Abstract
The innate immune response is highly conserved across all eukaryotes and has been studied in great detail in several model organisms. Hemocytes, the primary immune cell population in mosquitoes, are important components of the mosquito innate immune response, yet critical aspects of their biology have remained uncharacterized. Using a novel method of enrichment, we isolated phagocytic granulocytes and quantified their proteomes by mass spectrometry. The data demonstrate that phagocytosis, blood-feeding, and Plasmodium falciparum infection promote dramatic shifts in the proteomic profiles of An. gambiae granulocyte populations. Of interest, large numbers of immune proteins were induced in response to blood feeding alone, suggesting that granulocytes have an integral role in priming the mosquito immune system for pathogen challenge. In addition, we identify several granulocyte proteins with putative roles as membrane receptors, cell signaling, or immune components that when silenced, have either positive or negative effects on malaria parasite survival. Integrating existing hemocyte transcriptional profiles, we also compare differences in hemocyte transcript and protein expression to provide new insight into hemocyte gene regulation and discuss the potential that post-transcriptional regulation may be an important component of hemocyte gene expression. These data represent a significant advancement in mosquito hemocyte biology, providing the first comprehensive proteomic profiling of mosquito phagocytic granulocytes during homeostasis blood-feeding, and pathogen challenge. Together, these findings extend current knowledge to further illustrate the importance of hemocytes in shaping mosquito innate immunity and their principal role in defining malaria parasite survival in the mosquito host.
Insect immune cells, or hemocytes, are central to cellular and humoral immune responses, mediating the engulfment of bacteria via phagocytosis and the production of humoral components that sequester and/or kill invading pathogens (1–3). In contrast to the highly specialized immune cells characterized in mammalian systems, only three distinct classes of hemocytes have been described in mosquitoes that are primarily distinguished by morphology and limited biochemical characterization (4). Mosquito granulocytes are professional phagocytes similar to vertebrate macrophages that mediate immune activation through the production of humoral defense factors. Oenocytoids are thought to have a primary role in the production of melanin, and have been implicated in wound healing and pathogen killing in other insect systems (1–3). Lastly, prohemocytes are thought to serve as hematopoietic progenitors that differentiate to produce other hemocyte cell populations (5), or as recently proposed, may simply represent a smaller form of granulocytes (6).
Our current knowledge of mosquito hemocytes is still in its infancy, thereby leaving much of the fundamental aspects of their biology unknown (1, 4). Much of our knowledge has focused on circulating hemocytes because of their ease of isolation, yet a second population of sessile hemocytes remains attached to mosquito tissues that also contributes to the immune response and removal of pathogens (6, 7). Transcriptional analysis has been performed on a heterogeneous population of circulating mosquito hemocytes to measure their response in the context of either bacterial or Plasmodium parasite challenge and reveal distinct transcriptional profiles according to pathogen challenge and temporal patterns following infection (8, 9). In addition, Pinto et al. also identified several hemocyte factors that modulate parasite success (8).
Building on these studies, we report on the comprehensive proteomic analysis of circulating phagocytic granulocytes in response to blood feeding and infection in the African malaria vector, Anopheles gambiae. Using a novel technique to isolate granulocytes from other mosquito blood cells, we enriched and subsequently performed mass spectrometry-based, relative quantitative proteomics analyses on mosquito phagocytic granulocytes. The comparisons of the proteomic profiles to existing transcriptome data revealed new insights into hemocyte gene regulation, as well as identified candidate markers to distinguish hemocytes in future immunological studies. Together, our data further implicate the role of hemocytes as determinants of malaria parasite survival and suggest that these immune cells may be important components of an anticipatory response that may prime the mosquito immune system following blood-feeding. These data provide a rich community resource for mosquito hemocyte biology and provide important new information about gene expression in a granulocyte-enriched cell population of mosquito immune cells.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
The objective of this experiment was to examine the mosquito hemocyte proteome in response to blood-feeding and malaria parasite challenge. Because of the heterogeneous mosquito hemocyte cell population, we performed a novel method of enrichment using magnetic beads to enrich for phagocytic granulocytes. Using this technique, we compare the proteomes of nonselected hemocytes and granulocyte-enriched samples, as well as compare granulocyte-enriched samples across experimental treatments (naïve sugar-fed, blood-fed, and Plasmodium falciparum-infected samples). For each sample, 150 mosquitoes were perfused to collect either nonselected or granulocyte-enriched samples for each experimental condition using three independent biological replicates. Cell samples were lysed to release their protein contents and then separated by electrophoresis on a SDS-gel. Coomassie blue staining was performed to estimate protein quantities of equally loaded samples using densitometry analysis using a uniform protein band of ∼30kDa to verify that samples did not display significant differences in loading. Protein samples were then excised from the gel, processed, and used for LC-MS/MS analysis.
Protein identifications were performed by searching the raw data against an Anopheles gambiae protein database (VectorBase https://www.vectorbase.org, version 3.7) and abundances were determined by label-free quantification using normalized spectral counting with Scaffold software. Protein abundance data was used to compare samples across experimental treatments with significant differences determined by a Student t test.
Mosquito Rearing
The Keele strain of Anopheles gambiae was originally isolated for its high susceptibility to Plasmodium as previously described (10), and only rarely mounts a melanization response to malaria parasites. Mosquitoes were maintained at 27 °C and 80% relative humidity with a 14/10 h light/dark cycle. Larvae were reared in distilled water on a diet of fish food and cat food pellets, whereas adults were maintained on a 10% sucrose solution. Adult mosquitoes were housed in 8” × 8” steel cages with steel mesh sides to allow exposure to the light/dark cycles and a fabric sleeve for user access.
Blood Feeding and Plasmodium Infection
Approximately 5–7 day-old, physiologically matched, female An. gambiae mosquitoes were starved overnight prior to blood feeding. For P. falciparum infections, NF54 isolates of P. falciparum gametocyte cultures were obtained from the Johns Hopkins Malaria Research Institute Parasite Core facility and the gametocytemia was determined by microscopy analysis of Giemsa stained thin blood smears. Before feeding, gametocyte samples were diluted to 0.3% gametocytemia with human RBCs as previously described (11, 12). For all blood feedings (noninfected and P. falciparum-infected), serum was exchanged in all blood samples with heat inactivated human serum to a 45% hematocrit. Using artificial membrane feeders maintained by a circulating water bath, mosquitoes were fed on either noninfected human blood or P. falciparum-infected blood and maintained at 25 °C and 80% relative humidity under standard insectary conditions until hemocyte collection ∼48 h after feeding.
Magnetic Bead Injection and Hemocyte Collection
The following method was developed and optimized using several isolations from sugar-fed controls to ensure reproducible quantities of hemocyte samples are used for proteomics analyses. To collect circulating granulocytes, mosquitoes were cold anesthetized and individual mosquitoes were injected with 0.2 μl (2 mg/ml) of a suspension of 1–2 μm diameter MagnaBind Carboxyl Derivatized Beads (Thermo Scientific). For magnetic bead granulocyte-enriched samples, mosquitoes of each sample treatment (sugar-fed, blood-fed, or Plasmodium-infected) were injected with magnetic beads, ∼48 h after blood meal for the blood-fed and P. falciparum-infected samples. Based on previous reports (11, 13), P. falciparum ookinete invasion of the mosquito midgut occurs between 24–30 h, much later than the ∼18 h time point observed when using rodent malaria models. Age-matched samples were collected at the ∼48 h time point to measure the effects of P. falciparum ookinete invasion on hemocyte samples, with the appropriate noninfected blood meal serving as a control. Following injection, mosquitoes were returned to insectary conditions for 2 h. Hemocytes from both noninjected and magnetic bead-injected mosquitoes were collected using a perfusion method similar to those previously described (5, 6). Briefly, a small perforation was made in the abdomen of a cold anesthetized mosquito and ∼5 μl of anticoagulant buffer (70% Schneider's Insect medium, and (30% citrate buffer 98 mm NaOH, 186 mm NaCl, 1.7 mm EDTA, 41 mm citric acid; pH 4.5)) containing a protease inhibitor mixture (Sigma; P8340) was injected into the thorax, causing perfusion of the circulating hemolymph through the abdominal perforation. Exactly 150 mosquitoes were perfused to reproducibly collect equivalent hemocyte samples for each experimental condition. Perfusate was collected in a siliconized tube and kept on wet ice for the remainder of the collection process. Hemolymph perfusate from magnetic bead injected mosquitoes was placed on a collection device made from a 1/4 × 1/2 inch cylindrical Neodymium alloy magnet with a pull force of ∼8.64 pounds for 10 min at 4 °C to enrich for granulocytes that have phagocytosed the magnetic beads. To wash the granulocyte-enriched cell pellets, samples were resuspended in fresh 1× PBS with protease inhibitor mixture on ice and repeated twice prior to the collection of the final cell fractions. Additional samples from mosquitoes without magnetic bead enrichment (total hemocyte population) were centrifuged at 2000 × g for 5 mins to pellet the total hemocyte cell fraction.
Microscopy
To verify that specific hemocyte populations were responsible for magnetic bead uptake, hemocytes were examined by bright-field, phase-contrast, or fluorescence microscopy as previously described (6, 7). Hemocytes and fat body cells were stained with tubulin or CM-DiI as previously described (6, 7), or using a Drosophila Notch intracellular domain antibody (Developmental Studies Hybridoma Bank, Iowa City, Iowa; C17.9C6) at a 5 μg/ml dilution.
Protein Extraction
To extract total protein from the perfused hemocyte samples, the cellular pellet obtained from 150 mosquitoes was dissolved in 30 μl SDT-lysis buffer composed of 4% (w/v) SDS, 100 mm Tris/HCl, 0.1 m DTT, pH 7.6, and then boiled at 95 °C for 5 min.
Multilane Combined In-gel Digestion (MLCID)1
For SDS-PAGE, each lane was loaded with equivalent amounts of hemocyte protein lysates. After resolving on a 4–20% precast gradient gel (BioRad, Hercules, CA), the proteins were stained with Coomassie (Gel-Code Blue). Coomassie blue staining was used to estimate that equivalent levels of protein were loaded in each lane/replicate study, because direct quantification of the extremely small amounts of hemocytes would result in protein loss. To estimate the protein quantities and compare each independent biological replicate, the Coomassie stained gel images were analyzed using the quantitative densitometry analysis package in the Odyssey Image StudioTM software (Li-COR). Using a representative, uniform, singular protein band (∼30 kDa) that was common across all samples, the fluorescence signal was calculated as integrated intensities or K Counts/mm2 for each sample. These measurements provided an estimation that relatively equivalent amounts of hemocyte protein were reproducibly acquired and loaded into the gel prior to gel excision and downstream LC-MS/MS analysis. For gel band excision, clean and sterile razors were used to separate individual sample lanes and further cut into eight identical slices in a process-protection hood. These slices were further cut into 1 × 1 mm pieces prior to de-staining, reduction and alkylation, tryptic digestion and peptide extraction (14). The extracted peptides were lyophilized by speed-vac and re-suspended in 2% acetonitrile, 97.9% water, and 0.1% formic acid buffer for LC-MS/MS analysis.
LC-MS/MS
Following in-gel digestion, tryptic peptides from each biological replicate were individually analyzed. Half of each sample was injected onto an Agilent LC-MS system consisting of a 1200 LC system coupled to a 6520 Q-TOF via an HPLC Chip Cube interface. The samples were trapped and analyzed using an Agilent Polaris-HR-Chip-3C18 chip (360 nL, 180 Å C18 trap with a 75 μm i.d., 150 mm length, 180 Å C18 analytical column). Peptides were loaded onto the enrichment column by autosampler using 97% solvent A (0.1% formic acid in water) and 3% solvent B (0.1% formic acid in 90% acetonitrile) at a flow rate of 2 μl/min. Elution of peptides from the analytical column was performed using a gradient starting at 97% A at 300 nL/min. The mobile phase was 3–10% B for 4 min, 10–35% B for 56 min, 35–99% for 2 min, and maintained at 99% B for 6 min, followed by re-equilibration of the column with 3% B for 10 min. Data dependent (autoMS2) mode was used for MS acquisition by Agilent 6520 Q-TOF at 2 GHz. Precursor MS spectra were acquired from m/z 315 to 1700 and the top four peaks were selected for MS/MS analysis. Product scans were acquired from m/z 50 to 1700 at a scan rate of 1.5/second. A medium isolation width (∼4 amu) was used, and a collision energy of slope 3.9 V/100 Da with a 2.9 V offset was applied for fragmentation. A dynamic exclusion list was applied, with precursors excluded of 0.50 min after two MS/MS spectrum was acquired.
Mass Spectrometry Data Search and Analysis
All the LC-MS/MS raw data were converted to Mascot generic Format (.mgf) by Agilent MassHunter Qualitative Analysis B.04.00. Mascot version 2.4.1, OMSSA version 2.1.9 and X!Tandem version CYCLONE 2010.12.01.1 were used to search the Anopheles gambiae version 3.7 (14,667 sequences) protein FASTA sequence database for peptide sequence assignments using the following parameters: precursor ion mass tolerance of 50 ppm and a fragment ion mass tolerance of 0.2 daltons. Peptides were searched using fully tryptic cleavage constraints and up to two internal cleavages sites were allowed for tryptic digestion. Fixed modifications consisted of carbamidomethylation of cysteine. Variable modifications considered were oxidation of methionine residues. All the searched results were exported and then imported into the Scaffold software (Version 4.3.4, Proteome Software) for curation, label-free quantification analysis, and visualization. Scaffold's normalized spectral counting was employed to compare relative protein abundance between nonselected hemocytes (sugar-fed) and magnetic-bead enriched granulocytes (sugar-fed, blood-fed, and Plasmodium-infected) cell samples in each experiment as the basis for normalization of the spectral counts for all other LC-MS/MS data in that experiment. Scaffold calculates the spectrum count quantitative value by normalizing spectral counts across an experiment. The process of calculating normalized spectral counts is as follows: (1) Scaffold takes the sum of all the Total Spectrum Counts for each MS sample; (2) The sums are then scaled to the same level; and (3) Scaffold then applies the scaling factor for each sample to each protein group to produce an output with a normalized quantitative value. Peptide false discovery rate (FDR) was calculated as the sum of the Exclusive Spectrum Counts of decoy proteins divided by the sum of the Exclusive Spectrum Counts of target proteins, converted to a percentage. Protein FDR is the number of decoy proteins D divided by the number of target proteins T: FDR = D/T, expressed as a percentage. This approach assumes that decoy proteins can be filtered out of the considered protein list and that the user is interested in the FDR value of the remaining proteins in the list or target proteins T. Overall, protein false discovery rates of less than 1% and peptide false discovery rates of less than 0.1% were obtained with Scaffold filters, and each protein has ≥ 2 unique peptides.
Data Access
Our data meet all the standards regarding the Minimum Information About a Proteomics Experiment (MIAPE), and data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository (15) with the data set identifier PXD001507.
Gene-expression Analyses
Approximately 50 sugar-fed mosquitoes were perfused with anticoagulant buffer directly into TRIzol reagent (Invitrogen-Life Technologies, Waltham, MA) to obtain unselected hemocyte samples for RNA isolation. For granulocyte-enriched samples, mosquitoes of each sample treatment (sugar-fed, blood-fed, or Plasmodium-infected) were injected with magnetic beads (∼48 h after blood meal for the blood-fed and P. falciparum -infected samples) and allowed to recover under insectary conditions as described above. After perfusion, granulocyte enrichment, and washing, TRIzol reagent was added for RNA isolation. Total RNA was obtained using the Direct-zol RNA Mini kit (Zymo Research) according to the manufacturer's protocol. cDNA was prepared using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer's protocol and used for quantitative real-time PCR as previously described (11). Hemocyte gene expression was determined using gene-specific primers and normalized to levels of ribosomal protein S7 (rpS7). All qRT-PCR primers are listed in supplemental Table S1.
Multiple Coinertia Analysis (MCIA)
MCIA is an integrative analysis method that can be applied to multiple (OMICS) data sets simultaneously (16, 17). The data sets are matrices in which the number of features, typically stored in the rows, is much larger than the number of samples, typically stored in the columns. MCIA can be applied to multiple data sets that have matched features or matched samples. In our case, MCIA was applied to transcriptome and proteome data sets with the most optimally matched samples acquired from independent studies. In this analysis, the features refer to genes and proteins and a sample is computed as the log fold change between two treatments in a comparison between SF, BF and PF. This log fold change is computed for both genes and proteins.in both data sets.
MCIA finds maximally covariant axes, which allow the simultaneous projection of all features and samples in the same hyperspace. MCIA is a dimension reduction technique that maximizes the covariance between the data sets and the reference space. The MCIA axis selection starts with a one table ordination method such as principal component analysis, correspondence analysis or, as in this case, nonsymmetrical correspondence analysis. Afterward, the MCIA axes that maximize the squared covariance between scores of each data set projected on the synthetic axes are computed. The samples that share similar trends will group together in the MCIA space. Additionally, features can be projected into the MCIA space. Features highly expressed in a sample will be projected in the direction of that sample. The strength of association of a feature to a sample is directly proportional to the distance of the feature from the origin of the plot. The overall correlation between the two data sets is measured with the RV-coefficient, which is a generalization of the squared Pearson correlation coefficient (18). Here, a modified version was used, which overcomes the dimensionality bias of the original RV coefficient (19). The modified RV-coefficient ranges between –1 and 1. Its interpretation is similar to that of a correlation coefficient. A RV-coefficient of zero means there is no costructure between the two data sets. The higher the costructure between the data sets is, the higher the absolute value of the RV-coefficient.
There are two published anopheline hemocyte transcriptome data sets available (8, 9); however, the two studies used different microarray platforms. Merging the two data sets would artificially reduce the transcriptome data that can be used for MCIA to only the subset of transcripts that was measured in both analyses. This would limit the utility and primary advantage of a MCIA approach, i.e. avoid the need to subset the data sets for the analysis. As such, we decided to focus on the Pinto et al. (8) data set, which provided all the necessary matching transcript data for the MCIA comparison to the granulocyte proteome.
Cluster and Functional Analyses of Protein Data Sets
Averaged normalized spectral counts from each group were imported into Cluster 3.0 (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm) for analysis. An arbitrary cutoff of at least one sample mean of greater than 2.0 normalized spectra was used to filter the data. This led to the inclusion of 878 of 1128 total proteins, eliminating undifferentiated data from downstream analyses. Data were centered by median and normalized on a gene-wise basis. Hierarchical clustering was performed on both genes and data set using standard centered correlation analyses and average-linkage clustering. Following the identification of major clusters, gene IDs within each cluster were classified based on gene ontology to identify the functional categories of proteins for comparisons between clusters as previously described (20). We performed a conserved domain analysis for the 177 unknown or hypothetical genes that were analyzed in this study to further improve the genome annotation. For 125/177 unknowns, we found at least one significant domain hit, which can give a hint to the putative function of these genes (supplemental Table S2).
dsRNA Synthesis and Gene Silencing
For each candidate phagocyte gene to be analyzed, PCR products were amplified to create plasmid constructs for dsRNA production as previously described (11, 44). All PCR products were subcloned into a pJet1.2 vector using the CloneJet PCR cloning kit (Thermo Scientific) and used as a template for amplification to amplify T7-PCR products. T7 PCR amplicons were purified with the DNA Clean and Concentrator (Zymo Research, Irvine, CA) and resultant templates were used for dsRNA production using the MEGAscript RNAi kit (Life Technologies, Waltham, MA). Primers used for amplification and the production of T7-PCR templates are listed in supplemental Table S3. dsRNA products were used for injections as previously described (11). Surviving mosquitoes were then challenged with P. falciparum-infected blood 2 days post-injection and maintained at 25 °C. To determine the efficiency of dsRNA-mediated silencing, nonfed mosquitoes were collected from each candidate gene experiment and the effects of silencing were measured in whole mosquitoes by qRT-PCR and compared with GFP-silenced control mosquitoes. Primers used for knockdown validation are listed in supplemental Table S3. To determine the effects of gene-silencing on P. falciparum development, dsRNA-injected mosquitoes were maintained for 8 days at 25° and 80% humidity following infection. Oocyst numbers we determined by dissecting midguts in 1× PBS, staining in 0.2% mercurochrome, and then counting on a compound microscope.
VectorBase Gene Accession Numbers
VectorBase gene identifiers of genes featured in our analysis by order of appearance in the text: CTL4, AGAP005335; LYSC1, AGAP007347; DEF1, AGAP011294; HPX2, AGAP009033; TEP1, AGAP010815; SCRBQ2, AGAP010133; LRIM16A, AGAP028028; LRIM15, AGAP007045; SCRASP1, AGAP005625; VCP-like, AGAP007505; pretaporter, AGAP000044; Ras homology member A, AGAP005160; Ras-related, AGAP004559; Rab1A, AGAP004146; PPO2, AGAP006258; PPO4, LRIM1, AGAP006348; APL1C, AGAP007033; AGAP004981; LRIM9, AGAP007453; 60s ribosomal protein L14, AGAP005991; 60s ribosomal protein L36a, AGAP003538.
RESULTS
Magnetic Bead-based Isolation of the Phagocytic Hemocyte Populations
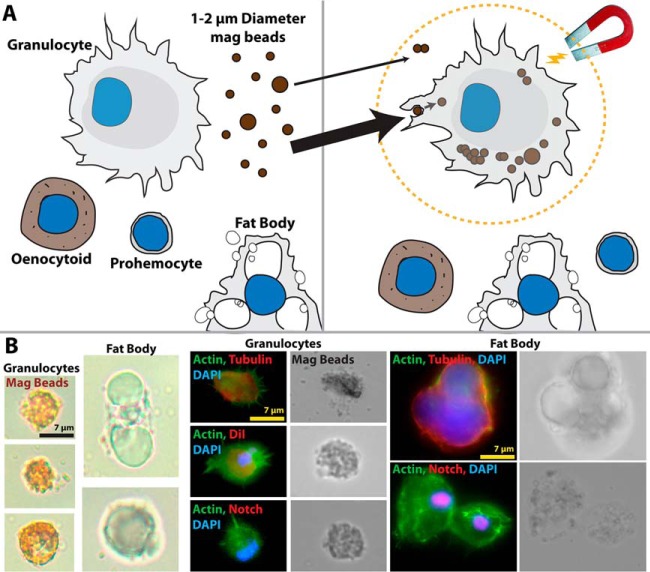
Several methodologies have been used to isolate mosquito hemocytes (4–9, 21). These rely on slight differences in perfusion techniques and isolation methods for the collection of complete hemocoel samples from individual insects. However, these methods often encounter complications such as variability in hemocyte numbers (1) and possible contamination by mosquito fat body cells, extraneous cellular debris, or bacteria (4). Based on the ability of granulocytes to undertake phagocytosis (22); we hypothesized that a highly enriched population of circulating granulocytes could be isolated by perfusion and magnetic purification following the phagocytic uptake of carboxylate-coated magnetic beads. Following this methodology (Fig. 1A), we were able to effectively purify circulating phagocytic granulocytes, enabling proteomic analysis on these enriched hemocyte populations. Microscopy confirmed that mosquito granulocytes efficiently phagocytosed magnetic beads and that perfused fat body cells did not associate with beads (Fig. 1B), demonstrating the specificity of our approach. These visible differences in morphology and phagocytic ability were further validated by immunostaining (Fig. 1B) using previously described hemocyte markers, tubulin and CM-DiI (6, 7) to differentiate between the staining pattern of phagocytic granulocytes and those of fat body cells. Additional experiments using a Notch antibody highlight differences between cell types, with Notch expression detected in fat body nuclei but absent from perfused granulocytes (Fig. 1B). In summary, these data suggest that our method of enrichment specifically targets the phagocytic granulocyte population of mosquito blood cells, thus enabling the advanced proteomic characterization of these relatively undescribed mosquito blood cells.
Fig. 1.
Purification of granulocytes using magnetic microbeads. A, Graphical overview of granulocyte separation by phagocytosis of magnetic beads. Other hemocyte cell types and common contaminants in hemocoel perfusate are not involved in phagocytosis, enabling purification of a highly enriched granulocyte population. B, Light, Phase-contrast and Fluorescent microscopy were used to verify the uptake of magnetic beads by granulocytes, but not fat body, the main contaminant present in such samples. Cell stains and conserved Drosophila antibodies were used to further morphologically differentiate the two cell types.
Proteomic Profiling of Phagocytic Granulocyte Populations in the Context of Homeostasis and Pathogen Challenge
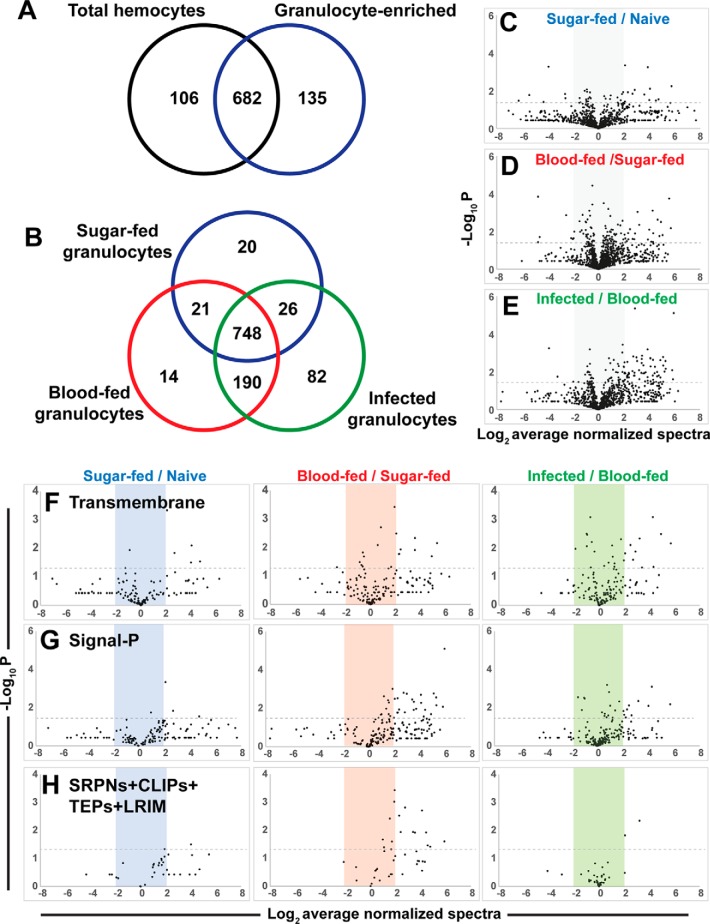
Using our technical advancement to enrich for phagocytic granulocyte populations, we performed proteomic analysis on circulating mosquito hemocyte populations to determine the effects of granulocyte enrichment (phagocytosis), blood feeding, and malaria parasite infection (supplemental Table S4). Initial comparisons were made between nonselected sugar-fed hemocytes (all cell types) and enriched sugar-fed phagocytic granulocytes (Fig. 2A). Additional experiments compared granulocyte-enriched cell populations 48 h after blood-feeding or P. falciparum infection to examine the effects of ookinete invasion (Fig. 2B). Across all samples, a total of 1128 proteins were identified, with 748 identified in all granulocyte-enriched samples (Fig. 2B). We further tested our estimated equivalence in protein loading using normalized spectral counts to perform label-free relative quantification. Average R2 values from pair-wise spectral count correlations indicate a high degree of similarity between the three independent biological replicates (supplemental Fig. S1). Given the difficulty in working with hemocyte samples and their small protein yield, our goal was to identify potential differences in protein abundance across sample treatments that in turn suggest hemocyte responses to granulocyte-enrichment, blood-feeding, and malaria parasite infection.
Fig. 2.
Proteomic analyses of phagocytic hemocyte populations. Venn diagram comparisons of protein identities from three independent biological replicates of mosquito hemocyte proteomes following granulocyte enrichment from total hemocyte populations (A) or comparing granulocyte proteomes in response to feeding status (naïve sugar-fed, blood-feeding, or Plasmodium infection) (B) are shown. Volcano plots of label-free relative quantitative analyses of protein abundance by average normalized spectral counts from three biological replicates (C–E). The three graphs depict total proteins from each sample versus the appropriate reference sample according to feeding status. Levels of predicted transmembrane proteins (F), secreted proteins (G), and a combination of immune gene families (SRPNs, CLIPs, TEPs and LRIMs) (H) were measured across each of the respective treatments (phagocytosis, blood-feeding, and infection). All values are depicted as the Log2 average of normalized spectra, whereas significance (p value) is measured as the -Log10. Dotted lines depict significance with a p value cutoff of 0.05.
Comparisons across proteomes illustrate the effects of granulocyte enrichment (Fig. 2C), blood feeding (Fig. 2D), and P. falciparum infection (Fig. 2E). Of interest, we detected widespread changes in relative protein abundance (both positive and negative) following granulocyte enrichment, yet the effects of blood feeding and parasite infection are largely positive (Figs. 2C–2E). Similar observations were also detected in proteins with predicted transmembrane domains (Fig. 2F) or predicted secreted proteins (Fig. 2G). To address components of mosquito immunity identified in our analysis, immune genes belonging to serine protease inhibitor (SRPNs), clip-domain serine protease (CLIPs), thioester protein (TEPs), or leucine rich-repeat immune protein (LRIMs) gene families were followed across each of the experimental conditions (Fig. 2H). Notably, members of these immune gene families were positively influenced by phagocytosis (granulocyte enrichment) and blood feeding, but showed little response to malaria parasite infection (Fig. 2H). Additional analyses of the Ras superfamily of small GTPases that have been previously implicated in cell proliferation and hemocyte activation (23) were down-regulated in enriched sugar-fed granulocyte populations, whereas blood-feeding and Plasmodium infection produced increased levels of protein abundance (supplemental Fig. S2).
Identification of Candidate Proteins Enriched in Response to Phagocytosis, Blood-feeding, and Parasite Challenge
To identify those proteins with significant changes in relative abundance across experimental samples, strict filtering criteria were applied to consider only statistically significant (p < 0.01) proteins with normalized spectral counts showing greater than 2-fold enrichment. Functional classifications were used to further define the 57 proteins identified according to these requirements (Table I). A complete list of all statistically significant (p < 0.05) proteins is listed in supplemental Table S5 for each experimental treatment.
TABLE 1. Protein enrichment in mag-bead-enriched hemocytes.
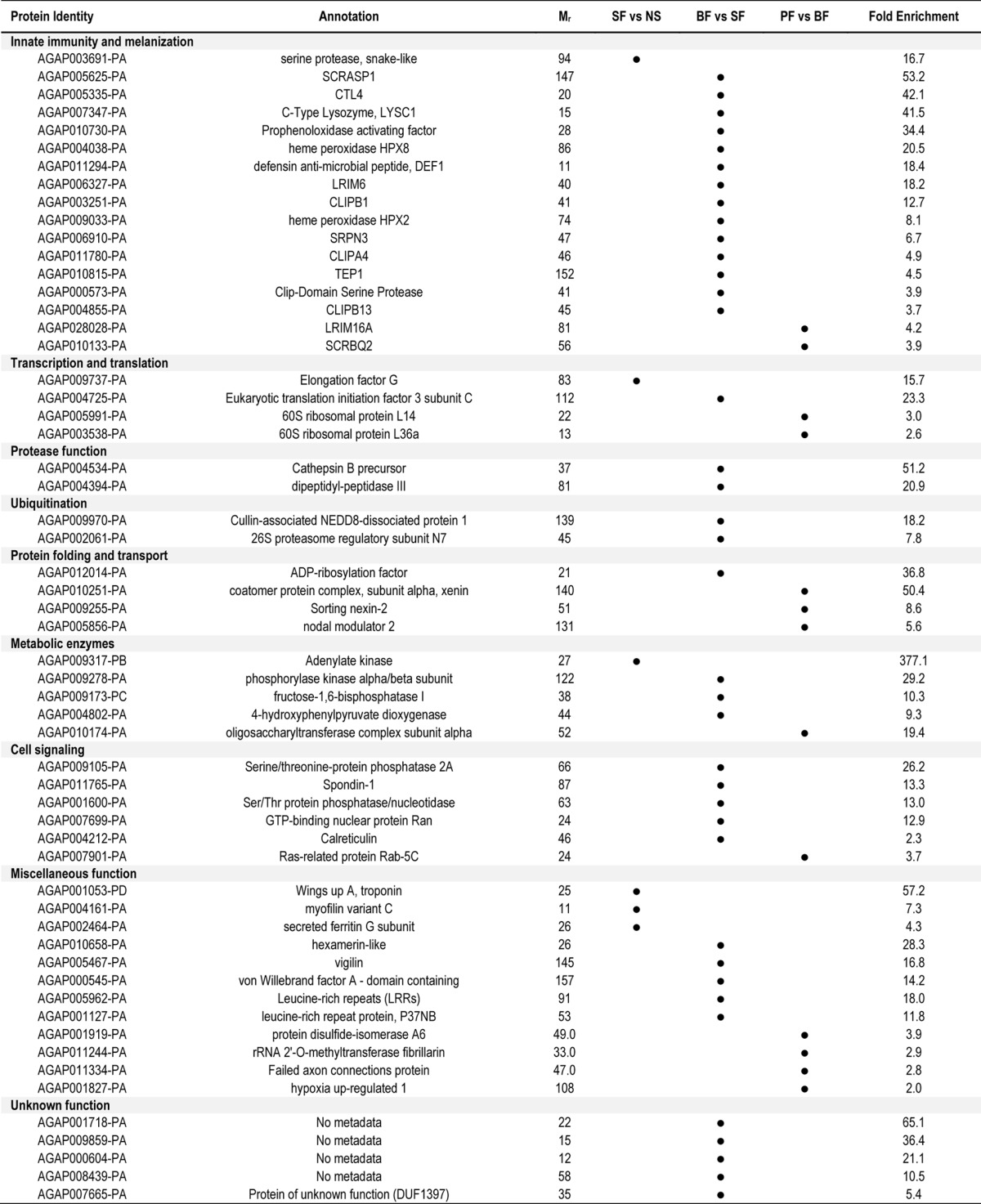
Six proteins appeared to be enriched following granulocyte enrichment (sugar-fed (SF) granulocyte-enriched samples versus nonselected (NS) hemocytes; Table I). However, the small sample size precludes any further conclusions regarding the concerted function of these proteins (Table I). As a result, it is unclear if the proteins identified are indicative of proteins expressed specifically in granulocyte populations or in response to phagocytosis.
Blood-feeding (blood-fed (BF) granulocyte-enriched samples versus sugar-fed (SF) granulocyte-enriched samples) produced the largest group of enriched proteins identified in our analysis (Table I). Approximately one-third of these proteins (14 of 38) are components of the mosquito innate immune response (Table I), despite the absence of parasite challenge in the noninfected blood samples. Several well-described genes influencing Plasmodium development were enriched in this group, including C-type lectin 4, CTL4 (24), lysozyme c-1, LYSC1 (25), defensin 1, DEF1 (26, 27), heme peroxidase 2, HPX2 (28), and thioester protein 1, TEP1 (29–31). In addition, several other proteins featured prominently with presumed functions in cell signaling and metabolism. This suggests that blood-feeding triggers extensive changes in phagocyte populations, and is supported by evidence that blood feeding promotes pervasive changes to mosquito hemocyte populations (9, 23, 32). The remaining proteins were distributed across several presumed biological roles including proteolysis, ubiquitination, metabolic enzymes, cell signaling molecules, and those of unknown function (Table I).
In contrast with the large number of immune components enriched with a noninfectious blood meal, only two of the 13 proteins enriched following Plasmodium infection have presumed roles in the immune response (Table I). Among these two proteins, SCRBQ2 has been previously implicated as an agonist of Plasmodium development (33), whereas LRIM16A is a yet undescribed member of a family of leucine-rich repeat proteins that are closely connected to the mosquito immune system (34). Additional proteins with presumed roles in protein folding and vesicular transport may represent a dramatic cellular reorganization or intracellular communication in response to parasite infection (Table I).
Comparisons Between Phagocyte Proteome Profiles and Published Hemocyte Transcriptomes
To compare our granulocyte-enriched proteomes (SF, BF, PF) (supplemental Table S4) with existing hemocyte transcriptional profiles (8), we performed MCIA (16, 17) to determine the correlation between protein and transcript data. Granulocyte protein samples were directly compared with transcriptional profiles produced by Pinto et al. (8) of nonselected hemocytes from naïve sugar-fed mosquitoes, 24 h after feeding with a noninvasive CTRP mutant Plasmodium berghei (comparable to a noninfectious blood meal), or 24 h post-infection with wild-type P. berghei (8). Despite differences in sample collection, sample time points, and the species of malaria parasite used, our MCIA analysis revealed a high level of similarity between our enriched granulocyte protein profiles and the previously published hemocyte transcriptomes (8) (RV-coefficient of 0.958, p = 0.023) (supplemental Fig. S3A). Additional comparisons were also performed to examine subpopulations of immune-specific (supplemental Fig. S3B) and proliferation-specific (supplemental Fig. S3C) protein profiles.
The MCIA results argue that the greatest degree of post-transcriptional regulation (increased length of the line between comparisons) occurs after an infectious blood meal (PFvSF) (supplemental Fig. S3). Transcript and protein profiles were most similar when parasite infection is compared with a noninfectious blood-meal (PFvBF) (supplemental Fig. S3, supplemental Table S6). This suggests that blood-feeding (independent of infection status) promote considerable differences in the levels of transcript and protein expression. In addition, the immune-specific proteome (supplemental Fig. S3B) displayed the highest levels of post-transcriptional regulation (based on the pair-wise RV-coefficient), implying that components of the mosquito immune response are more likely to undergo translational regulation in agreement with previous studies (35).
Based on these findings, we examined a subset of hemocyte transcripts displaying greater than 2-fold transcript up-regulation (8) and proteins that were enriched in each of our granulocyte samples (Table I) to determine if gene regulation occurs at either the transcript or protein level. Correlations between the log fold-change from the gene expression data (8) and phagocytic granulocyte protein abundance were low (PFvBF = 0.23; BFvSF = 0.10; PFvSF = 0.22) across all experimental samples (supplemental Table S4). However, when the direction of the fold-change is considered, we observed higher levels of concordance between gene and protein expression (PFvBF = 0.54: BFvSF = 0.54; PFvSF = 0.67) (supplemental Table S7), inferring that transcript and protein abundance are much different measurements of granulocyte gene regulation.
To further explore differences between transcript and protein levels, we examined a subset of proteins that featured prominently in our enrichment (Table I, supplemental Table S5), MCIA analyses (supplemental Fig. S3, supplemental Table S6), or qRT-PCR (supplemental Fig. S4). Several patterns emerged, including transcript levels that closely correlate with protein abundance, small differences in transcript levels resulting in large changes in protein abundance, inverse relationships between mRNA/protein levels, and evidence of post-transcriptional regulation (supplemental Fig. S4). In addition, similar expression profiles are shared among granulocyte proteins responsive to blood-feeding, suggesting that they are induced by related mechanisms (supplemental Fig. S4B). This contrasts those genes responsive to Plasmodium infection, which display the greatest discordance (opposite trends) between transcript and protein expression (supplemental Fig. S4C). Although speculative because of the small sample size, these patterns of regulation corresponding with feeding status warrant further study to examine the impact of blood-feeding and parasite infection on granulocyte gene expression.
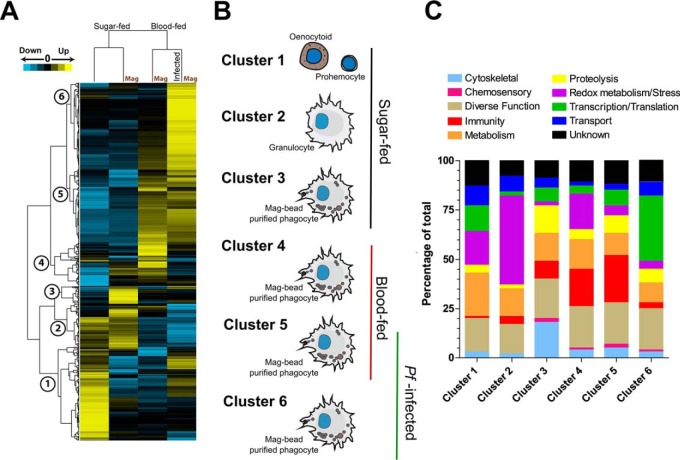
Cluster Analyses Reveals Distinct Hemocyte Protein Profiles Indicative of Cell Type and Feeding Status
Cluster analysis was performed to better understand the association of proteins present in our enriched phagocyte populations across the sample treatments, resulting in six protein clusters indicative of selection and feeding status (Fig. 3A). Clusters 1 to 3 comprise proteins with the highest abundance in sugar-fed hemocytes, whereas clusters 4 to 6 demonstrate responses to blood-feeding and infection (Fig. 3B). Proteins that identified with each cluster (supplemental Table S8) were further classified based on predictive gene function for comparative analysis (Fig. 3C). We identified distinct protein signatures that are associated with hemocyte subtypes (prohemocytes and oenocytoids versus granulocytes) from total and enriched cell samples. In agreement with the cluster analysis, predictive gene ontologies suggest that the cell types associated with each respective cluster have distinct molecular functions (Fig. 3C). These data reveal clear differences between nonselected cell populations (prohemocytes and oenocytoids; Cluster 1) and those observed for granulocyte populations in other clusters (Clusters 2–6, Figs. 3A and 3B). These nonselected hemocyte populations had the highest abundance of proteins involved in cell metabolism and energy transport, distinct from the molecular profiles of the granulocyte-enriched populations identified in other clusters (Fig. 3C). However, because of the presence of both prohemocytes and oenocytoids in the unselected cell population, we are currently unable to assign further roles to the contributions of these hemocyte subpopulations.
Fig. 3.
Clustering analyses reveals expression patterns indicative of feeding status and infection. Proteomic data from each sample treatment was clustered into six distinct coexpression clusters based on protein abundance (A). The analysis clearly defined feeding status (sugar- or blood-fed) into two distinct clades. B, Visual representations display information regarding cell type and feeding status for each cluster group. Proteins associated with each cluster reveal distinct distributions of molecular function groups (C).
In contrast, we provide significant, new insight into the biology of phagocytic granulocytes and their protein composition in response to phagocytosis, blood-feeding, and Plasmodium infection. These profiles are distinct from the proteins in Cluster 2 that likely represent granulocyte-specific populations, independent of magnetic bead selection (Fig. 3). Based on the cluster and gene ontology analysis, we identify specific responses to phagocytosis (Cluster 3), the effects of a naïve blood meal (Cluster 4), and conserved responses independent of infection status (Cluster 5; Fig. 3). Plasmodium-specific responses are represented by Cluster 6 (Fig. 3).
Candidate Gene Silencing Reveals the Importance of Phagocytic Granulocytes as Modulators of Malaria Parasite Development
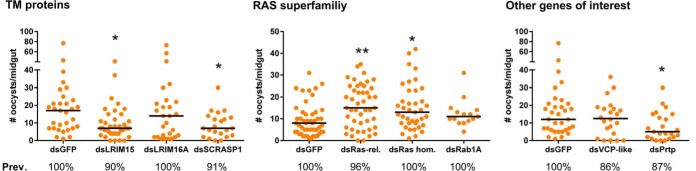
To determine the functional role of several phagocyte proteins in malaria parasite survival, eight candidate genes featuring prominently in our analyses were evaluated for their effects on parasite numbers following dsRNA-mediated gene silencing. These targets were chosen for their presumed roles as cell surface-expressed transmembrane proteins (LRIM15, LRIM16A, SCRASP1), implications in the immune response (LRIM15, LRIM16A, SCRASP1, VCP-like, Prtp), or cell proliferation (Ras-homology member A, Ras-related, Rab1A). Seven of the eight candidate genes were efficiently silenced following dsRNA injection (ranging between 39–72%) when measured by qRT-PCR (supplemental Fig. S5). The remaining gene, VCP-like, proved refractory to gene-silencing under our experimental conditions (supplemental Fig. S5), and had no effect on parasite numbers in the absence of silencing as expected (Fig. 4).
Fig. 4.
Gene-silencing of candidate phagocyte genes reveals agonistic and antagonistic roles in malaria parasite survival. Candidate phagocyte genes were tested by dsRNA-mediated gene silencing and evaluated for their effects on P. falciparum oocyst numbers. Candidate genes are displayed by predicted function as transmembrane (TM) proteins, members of the RAS superfamily, or as other predicted immune genes of interest. For all experiments, each dot represents the number of parasites on an individual midgut 8 days after infection. Oocyst numbers were pooled from three or more independent biological replicates for each candidate gene. Median oocyst numbers from each experimental condition are denoted by the horizontal red line. Statistical analysis was performed using Kruskal-Wallis with a Dunn's post-test to determine significance. Asterisks denote significance (*; p < 0.05; **, p < 0.01). ns, not significant.
For the seven genes that displayed knockdowns by dsRNA-mediated silencing, five candidate genes produced significant mosquito phenotypes that influenced Plasmodium survival (Fig. 4). Silencing of LRIM15, SCRASP1, and pretaporter (Prtp) significantly decreased parasite numbers (Fig. 4), suggesting that these proteins have protective roles that aid Plasmodium development. In contrast, silencing either Ras-related or Ras-homology member A increased parasite numbers (Fig. 4), arguing that the signaling cascades initiated by these signaling molecules limit Plasmodium development. LRIM16A and Rab1A had no measurable effects on parasite survival in our experiments (Fig. 4). Together, these data demonstrate that the granulocyte proteins identified in this study can have profound positive or negative effects on malaria parasite survival in the mosquito host, highlighting the specific role of these immune cells in the mosquito immune response (summarized in Fig. 5).
Fig. 5.
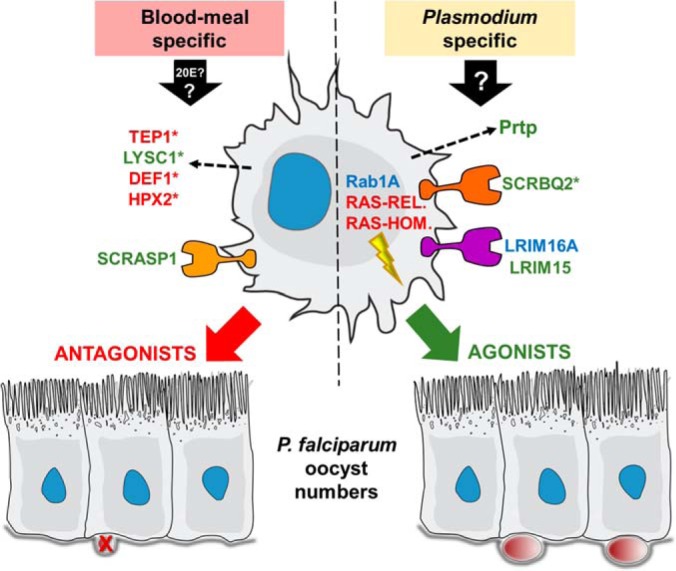
Summary of phagocyte proteins on Plasmodium development. Phagocyte proteins induced by blood-feeding (possibly by 20E or other blood-meal induced factors), or as a result of unknown stimuli following Plasmodium infection, can significantly influence malaria parasite development in the mosquito. Seven novel proteins were examined in this study that when silenced behaved as agonists (green), antagonists (red), or did not influence parasite development (blue). Five additional proteins, with previously described effects on Plasmodium numbers, also featured prominently in our analysis (denoted by asterisks). Predicted roles are depicted for these phagocyte proteins as cell surface receptors, intracellular signaling, or secreted function.
DISCUSSION
Most of our current knowledge of hemocyte biology stems from experiments performed in other insect species, leaving several important aspects of mosquito hemocyte biology unexplored. Unlike commonly studied Drosophila or lepidopteran insects, mosquitoes require a blood meal to complete their life cycle, exposing them to blood meal-derived molecules and pathogens that influence their physiology. Therefore, mosquito hemocyte biology is unique and can serve as a model for other blood-feeding insects. To better understand mosquito hemocyte biology, we performed a comprehensive proteomic analysis of hemocytes in naïve, blood-fed, or Plasmodium-infected An. gambiae. Using a novel “low-tech” strategy to enrich for highly purified phagocytic granulocyte cell populations, we could overcome the lack of genetic tools and cellular markers for hemocytes to enrich for specific hemocyte populations. This enabled the characterization of a highly purified granulocyte-enriched population by proteomic analysis, simplifying the interpretation of the results to a specific hemocyte subtype to examine their cellular context to feeding status and infection.
Based on this enrichment, we identify important components of granulocyte biology in the context of nutritional- and infection-based stimuli. Profiles of sugar-fed granulocytes (Cluster 2) suggest that proteins involved in redox metabolism are a large component of their biology, likely involved in the production of reactive oxygen species used in cell signaling events or killing invading pathogens (36). However, when examining the specific responses to phagocytosis (Cluster 3), it does not appear as redox metabolism proteins are directly activated in response to the “pathogen.” Although these proteins may have been rapidly turned over before our sampling window, we find that cytoskeletal components may have an important role in phagocytosis (Fig. 3, supplemental Table S8) in agreement with previous transcriptional studies in mosquito hemocytes (9) and conserved roles in other experimental systems (37, 38). This might reflect adaptations to an increase in cellular volume or structural rearrangements associated with phagosome maturation and lysosome fusion events that accompany pathogen destruction (39). Additional proteins linked to immune activation and the melanization cascade also clustered with the phagocytic response, including two prophenoloxidase proteins (PPO2 and PPO4), in agreement with previous reports that suggest that melanin deposition is a critical step in phagocytosis (7, 22, 40). However, because of the brown color of the magnetic beads in cells undergoing phagocytosis (Fig. 1), the deposition of melanin on the bead surface is not easily distinguished.
In mosquitoes, our understanding of prophenoloxidase (PPO) expression has been limited, yet often correlated with oenocytoid cell populations based on PPO expression in the equivalent cell type (crystal cells) in Drosophila (42). Although a subset of the 10 An. gambiae PPOs may be exclusively expressed in oenocytoid populations, our results argue that mosquito PPOs are also expressed in granulocyte populations. This is further supported by PPO6 staining in An. gambiae granulocytes (4, 23, 43), as well as the presence of PPO-positive granulocytes in other mosquito species, moths, and houseflies (44, 45). Together, these results suggest new possible roles for PPOs in mosquito granulocytes and mosquito innate immunity.
The results of our study also provide new perspectives into the regulation of the mosquito innate immune response. Large numbers of immune components were regulated by blood-feeding alone, suggesting that immune activation is largely independent of malaria parasite infection. Among these proteins, many have well-characterized roles in influencing Plasmodium survival and clearance, including TEP1, LRIM1, and APL1, which are core components of a complement-like immune response (30, 31, 41). Previous work has suggested that changes in mosquito physiology associated with blood-feeding may strongly influence mosquito immunity in the absence of infection (23, 32, 43, 46). This is supported by the induction of LRIM9 following 20-hydroxyecdysone (20E) injection (46) and the influence of 20E on prophenoloxidase expression in mosquito cell lines (47). LRIM9 and multiple prophenoloxidases were identified in our phagocyte proteomes displaying increased levels of protein abundance after a noninfected blood meal. Therefore, it is suggestive that hormonal changes associated with blood-feeding may influence mosquito granulocytes in the absence of infection. It has recently been proposed that physiological signals produced in response to blood-feeding comprise a preemptive or anticipatory response, to combat blood-borne pathogens (46). Our findings provide support that these physiological signals promote an anticipatory immune response in mosquito granulocytes.
Plasmodium infection also initiated dramatic changes to the hemocyte proteome beyond the changes identified with blood-feeding alone. However, very few proteins of immune function were specifically associated with parasite infection. In contrast, infection promoted marked increases in the translation machinery, most notably in 40S and 60S ribosomal protein subunits and translation initiation factors including two 60s ribosomal subunits (L14 and L36a) that were enriched following Plasmodium infection (Table I, supplemental Table S8). These data suggest that Plasmodium infection broadly increases mRNA translation and protein synthesis in granulocytes, in agreement with previously published results examining mosquito midgut responses to infection (35, 48). This may result from changes in metabolic activity in response to parasite infection that promote granulocyte activation and/or differentiation, and is supported by recent evidence that ookinete traversal of the midgut promotes hemocyte differentiation and increased granulocyte numbers (49).
Both blood-feeding and parasite infection have dramatic impacts on mosquito physiology, with a primary function to convert the blood meal into a nutrient source for egg production (50), yet there is a delicate balance between reproduction and diverting resources toward an immune response to limit malaria parasite infection (51). These physiological changes also influence mosquito hemocyte populations, as blood-feeding promotes an increase in hemocyte numbers (9, 23, 32), whereas Plasmodium infection promotes hemocyte differentiation (5, 47, 52). In agreement with these observations, we noted increases in the Ras family of small GTPases in response to blood-feeding and infection, and demonstrate that two Ras components (Ras-related and Ras-homology member A) have integral roles in shaping parasite numbers in the mosquito host. Ras superfamily members have described roles in the regulation of cell growth and proliferation in metazoans, and over-expression of Ras85D results in dramatic increases of Drosophila hemocyte cell numbers (53, 54). In addition, Bryant et al. (23) suggest that Ras-MAPK signaling contributes to blood-meal induced hemocyte activation, similar to the signaling pathways that define macrophage (55) and Drosophila hemocyte activation (54, 56). While the precise mechanism in which these Ras-like genes function have yet to be determined, these signaling components may regulate the respective hemocyte proliferation or differentiation responses produced as a result of blood feeding and ookinete invasion (9, 23, 32, 47, 50).
Additional gene silencing experiments illustrate that hemocyte proteins can also positively influence Plasmodium survival. In contrast to most previously described mosquito genes that manipulate malaria parasite numbers, few genes have been identified that confer protective effects on parasite growth (57). Of these examples, several contribute to parasite melanization (24, 58), a phenotype not typically seen in the Keele strain of An. gambiae that was used in our infection experiments with P. falciparum. Two of the three Plasmodium agonists identified in our knockdown experiments are putative cell surface proteins, as well as another previously described putative cell surface protein SCRBQ2 (33) that was identified in our enriched phagocyte samples that similarly reduce oocyst numbers when silenced (Fig. 5), suggesting that hemocyte surface proteins may be important regulators of mosquito immunity.
LRIM15 is a poorly described member of the leucine-rich repeat immune (LRIM) protein family containing a transmembrane domain and described function as a cell-surface marker (34, 59). LRIM family members have been previously implicated in the immune response (34, 59) and serve important roles as components of the mosquito complement-like system (30, 31). Family members contain signal peptide sequences that mediate the secretion of these proteins to the hemolymph with the exception of the two LRIMs containing transmembrane domains examined in our study, LRIM15 and LRIM16A, suggesting that these two proteins may have much different functions. As evident by our gene-silencing experiments, only LRIM15 is an important effector of mosquito immunity. We also demonstrate that a putative scavenger-receptor, SCRASP1, is an important modulator of Plasmodium development. SCRASP1 is of undescribed function, yet uniquely contains predicted chitin-binding, serine protease, low-density lipoprotein receptor, and a transmembrane domain that confer its function. Similar to the reduced numbers of malaria parasites produced when SCRBQ2 was silenced (33), the silencing of SCRASP1 also led to decreased oocyst numbers, arguing that these scavenger receptors may influence malaria parasite development by comparable mechanisms. Highly enriched after blood-feeding alone, SCRASP1 may contribute to pre-emptive immune responses activated by the hormonal changes that accompany blood feeding as described above, distinct from the Plasmodium-specific enrichment of SCRBQ2. Although the putative functions for LRIM15, SCRASP1, and SCRBQ2 remain unknown, their concerted role as cell surface receptors suggest that they may act as immuno-suppressors that when silenced, increase innate immune signaling in mosquito hemocyte populations.
In contrast, the presumed role of Prtp in mosquitoes suggests that its protective effects on parasite numbers may occur through different mechanisms. Prtp contains two thioredoxin-like domains that may be involved in redox metabolism (60), inferring that the loss of Prtp by dsRNA-mediated gene silencing may be attributed to increased levels of reactive oxygen species (ROS) and is supported by evidence that thioredoxins protect cells from ROS-mediated cytotoxicity in orthologous systems (60). This argues that silencing may create a toxic cellular environment in the mosquito host that limits parasite numbers. Alternatively, based on previous studies in Drosophila embybros, Prtp has been shown to interact with Draper to influence the phagocytosis of apoptotic cells (61). Induced following Plasmodium infection, Prtp may be involved in a wound healing response and clearance of apoptotic cells following ookinete invasion, although at this time it is unclear how this might influence parasite numbers.
The results of our gene silencing experiments, combined with the previously published roles of several proteins identified in our study, further illustrate the role of mosquito hemocytes as important modulators of parasite development (Fig. 5). However, because of the systemic nature of dsRNA-mediated silencing, we cannot fully exclude that these candidate proteins might have roles in other mosquito tissues. Although further work is required to explore the roles of phagocyte proteins in anti-Plasmodium immunity, the identification of TEP1 and other secreted proteins argue that hemocyte-derived hemolymph factors are important components of the “early-phase” response responsible for ookinete killing. Alternatively, some of the phagocyte proteins identified in our study may also contribute to the yet uncharacterized “late-phase” immune responses that mediate oocyst survival (49). With functional roles as cell surface receptors, secreted proteins, and intracellular signaling molecules, these findings demonstrate that multiple; temporally coordinated events contribute to hemocyte immune responses in the mosquito host.
Hemocytes have a primary role in mosquito host defenses to extracellular and invading pathogens by initiating both cellular and humoral immune responses (1, 6–8). Evidence suggests that the presence of commensal bacteria in the mosquito midgut can shape basal immune levels, determine immune priming, and can modulate the development of Plasmodium parasites in the mosquito host (41, 62). Shortly after blood ingestion, the resident microbiota undergoes a dramatic proliferation that likely produces an immune response in the absence of an infectious blood meal. Therefore, it is possible that bacterial titers in the mosquito midgut could influence immune responses produced by hemocytes. However, the effects of the microbiome were not addressed in our current study, and therefore we cannot exclude the role of commensal bacteria on our phagocyte proteomes. This is of interest because of the putative roles of bacteria in shaping the anticipatory immune response between comparisons of our sugar-fed and blood-fed phagocyte proteomes. However, the up-regulation of LRIM9 after blood feeding (an immune component that grouped with our blood-responsive clustering analyses and responsive to 20E) is independent of the endogenous midgut microflora (46), arguing support for blood-meal induced components in our analyses. More importantly, proteome comparisons between phagocyte samples from blood-fed and Plasmodium-infected mosquitoes encounter similar changes in their microflora following blood-feeding, minimizing the effects of the microbiota in our analysis.
Mosquito hemocytes contribute to pathogen clearance by phagocytosis (7, 22, 40) and are presumed to have a major role in the secretion of proteins into the hemolymph (29–31, 59, 63). An initial characterization of mosquito hemolymph components identified 26 proteins (64), of which approximately two-thirds (17/26) were identified in our proteomic data. In addition, several well-described humoral immune components such as TEP1, leucine-rich repeat immune protein 1 (LRIM1), and Anopheles Plasmodium-responsive leucine-rich repeat 1, APL1 (30, 31) were identified in our proteomic analysis, providing confirmation that several key modulators of the immune response are produced at least in part by mosquito granulocytes. Together, this argues that many immune-related genes measured in studies that focused on midgut immunity may actually be derived from hemocytes, possibly from granulocytes attached to the midgut basal lamina. Hemocyte attachment to the midgut basal lamina has been previously suggested to account for midgut TEP1 expression (29, 65) and likely accounts for the detection of several more hemocyte genes identified in this study. Therefore, our proteomic analysis should be a valuable resource to delineate the contributions of hemocytes to the immune responses that determine Plasmodium development in the mosquito host.
Our analysis of hemocyte protein profiles provides a strong foundation for the advanced study of mosquito hemocyte biology, and identifies several candidate surface markers of phagocytic granulocyte populations to enable their future study. We identify 177 proteins with predicted transmembrane domains, including SCRBQ2, SCRASP1, LRIM15, and LRIM16A that feature prominently in our analysis. Previous studies have demonstrated that LRIM15 is expressed on the surface of transfected Sf9 cells (59), adding further support and validation for this list of putative surface markers. In addition, we also identified several genes in our hemocyte proteomes that have been previously proposed as hemocyte-subtype specific protein markers (8). Collectively, these putative markers may be used in combination to select and further profile hemocyte populations in future experiments.
Integrating proteomic data with existing hemocyte transcriptional data, our MCIA analysis sheds significant insight into hemocyte gene regulation. Comparisons across transcript/protein data sets and feeding status suggest that the highest level of post-transcriptional regulation in phagocytic granulocytes occurs following Plasmodium infection and is most pronounced with components of the mosquito innate immune response. Our qRT-PCR analysis comparing transcript levels and protein abundance for a limited subset of genes identified in the phagocyte proteomes further supports this statement, with the majority of immune proteins enriched following Plasmodium infection displaying patterns of post-transcriptional gene regulation. This is supported by evidence demonstrating the increased translation of immune genes in the mosquito midgut following P. falciparum infection, with little change in mRNA expression levels (35). Together, this would suggest that post-transcriptional regulation serves an important role in the regulation of the mosquito immune response. Although it requires further investigation, it is speculative that granulocytes (and possibly other mosquito tissues) may store select transcripts to facilitate quick responses to stimuli following pathogen infection.
The multicomponent analysis also highlights differences in gene regulation at the transcript and protein level. Although the previously reported hemocyte transcriptomes displayed strong similarities with our proteomic data by MCIA analysis, closer examination of transcript and protein expression levels by log fold change highlight differences in these two measurements of gene function. When a subset of genes was examined by qRT-PCR, most transcripts display modest changes in expression, generally less than 5 fold between sample treatments. In contrast, in some instances we observe dramatic changes in protein abundance including ∼400-fold change in protein abundance for adenylate kinase, and ∼20–40-fold differences for snake-like, CTL4, and DEF1. These data highlight important differences in the regulation of transcript and protein profiles, especially when previous descriptions of mosquito hemocyte gene regulation have solely focused on transcript abundance.
In summary, we provide the first integrative-OMICS analysis of phagocytic granulocytes in An. gambiae and demonstrate specific changes in protein abundance that corresponds with granulocyte enrichment, blood-feeding, and Plasmodium infection. Using MCIA and qRT-PCR analysis, we provide insight into patterns of hemocyte gene regulation at the transcript and protein levels, arguing that post-transcriptional regulation is an important aspect of mosquito hemocyte biology. In addition, our findings offer a new perspective of the mosquito immune system, providing strong evidence that hemocytes are primary components of an anticipatory immune response that primes mosquito immunity upon blood-feeding. These physiological changes, likely mediated through the effects of mosquito hormones or blood components, dramatically shape mosquito immunity. We further establish the involvement of several candidate phagocyte proteins by gene-silencing experiments, demonstrating that mosquito hemocyte can have positive or negative influences on malaria parasite development in the mosquito host. Together, these findings represent a significant advancement in An. gambiae hemocyte biology and provide important new insights into the integral roles of granulocytes in shaping mosquito physiology and immune responses to malaria parasites.
Supplementary Material
Acknowledgments
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions: R.C.S., J.G.K., C.B., G.G.T., and R.R.D. conceptualized the study. R.C.S., J.G.K., D.T., O.A.Z., G.G.T., and R.R.D. designed the experiments. R.C.S., J.G.K., D.T., and O.A.Z. performed the experiments. All authors contributed to data analysis, writing, and final preparation of the manuscript text and figures.
* The work was supported in part by the Bloomberg Family Foundation through the Johns Hopkins Malaria Research Institute (to RRD) and the Austrian BMWFJ, BMVIT, SFG, Standortagentur Tirol, and ZIT through the Austrian FFG-COMET-Funding Program FFG grant #824186 (to GGT). Additional support was provided by a JHMRI Postdoctoral fellowship (to RCS).
 This article contains supplemental material.
This article contains supplemental material.
To facilitate the review of the mass spectral data, we have uploaded the files to PRoteomics IDEntifications database (PRIDE). The data can be accessed through the following reviewer account details:
Username: reviewer62532@ebi.ac.uk
Password: IcSSRtVQ
The authors declare that there are no competing interests that may be perceived to influence the conduct or analysis of the data.
1 The abbreviations used are:
- MLCID
- multilane combined in-gel digestion
- FDR
- false discovery rate
- MCIA
- multiple coinertia analysis.
REFERENCES
- 1. Hillyer J. F., and Strand M. R. (2014) Mosquito hemocyte-mediated immune responses. Curr. Opin. Insect. Sci. 3, 14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Honti V., Csordás G., Kurucz É Márkus R, and Andó I. (2014) The cell-mediated immunity of Drosophila melanogaster: hemocyte lineages, immune compartments, microanatomy and regulation. Dev. Comp. Immunol. 42, 47–56 [DOI] [PubMed] [Google Scholar]
- 3. Wang L., Kounatidis I., and Ligoxygakis P. (2014) Drosophila as a model to study the role of blood cells in inflammation, innate immunity and cancer. Front. Cell Infect. Microbiol. 3, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castillo J. C., Robertson A. E., and Strand M. R. (2006) Characterization of hemocytes from the mosquitoes Anopheles gambiae and Aedes aegypti. Insect. Biochem. Mol. Biol. 36, 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodrigues J., Brayner F. A., Alves L. C., Dixit R., and Barillas-Mury C. (2010) Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 329, 1353–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. King J. G., and Hillyer J. F. (2013) Spatial and temporal in vivo analysis of circulating and sessile immune cells in mosquitoes: hemocyte mitosis following infection. BMC Biol. 11, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. King J. G., and Hillyer J. F. (2012) Infection-induced interaction between the mosquito circulatory and immune systems. PLoS Pathog. 8, e1003058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pinto S. B., Lombardo F., Koutsos A. C., Waterhouse R. M., McKay K., An C., Ramakrishnan C., Kafatos F. C., and Michel K. (2009) Discovery of Plasmodium modulators by genome-wide analysis of circulating hemocytes in Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 106, 21270–21275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baton L., Robertson A., Warr E., Strand M., and Dimopoulos G. (2009) Genome-wide transcriptomic profiling of Anopheles gambiae hemocytes reveals pathogen-specific signatures upon bacterial challenge and Plasmodium. BMC Genom. 13, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hurd H., Taylor P. J., Adams D., Underhill A., and Eggleston P. (2005) Evaluating the costs of mosquito resistance to malaria parasites. Evolution 59, 2560–2572 [PMC free article] [PubMed] [Google Scholar]
- 11. Smith R. C., Eappen A., Radtke A., and Jacobs-Lorena M. (2012) Regulation of anti-Plasmodium immunity by a LITAF-like transcription factor in the malaria vector Anopheles gambiae. PLoS Pathog. 8: e1002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eappen A. G., Smith R. C., and Jacobs-Lorena M. (2013) Enterobacter-activated mosquito immune responses to Plasmodium involve activation of SRPN6 in Anopheles stephensi. PLoS ONE 8, e62937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramphul U.N., Garver L.S., Molina-Cruz A., Canepa G.E., and Barillas-Mury C.. Plasmodium falciparum evades mosquito immunity by disrupting JNK-mediated apoptosis of invaded midgut cells. Proc. Natl. Acad. Sci. U.S.A. 112(5), 1273–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tao D., Ubaida-Mohien C., Mathias D., King J., Pastrana-Mena R., Tripathi A., Goldowitz I., Graham D., Moss E., Marti M., and Dinglasan R. R. (2014) Sex-partitioning of the Plasmodium falciparum stage V gametocyte proteome provides insight into falciparum-specific cell biology. Mol. Cell. Proteomics 13, 2705–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meng C., Kuster B., Culhane A. C., Gholami AM:. (2014) A multivariate approach to the integration of multi-omics data sets. BMC Bioinformatics 15, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tomescu OA, Mattanovich D, Thallinger GG: Integrative omics analysis. A study based on Plasmodium falciparum mRNA and protein data. BMC Syst. Biol. 2014, 8 Suppl 2:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robert P., and Escoufier Y. (1976) A unified tool for linear multivariate statistical methods:The RV-coefficient. Appl. Stat. – J. Roy. St. C. 25, 8 [Google Scholar]
- 19. Smilde A. K., Kiers H. A., Bijlsma S., Rubingh C., and Van Erk M. (2009) Matrix correlations for high-dimensional data: the modified RV-coefficient. Bioinformatics 25, 401–405 [DOI] [PubMed] [Google Scholar]
- 20. Mendes A. M., Awono-Ambene P. H., Nsango S. E., Cohuet A., Fontenille D., Kafatos F. C., Christophides G. K., Morlais I., and Vlachou D. (2011) Infection intensity-dependent responses of Anopheles gambiae to the African malaria parasite Plasmodium falciparum. Infect. Immun. 79, 4708–4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abraham E., Pinto S., Ghosh A., Vanlandingham D., Budd A., Higgs S., Kafatos F., Jacobs-Lorena M., and Michel K. (2005) An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 102, 16327–16332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hillyer J., Schmidt S., and Christensen B. (2003) Rapid phagocytosis and melanization of bacteria and Plasmodium sporozoites by hemocytes of the mosquito Aedes aegypti. J. Parasitol. 89, 62–69 [DOI] [PubMed] [Google Scholar]
- 23. Bryant W. B., and Michel K. (2014) Blood feeding induces hemocyte proliferation and activation in the African malaria mosquito, Anopheles gambiae Giles. J. Exp. Biol. 217, 1238–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osta M. A., Christophides G. K., and Kafatos F. C. (2004) Effects of mosquito genes on Plasmodium development. Science 303, 2030–2032 [DOI] [PubMed] [Google Scholar]
- 25. Kajla M. K., Shi L., Li B., Luckhart S., Li J., and Paskewitz S. M. (2011) A new role for an old antimicrobial: lysozyme c-1 can function to protect malaria parasites in Anopheles mosquitoes. PLoS ONE 6, e19649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dimopoulos G., Richman A., Muller H.-M., and Kafatos F. C. (1997) Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl. Acad. Sci. USA 94, 11508–11513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luna C., Hoa N. T., Lin H., Zhang L., Nguyen H. L., Kanzok S. M., and Zheng L. (2006) Expression of immune responsive genes in cell lines from two different Anopheline species. Insect Mol. Biol. 15, 721–729 [DOI] [PubMed] [Google Scholar]
- 28. Oliveira G. D. A., Lieberman J., and Barillas-Mury C. (2012) Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science 335, 856–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blandin S., Shiao S. H., Moita L. F., Janse C. J., Waters A. P., Kafatos F. C., and Levashina E. A. (2004) Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116, 661–670 [DOI] [PubMed] [Google Scholar]
- 30. Povelones M., Waterhouse R. M., Kafatos F. C., and Christophides G. K. (2009) Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 324, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fraiture M., Baxter R. H., Steinert S., Chelliah Y., Frolet C., Quispe-Tintaya W., Hoffmann J. A., Blandin S., and Levashina E. A. (2009) Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe 5, 273–284 [DOI] [PubMed] [Google Scholar]
- 32. Castillo J., Brown M. R., and Strand M. R. (2011) Blood Feeding and Insulin-like Peptide 3 Stimulate Proliferation of Hemocytes in the Mosquito Aedes aegypti. PLoS Pathog. 7, e1002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. González-Lázaro M., Dinglasan R. R., Hernández-Hernández L., Rodríguez M. H., Laclaustra M., Jacobs-Lorena M., and Flores-Romo L. (2009) Anopheles gambiae Croquemort SCRBQ2, expression profile in the mosquito and its potential interaction with the malaria parasite Plasmodium berghei. Insect Biochem. Mol. Biol. 39, 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Waterhouse R. M., Povelones M., and Christophides G. K. (2010) Sequence-structure-function relations of the mosquito leucine-rich repeat immune proteins. BMC Genomics 11, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mead E. A., Li M., Tu Z., and Zhu J. (2012) Translational regulation of Anopheles gambiae mRNAs in the midgut during Plasmodium falciparum infection. BMC Genomics 13, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dupré-Crochet S., Erard M., and Nüβe O. (2013) ROS production in phagocytes: why, when, and where? J. Leukoc. Biol. 94, 657–670 [DOI] [PubMed] [Google Scholar]
- 37. May R. C., and Machesky L. M. (2001) Phagocytosis and the actin cytoskeleton. J. Cell Sci. 114, 1061–1077 [DOI] [PubMed] [Google Scholar]
- 38. Coppolino M. G., Krause M., Hagendorff P., Monner D. A., Trimble W., Grinstein S., Wehland J., and Sechi A. S. (2001) Evidence for a molecular complex consisting of Fyb/SLAP, SLP-76, Nck, VASP and WASP that links the actin cytoskeleton to Fcgamma receptor signalling during phagocytosis. J. Cell Sci. 114, 4307–4318 [DOI] [PubMed] [Google Scholar]
- 39. Lemaitre B., and Hoffmann J. (2007) The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]
- 40. Hillyer J. F., Schmidt S. L., and Christensen B. M. (2003) Hemocyte-mediated phagocytosis and melanization in the mosquito Armigeres subalbatus following immune challenge by bacteria. Cell Tissue Res. 313, 117–127 [DOI] [PubMed] [Google Scholar]
- 41. Smith R. C., Vega-Rodríguez J., and Jacobs-Lorena M. (2014) The Plasmodium bottleneck : malaria parasite losses in the mosquito vector. Mem. Inst. Oswaldo Cruz 109, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dudzic J. P., Kondo S., Ueda R., Bergman C. M., and Lemaitre B. (2015) Drosophila innate immunity: regional and functional specialization of prophenoloxidases. BMC Biol. 13, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bryant W. B., and Michel K. (2015) Anopheles gambiae hemocytes exhibit transient states of activation. Dev. Comp. Immunol. 55, 119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z., Lu A., Li X., Shao Q., Beerntsen B. T., Liu C., Ma Y., Huang Y., Zhu H., and Ling E. (2011) A systematic study on hemocyte identification and plasma prophenoloxidase from Culex pipiens quinquefasciatus at different developmental stages. Exp. Parasitol. 127, 135–141 [DOI] [PubMed] [Google Scholar]
- 45. Ling E., and Yu X. Q. (2005) Prophenoloxidase binds to the surface of hemocytes and is involved in hemocyte melanization in Manduca sexta. Insect Biochem. Mol. Biol. 35, 1356–1366 [DOI] [PubMed] [Google Scholar]
- 46. Upton L. M., Povelones M., and Christophides G. K. (2015) Anopheles gambiae Blood Feeding Initiates an Anticipatory Defense Response to Plasmodium berghei. J. Innate Immun. 7, 74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Müller H. M., Dimopoulos G., Blass C., and Kafatos F. C. (1999) A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J. Biol. Chem. 274, 11727–11735 [DOI] [PubMed] [Google Scholar]
- 48. Wang B., Pakpour N., Napoli E., Drexler A., Glennon E. K., Surachetpong W., Cheung K., Aguirre A., Klyver J. M., Lewis E. E., Eigenheer R., Phinney B. S., Giulivi C., and Luckhart S. (2015) Anopheles stephensi p38 MAPK signaling regulates innate immunity and bioenergetics during Plasmodium falciparum infection. Parasit. Vectors 8, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith R. C., Barillas-Mury C., and Jacobs-Lorena M. (2015) Hemocyte differentiation mediates the late-phase immune response against Plasmodium in Anopheles gambaie. Proc. Natl. Acad. Sci. U.S.A. 112, E3412–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kokoza V. A., Martin D., Ahmed A., Mienaltowski M. J., Morton C. M., and Raikhel A. S. (2001) Transcriptional regulation of the mosquito vitellogenin gene via a blood meal-triggered cascade. Gene 274, 47–65 [DOI] [PubMed] [Google Scholar]
- 51. Rono M. K., Whitten M. M., Oulad-Abdelghani M., Levashina E. A., and Marois E. (2010) The major yolk protein vitellogenin interferes with the anti-plasmodium response in the malaria mosquito Anopheles gambiae. PLos Biol. 8, e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ramirez J. L., Garver L. S., Brayner F. A., Alves L. C., Rodrigues J., Molina-Cruz A., and Barillas-Mury C. (2014) The Role of Hemocytes in Anopheles gambiae Antiplasmodial Immunity. J. Innate Immun. 6, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Asha H., Nagy I., Kovacs G., Stetson D., and Ando I. (2003) Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zettervall C. J., Anderl I., Williams M. J., Palmer R., Kurucz E., Ando I., and Hultmark D. (2004) A directed screen for genes involved in Drosophila blood cell activation. Proc. Natl. Acad. Sci. USA 101, 14192–14197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cook A. D., Braine E. L., and Hamilton J. A. (2004) Stimulus-dependent requirement for granulocyte-macrophage colony-stimulating factor in inflammation. J. Immunol. 173, 4643–4651 [DOI] [PubMed] [Google Scholar]
- 56. Sinenko S. A., Shim J., and Banerjee U. (2011) Oxidative stress in the haematopoietic niche regulates the cellular immune response in Drosophila. EMBO Rep. 13, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sreenivasamurthy S. K., Dey G., Ramu M., Kumar M., Gupta M. K., Mohanty A. K., Harsha H. C., Sharma P., Kumar N., Pandey A., Kumar A., and Prasad T. S. (2013) A compendium of molecules involved in vector-pathogen interactions pertaining to malaria. Malar J. 12, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Michel K., Budd A., Pinto S., Gibson T. J., and Kafatos F. C. (2005) Anopheles gambiae SRPN2 facilitates midgut invasion by the malaria parasite Plasmodium berghei. EMBO Rep. 6, 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Midega J., Blight J., Lombardo F., Povelones M., Kafatos F., and Christophides G. K. (2013) Discovery and characterization of two Nimrod superfamily members in Anopheles gambiae. Pathog. Glob. Health 107, 463–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shao L. E., Tanaka T., Gribi R., and Yu J. (2002) Thioredoxin-related regulation of NO/NOS activities. Ann. N.Y. Acad. Sci. 962, 140–150 [DOI] [PubMed] [Google Scholar]
- 61. Kuraishi T., Nakagawa Y., Nagaosa K., Hashimoto Y., Ishimoto T., Moki T., Fujita Y., Nakayama H., Dohmae N., Shiratsuchi A., Yamamoto N., Ueda K., Yamaguchi M., Awasaki T., and Nakanishi Y. (2009) Pretaporter, a Drosophila protein serving as a ligand for Draper in the phagocytosis of apoptotic cells. EMBO J. 28, 3868–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dong Y., Manfredini F., and Dimopoulos G. (2009) Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 5, e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frolet C., Thoma M., Blandin S., Hoffmann J. A., and Levashina E. A. (2006) Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity 25, 677–685 [DOI] [PubMed] [Google Scholar]
- 64. Paskewitz S. M., and Shi L. (2005) The hemolymph proteome of Anopheles gambiae. Insect Biochem. Mol. Biol. 35, 815–824 [DOI] [PubMed] [Google Scholar]
- 65. Vlachou D., Schlegelmilch T., Christophides G. K., and Kafatos F. C. (2005) Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr. Biol. 15, 1185–1195 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.