Abstract
Several chronic inflammatory liver diseases, e.g. chronic hepatitis B or C viral infection and steatohepatitis, have been shown to predispose to the development of hepatocellular carcinoma (HCC). In patients with chronic liver disease, interleukin-6 (IL-6) serum levels are elevated and increase even more when HCC develops. However, the impact and regulatory mechanisms of IL-6 signaling during hepatocarcinogenesis are still poorly defined. Here, we could show that gene expression profiling of patients with chromosome 8p loss correlated with increased IL-6 signaling. In addition, the chromosome 8p tumor suppressor genes SH2D4A (Src homology 2 domain containing 4A) and SORBS3 (Sorbin and Src homology 3 domain containing 3) together exerted greater inhibition of cell growth and clonogenicity compared to a single gene. Overexpression of SH2D4A and SORBS3 in HCC cells led to decreased IL-6 target gene expression and reduced Signal Transducer and Activator of Transcription 3 (STAT3) signaling. In situ and in vitro co-immunoprecipitation assays revealed that SH2D4A directly interacts with STAT3 thereby retaining STAT3 in the cytoplasm and inhibiting STAT3 transcriptional activity. On the other hand, SORBS3 co-activated estrogen receptor α (ERα) signaling leading indirectly to repression of STAT3 signaling. In human HCC tissues, SH2D4A was positively associated with infiltrating regulatory and cytotoxic T cell populations suggesting distinct immunophenotypes in HCC subgroups with chromosome 8p loss. Thus, the genetically linked tumor suppressors SH2D4A and SORBS3 functionally cooperate to inhibit STAT3 signaling in HCC.
Conclusion
The chromosome 8p tumor suppressor genes SORBS3 and SH2D4A are physically and functionally linked and provide a molecular mechanism of inhibiting STAT3-mediated IL-6 signaling in HCC cells.
Keywords: Liver cancer, Interleukin-6, STAT3, tumor suppression, tumor microenvironment
Hepatocellular carcinoma (HCC) is the most common type of liver cancer, which is the second-leading cause of cancer-related mortality worldwide (1). In contrast to the decreasing overall cancer-related death rate, liver cancer incidence and mortality rates have been projected to increase until 2030 due to demographic shifts (2). Underlying liver diseases such as chronic infection with hepatitis B (HBV) and C viruses (HCV) and other inflammatory liver diseases such as alcoholic and non-alcoholic steatohepatitis contribute to the development of HCC, making HCC a paradigm for inflammation and virus-induced cancer (3, 4). It has been shown that tumor cells grow in a complex microenvironment of tumorous and non-tumorous cells, cellular components, and secreted small molecules (3, 5, 6). Tumor cells are not only effective in escaping from immune mediated eradication but they can also induce tumor promoting factors in the tumor microenvironment (7). This interplay of tumor cells with components of their microenvironment can activate several molecular pathways, some of which may enhance tumor progression, leading to metastasis and poor outcome (8, 9). Therefore, better understanding of the principles underlying tumor development and tumor-microenvironment interaction is critical to devise novel treatment modalities. Multiple studies in HCC and other cancer types have demonstrated the critical involvement of inflammation, especially of interleukin-6 (IL-6) signaling during tumorigenesis and metastasis (3, 7, 10). High serum IL-6 levels have been found to be associated with rapid progression from chronic viral hepatitis to HCC in HBV- and HCV-positive patients (11, 12). In addition, IL-6 has been shown to be involved in gender disparity of liver cancer (13, 14). Ablation of IL-6 expression leads to lower HCC incidence in DEN-induced hepatocarcinogenesis in male mice, whereas, female mice are protected by estrogen-mediated inhibition of IL-6 secretion (14). However, it is still unclear how IL-6 signal transduction is modulated during hepatocarcinogenesis.
Like other solid tumors, HCC is heterogeneous in terms of its clinical presentation and genomic and transcriptomic patterns due to genomic instability (4, 15, 16). Most tumors, including HCC, harbor a large number of genomic aberrations, but only few genes are likely to drive tumor development and progression. In addition, numerous recurrent hemizygous deletions, such as the deletion of large genomic regions on chromosome 8p, suggest a selective advantage of loss of multiple tumor suppressor genes (TSGs) over loss of a single gene to these tumor cells (17, 18). Furthermore, a genetic screen of TSGs revealed that inactivation of multiple chromosome 8p TSGs increases tumor development in a mouse model (18). Thus, these results suggest that the aberrant expression of multiple oncogenes and TSGs might have a stronger tumorigenic effect compared to a single gene. Therefore, loss of multiple TSGs, which are physically linked by their genomic location, may result in their functional cooperation.
Using an integrative genomic approach, we have addressed the functional impact of chromosome 8p loss, which is observed in 45% of HCC patients and associated with poor outcome (19). Integration of gene expression and somatic copy number profiles led to the identification of a 10-gene signature associated with chromosome 8p that predicted the outcome in two independent HCC cohorts (19). This gene signature contained six potential TSGs on chromosome 8p, the bona fide TSG DLC1 (Deleted in Liver Cancer 1) and two novel TSGs, namely SH2D4A (Src Homology 2 Domain Containing 4A) and SORBS3 (Sorbin and Src Homology 3 Domain Containing 3, encoding the protein Vinexin) (19).
In the present study, we analyzed the molecular mechanisms underlying SH2D4A and SORBS3 tumor suppressive function. We were able to show that SH2D4A and SORBS3 represent two tumor suppressor genes that cooperate to inhibit IL-6 mediated STAT3 (Signal Transducer and Activator of Transcription 3) signaling in hepatocarcinogenesis. In addition, SH2D4A expression levels are associated with tumor infiltrating T lymphocyte populations. Thus, these data demonstrate a link between the tumor’s genotype and the tumor-microenvironment in distinct HCC subgroups suggesting a benefit of combined genotype- and immunophenotype-specific therapies.
Experimental Procedures
Tissue microarray (TMA) and human subjects
The tissue microarray contained two representative areas (core diameter: 0.6 mm) of 127 HCCs. Cases were surgically resected between 2006 and 2011 at the University Hospital of Heidelberg and histologically classified according to established criteria by two experienced pathologists (P.S., N.W). The study was approved by the institutional ethics committee (application no. 206/05) of the Medical Faculty of Heidelberg University and supported by the tissue bank of the National Center for Tumor Diseases (NCT, Heidelberg, Germany).
Gene expression analysis and human subjects
Our study used a published Affymetrix U133A2.0 gene expression data set derived from 247 HCC patients as described previously (Gene Expression Omnibus accession number GSE14520) (20). Patient samples of this data set were obtained with informed consent from patients at the Liver Cancer Institute (LCI) and Zhongshan Hospital (Fudan University, Shanghai, China).
Statistical analysis
If not otherwise indicated, Student's t-test (two-tailed) was used for statistical analysis of comparative data between different groups. A value of p < 0.05 was considered to be statistically significant. Data are presented as mean ± SD (standard deviation) or mean ± SEM (standard error of the mean). Statistical analyses were performed using GraphPad Prism 6 for Windows software.
Additional experimental procedures are included in the Supplemental Information.
Results
SORBS3 and SH2D4A reduce proliferation and colony formation of HCC cells
Due to the genomic co-localization of SORBS3 and SH2D4A on chromosome 8p and their frequent co-deletion in human HCCs, we sought to test if SORBS3 and SH2D4A functionally interact in their tumor suppressive activity. Both SORBS3 and SH2D4A are adaptor proteins that appear to lack enzymatic activity. SH2D4A contains one SH2 (Src Homology 2) domain that might allow for binding of SH2D4A to phosphorylated tyrosine residues (Figure S1A). In the human embryonic kidney cell line HEK, it has been demonstrated that SH2D4A inhibits cell proliferation (21). SORBS3 on the other hand has one SoHo (Sorbin-homology) and three SH3 (Src Homology 3) domains which suggests a role in the cytoskeleton or the modulation of kinase activity (Figure S1A). Further, it has been shown that SORBS3 protein may be expressed in two different isoforms, SORBS3α and SORBS3β, of which only SORBS3α contains the SoHo domain ((22) and Figure S1A). SORBS3β has been shown to inhibit cell spreading, migration and anchorage-independent growth in the prostate carcinoma line LNCaP (23). However, the role of combined loss of SORBS3 and SH2D4A has not yet been characterized in HCC.
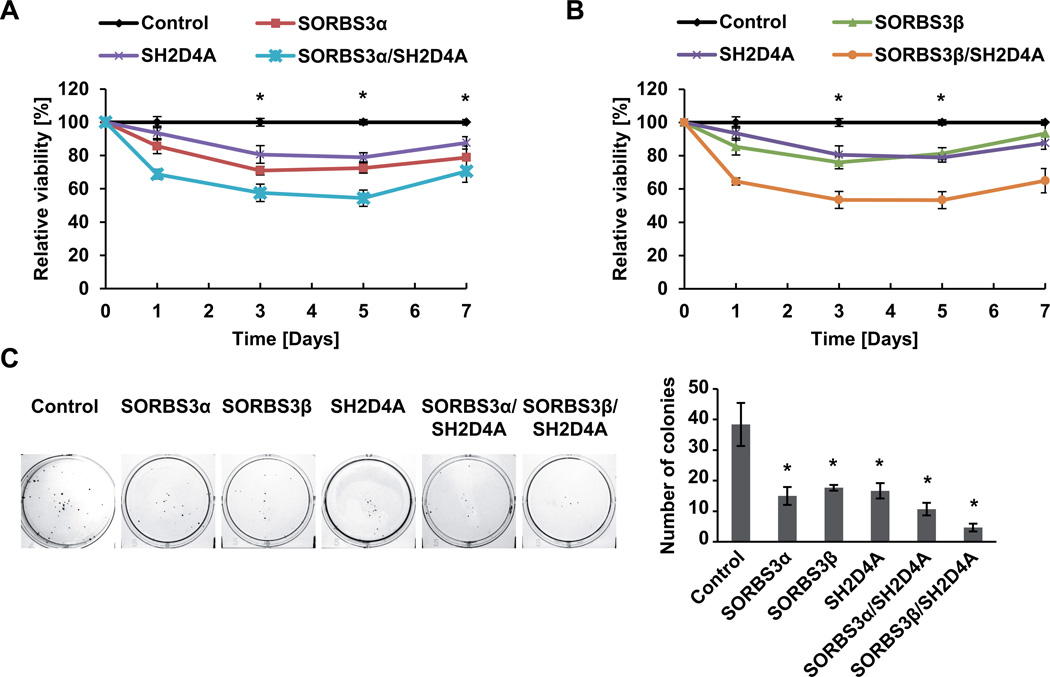
Western blot and semi-quantitative real-time RT-PCR analysis of SORBS3α, SORBS3β and SH2D4A showed that SORBS3α and SORBS3β are expressed at varying levels in HCC cell lines and SV40 large T antigen transformed normal liver THLE-2 cells (Figure S1B). In addition, mRNA levels of SORBS3 and SH2D4A positively correlated across different cell lines (Figure S1C and D). To test the functional interaction of SORBS3 and SH2D4A, we compared the activity of the two isoforms of SORBS3, SORBS3α and SORBS3β, alone or together with SH2D4A. Proliferation assays in the HepG2 cell line expressing low levels of endogenous SORBS3 and SH2D4A showed that transient overexpression of SORBS3α or SH2D4A alone was able to significantly reduce cell proliferation (p < 0.001; Figure 1A). Similarly, the shorter isoform SORBS3β was also able to significantly inhibit cell proliferation (p < 0.001; Figure 1B). Interestingly, overexpression of SH2D4A together with SORBS3α or SORBS3β demonstrated collaborative effects of these two TSGs resulting in significantly decreased proliferation compared to a single TSG (p < 0.001; Figure 1A and B). Similar results were obtained in the HuH1 cell line for SORBS3α and SH2D4A, whereas, the effect of SORBS3β was cell line dependent (Figure S2A and B). The analysis of clonogenic cell growth showed that SORBS3α, SORBS3β or SH2D4A alone reduced the clonogenic potential of HCC cells and that SORBS3α and SORBS3β similarly collaborated with SH2D4A to diminish colony formation in HepG2 and HuH1 cells (Figures 1C and S2C). Thus, the chromosome 8p TSGs SH2D4A and SORBS3 appear to functionally collaborate to inhibit cell proliferation and clonogenic tumor cell growth in HCC cells.
Figure 1. SH2D4A and SORBS3 cooperate to inhibit tumor cell proliferation and colony formation.
(A) Cell proliferation assay showing the relative viable cell number of HepG2 cells transiently transfected with empty vector control, SORBS3α, SH2D4A alone or together as indicated. (B) Cell proliferation assay showing the relative cell number of HepG2 cells transiently transfected with empty vector control, SORBS3β, SH2D4A alone or together. Data represent mean ± SEM normalized to transfection control at each time point of three independent experiments. * p < 0.05 of two-tailed Mann-Whitney U test of control vs. overexpression groups. (C) Representative images of colony formation assay of HepG2 cells transfected with empty vector control, SORBS3α, SORBS3β, SH2D4A alone or together as indicated (left panels). The right panel shows the statistical representation of the number of colonies in three replicates. One out of three independent experiments is shown. Data represent mean ± SD; * p < 0.05 of two-sided Mann-Whitney U test.
SORBS3 and SH2D4A expression lead to decreased IL-6 signaling
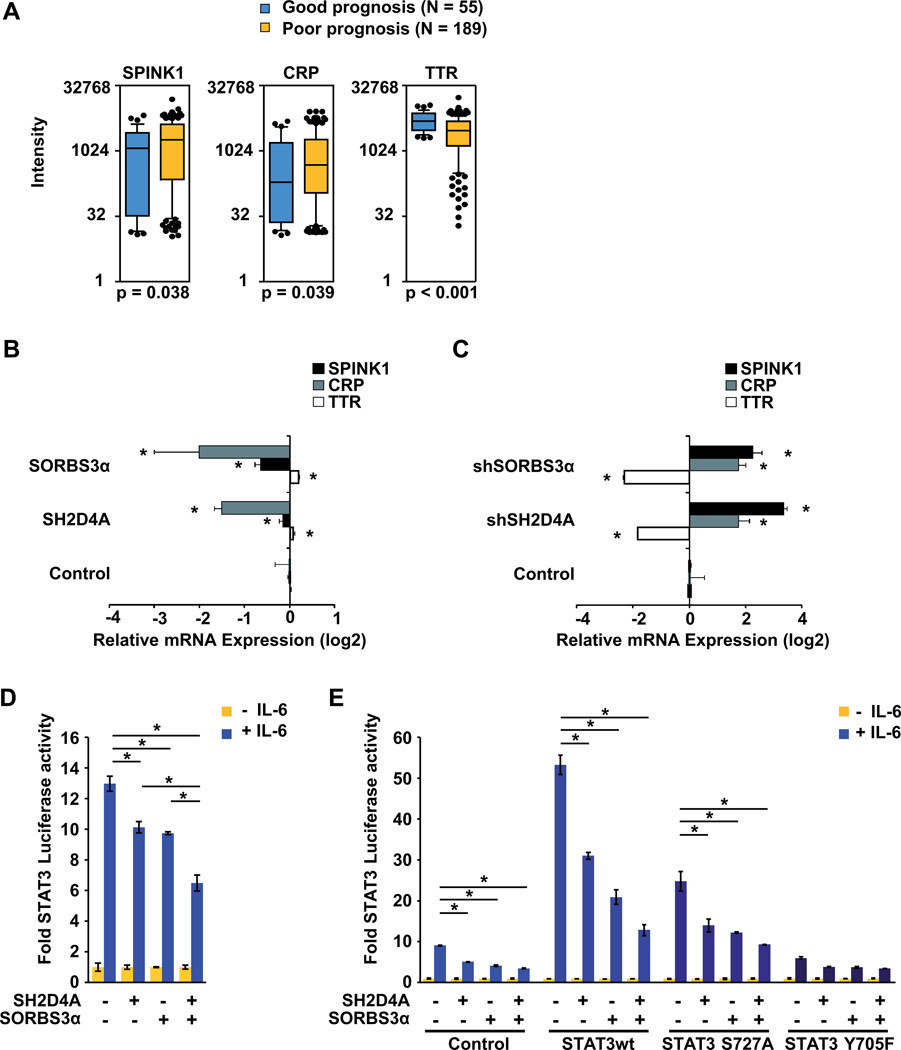
To identify pathways that may be repressed by SORBS3 and SH2D4A, we compared the gene expression profiles of patients with SORBS3 and SH2D4A gene expression classified as good prognosis group by the chromosome 8p-associated gene signature to patients with downregulation of SORBS3 and SH2D4A gene expression classified as poor prognosis group (19). Therefore, we used the previously published Affymetrix microarray data for class comparison of the good and poor outcome groups and pathway analysis (19). We found that the classical IL-6 target genes TTR, SPINK1, and CRP (24) are differentially expressed in HCC patients with good versus poor prognosis. SPINK1 and CRP were more than two-fold upregulated in HCC patients with loss of chromosome 8p signature and poor prognosis (Figure 2A and Supplemental Table S1). TTR which is repressed by IL-6 signaling showed the opposite effect by significant downregulation in the poor prognosis group. These data suggest that expression of the chromosome 8p signature, including SORBS3 and SH2D4A, is associated with IL-6 signaling which is increased in the poor outcome group.
Figure 2. Chromosome 8p tumor suppressor genes SH2D4A and SORBS3 cooperate to inhibit IL-6 signaling.
(A) Expression of IL-6 target genes SPINK1, CRP and TTR obtained by Affymetrix microarray profiling in HCC tumor tissues of patients with (poor prognosis) or without (good prognosis) expression of the chromosome 8p gene signature which is associated with poor outcome. (B) Expression of IL-6 target genes SPINK1, CRP and TTR measured by semi-quantitative RT-PCR in HuH7 cells upon lentivirus-mediated SORBS3, SH2D4A or Control (eGFP) shRNA knockdown. (C) Expression of the IL-6 target genes measured by semi-quantitative RT-PCR in Hep3B cells transfected with SORBS3, SH2D4A or empty vector control, respectively. (D) Luciferase assay in HepG2 cells measuring endogenous STAT3 transcriptional activity using STAT3 binding elements co-transfected with control vector, SH2D4A or SORBS3α expression vector in the presence or absence of 20 ng/ml IL-6 for 2 h. (E) Luciferase assay measuring STAT3 transcriptional activity of HuH1 cells co-transfected with control vector, SH2D4A, SORBS3α, STAT3wt, STAT3 S727A and STAT3 Y705F expression vectors in the presence or absence of 20 ng/ml IL-6 for 2 h, as indicated. Renilla luciferase was used as internal transfection control and for normalization. Data were normalized to corresponding IL6 unstimulated cells (mean ± SD; *p < 0.05).
To test whether SORBS3α and SH2D4A were directly involved in the inhibition of IL-6 signaling of patients with good outcome, we expressed these two proteins in HCC cell lines and found that both, SORBS3α and SH2D4A, decreased the expression of SPINK1 and CRP, which are activated by IL-6 signaling, and increased TTR expression, which is repressed by IL-6 signaling (Figure 2B, Figure S3A). Consistently, knockdown of SORBS3 and SH2D4A led to the opposite effect (Figure 2C, Figure S3B). Thus, SORBS3 and SH2D4A expression led to a phenotype consistent with inhibition of endogenous gene expression of IL-6 target genes and recapitulated the phenotype found in human HCC gene expression profiles.
Since IL-6 binding to its receptor leads to activation of Janus Kinases (JAK) 1 and 2 that phosphorylate and activate STAT3, we sought to examine whether SORBS3 and SH2D4A could inhibit IL-6 target gene expression by inhibition of STAT3 signaling (25). Therefore, we used a luciferase reporter construct containing five copies of the sis-inducible element (SIE; STAT3 binding site) in the two cell lines HepG2 and HuH1 which contain low levels of SORBS3 and SH2D4A. In the presence of IL-6, endogenous STAT3 transcriptional activity was 10- to 14-fold increased, whereas, the reconstitution of SORBS3α or SH2D4A significantly reduced the STAT3 activity in both HCC cell lines (Figure 2D and E). This effect was further increased through co-expression of SORBS3α and SH2D4A showing collaborative effects of both proteins (Figure 2D and E). It has been shown previously that IL-6 stimulation leads to STAT3 phosphorylation at Tyrosine 705 (Y705) and Serine 727 (S727) which have been suggested to be required for dimerization and maximal transcriptional activity of STAT3 (26, 27). Thus, we co-transfected wild type STAT3 (STAT3wt), STAT3 S727A, or STAT3 Y705F and found that inhibition of STAT3 activity by SH2D4A and SORBS3 was independent of STAT3 S727 phosphorylation (Figure 2E). Taken together, these results suggest that both SORBS3 and SH2D4A act together to inhibit IL-6 target gene expression through inhibition of STAT3 functional activity.
SH2D4A directly interacts with STAT3 and inhibits its translocation to the nucleus
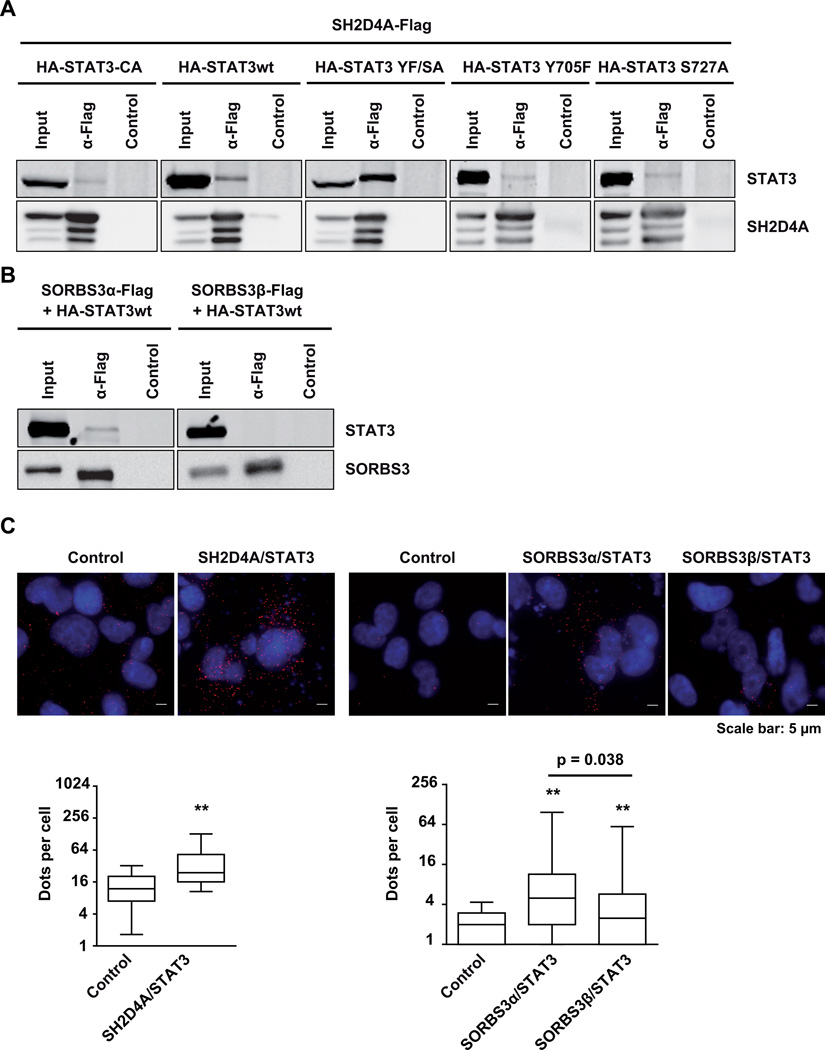
To dissect the molecular mechanisms underlying inhibition of STAT3 signaling by SH2D4A and SORBS3, we performed co-immunoprecipitation (co-IP) and in situ proximity ligation assays (PLA) and found that SH2D4A directly bound to STAT3 protein. To further characterize the interaction of SH2D4A and STAT3, wild type STAT3 (STAT3wt), constitutively active STAT3 (STAT3-CA, double mutant STAT3 C661A and C663N), mutant STAT3 Y705F, STAT3 S727A, and the inactive double mutant STAT3 Y705F and S727A (STAT3 YF/SA) were compared for their binding to SH2D4A. We found that STAT3 YF/SA exhibited the strongest binding affinity to SH2D4A, whereas, STAT3wt, STAT3-CA and single phospho-STAT3 mutants showed weaker binding (Figure 3A). Interestingly, SH2D4A binding affinity might be strongest for unphosphorylated, inactive STAT3. In addition, we also tested whether SORBS3 directly interacts with STAT3. In co-IP experiments we observed binding of SORBS3α but not SORBS3β to STAT3 (Figure 3B). Next, we performed PLA, also named In-cell co-IP, in HLF cells and observed distinct signals suggesting a direct interaction of SH2D4A and STAT3 in situ. Quantification of dots per cell, each displaying a protein-protein interaction, revealed significantly higher signal numbers in the transfected cells compared to the controls (Figure 3C). SORBS3α and SORBS3β also showed direct interaction with STAT3, however, the binding was significantly less for SORBS3β compared to SORBS3α. Thus, these data indicate that SH2D4A and SORBS3 directly interact with STAT3 protein in vitro and in situ.
Figure 3. SH2D4A and SORBS3α proteins directly interact with STAT3.
(A) HEK293T cells were transiently co-transfected with N-terminally HA-tagged STAT3-CA, STAT3wt, STAT3 YF/SA, STAT3 Y705F or STAT3 S727A and C-terminally Flag-tagged SH2D4A. After anti-Flag immunoprecipitation, samples were separated by SDS–polyacrylamide gel electrophoresis and subjected to Western blot analysis with anti-HA and anti-SH2D4A antibodies. One representative experiment out of three with similar outcome is shown. (B) HEK293T cells were transiently co-transfected with N-terminally HA-tagged STAT3wt and C-terminally Flag-tagged SORBS3α or SORBS3β, respectively. Proteins were immunoprecipitated with anti-Flag antibodies and immunoblotted with anti-HA and anti-SORBS3 antibodies. One representative experiment out of three with similar outcome is shown. (C) Representative images of proximity ligation assays (PLA) of HLF control cells or cells transiently transfected with SH2D4A, SORBS3α or SORBS3β and STAT3. PLA signals are shown in red; cell nuclei were stained with DAPI in blue. Quantitative representation of PLA dots per cell (log) by boxplots with median and 25th to 75th percentile. Whiskers represent 5–95% confidence intervals. N≥50 cells; P-values were obtained by two-sided Mann-Whitney U test. ** p < 0.0001 vs. control.
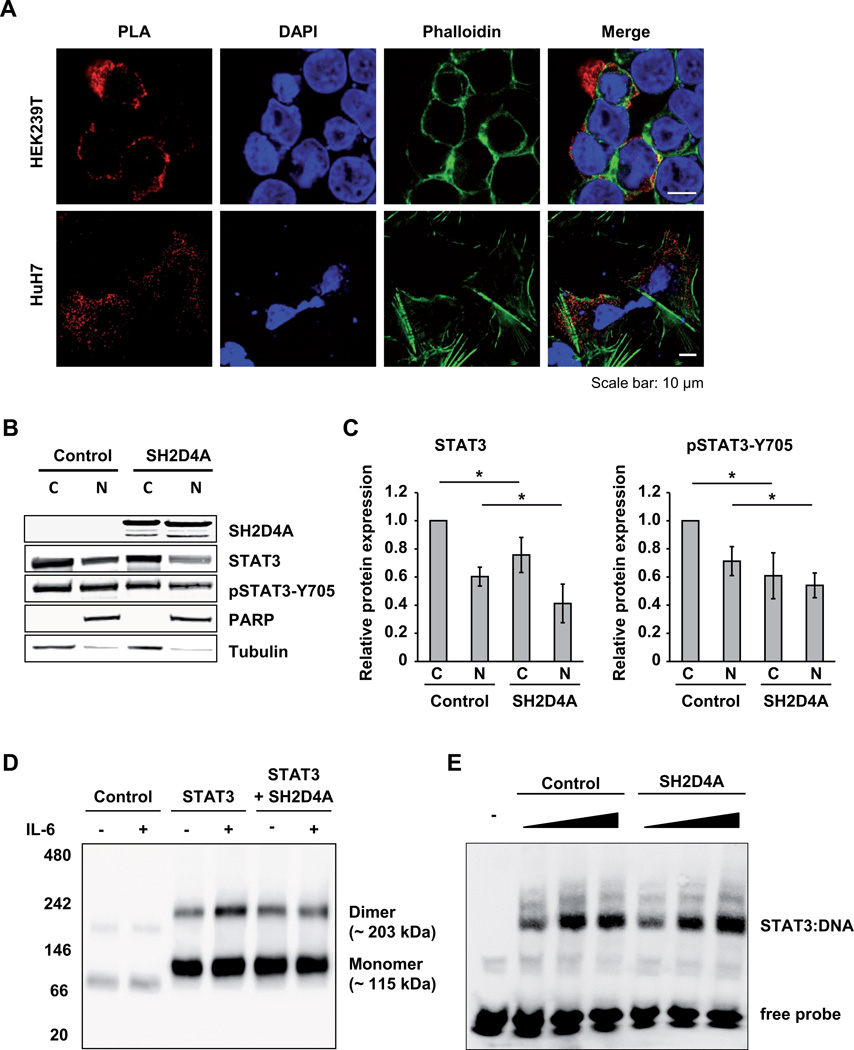
Upon activation by IL-6, STAT3 is phosphorylated by JAK1 or JAK2, forms dimers, and translocates to the nucleus where it acts as a transcription factor and an oncogene in HCC (25, 28). Interestingly, overexpression of SORBS3 and SH2D4A in HepG2 cells showed no effect on the phosphorylation of JAK1, JAK2 and STAT3 upon IL-6 stimulation (Figure S4). Next, we asked the question whether SH2D4A binding to STAT3 may lead to the retention of STAT3 in the cytoplasm. Confocal imaging of PLA showed that the STAT3-SH2D4A complexes were predominantly localized to the cytoplasm (Figure 4A). The SH2D4A-STAT3 interaction was not altered when the cells were treated with IL-6 (Figure S5). To further analyze STAT3 localization, we also performed nuclear and cytoplasmic fractionation of control cells that do not express endogenous SH2D4A and SH2D4A-transfected HEK293T cells. Western blotting of the cytoplasmic and nuclear fractions revealed that expression of SH2D4A significantly reduced total STAT3 and at Y705 phosphorylated STAT3 in the nucleus and cytoplasm (Figure 4B and C). In addition, we performed blue native polyacrylamide gel electrophoresis (BN-PAGE) under non-denaturing conditions to analyze the formation of STAT3 dimers in the presence and absence of SH2D4A. Consistent with previous studies, GFP-tagged STAT3 preformed dimers in the absence of IL-6 (29, 30) and upon IL-6 stimulation STAT3 dimerization significantly increased (Figure 4D). Expression of SH2D4A reduced the amount of STAT3 dimer formation after IL-6 stimulation to preformed STAT3-dimer level which was not further decreased by SH2D4A (Figure 4D). Thus, SH2D4A directly binds to STAT3 protein in the cytoplasm leading to cytoplasmic retention and inhibition of IL-6 stimulation-driven dimerization of STAT3.
Figure 4. SH2D4A leads to the retention of STAT3 in the cytoplasm and blocks STAT3 dimerization.
(A) Confocal imaging of proximity ligation assays (PLA) in HEK293T and HuH7 cells transfected with SH2D4A and STAT3. PLA dots are shown in red; Cell nuclei were stained with DAPI in blue; F-actin was stained with Phalloidin in green. (B) Western blot analysis of nuclear and cytoplasmic extracts of HEK293T control or SH2D4A transfected cells with indicated antibodies. A representative experiment out of four with similar outcome is shown. (C) Quantitative analysis of pan-STAT3 and Y705-phosphorylation-specific anti-pSTAT3 antibody in four independent cell fractionation experiments. Data are presented as mean ± SD derived from four biological replicates normalized to cytoplasmic control fractions. * p < 0.05 of two-sided Mann-Whitney U test. (D) HEK293T cells were transfected with vector control, GFP-STAT3 and SH2D4A-Flag, and treated with 20 ng/ml IL-6 or left untreated. Protein lysates were analyzed by Blue native polyacrylamide gel electrophoresis (BN-PAGE) followed by Western blotting with anti-STAT3 antibody. One out of three independent experiments is shown. (E) Electrophoretic mobility shift assay (EMSA) of nuclear extracts of HepG2 cells transfected with empty control vector or SH2D4A-Flag, and stimulated with 20 ng/ml IL-6 for 20 min. Protein lysates (1 µg, 2.5 µg and 5 µg) were incubated with biotin end-labeled STAT1/STAT3 oligonucleotides (m67SIE) and separated on a native polyacrylamide gel. The DNA-protein complexes were detected using streptavidin-horseradish peroxidase conjugate and chemiluminescent substrate.
In recent studies the STAT family member STAT1 has also been shown to play a critical role in liver tumorigenesis (31). Therefore, we performed co-IP experiments to test if SORBS3 and SH2D4A also could bind to STAT1. Similarly to STAT3, we observed binding of SH2D4A and SORBS3α but not SORBS3β to STAT1 (Figure S6A and B). Next we sought to test if STAT1 activity is affected by SH2D4A or SORBS3 binding. Therefore, we applied luciferase assays using a reporter construct containing 4× interferon-gamma activated sites (GAS elements). However, we could not observe any activation of this reporter upon IL-6 stimulation, and SORBS3 and SH2D4A did not have any effect on STAT1-luciferase reporter activity (Figure S6C). As a control, we stimulated the cells with interferon-γ (IFNγ) and found that luciferase activity was induced more than 14-fold. In contrast to STAT3, SH2D4A and SORBS3α but not SORBS3β increased STAT1 activity (Figure S6D). Thus, we were able to show that SH2D4A and SORBS3α both interact with STAT1 and can increase IFNγ/STAT1signaling. These results are consistent with previous reports suggesting that STAT1 mostly triggers anti-proliferative and pro-apoptotic responses, whereas, STAT3 promotes cell survival and motility and is considered as an oncogene (31).
SORBS3α but not SORBS3β directly interacts with ERα
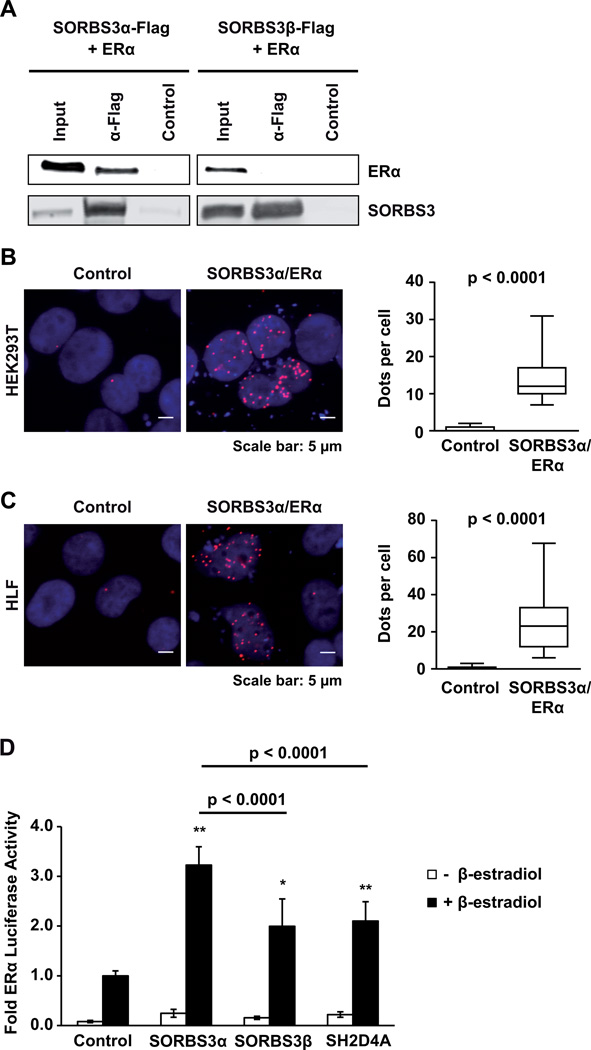
Next, we aimed to elucidate the functional role of SORBS3 in the inhibition of IL-6 signaling. It has been shown previously in COS-7 cells that SORBS3α binds to ERα and modulates ERα function through phosphorylation (32). In addition, ERα signaling inhibits STAT3 through direct binding in vitro and in vivo in breast cancer cells (33). Thus, we analyzed whether SORBS3 functions as a co-activator of ERα signaling to inhibit STAT3 in HCC cells. First, we performed co-IP experiments and found that SORBS3α but not SORBS3β directly bound to ERα in vitro (Figure 5A). These results were confirmed by PLA which showed that SORBS3α directly interacts with ERα in situ (Figure 5B and C). To functionally validate the enhancement of ERα activity by SORBS3, we performed luciferase reporter assays. Using a reporter construct containing tandem estrogen response elements (ERE), we confirmed that SORBS3α increased ERα signaling 3.2-fold in the presence of β-estradiol (p < 0.0001; Figure 5D). In contrast, SORBS3β and SH2D4A only slightly elevated ERα signaling by two-fold (Figure 5D). Taken together, these results show that rather SORBS3α than SORBS3β or SH2D4A enhances ERα signaling which might in turn inhibit STAT3 signaling.
Figure 5. Direct interaction of SORBS3 with ERα.
(A) HEK293T cells were transiently co-transfected with SORBS3α-Flag (left) or SORBS3β-Flag (right) together with ERα. After anti-Flag immunoprecipitation, samples were separated by SDS–PAGE and subjected to Western blot analysis with anti-ERα-reactive and anti-Flag antibodies. One representative experiment out of three with similar outcome is shown. (B) Representative images of proximity ligation assays (PLA) in HEK293T and in (C) HLF control cells or cells transfected with SORBS3α and ERα (left panel). Quantitative representation of PLA dots per cell by boxplots with whiskers representing 5–95% confidence intervals (right panel). P-values were obtained by two-tailed Mann-Whitney U test. (D) Luciferase assay analyzing ERα signaling activity using a 3× ERE-luciferase reporter construct in HuH7 cells expressing ERα and control vector, SORBS3α, SORBS3β or SH2D4A, stimulated with or without 10 nM β-estradiol for 24 h, as indicated. Data represent mean ± SD of three independent biological experiments. Data were normalized to Renilla and the stimulated transfection control. * p < 0.001; ** p < 0.0001 vs. β-estradiol stimulated control using two-tailed Mann-Whitney U test.
SH2D4A expression is associated with tumor infiltrating cells in human HCC tumor tissues
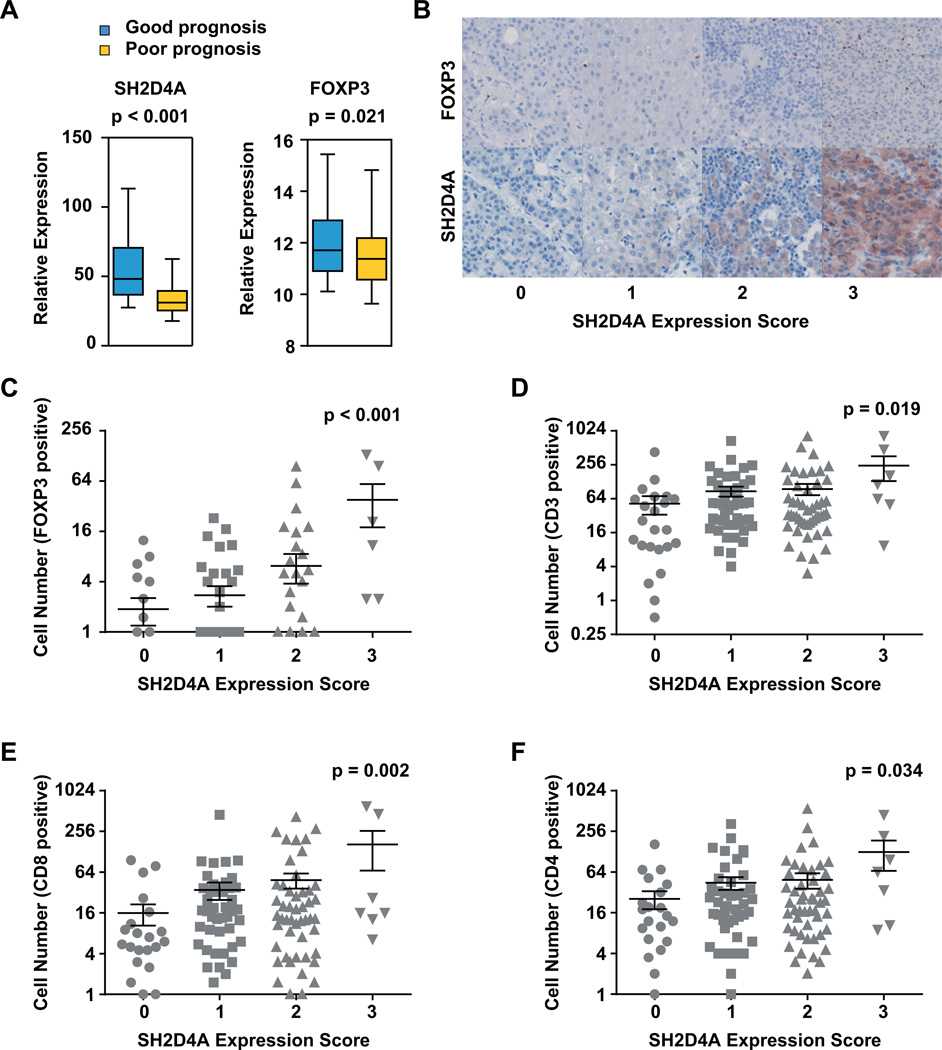
Immune cell infiltrations play an important role in the progression of many solid tumor entities. In addition, there is a close crosstalk including multiple feedback mechanisms between immune cells and tumor cell signaling (31). Specifically, T cell infiltration is induced by STAT3 inhibition in tumor cells (34). Thus, we analyzed if chromosome 8p tumor suppressor genes were associated with alterations of the tumor microenvironment. First, we compared the expression of T cell markers in gene expression profiles of HCCs with (good outcome) or without (poor outcome) chromosome 8p gene signature. We found that FOXP3 exhibited significantly lower expression in HCC patients with poor outcome (Figure 6A). Other T cell markers including CD3E, CD3G, CD8A, CD8B, and CD4 did not show differential expression, while CD3D was slightly upregulated on mRNA level (Figure S7). In addition, FOXP3 expression was positively correlated with SH2D4A expression in patients with poor but not good outcome (Figure S7B and C). Therefore, we sought to visually characterize the immune cell infiltrates in human HCCs by immunohistochemical staining of tumor tissues using different T cell markers. Consistently with the gene expression analysis, we found the number of FOXP3-positive regulatory T cells to significantly correlate with increased SH2D4A expression in HCC cells (Figure 6B and C). Also, CD3-, CD4- and CD8-positive T cell infiltrations were higher in HCC tumors with high SH2D4A expression (Figure 6D–F). In contrast, CD68-positive macrophages, NASDCL-positive granulocytes and CD20-positive B cells did not show differential infiltration patterns in HCC patient groups with distinct SH2D4A expression levels (Figure S8). Thus, high SH2D4A expression in HCC tumor cells is associated with tumor infiltration of FOXP3-positive regulatory T cells and CD8-positive cytotoxic T cells.
Figure 6. SH2D4A expression is associated with tumor infiltration of regulatory and cytotoxic T cells.
(A) Expression of SH2D4A and FOXP3 obtained by Affymetrix gene expression profiling of HCC tumor tissues of patients in the poor or good prognosis patient subgroups. (B) Representative images of FOXP3 and SH2D4A immunohistochemical staining in HCC tumor tissues of a tissue microarray (TMA). Representative images are shown for four HCC patients with different expression levels of SH2D4A (Score 0, 1, 2 or 3). (C) Quantitative analysis of FOXP3-positive, (D) CD3-positive, (E) CD8-positive and (F) CD4-positive T cells in HCC patients groups without SH2D4A (Score 0), with low (Score 1), moderate (Score 2) or high (Score 3) SH2D4A expression. Data represent mean ± SEM; p-value obtained by ANOVA analysis of variance.
Discussion
It is well established that the tumor microenvironment can influence tumor growth and disease progression mainly through secretion of cytokines, chemokines, growth factors and reactive oxygen and nitrogen species. However, it is still unclear which role genomic changes that are specific to tumor cells are playing in the crosstalk between tumor cells and the tumor microenvironment. Many studies in HCC and other solid tumors have demonstrated an oncogenic mechanism of activated IL-6/STAT3 signaling (31, 35). In line with these findings, hepatocyte-specific STAT3 knockout mice are resistant to liver tumorigenesis induced by diethyl-nitrosamine (DEN) which leads to minimal chronic liver inflammation and fibrosis (28). Furthermore, high levels of phosphorylated STAT3 protein correlate with poor outcome in HCC patients (36). However, recent studies in prostate cancer and KRAS mutant lung tumors suggest that STAT3 may also act as a tumor suppressor challenging the current discussion on therapeutic benefit or risk of IL-6/STAT3 inhibition (37, 38). In the chronic carbon tetrachloride (CCl4)-induced liver fibrosis mouse model, STAT3 also exhibits hepatoprotective functions by prevention of hepatic damage and fibrosis ultimately leading to reduced liver tumor formation (28). Thus, STAT3’s function is tightly regulated and depends on cell type and cellular microenvironment.
In this study, we demonstrate that the chromosome 8p tumor suppressor genes SH2D4A and SORBS3 cooperate to inhibit HCC tumor cell growth and proliferation. This finding is in concordance with the finding that loss of chromosome 8p is frequent in HCC but no focal deletions exist on chromosome 8p and that consequently SORBS3 and SH2D4A are typically co-deleted in HCC (18, 19). Interestingly, our data revealed that both genes are associated with repression of IL-6 signaling in HCC tumor tissues. Functional studies showed that both genes synergize to inhibit IL-6 mediated STAT3 signaling. SH2D4A directly binds to STAT3 protein in the cytoplasm, leading to reduced levels of STAT3 in the nucleus and inhibition of STAT3-dimerization upon IL-6 stimulation. However, STAT3 and phospho-STAT3 protein levels in whole cell lysates were not altered upon SH2D4A or SORBS3 expression. It has been shown previously that STAT3 protein is found in lipid rafts and in the early endosomal fraction of Hep3B cells (39, 40). In contrast, the fractionation method applied in our study does not isolate proteins in endoplasmic vesicles nor proteins bound to the cell membrane. Therefore, we assume that the observed differences in STAT3 and phospho-STAT3 protein levels in the cytoplasmic and nuclear fractions could be due to a shift of STAT3 protein localization upon SH2D4A expression. The inhibition of IL-6 signaling by SH2D4A may provide a potential mechanism of tumor suppression in chronic liver injury and inflammation. Loss of SH2D4A during tumorigenesis might enhance IL-6/STAT3 signaling in the subgroup of HCC patients with loss of chromosome 8p gene signature and poor prognosis. Monoclonal antibodies specifically binding IL-6 (siltuximab) or IL-6R (tocilizumab) have been shown to be well tolerated and effective in reducing STAT3 phosphorylation and expression of IL-6 target genes in patients (41). Thus, inhibition of IL-6 or IL-6R might be a therapeutic option for HCC patients with chromosome 8p loss.
A recent study demonstrated that gp130 signaling in the liver and intestine led to activation of the YAP pathway independent of STAT3 (42). Supporting our observations of SH2D4A and SORBS3 inhibition of STAT3, we did not detect any effect on YAP phosphorylation and target gene expression in HCC cells (Figure S9). However, we found that SORBS3 binds and activates ERα which has been shown to inhibit IL-6/STAT3 signaling. Epidemiological studies in HCC-high risk patient groups have observed a gender bias with a two- to three-fold increased incidence in male versus female patients suggesting a protective effect of estrogen in patients with underlying chronic liver disease (43). Furthermore, the inhibition of IL-6/STAT3 signaling by estrogen/ERα signaling has also been functionally demonstrated in vivo. Estradiol treatment of male mice reduced liver injury in IL-6-treated mice suggesting that estrogen/ERα signaling may also attenuate signaling downstream of IL-6 (14). In breast cancer cells ERα signaling efficiently inhibits IL-6/STAT3 signaling by direct protein-protein interaction of ERα and STAT3 leading to repression of STAT3 transcriptional activity (33). In multiple myeloma on the other hand, ERα signaling leads to activation of PIAS3 expression which binds to and inhibits STAT3 signaling (44). A recent study in HCC cell lines and tumor tissues suggested that ERα could repress STAT3 activity by increased expression of protein tyrosine phosphatase receptor type O (PTPRO) (45). PTPRO dephosphorylated STAT3 at Y705 and S727 resulting in diminished STAT3 signaling. These studies suggest that the inhibitory function of ERα signaling on STAT3 is common in multiple tumor entities. However, the molecular mechanisms leading to reduced STAT3 transcriptional activity may be diverse. We found that SORBS3 repressed STAT3 transcriptional activity and functioned as a co-activator of ERα signaling in HCC cells. Thus, co-activation of ERα signaling by SORBS3 may enhance the inhibitory effect of ERα on STAT3 in hepatocytes during chronic inflammatory liver disease.
Tumor infiltrating T lymphocytes are present in multiple tumor entities in various quantities and their presence is associated with better outcome possibly explained by immune surveillance of tumor growth. High levels of CD8-positive T cells in the tumor, but not in the distant normal tissue of HCC patients, have been found to be an independent predictor of good prognosis (46). Although most tumor cells are believed to express antigens that can be recognized by CD8-positive T cells, the antitumor effect of CD8-positive cytotoxic T cells is limited and exhausts over time (47). A mouse model of NRasG12V-induced senescent hepatocytes demonstrated that CD4 T cell-mediated immune clearance of pre-malignant senescent cells is crucial to suppress liver cancer development (48). Therefore, both CD8-positive cytotoxic and CD4-positive regulatory T cells are required for immune surveillance and it is conceivable that efficient activation of cytotoxic and regulatory T cells may improve immunotherapy of HCC. In the present study, we found in gene expression microarray data that the chromosome 8p 10-gene signature including SH2D4A is associated with altered FOXP3 gene expression (figure 6A). To analyze whether these differences originate from infiltrating FOXP3-positive T cells, we chose to use immunohistochemical analysis on formalin-fixed paraffin-embedded (FFPE) tissue. Since FFPE tissue is not anymore available for the Affymetrix cohort, we used the 127 HCC patient cohort from Heidelberg and could confirm that high levels of SH2D4A protein are associated with high numbers of FOXP3-positive T cells. Hence, we found in both cohorts an association of SH2D4A expression and FOXP3 expression. The utilization of two independent cohorts might bare intrinsic confounders. However, in our previous study we showed that the prognostic value of the chromosome 8p 10-gene signature is independent of etiology and clinical staging in HCC (19).
Taken together, in this study we showed that SH2D4A and SORBS3 are not only physically linked on chromosome 8p but also functionally interact to inhibit STAT3 transcriptional activity in HCC. In addition, we found a correlation of low SH2D4A protein expression with decreased infiltration of CD4-, CD8- and FOXP3-positive T cells in human HCC tissues. This suggests a decrease of immune surveillance in patients with low expression of the chromosome 8p gene SH2D4A. In conclusion, the combination of genotype-specific HCC therapy combined with immunotherapy may aid in the development of more effective HCC treatment modalities.
Supplementary Material
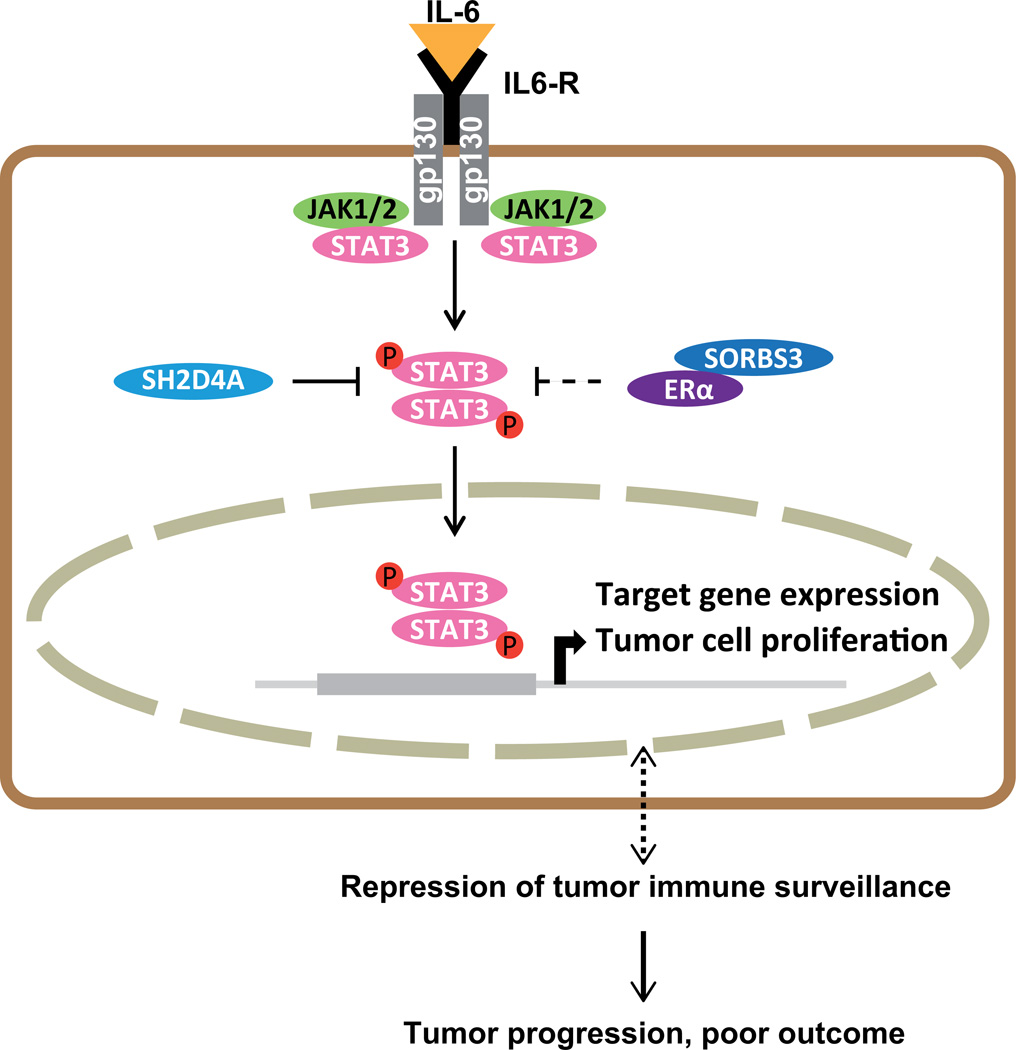
Figure 7. Schematic model of SH2D4A and SORBS3 mediated repression of STAT3 signaling.
IL-6 binds to its receptor which is constituted by IL6R and gp130. Upon IL-6 binding JAK1/2 and STAT3 are activated through phosphorylation, phosphorylated STAT3 forms dimers, translocates to the nucleus and activates target gene expression. SH2D4A physically binds STAT3 leading to retention of STAT3 in the cytoplasm and thus, inhibition of STAT3 signaling. SORBS3 might function indirectly through binding of ERα also leading to inhibition of STAT3 signaling.
Acknowledgments
Financial Support: P.S. was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG, SFB/TRR77, Z2). S.R. was support by DFG grant RO 4673, the Olympia-Morata Program, Brigitte-Schiebenlange Fellowship and NCT Heidelberg School of Oncology Fellowship. X.W.W. was supported by the intramural research program of the Center for Cancer Research, the U.S. National Cancer Institute.
List of Abbreviations
- BN-PAGE
Blue native polyacrylamide gel electrophoresis
- co-IP
co-immunoprecipitaion
- CRP
C-reactive protein
- CTGF
Connective Tissue Growth Factor
- DLC1
Deleted in Liver Cancer 1
- ERα
estrogen receptor α
- ERE
estrogen response element
- FFPE
formalin-fixed paraffin-embedded
- FOXM1
Forkhead box M1
- FOXP3
Forkhead box P3
- GAS
interferon-gamma activated site
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HCC
hepatocellular carcinoma
- IL-6
interleukin-6
- IL6R
interleukin-6 receptor
- IFNγ
interferon-γ
- JAK
Janus Kinases
- PEI
polyethylenimine
- PLA
proximity ligation assay
- RT-PCR
reverse transcription-polymerase chain reaction
- SD
standard deviation
- SEM
standard error of the mean
- SH2
Src Homology 2
- SH3
Src Homology 3
- SH2D4A
Src Homology 2 Domain Containing 4A
- SIE
sis-inducible element
- SoHo
Sorbin-homology
- SORBS3
Sorbin and Src Homology 3 Domain Containing 3
- SPINK1
Serine Peptidase Inhibitor, Kazal Type 1
- STAT
Signal Transducer and Activator of Transcription
- TSG
tumor suppressor gene
- TTR
Transthyretin
- wt
wild type
Footnotes
Disclosures: Nothing to report.
Contributor Information
Carolin Ploeger, Email: Carolin.Ploeger@med.uni-heidelberg.de.
Nina Waldburger, Email: Nina.Waldburger@med.uni-heidelberg.de.
Angelika Fraas, Email: Angelika.Fraas@med.uni-heidelberg.de.
Benjamin Goeppert, Email: Benjamin.Goeppert@med.uni-heidelberg.de.
Stefan Pusch, Email: s.pusch@Dkfz-Heidelberg.de.
Kai Breuhahn, Email: Kai.Breuhahn@med.uni-heidelberg.de.
Xin Wei Wang, Email: xw3u@nih.gov.
Peter Schirmacher, Email: Peter.Schirmacher@med.uni-heidelberg.de.
Stephanie Roessler, Email: Stephanie.Roessler@med.uni-heidelberg.de.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 5.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 7.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 9.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grivennikov S, Karin M. Autocrine IL-6 signaling: a key event in tumorigenesis? Cancer Cell. 2008;13:7–9. doi: 10.1016/j.ccr.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa H, Maeda S, Yoshida H, Tateishi R, Masuzaki R, Ohki T, Hayakawa Y, et al. Serum IL-6 levels and the risk for hepatocarcinogenesis in chronic hepatitis C patients: an analysis based on gender differences. Int J Cancer. 2009;125:2264–2269. doi: 10.1002/ijc.24720. [DOI] [PubMed] [Google Scholar]
- 12.Wong VW, Yu J, Cheng AS, Wong GL, Chan HY, Chu ES, Ng EK, et al. High serum interleukin-6 level predicts future hepatocellular carcinoma development in patients with chronic hepatitis B. Int J Cancer. 2009;124:2766–2770. doi: 10.1002/ijc.24281. [DOI] [PubMed] [Google Scholar]
- 13.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 15.Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, Tsuji S, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 16.Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, Caldas C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solimini NL, Xu Q, Mermel CH, Liang AC, Schlabach MR, Luo J, Burrows AE, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue W, Kitzing T, Roessler S, Zuber J, Krasnitz A, Schultz N, Revill K, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci U S A. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966. e912. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Li W, Lu J, Liu H, Li Y, Zhao Y. SH2D4A regulates cell proliferation via the ERalpha/PLC-gamma/PKC pathway. BMB Rep. 2009;42:516–522. doi: 10.5483/bmbrep.2009.42.8.516. [DOI] [PubMed] [Google Scholar]
- 22.Kioka N, Sakata S, Kawauchi T, Amachi T, Akiyama SK, Okazaki K, Yaen C, et al. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J Cell Biol. 1999;144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizutani K, Ito H, Iwamoto I, Morishita R, Deguchi T, Nozawa Y, Asano T, et al. Essential roles of ERK-mediated phosphorylation of vinexin in cell spreading, migration and anchorage-independent growth. Oncogene. 2007;26:7122–7131. doi: 10.1038/sj.onc.1210512. [DOI] [PubMed] [Google Scholar]
- 24.Rebouissou S, Amessou M, Couchy G, Poussin K, Imbeaud S, Pilati C, Izard T, et al. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457:200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 26.Aziz MH, Hafeez BB, Sand JM, Pierce DB, Aziz SW, Dreckschmidt NE, Verma AK. Protein kinase Cvarepsilon mediates Stat3Ser727 phosphorylation, Stat3-regulated gene expression, and cell invasion in various human cancer cell lines through integration with MAPK cascade (RAF-1, MEK1/2, and ERK1/2) Oncogene. 2010;29:3100–3109. doi: 10.1038/onc.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin HR, Kim HJ, Kim JY, Hurt EM, Klarmann GJ, Kawasaki BT, Duhagon Serrat MA, et al. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 2008;68:7736–7741. doi: 10.1158/0008-5472.CAN-08-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Lafdil F, Wang L, Park O, Yin S, Niu J, Miller AM, et al. Hepatoprotective versus oncogenic functions of STAT3 in liver tumorigenesis. Am J Pathol. 2011;179:714–724. doi: 10.1016/j.ajpath.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogt M, Domoszlai T, Kleshchanok D, Lehmann S, Schmitt A, Poli V, Richtering W, et al. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J Cell Sci. 2011;124:900–909. doi: 10.1242/jcs.072520. [DOI] [PubMed] [Google Scholar]
- 30.Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 31.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tujague M, Thomsen JS, Mizuki K, Sadek CM, Gustafsson JA. The focal adhesion protein vinexin alpha regulates the phosphorylation and activity of estrogen receptor alpha. J Biol Chem. 2004;279:9255–9263. doi: 10.1074/jbc.M312160200. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto T, Matsuda T, Junicho A, Kishi H, Saatcioglu F, Muraguchi A. Cross-talk between signal transducer and activator of transcription 3 and estrogen receptor signaling. FEBS Lett. 2000;486:143–148. doi: 10.1016/s0014-5793(00)02296-1. [DOI] [PubMed] [Google Scholar]
- 34.Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 35.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15:79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang CH, Xu GL, Jia WD, Li JS, Ma JL, Ren WH, Ge YS, et al. Activation of STAT3 signal pathway correlates with twist and E-cadherin expression in hepatocellular carcinoma and their clinical significance. J Surg Res. 2012;174:120–129. doi: 10.1016/j.jss.2010.10.030. [DOI] [PubMed] [Google Scholar]
- 37.Grabner B, Schramek D, Mueller KM, Moll HP, Svinka J, Hoffmann T, Bauer E, et al. Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nat Commun. 2015;6:6285. doi: 10.1038/ncomms7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pencik J, Schlederer M, Gruber W, Unger C, Walker SM, Chalaris A, Marie IJ, et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat Commun. 2015;6:7736. doi: 10.1038/ncomms8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sehgal PB, Guo GG, Shah M, Kumar V, Patel K. Cytokine signaling: STATS in plasma membrane rafts. J Biol Chem. 2002;277:12067–12074. doi: 10.1074/jbc.M200018200. [DOI] [PubMed] [Google Scholar]
- 40.Shah M, Patel K, Mukhopadhyay S, Xu F, Guo G, Sehgal PB. Membrane-associated STAT3 and PY-STAT3 in the cytoplasm. J Biol Chem. 2006;281:7302–7308. doi: 10.1074/jbc.M508527200. [DOI] [PubMed] [Google Scholar]
- 41.Rossi JF, Lu ZY, Jourdan M, Klein B. Interleukin-6 as a therapeutic target. Clin Cancer Res. 2015;21:1248–1257. doi: 10.1158/1078-0432.CCR-14-2291. [DOI] [PubMed] [Google Scholar]
- 42.Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, Wang K, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maheshwari S, Sarraj A, Kramer J, El-Serag HB. Oral contraception and the risk of hepatocellular carcinoma. J Hepatol. 2007;47:506–513. doi: 10.1016/j.jhep.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Wang LH, Yang XY, Mihalic K, Xiao W, Li D, Farrar WL. Activation of estrogen receptor blocks interleukin-6-inducible cell growth of human multiple myeloma involving molecular cross-talk between estrogen receptor and STAT3 mediated by co-regulator PIAS3. J Biol Chem. 2001;276:31839–31844. doi: 10.1074/jbc.M105185200. [DOI] [PubMed] [Google Scholar]
- 45.Hou J, Xu J, Jiang R, Wang Y, Chen C, Deng L, Huang X, et al. Estrogen-sensitive PTPRO expression represses hepatocellular carcinoma progression by control of STAT3. Hepatology. 2013;57:678–688. doi: 10.1002/hep.25980. [DOI] [PubMed] [Google Scholar]
- 46.Brunner SM, Rubner C, Kesselring R, Martin M, Griesshammer E, Ruemmele P, Stempfl T, et al. Tumor-infiltrating, interleukin-33-producing effector-memory CD8(+) T cells in resected hepatocellular carcinoma prolong patient survival. Hepatology. 2015;61:1957–1967. doi: 10.1002/hep.27728. [DOI] [PubMed] [Google Scholar]
- 47.Li KK, Adams DH. Antitumor CD8+ T cells in hepatocellular carcinoma: present but exhausted. Hepatology. 2014;59:1232–1234. doi: 10.1002/hep.26779. [DOI] [PubMed] [Google Scholar]
- 48.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.