Abstract
Protein post-translational methylation has been reported to occur in archaea, including members of the genus Sulfolobus, but has never been characterized on a proteome-wide scale. Among important Sulfolobus proteins carrying such modification are the chromatin proteins that have been described to be methylated on lysine side chains, resembling eukaryotic histones in that aspect. To get more insight into the extent of this modification and its dynamics during the different growth steps of the thermoacidophylic archaeon S. islandicus LAL14/1, we performed a global and deep proteomic analysis using a combination of high-throughput bottom-up and top-down approaches on a single high-resolution mass spectrometer. 1,931 methylation sites on 751 proteins were found by the bottom-up analysis, with methylation sites on 526 proteins monitored throughout three cell culture growth stages: early-exponential, mid-exponential, and stationary. The top-down analysis revealed 3,978 proteoforms arising from 681 proteins, including 292 methylated proteoforms, 85 of which were comprehensively characterized. Methylated proteoforms of the five chromatin proteins (Alba1, Alba2, Cren7, Sul7d1, Sul7d2) were fully characterized by a combination of bottom-up and top-down data. The top-down analysis also revealed an increase of methylation during cell growth for two chromatin proteins, which had not been evidenced by bottom-up. These results shed new light on the ubiquitous lysine methylation throughout the S. islandicus proteome. Furthermore, we found that S. islandicus proteins are frequently acetylated at the N terminus, following the removal of the N-terminal methionine. This study highlights the great value of combining bottom-up and top-down proteomics for obtaining an unprecedented level of accuracy in detecting differentially modified intact proteoforms. The data have been deposited to the ProteomeXchange with identifiers PXD003074 and PXD004179.
Living organisms are classified into three domains of life, Bacteria, Eukarya and Archaea. Archaea inhabit highly diverse environments, ranging from marine waters and soil to the human gut. However, they are perhaps best known for their ability to sustain harsh physicochemical conditions, which are too extreme for the growth of bacteria and eukaryotes. Indeed, archaea can thrive in the environments where the temperature exceeds 100 °C (up to 121 °C; (1)) and pH drops below 1 (2).
Like in eukaryotes and bacteria, protein post-translational modifications (PTMs)1 play a crucial role in the functioning of archaeal cells. More and more studies are dedicated to archaeal PTMs (3, 4), and several PTMs have already been described: proteolytic processing, methylation, acetylation, phosphorylation, ADP-ribosylation, glycosylation (5), and modification with ubiquitin-like proteins (6). Nonetheless, thus far, most of the studies on archaeal PTMs have focused on single proteins or protein complexes, with only few of them using proteome-wide approaches (7, 8). Therefore, the global extent of PTMs and their dynamics in the course of cell cycle remain largely unexplored.
Although the subcellular organization of archaea is generally similar to that of bacteria (both lack nucleus and intracellular compartments), some molecular machineries responsible for the key aspects of archaeal cell biology closely resemble the corresponding systems of eukaryotes. Members of the phylum Crenarchaeota, including the Sulfolobus species, encode a distinct set of chromatin proteins that do not belong to the histone family and have no equivalent in either eukaryotes or bacteria. However, reversible acetylation at a specific lysine residue has been reported in the case of the crenarchaeal chromatin protein Alba (9), in a manner resembling eukaryotic histone acetylation. Moreover, deacetylation of Alba by the eukaryotic-like Sir2 deacetylase has been shown to increase its DNA binding affinity (hence the name: acetylation lowers binding affinity), indicating that PTMs in archaea may play an important regulatory role, similar to that in eukaryotes. In hyperthermophiles, PTMs, specifically methylation, also appear to play a significant role in increasing protein thermostability (10, 11). Indeed, a number of experimental studies have shown that protein methylation in hyperthermophilic archaea significantly increases resistance to heat denaturation and aggregation compared with the unmodified recombinant counterpart (10, 11, 12–14). Although the basis of this phenomenon is not well understood, it has been suggested that methylation at sites located on the accessible protein surface might modulate the intra- and inter- molecular interactions by changing the local hydrophobicity and surface charge (13).
Top-down proteomics (15), which implies the analysis of complex protein samples without prior enzymatic digestion, emerged recently as a powerful method for unambiguous characterization of intact proteoforms (16) bearing multiple PTMs in various combinations (17, 18). With the capabilities of the latest high-resolution tandem mass spectrometry systems, it is now possible to envision an ambitious goal: perform high-throughput discovery of proteoforms on a proteome-wide scale (19), analyzing both qualitative (20) and quantitative (21, 22) changes at the proteoform level. This makes top-down proteomics a complementary approach to bottom-up proteomics, which offers a tremendous depth of analysis, but lacks information on intact proteoforms. Top-down proteomics has been used to characterize heavily modified histone proteoforms (23). In this study we show that it can be also successfully applied to study post-translationally modified archaeal chromatin proteins, which have comparable molecular weights (6–11 kDa).
In the present study, we combined bottom-up and top-down proteomics approaches to monitor the proteome-wide post-translational modifications in the hyperthermoacidophilic archaeon Sulfolobus islandicus during three different cell growth stages, including the early-exponential, mid-exponential, and stationary stage. We used bottom-up proteomics to perform relative quantitation of proteins at different stages and to achieve high proteome coverage for PTM investigation, whereas high-throughput top-down analysis allowed us to characterize the post-translationally modified proteoforms and their dynamics throughout the cell growth. Our results reveal abundant N-terminal protein acetylation and massive methylation of the S. islandicus proteome that becomes more pronounced in the stationary phase of the cell growth. This tendency is also observed in the case of several chromatin proteins, including Alba, which are progressively methylated in the course of the cell growth, revealing potential parallel with chromatin remodeling in eukaryotes.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
Early exponential (OD600 of 0.15) and stationary stage [OD600 of 1.2] were selected for the study, as well as one time point in between at OD600 = 0.6 (supplemental Fig. S1 in the file ESI 1). Samples on each stage were prepared in three biological replicates to allow a basic assessment of reproducibility and reliability of the experimental findings. Each biological replicate was analyzed in three technical replicates for the label-free quantitation.
Cell Culture and Lysis
Sulfolobus islandicus strain LAL14/1 (Taxonomy ID 1241935) cultures were grown at 78 °C in rich medium containing 0.2% (w/v) tryptone, 0.1% (w/v) sucrose and 0.1% (w/v) yeast extract. The pH was adjusted to pH 3.5 with H2SO4 (24). Growth was monitored spectrophotometrically at 600 nm, and a doubling time of ∼11 h was obtained.
Samples collected at the “Early” (OD600 of 0.15), “Mid” (OD600 of 0.6) and stationary (“Late”) [OD600 of 1.2] growth stages in three biological replicates on each stage. Cells were harvested and washed three times with the sample buffer (20 mm Tris acetate, pH 6 and complete protease inhibitor mixture).
Cells were resuspended in the sample buffer and disrupted by French press (3 passages of 800 psi). Unbroken cells were removed by low-speed centrifugation for 20 min at 2000 × g at 4 °C.
Subcellular Fractionation
Cell membranes were separated from the cytosolic fraction by ultracentrifugation at 100,000 × g at 4 °C for 45 min. Cytosolic proteins in the supernatant were precipitated with ice cold acetone (80% final concentration), incubated for 1 h at −20 °C and pelleted at 15,000 × g for 15 min at 4 °C.
Protein Fractionation
Protein concentration was determined using micro BCA assay kit (Thermo Scientific, Rockford, IL) and NanoDrop 2000 spectrophotometer (Thermo Scientific).
For strong anion exchange (SAX) protein fractionation, 1 mg of the precipitated cytosolic sample was redissolved in 100 μl of 10 mm TrisHCl buffer (pH 8.0), the sample was split into three equal parts to avoid column overload, and each was separated on an ÄKTAmicro chromatography system (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) using MonoQ PC 1.6/5 SAX column (GE Healthcare Life Sciences, Sweden). For each part, the flow-through fraction and 10 eluted fractions were collected. After injection, the column was washed by 10 column volumes (CV) of solvent A, then eluted at 0.1 ml/min with gradient of 0% to 100% of solvent B in 40 CV (Sol. A. - 10 mm TrisHCl buffer pH 8.0, Sol. B. - 0.5 m NaCl, 10 mm TrisHCl buffer pH 8.0). Replicates of each fraction were combined, and the resulting 11 fractions (except flow-through) were exchanged 3 times to 10 mm TrisHCl, pH 8.0 using Amicon Ultra 0.5 ml 3K centrifugal filters (Merck Millipore, Tulagreen, Ireland). The resulting fractions were dried on Speedvac and stored at −20 °C.
The GELFrEE (Expedeon Protein Discovery and Analysis, San Diego, CA) separation (25) was accomplished using a 12% Tris-Acetate GELFrEE cartridge in full accordance with manufacturer's instructions. Briefly, 1 mg of each sample was separated into 4 fractions (fr.) during 1 h at 50 V (fr. 1), 10 min 35 s at 50 V (fr. 2), 4 min 47 s at 85 V (fr. 3) and 6 min 6 s at 85 V (fr. 4). Each fraction was exchanged 4 times with 50 mm ammonium bicarbonate and simultaneously concentrated to 40 μl using Amicon Ultra 0.5 ml 10 K centrifugal filters (Merck Millipore, Tulagreen, Ireland) and the resulting solution was subjected to methanol/water/chloroform precipitation as described previously (26). The precipitate was dried on Speedvac and stored at −20 °C.
Western Blot
Cytosolic fractions from the three growth stages (50 μg of proteins) were loaded onto a NuPAGE 4–12% polyacrylamide Bis-Tris gel (Thermo Scientific) using NuPAGE-MES SDS running buffer (Thermo Scientific). Proteins were visualized using Coomassie-based Instant Blue (Expedeon Protein Discovery and Analysis) staining. For Western hybridization, proteins were transferred onto a PVDF membrane. Acetylated proteins were visualized using a 1:250 dilution of the anti-acetyllysine rabbit antibody (SAB5200090, Sigma-Aldrich, Saint Louis, MO) and peroxidase-coupled anti-rabbit IgG produced in goat (Sigma-Aldrich) and the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) on Amersham Biosciences Hyperfilm ECL (GE Healthcare). Acetylated BSA (B-2518, Sigma-Aldrich), S. solfataricus and E. coli cytosolic proteins and S. cerevisiae protein extracts were used as positive controls.
Digestion and Peptide Fractionation E. coli
Tryptic digest was purchased from Waters (Milford, MA). One hundred micrograms of each S. islandicus sample was dissolved in 50 mm ammonium bicarbonate (pH 8.0), 8 m urea, reduced with 10 mm dithiothreitol for 30 min at 56 °C, alkylated with 15 mm iodoacetamide at room temperature in the dark for 30 min, then digested at 37 °C in 8 m urea using sequencing grade Lys-C (Promega, Madison, WI) at 1:100 enzyme/protein ratio for 4 h and with sequencing grade modified trypsin (Promega, Madison, WI) in 1 m urea at 37 °C overnight. The resulting digested sample was purified using C18 Sep-Pak cartridges (Waters, Milford, MA), dried on Speedvac and stored at −20 °C.
In-gel digestion was performed with trypsin as described previously (27).
SCX peptide fractionation was performed on the Late stage sample replicate 1 using SCX SpinTips (Protea Biosciences, Morgantown, WV) according to the manufacturer's instructions. Purified peptide sample was redissolved in loading buffer (5 mm ammonium formate (AF), pH 3.64, 20% acetonitrile) and loaded 3 times on the SCX tip, unbound fraction (“flowthrough”) was collected, as well 5 eluted fractions: 50, 100, 250 and 500 mm AF, pH 3.6, 10% acetonitrile and 1 m AF, pH 6.5. The resulting fractions were evaporated on Speedvac and stored at −20 °C.
Bottom-up LC-MS Analysis
A nanoflow ultra-high pressure liquid chromatography system consisted of Thermo Scientific Dionex NCS-3500RS NANO pump unit and WPS-3000TPL RS autosampler (both - Dionex Softron GmbH, Germering, Germany) was used for all LC-MS experiments. Solvent A consisted of 0.1% aqueous formic acid, solvent B consisted of 80% acetonitrile with 0.1% of formic acid. Mass spectrometry was performed on the ETD-enabled Orbitrap Fusion Tribrid instrument (Thermo Scientific, San Jose, CA) with Easy-Spray nano-ESI source (Thermo Scientific).
C18 μ-precolumn cartridge (300 μm i.d. × 5 mm, C18 PepMap 100 particles with 5 μm size and 100 Å pores, PN 160454) and Easy-Spray PepMap C18 analytical column (75 μm i.d. x 50 cm, 1.9 μm 100 Å particles, PN ES803) were purchased from Thermo Scientific. Two micrograms (5 μg for SCX-fractionated peptides) of a digested sample was trapped on a precolumn for 3 min at 20 μl/min of 100% solvent A, then eluted at 300 nl/min onto the analytical column using the following gradient: 1% B at 3 min, 8% B at 5 min, 35% B at 70 min, 50% B at 120 min, 90% B at 125–130 min.
All data-dependent LC-MS runs were performed with MS1 scan resolution 60,000, MS2 resolution 15,000. For label-free quantitation, the HCD-only runs were acquired. Separate ETD-only runs were acquired for the PTM characterization. “Top time” data-dependent settings with duty cycle time of 0.7 s for label-free quantitation HCD runs and 2 s for ETD runs were used. Precursor selection range was 330–1530 for HCD and 330–1000 for ETD. Precursors with the charge states 2–24 were selected for HCD and 3–24 were selected for ETD. Quadrupole isolation window was 1.4. Dynamic exclusion was set to 60 s with the mass tolerance of ± 10 ppm. HCD collision energy was set to 28. ETD reaction time was 70 ms and CID supplementary activation at collision energy of 20 was used. Maximum injection time was 50 ms for HCD and 200 ms for ETD. For more detailed MS method description see the Supplementary Methods in the file ESI 1. HCD runs for label-free quantitation were performed in 3 technical replicates for each biological replicate.
Top-down LC-MS Analysis
The same equipment and eluents as the ones described for bottom-up analysis were used for top-down LC-MS experiments.
C4 μ-precolumn cartridge (300 μm i.d. × 5 mm, C4 PepMap300 particles with 5 μm size and 300 Å pores) and Easy-Spray PepSwift monolithic reversed-phase PS-DVB analytical column (200 μm i.d. × 25 cm) were purchased from Thermo Scientific. After loading a protein sample onto a precolumn for 3 min at 20 μl/min of 100% solvent A, different elution gradients at 1 μl/min flow rate were used: for SAX fractions - 5% B at 3 min, 10% B at 5 min, 60% B at 125 min, 90% B at 128–130 min; for GELFrEE fraction 1 - 5% B at 3 min, 10% B at 5 min, 40% B at 35 min, 60% B at 40 min, 90% B at 42–46 min; for GELFrEE fractions 2–4 - 5% B at 3 min, 25% B at 5 min, 50% B at 125 min, 90% B at 126–130 min.
Orbitrap Fusion was operating in the intact protein mode. For each SAX or GELFrEE fraction, an HCD-only LC-MS run and an ETD-only LC-MS run were acquired. MS1 scans were performed at 120,000 resolution (60,000 for GELFrEE fraction 1) with 2–5 microscans per scan. Data-dependent selection was performed on top 3 most intense precursor ions with 90 s dynamic exclusion time. HCD collision energies of 20 to 25 and 1 microscan for were used for HCD runs, ETD reaction time 4 to 7 ms and 2–3 microscans per scan were used for ETD runs. MS/MS resolution was 120,000 (60,000 resolution was used for GELFrEE fraction 1). Alba1-containing SAX fraction was also rerun with 240,000 MS/MS resolution and 4 MS/MS microscans to increase fragmentation spectra quality. For more detailed MS method description see file ESI 1.
Data Analysis
Fasta files for the E. coli strain K12 (reference proteome, version from May 2016, 4306 entries) and S. islandicus LAL14/1 (December 2014, 2591 entry) were downloaded from Uniprot, common contaminants database was furnished with MaxQuant. Andromeda search engine in MaxQuant 1.5.2.8 (28) was used for protein identification and label-free relative quantitation. Briefly, Acetyl (protein N terminus) and Oxidation (M) and Methylation (K) were used as variable modifications, trypsin/P was used as an enzyme with maximum of 2 missed cleavages allowed, precursor and fragment tolerances were 5 ppm and 18 ppm, respectively, peptide and protein identification FDRs were set to 1%, for detailed settings see file ESI 1. For the stagewise data sets with HCD and ETD data, the FDRs were calculated jointly for HCD and ETD files of a biological replicate. For gel band identifications, Acetyl (protein N terminus), Acetyl (K), Oxidation (M) and Methylation (K) were set as variable modifications. Methylation sites obtained with Andromeda database search were filtered using home-made scripts. A K-methylation site was considered confident if it had a continuous fragment ion series localizing the site, i.e. having the fragment ion series continuing on both sides of the lysine (or at one, if it is a C-terminal lysine). Home-made scripts were written on Python 3.3.2 using Pyteomics package (29). Unless stated otherwise, we use the term methylation interchangeably with the term monomethylation throughout this article.
For label-free quantitation, the averaging of technical replicates, intensity normalization and retention time alignment (2 min window) were performed automatically in MaxQuant 1.5.2.8 using the MaxLFQ algorithm (30), unique and razor peptides were used for calculation. The MaxLFQ-normalized values were loaded into Perseus 1.5.4.0, were log2-transfromed, the proteins used for quantitation must contain 3 valid values at at least 1 growth stage. The missing values were imputed from a normal distribution (shift 1.8, width 0.3). Two-sided Student t-tests were performed on different groups, permutation-based FDR was calculated (with S0 parameter of 1) and the proteins with q-values of 0.01 or less were considered differentially expressed between conditions. For quantification of K-monomethylated peptides, the nonnormalized peptide intensities were loaded into Perseus 1.5.4.0, the peptides used for quantitation must contain 3 valid values at at least 1 growth stage. Intensities were normalized on the median in each sample, then log2-transformed. The missing values were imputed from a normal distribution (shift 1.8, width 0.3). Two-sided Student t-tests were performed on different groups, permutation-based FDR was calculated (with S0 parameter of 2).
ProSight PC 3 SP 1 (Thermo Scientific) was used for top-down database search. For detailed search parameters see in ESI 1. Briefly, raw files were deconvoluted using Xtract and subjected to a four-stage database search consisted of “Absolute Mass” search with 4.08 Da precursor tolerance, then 3 kDa precursor tolerance, then “Biomarker” search with 10 ppm precursor tolerance and “Absolute Mass” search with 250 kDa precursor tolerance, fragment ion tolerance was set to 3 or 4 ppm. S. islandicus LAL14/1 TrEMBL database of 2,591 entries was downloaded in Uniprot flatfile format and imported with full PTM annotation, N-terminal acetylation and methionine excision, yielding a database of 10,442 entries. According to the article from Catherman et al. (31) FDR cutoff of 0.01 corresponded to a ProSightPC p-score cutoff of 4.7 × 10−9, which gives an E-value cutoff of 5 × 10−5 on the database of 10,442 entries. ProSightPC E-value cutoff of 5 × 10−5 was thus used to filter the confident identification results. Annotated top-down spectra from ProSightPC search output can be viewed in MS-Viewer (32) on the Protein Prospector public website (prospector.ucsf.edu): search key dk458i4qtb for SAX ETD data, rvs63gz69o for SAX HCD data, vmhsk5c8pb for GELFrEE ETD data and wgvkcvlyeg for GELFrEE HCD data.
To quantify the relative abundancies of Alba1 and Cren7 proteoforms, for each biological replicate in two technical repeats, extracted ion chromatograms (XIC) were calculated in Xcalibur 2.2 (Thermo Scientific) with 10 ppm accuracy for 4–5 charge states per proteoform. Then, peak areas for methylated proteoform in each file were divided by the nonmethylated proteoform XIC area in the respective chromatogram, the results were summarized and averaged (see the file ESI 10). Welch t-tests were performed using standard function of R 3.1.2.
RESULTS
Label-Free Relative Quantitation of Protein Expression Between Different Growth Stages by Bottom-Up Proteomics
Cultures of S. islandicus cells were harvested at three different time points corresponding to the early-exponential (throughout the text referred to as “Early”), mid-exponential (“Mid”), and stationary (“Late”) growth phases (supplemental Fig. S1) in three biological replicates. Cytosolic fraction of each cell lysate was digested in solution and analyzed using a 2 h chromatography gradient and an Orbitrap Fusion in data-dependent mode with HCD fragmentation. This single-shot approach allowed us to identify 1529 proteins at FDR 0.01 (out of 2591 sequences in the S. islandicus LAL14/1 protein database) and to quantify 1335 proteins with nonzero intensities for all three biological replicates at least at one time point. To assess the suitability of the data for the label-free quantitation, coefficients of variability (CVs) were calculated for technical replicates on the nonlogarithmized, nonnormalized peptide intensities. The determined median (11–20%) and mean (16–23%) CV levels for different biological samples were considered acceptable for label-free quantification. The details of the calculation are provided in ESI 2.
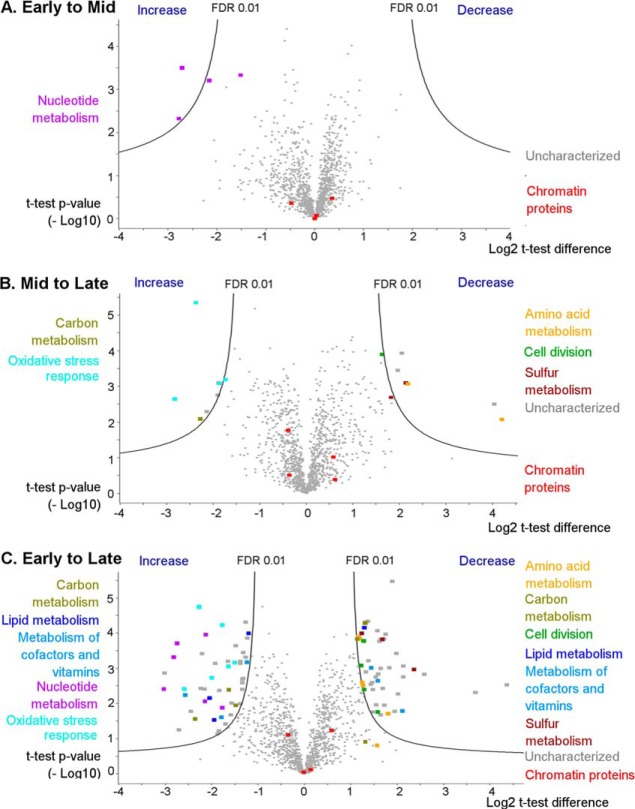
Fig. 1 represents the volcano plots corresponding to the relative changes in protein abundances between Early and Mid (Fig. 1A), Mid and Late (Fig. 1B) as well as Early and Late (Fig. 1C) time points. An FDR of 0.01 and S0 parameter of 1 were chosen as the threshold values to filter statistically significant changes in Perseus. Spreadsheet containing the intensity data for all the proteins, as well as lists of differentially expressed proteins, can be found in the ESI 3 file.
Fig. 1.
LFQ-based volcano plots showing the protein expression level changes between the growth stages of S. islandicus. A, Early to Mid stage, B, Mid to Late stage, C, Early to Late stage. Black solid lines represent the significance threshold at FDR 0.01 and S0 = 1. Color-coded functions are shown for some of the differentially expressed protein functional groups. Four DNA-binding proteins are also highlighted in red: Alba1, Alba2, proteins Sul7d1 and Sul7d2 (quantified together), and Cren7 protein.
As evidenced from Fig. 1A, the vast majority of proteins shows insignificant differences in expression level between the Early and Mid growth phases. The two exceptions are phosphoribosylformylglycinamidine synthase subunit PurQ (Uniprot accession M9U759) and phosphoribosylaminoimidazole-succinocarboxamide synthase (M9UE35), which are significantly up-regulated (4.5–7-fold increase) at Mid compared with the Early time point. Both proteins participate in purine biosynthesis (33), pointing toward a more active nucleotide metabolism during the transition from the early- to mid-exponential growth phase.
Changes from the Mid to Late sampling points are more pronounced (see Fig. 1B), with 13 proteins being differentially expressed at FDR 0.01 and 106 at FDR 0.05 (ESI 3 file). Among them, four proteins involved in oxidative stress response showed 3- to 8-fold increase in abundance at the Late time point. The latter observation is consistent with the studies in bacteria showing that development of cellular resistance to oxidative stress is a classical trait of the stationary phase, when reactive oxygen species might be particularly harmful for nondividing cells (34, 35). Not surprisingly, proteins involved in cell division, ribosome biogenesis, tRNA biogenesis are down-regulated, when comparing Mid growth stage to the stationary Late stage.
Proteomic changes between the actively growing and stationary S. islandicus cells are the most prominent when comparing Early and Late time points (Fig. 1C), with 93 proteins found to be significantly differentially expressed at FDR 0.01. Fifteen enzymes participating in amino acid (8 proteins), carbon (3 proteins) and sulfur (3 proteins) metabolism are found to be significantly down-regulated (2.5- to 8.9-fold decrease in abundance), as are four cell division-associated proteins, including Vps4 and ESCRT-III (2.5–3.5-fold decrease). By contrast, 6 proteins involved in nucleotide metabolism are significantly up-regulated (3.2–8-fold increase), in accordance with the trend observed between Early and Mid time points. Other proteins that increase in abundance include five oxidative stress response proteins (2.1- to 6-fold), two heterodisulfide reductase subunits (3- to 6-fold increase), three lipid metabolism proteins (2.3- to 4-fold increase) and two carbohydrate metabolism-associated proteins (Fig. 1C).
Despite reported growth phase-dependent changes in the organization of the Sulfolobus nucleoid (36, 37), the major chromatin proteins, namely Alba1 (M9UDF6), Alba2 (M9U6G3), 7-kDa DNA-binding proteins Sul7d1 (M9UD85) and Sul7d2 (M9UII2) as well as Cren7 (M9U8W4), do not significantly change their relative abundances across the three sampling time points.
Characterization of Protein N-Terminal Acetylation by Bottom-Up Proteomics
The shotgun analysis of the Lys-C/trypsin digests of S. islandicus on the Orbitrap Fusion allowed us to routinely identify around 1,500 proteins, which corresponds to almost 60% of the whole theoretical proteome. To achieve a better proteome coverage depth, peptides were prefractionated before the LC-MS/MS analysis. To this end, one sample at the Late stage (as the amount of available material at the Late stage is maximal) was digested with Lys-C and trypsin and subjected to fast tip-based strong cation exchange (SCX) fractionation into 6 fractions, which were analyzed with separate HCD and ETD LC-MS/MS methods. The fractionation proved rather efficient: 11,602 out of 18,227 peptides (63.6%) were identified only in one fraction, and 4,396 additional peptides (24.1%) were identified in 2 fractions. However, SCX prefractionation increased the number of identified proteins only to 1,618 (FDR 0.01), i.e. by 5% compared with single-shot LC-MS.
Additionally, we reused the files from the LFQ data set (HCD fragmentation) and acquired a supplementary shotgun run with ETD fragmentation for each biological replicate on the unfractionated samples of each growth stage. The resulting HCD+ETD shotgun data sets for each biological replicate were then subjected to database search with PTMs of interest set as variable modifications.
Global identification of protein lysine N6-acetylation is notoriously difficult because of its dynamic property and rather low abundance (38–40). Indeed, searches for lysine side chain acetylation yielded no occurrences in our data set, suggesting that special enrichment steps might be required for detection of this modification. Thus, lysine N6-acetylation was not further considered in this study.
Similar to eukaryotes, archaeal proteins can also be acetylated at the alpha-amino group of the N-terminal amino acid (41). Our data set contained N-terminal peptides of 372 different proteins. In nearly half of them (180; 48%) the N-terminal methionine is removed. One third of all detected protein N termini (127; 34%) were found to be acetylated (ESI 4). Among the N-terminally acetylated proteins, 91 have their N-terminal methionine removed. Serine and glutamate residues in the second and third position, respectively, strongly facilitate N-terminal methionine excision and subsequent acetylation (see supplemental Fig. S3 in the file ESI 1). Additionally, acetylation occurs frequently at the N-terminal methionine itself when a glutamate residue is situated in the second position. Other terminal amino acids that undergo acetylation after the N-terminal methionine removal include alanine (11 occurrences), threonine (7), glutamate (1), and valine (1). Acetylation of the latter two amino acids at the N-terminal position has not been previously described in archaea (42).
Ubiquitous Lysine Methylation Revealed by Bottom-Up Proteomics
It has been previously shown that certain Sulfolobus proteins are methylated (unless stated otherwise, monomethylation is meant) on lysine residues (43). This methylation might play a role in thermal stabilization of these proteins (44) (12), which is highly important for cell functioning of hyperthermophilic archaea. Modern proteomic techniques recently allowed the characterization of lysine methylation sites on several Sulfolobus proteins. For example, 20, 21, and 26 methylation sites were identified on different subunits of the DNA-directed RNA polymerases (DdRp) from S. shibatae, S. solfataricus and S. acidocaldarius, respectively (11) (45). However, a proteome-wide analysis of lysine methylation has not been performed thus far on any archaeon.
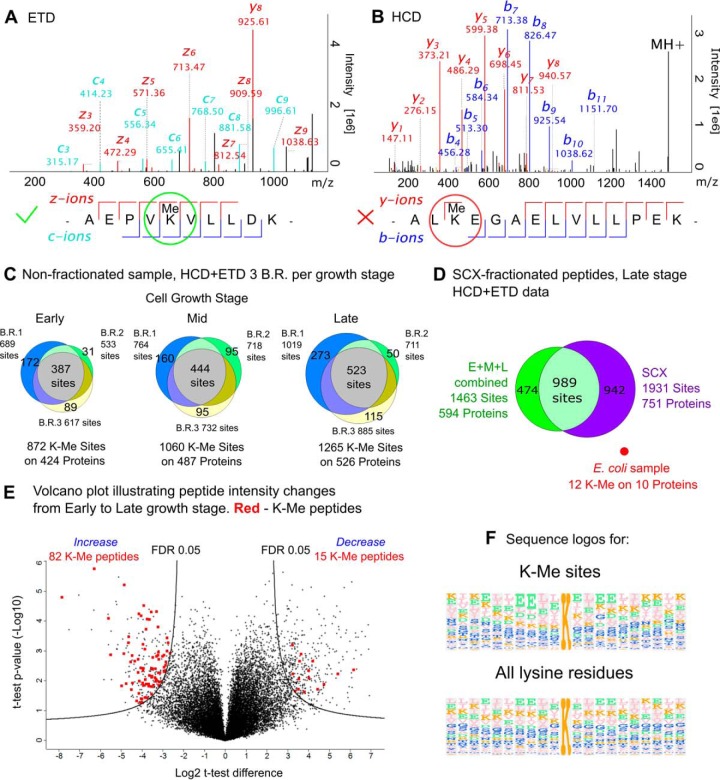
Searching the SCX HCD+ETD data set for lysine side-chain monomethylation revealed a staggering number of 2,518 sites on 872 proteins, i.e. more than half of the total 1,623 identified proteins were found to be methylated. To increase the confidence of methylation localization we decided to filter the results based on the fragmentation quality of the corresponding spectra (Fig. 2A, 2B). A methylation site was considered confident only if b, c, y, or z-ion series in a fragmentation spectrum was continuous on both sides of the methylated lysine residue, i.e. ions corresponding to fragmentation at N- and C termini of a methylated lysine were registered in the same spectrum. For example, the spectrum shown in Fig. 2A contains continuous c, y, and z-series, thus the methylation site was considered confident, whereas the ion series in Fig. 2B are discontinuous around the lysine methylation site and, accordingly, was not taken into account. This approach filters out low quality spectra, increasing the confidence of identification. It also introduces a certain degree of modification site localization to distinguish lysine methylation from glutamate side-chain O-methylation, which is also known to occur in archaea (46, 47), although not in Sulfolobus species. After such filtering, we obtained 1,931 confident lysine methylation sites on 751 proteins (Fig. 2D, ESI 5 file). In comparison, recent analysis of PTMs in E. coli identified only 84 methylated lysine residues in 64 different proteins (48) in an extensive study employing OFFGEL peptide prefractionation before LC-MS analysis. Analysis of the trypsin digest of E. coli cell lysate using the same LC-MS/MS setup and data processing workflow as for the S. islandicus proteome, gave even lower number of methylation sites (12 sites on 10 proteins out of 4305 encoded proteins; see file ESI6), emphasizing a major difference with respect to proteome methylation in the two organisms (Fig. 2D).
Fig. 2.
A and B - representative spectra of the methylated peptides from S. islandicus Lys-C/trypsin digest, corresponding fragmentation maps are shown below the spectra. A, spectrum (ETD) contains the continuous ion series on both sides of the methylated lysine residue, thus the methylated lysine site is filtered in; B, (HCD) fragment ion series is discontinuous, hence methylated lysine residue discarded; C, Venn diagrams show the number of the found methylation sites per growth stage (circles are given for each biological replicate B.R. at the corresponding growth stage); D, Venn diagram, showing the combined number of sites and methylated proteins for the three growth stages, and well as for the SCX fractionated late growth stage sample; E, volcano plot of the normalized peptide intensity changes between Early and Late stages. Significantly changing K-Me peptides are show in red, significance threshold at FDR 0.05 and S0 = 2; F, sequence logos, showing the frequency of the amino acid residue occurrences in the proximity of a methylated lysine (left) or in proximity to any lysine residue (right).
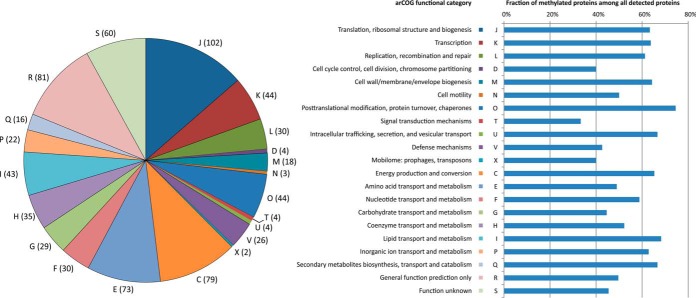
To gain insight into the specificity of lysine methylation in hyperthermophilic archaea, we generated a sequence logo for all identified K-methylated sites and compared it with that of all lysine residues in the corresponding proteins (Fig. 2F). The two sequence logos look strikingly similar, confirming that the involved methyltransferase lacks sequence specificity. The methylated proteins belong to all 21 functional categories defined in the new implementation of the archaeal clusters of orthologous groups (arCOGs) (49, 50). Over 70% of all detected proteins in the “Post-translational modification, protein turnover, chaperones (O)” category are found to be methylated (Fig. 3). By contrast, in some of the relatively well populated functional categories (i.e. with more than 60 detected proteins), such as “Carbohydrate transport and metabolism (G)” and “Defense mechanisms (V; e.g. components of the CRISPR-Cas system),” just above 40% of the proteins are methylated. Nevertheless, the average level of methylation for detected proteins across all functional categories is 58.8%, suggesting that there is little specificity toward proteins of any particular functional category. No difference in molecular weight has been observed between the methylated and nonmethylated proteins (data not shown). However, the calculated pI values for the nonmethylated proteins have a clear maximum around pI 9.5, which is not well pronounced for the proteins that are found to be methylated (see supplemental Fig. S5 in the file ESI 1). The reasons for such tendency remain unclear.
Fig. 3.
Functional annotation of the identified methylated proteins. Pie-chart on the left shows the classification of identified methylated proteins according to functional categories, as defined in the archaeal clusters of orthologous groups (arCOG) (46, 47). The numbers in parenthesis indicate the exact number of identified methylated proteins in each category. The categories are denoted by capital letters and the key is provided in the right panel. Horizontal bars show the proportion of methylated proteins among all detected proteins in each functional category.
The comparison of the extent of lysine methylation between different growth phases shows that the total number of identified methylation sites tends to increase from the Early to Mid and ultimately to the Late stage (Fig. 2C). About 42–44% of all the sites being found in all three biological replicates for each stage (termed confident sites). Some confident methylation sites are found at one growth stage, but are absent in all three biological replicates at a different stage. For example, there are 13 sites that are reproducibly found at the Early stage, but absent at the Late stage (see file ESI 5). These 13 sites include K-88 and 167 sites for aspartokinase (Uniprot accession M9U961) and K-109 and 182 sites on phosphoribosyltransferase (M9U3M0). However, these proteins are downregulated at the Late stage according to LFQ data, hence the absence of any confident methylation sites could be attributed to the low abundance of the corresponding tryptic peptides. By contrast, 60 confident sites, which emerge at the Late stage, are absent at the Early stage. For instance, the isocitrate dehydrogenase (M9UDB0) has three sites out of nine emerging at the Late stage, although the protein total LFQ intensity remains stable throughout the growth stages. In 32 other cases the number of methylation sites increased at the Late stage without a noticeable increase of the LFQ abundance of the corresponding protein.
Peptide intensity values from the reprocessed LFQ data set were normalized according to the median value of the sample and t-tests were performed to compare the peptide abundances across growth stages. Because of variability in peptide intensity and high number of peptides, which inevitably result in the elevated multiple testing error probability, only 4 K-monomethylated peptides passed the FDR threshold of 0.01. After relaxing the FDR threshold to 0.05 (S0 = 2), 625 peptides out of 18,369 were found to significantly change in abundance from Early to Late stage (see Fig. 2E and the ESI file 7). Among them, 15 K-methylated peptides significantly decrease in their abundance, including 4 that do not show the corresponding change at the protein level. At the same time, 82 K-methylated peptides significantly increase in their abundance from Early to Late stage, including 53 cases where there was no such increase at the protein level (ESI file 3). The abundance of methylated sites K-53 and K-61 of the Sul7d DNA-binding proteins were found to be significantly increased at the Late stage. The other cases include metabolic enzymes and ribosomal proteins, with no obvious functional clustering between them.
Chromatin-associated proteins, with the exception of Alba1, were also found to be methylated in the shotgun proteomic data set (see Table I). Lysine methylation on Alba1 has not been found in the LFQ data set, but the in-gel digestion of the 12 kDa band from an SDS-PAGE gel (see supplemental Fig. S2 in ESI 1) allows for the confident identification of the lysine methylation site on K-16 (see supplemental Fig. S4 in ESI 1). Alba2 protein was found to be methylated at K-4, which has not been reported previously. Cren7 protein is methylated at lysine residues 11, 16, 24, 31, 42. The genome of the S. islandicus strain LAL14/1 contains two closely related (95% sequence identity) 7-kDa DNA-binding proteins, which we call Sul7d1 and Sul7d2 (Table I). Nonunique peptides, shared by both Sul7d proteins, are methylated on lysine residues 28, 40, 49, 53, 61, 63, and 64, whereas the peptide unique to Sul7d1 shows an additional methylation on lysine 5 and/or 7. Sul7d methylation has been recently investigated by Niu et al. in S. islandicus REY15A (Sul7d from strain REY15A has an identical sequence to Sul7d1 from strain LAL14/1) (51). Our study confirms the Sul7d methylation pattern determined in S. islandicus REY15A and reveals two additional sites, K-28 and K-40, which were not identified in the previous report. When comparing the different growth stages, Alba2 methylation is not detected at the Early stage, but emerges at the Mid and Late time points. The same is observed for the methylation at K-49 of the Sul7d proteins. When the label-free quantitation at the peptide level is considered, a clear tendency toward higher abundance of the K-Me peptides for Cren7 and Sul7d proteins is readily apparent (see file ESI 8). The peptides, corresponding to the K-53 and K-61 sites of Sul7d and K-11, 16, 24, and 31 of Cren7 protein pass the t test p value threshold of 0.05 when comparing the abundances between the Early and Late growth stages (see ESI 8). The difference for Sul7d methylated peptides remains significant even after the multiple-testing FDR correction with the threshold of 0.05 (ESI 7). This suggests that at least 3 of the 5 chromatin proteins conform to the same tendency of increased methylation level from the Early toward the Late stage.
Table I. Lysine methylation, identified on S. islandicus DNA/RNA-binding proteins by bottom-up approach with HCD and ETD fragmentation. Methylated residues are underlined and marked in bold.
| Uniprot accession | Protein description | Methylation sites | Sequence |
|---|---|---|---|
| M9UDF6 | Alba1 protein | 16* | SSGTPTPSN VVLIGKKPVM NYVLAALTLL |
| NQGVSEIVIK ARGRAISKAV DTVEIVRNRF | |||
| LPDKIEIKEI RVGSQVVTSQ DGRQSRVSTI EIAIRKK | |||
| M9U6G3 | Alba2 protein | 4 | TEKLNEIVV RKTKNVEDHV LDVIVLFNQG |
| IDEVILKGIG REISKAVDVY NSLKDRLGDG | |||
| IQLVNVQTGS EVRDRRRISY ILLRLKRVY | |||
| M9U8W4 | Chromatin protein Cren7 | 11, 16. 24, 31, 42 | SSGKKAVKV KTPAGKEAEL VPEKVWALAP |
| KGRKGVKIGL FKDPETGKYF RHKLPDDYPI | |||
| M9UD85 | 7-kDa DNA-binding protein (Sul7d1) | 5, 7, 28, 40, 49, 53, 61, 63.64 | TTVKFKYKG EEKQVDTSKI KKVWRVGKMI |
| SFTYDEGGGK TGRGAVSEKD APKELLQMLEKQKK | |||
| M9UII2 | 7-kDa DNA-binding protein (Sul7d2) | 28, 40, 49, 53, 61, 63.64 | VTVKFKYKG EDKEVDTSKI KKVWRVGKMI |
| SFTYDEGGGK TGRGAVSEKD APKELLQMLEKQKK |
* - only found in the in-gel digested 12 kDa SDS-PAGE band
As mentioned above, several studies have investigated the methylation state of the multisubunit DNA-directed RNA-polymerase (DdRp) in different Sulfolobus species. Our data set also contains the information on 24 methylation sites distributed over 9 subunits of the DdRp in S. islandicus (supplemental Table S1 in file ESI 1). This result is comparable to the 21 sites/9 subunits identified by Botting et al. in S. solfataricus (11) as well as 20 sites/13 subunits in S. shibatae and 26 sites/13 subunits in S. acidocaldarius identified recently by Azkargorta et al. (45). The methylation sites are not fully conserved between the mentioned studies and our report: 12 sites from our data set have not been reported previously. This observation might further point toward low sequence specificity of the lysine methyltransferase in action. However, it cannot be excluded that the observed variation arises from the technical differences in the experimental setup between the respective analyses.
Interestingly, our results indicate that the chromatin-associated proteins show no significant change in their relative abundance at the protein level across the three sampling time points, but Cren7 and Sul7d show a tendency toward increased lysine methylation at the Late stage. When considering PTMs, important changes in the PTM profiles of the chromatin proteins might occur at the proteoform level. To explore this possibility, we performed the proteome-wide top-down analysis.
Proteoform-Level Characterization by High-Throughput Top-Down Analysis
Unlike bottom-up mass spectrometry, top-down analysis allows for the characterization of intact proteoforms containing combinations of PTMs, rather than separate modification sites. Top-down is also useful for the characterization of unexpected PTMs, because modified forms often coelute during chromatography and can be easily linked together. One should note that the sensitivity of top-down approaches is not (yet) as good as the one for the bottom-up and that the analysis is restricted to proteins with low molecular mass (generally < 30 kDa), which can introduce a bias in the analysis. However, analysis of the intact proteoforms by top-down approaches provides valuable complementary information to that obtained by bottom-up techniques.
Top-down proteomics has already proven to be a useful tool to study archaeal proteins (52, 53). However, thus far, it has not been applied to Sulfolobus species.
Strong anion exchange (SAX) chromatography has been reported by Bunger et al. (54) to be an efficient prefractionation method that can be coupled with subsequent RPLC-MS top-down analysis. Using an LTQ XL ion trap mass spectrometer, Bunger et al. (54) identified 322 proteoforms of 174 proteins of E. coli. We therefore performed a SAX separation of 1 mg of one of the S. islandicus cytosolic preparations (Late stage replicate 1) into 11 fractions, including flow-through and 10 eluted fractions. All fractions were further analyzed on the Orbitrap Tribrid Fusion, using data-dependent methods with HCD or ETD fragmentation. ProSightPC was used for the database search, allowing the characterization of intact unmodified proteoforms, modified proteoforms and proteolytic fragments. Fig. 4 summarizes the results for the 2D SAX-RPLC-MS global top-down analysis.
Fig. 4.
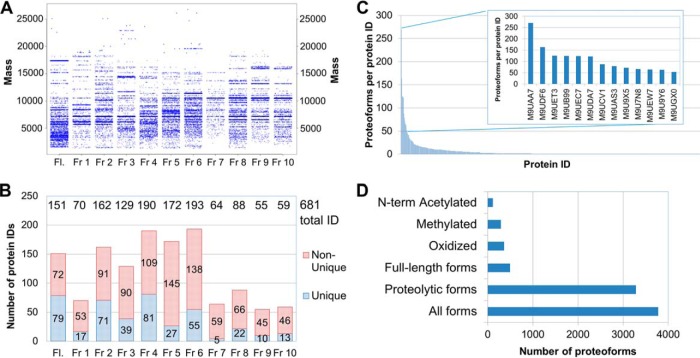
Summary of the top-down LC-MS/MS analysis of the SAX-prefractionated S. islandicus cytosolic sample: A, distributions of the experimental intact protein masses by fraction; B, numbers of protein IDs, identified in each SAX fraction, including a number of the unique IDs, found exclusively in the respective fraction, and nonunique IDs, which were also found in one or more other fractions; C, distribution of the proteoforms found per protein ID; D, distribution of the most frequent types of identified proteoforms. Fl, flow-through fraction.
Fig. 4A exemplifies the SDS-PAGE-like representation of the experimental intact charge-deconvoluted masses measured in the SAX top-down data set. The maximal measured mass is around 28 kDa. In S. islandicus LAL14/1, 1351 proteins (52% of all the proteins) are lighter than 28 kDa. Collectively, in the 11 fractions, 3,978 proteoforms of 681 unique protein IDs (see Fig. 4B) are identified with an E-value cutoff of 5 × 10−5 (see the list in the file ESI 9). As shown in Fig. 4B, 61 to 208 unique protein IDs are found in each fraction, with fractions 4–6 being the largest in terms of unique IDs. Most proteins can be found in more than one fraction, which in some cases is caused by the presence of several proteoforms, e.g. different proteolytic fragments, and in other cases—by insufficient separation. The 681 detected unique proteins correspond to more than 40% of the proteins identified in the same S. islandicus sample by bottom-up approach and to 26% of the total number of encoded proteins. For comparison, 563 unique IDs (12% of the encoded proteins) and >1,600 proteoforms have been identified in the recent study by Ansong et al. (20) for another prokaryotic organism, enterobacterium Salmonella typhimurium. However, in that study the authors utilized 5% FDR cutoff and a different software (MS-Align+) for database search, whereas the present study assumes an FDR cut-off of 1%.
Abundant cytosolic proteins are represented by a large number of different proteoforms. For instance, there are 270 forms for peroxiredoxin (M9UAA7, see Fig. 4C). Proteolytic fragments represent ca. 3483 (∼87%) of all the identified proteoforms (Fig. 4D), despite the use of a protease inhibitor mixture during sample preparation. N-terminal methionine cleavage and N-terminal acetylation (113 N-terminal acetylation occurrences) were also frequent and easily characterized by the top-down approach (Fig. 4D). As discussed in a recent paper by Fortelny et al. (55), about 90% of N-terminal processing events in the human proteome remain unexplained, and this percentage may be even higher in archaea, given their largely undiscovered protein processing pathways.
Massive methylation is also observed in the top-down data set, with 292 potentially methylated proteoforms (Fig. 4D). For 85 methylated forms of 54 proteins it is possible to locate the most plausible methylation sites using high-resolution MS/MS data (see supplemental Table S2 in the file ESI 1 for data on all the 85 methylated proteoforms). Methylated proteoforms are in the 5.7 to 19.5 kDa mass range. Among those were ribosomal proteins L14e and L29P, transcriptional regulators, DNA-directed RNA polymerase subunits H and L and peroxiredoxin. Some of the characterized proteins bear up to 5 methyl groups on the same polypeptide (prefoldin subunit β [M9U9D6] and acylphosphatase [M9UDL6]). Lower sensitivity of the top-down analysis as compared with the bottom-up approach is the most plausible explanation for a lower number of methylation sites detected in the top-down mode.
Cysteine-containing putative redox proteins M9U7N8 and M9UCV1 appear to bear methylthio groups (-SMe, 45.99 Da) on their cysteine-18 and cysteine-17, respectively (see supplemental Fig. S6 in the file ESI 1). This may be a result of cellular oxidative stress or, if the methylthio group transfer is reversible, a specific metabolic form of these redox proteins.
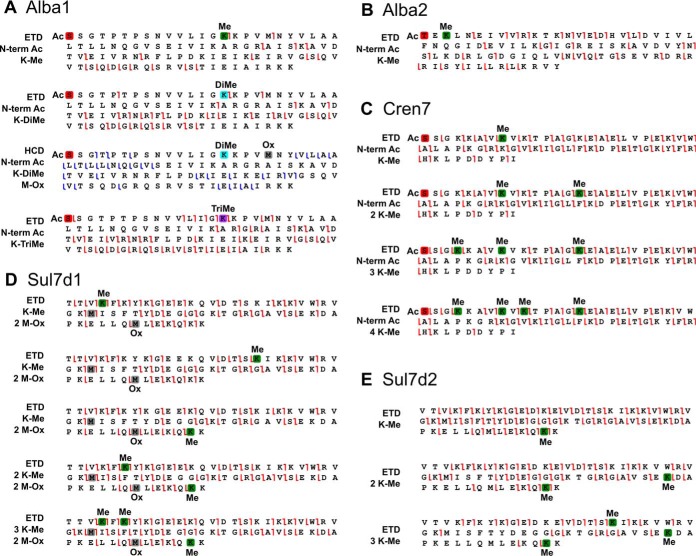
Small and basic chromatin proteins are particularly suitable for top-down approaches. All of the five chromatin proteins are identified as two or more proteoforms. Fragmentation maps for the characterized proteoforms based on ETD and HCD experiments are represented in Fig. 5. ETD yielded high sequence coverage and site location confidence. Notably, for the chromatin protein Cren7, we identified proteoforms with up to 4 methylations. The MS/MS data indicates that the most plausible locations for these modifications are in the N-terminal part of the sequence between Lys-5 and 11; by contrast, the bottom-up data suggested additional methylation sites on Lys-11 to 42 (see Table I). Top-down and bottom-up approaches are therefore complementary in this case, because top-down provides the exact number of simultaneously methylated lysine residues and the preferential modification locations, whereas bottom-up reveals minor methylation locations. This is in accordance with a previous report (51) showing that Cren7 is methylated on many residues, however the top-down analysis allows us to suggest that the N-terminal part is methylated preferentially.
Fig. 5.
Fragmentation maps, showing localization of methylations and acetylations on the main S. islandicus methylated chromatin protein forms by ETD or HCD MS/MS. A, Alba1 proteoforms; B, Alba2 proteoforms; C, Cren7 proteoforms; D, Sul7d1 proteoforms, E, Sul7d2 proteoforms. Red box represents N-terminal acetylation; green box, lysine methylation; light blue box, lysine dimethylation; violet box, lysine trimethylation; gray box, methionine monooxidation.
Sul7d1 and Sul7d2 proteins are 95% identical to each other and the mass difference between the two proteins is 15 Da. Consequently, they can be partially separated on the PS-DVB HPLC column. For both proteins, proteoforms with up to 3 methylations are characterized (Fig. 5). Surprisingly, despite high sequence similarity, the proteins show a difference in their methylation pattern: Sul7d1 tends to be methylated at its N- and C-terminal extremities, whereas Sul7d2 is preferentially methylated in its C-terminal part and at Lys-17. These differences may be attributed to different DNA binding modes for these proteins or unequal availability to the methyltransferase in the cytosol. Alba2 methylation at Lys-4 is also confirmed by top-down analysis.
Analysis of Alba1 Methylation by Top-Down Proteomics
Unlike in the bottom-up analysis, the Alba1 protein is one of the most abundant proteins in the top-down total ion chromatograms, which enables the facile characterization of its proteoforms. Alba1 is found to be acetylated at the N-terminal serine and occasionally oxidized on Met-20. The previous report states that Alba1 is not methylated in vivo and even in vitro under action of the promiscuous Sulfolobus protein methyltransferase aKMT4, which is known to methylate other chromatin proteins (51). However, our results unequivocally show that Alba1 is methylated at K-16. Interestingly, the same residue has been previously found to be acetylated in vivo (9). Importantly, acetylation at this position modulates the DNA-binding affinity of Alba. Strikingly, Lys-16 is not only monomethylated, but can also be di- and trimethylated (Fig. 5). The HCD fragmentation confirms that an acetyl group is indeed on the N terminus of the protein and suggests that methyl groups are on Lys-16 or Lys-17. By contrast, ETD data confidently places both methyl groups on Lys-16, i.e. Lys-16 is dimethylated. In addition, a proteoform with a nominal ΔM of 42 Da was found, which could correspond to the expected lysine acetylation. However, the high-resolution, high mass accuracy ETD data indicate that Lys-16 is trimethylated (ΔM = 42.0469 Da) rather than acetylated (ΔM = 42.0106 Da). At 3 ppm matching tolerance, 40 fragments match to the acetylated form of the protein and 56 fragments match to the trimethylated form (see supplemental Fig. S7 in ESI 1). To investigate the acetylation state of Alba we performed Western blot analysis using anti-acetyllysine primary antibody (supplemental Fig. S2). We analyzed cell extracts from the Early, Mid, and Late sampling points, whereas extracts from E. coli, Saccharomyces cerevisiae and S. solfataricus (previously shown to contain acetylated Alba protein) were used as positive controls. Prominent protein bands were cut from the SDS-PAGE gel, in-gel digested and subjected to LC-MS analysis. In-gel digestion of the 12kDa band was dominated by the Alba1 protein. The anti-acetyllysine antibodies revealed the acetylation in the band corresponding to Alba1 in both S. islandicus and S. solfataricus (supplemental Fig. S2). Collectively, LC-MS and the Western blot analyses strongly suggest that Alba proteins in S. islandicus LAL14/1 are both methylated and acetylated on the lysine side chains in vivo, although acetylation appears to be less pronounced (and hence undetected by LC-MS) compared with methylation. For the three methylated Alba1 proteoforms, the complementary pair of c15 and z81-ions in ETD spectra suggests that the methylation is located on Lys-16, whereas the neighboring Lys-17 appears to be unmodified. To date, lysine dimethylation and trimethylation have not been reported to occur in archaea. These modifications constitute a direct analogy between Alba1 and eukaryotic histones.
Growth Stage-Dependent Dynamics of Methylated Proteoforms by Top-Down Analysis
SAX prefractionation of the proteins at the Late culture growth stage allowed us to achieve a high proteome coverage in top-down mode with 681 unique identifications. However, this strategy requires a large number of LC-MS runs and is not suitable for comparing the three growth stages in three replicates. We found a good compromise by using a GELFrEE molecular weight-based separation, instead of a SAX fractionation. Each cytosolic sample was separated into four fractions in the range of 5–25 kDa (supplemental Fig. S8 in the file ESI 1), representing the “light” part of the proteome, which is the most suitable for a high-resolution top-down analysis on an Orbitrap Fusion. The first fraction was therefore found to be enriched in the smallest proteins, such as Sul7d and Cren7, the second and third contained the Alba proteins along with other proteins of a similar weight.
In accordance with the results of the bottom-up analysis, top-down approach also revealed the tendency toward an increase of the relative abundance of methylated proteoforms at the later growth stages (see supplemental Fig. S9 in ESI 1).
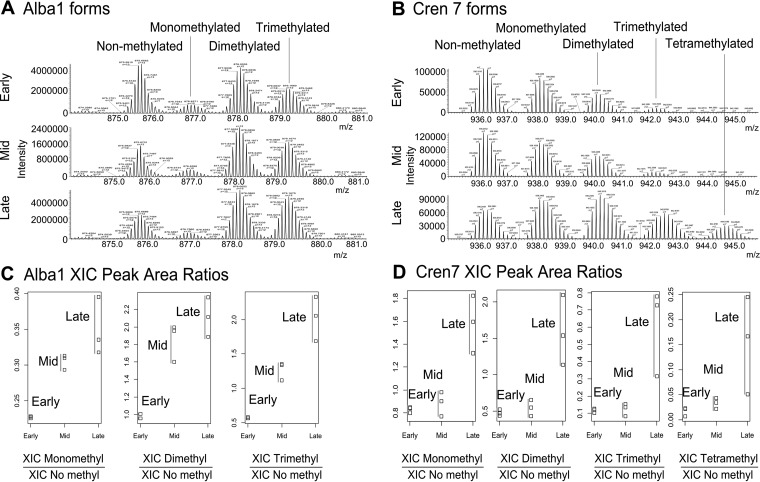
For example, in the case of Alba1, Cren7 (see spectra in Fig. 6A and 6B, respectively), Sul7d proteins, small ribonucleoprotein (M9U8A0), ribosomal proteins L7Ae and L12 as well as the NT domain-containing protein, the relative intensity of the signals corresponding to the methylated forms compared with those of the nonmethylated counterparts clearly increased from the Early growth stage to the Mid and the Late stage. By contrast, such changes in the relative abundances are not observed for the proteoforms of Alba2 and a transcriptional regulator (M9UEC7) (see supplemental Fig. S9). Interestingly, these proteins were not found to be differentially expressed between the different growth stages according to the LFQ data. This means that despite the same overall expression level, these proteins are more heavily methylated at the Late growth stage compared with the Mid and Early stages. This correlates with the increased number of identified methylation sites in the bottom-up experiments (Fig. 2).
Fig. 6.
Relative quantification of Alba1 and Cren7 proteoforms by top-down experiments. A, averaged spectra of Alba1 proteoforms at three growth stages; B, averaged spectra of Cren7 proteoforms at three growth stages; C, relative abundances of the Alba1 methylated forms at three growth stages; D, relative abundances of the Cren7 methylated forms at three growth stages. Peak areas on the extracted ion chromatograms (XIC) for each form are related to XIC peak area of nonmethylated form in the corresponding chromatogram.
To test the significance of the observed effect, we chose Alba1 and Cren7 to perform a relative proteoform quantitation, based on the relative XIC areas. The results are summarized on the dot plots in Fig. 6. The details of the calculations can be found in the file ESI 10. As shown in Fig. 6A, the relative abundance of methylated Alba1 forms compared with the nonmethylated counterpart rises drastically from the Early to Mid and Late time points, from 1 to 2 for the dimethylated proteoform and from 0.6 to 2 for the trimethylated form. These results are consistent for all three biological replicates. Welch t test p values were calculated on the ln-transformed relative intensity values to check the significance of the changes between growth stages (see supplemental Table S3 in ESI 1). The changes for mono-, di-, and trimethylated forms appear to be significant with p values < 0.05 for the comparisons between Early and Mid (p values 3.3 × 10−3 - 8.5 × 10−3) as well as between Early and Late stage (p values 3.7 × 10−3 - 2.3 × 10−2). By contrast, for the Cren7 protein the differences between Early and Mid stages turned out to be insignificant, whereas the comparison of the relative methylated proteoform abundances between Early and Late stages reveals a significant rise with the Welch t test p values of 0.016– 0.027 (see Fig. 6 above and supplemental Table S4 in ESI 1). Hence, we conclude that for both Alba1 and Cren7 there is a significant increase in the relative abundance of methylated proteoforms from the Early to the Late stage of cell growth, which strongly confirms and complements the bottom-up data, where we were able to conclude a statistically significant increase of the methylated peptide abundance for the Sul7d proteins.
DISCUSSION
We report here a comprehensive study of the proteomic landscape of the thermoacidophilic archaeon Sulfolobus islandicus during cell growth. Bottom-up and top-down proteomics approaches provided complementary information and shed light not only on the differential abundance of proteins at the three different growth phases but also on the nature and dynamics of post-translational protein modifications in this archaeon. We paid a particular attention to a subset of small proteins that are abundant in the cell and involved in structuring of the Sulfolobus chromatin.
Previous studies focusing on the nucleoid organization in different Sulfolobus species revealed that the nucleoids were highly structured and differentially distributed in the cell interior, depending on the cell growth phase (36, 37). These observations along with the changes in the kinetics of cell division at different growth phases (supplemental Fig. S1) imply a major shift in the metabolic activity as well as subcellular organization of Sulfolobus cells, which might be accompanied by changes in protein content and/or their post-translational modification. To date, only a few quantitative proteomic studies of Sulfolobus species (56) have been reported. To assess the variations in relative abundance of Sulfolobus proteins during different stages of the cell growth, we performed a proteome-wide label-free relative quantitation (LFQ) (57) using an Orbitrap Fusion mass spectrometer (58). The results of this analysis were expected to serve a dual purpose: (1) provide novel information on the proteome dynamics during the growth of a hyperthermophilic archaeon; (2) serve as a basis for the subsequent PTM and proteoform analyses.
Proteome-wide label-free relative quantitation at the peptide level allowed us to gain insight into the changes in protein expression levels at different growth stages, known to be linked to changes in metabolic activity and architecture of the nucleoid (36, 37). A total of 93 proteins were found to significantly change their relative abundances between the early-exponential and the stationary growth phases (FDR 0.01). Some of the proteins increased in abundance, whereas others were significantly diminished. Among the proteins that became more abundant in the stationary phase, we found proteins involved in the oxidative stress response and nucleotide metabolism, whereas proteins involved in various metabolic processes, such as enzymes implicated in metabolism of amino acid, carbon, and sulfur, were found to be significantly down-regulated. Not unexpectedly, proteins participating in cell division, tRNA and ribosome biogenesis also became less abundant as the cell culture reached the stationary phase. The results obtained are fully consistent with the physiological response of the cell population toward the reduced availability of the resources in the growth medium, which manifests in the reduced rate of cell growth and diminished metabolic activity.
Strikingly, there was no change in abundance at the protein level for any of the five chromatin proteins: Alba1, Alba2, Cren7, Sul7d1, and Sul7d2. Several in vitro studies have shown that Alba1 and Alba2 bind DNA as dimers and that binding affinities as well as effects on the chromatin folding are different for homo- and heterodimers (59, 60). Consequently, it has been suggested that differential expression of Alba1 and Alba2 would change the ratio of Alba homo- and hetero-dimers within the cell thereby providing means to regulate the global gene expression (59–61). Specifically, because heterodimers exhibit weaker dimer-dimer interactions compared with Alba1 homodimer, increase in the level of heterodimers would make the DNA more accessible for transcription (61). Our results demonstrate that the ratio between different chromatin proteins, including both Alba proteins, is rather constant throughout the growth stages, suggesting that, at least in S. islandicus, nucleoid organization and gene expression are likely to be regulated by a mechanism independent of the differential expression of chromatin proteins.
One of the most striking results of our analysis is the unprecedented extent of lysine methylation in S. islandicus, with up to 1,931 methylation sites on 731 proteins, which were found using bottom-up proteomics approach. Protein methylation in Sulfolobus is believed to be one of the adaptations to high temperature and its ubiquity is unique in comparison with mesophilic bacteria or eukaryotes (11, 44). The latter possibility is consistent with our observation that methylation is independent of the functional category to which the protein belongs—proteins from all 21 arCOG functional categories were found to be methylated, with roughly 50% of proteins from each category containing this PTM. Furthermore, comparison of the methylation profiles at different growth phases revealed that methylation of the proteome becomes considerably more pronounced as the cell culture approaches stationary phase. Consistent with the ubiquity of methylation, our analysis also shows a complete lack of sequence specificity for the involved lysine methyltransferase. Although it was beyond the scope of this work to pinpoint the exact enzyme(s) responsible for the modification, the previously described aKMT4 methyltransferase, which remains the only characterized lysine methyltransferase in Sulfolobus (62), represents the most likely candidate. Indeed, the latter protein is conserved in organisms of the phylum Crenarcheota and has been shown to have a very relaxed specificity (62). According to our LFQ data set, this methyltransferase (M9U9Y9) is abundant in S. islandicus and its abundance is stable throughout the growth phases. Notably both top-down and bottom-up analyses showed that the aKMT4 methyltransferase is itself methylated (K-15, K-25, and K-74), consistent with the previous report (51).
Although the bulk methylation is likely to represent a specific adaptation of hyperthermophiles to high temperatures, some of the methylations might also play other, sometimes regulatory, roles, like in the case of eukaryotic histones. By analogy, this might be the case for chromatin proteins. High-throughput top-down analysis of S. islandicus cytosolic proteins enabled the comprehensive characterization of 85 methylated forms of 54 proteins of up to 19.5 kDa, including chromatin proteins. Very interestingly, the top-down approach brought a unique level of accuracy for the identified proteins, highlighting changes in PTMs during cell growth that could not be monitored by the classical bottom-up approach. In addition, the number of proteoforms identified in this study is the largest ever described for prokaryotic cells. This was made possible by the use of a simple fractionation step before the LC-MS analysis and the use of a combination of fragmentation techniques such as HCD and ETD. We found, for the first time, that S. islandicus Alba1 and Alba2 proteins are methylated, and Alba1 may exist under mono-, di-, and trimethylated proteoforms. To the best of our knowledge, lysine dimethylation and trimethylation have never been previously reported in Sulfolobus. Importantly, the methylation in Alba1 protein occurs at Lys-16, the residue that is located at the protein-DNA interface. Previous reports have shown that acetylation of Alba1 from S. solfaricus at Lys-16 lowers the DNA-binding affinity of the protein. Methylation is expected to change the hydrophobic and steric properties of Lys-16 of Alba1 in S. islandicus and thus might also modulate the protein-DNA interactions. These observations further strengthen the previously noticed analogy between Alba and eukaryotic histones (63–65). Because Alba1 bears dimethylation and trimethylation and it is not modified upon aKMT4 even in vitro in closely related Sulfolobus strains (51), we hypothesize that another methyltransferase may be involved in Alba1 methylation in S. islandicus LAL14/1.
Why does the proteome become more methylated at the later stages of the cell growth? We hypothesize that the protein turnover rate and availability to the methyltransferase may play a major role. If we assume the constant rate of any given protein methylation in the cytosol, then during active cell growth the proteins are synthesized at high rate in the dividing cells, constantly diluting the methylated protein pool with the “fresh” nonmethylated proteoforms. When the protein turnover slows down, an average methylation reaction time for each protein copy increases and the extent of methylation rises. Proteins with limited availability to the solvent are not expected to show much increase in methylation because of the slow overall reaction rate. Also, proteins with high turnover will be unaffected by methylation increase as their short half-life may remain unchanged in actively growing and stationary phase cells. Another possible cause is the higher degradation rate of the nonmethylated proteoforms as methylation has been shown to increase protein thermostability (11): in this case nonmethylated forms are depleted faster, than methylated ones and this effect should be more pronounced in the stationary stage, because the overall protein synthesis rate is lower. However, the details, exact mechanism and consequences of the increase in the extent of lysine methylation in S. islandicus are subjects for future research.
Although acetylation of the lysine side chains in S. islandicus was not detected by LC-MS in the present study, we found that N-terminal protein acetylation is rather prevalent. More than one third of the 372 detected N-terminal peptides were acetylated. This result is consistent with the previous smaller-scale analysis reported by Mackay et al. (42) that showed that 17 out of 26 analyzed N-terminal peptides from S. solfataricus were acetylated. More generally, the extent of N-terminal acetylation appears to vary across archaeal species (41). For example, in halophilic archaea Halobacterium salinarum and Natronomonas pharaonis, 14% to 19% of proteins are N-terminally acetylated (66), whereas in another haloarchaeon, Haloferax volcanii, 29% of the analyzed proteins carry this modification (67). By contrast, it is believed that N-terminal acetylation in methanogenic archaea is very rare or does not occur at all (41). N-terminal protein acetylation in S. islandicus appears to be rather promiscuous—although serine residue is the most prevalent substrate, acetylated N-terminal methionine, alanine, threonine, glutamate, and valine could also be detected, albeit at different frequencies. Biochemical and structural characterization of the N-terminal acetyltransferase of S. solfataricus (96% identical to the homolog in S. islandicus) revealed the basis of the relaxed substrate specificity and showed that the archaeal enzyme is homologous to the eukaryotic counterpart (68). Notably, three of the five chromatin proteins, specifically Alba1, Alba2, and Cren7, are N-terminally acetylated. However, the implications of this modification remain unclear.
To sum up, this study shows that although bottom-up proteomics allows a deep analysis of proteomes, the high complexity and variability of the data complicates the discovery of subtle proteomic differences because of change in post-translational modifications. Top-down approach allows to reduce the sample complexity and to conveniently analyze proteins at the proteoform level. The bottom-up analysis enables comparing the intensities of different peptides, which increases the breadth of the results. The high number of peptides also allows for the multiple-testing error probability, addressed using FDR correction, which inevitably hides some of the results below the significance threshold. By contrast, the top-down approach provides the opportunity to compare the signal intensities of the proteoforms from the same spectra, which reduces the variability and facilitates the comparison of the proteoforms. Bottom-up and top-down proteomics approaches are therefore highly complementary, and as shown in this study, can be performed on the same mass spectrometer, in a routine fashion: all the consumables used here are commercially available and all the methods can be directly transferred to another instrument without much effort.
Associated Content
Supplementary file ESI 1: supplementary experimental procedures, supplementary figures and tables. File ESI 2: technical variability between the technical replicates in the bottom-up LFQ data set. File ESI 3: output tables for the bottom-up LFQ (protein level data). File ESI 4: N-terminal acetylation sites. File ESI 5: lysine monomethylation sites identified from the bottom-up data for S. islandicus. File ESI 6: lysine monomethylation sites identified from the bottom-up data for E. coli. File ESI 7: peptide-level abundance changes between the Early and Late growth stages, including the K-Me peptides. File ESI 8: peptide-level abundance changes between the Early and Late growth stages for the 5 chromatin proteins. File ESI 9: the proteoforms identified in the SAX-fractionated sample using top-down approach. File ESI 10: summary of the Alba1 and Cren7 proteoforms relative quantitation by top-down. The raw data files have been deposited to the ProteomeXchange with identifiers PXD003074 and PXD004179. Annotated top-down spectra from ProSightPC search output can be viewed in MS-Viewer (32) on the Protein Prospector public website (prospector.ucsf.edu): search key dk458i4qtb for SAX ETD data, rvs63gz69o for SAX HCD data, vmhsk5c8pb for GELFrEE ETD data and wgvkcvlyeg for GELFrEE HCD data.
Supplementary Material
Footnotes
Author contributions: E.V., M.K., D.P. and J.C.R. designed the research; E.V. and E.R. performed the research; E.V., E.R. and M.K. treated the data; E.V., E.R., M.K., J.C.R. wrote the manuscript.
*
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PTM
- post translational modification
- AF
- ammonium formate
- ETD
- electron-transfer dissociation
- GELFrEE
- gel elution liquid fraction entrapment electrophoresis
- HCD
- higher-energy collision-induced dissociation
- LFQ
- label-free quantification
- PS-DVB
- polystyrene-divinylbenzene co-polymer
- SAX
- strong anion exchange
- SCX
- strong cation exchange.
REFERENCES
- 1. Kashefi K., and Lovley D. R. (2003) Extending the Upper Temperature Limit for Life. Science 301, 934. [DOI] [PubMed] [Google Scholar]
- 2. Schleper C., Puehler G., Holz I., Gambacorta A., Janekovic D., Santarius U., Klenk H. P., and Zillig W. (1995) Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J. Bacteriol. 177, 7050–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eichler J., and Adams M. W. W. (2005) Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69, 393–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eichler J., and Maupin-Furlow J. (2013) Post-translation modification in Archaea: lessons from Haloferax volcanii and other haloarchaea. FEMS Microbiol. Rev. 37, 583–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jarrell K. F., Ding Y., Meyer B. H., Albers S. V, Kaminski L., and Eichler J. (2014) N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol. Mol. Biol. Rev. 78, 304–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Humbard M. A. Miranda H. V., Lim J. M, Krause D. J., Pritz J. R., Zhou G., Chen S., Wells L., and Maupin-Furlow J. a (2010) Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Esser D., Pham T. K., Reimann J., Albers S. V., Siebers B., and Wright P. C. (2012) Change of carbon source causes dramatic effects in the phospho- proteome of the Archaeon Sulfolobus solfataricus. J. Proteome Res. 11, 4823–4833 [DOI] [PubMed] [Google Scholar]
- 8. Palmieri G., Balestrieri M., Peter-Katalinić J., Pohlentz G., Rossi M., Fiume I., and Pocsfalvi G. (2013) Surface-exposed Glycoproteins of Hyperthermophilic Sulfolobus solfataricus P2 Show a Common N-Glycosylation Profile. J. Proteome Res. 12, 2779–2790 [DOI] [PubMed] [Google Scholar]
- 9. Bell S. D., Botting C. H., Wardleworth B. N., Jackson S. P., and White M. F. (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296, 148–151 [DOI] [PubMed] [Google Scholar]
- 10. Maras B., Valiante S., Chiaraluce R., Consalvi V., Politi L., De Rosa M., Bossa F., Scandurra R., and Barra D. (1994) The amino acid sequence of glutamate dehydrogenase fromPyrococcus furiosus, a hyperthermophilic archaebacterium. J. Protein Chem. 13, 253–259 [DOI] [PubMed] [Google Scholar]
- 11. Botting C. H., Talbot P., Paytubi S., and White M. F. (2010) Extensive lysine methylation in hyperthermophilic crenarchaea: potential implications for protein stability and recombinant enzymes. Archaea 2010, 106341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Febbraio F., Andolfo A., Tanfani F., Briante R., Gentile F., Formisano S., Vaccaro C., Scirè A., Bertoli E., Pucci P., and Nucci R. (2004) Thermal stability and aggregation of sulfolobus solfataricus beta-glycosidase are dependent upon the N-epsilon-methylation of specific lysyl residues: critical role of in vivo post-translational modifications. J. Biol. Chem. 279, 10185–10194 [DOI] [PubMed] [Google Scholar]
- 13. Xia Y., Niu Y., Cui J., Fu Y., Chen X. S., Lou H., and Cao Q. (2015) The helicase activity of hyperthermophilic archaeal MCM is enhanced at high temperatures by lysine methylation. Front. Microbiol. 6, 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baumann H., Knapp S., Lundback T., Ladenstein R., and Hard T. (1994) Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nat. Struct. Mol. Biol. 1, 808–819 [DOI] [PubMed] [Google Scholar]
- 15. Kelleher N. L. (2004) Top-Down Proteomics. Anal. Chem. 76, 196A–20A [PubMed] [Google Scholar]
- 16. Smith L. M., and Kelleher N. L. (2013) Proteoform: a single term describing protein complexity. Nat. Meth. 10, 186–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gault J., Malosse C., Machata S., Millien C., Podglajen I., Ploy M. C, Costello C. E., Duménil G., and Chamot-Rooke J. (2014) Complete posttranslational modification mapping of pathogenic Neisseria meningitidis pilins requires top-down mass spectrometry. Proteomics 14, 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gault J., Ferber M., Machata S., Imhaus A. F, Malosse C., Charles-Orszag A., Millien C., Bouvier G., Bardiaux B., Péhau-Arnaudet G., Klinge K., Podglajen I., Ploy M. C., Seifert H. S., Nilges M., Chamot-Rooke J., and Duménil G. (2015) Neisseria meningitidis Type IV Pili composed of sequence invariable pilins are masked by multisite glycosylation. PLoS Pathog. 11, e1005162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tran J. C., Zamdborg L., Ahlf D. R., Lee J. E., Catherman A. D., Durbin K. R., Tipton J. D., Vellaichamy A., Kellie J. F., Li M., Wu C., Sweet S. M., Early B. P., Siuti N., LeDuc R. D., Compton P. D., Thomas P. M., and Kelleher N. L. (2011) Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ansong C., Wu S., Meng D., Liu X., Brewer H. M., Deatherage Kaiser B. L., Nakayasu E. S., Cort J. R., Pevzner P., Smith R. D., Heffron F., Adkins J. N., and Pasa-Tolic L. (2013) Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proc. Natl. Acad. Sci. 110, 10153–10158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu S., Brown J. N., Tolic N., Meng D., Liu X., Zhang H., Zhao R., Moore R. J., Pevzner P., Smith R. D., and Pasa-Tolic L. (2014) Quantitative analysis of human salivary gland-derived intact proteome using top-down mass spectrometry. Proteomics 14, 1211–1222 [DOI] [PubMed] [Google Scholar]
- 22. Ntai I., Kim K., Fellers R. T., Skinner O. S., Smith A. D., Early B. P., Savaryn J. P., LeDuc R. D., Thomas P. M., and Kelleher N. L. (2014) Applying label-free quantitation to top down proteomics. Anal. Chem. 86, 4961–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moradian A., Kalli A., Sweredoski M. J., and Hess S. (2014) The top-down, middle-down, and bottom-up mass spectrometry approaches for characterization of histone variants and their post-translational modifications. Proteomics 14, 489–497 [DOI] [PubMed] [Google Scholar]
- 24. Jaubert C., Danioux C., Oberto J., Cortez D., Bize A., Krupovic M., She Q., Forterre P., Prangishvili D., and Sezonov G. (2013) Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 3, 130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tran J. C., and Doucette A. A. (2008) Gel-Eluted Liquid Fraction Entrapment Electrophoresis: An Electrophoretic Method for Broad Molecular Weight Range Proteome Separation. Anal. Chem. 80, 1568–1573 [DOI] [PubMed] [Google Scholar]
- 26. Botelho D., Wall M. J., Vieira D. B., Fitzsimmons S., Liu F., and Doucette A. (2010) Top-Down and Bottom-Up Proteomics of SDS-Containing Solutions Following Mass-Based Separation. J. Proteome Res. 9, 2863–2870 [DOI] [PubMed] [Google Scholar]
- 27. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 29. Goloborodko A., Levitsky L., Ivanov M., and Gorshkov M. V. (2013) Pyteomics—a Python Framework for Exploratory Data Analysis and Rapid Software Prototyping in Proteomics. J. Am. Soc. Mass Spectrom. 24, 301–304 [DOI] [PubMed] [Google Scholar]
- 30. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Catherman A. D., Durbin K. R., Ahlf D. R., Early B. P., Fellers R. T., Tran J. C., Thomas P. M., and Kelleher N. L. (2013) Large-scale top-down proteomics of the human proteome: membrane proteins, mitochondria, and senescence. Mol. Cell. Proteomics 12, 3465–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baker P. R., and Chalkley R. J. (2014) MS-Viewer: a web based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown A. M., Hoopes S. L., White R. H., and Sarisky C. A. (2011) A purine biosynthesis in archaea: variations on a theme. Biol. Direct 6, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schnell S., and Steinman H. M. (1995) Function and stationary-phase induction of periplasmic copper-zinc superoxide dismutase and catalase/peroxidase in Caulobacter crescentus. J. Bacteriol. 177, 5924–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nair S., and Finkel S. E. (2004) Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186, 4192–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Poplawski A., and Bernander R. (1997) Nucleoid structure and distribution in thermophilic Archaea. J. Bacteriol. 179, 7625–7630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hjort K., and Bernander R. (1999) Changes in cell size and DNA content in Sulfolobus cultures during dilution and temperature shift experiments. J. Bacteriol. 181, 5669–5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550 [DOI] [PubMed] [Google Scholar]
- 39. Svinkina T., Gu H., Silva J. C., Mertins P., Qiao J., Fereshetian S., Jaffe J. D., Kuhn E., Udeshi N. D., and Carr S. A. (2015) Deep, Quantitative Coverage of the Lysine Acetylome Using Novel Anti-acetyl-lysine Antibodies and an Optimized Proteomic Workflow. Mol. Cell. Proteomics 14, 2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang K., Tian S., and Fan E. (2013) Protein lysine acetylation analysis: current MS-based proteomic technologies. Analyst 138, 1628–1636 [DOI] [PubMed] [Google Scholar]
- 41. Soppa J. (2010) Protein acetylation in archaea, bacteria, and eukaryotes. Archaea 2010, 820681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mackay D. T., Botting C. H., Taylor G. L., and White M. F. (2007) An acetylase with relaxed specificity catalyses protein N-terminal acetylation in Sulfolobus solfataricus. Mol. Microbiol. 64, 1540–1548 [DOI] [PubMed] [Google Scholar]
- 43. Ammendola S., Raia C. A., Caruso C., Camardella L., D'Auria S., De Rosa M., and Rossi M. (1992) Thermostable NAD+-dependent alcohol dehydrogenase from Sulfolobus solfataricus: gene and protein sequence determination and relationship to other alcohol dehydrogenases. Biochemistry 31, 12514–12523 [DOI] [PubMed] [Google Scholar]
- 44. Maras B., Consalvi V., Chiaraluce R., Politi L., De Rosa M., Bossa F., Scandurra R., and Barra D. (1992) The protein sequence of glutamate dehydrogenase from Sulfolobus solfataricus, a thermoacidophilic archaebacterium. Eur. J. Biochem. 203, 81–87 [DOI] [PubMed] [Google Scholar]
- 45. Azkargorta M., Wojtas M. N., Abrescia N. G. A., and Elortza F. (2014) Lysine methylation mapping of Crenarchaeal DNA-directed RNA polymerases by collision-induced and electron-transfer dissociation mass spectrometry. J. Proteome Res. 13, 2637–2648 [DOI] [PubMed] [Google Scholar]
- 46. Perazzona B., and Spudich J. L. (1999) Identification of Methylation Sites and Effects of Phototaxis Stimuli on Transducer Methylation in Halobacterium salinarum. J. Bacteriol. 181, 5676–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koch M. K., Staudinger W. F., Siedler F., and Oesterhelt D. (2008) Physiological sites of deamidation and methyl esterification in sensory transducers of Halobacterium salinarum. J. Mol. Biol. 380, 285–302 [DOI] [PubMed] [Google Scholar]
- 48. Schmidt A., Kochanowski K., Vedelaar S., Ahrne E., Volkmer B., Callipo L., Knoops K., Bauer M., Aebersold R., and Heinemann M. (2016) The quantitative and condition-dependent Escherichia coli proteome. Nat Biotech 34, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Makarova K. S., Sorokin A. V., Novichkov P. S., Wolf Y. I., and Koonin E. (2007) V Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol. Direct 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Makarova K. S., Wolf Y. I., and Koonin E. V. (2015) Archaeal clusters of orthologous genes (arCOGs): An update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life 5, 818–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Niu Y., Xia Y., Wang S., Li J., Niu C., Li X., Zhao Y., Xiong H., Li Z., Lou H., and Cao Q. (2013) A prototypic lysine methyltransferase 4 from archaea with degenerate sequence specificity methylates chromatin proteins Sul7d and Cren7 in different patterns. J. Biol. Chem. 288, 13728–13740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Forbes A. J., Patrie S. M., Taylor G. K., Kim Y. B, Jiang L., and Kelleher N. L. (2004) Targeted analysis and discovery of posttranslational modifications in proteins from methanogenic archaea by top-down MS. Proc. Natl. Acad. Sci. U.S.A. 101, 2678–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ferguson J. T., Wenger C. D., Metcalf W. W., and Kelleher N. L. (2009) Top-down proteomics reveals novel protein forms expressed in methanosarcina acetivorans. J. Am. Soc. Mass Spectrom. 20, 1743–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bunger M. K., Cargile B. J., Ngunjiri A., Bundy J. L., and Stephenson J. L. (2008) Automated proteomics of E. coli via top-down electron-transfer dissociation mass spectrometry. Anal. Chem. 80, 1459–1467 [DOI] [PubMed] [Google Scholar]
- 55. Fortelny N., Pavlidis P., and Overall C. M. (2015) The path of no return-Truncated protein N-termini and current ignorance of their genesis. Proteomics 15, 2547–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kort J. C., Esser D., Pham T. K., Noirel J., Wright P. C., and Siebers B. (2013) A cool tool for hot and sour Archaea: Proteomics of Sulfolobus solfataricus. Proteomics 13, 2831–2850 [DOI] [PubMed] [Google Scholar]
- 57. Bantscheff M., Schirle M., Sweetman G., Rick J., and Kuster B. (2007) Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 58. Hebert A. S., Richards A. L., Bailey D. J., Ulbrich A., Coughlin E. E., Westphall M. S., and Coon J. J. (2014) The one hour yeast proteome. Mol. Cell. Proteomics, 13, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jelinska C., Conroy M. J., Craven C. J., Hounslow A. M., Bullough P. A., Waltho J. P., Taylor G. L., and White M. F. (2005) Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging. Structure 13, 963–971 [DOI] [PubMed] [Google Scholar]
- 60. Laurens N., Driessen R. P., Heller I., Vorselen D., Noom M. C., Hol F. J., White M. F., Dame R. T., and Wuite G. J. (2012) Alba shapes the archaeal genome using a delicate balance of bridging and stiffening the DNA. Nat Commun 3, 1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Driessen R. P., and Dame R. T. (2013) Structure and dynamics of the crenarchaeal nucleoid. Biochem. Soc. Trans. 41, 321–325 [DOI] [PubMed] [Google Scholar]
- 62. Chu Y., Zhang Z., Wang Q., Luo Y., and Huang L. (2012) Identification and characterization of a highly conserved crenarchaeal protein lysine methyltransferase with broad substrate specificity. J. Bacteriol. 194, 6917–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao K., Chai X., and Marmorstein R. (2003) Structure of a Sir2 Substrate, Alba, Reveals a Mechanism for Deacetylation-induced Enhancement of DNA Binding. J. Biol. Chem. 278, 26071–26077 [DOI] [PubMed] [Google Scholar]
- 64. Sandman K., and Reeve J. N. (2005) Archaeal chromatin proteins: different structures but common function? Curr. Opin. Microbiol. 8, 656–661 [DOI] [PubMed] [Google Scholar]
- 65. Peeters E., Driessen R. P. C., Werner F., and Dame R. T. (2015) The interplay between nucleoid organization and transcription in archaeal genomes. Nat. Rev. Micro. 13, 333–341 [DOI] [PubMed] [Google Scholar]
- 66. Aivaliotis M., Gevaert K., Falb M., Tebbe A., Konstantinidis K., Bisle B., Klein C., Martens L., Staes A., Timmerman E., Van Damme J., Siedler F., Pfeiffer F., Vandekerckhove J., and Oesterhelt D. (2007) Large-scale identification of N-terminal peptides in the halophilic Archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res. 6, 2195–2204 [DOI] [PubMed] [Google Scholar]
- 67. Kirkland P. A., Humbard M. A., Daniels C. J., and Maupin-Furlow J. A. (2008) Shotgun Proteomics of the Haloarchaeon Haloferax volcanii. J. Proteome Res. 7, 5033–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chang Y. Y., and Hau C. H. (2015) Structural Basis for Substrate-specific Acetylation of Nα-acetyltransferase Ard1 from Sulfolobus solfataricus. Sci. Rep. 5, 8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.