusion. Stroke. 2012;43:3371–3374. doi: 10.1161/STROKEAHA.112.663864. [DOI] [PubMed] [Google Scholar]
- 61.Smith K.R., Leventer R.J., Mackay M.T., Pope K., Gillies G., Delatycki M.B., Amor D.J., Bahlo M., Lockhart P.J. Identification of a novel RNF213 variant in a family with heterogeneous intracerebral vasculopathy. Int. J. Stroke. 2014;9:E26–E27. doi: 10.1111/ijs.12306. [DOI] [PubMed] [Google Scholar]
- 62.Bang O.Y., Ryoo S., Kim S.J., Yoon C.H., Cha J., Yeon J.Y., Kim K.H., Kim G.M., Chung C.S., Lee K.H. Adult Moyamoya Disease: A Burden of Intracranial Stenosis in East Asians? PLoS ONE. 2015;10:e0130663. doi: 10.1371/journal.pone.0130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Z., Jiang H., Zhang L., Xu X., Zhang X., Kang Z., Song D., Zhang J., Guan M., Gu Y. Molecular analysis of RNF213 gene for moyamoya disease in the Chinese Han population. PLoS ONE. 2012;7:e48179. doi: 10.1371/journal.pone.0048179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moteki Y., Onda H., Kasuya H., Yoneyama T., Okada Y., Hirota K., Mukawa M., Nariai T., Mitani S., Akagawa H. Systematic Validation of RNF213 Coding Variants in Japanese Patients With Moyamoya Disease. J. Am. Heart Assoc. 2015;4:4. doi: 10.1161/JAHA.115.001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee M.J., Chen Y.F., Fan P.C., Wang K.C., Wang K., Wang J., Kuo M.F. Mutation genotypes of RNF213 gene from moyamoya patients in Taiwan. J. Neurol. Sci. 2015;353:161–165. doi: 10.1016/j.jns.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 66.Koizumi A., Kobayashi H., Hitomi T., Harada K.H., Habu T., Youssefian S. A new horizon of moyamoya disease and associated health risks explored through RNF213. Environ. Health Prev. Med. 2016;21:55–70. doi: 10.1007/s12199-015-0498-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott R.M., Smith E.R. Moyamoya disease and moyamoya syndrome. N. Engl. J. Med. 2009;360:1226–1237. doi: 10.1056/NEJMra0804622. [DOI] [PubMed] [Google Scholar]
- 68.Kessler K., Borges L.F., Ho-Tin-Noé B., Jondeau G., Michel J.B., Vranckx R. Angiogenesis and remodelling in human thoracic aortic aneurysms. Cardiovasc. Res. 2014;104:147–159. doi: 10.1093/cvr/cvu196. [DOI] [PubMed] [Google Scholar]
- 69.Hoh B.L., Hosaka K., Downes D.P., Nowicki K.W., Wilmer E.N., Velat G.J., Scott E.W. Stromal cell-derived factor-1 promoted angiogenesis and inflammatory cell infiltration in aneurysm walls. J. Neurosurg. 2014;120:73–86. doi: 10.3171/2013.9.JNS122074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li B., Li F., Chi L., Zhang L., Zhu S. The expression of SPARC in human intracranial aneurysms and its relationship with MMP-2/-9. PLoS ONE. 2013;8:e58490. doi: 10.1371/journal.pone.0058490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Imai Y., Sato K., Ishikawa T., Comerford A., David T., Yamaguchi T. ATP transport in saccular cerebral aneurysms at arterial bends. Ann. Biomed. Eng. 2010;38:927–934. doi: 10.1007/s10439-009-9864-1. [DOI] [PubMed] [Google Scholar]
- 72.Power A.S., Pham T., Loiselle D.S., Crossman D.H., Ward M.L., Hickey A.J. Impaired ADP channeling to mitochondria and elevated reactive oxygen species in hypertensive hearts. Am. J. Physiol. Heart Circ. Physiol. 2016;310:H1649–H1657. doi: 10.1152/ajpheart.00050.2016. [DOI] [PubMed] [Google Scholar]
- 73.Vanhoutte P.M., Zhao Y., Xu A., Leung S.W. Thirty Years of Saying NO: Sources, Fate, Actions, and Misfortunes of the Endothelium-Derived Vasodilator Mediator. Circ. Res. 2016;119:375–396. doi: 10.1161/CIRCRESAHA.116.306531. [DOI] [PubMed] [Google Scholar]
- 74.van ’t Hof F.N., Kurki M.I., Kleinloog R., de Bakker P.I., von und zu Fraunberg M., Jääskeläinen J.E., Gaál E.I., Lehto H., Kivisaari R., Laakso A. Genetic risk load according to the site of intracranial aneurysms. Neurology. 2014;83:34–39. doi: 10.1212/WNL.0000000000000547. [DOI] [PubMed] [Google Scholar]
- 75.Kurki M.I., Gaál E.I., Kettunen J., Lappalainen T., Menelaou A., Anttila V., van ’t Hof F.N., von Und Zu Fraunberg M., Helisalmi S., Hiltunen M. High risk population isolate reveals low frequency variants predisposing to intracranial aneurysms. PLoS Genet. 2014;10:e1004134. doi: 10.1371/journal.pgen.1004134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.