Abstract
Intracellular GlcNAcylation of Ser and Thr residues is a well-known and widely investigated post-translational modification. This post-translational modification has been shown to play a significant role in cell signaling and in many regulatory processes within cells. O-GlcNAc transferase is the enzyme responsible for glycosylating cytosolic and nuclear proteins with a single GlcNAc residue on Ser and Thr side-chains. Here we report that the same enzyme may also be responsible for S-GlcNAcylation, i.e. for linking the GlcNAc unit to the peptide by modifying a cysteine side-chain. We also report that O-GlcNAcase, the enzyme responsible for removal of O-GlcNAcylation does not appear to remove the S-linked sugar. Such Cys modifications have been detected and identified in mouse and rat samples. This work has established the occurrence of 14 modification sites assigned to 11 proteins unambiguously. We have also identified S-GlcNAcylation from human Host Cell Factor 1 isolated from HEK-cells. Although these site assignments are primarily based on electron-transfer dissociation mass spectra, we also report that S-linked GlcNAc is more stable under collisional activation than O-linked GlcNAc derivatives.
Protein glycosylation is one of the most frequently occurring post-translational modifications. The generic term “glycosylation” includes a diverse set of modifications featuring different sugar units bound to amino acid side-chains (1). Based on the atom through which the peptide-saccharide linkage is established, C-, N-, O-, and S-glycosylation can be distinguished. C-glycosylation is a mannose modification on the side-chain of Trp (2). N-glycans, which feature a common core structure of GlcNAc2Man3 elongated with a variety of different moieties, are linked to Asn side-chains. This modification occurs within the consensus motif NX(S/T/C), where X cannot be Pro (3, 4). O-glycoproteins belong to two distinct categories: GlcNAcylation of Ser and Thr residues occurs within the cytoplasm or nucleus (3, 5); proteins destined for secretion or presentation on the cell membrane are modified in the ER and the Golgi, where GalNAc, Xyl, Fuc, Glc, or Man may be directly attached onto hydroxy amino acid side-chains (3). With the introduction of better analytical tools our knowledge about protein glycosylation is constantly expanding. Nonconsensus motif N-glycosylation has been reported (6–9) and O-glycosylation of Tyr residues has recently been discovered (9, 10). S-glycosylation is also a new discovery in prokaryotes: the sugar modification of Cys side-chains has been reported in Bacillus subtilis (S-glucosylation) and in Lactobacillus plantarum (S-GlcNAcylation) (11–13). Most of these discoveries are the result of mass spectrometric analysis of glycopeptides found in proteolytic digests of protein mixtures. It has become common place to use high mass accuracy collision-induced dissociation (CID)1 techniques to generate mass spectra of these substances, but the alternative non-ergodic technique, electron-transfer dissociation (ETD), has unique advantages for the assignment of sugar/protein linkage sites. The CID strategy yields extensive information about the structural nature of glycans in the observation of nonreducing end oxonium (B) ions and reducing end (Y) fragments (14) in the mass spectra, both series formed by glycosidic bond cleavages (15, 16). In addition, beam-type CID (in Thermo instruments termed “higher-energy C-trap dissociation,” i.e. HCD) usually induces sufficient peptide fragment ions to identify the peptide (17). However, as noted above because of the highly labile nature of the glycosidic bond upon collisional activation, information required for glycan site-localization is typically lost. This drawback may not be a problem for studies of N-glycosylation because of its consensus motif, but it is most certainly an obstacle in those cases where consensus motifs do not exist, i.e. in O-GlcNAcylation and extracellular O-glycosylation. The ETD process triggers peptide backbone cleavages without internal energy randomization; thus, preserving the side-chains intact, including the fragile oligosaccharide structures attached (9, 17–21). Thus, the information required to assign modification sites is usually present in ETD spectra. There are search engines, for example Protein Prospector and Byonic, that automatically perform the spectral assignments, but an array of potential glycans and the amino acids to be modified have to be specified prior to the database search (9, 22, 23). Automated interpretation of glycopeptide mass spectra is still in its infancy, therefore, careful inspection of such search results is recommended. Human intervention may help to identify misassignments as well as flag new, unexpected structural clues. In this article we report that S-linked glycosylation occurs in mammals, including humans. The side-chains of Cys residues are modified by a single GlcNAc, and O-GlcNAc transferase (OGT) (24, 25), the enzyme required for intracellular O-GlcNAcylation, is responsible for it.
EXPERIMENTAL PROCEDURES
Purification and Proteolytic Digestion of Mouse Synaptic Membranes
Isolation and proteolytic digestion of the mouse synaptosome was performed as described previously (20). Briefly, brains (minus cerebellum) from several adult C57BL/6J mice were harvested and immediately frozen in liquid nitrogen. Tissue was homogenized in sucrose buffer in the presence of 20 μm PUGNAc and phosphatase inhibitors and cleared by centrifugation. The membrane containing fraction was layered on a sucrose density gradient and fractionated by centrifugation. Synaptic membranes were collected at the 1.0–1.2 M interface. The synaptosome fraction (10 mg) was denatured in 6 m guanidine hydrochloride containing 20 μm PUGNAc and 6× phosphatase inhibitor mixture (I and III Sigma); samples were reduced with 2 mm Tris(2-carboxyethyl)phosphine hydrochloride at 57 °C for 1 h and alkylated with 4.2 mm iodoacetamide in the dark for 45 min at room temperature, then diluted 6-fold and incubated with 5.0% (w/w) TPCK treated trypsin (ThermoFisher Scientific, Rockford, IL) at 37 °C for 18 h.
Lectin Weak Affinity Chromatography
Desalted synaptosome tryptic peptides were resuspended in 600 μl LWAC buffer (100 mm Tris pH 7.5, 150 mm NaCl, 10 mm MgCl2, 10 mm CaCl2, 5% acetonitrile) and 100 μl were run over a 2.0 × 250 mm POROS-WGA column at 100 μl/min under isocratic conditions with LWAC buffer and eluted with a 100 μl injection of 40 mm GlcNAc. Glycopeptides from the rightmost 10% of the flow-through peak through the 40 mm GlcNAc peak were collected inline on a Luna 10u C18 30 × 4.6 mm column (Phenomenex, Torrance, CA). Glycopeptides collected from all 10 mg of total peptides were eluted with 50% acetonitrile 0.1% formic acid in a single 500 μl fraction. Glycopeptides from the eluate were subsequently enriched twice more for a total of three enrichment steps.
High pH Reverse Phase Chromatography
LWAC enriched glycopeptides were further fractionated by high pH RP chromatography with a 1.0 × 100 mm Gemini 3μ C18 column (Phenomenex). Peptides were loaded onto the column in 100% buffer A (20 mm HCOONH4, pH 10). Buffer B consisted of buffer A with 50% acetonitrile. A gradient of 1% B to 21% B over 1.1 ml, to 62% B over 5.4 ml, and then directly to 100% B with a flow rate of 80 μl/min was used and 24 fractions were collected.
In Vitro GlcNAcylation and Deglycosylation
Peptides MCAALNSMDQYGGR, QKAPFPATCEAPSR, LDFGQGSGSPVCLAQVK, AVCCDMVYKLPFGR, QKAPFPAACEAPAR, LDFGQGAGAPVCLAQVK were synthesized by Elim Biopharmaceuticals (Hayward, CA). O-GlcNAc standard peptide, TAPTS(GlcNAc)TIAPG was purchased from ThermoFisher Scientific. Recombinant O-GlcNAc transferase expressed and purified from E. coli (30 μm) and synthetic peptide (33 μm) were combined with 30 μm UDP-GlcNAc in 50 mm Tris-HCl, 12.5 mm MgCl2, 1 mm DTT for 18 h at 37 °C. Reactions were stopped with the addition of formic acid to 0.3% (v/v) and desalted prior to MS analysis. As a control the above reactions were also carried out without addition of OGT.
Cys-GlcNAc modified peptides produced with OGT under the above reaction conditions were separated from the enzyme using a 10 K Amicon Ultra spin filter. Peptides were then desalted, dried, and resuspended in 20 mm HEPES pH 7.2 with or without 7 nm recombinant O-GlcNAcase from Streptococcus pyogenes (Prozomix, Haltwhistle, UK). The activity of the glycosidase was also tested with the standard O-GlcNAcylated peptide.
Mass Spectrometry Analysis
All samples were analyzed on an LTQ-Orbitrap Velos mass spectrometer (Thermo) equipped with a nano-Acquity UPLC system (Waters, Milford, MA). Peptides were fractionated on a 15 cm × 75 μm ID 3 μm C18 EASY-Spray column using a linear gradient, from 2–35% solvent B developed in 60 min. Mass measurements were performed in the Orbitrap. The three most abundant multiply charged ions were computer-selected for HCD as well as ETD analysis. The trigger intensity was set to 2000. Supplemental activation was enabled. The ETD fragments were measured in the linear trap and the HCD fragments were analyzed in the Orbitrap. Each sample was injected twice. In the first analysis, precursor ion selection was restricted to 2+ ions, in the second analysis doubly charged precursors were excluded. Raw files have been uploaded to MassIVE under the accession# MSV000079722. Peaklists were extracted using Proteome Discoverer 1.4. ETD and HCD data was searched against the UniProt Mus musculus database (73,955 entries, downloaded June 6, 2013) (and concatenated with a randomized sequence for each entry) using Protein Prospector (v5.10.15). Cleavage specificity was set as tryptic allowing for 2 missed cleavages. Carbamidomethylation of Cys, acetylation of protein N termini, oxidation of Met, cyclization of N-terminal Gln, and HexNAc modification of Ser, Thr, and Asn, and a mass modification of 203–203.1 Da of Cys were set as variable modifications. Three modifications per peptide were permitted. The required mass accuracy was 20 ppm for precursor ions, and 30 ppm or 0.8 Da for HCD and ETD fragments, respectively. Spectra identified as representing peptides featuring a Cys-modification close to the exact mass of HexNAc (203.0794) with a minimum score of 22, a maximum E value of 0.05 and with a SLIP (site localization) score of 5 were tentatively accepted, and carefully scrutinized. A new mouse synaptosome mixture was analyzed after glycopeptide enrichment in LC/MS EThcD mode on an Orbitrap Fusion Lumos Tribrid Mass Spectrometer in 1 h gradient fractionation/data acquisition. In this experiment both the precursor and fragment ions were measured in the Orbitrap. Thus, the database searches were performed as described above but with a 10 ppm and 20 ppm mass accuracy requirement for the precursors and fragments, respectively. This search was performed with Protein Prospector v5.14.20 and the CysGlcNAcylation was accurately defined. All other parameters remained the same. When generating the XIC chromatogram for the comparison of the relative amounts of S- and O-GlcNAcylated sequences the monoisotopic masses ±20 ppm were extracted from the MS1 data.
RESULTS
Glycopeptide enrichment studies were conducted with a mouse synaptosome tryptic digest using lectin affinity chromatography with wheat germ agglutinin as described previously (20). The resulting mixtures were further fractionated by high pH reverse phase liquid chromatography and subjected to LC/MS/MS analyses. Precursor ions were fragmented with beam-type collisional activation (“higher energy C-trap dissociation,” HCD) as well as electron-transfer dissociation (ETD). Manual inspection of particular spectra revealed that in certain instances the spectral assignments would be more convincing if the HexNAc-modification of Cys-residues was considered. A database search was then performed with a 203–203.1 Da variable modification on Cys side-chains and a significant number of Cys-modified sequences were identified with high confidence (Table I, supplemental Table S1). The ETD spectra in question were carefully evaluated along with their corresponding HCD spectra. In all rigorous cases the interpretation of the ETD spectra unambiguously established the Cys as the site of modification (Fig. 1, supplemental Table S1; supplemental Figs. S1–S10). The single modifying HexNAc was assigned as GlcNAc based on its characteristic fragment ions (Fig. 2A, supplemental Figs. S1–S17, S26) (26). Interestingly, S-linked GlcNAcylation seems to be more stable than O-linked GlcNAcylation as illustrated with the fragmentation of an O-GlcNAcylated peptide of identical amino acid sequence (Fig. 2B). It was also observed that the ETD spectra of these S-GlcNAcylated peptides usually featured significant S-GlcNAc losses from the charge-reduced precursor ions (Fig. 1, supplemental Figs. S1–S17, S26). Additional database searches with rat sciatic nerve (SN) and mouse embryonic stem cell (ESC) glycopeptide datasets, where the sample preparation, glycopeptide enrichment, and mass spectrometry analysis were performed in a similar manner, yielded 7 S-linked GlcNAcylated peptides (Table I, supplemental Table S1, supplemental Figs. S11–S17). One of these peptides, NAC(GlcNAc)IAPAAFSGQPQK from MLX-interacting protein was also found in the synaptosome data set, whereas a homologous sequence was present among the rat glycopeptides: SAC(GlcNAc)IAPAAFTGQPQK of Protein Mlxip. In addition, identical Protein Fam222b S-linked GlcNAcylated peptides were identified from the mouse ESC and rat SN samples (Table I, supplemental Table S1, supplemental Figs. S12 and S17). The S-linked GlcNAc peptides were reproducibly enriched and detected from a recent additional mouse synaptosome preparation. The sample was analyzed after glycopeptide enrichment but without further fractionation. In this new study three bassoon peptides, LDFGQGSGSPVC(GlcNAc)LAQVK, FPFGSSC(GlcNAc)TGTFHPAPSAPDK, and QKAPFPATC(GlcNAc)EAPSR were identified (supplemental Figs. S18–S20). Quite a few of the proteins detected with this new modification have been reported previously as being O-GlcNAcylated (18, 20, 21). Sequences identified as S-GlcNAc-modified were also observed O-GlcNAcylated within the same synaptosome data sets (see supplemental Table S1, supplemental Figs. S21–S24). We attempted to assess the relative amounts of S-and O-linked modifications on the same sequence using the LC/MS data set recorded from the unfractionated synaptosome glycopeptides. Peptide FPFGSSCTGTFHPAPSAPDK was identified both S- and O-GlcNAcylated, residues 1918 and 1919 respectively, under these circumstances, and based on the extracted ion chromatograms and relative peak-intensities the two variants were present approximately in the same relative amounts (supplemental Fig. S25); for the other two sequences only the S-GlcNAcylated peptide was detected. To assess whether such modification occurs in humans, we also reanalyzed glycopeptide data sets obtained from human tissue culture samples, and found that recombinant Host Cell Factor 1 isolated from HEK cells (18) was Cys-GlcNAcylated at position 1139 (supplemental Fig. S26). With these assignments in hand we hypothesized that OGT may be flexible concerning the sugar acceptor, and thus, Cys-GlcNAcylation may be part of its repertoire.
Table I. S-GlcNAcylated mammalian glycopeptides identified in our datasets.
| Protein name | Cys | Peptide | Data set |
|---|---|---|---|
| Bassoon | 1775 | LDFGQGSGSPVC(GlcNAc)LAQVK | Mouse syn |
| Bassoon | 1918 | FPFGSSC(GlcNAc)TGTFHPAPSAPDK | Mouse syn |
| Bassoon | 2107 | MC(GlcNAc)AALNSMDQYGGR | Mouse syn |
| Bassoon | 2489 | QKAPFPAT(GlcNAc)C(GlcNAc)EAPSR | Mouse syn |
| Bassoon | 2489 | QKAPFPATC(GlcNAc)EAPSR | Mouse syn |
| Piccolo | 2870 | AVC(Carbamidomethyl)C(GlcNAc)DMVYKLPFGR | Mouse syn |
| Ankyrin-2 | 2906 | IQTDTC(GlcNAc)HSTVVHSPEVYSVIIR | Mouse syn |
| Serine/threonine-protein kinase SIK3 | 1289 | YPATC(GlcNAc)VTDIMLSHK | Mouse syn |
| Trafficking kinesin-binding protein 1 | 695 | VTPSLNSAPAPAC(GlcNAc)SSTSHLK | Mouse syn |
| MLX-interacting protein | 579 | NAC(GlcNAc)IAPAAFSGQPQK | Mouse syn |
| Dihydropyrimidinase-related protein 2 | 504 | GLYDGPVC(GlcNAc)EVSVTPK | Rat SN |
| Protein Fam222b | 288 | AGISTTSVC(GlcNAc)EGQIANPSPISR | Rat SN |
| Glucocorticoid receptor | 322 | LGPVYC(GlcNAc)QASFSGTNIIGNK | Rat SN |
| Protein Mlxip | 579 | SAC(GlcNAc)IAPAAFTGQPQK | Rat SN |
| YEATS domain-containing protein 2 | 980 | TPGQPQVC(GlcNAc)VSQATMATC(Carbamidomethyl)K | Mouse ESC |
| MLX-interacting protein | 579 | NAC(GlcNAc)IAPAAFSGQPQK | Mouse ESC |
| Protein Fam222b | 288 | AGISTTSVC(GlcNAc)EGQIANPSPISR | Mouse ESC |
| Host Cell Factor 1 | 1139 | RAC(GlcNAc)AAGTPAVIR | HEK cells |
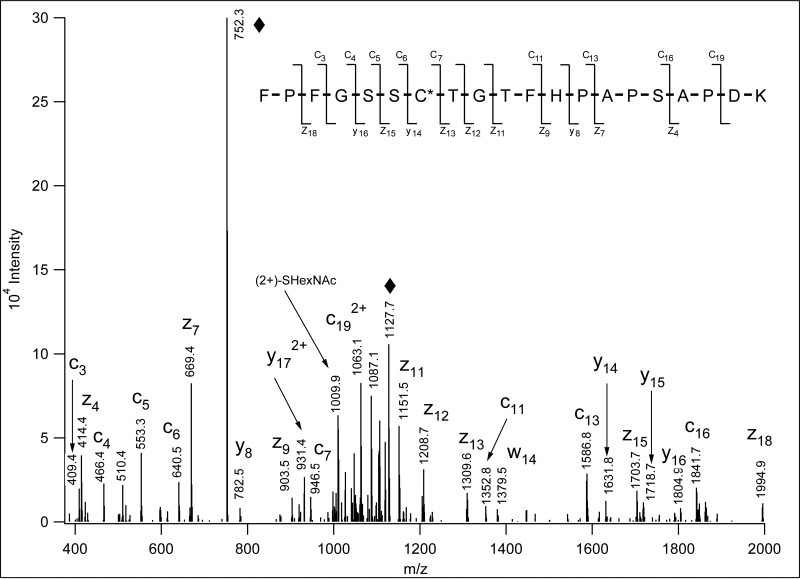
Fig. 1.
ETD spectrum of m/z 752. 0078(3+) identified as Bassoon 1912FPFGSSC(HexNAc)TGTFHPAPSAPDK1931. From these data the identity of the sugar unit cannot be determined, but the modification site assignment is unambiguous. “♦” labels the precursor ion and its charge-reduced form. The insert shows the bond cleavages detected. The modified residue is labeled with an asterisk.
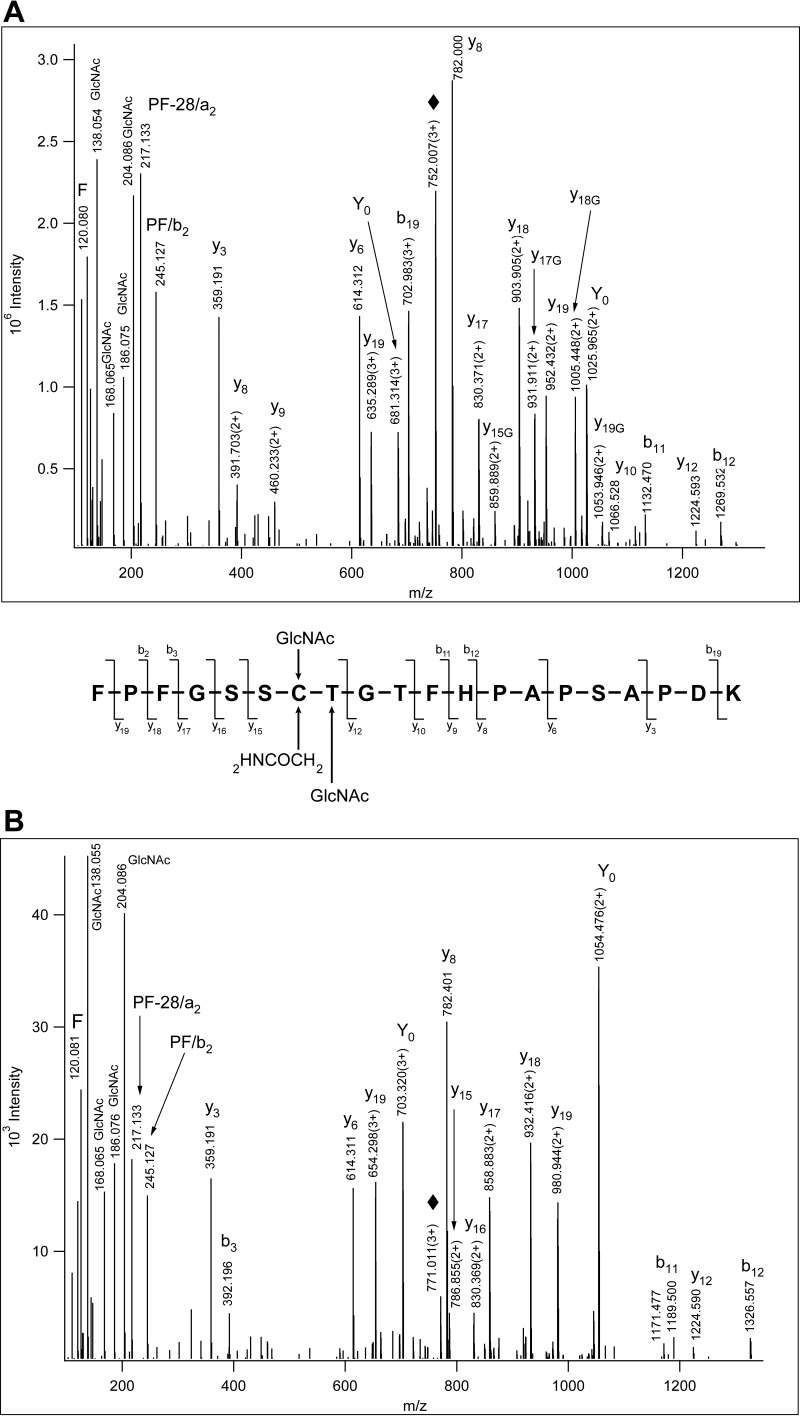
Fig. 2.
Comparison of S- and O-GlcNAcylation of a single peptide. A, HCD spectrum of 1912FPFGSSC(HexNAc)TGTFHPAPSAPDK1931. From the fragmentation pattern of the m/z 204.087 oxonium ion the modifying sugar was identified as GlcNAc (26). “♦” labels the precursor ion. Y0 stands for the peptide after the gas-phase GlcNAc elimination (Nomenclature (11)). Peptide fragments that retained the GlcNAc feature a 'G' next to their position number. Combined fragmentation pattern for A and B is shown. B, HCD spectrum of m/z 771.0135(3+) that was identified from ETD data as Bassoon's 1912FPFGSSC(Carbamidomethyl)T(HexNAc)GTFHPAPSAPDK1931. The glycosylation site cannot be determined, but the fragmentation pattern of the m/z 204.087 oxonium ion identifies the modifying sugar as GlcNAc (26). No glycosylated fragments were detected. “♦” labels the precursor ion.
In vitro Glycosylation and Deglycosylation
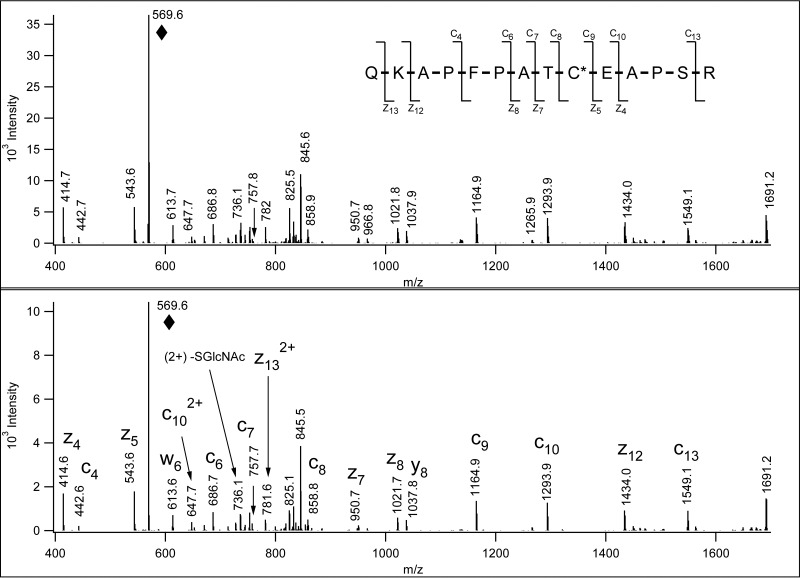
In vitro glycosylation experiments were performed with selected synthetic peptides MCAALNSMDQYGGR, QKAPFPATCEAPSR, LDFGQGSGSPVCLAQVK, AVCCDMVYKLPFGR, and recombinant OGT. Once the reaction mixture was analyzed by LC/MS/HCD and ETD analysis it became obvious that the enzyme indeed modified two of the synthetic peptides, QKAPFPATCEAPSR, and LDFGQGSGSPVCLAQVK, and the GlcNAc was linked to the Cys residue. Fig. 3 shows that the ETD spectrum of the in vitro modified QKAPFPATC(GlcNAc)EAPSR is identical to that acquired from the glycopeptide isolated from the mouse synaptosome. We also performed these experiments with versions of the above peptides with Ala for Ser/Thr substitutions and again found that recombinant OGT was able to modify Cys residues (supplemental Figs. S27 and S28). Supplemental Fig. S1B shows the HCD and ETD data for the other in vitro S-GlcNAcylated sequence. In the control mixture where no enzyme was added Cys-GlcNAc-modification was not detected.
Fig. 3.
Comparison of in vivo and in vitro S-GlcNAcylation. ETD data of the synthetic peptide in vitro S-GlcNAcylated by O-GlcNAc transferaseglycosylated (lower panel) are identical to the ETD data acquired from the S-GlcNAcylated peptide identified in the mouse synaptosome sample (upper panel). “♦” labels the precursor ion.
The addition and removal of GlcNAc from Ser and Thr residues is each performed by a single enzyme, OGT and O-GlcNAcase (OGA), respectively (27, 28). Although our data shows that OGT is capable of modifying Cys residues with GlcNAc, we also examined whether or not OGA could remove this S-linked modification. In vitro OGT reactions were carried out as above; peptides were separated from OGT, and then incubated with or without OGA. Unfortunately, in both reactions there is a mixture of modified and unmodified peptides. Ratios of these peptides clearly show that although OGA was capable of deglycosylating the standard O-GlcNAcylated peptide (supplemental Figs. S29A, S29B, S29E), it was unable to remove the sugar from the S-linked GlcNAc peptide (supplemental Figs. S29C, S29D, S29F). This observation was consistent with earlier findings and the proposed mechanism of GlcNAc hydrolysis by the enzyme (29).
DISCUSSION
We discovered GlcNAcylation of Cys side-chains on a series of intracellular proteins. ETD analysis was essential for the identification of the modified residues and sites described here. The ETD spectra of S-linked GlcNAcylation feature the expected fragmentation caused by side-chain loss, based on a similar side chain loss from alkyl-Cys residues following electron-capture (30). Upon collisional activation S-GlcNAcylated sequences undergo gas-phase deglycosylation similar to O-linked glycopeptides (16, 17, 31, 32). However, when identical O- and S-modified sequence ion series are compared, the latter ones frequently retain the GlcNAc, something rarely observed for their O-linked counterparts (31, 33). Some CID data even contain sufficient information for the unambiguous assignment of S-GlcNAcylation.
S-GlcNAcylation has been observed in some proteins that have previously been reported to be O-GlcNAc modified on Ser and Thr residues (supplemental Table S1) (18, 20, 21). Based on our in vitro experiments the O-GlcNAc transferase that glycosylates the hydroxy amino acids modifies Cys residues as well, and in certain sequence stretches Cys may be preferred. This behavior is not entirely unique. The glycosyltransferase responsible for the S-glycosylation in prokaryote Bacillus subtilis is also able to perform O-glycosylation (34). Our data also suggests, however, that OGA is not able to remove GlcNAc from Cys residues. Further experimentation will be necessary to assess whether Cys-GlcNAc is a dynamic modification removed by an enzyme other than OGA. Based on our very limited data set we believe that the modification may be conserved among species. Identical sequences (AGISTTSVC(GlcNAc)EGQIANPSPISR representing protein Fam222b) and homolog peptide pair (NAC(GlcNAc)IAPAAFSGQPQK and SAC(GlcNAc)IAPAAFSGQPQK from Protein Mlxip) were detected in mouse and rat samples, respectively (see Table I).
The interplay of different post-translational modifications on numerous proteins has been reported (35, 36), and is documented for phosphorylation and GlcNAcylation in some cases (15, 20, 26, 27, 37, 38). Both S- and O-GlcNAcylation were detected on the same peptide sequences in some synaptosome glycoproteins but generally were not found concurrently within the same peptide. Based on these results we may speculate that S-GlcNAcylation may prevent/replace O-GlcNAcylation in the proximity of the modified Cys. However, the biological significance of S-GlcNAcylation and potential interactions with other modifications remains to be investigated.
Supplementary Material
Acknowledgments
We thank Krista Kaasik for technical assistance with the synaptosome preparation, Jeffrey Milbrandt (Washington University, Medical School, St. Louis) for the rat sciatic nerve sample, Sam Myers for the Host Cell Factor 1 data set, Nilanjana Chatterjee for providing recombinant OGT, and Joel Hrit and Barbara Panning for providing the mouse embryonic data set.
Footnotes
Author contributions: J.C.M., A.L.B., and K.F.M. designed research; J.C.M. performed research; J.C.M. and K.F.M. analyzed data; J.C.M. and K.F.M. wrote the paper.
* This work was supported by NIH grant NIGMS 8P41GM103481, the Howard Hughes Medical Institute and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CID
- Collision-induced dissociation
- ETD
- Electron-transfer dissociation
- GlcNAc
- N-acetylglucosamine
- HCD
- higher-energy C-trap dissociation
- HexNAc
- N-acetylhexosamine
- LWAC
- Lectin weak affinity chromatography
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase.
REFERENCES
- 1. Moremen K. W., Tiemeyer M., and Nairn A. V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Furmanek A., and Hofsteenge J. (2000) Protein C-mannosylation: facts and questions. Acta Biochim. Pol. 47, 781–789 [PubMed] [Google Scholar]
- 3. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., and Etzler M. E., editors. Essentials of Glycobiology. 2nd edition Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009 [PubMed] [Google Scholar]
- 4. Pless D. D., and Lennarz W. J. (1977) Enzymatic conversion of proteins to glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 74, 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hart G. W. (1997) Dynamic o-linked glycosylation of nuclear and cytoskeletal proteins. Annu. Rev. Biochem. 66, 315–335 [DOI] [PubMed] [Google Scholar]
- 6. Valliere-Douglass J. F., Eakin C. M., Wallace A., Ketchem R. R., Wang W., Treuheit M. J., and Balland A. (2010) Glutamine-linked and non-consensus asparagine-linked oligosaccharides present in human recombinant antibodies define novel protein glycosylation motifs. J. Biol. Chem. 285, 16012–16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zielinska D. F., Gnad F., Wiśniewski J. R., and Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 8. Zhang H., Huang R. Y., Jalili P. R., Irungu J. W., Nicol G. R., Ray K. B., Rohrs H. W., and Gross M. L. (2010) Improved mass spectrometric characterization of protein glycosylation reveals unusual glycosylation of maize-derived bovine trypsin. Anal. Chem. 82, 10095–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trinidad J. C., Schoepfer R., Burlingame A. L., and Medzihradszky K. F. (2013) N- and O-glycosylation in the murine synaptosome. Mol. Cell. Proteomics 12, 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Halim A., Brinkmalm G., Ruetschi U., Westman-Brinkmalm A., Portelius E., Zetterberg H., Blennow K., Larson G., and Nilsson J. (2011) Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc. Natl. Acad. Sci. U.S.A. 108, 11848–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oman T. J., Boettcher J. M., Wang H., Okalibe X. N., and van der Donk WA. (2011) Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat. Chem. Biol. 7, 78–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stepper J., Shastri S., Loo T. S., Preston J. C., Novak P., Man P., Moore C. H., Havlíček V., Patchett M. L., and Norris G. E. (2011) Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett. 585, 645–650 [DOI] [PubMed] [Google Scholar]
- 13. Venugopal H., Edwards P. J., Schwalbe M., Claridge J. K., Libich D. S., Stepper J., Loo T., Patchett M. L., Norris G. E., and Pascal S. M. (2011) Structural, dynamic, and chemical characterization of a novel Sglycosylated bacteriocin. Biochemistry. 50, 2748–2755 [DOI] [PubMed] [Google Scholar]
- 14. Domon B., and Costello C. E. (1988) A systematic nomenclature for carbohydrate fragmentations in FABMS/MS spectra of glycoconjugates. Glycoconj. J. 5, 397–409 [Google Scholar]
- 15. Medzihradszky K. F. (2005) Characterization of protein N-glycosylation. Methods Enzymol. 405, 116–138 [DOI] [PubMed] [Google Scholar]
- 16. Peter-Katalinic J. (2005) Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 405, 139–171 [DOI] [PubMed] [Google Scholar]
- 17. Darula Z., Chalkley R. J., Lynn A., Baker P. R., and Medzihradszky K. F. (2011) Improved identification of O-linked glycopeptides from ETD data with optimized scoring for different charge states and cleavage specificities. Amino Acids 41, 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chalkley R. J., Thalhammer A., Schoepfer R., and Burlingame A. L. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perdivara I., Petrovich R., Allinquant B., Alliquant B, Deterding L. J., Tomer K. B., and Przybylski M. (2009) Elucidation of O-glycosylation structures of the beta-amyloid precursor protein by liquid chromatography-mass spectrometry using electron transfer dissociation and collision induced dissociation. J. Proteome Res. 8, 631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., and Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Myers S. A., Daou S., Affar el B., and Burlingame A. (2013) Electron transfer dissociation (ETD): The mass spectrometric breakthrough essential for O-GlcNAc protein site assignments-a study of the O-GlcNAcylated protein Host Cell Factor C1. Proteomics 13, 982–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baker P. R., Trinidad J. C., and Chalkley R. J. (2011) Modification site localization scoring integrated into a search engine. Mol. Cell. Proteomics 10(7) doi: 10.1074/mcp.M111.008078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bern M., Kil Y. J., and Becker C. (2012) Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics Chapter 13:Unit13.20. doi: 10.1002/0471250953.bi1320s40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haltiwanger R. S., Blomberg M. A., and Hart G. W. (1992) Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 267, 9005–9013 [PubMed] [Google Scholar]
- 25. Kreppel L. K., Blomberg M. A., and Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 26. Halim A., Westerlind U., Pett C., Schorlemer M., Rüetschi U., Brinkmalm G., Sihlbom C., Lengqvist J., Larson G., and Nilsson J. (2014) Assignment of Saccharide Identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome Res. 13, 6024–6032 [DOI] [PubMed] [Google Scholar]
- 27. Dong D. L., and Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 [PubMed] [Google Scholar]
- 28. Gao Y., Wells L., Comer F. I., Parker G. J., and Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 [DOI] [PubMed] [Google Scholar]
- 29. Rao F. V., Dorfmueller H. C., Villa F., Allwood M., Eggleston I. M., van Aalten DM (2006) Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 5; 25, 1569–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chalkley R. J., Brinkworth C. S., and Burlingame A. L. (2006) Side-chain fragmentation of alkylated cysteine residues in electron capture dissociation mass spectrometry. J. Am. Soc. Mass Spectrom. 17, 1271–1274 [DOI] [PubMed] [Google Scholar]
- 31. Chalkley R. J., and Burlingame A. L. (2001) Identification of GlcNacylation sites of peptides and alpha-crystallin using Q-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 12, 1106–1113 [DOI] [PubMed] [Google Scholar]
- 32. Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., and Burlingame A. L. (2006) O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 33. Chalkley R. J., and Burlingame A. L. (2003) Identification of novel sites of O-N-acetylglucosamine modification of serum response factor using quadrupole time-of-flight mass spectrometry. Mol. Cell. Proteomics 2, 182–190 [DOI] [PubMed] [Google Scholar]
- 34. Wang H., Oman T. J., Zhang R., Garcia De Gonzalo C. V., Zhang Q., van der Donk W. A. (2014) The glycosyltransferase involved in thurandacin biosynthesis catalyzes both O- and S-glycosylation. J. Am. Chem. Soc. 136, 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Venne A. S., Kollipara L., and Zahedi R. P. (2014) The next level of complexity: crosstalk of posttranslational modifications. Proteomics. 14, 513–524 [DOI] [PubMed] [Google Scholar]
- 36. Hunter T. (2007) The Age of Crosstalk: Phosphorylation, ubiquitination, and beyond. Mol.. Cell. 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 37. Kaasik K., Kivimäe S., Allen J. J., Chalkley R. J., Huang Y., Baer K., Kissel H., Burlingame A. L., Shokat K. M., Ptáček L. J., and Fu Y. H. (2013) Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hart G. W. (2014) Three Decades of Research on O-GlcNAcylation - A Major Nutrient Sensor That Regulates Signaling, Transcription and Cellular Metabolism. Front. Endocrinol. 5, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.