Abstract
Sarpogrelate is an antiplatelet agent widely used to treat arterial occlusive diseases. Evaluation of platelet aggregation is essential to monitor therapeutic effects of sarpogrelate. Currently, no molecular signatures are available to evaluate platelet aggregation. Here, we performed comprehensive proteome profiling of platelets collected from 18 subjects before and after sarpogrelate administration using LC-MS/MS analysis coupled with extensive fractionation. Of 5423 proteins detected, we identified 499 proteins affected by sarpogrelate and found that they strongly represented cellular processes related to platelet activation and aggregation, including cell activation, coagulation, and vesicle-mediated transports. Based on the network model of the proteins involved in these processes, we selected three proteins (cut-like homeobox 1; coagulation factor XIII, B polypeptide; and peptidylprolyl isomerase D) that reflect the platelet aggregation-related processes after confirming their alterations by sarpogrelate in independent samples using Western blotting. Our proteomic approach provided a protein profile predictive of therapeutic effects of sarpogrelate.
Sarpogrelate, an antagonist of 5-hydroxytryptamine (5-HT1, also called serotonin) receptors (subtype 2A and 2B), has been widely used in the treatment of arterial occlusive diseases (1, 2). It is metabolized to racemic M-1, and both enantiomers of the M-1 also act as antagonists of the 5-HT receptors. Sarpogrelate inhibits the responses to 5-HT mediated by the 5-HT receptors, such as platelet aggregation, vasoconstriction, and vascular smooth muscle proliferation (3). Moreover, several previous studies have shown that sarpogrelate can prevent thrombosis in atherosclerotic diseases, although the detailed mechanism is unknown (4, 5). Also, sarpogrelate has been used to treat Asian patients with peripheral arterial diseases (6). A sarpogrelate-aspirin comparative clinical study in 1510 Japanese patients with cerebral infarction showed that sarpogrelate was not inferior to aspirin in prevention of recurrence of cerebral infarction while it reduced significantly bleeding events, compared with aspirin (7).
To monitor therapeutic effects of sarpogrelate, it is essential to assess platelet aggregation induced by 5-HT. Platelet aggregation is typically evaluated with platelet aggregometry, an optical densitometry method, which is rather insensitive to small platelet aggregates (8). Unlike potent agonists of thrombin and collagen, 5-HT by itself induces only the formation of small aggregates. Thus, the effect of sarpogrelate on platelet aggregation induced by 5-HT has been difficult to be evaluated using the conventional platelet aggregometry (9). Thus, there has been a significant need for an effective assay method that can be used to evaluate the effect of sarpogrelate.
A number of studies have shown that protein profiles can serve as effective measures for evaluation of drug responses (10–12). In these studies, the search for a protein profile indicative of the response of a drug was facilitated by the use of proteomic technologies. Platelets in blood play important roles in hemostatic or thrombotic mechanisms, inflammatory process, and cardiovascular pathogenesis. Platelets undergo considerable alterations by sarpogrelate. Moreover, because platelets are anucleated cells, protein profiles can provide more relevant information regarding the responses to sarpogrelate, compared with transcriptomic or genomic profiles. Several proteomic studies provided global proteome profiles of platelets under various conditions (13–16). For example, Burkhartet al. (13) and Qureshi et al. (16) identified 4191 and 1507 proteins, respectively, from human platelets in resting states. Also, several studies provided proteomes associated with responses of antiplatelet drugs. For example, Azcona et al. (17) compared platelet proteomes of type 2 diabetic patients with stable coronary ischemia after treatment of aspirin and clopidogrel + aspirin. As a proteome profile indicative of clopidogrel + aspirin, they proposed downregulated actin-binding protein isotypes 2 and 5 associated with cytoskeleton organization in patients treated with clopidogrel + aspirin, compared with aspirin treated patients. Lopez-Farre et al. (18) and Mateos-Caceres et al. (19) compared the proteomes of platelets from aspirin resistant and sensitive coronary ischemic patients and proposed, as proteome profiles indicative of aspirin resistance, up- and down-regulated proteins associated with platelet functions (cytoskeleton, energetic metabolism, oxidative stress, and cell survival) in aspirin resistant patients. However, to our knowledge, no proteomic analyses have been performed to identify a platelet proteome profile indicative of the response to sarpogrelate.
Here, we performed a comprehensive proteome profiling of platelets collected from 18 healthy subjects before and after administration of sarpogrelate. For each subject, platelets were isolated before and then again after drug administration to investigate the alteration in the platelet proteome by sarpogrelate. A sample preparation method involving filter-aided sample preparation (FASP) (20) was used to improve extraction of proteins including both soluble and membranous proteins. Moreover, an ultrahigh pressure liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) coupled with extensive fractionation was used to generate a comprehensive proteome of platelets. The platelet proteome obtained from LC-MS/MS analyses included 47,045 peptides for 5423 proteins. Of them, we identified hundreds of differentially expressed proteins (DEPs) in platelets by sarpogrelate, which are involved in cellular responses related to platelet activation and aggregation induced by 5-HT. Finally, of these proteins, we selected three proteins indicative of the response to sarpogrelate in platelets.
EXPERIMENTAL PROCEDURES
Sample Preparation
We collected two independent sets of platelet samples from healthy male Korean volunteers. The first set, including platelet samples from 18 subjects before and after administration of sarpogrelate, was used for the proteome profiling (supplemental Table S1A). The second set, including independent platelet samples from 5 subjects, which were not included in the first set, before and after sarpogrelate administration, was used to confirm the validity of the selected proteins by Western blotting (supplemental Table S1B). The eligibility was determined after a complete medical and laboratory examination. The exclusion criteria used were as follows: a history of cardiovascular, hepatic, renal, endocrine, gastrointestinal, hemorrhagic, or bleeding disorders; clinically significant findings on routine clinical laboratory testing or on physical examination; a history of hypersensitivity to sarpogrelate or other antiplatelet agents; and any medication history within 1 week prior to the enrollment in this study. Characteristics of the subjects in the two sets, such as age, body weight, and platelet counts, were summarized in supplemental Table S1. The subjects were admitted to the Clinical Trial Center at Samsung Medical Center on the day before the drug administration. All the subjects provided written informed consents, and the study protocol (SMCCPT-DP-1202) was approved by the institutional review board (IRB No. 2011–11-006). The food was standardized and strictly controlled during the whole study period. All volunteers were given 100 mg of sarpogrelate hydrochloride (Anplug 100 mg, Yuhan Corporation, Seoul, Korea) orally after an overnight fast. Blood samples were obtained before and 6 h after the drug administration.
Experimental Design and Statistical Rationale
For the proteomic analysis, we obtained a total of 18 pairs of platelet samples before and after the administration of sarpogrelate (supplemental Table S1A). The samples from 15 of 18 subjects were pooled and then used to generate a master accurate mass and time tag (AMT) database (DB). Next, the pairs of platelet samples for 14 of 18 subjects before (controls) and after the administration of sarpogrelate were used for LC-MS/MS profiling (supplemental Table S1A). According to power analysis (21) for one sample t test, which was used to identify differentially expressed proteins (DEPs; see below for justification), the sample size required to reliably detect an effect size of 1.3-fold with false negative rate (β) = 0.05 was 9. In the power analysis, the control distribution was defined as a normal distribution with× μ = 7.32 × 10−2 and σ = 0.76 estimated from the mean log2-fold changes of proteins in our data sets. Considering that our sample size (14 paired platelet samples) was larger than 9, our sample size is sufficient to detect the DEPs with 2-fold (≥ effect size of 1.3-fold used in power analysis), which was used as the cutoff to identify the DEPs in this study. For each of the 14 biological replicates, we performed three LC-MS/MS experiments as technical replicates, resulting in 42 data sets for the samples before (controls) and after sarpogrelate administration, respectively.
Platelet Preparation
Eight milliliters of whole blood from each subject was collected using Acid-Citrate-Dextrose (ACD) as anticoagulant and then transferred into new polypropylene tubes. Platelet-rich plasma (PRP) was prepared by centrifugation at 200 × g for 20 min at room temperature. Two thirds of the PRP were transferred into a new tube, washed, and isolated by centrifugation at 800 × g for 10 min. Isolated platelets were resuspended in Tyrode's Buffer containing 134 mm NaCl, 12 mm NaHCO3, 2.9 mm KCl, 0.34 mm Na2HPO4, 1 mm MgCl2, and 10 mm Hepes (pH 7.4). The platelet count was measured with a Coulter LH780 system (Beckman Coulter, Inc., Brea, CA). Because the protein contents of leukocytes are much higher than those of platelets, the contamination of platelets with leukocytes can affect significantly the quality of the measured platelet proteome. Thus, we checked the purity of our platelet samples and leukocyte contamination during our platelet preparation by flow cytometric analysis with CD45 and CD61 labeling, as previously described (22, 23), using the FACSCanto II Flow Cytometer (Becton Dickinson, San Jose, CA) and the Kaluza software (Beckman Coulter, Brea, CA). The flow cytometric assessment showed that our platelet samples contained 99.5–99.73% CD61 positive cells (platelets), but only 0.02–0.06% CD45 positive cells (leukocytes) (supplemental Fig. S1), which corresponded to 2–6 leukocytes per 1 × 104 platelets and also to 1–3% protein contamination based on the estimate previously described (24, 25). In this study, we used multiple centrifugation for platelet isolation and flow cytometric analysis to measure leukocyte contamination. In the four previous studies using these methods (16, 26–28), leukocyte contamination was in the range between 1 per 1 × 105 and 1 × 103 platelets. Thus, the leukocyte contamination in this study (2–6 per 1 × 104 platelets) was within this range, implying that the purity of platelet samples and leukocyte contamination were comparable to those in previous studies.
Protein Extraction and Digestion
Platelet samples were dissolved in 200 μl of lysis buffer (4% SDS in 0.1 m Tris-HCl, pH 7.6) by 1 min of vortexing. Samples were then lysed by sonication using hand-type sonicator (Q55 Sonicator, QSONICA, CT) for 10 s at 30 W on ice. This lysis step was repeated five times. Protein concentration of each platelet samples were determined by BCA assay (Pierce, Rockford, IL). All platelet proteins were digested using slightly modified Filter-Aided Sample Preparation (FASP) method as previously described (20). Briefly, the protein sample was reduced with the SDT buffer (4% SDS and 0.1 m DTT in 0.1 m Tris-HCl, pH 7.6) for 45 min at 37 °C and boiled for 10 min at 95 °C on a thermomixer (Thermomixer comfort, Eppendorf, Hamburg, Germany). Subsequently, it was sonicated for 10 min in bath sonicator (Power sonic 505, Hwashin Technology, Seoul, Korea) followed by centrifugation at 16,000 × g and 20 °C for 5 min. The supernatant was transferred to a membrane filter (YM-30, Millipore Corporation, Bedford, MA) and added 200 μl of 8 m urea in 0.1 m Tris-HCl (pH 8.5) to membrane filter. The protein sample on the membrane filter was centrifuged at 14,000 × g and 20 °C for 60 min to remove SDS. This wash step was repeated three times. Subsequently, reduced protein sample was alkylated on membrane filter with 100 μl of 50 mm iodoacetamide in 8 m urea for 25 min at room temperature in dark and followed by centrifugation at 14,000 × g for 30 min. The filter was washed with 200 μl of 8 m urea four times and again washed with 100 μl of 50 mm ammonium bicarbonate (pH 8.0) twice to buffer exchange. And then trypsin was added to the filter at an enzyme to protein ratio of 1:50 (w/w) and protein sample was digested at 37 °C for overnight. After the first digestion, the sample was digested with trypsin (1:100 w/w ratio) at 37 °C for 6 h again. Finally, the tryptic peptides were eluted by centrifugation at 14,000 × g and at 20 °C for 30 min and the filter was rinsed with 60 μl of 50 mm ammonium bicarbonate by centrifugation at 14,000 × g and at 20 °C for 20 min. The resulting peptide sample was dried by vacuum centrifugation and kept in −80 °C until the subsequent LC-MS/MS analysis.
Basic pH Reversed Phase Liquid Chromatography Fractionation (BRP fractionation)
Fifteen pairs of peptide samples (i.e. 15 before and 15 after sarpogrelate administration; supplemental Table S1A) were pooled and immediately fractionated into twelve fractions by high pH reverse-phase liquid chromatography fractionation method as previously described (29). The fractionation was performed using an Agilent 1260 Infinity HPLC system (Palo Alto, CA) equipped with an analytical column (Xbridge C18, 4.6 mm × 250 mm, 130 Å, 5 μm) and a guard column (Xbridge C18, 4.6 mm × 20 mm, 130 Å, 5 μm). For the peptide separation, a 115 min gradient was used at a flow rate of 500 μl/min: at 0% solvent B for 10 min, from 0% to 5% solvent B over 10 min, from 5 to 35% solvent B over 60 min, from 35 to 70% solvent B over 15 min, 70% solvent B for 10 min, from 70 to 0% solvent B over 10 min. Solvents A and B were 10 mm ammonium formate in water (pH 10) and 10 mm ammonium formate in 90% acetonitrile (ACN, pH 10), respectively. A total of 96 fractions were collected continuously into 96-well plate by a fraction collector (G1364C, Agilent, Palo Alto, CA) in every 1 min from 10 to 105 min. The 96 fractions were divided into four sections of early (from #1 to #24), first mid (from #25 to #48), second mid (from #49 to #72) and late (from #73 to #96) sections according to their elution time. The collected fractions were noncontiguously concatenated into 12 fractions by pooling two fractions from each section (i.e. #1-#13-#25-#37-#49-#61-#73-#85, #2-#14-#26-#38-#50-#62-#74-#86, … #12-#24-#36-#48-#60-#72-#84-#96). The concatenated fractions were dried by vacuum centrifugation and stored at −80 °C until further LC-MS/MS analysis.
LC-MS/MS Analysis
The 14 pairs of unfractionated peptide samples (supplemental Table S1A) and 12 mRP fractions were analyzed by a 7-T Fourier transform ion cyclotron resonance mass spectrometer (LTQ-FT, Thermo Electron, San Jose, CA) coupled with a modified nanoACQUITY UPLC system (Waters, Milford, MA). LC-MS/MS experiments were performed triplicate on each of 28 unfractionated peptide samples (i.e. a total of 84 LC-MS/MS experiments). The nanoACQUITY system was modified to dual-online ultra-high pressure liquid chromatography (DO-UHPLC) system for the high throughput analysis (30). Two capillary analytical columns (75 μm × 100 cm) and two solid phase extraction column (SPE) columns (150 μm × 3 cm) were manufactured in house with C18 bonded particles (Jupiter, 300 Å, 3 μm, Phenomenex, Torrence, CA) as previously described (31). Solvent A and B were 0.1% formic acid in water and 0.1% formic acid in ACN, respectively. A 240 min gradient was used at a flow rate of 300 nL/min: from 1 to 40% solvent B over 220 min, from 40 to 80% solvent B over 5 min, 80% solvent B for 10 min, and 1% solvent B for 5 min. The temperature of column was maintained at 60 °C (32). The electrospray voltage was 3.0 kV and the mass spectrometer was operated by data-dependent MS/MS mode. MS precursor scans (m/z range of 500–2,000 Th) were acquired at an automated gain control (AGC) target value of 1.0 × 106 and a resolution of 1.0 × 105 and maximum ion injection time (IT) of 1,000 ms. Up to 7 most abundant ions in a precursor scan were selected with isolation window of 3 Th with the exclusion duration of 30 s and fragmented using collision induced dissociation (CID) at normalized collision energy (NCE) of 35. The intensity threshold of initiating fragmentation was set to 500. Tandem mass spectra were acquired at an AGC target value of 1.0 × 104 and maximum IT of 100 ms.
Database Search for Peptide Identification
The MS/MS data from a total of 96 LC-MS/MS experiments (i.e. 86 for unfractionated samples and 12 for fractionated samples) were processed using iPE-MMR (33) for the refinement of precursor masses. The resultant MS/MS data were searched against uniprot human reference database (released September 2012; 69,391 entries) and the common contaminants (180 entries) using MS-GF+ search engine (V9012) (34). The mass tolerance for the precursor ion was set at 15 ppm and the number of tryptic termini was 1. Carbamidomethylation of cysteine was set to static modification. Oxidation of methionine and the carbamylation of N termini were set to variable modifications. Peptide-spectrum matches at the peptide-level false discovery rate (FDR) of 0.01 were used for further analyses. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD003613 and 10.6019/PXD003613 (35).
Label-free Peptide Quantification
An MS intensity based label-free quantitative analysis was performed to platelet LC-MS/MS data as previously described (12). Briefly, in an LC-MS/MS experiment, all MS features of a peptide in different charge states that have similar monoisotopic masses (i.e. within the mass tolerance of 10 ppm) but different LC elution times over a period of elution times were grouped into a unique mass class (UMC) (36). UMC mass was calculated as intensity weighted average of monoisotopic masses of all the components of each UMC. Sum intensity of a UMC was calculated by summing the spectral intensities of all MS components of a UMC. During the PE-MMR analysis, the precursor mass of the MS/MS data (i.e. mgf files) was matched to the UMC masses and replaced by the UMC masses. Then, MS/MS information was linked to the matched UMC. When the linked spectra resulted in peptide identification within FDR of 1% after database search and target-decoy analysis, the corresponding peptide ID was linked to the UMC and the sum intensity of the UMC was assigned to the peptide ID. In order to minimize the LC elution time variance of the same peptide across the different LC-MS/MS runs, normalized elution time (NET) utility was used to calculate the experimental NETs of the identified UMCs using UMC elution time (37). Briefly, we plotted calibration graphs between the UMC elution time of the peptide IDs (experimental value) from a LC-MS/MS experiment against the theoretical NET values of the peptide IDs from the NET utility program (i.e. using the peptide sequence). Then we used the resultant calibration function to calculate the experimental NET for all UMCs. A peptide ID of a UMC mass and an experimental NET was classified as an AMT. All AMTs from 96 LC-MS/MS experiments were used to construct an AMT DB (Oracle Database 10g Enterprise Edition, Release 10.2.0.1.0). It is an assembly of AMTs consisting of unique peptide sequences with the UMC mass and the experimental NET. For the multiply measured peptide, the average mass and the average NET were chosen for each AMT. After constructing the master AMT DB, unidentified UMCs (i.e. UMCs with no peptide IDs) were matched to the AMTs of the master AMT DB using mass and elution time with ±10 ppm of mass tolerance and ± 0.01 of NET tolerance. The resultant assigned UMCs from the 84 LC-MS/MS data sets (i.e. triplicate LC-MS/MS data sets of 14 pairs of peptide samples) were combined into an alignment table in which each row contained sum intensities of UMC for each of the identified peptide.
Identification of Differentially Expressed Proteins (DEPs)
The intensities of the aligned peptides were normalized using quantile-normalization methods (38). The information of the aligned peptides were rolled up to infer protein groups using bipartite graph analysis. For each protein group, the representative proteins (or protein isoforms) were selected as the ones with the largest number of peptides or unique peptides as previously described (39). When more than two proteins in a protein group had the same number of peptides, the protein with the higher sequence coverage was selected as the representative protein. The representative proteins of more than two peptide identifications were used for further analysis. For each subject, relative abundances of the representative proteins in individual protein groups were then computed from peptide intensities using the linear-programming formulation as previously described (40). To identify the DEPs, we first selected the sets of proteins that have more than two nonredundant peptides and were detected in more than 50% of all subjects to ensure their statistical power. For the selected proteins, we then performed one sample t test using log2-fold changes in all subjects to compute the significance of t values. The mean log2-fold changes of proteins were found to follow a normal distribution with μ = 7.32 × 10−2 and σ = 0.76 (p = 1.83 × 10−30, Two-sample Kolmogorov-Smirnov test), thus supporting the use of one sample t test. An empirical distribution of the null hypothesis that a protein is not differentially expressed was estimated by performing 100,000 random permutations of the samples and then by applying the Gaussian kernel density estimation method to t values resulting from the random permutations. FDRs of each protein for one sample t test were then calculated using Storey method (41). The DEPs were identified as the ones with FDR ≤ 0.1 and absolute log2-fold change ≥ 0.58 (1.5-fold). The use of the empirical distribution to compute FDR can reduce the problems caused by potential deviations from the underlying assumptions of one sample t test. To determine the FDR cutoff, we first identified three sets of DEPs using three different cutoff values of 0.01, 0.05, and 0.1, examined manually differential expression (up- or down-regulation) of the DEPs identified using each of the cutoff values, and then found that up- and down-regulation of the DEPs could be still clearly observed even using the FDR cutoff of 0.1. Thus, we used the FDR cutoff of 0.1 to include false negative DEPs without including a significant number of false positive DEPs for the following analyses (enrichment analysis of GOBPs and network analysis).
Enrichment Analysis
Enrichment analysis for a list of proteins (e.g., detected proteins or DEPs) was performed using DAVID software to identify GO biological processes (GOBPs) and GO cellular components (GOCCs) represented by the DEPs (42). The GOBPs and GOCCs enriched by a list of proteins were identified as the ones with the enrichment p value < 0.05 calculated by DAVID.
Network Analysis
To reconstruct a network model for the DEPs, we first selected a subset of the DEPs involved in the GOBPs related to platelet activation and aggregation (cell activation, vesicle-mediated transport, Golgi vesicle transport, protein transport, and coagulation). We then collected the protein-protein interactions among the selected DEPs from five protein-protein interactome databases: BIND (Biomolecular Interaction Network Database) (43), HPRD (Human Protein Reference Database) (44), BioGRID (Biological General Repository for Interaction Data sets) (45), MINT (Molecular INTeraction Database) (46), and STRING (search tool for recurring instances of neighboring genes) (47). Using Cytoscape (48), the network model was reconstructed for the selected DEPs based on their protein-protein interactions. The nodes in the network model were arranged based on the locations and relationships of the corresponding proteins in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (49). The nodes with no information in the KEGG pathway database were located based on their functions, locations, and/or relationships reported in the previous literature. Of the 99 proteins used for network modeling, 14 proteins with no information of functions, locations, or relationships were removed from the network model.
Partial Least Squares-Discriminant Analysis (PLS-DA)
We applied PLS-DA to evaluate whether the abundances of the five selected proteins can separate platelet samples obtained before sarpogrelate administration from those obtained after the administration (50). We generated an X-block data matrix (14 × 5) containing relative abundances of the 5 selected proteins and also a Y-block data matrix (14 × 1) containing group information (zeros and ones for before and after the administration). The data matrices were then auto-scaled for each column to have zero mean and unit standard deviation. PLS-DA was applied to the auto-scaled X- and Y-blocks. After the PLS-DA application, the three PLS latent variables (LV1–3) were selected after 10-fold cross-validations as previously described (51).
Western Blot
Briefly, 1 μg of the total protein from each sample was resolved on 4–12% NuPAGE gradient SDS-PAGE and then electrotransferred to a nitrocellulose membrane (Invitrogen, San Diego, CA). The blots were incubated overnight at 4 °C with primary antibodies against GDI1 (1:600 dilution; Abcam, Cambridge, UK), PPID (1:500 dilution, Abcam), CD36 (1:500 dilution; R&D Systems, Minneapolis, MN), F13B (1:500 dilution; ProSci, Atlanta, GA), HOOK2 (1:500 dilution; Abcam), and CUX1 (1:500 dilution; Abcam) after blocking with 5% skim milk in PBS. The blots were treated with a horseradish peroxidase-conjugated (HRP) secondary polyclonal antibody (1:5,000 dilution; Abcam), detected using an ECL Western blotting detection system (Amersham Bioscience, GE Healthcare Life Science), and then monitored with a LAS-4000 (Fuji Film, Tokyo, Japan). We measured β-actin levels as a loading control. The blots were then quantified with Fujifilm Multi Gauge version 3.0. The list of primary antibodies for the selected proteins is summarized in supplemental Table S2.
RESULTS
Comprehensive Profiling of Platelet Proteomes
We first carefully selected healthy male subjects (n = 18) with no history of vascular diseases or hypersensitivity to antiplatelet agents, as well as no abnormal findings from clinical laboratory testing (see Experimental Procedures). The characteristics of the subjects, such as ages, body weights, and platelet counts, are summarized in supplemental Table S1. From each subject, we isolated platelets before and six hours after the administration of sarpogrelate (100 mg) to investigate platelet proteomes affected by sarpogrelate. To improve the coverage of proteomes to include membranous proteins, we used the FASP (20) method for protein digestion after obtaining the lysates of platelets (see Experimental Procedures).
To extensively profile platelet proteomes, we generated a master accurate mass and time tag (AMT) database (DB). To this end, we first pooled the peptide samples from 15 of the 18 pairs of platelet samples (supplemental Table S1A) obtained before and after the administration of sarpogrelate, respectively, performed high pH reverse phase fractionation of the pooled peptides into 12 fractions, and then analyzed these fractions using ultra-high pressure LC-MS/MS (Fig. 1A, left column). This resulted in 12 LC-MS/MS data sets (supplemental Fig. S2). In addition, we performed triplicate experiments on 14 of 18 pairs of the platelet samples (supplemental Table S1A), resulting in 84 (14 × 2 × 3) LC-MS/MS data sets (Fig. 1A, right column; supplemental Fig. S3). A total of 96 (12 + 84) LC-MS/MS data sets from the fractionation and triplicate experiments were used to construct a master AMT DB of platelet proteome.
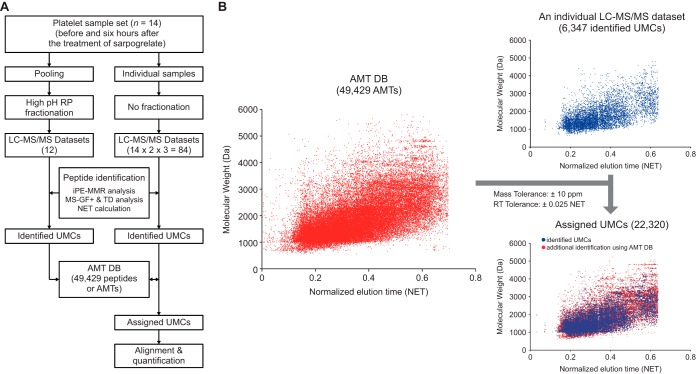
Fig. 1.
The master AMT DB for the platelet proteome. A, The overall scheme of AMT DB construction. A total of 96 data sets were generated from LC-MS/MS analyses. For individual data sets, UMCs were assigned with peptide IDs (identified UMCs) that were identified by iPE-MMR analysis followed by MS-GF+ search in the target-decoy (TD) setting. The identified UMCs were compiled into an AMT DB. B, Utilization of the AMT DB to assign peptide IDs to unidentified UMCs. The 49,429 AMTs (magenta dots) in the AMT DB are visualized in a 2D (NET and molecular weight) scatter plot (left). For a LC-MS/MS data set, the 6347 identified UMCs (blue dots) are shown in the upper right scatter plot. By matching unidentified UMCs in this data set with AMTs in the DB using the indicated mass and NET tolerances, a subset of unidentified UMCs were assigned with peptide IDs of the AMTs matched with them. These matched UMCs (red dots) are shown in the bottom right scatter plot.
We first generated unique mass classes (UMCs) of peptide MS features from individual data sets using iPE-MMR analysis (33) and then assigned both peptide IDs and normalized elusion times (NETs) to UMCs (identified UMCs) after target-decoy MS-GF+ search (FDR < 0.01) and NET calculation. All identified UMCs from the 96 data sets were compiled into the master AMT DB (see Experimental Procedures; supplemental Table S3), which comprised 49,429 peptides (or AMTs; dots in Fig. 1B, left). For each of the 84 LC-MS/MS data sets, we then used the master AMT DB to assign peptide IDs to unidentified UMCs by matching them with AMTs using the mass and NET tolerances (10 ppm and 0.025 NET, respectively; Fig. 1B, right). Of the 49,429 peptides in the master AMT DB, we selected 47,045 peptides that cover 5423 platelet proteins with two or more sibling peptides and then used them for protein quantitation in 84 LC-MS/MS data sets (supplemental Tables S4 and S5).
Characteristics of Platelet Proteome Profiles
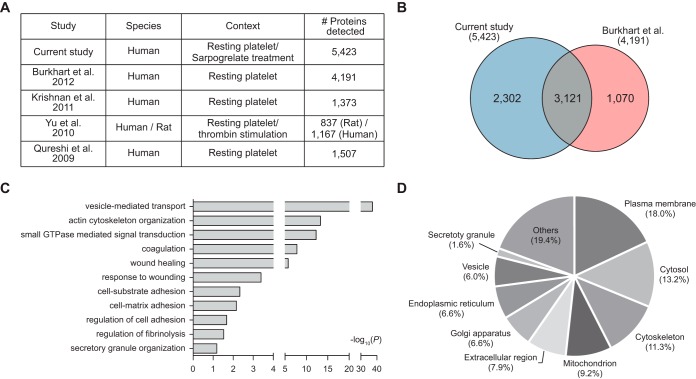
To assess the comprehensiveness of our platelet proteome, we first compared our proteome (5,423 proteins) with the platelet proteomes previously reported (Fig. 2A). Four previous studies profiled the proteomes of resting platelets and then identified 4191 (13), 1373 (14), 837 (15), and 1507 (16) platelet proteins, respectively. Of these studies, Burkhart et al. (13) provided the largest platelet proteome including 4191 proteins by profiling the proteomes of resting platelets collected from four individuals using LC-MS/MS analysis after isobaric tag for relative and absolute quantitation (iTRAQ) labeling. The comparison of our proteome with the largest previous platelet proteome showed that of the 4191 proteins previously reported, 3121 (74.5% of 4191) were also detected in our proteome (Fig. 2B). Additionally, our proteome provided 2302 (42.5% of 5423) proteins not detected in the previous platelet proteome. These data indicate that our proteome can serve as one of comprehensive proteomes of platelets.
Fig. 2.
Comprehensive profiles of platelet proteomes. A, Previous platelet proteomes measured from resting platelet cells or after thrombin stimulation. The numbers of detected proteins in individual studies are shown. B, Venn diagram showing the relationships of our platelet proteome with the largest previous platelet proteome. C, Cellular processes (GOBPs) in which the identified platelet proteins are mainly involved. The bars represent -log10(P) where P represents the significance of each GOBP being enriched by the platelet proteins. The p value was computed by the DAVID software. D, Relative proportions of cellular components (GOCCs) in which the measured platelet proteins are mainly localized.
We then examined cellular processes represented by the 5423 platelet proteins by performing enrichment analysis of GOBPs using DAVID software (42). The cellular processes significantly represented by the platelet proteome included the processes related to major functions of platelets: adhesion to the interrupted endothelium (cell-substrate and cell-matrix adhesions and regulation of cell adhesion); activation involving shape changes and granule secretion (actin cytoskeleton organization, small GTPase mediated signal transduction, and vesicle-mediated transport); and aggregation (coagulation, wound healing, and regulation of fibrinolysis) (Fig. 2C; supplemental Table S6A). Moreover, the enrichment analysis of GOCCs revealed that a majority of the platelet proteins were localized in cellular components related to secretory granule that has been shown to be associated with platelet activation and aggregation (endoplasmic reticulum, Golgi apparatus, vesicle, secretory granule, and extracellular region) (28.8%), as well as plasma membrane (18.0%); cytosol (13.2%); cytoskeleton (11.3%); and mitochondrion (9.2%) (Fig. 2D; supplemental Table S6B). Specifically, our proteome included platelet granular proteins including 44 platelet α-granule and 5 platelet δ-granule proteins (secretory granule with 1.6% in Fig. 2D). These data indicate that our platelet proteome can provide a broad spectrum of information related to cellular processes occurring during platelet activation and aggregation in diverse cellular compartments.
Identification of Platelet Proteins Affected by Sarpogrelate
To identify platelet proteins whose abundances were affected by sarpogrelate, we first aligned each of 47,045 peptides for the 5423 proteins in the 84 LC-MS/MS data sets (42 data sets for platelets collected before administration of sarpogrelate and the other half for those after sarpogrelate administration) and then determined the ratios of abundances of the 5423 platelet proteins measured before and after the administration of sarpogrelate in individual samples using the linear programming method previously reported (see Experimental Procedures) (40). By applying t test to the protein ratios in 14 samples, we then identified 499 differentially expressed proteins (DEPs; FDR ≤ 0.1 from t test) before and after the administration of sarpogrelate (see Experimental Procedures; supplemental Table S7). Of the 499 DEPs, 348 proteins were up-regulated by the administration of sarpogrelate, whereas 151 were downregulated.
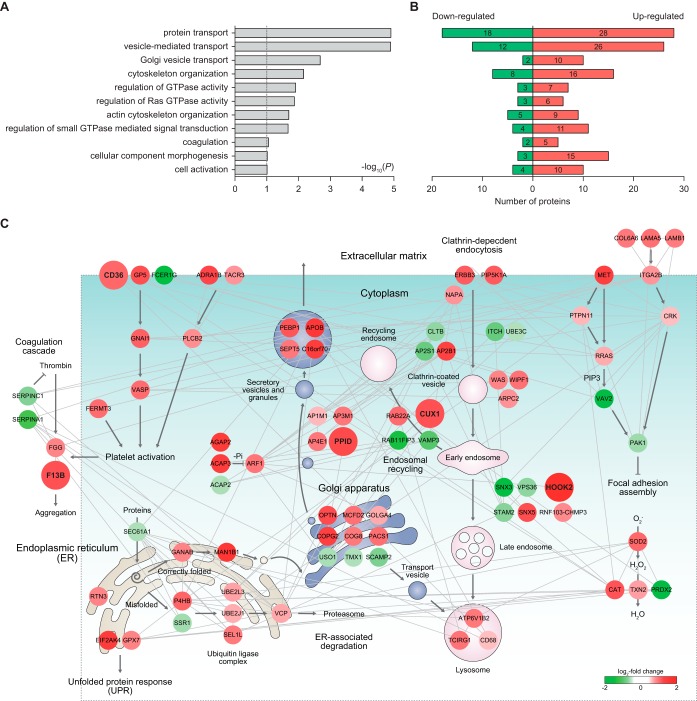
To understand the association of the DEPs with functions of sarpogrelate in platelets, we identified cellular processes represented by the 499 DEPs in platelets using DAVID software. This analysis revealed that the DEPs were mainly involved in the processes related to platelet activation and aggregation (cell activation, cell component morphogenesis, vesicle-mediated transport/Golgi vesicle transport/protein transport, actin cytoskeleton organization, coagulation, and regulation of small GTPase mediated signal transduction) (Fig. 3A;supplemental Table S8), consistent with the effect of sarpogrelate on platelet activation and aggregation involving shape changes and granule secretion (5, 52, 53). Interestingly, these processes were represented by both the up- and down-regulated proteins (Fig. 3B), suggesting that the up- and downregulated proteins by sarpogrelate collectively act to define the function of sarpogrelate in the processes related to platelet activation and aggregation, such as coagulation and granule-mediated transports.
Fig. 3.
Network models delineating cellular processes affected by sarpogrelate. A, Cellular processes (GOBPs) represented by the differentially expressed proteins (DEPs) between platelets before and after administration of sarpogrelate. The bars (A) represent -log10(P) where P represents the significance of each GOBP being enriched by the DEPs. B, Numbers of the DEPs involved in the GOBPs. The numbers of up- and down-regulated proteins involved in each GOBP were separately shown. C, A network model describing the interactions of the DEPs involved in the GOBPs related to platelet activation and aggregation. Node colors represent the increase (red) and the decrease (green) in the abundance after administration of sarpogrelate. The color bar denotes the gradient of log2-fold changes. Edges represent protein-protein interactions (gray) collected from five interactome databases (see Experimental Procedures). The arrows denote the reactions between the proteins, vesicle trafficking information, and flows of intracellular signaling obtained from the KEGG pathway database. The repression symbols denote suppressive reactions between the proteins obtained from the KEGG pathway database. See Experimental Procedures for further details regarding network modeling.
To explore collective functions of the DEPs in the aforementioned processes affected by sarpogrelate, we reconstructed a network model describing the interactions among the DEPs. In this network modeling, we focused on the DEPs involved in the processes relevant to granule secretion during platelet activation (cell activation, vesicle-mediated transport, Golgi vesicle transport, and protein transport); and platelet aggregation (coagulation) (Fig. 3C). First, the network model showed the alteration of the pathways associated with platelet activation (CD36-GNAI1-VASP, ADRA1B-PLCB2, and FERMT3 pathways) and a thrombin-coagulation pathway for platelet aggregation involving FGG and F13B, suggesting the effect of sarpogrelate on these pathways associated with platelet activation and aggregation. Second, the network model showed the alteration of 1) Golgi-secretory granule trafficking involving COPG2, PPID, ACAP2/3-ARF1, and SEPT5 DEPs and 2) the endosomal network for vesicle trafficking involving PIP5K1A, AP2B1, CUX1, HOOK2, and ATP6V1B2 DEPs. The DEPs are involved in the components in these trafficking networks, suggesting the effect of sarpogrelate on the trafficking networks associated with granule secretion during platelet activation (Fig. 3C). The collective alteration of these processes further indicates that the processes related to platelet activation and aggregation are highly integrated and their collective contribution of these processes to the effects of sarpogrelate on platelet activation and aggregation, consistent with the previous findings (54–56).
A Proteome Profile Representing Antiplatelet Functions of Sarpogrelate
The network model above showed the alteration of the pathways associated platelet activation and the thrombin-coagulation pathway associated with platelet aggregation, as well as Golgi-secretory granule and the endosomal networks for vesicle trafficking associated with granule secretion during platelet activation (Fig. 3C). Thus, to select a proteome profile that can collectively represent the functions of sarpogrelate in platelets, we focused on the DEPs that can represent these pathways and trafficking networks. The up-regulated proteins can be preferred as biomarker candidates because of the easiness to detect their alterations, compared with the downregulated proteins (57–59). Moreover, the DEPs with large fold-changes can be further preferred for the increased accuracy in detecting their alterations. Based on these preference criteria, we selected the following five up-regulated proteins five up-regulated proteins for which (1) the average of fold-changes in 14 samples was larger than 2, and (2) the number of samples having fold-changes >1.5 should be larger than half of 14 samples, as a proteome profile that represents the pathways and trafficking networks associated with platelet activation and aggregation in the network model (supplemental Fig. S4): (1) CD36 for platelet activation pathways, (2) F13B for the thrombin-coagulation pathway, (3) PPID for the Golgi-secretory granule network, and (4–5) CUX1 and HOOK2 for the endosomal network.
Validation of the Selected Protein Profile
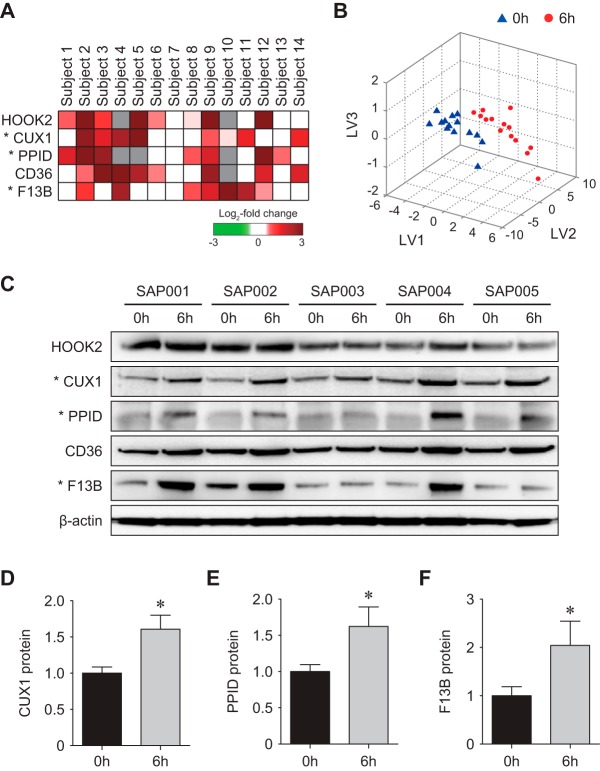
Based on the network model, the five proteins were selected as a protein profile that can represent the effect of sarpogrelate on platelet activation and aggregation. These proteins showed up-regulation in 14 subjects used for LC-MS/MS analysis by sarpogrelate (Fig. 4A). For the five selected proteins, we first examined whether they are capable of distinguishing platelet samples collected before and after administration of sarpogrelate using partial-least square-discriminant analysis (PLS-DA; Fig. 4B) (12). The PLS-DA analysis showed that all platelet samples obtained after the administration of sarpogrelate (red dots in Fig. 4B) were correctly separated from those before the administration (blue triangles in Fig. 4B) using three latent variables (LV1–3; supplemental Table S9; Experimental Procedures).
Fig. 4.
Validation of the selected proteins. A, A heat map showing differential expression of the selected candidate proteins. Colors represent up-regulation (red) and down-regulation (green) by sarpogrelate in 14 subjects. Gray denotes that the proteins are not detected in the corresponding subjects. The color bar denotes the gradient of log2-fold changes. B, 3-d PLS-DA score plot showing that the five selected proteins can achieve clear separation between platelet samples before (triangles) and after (circles) sarpogrelate administration. LV1–3 represent the first, second, and third latent variables obtained by PLS-DA. The coordinates of each sample in the 3-d space were computed by projecting the amounts of the five proteins onto the LV1–3. C–F, Western blotting results (C) and quantitative analysis (D–F) of the selected proteins in the independent set of platelet samples. Up-regulation of F13B, CUX1, and PPID in platelet samples treated with sarpogrelate were confirmed. The data are shown as mean ± S.D. *p < 0.05 by Student's t test.
To test the validity of the five selected proteins, we next collected the platelets from an independent set of five subjects based on the same criteria (see Experimental Procedures) employed for the collection of the samples at the discovery phase using LC-MS/MS analysis (supplemental Table S1). For these new samples, using Western blotting, we then tested differential expression of the five selected proteins measured by LC-MS/MS analysis. Of the five proteins, we finally selected the three proteins (CUX1, PPID, and F13B) that showed the changes in the independent samples consistent to those shown in the LC-MS/MS data (Fig. 4C–F). These data indicate that the three proteins can serve as a protein profile that represents the effects of sarpogrelate in platelets.
DISCUSSION
Sarpogrelate, as an antagonist of 5-HT receptors, has been widely used to treat the diseases involving platelet aggregation induced by 5-HT. However, there are no assay methods that can be used to monitor the effect of sarpogrelate on platelet aggregation. In this study, we thus searched for a protein profile that can be used to evaluate platelet aggregation. In this search, we used the following approach that involves (1) a comprehensive proteome profiling of platelets with generation of a master AMT DB and ultra-high pressure LC-MS/MS analysis; (2) identification of the DEPs before and after administration of sarpogrelate; (3) selection of a protein profile that can represent cellular processes associated with platelet aggregation and activation by analyzing the network model of the DEPs involved in such cellular processes; and (4) validation of the selected protein profile in independent platelet samples using Western blotting. Using this approach, we identified a protein profile composed of three up-regulated proteins involved in the following platelet aggregation-related processes: F13B, PPID, and CUX1 involved in the thrombin-coagulation pathway, ER-Golgi-secretory granule network, and the endosomal network, respectively.
A number of previous studies have shown associations of the three proteins selected in this study with pathophysiology of platelets under various conditions: (1) Alteration of serum F13B level caused by homozygous or compound heterozygous mutation in the F13B gene was linked to autosomal recessive hematologic disorder characterized by increased bleeding and poor wound healing (60, 61); and (2) PPID was reported as an essential regulator of the mitochondrial permeability transition pore that plays a critical role in platelet activation, and murine platelet activation responses thus showed the correlations with alterations of PPID amounts (62). However, the association of CUX1 with platelet functions has not been previously reported, although it plays important roles in the endosomal network for vesicle trafficking, indicating its potential role in granule secretion during platelet activation. These data suggest the validity of the selected proteins as a protein profile that can represent platelet aggregation. However, none of these proteins have been reported to be altered in their protein levels by sarpogrelate, suggesting the novelty of the three protein panel.
Platelets are anucleated cells, and the proteome data thus provide the more relevant information regarding functions of platelets, compared with genomic data. A number of studies have profiled the proteomes of platelets. The four studies (13–16) provided the proteomes of platelets in resting states. Although they could have served as useful resources for various studies of platelets, none of them have performed comparative proteomic analysis, thus providing no lists of DEPs related to platelet functions (e.g. the DEPs related to platelet activation and aggregation identified in this study). All of the three proteins selected as a protein profile indicative of platelet aggregation were detected in at least one of the previous studies. However, our study reported for the first time differential expression of the three selected proteins in platelets by sarpogrelate, thereby suggesting their potential use as the indicators of the effect of sarpogrelate on platelet aggregation in clinical settings.
Sarpogrelate was shown to negatively regulate platelet activation and aggregation (5). In this study, to investigate the proteins affected by sarpogrelate, we treated sarpogrelate on platelets obtained from healthy subjects. Because of the inhibitory effect of sarpogrelate on platelet aggregation, the proteins involved in the processes associated with platelet activation and aggregation are expected to be down-regulated by sarpogrelate. Unexpectedly, however, many of these proteins were up-regulated at 6 hours after treatment of sarpogrelate (Fig. 3B). Previously, Volpi et al. (63) profiled platelet proteomes at two different time points (12 and 24 h) after treatment of clopidogrel, an antiplatelet agent, and found up- and down-regulation of the proteins affected by clopidogrel at the different time points. For example, the abundances of the proteins (PFN1, MTPN, and MYL9) associated with actin organization were increased at 12 h after the loading dose of clopidogrel, but decreased below to the basal level at 24 h after stenting. This may account for the up-regulation of the proteins affected by sarpogrelate (e.g. proteins associated with actin organization) at 6 h after the treatment. Also, the secretion of 5-HT stored in platelets is not perturbed in healthy subjects, unlike the activated platelets under pathological conditions. Thus, the action of sarpogrelate can be different in platelets obtained from healthy subjects, which may lead to the up-regulation of the proteins affected by sarpogrelate. Detailed studies that examine temporal expression of the selected proteins in platelets from healthy subjects and activated platelets under pathological conditions can be carried out to understand how these proteins reflect the effects of sarpogrelate on platelet activation and aggregation. Moreover, the clinical implications of the three selected proteins can be further tested with a larger number of subjects. In addition, longitudinal studies can be designed to further demonstrate the nature of dynamic changes of the proposed protein profile after administration of sarpogrelate. Furthermore, more detailed functional studies can be carried out to understand the mechanistic contributions of the selected protein profiles to platelet aggregation.
In addition to the three proteins selected in this study, our approach provided a comprehensive list of DEPs related to platelet aggregation, thus extending extensively the current list identified by the conventional small-scale experiments or approaches. For example, deficiency or inhibition of growth arrest-specific gene 6 (GAS6), peptidylprolyl isomerase A (PPIA), serglycin (SRGN), and secreted protein, acidic, cysteine-rich (SPARC) were reported to be associated with thrombosis, inflammation, atherosclerosis, and metastasis, respectively (64–70). Thus, the list of DEPs can serve as a comprehensive resource to biologists who study the cellular processes associated with platelet aggregation (Fig. 3A). Moreover, the network model can provide a basis for understanding of the interplays of the cellular processes to collectively define platelet activation and aggregation induced by 5-HT (Fig. 3C). The GOBP enrichment analysis (supplemental Table S8) further showed that protein homeostasis-related processes (e.g. protein synthesis, protein folding, protein transport, and protein degradation) could be associated with platelet aggregation induced by 5-HT. This can suggest that protein homeostasis can serve as a potential measure to monitor the effect of sarpogrelate on the 5-HT-induced platelet aggregation. Moreover, the inhibition of protein homeostasis can be proposed as alternative targets of antiplatelet agents to the 5-HT receptors. In summary, our approach successfully identified a protein profile that can be used to monitor the effect of sarpogrelate on platelet aggregation induced by 5-HT, and the protein profile can provide new insights into the mechanism associated with functions of sarpogrelate in platelets.
Supplementary Material
Footnotes
Author contributions: D.H., Sang-Won Lee, and Soo-Youn Lee designed research; H.L., S.C., J.P., J.B., E.G., S.K., D.H., Sang-Won Lee, and Soo-Youn Lee performed research; H.L., S.C., H.K., D.H., Sang-Won Lee, and Soo-Youn Lee analyzed data; H.L., S.C., D.H., Sang-Won Lee, and Soo-Youn Lee wrote the paper.
* This work was supported by grants from the Multi-omics Research Program (NRF-2012M3A9B9036675), Institute for Basic Science (IBS-R013-G1-2015-a00), and NRF-2014M3C7A1046047 funded by the Korean Ministry of Science, ICT & Future Planning, as well as a grant of Korea Health Industry Development Institute (HI13C2098) funded by the Ministry of Health & Welfare, Republic of Korea.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- 5-HT
- 5-hydroxytryptamine
- FASP
- filter-aided sample preparation
- FTICR
- Fourier transform ion cyclotron resonance mass spectrometer
- iPE-MMR
- integrated post experiment monoisotopic mass refinement
- UMC
- unique mass class
- AMT DB
- accurate mass and time tag database
- NET
- normalized elution time
- FDR
- false discovery rate
- ESI
- electrospray ionization
- DEP
- differentially expressed protein
- GOBP
- Gene Ontology Biological Process
- GOCC
- Gene Ontology Cellular Component
- GOMF
- Gene Ontology molecular function.
REFERENCES
- 1. Miyazaki M., Higashi Y., Goto C., Chayama K., Yoshizumi M., Sanada H., Orihashi K., and Sueda T. (2007) Sarpogrelate hydrochloride, a selective 5-HT2A antagonist, improves vascular function in patients with peripheral arterial disease. J. Cardiovasc. Pharmacol. 49, 221–227 [DOI] [PubMed] [Google Scholar]
- 2. Saini H. K., Takeda N., Goyal R. K., Kumamoto H., Arneja A. S., and Dhalla N. S. (2004) Therapeutic potentials of sarpogrelate in cardiovascular disease. Cardiovasc. Drug Rev. 22, 27–54 [DOI] [PubMed] [Google Scholar]
- 3. Doggrell S. A. (2004) Sarpogrelate: cardiovascular and renal clinical potential. Expert Opin. Investig. Drugs 13, 865–874 [DOI] [PubMed] [Google Scholar]
- 4. Hayashi T., Sumi D., Matsui-Hirai H., Fukatsu A., Arockia Rani P. J., Kano H., Tsunekawa T., and Iguchi A. (2003) Sarpogrelate HCl, a selective 5-HT2A antagonist, retards the progression of atherosclerosis through a novel mechanism. Atherosclerosis 168, 23–31 [DOI] [PubMed] [Google Scholar]
- 5. Nishihira K., Yamashita A., Tanaka N., Kawamoto R., Imamura T., Yamamoto R., Eto T., and Asada Y. (2006) Inhibition of 5-hydroxytryptamine receptor prevents occlusive thrombus formation on neointima of the rabbit femoral artery. J. Thromb. Haemost. 4, 247–255 [DOI] [PubMed] [Google Scholar]
- 6. Matsuo H., and Shigematsu H. (2008) Effects of the 5-HT2A Antagonist Sarpogrelate on Walking Ability in Patients with Intermittent Claudication as Measured Using the Walking Impairment Questionnaire. Ann. Vasc. Dis. 1, 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shinohara Y., Nishimaru K., Sawada T., Terashi A., Handa S., Hirai S., Hayashi K., Tohgi H., Fukuuchi Y., Uchiyama S., Yamaguchi T., Kobayashi S., Kondo K., Otomo E., Gotoh F., and Group S. A. S. (2008) Sarpogrelate-Aspirin Comparative Clinical Study for Efficacy and Safety in Secondary Prevention of Cerebral Infarction (S-ACCESS): A randomized, double-blind, aspirin-controlled trial. Stroke 39, 1827–1833 [DOI] [PubMed] [Google Scholar]
- 8. Thompson N. T., Scrutton M. C., and Wallis R. B. (1986) Particle volume changes associated with light transmittance changes in the platelet aggregometer: dependence upon aggregating agent and effectiveness of stimulus. Thromb. Res. 41, 615–626 [DOI] [PubMed] [Google Scholar]
- 9. Qi R., Ozaki Y., Satoh K., Kurota K., Asazuma N., Yatomi Y., and Kume S. (1996) Quantitative measurement of various 5-HT receptor antagonists on platelet activation induced by serotonin. Thromb. Res. 81, 43–54 [DOI] [PubMed] [Google Scholar]
- 10. Zhao L., Lee B. Y., Brown D. A., Molloy M. P., Marx G. M., Pavlakis N., Boyer M. J., Stockler M. R., Kaplan W., Breit S. N., Sutherland R. L., Henshall S. M., and Horvath L. G. (2009) Identification of candidate biomarkers of therapeutic response to docetaxel by proteomic profiling. Cancer Res. 69, 7696–7703 [DOI] [PubMed] [Google Scholar]
- 11. Ma Y., Ding Z., Qian Y., Shi X., Castranova V., Harner E. J., and Guo L. (2006) Predicting cancer drug response by proteomic profiling. Clin. Cancer Res. 12, 4583–4589 [DOI] [PubMed] [Google Scholar]
- 12. Hyung S. W., Lee M. Y., Yu J. H., Shin B., Jung H. J., Park J. M., Han W., Lee K. M., Moon H. G., Zhang H., Aebersold R., Hwang D., Lee S. W., Yu M. H., and Noh D. Y. (2011) A serum protein profile predictive of the resistance to neoadjuvant chemotherapy in advanced breast cancers. Mol. Cell. Proteomics 10, M111.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burkhart J. M., Vaudel M., Gambaryan S., Radau S., Walter U., Martens L., Geiger J., Sickmann A., and Zahedi R. P. (2012) The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood 120, e73–e82 [DOI] [PubMed] [Google Scholar]
- 14. Krishnan S., Gaspari M., Della Corte A., Bianchi P., Crescente M., Cerletti C., Torella D., Indolfi C., de Gaetano G., Donati M. B., Rotilio D., and Cuda G. (2011) OFFgel-based multidimensional LC-MS/MS approach to the cataloguing of the human platelet proteome for an interactomic profile. Electrophoresis 32, 686–695 [DOI] [PubMed] [Google Scholar]
- 15. Yu Y., Leng T., Yun D., Liu N., Yao J., Dai Y., Yang P., and Chen X. (2010) Global analysis of the rat and human platelet proteome - the molecular blueprint for illustrating multi-functional platelets and cross-species function evolution. Proteomics 10, 2444–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Qureshi A. H., Chaoji V., Maiguel D., Faridi M. H., Barth C. J., Salem S. M., Singhal M., Stoub D., Krastins B., Ogihara M., Zaki M. J., and Gupta V. (2009) Proteomic and phospho-proteomic profile of human platelets in basal, resting state: insights into integrin signaling. PLoS ONE 4, e7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Azcona L., Lopez Farre A. J., Jimenez Mateos-Caceres P., Segura A., Rodriguez P., Modrego J., Zamorano-Leon J. J., and Macaya C. (2012) Impact of clopidogrel and aspirin treatment on the expression of proteins in platelets from type-2 diabetic patients with stable coronary ischemia. J. Pharm. Sci. 101, 2821–2832 [DOI] [PubMed] [Google Scholar]
- 18. Lopez-Farre A. J., Mateos-Caceres P. J., Sacristan D., Azcona L., Bernardo E., de Prada T. P., Alonso-Orgaz S., Fernandez-Arquero M., Fernandez-Ortiz A., and Macaya C. (2007) Relationship between vitamin D binding protein and aspirin resistance in coronary ischemic patients: a proteomic study. J. Proteome Res. 6, 2481–2487 [DOI] [PubMed] [Google Scholar]
- 19. Mateos-Caceres P. J., Macaya C., Azcona L., Modrego J., Mahillo E., Bernardo E., Fernandez-Ortiz A., and Lopez-Farre A. J. (2010) Different expression of proteins in platelets from aspirin-resistant and aspirin-sensitive patients. Thromb. Haemost. 103, 160–170 [DOI] [PubMed] [Google Scholar]
- 20. Wisniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 21. Skates S. J., Gillette M. A., LaBaer J., Carr S. A., Anderson L., Liebler D. C., Ransohoff D., Rifai N., Kondratovich M., Tezak Z., Mansfield E., Oberg A. L., Wright I., Barnes G., Gail M., Mesri M., Kinsinger C. R., Rodriguez H., and Boja E. S. (2013) Statistical design for biospecimen cohort size in proteomics-based biomarker discovery and verification studies. J. Proteome Res. 12, 5383–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Catellier D. J., Aleksic N., Folsom A. R., and Boerwinkle E. (2008) Atherosclerosis Risk in Communities (ARIC) Carotid MRI flow cytometry study of monocyte and platelet markers: intraindividual variability and reliability. Clin. Chem. 54, 1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park S. H., Park C. J., Kim M. J., Han M. Y., Lee B. R., Cho Y. U., and Jang S. (2014) The Sysmex XN-2000 hematology autoanalyzer provides a highly accurate platelet count than the former Sysmex XE-2100 system based on comparison with the CD41/CD61 immunoplatelet reference method of flow cytometry. Ann. Lab. Med. 34, 471–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Birschmann I., Mietner S., Dittrich M., Pfrang J., Dandekar T., and Walter U. (2008) Use of functional highly purified human platelets for the identification of new proteins of the IPP signaling pathway. Thromb. Res. 122, 59–68 [DOI] [PubMed] [Google Scholar]
- 25. Trichler S. A., Bulla S. C., Thomason J., Lunsford K. V., and Bulla C. (2013) Ultra-pure platelet isolation from canine whole blood. BMC Vet. Res. 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pignatelli P., Sanguigni V., Lenti L., Ferro D., Finocchi A., Rossi P., and Violi F. (2004) gp91phox-dependent expression of platelet CD40 ligand. Circulation 110, 1326–1329 [DOI] [PubMed] [Google Scholar]
- 27. Panes O., Matus V., Saez C. G., Quiroga T., Pereira J., and Mezzano D. (2007) Human platelets synthesize and express functional tissue factor. Blood 109, 5242–5250 [DOI] [PubMed] [Google Scholar]
- 28. Lopez-Farre A. J., Zamorano-Leon J. J., Azcona L., Modrego J., Mateos-Caceres P. J., Gonzalez-Armengol J., Villarroel P., Moreno-Herrero R., Rodriguez-Sierra P., Segura A., Tamargo J., and Macaya C. (2011) Proteomic changes related to “bewildered” circulating platelets in the acute coronary syndrome. Proteomics 11, 3335–3348 [DOI] [PubMed] [Google Scholar]
- 29. Wang Y., Yang F., Gritsenko M. A., Wang Y., Clauss T., Liu T., Shen Y., Monroe M. E., Lopez-Ferrer D., Reno T., Moore R. J., Klemke R. L., Camp D. G. 2nd, and Smith R. D. (2011) Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11, 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee H., Lee J. H., Kim H., Kim S. J., Bae J., Kim H. K., and Lee S. W. (2014) A fully automated dual-online multifunctional ultrahigh pressure liquid chromatography system for high-throughput proteomics analysis. J. Chromatogr. A 1329, 83–89 [DOI] [PubMed] [Google Scholar]
- 31. Lee J. H., Hyung S. W., Mun D. G., Jung H. J., Kim H., Lee H., Kim S. J., Park K. S., Moore R. J., Smith R. D., and Lee S. W. (2012) Fully automated multifunctional ultrahigh pressure liquid chromatography system for advanced proteome analyses. J. Proteome Res. 11, 4373–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hyung S. W., Kim M. S., Mun D. G., Lee H., and Lee S. W. (2011) The effect and potential of using a temperature controlled separation column with ultra-high pressure microcapillary liquid chromatography/tandem mass spectrometry on proteomic analysis. Analyst 136, 2100–2105 [DOI] [PubMed] [Google Scholar]
- 33. Jung H. J., Purvine S. O., Kim H., Petyuk V. A., Hyung S. W., Monroe M. E., Mun D. G., Kim K. C., Park J. M., Kim S. J., Tolic N., Slysz G. W., Moore R. J., Zhao R., Adkins J. N., Anderson G. A., Lee H., Camp D. G. 2nd, Yu M. H., Smith R. D., and Lee S. W. (2010) Integrated post-experiment monoisotopic mass refinement: an integrated approach to accurately assign monoisotopic precursor masses to tandem mass spectrometric data. Anal. Chem. 82, 8510–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim S., and Pevzner P. A. (2014) MS-GF plus makes progress towards a universal database search tool for proteomics. Nat. Commun. 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shin B., Jung H. J., Hyung S. W., Kim H., Lee D., Lee C., Yu M. H., and Lee S. W. (2008) Postexperiment monoisotopic mass filtering and refinement (PE-MMR) of tandem mass spectrometric data increases accuracy of peptide identification in LC/MS/MS. Mol. Cell. Proteomics 7, 1124–1134 [DOI] [PubMed] [Google Scholar]
- 37. Jaitly N., Monroe M. E., Petyuk V. A., Clauss T. R., Adkins J. N., and Smith R. D. (2006) Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal. Chem. 78, 7397–7409 [DOI] [PubMed] [Google Scholar]
- 38. Bolstad B. M., Irizarry R. A., Astrand M., and Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 39. Zhang B., Chambers M. C., and Tabb D. L. (2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dost B., Bandeira N., Li X., Shen Z., Briggs S. P., and Bafna V. (2012) Accurate mass spectrometry based protein quantification via shared peptides. J. Comput. Biol. 19, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Storey J. D., and Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang, da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 43. Bader G. D., Betel D., and Hogue C. W. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 31, 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peri S., Navarro J. D., Kristiansen T. Z., Amanchy R., Surendranath V., Muthusamy B., Gandhi T. K., Chandrika K. N., Deshpande N., Suresh S., Rashmi B. P., Shanker K., Padma N., Niranjan V., Harsha H. C., Talreja N., Vrushabendra B. M., Ramya M. A., Yatish A. J., Joy M., Shivashankar H. N., Kavitha M. P., Menezes M., Choudhury D. R., Ghosh N., Saravana R., Chandran S., Mohan S., Jonnalagadda C. K., Prasad C. K., Kumar-Sinha C., Deshpande K. S., and Pandey A. (2004) Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 32, D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stark C., Breitkreutz B. J., Reguly T., Boucher L., Breitkreutz A., and Tyers M. (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chatr-aryamontri A, Ceol A., Palazzi L. M., Nardelli G., Schneider M. V., Castagnoli L., and Cesareni G. (2007) MINT: the Molecular INTeraction database. Nucleic Acids Res. 35, D572–D574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., and Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanehisa M., Goto S., Sato Y., Furumichi M., and Tanabe M. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Geladi P., and Kowalski B. R. (1986) Partial Least-Squares Regression - a Tutorial. Anal. Chim. Acta 185, 1–17 [Google Scholar]
- 51. Yun S. J., Park J. W., Choi I. J., Kang B., Kim H. K., Moon D. W., Lee T. G., and Hwang D. (2011) TOFSIMS-P: a web-based platform for analysis of large-scale TOF-SIMS data. Anal. Chem. 83, 9298–9305 [DOI] [PubMed] [Google Scholar]
- 52. Nagatomo T., Rashid M., Abul Muntasir H., and Komiyama T. (2004) Functions of 5-HT2A receptor and its antagonists in the cardiovascular system. Pharmacol. Ther. 104, 59–81 [DOI] [PubMed] [Google Scholar]
- 53. Walther D. J., Peter J. U., Winter S., Holtje M., Paulmann N., Grohmann M., Vowinckel J., Alamo-Bethencourt V., Wilhelm C. S., Ahnert-Hilger G., and Bader M. (2003) Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell 115, 851–862 [DOI] [PubMed] [Google Scholar]
- 54. Golebiewska E. M., and Poole A. W. (2015) Platelet secretion: From haemostasis to wound healing and beyond. Blood Rev. 29, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blair P., and Flaumenhaft R. (2009) Platelet alpha-granules: basic biology and clinical correlates. Blood Rev. 23, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rendu F., and Brohard-Bohn B. (2001) The platelet release reaction: granules' constituents, secretion and functions. Platelets 12, 261–273 [DOI] [PubMed] [Google Scholar]
- 57. Beer L. A., Wang H., Tang H. Y., Cao Z. J., Chang-Wong T., Tanyi J. L., Zhang R. G., Liu Q., and Speicher D. W. (2013) Identification of Multiple Novel Protein Biomarkers Shed by Human Serous Ovarian Tumors into the Blood of Immunocompromised Mice and Verified in Patient Sera. Plos One 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kulasingam V., and Diamandis E. P. (2008) Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat. Clin. Pract. Oncol. 5, 588–599 [DOI] [PubMed] [Google Scholar]
- 59. Finlay J. A., Klee E. W., McDonald C., Attewell J. R., Hebrink D., Dyer R., Love B., Vasmatzis G., Li T. M., Beechem J. M., and Klee G. G. (2006) A systematic method for selection of promising serum protein biomarkers to improve prostate cancer (PCa1) detection. Clin. Chem. 52, 2159–2162 [DOI] [PubMed] [Google Scholar]
- 60. Hashiguchi T., Saito M., Morishita E., Matsuda T., and Ichinose A. (1993) Two genetic defects in a patient with complete deficiency of the b-subunit for coagulation factor XIII. Blood 82, 145–150 [PubMed] [Google Scholar]
- 61. Ichinose A., Izumi T., and Hashiguchi T. (1996) The normal and abnormal genes of the a and b subunits in coagulation factor XIII. Semin. Thromb. Hemost. 22, 385–391 [DOI] [PubMed] [Google Scholar]
- 62. Jobe S. M., Wilson K. M., Leo L., Raimondi A., Molkentin J. D., Lentz S. R., and Di Paola J. (2008) Critical role for the mitochondrial permeability transition pore and cyclophilin D in platelet activation and thrombosis. Blood 111, 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Volpi E., Giusti L., Ciregia F., Da Valle Y., Giannaccini G., Berti S., Clerico A., and Lucacchini A. (2012) Platelet proteome and clopidogrel response in patients with stable angina undergoing percutaneous coronary intervention. Clin. Biochem. 45, 758–765 [DOI] [PubMed] [Google Scholar]
- 64. Maree A. O., Jneid H., Palacios I. F., Rosenfield K., MacRae C. A., and Fitzgerald D. J. (2007) Growth arrest specific gene (GAS) 6 modulates platelet thrombus formation and vascular wall homeostasis and represents an attractive drug target. Curr. Pharm. Des. 13, 2656–2661 [DOI] [PubMed] [Google Scholar]
- 65. Angelillo-Scherrer A., de Frutos P., Aparicio C., Melis E., Savi P., Lupu F., Arnout J., Dewerchin M., Hoylaerts M., Herbert J., Collen D., Dahlback B., and Carmeliet P. (2001) Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nat. Med. 7, 215–221 [DOI] [PubMed] [Google Scholar]
- 66. Foley J. H., and Conway E. M. (2013) Gas6 gains entry into the coagulation cascade. Blood 121, 570–571 [DOI] [PubMed] [Google Scholar]
- 67. Elvers M., Herrmann A., Seizer P., Munzer P., Beck S., Schonberger T., Borst O., Martin-Romero F. J., Lang F., May A. E., and Gawaz M. (2012) Intracellular cyclophilin A is an important Ca2+ regulator in platelets and critically involved in arterial thrombus formation. Blood 120, 1317–1326 [DOI] [PubMed] [Google Scholar]
- 68. Woulfe D. S., Lilliendahl J. K., August S., Rauova L., Kowalska M. A., Abrink M., Pejler G., White J. G., and Schick B. P. (2008) Serglycin proteoglycan deletion induces defects in platelet aggregation and thrombus formation in mice. Blood 111, 3458–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Clezardin P., Malaval L., Morel M. C., Guichard J., Lecompte T., Trzeciak M. C., Dechavanne M., Breton-Gorius J., Delmas P. D., and Kaplan C. (1991) Osteonectin is an alpha-granule component involved with thrombospondin in platelet aggregation. J. Bone Miner. Res. 6, 1059–1070 [DOI] [PubMed] [Google Scholar]
- 70. Stenner D. D., Tracy R. P., Riggs B. L., and Mann K. G. (1986) Human platelets contain and secrete osteonectin, a major protein of mineralized bone. Proc. Natl. Acad. Sci. U.S.A. 83, 6892–6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.