Abstract
Yin-deficiency-heat (YDH) syndrome is a concept in Traditional Chinese Medicine (TCM) for describing subhealth status. However, there are few efficient diagnostic methods available for confirming YDH syndrome. To explore the novel method for diagnosing YDH syndrome, we applied iTRAQ to observe the serum protein profiles in YDH syndrome rats and confirmed protein levels by ELISA. A total of 92 differentially expressed proteins (63 upregulated proteins and 29 downregulated proteins), which were mainly involved in complement and coagulation cascades and glucose metabolism pathway, were identified by the proteomic experiments. Kininogen 1 (KNG1) was significantly increased (p < 0.0001), while apolipoprotein C-III (APOC3, p < 0.005) and paraoxonase 1 (PON1, p < 0.001) were significantly decreased in the serum of YDH syndrome rats. The combination of KNG1, APOC3, and PON1 constituted a diagnostic model with 100.0% sensitivity and 85.0% specificity. The results indicated that KNG1, APOC3, and PON1 may act as potential biomarkers for diagnosing YDH syndrome. KNG1 may regulate cytokines and chemokines release in YDH syndrome, and the low levels of PON1 and APOC3 may increase oxidative stress and lipolysis in YDH syndrome, respectively. Our work provides a novel method for YDH syndrome diagnosis and also provides valuable experimental basis to understand the molecular mechanism of YDH syndrome.
1. Introduction
Yin-deficiency-heat (YDH) syndrome, also known as the pathological condition “internal heat due to Yin deficiency,” is a common subhealth status in Traditional Chinese Medicine (TCM). YDH syndrome occurs frequently in individuals with Yin deficiency constitution, which is the fourth most common pathological constitution in general population [1]. Nowadays, YDH syndrome presents a great challenge in China, prevailing especially among white collar workers and college students [2]. YDH syndrome among individuals aged between 15 and 34 years has been shown to be more common than among other age groups [1]. Individuals with YDH syndrome present with deterioration in physical function, fatigue, weakness, emaciation, five-center (the palms, soles, and chest) heat, and tidal fever, which negatively affect the quality of life and the work productivity.
YDH syndrome could be relieved by Chinese herbal compound, and if diagnosed and treated promptly, it can be prevented from developing into disease. However, as a subhealth status without any obvious manifestations, the diagnosis of YDH syndrome has always been a tough problem. Currently, the traditional clinical diagnosis of YDH syndrome is based on subjective observation, which depends heavily on the clinical experience or knowledge of practitioner in TCM. In addition, questionnaires are commonly used to identify YDH syndrome, whereas the questions which are ambiguous or difficult-to-distinguish may lead to the bias in the test. The current methods for the diagnosis of YDH syndrome are lack of scientific basis. Therefore, novel methods suitable for YDH syndrome diagnosis are urgently needed.
Given the noninvasiveness and easy accessibility, serum is frequently used to screen diagnostic biomarkers for many diseases. Furthermore, serum contains approximately 10,000 proteins from cells and tissues [3], and the alteration of protein level sensitively reflects different diseases or pathological conditions. Former studies have successfully identified serum biomarkers for the diagnosis of breast carcinoma [4], colorectal cancer hepatic metastasis [5], and pathological characteristics of pulmonary TB with TCM syndromes [6]. In this study, we applied iTRAQ labeling coupled with two-dimensional liquid chromatography-tandem mass spectrometry (2D LCMS/MS) to investigate differentially expressed proteins in the serum of YDH syndrome rats. Further bioinformatics analysis revealed that differentially expressed proteins KNG1, APOC3, and PON1 were closely related to YDH syndrome and may act as potential biomarkers for diagnosis of YDH syndrome. Our results suggest the potential role of KNG1, APOC3, and PON1 in the diagnosis of YDH syndrome and provide important insights to understand the molecular mechanism of YDH syndrome.
2. Materials and Methods
2.1. Experimental Design
The schematic of the experimental workflow is shown in Figure 1. Serum samples from YDH syndrome rats and normal rats were subjected to iTRAQ-coupled 2D LC-MS/MS analysis. Bioinformatics approaches were used to explore the function of differentially expressed proteins. The expression levels of proteins were measured by ELISA.
Figure 1.
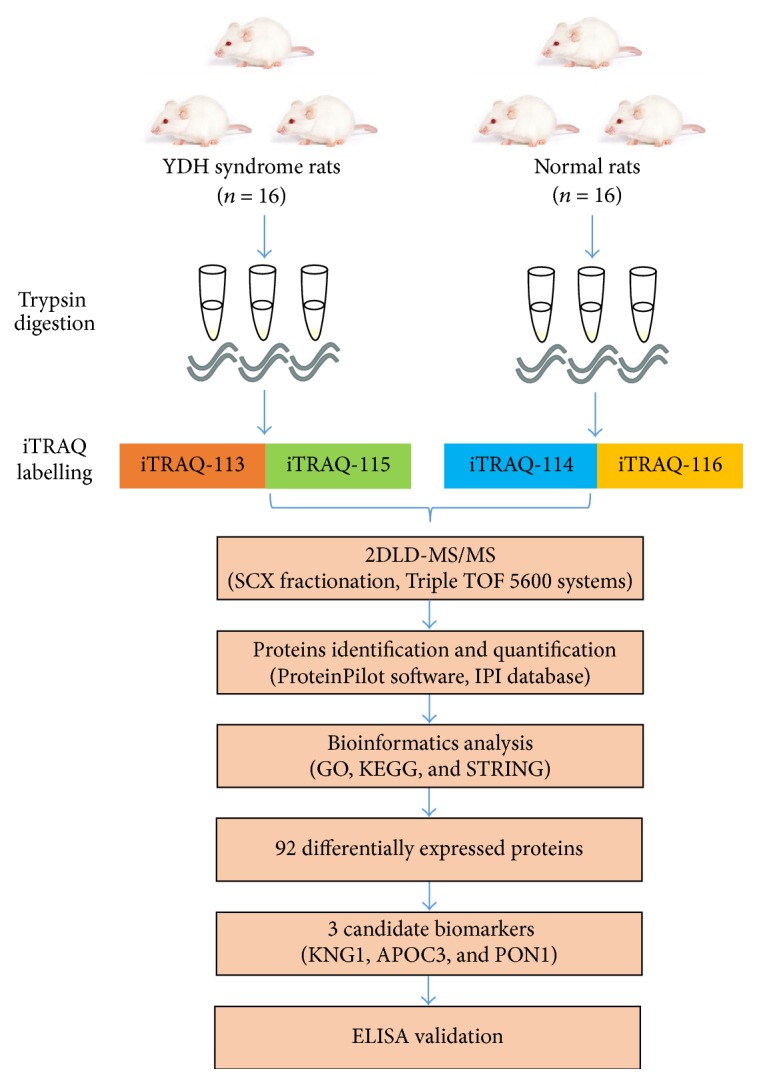
Schematic of the workflow design based on iTRAQ-2DLC-MS/MS analysis of YDH-related proteins in rats.
2.2. Animals and Treatment
SPF female Sprague-Dawley rats, weighing 180–220 g, were purchased from the Experimental Animal Center of Zhejiang Province (China). All rats were housed at a constant temperature of 23 ± 1°C and a 12 h light/dark cycle with free access to water and standard rat diet. All experimental procedures were approved by the Zhejiang University Institutional Animal Care and Use Committee. The pungent and hot Chinese herbs, Fuzi (Radix Aconiti Praeparata), Ganjiang (Rhizoma Zingiberis), and Rougui (Cinnamomum cassia) were purchased from the Hangzhou Hu Qingyu Drugstore (China). These herbs with warm nature consume Yin fluid, which is commonly used in TCM to induce YDH syndrome animal models [7]. Equal amounts of Fuzi, Ganjiang, and Rougui were mixed, decocted, and concentrated to a final concentration of 4 g/mL according to the previously described method [8]. After acclimation for 7 days, rats were randomly divided into the model group (herbal decoction, 2 mL/100 g/d, by gastrogavage) and the control group (sterile saline solution, 2 mL/100 g/d, by gastrogavage). All rats were sacrificed after 3 weeks of treatment, and serum samples were collected and stored at −80°C. Thymus, spleen, and adrenal gland were immediately removed and weighed. The anesthetized rats were euthanized by cervical dislocation.
2.3. iTRAQ-2D LC-MS/MS
Sample pooling is commonly used in proteomic studies to increase the precision and accuracy of the results [9]. Serum samples from the model group (n = 16) and the control group (n = 16) were subjected to iTRAQ labeling according to the manufacturer's instructions (Applied Biosystems, Foster city, CA, USA), and 16 samples in each group were randomly separated into two subgroups to establish two biological replicates, so 4 pooling samples were generated. To enrich the low abundant protein, Agilent MARS-14 column (Agilent Technologies, Santa Clara, CA, USA) was used to remove 14 highly abundant proteins, including albumin, IgG, antitrypsin, IgA, transferring, haptoglobin, fibrinogen, alpha 2-macroglobulin, alpha 1-acid glycoprotein, IgM, apolipoprotein AI, apolipoprotein AII, complement C3, and transthyretin, and the eluent of protein samples were quantified by the Bradford method. The protein samples (100 μg) of each group were reduced, alkylated, and digested with trypsin (Sigma, St. Louis, MO, USA) overnight at 37°C. Then, protein samples from the control group were labeled with reporter tags 114 and 116 and protein samples from the YDH model group were labeled with reporter tags 113 and 115. The labeled peptides were pooled and desalted and then dried by a rotation vacuum concentrator (Christ RVC 2-25, Christ, Osterode, Germany).
The iTRAQ-labeled peptides were fractionated by strong cation exchange (SCX) liquid chromatography using a polysulfoethyl column (2.1 × 100 mm, 5 μm, 200 Ǻ; Nest Group, Inc., Southborough, MA, USA). A gradient of buffer A (10 mM KH2PO4, 25% ACN, pH 2.6) and buffer B (350 mM KCl, 10 mM KH2PO4, 25% ACN, pH 2.6) was used to elute the peptides adsorbed on the column. The concentrations of the eluted peptides were monitored by measuring the absorbance at 214 nm [10]. A total of ten SCX fractions were collected and concentrated. Each fraction was dissolved and subjected to Reverse-phase LC fractionation. The LC elution was subsequently analyzed on a Triple TOF 5600 system (Applied Biosystems) in duplicate as two technical replicates. In an information-dependent acquisition mode (IDA), the survey scans were acquired from 400 to 1500 m/z with up to 20 most intense multiply charged ions selected for MS/MS analysis [11]. The product ion spectra were accumulated in the 100–2000 m/z to enhance the intensities of the iTRAQ reporter ions (113, 114, 115, and 116 m/z) for quantification. The relative abundance of the peptides and proteins was reflected by the ratio of the peak area of the iTRAQ reporter ions intensities [12].
The data of MS/MS spectra were searched against the IPI rat database (IPI.rat.v3.69, 39 578 entries) with ProteinPilot™ 2.0.1 software (Applied Biosystems) [13]. ProteinPilot was searched with a fragment ion mass tolerance of 0.050 Da and a parent ion tolerance of 10.0 ppm. Scaffold (version Scaffold_4.6.2, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could establish an FDR less than 1.0% by the Scaffold Local FDR algorithm and contained at least 2 identified peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Scaffold Q+ (version Scaffold_4.6.2, Proteome Software Inc., Portland, OR) was used to quantitate Label Based Quantitation (iTRAQ, TMT, SILAC, etc.) peptide and protein identifications. Normalization was performed iteratively (across samples and spectra) on intensities, as described in Statistical Analysis of Relative Labeled Mass Spectrometry Data from Complex Samples Using ANOVA [14]. Medians were used for averaging. Spectra data were log-transformed, pruned of those matched to multiple proteins, and weighted by an adaptive intensity weighting algorithm. p values less than 0.05 in each independent iTRAQ experiment and the fold-changes ratio lower than 0.8 or higher than 1.25 were considered as significant [15, 16].
2.4. Bioinformatics Analysis
We annotated function or feature of proteins from several different categories, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) software. The molecular function, cellular component, and biological process were analyzed by searching GO database (http://geneontology.org/). The signal pathways of proteins were annotated by KEGG pathway database (http://www.genome.jp/kegg/mapper.html). The interaction networks of the identified proteins were analyzed by STRING software (http://string-db.org/).
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
Kininogen 1 (KNG1) Rat ELISA Kit (Abcam, detection limit 4.97 ng/mL), Apoc3 (Rat/Mouse) ELISA Kit (Abnova, detection limit 0.3 g/mL), and rat paraoxonase 1 (PON1) ELISA Kit (Cusabio Biotech, Wuhan, Hubei, China; detection limit 15.6 U/mL) were used to measure the expression levels of proteins. Briefly, serum samples were incubated in microtiter wells with appropriate dilution according to the manufacturer's instructions. After discarding the solution and washing, biotinylated antibody, HRP-streptavidin, substrate reagent, and stop solution were sequentially added to the wells. Finally, absorbance was read at wavelength of 450 nm immediately. The expression levels of the proteins were calculated by a four-parameter logistic curve (Microplate manager 6 software, Bio-Rad).
2.6. Statistical Analysis
Experimental data were presented as mean ± standard deviation (SD). Statistical analyses were performed with SPSS software, version 18.0 (SPSS, Chicago, IL, USA) and p value of less than 0.05 was considered statistically significant. Nonparametric analyses were performed using the Mann–Whitney U test. Receiver operating characteristic (ROC) curves and logistic regression models were performed using MedCalc Software (Version 12.4.2.0, Belgium).
3. Results
3.1. Characteristics of YDH Syndrome Rats
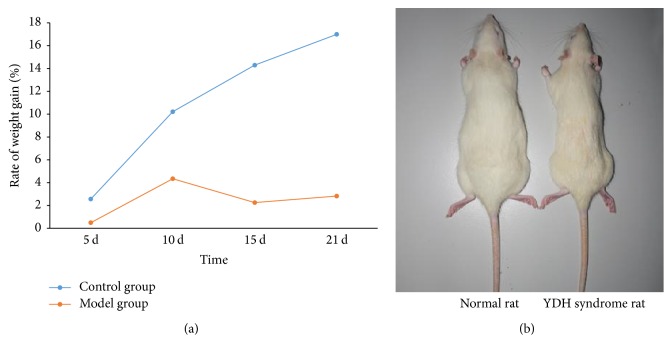
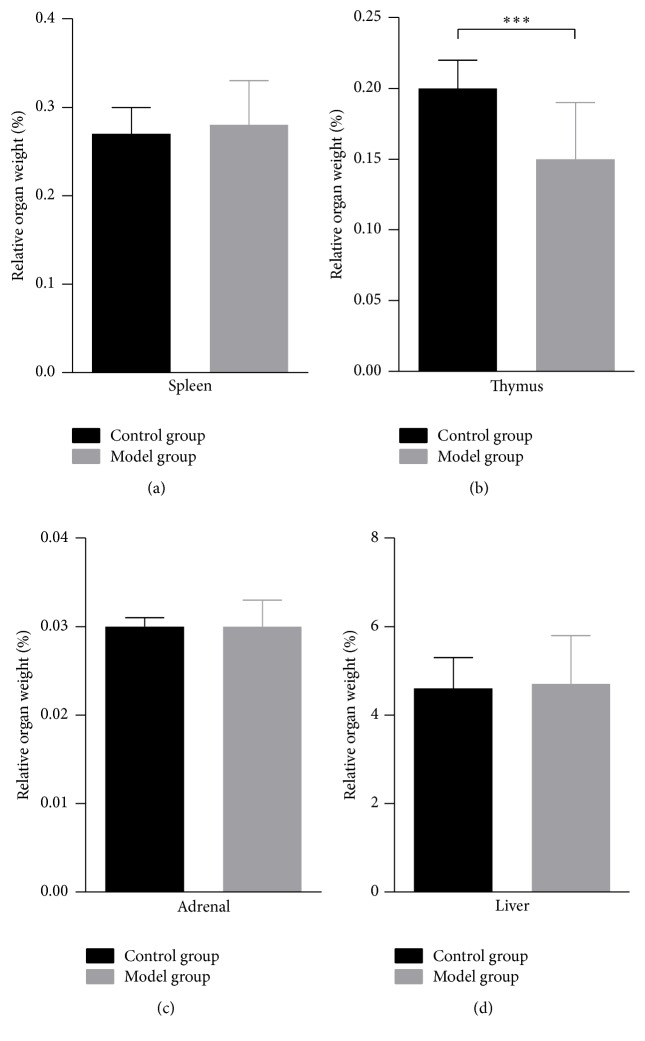
Compared with the normal group, rats in the model group showed obvious YDH symptoms, including dry hair, restlessness, dry stool, and weight loss. The rate of weight gain in the YDH syndrome rats was consistently lower than that of the control group (Figure 2(a)). At the end of the third week, rats in the YDH model group appeared much smaller than the normal rats (Figure 2(b)), and the relative thymus weight decreased significantly in the model group compared with the normal group (p < 0.001, Figure 3(b)). However, the relative weights of the spleen, adrenal gland, and liver showed no remarkable changes (Figures 3(a), 3(c), and 3(d)).
Figure 2.
(a) The rate of weight gain in the normal rats and the YDH syndrome rats at each time point. (b) Gross appearance of the YDH syndrome rat compared to the normal rat at the end of the experiment.
Figure 3.
The comparison of relative organs weight between the normal rats and YDH syndrome rats. Values are means ± SD. p value of less than 0.05 indicates statistical significance using the Mann–Whitney U test. ∗∗∗ p < 0.001. (a) The relative weight of the spleen in each group. (b) The relative weight of the thymus in each group. (c) The relative weight of the adrenal gland in each group. (d) The relative weight of the liver in each group.
3.2. Proteomic and Bioinformatics Analysis
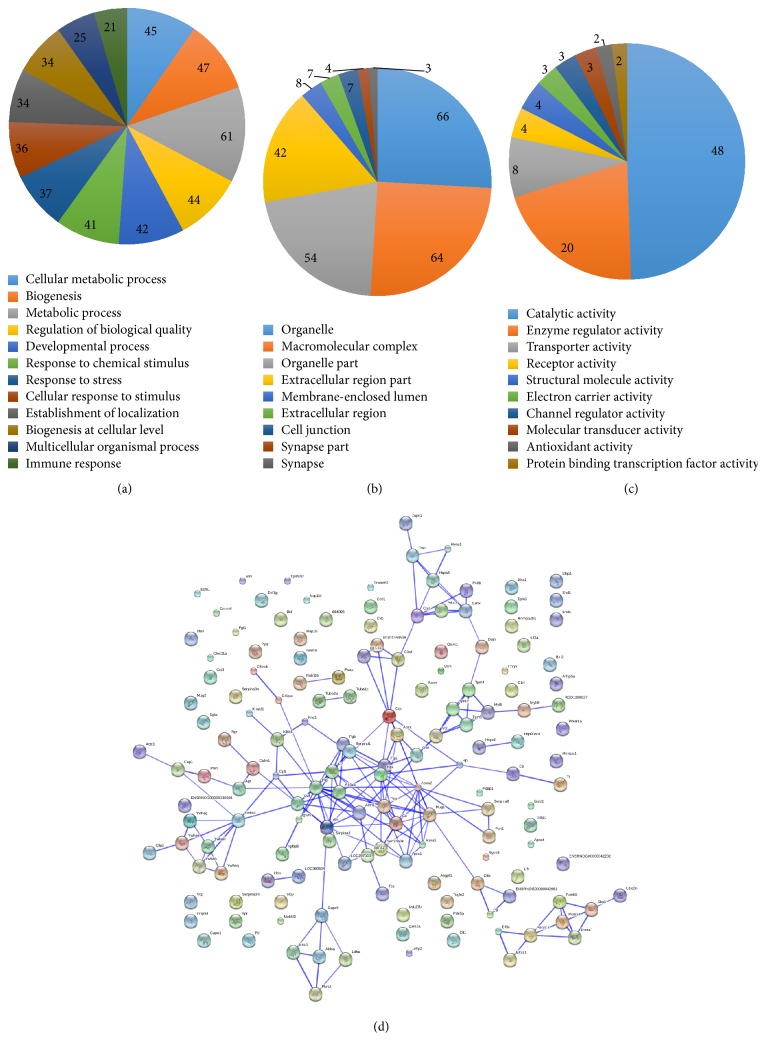
The intensity distribution for all channels was shown in Supplementary Figure 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/5176731, and higher values represent better signal intensities. The deviation of the MS data was less than 2.5 ppm, indicating a high degree of accuracy (Supplementary Figure 2). The distribution of discriminant score calculated by Scaffold software in PeptideProphet algorithm was shown in Supplementary Figure 3, and the channel signals before and after the normalization of the channel distribution were shown in Supplementary Figure 4. A total of 165 differentially expressed proteins were identified in the serum of YDH syndrome rats using iTRAQ-2D LC-MS/MS, 127, and 130 proteins in two replicates, respectively. 92 differentially expressed proteins were screened in both replicates, including 63 upregulated proteins (>1.25-fold, p < 0.05, Table 1) and 29 downregulated proteins (<0.8-fold, p < 0.05, Table 2). GO annotation was used to analyze the cellular component, molecular function, and participation in biological processes of these 92 distinct proteins. In the biological processes analysis, we found that a majority of differentially expressed proteins were associated with metabolic process (48, 36%; Figure 4(a)), indicating that metabolic abnormalities were closely related to the YDH syndrome. Cellular component analysis revealed that the differentially expressed proteins were mainly localized to the organelle (66, 26%), macromolecular complex (64, 25%), organelle part (54, 21%), and extracellular region part (42, 16%; Figure 4(b)). Function-based enrichment analysis indicated that most of these proteins played important roles in catalyzing (48, 50%) and enzyme regulating (20, 21%; Figure 4(c)). KEGG pathway annotation revealed a greater relative abundance of proteins involved in immune system (especially in complement and coagulation cascades, Table 3) and carbohydrate metabolism (especially in glycolysis and gluconeogenesis, Table 4), indicating that the YDH syndrome was closely related to the abnormality of immunoreaction and glycometabolism. Furthermore, STRING analysis showed that most of the differentially expressed proteins interacted with each other (Figure 4(d)).
Table 1.
Upregulated serum proteins in YDH syndrome group quantified by iTRAQ-based proteomics and the ratios to the control group.
| Protein ID | Gene name | Protein name | M/C (114/113) | M/C (116/115) | ||||
|---|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Mean | Run 1 | Run 2 | Mean | |||
| P62161 | Calm1 | Calmodulin | 8.09 | 16.75 | 12.42 | 12.82 | 6.08 | 9.45 |
| P62260 | Ywhae | 14-3-3 protein epsilon | 2.68 | 6.31 | 4.49 | 15.28 | 14.59 | 14.93 |
| Q6AYZ1 | Tuba1c | Tubulin alpha-1C chain | 9.29 | 4.79 | 7.04 | 3.91 | 17.22 | 10.56 |
| P45592 | Cfl1 | Cofilin-1 | 4.37 | 3.44 | 3.90 | 11.07 | 13.93 | 12.50 |
| P01048 | Map1 | T-kininogen 1 | 5.50 | 4.57 | 5.03 | 16.29 | 5.65 | 10.97 |
| P09006 | Serpina3n | Serine protease inhibitor A3N | 2.99 | 2.86 | 2.92 | 11.91 | 13.18 | 12.55 |
| P08932 | — | T-kininogen 2 | 3.91 | 4.79 | 4.35 | 14.72 | 6.14 | 10.43 |
| Q62812 | Myh9 | Myosin-9 | 7.11 | 9.55 | 8.33 | 6.79 | 6.03 | 6.41 |
| P18418 | Calr | Calreticulin | 4.13 | 2.75 | 3.44 | 8.55 | 9.73 | 9.14 |
| Q66HD0 | Hsp90b1 | Endoplasmin | 3.02 | 6.25 | 4.64 | 13.55 | 2.03 | 7.79 |
| P34058 | Hsp90ab1 | Heat shock protein HSP 90-beta | 4.25 | 9.64 | 6.94 | 5.25 | 5.50 | 5.37 |
| P68511 | Ywhah | 14-3-3 protein eta | 3.84 | 1.46 | 2.65 | 13.68 | 5.06 | 9.37 |
| P04276 | Gc | Vitamin D-binding protein | 6.61 | 4.45 | 5.53 | 5.65 | 6.03 | 5.84 |
| Q62636 | Rap1b | Ras-related protein Rap-1b | 3.19 | 2.58 | 2.89 | 8.71 | 7.66 | 8.18 |
| Q9Z1P2 | Actn1 | Alpha-actinin-1 | 2.99 | 2.63 | 2.81 | 6.79 | 8.79 | 7.79 |
| P09495 | Tpm4 | Tropomyosin alpha-4 chain | 2.81 | 2.51 | 2.66 | 6.14 | 9.12 | 7.63 |
| Q6PCT3 | Tpd52l2 | Tumor protein D54 | 1.71 | 6.31 | 4.01 | 4.41 | 7.87 | 6.14 |
| P11980 | Pkm | Pyruvate kinase isozymes M1/M2 | 1.82 | 2.56 | 2.19 | 11.27 | 4.57 | 7.92 |
| Q08163 | Cap1 | Adenylyl cyclase-associated protein 1 | 3.22 | 3.94 | 3.58 | 4.17 | 8.39 | 6.28 |
| P11598 | Pdia3 | Protein disulfide-isomerase A3 | 11.17 | 3.16 | 7.17 | 2.13 | 2.27 | 2.20 |
| P16086 | Sptan1 | Spectrin alpha chain | 2.38 | 3.16 | 2.77 | 5.92 | 6.92 | 6.42 |
| P62963 | Pfn1 | Profilin-1 | 3.13 | 2.31 | 2.72 | 4.79 | 7.45 | 6.12 |
| Q68FR2 | Bin2 | Bridging integrator 2 | 2.25 | 2.17 | 2.21 | 6.03 | 6.08 | 6.05 |
| Q63610 | Tpm3 | Tropomyosin alpha-3 chain | 2.21 | 2.70 | 2.46 | 5.30 | 6.25 | 5.77 |
| P06866 | Hp | Haptoglobin | 7.11 | 3.50 | 5.31 | 2.75 | 3.05 | 2.90 |
| P08934 | Kng1 | Kininogen-1 | 5.86 | 4.88 | 5.37 | 1.60 | 2.83 | 2.22 |
| B0BNA5 | Cotl1 | Coactosin-like protein | 2.56 | 2.25 | 2.40 | 5.65 | 4.53 | 5.09 |
| Q63081 | Pdia6 | Protein disulfide-isomerase A6 | 3.02 | 4.21 | 3.61 | 4.29 | 3.44 | 3.86 |
| P85972 | Vcl | Vinculin | 3.56 | 3.08 | 3.32 | 4.09 | 3.94 | 4.02 |
| P04642 | Ldha | L-lactate dehydrogenase A chain | 4.41 | 1.67 | 3.04 | 6.61 | 1.82 | 4.21 |
| P63102 | Msfs1 | 14-3-3 protein zeta/delta | 2.81 | 1.84 | 2.32 | 5.20 | 4.53 | 4.86 |
| B2GUZ5 | Capza1 | F-actin-capping protein subunit alpha-1 | 2.23 | 1.72 | 1.98 | 3.73 | 6.49 | 5.11 |
| P46462 | Vcp | Transitional endoplasmic reticulum ATPase | 2.99 | 2.40 | 2.70 | 2.65 | 5.45 | 4.05 |
| P20767 | — | Ig lambda-2 chain C region | 2.25 | 2.05 | 2.15 | 2.58 | 6.55 | 4.56 |
| Q5XFX0 | Tagln2 | Transgelin-2 | 3.63 | 2.07 | 2.85 | 2.81 | 4.37 | 3.59 |
| Q4V7E8 | Lrrfip2 | Flightless-interacting protein 2 | 2.33 | 1.96 | 2.15 | 4.06 | 3.98 | 4.02 |
| Q63617 | Hyou1 | Hypoxia upregulated protein 1 | 1.79 | 5.97 | 3.88 | 1.75 | 2.58 | 2.17 |
| Q62930 | C9 | Complement component C9 | 3.63 | 2.83 | 3.23 | 2.25 | 3.25 | 2.75 |
| Q5XI73 | Arhgdia | Rho GDP-dissociation inhibitor 1 | 1.74 | 2.99 | 2.37 | 2.96 | 4.02 | 3.49 |
| P04797 | Gapdh | Dehydrogenase | 1.77 | 1.51 | 1.64 | 3.50 | 4.92 | 4.21 |
| Q07009 | Capn2 | Calpain-2 catalytic subunit | 1.28 | 3.60 | 2.44 | 3.80 | 2.65 | 3.23 |
| Q5U211 | Snx3 | Sorting nexin-3 | 1.91 | 4.25 | 3.08 | 1.60 | 3.22 | 2.41 |
| Q63514 | C4bpa | C4b-binding protein alpha chain | 2.99 | 2.49 | 2.74 | 2.88 | 2.31 | 2.60 |
| Q64119 | Myl6 | Myosin light polypeptide 6 | 1.72 | 2.27 | 2.00 | 3.60 | 2.68 | 3.14 |
| P97571 | Capn1 | Calpain-1 catalytic | 3.73 | 2.15 | 2.94 | 2.11 | 1.61 | 1.86 |
| P20760 | Igg-2a | Ig gamma-2A chain C region | 1.50 | 1.84 | 1.67 | 2.63 | 3.22 | 2.93 |
| P06761 | Hspa5 | 78 kDa glucose-regulated protein | 1.56 | 1.46 | 1.51 | 3.53 | 2.56 | 3.05 |
| Q63515 | C4bpb | C4b-binding protein beta chain | 3.60 | 2.09 | 2.84 | 1.56 | 1.61 | 1.59 |
| Q7M0E3 | Dstn | Destrin | 3.16 | 1.29 | 2.23 | 2.61 | 1.58 | 2.10 |
| Q9EPH8 | Pabpc1 | Polyadenylate-binding protein 1 | 1.41 | 1.80 | 1.60 | 3.40 | 2.00 | 2.70 |
| P63018 | Hspa8 | Heat shock cognate 71 kDa protein | 1.60 | 2.19 | 1.89 | 2.63 | 2.17 | 2.40 |
| P04785 | P4hb | Protein disulfide-isomerase | 2.25 | 2.01 | 2.13 | 1.82 | 2.49 | 2.15 |
| Q9EQX9 | Ube2n | Ubiquitin-conjugating enzyme E2 N | 2.86 | 1.58 | 2.22 | 1.87 | 2.15 | 2.01 |
| P06399 | Fga | Fibrinogen alpha chain | 4.37 | 1.43 | 2.90 | 1.33 | 1.28 | 1.31 |
| P01835 | — | Ig kappa chain C region | 2.01 | 1.57 | 1.79 | 2.03 | 2.70 | 2.37 |
| P12346 | Tf | Serotransferrin | 1.60 | 1.91 | 1.75 | 2.54 | 2.03 | 2.28 |
| Q9JLT6 | Bid | BH3-interacting domain death agonist | 1.36 | 1.64 | 1.50 | 1.69 | 3.05 | 2.37 |
| P68101 | Eif2s1 | Eukaryotic translation initiation factor | 1.66 | 1.26 | 1.46 | 3.16 | 1.46 | 2.31 |
| P35213 | Ywhab | 14-3-3 protein beta/alpha | 1.85 | 1.28 | 1.57 | 2.65 | 1.49 | 2.07 |
| Q91ZN1 | Coro1a | Coronin-1A | 1.64 | 1.37 | 1.51 | 2.36 | 1.69 | 2.02 |
| P04906 | Gstp1 | Glutathione S-transferase P | 1.33 | 1.47 | 1.40 | 2.51 | 1.67 | 2.09 |
| P14272 | Klkb1 | Plasma kallikrein | 2.11 | 1.96 | 2.03 | 1.27 | 1.42 | 1.34 |
| P07335 | Ckb | Creatine kinase B-type | 1.41 | 1.28 | 1.34 | 1.57 | 1.38 | 1.48 |
M, model group; C, control group; 114/113 and 116/115, two biological replicates; Run 1 and Run 2, two technical replicates.
Table 2.
Downregulated serum proteins in YDH syndrome group quantified by iTRAQ-based proteomics and the ratios to the control group.
| Protein ID | Gene name | Protein name | M/C (114/113) | M/C (116/115) | ||||
|---|---|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Mean | Run 1 | Run 2 | Mean | |||
| P02091 | Hbb | Hemoglobin subunit beta-1 | 0.71 | 0.70 | 0.71 | 0.67 | 0.70 | 0.68 |
| P05545 | Serpina3k | Serine protease inhibitor A3K | 0.66 | 0.64 | 0.65 | 0.65 | 0.70 | 0.68 |
| Q6IE52 | Mug2 | Murinoglobulin-2 | 0.67 | 0.75 | 0.71 | 0.42 | 0.73 | 0.58 |
| P15978 | RT1-Aw2 | Class I histocompatibility antigen | 0.73 | 0.61 | 0.67 | 0.58 | 0.60 | 0.59 |
| P36953 | Afm | Afamin | 0.54 | 0.45 | 0.49 | 0.74 | 0.69 | 0.72 |
| Q62894 | Ecm1 | Extracellular matrix protein 1 | 0.77 | 0.55 | 0.66 | 0.52 | 0.51 | 0.52 |
| O88201 | Clec11a | C-type lectin domain family 11 member A | 0.77 | 0.53 | 0.65 | 0.32 | 0.72 | 0.52 |
| P55314 | C8b | Complement component C8 beta chain | 0.74 | 0.74 | 0.74 | 0.37 | 0.47 | 0.42 |
| P14630 | Apom | Apolipoprotein M | 0.64 | 0.44 | 0.54 | 0.53 | 0.60 | 0.57 |
| P35859 | Igfals | Insulin-like growth facto | 0.50 | 0.58 | 0.54 | 0.44 | 0.58 | 0.51 |
| P14046 | A1i3 | Alpha-1-inhibitor 3 | 0.48 | 0.56 | 0.52 | 0.67 | 0.34 | 0.50 |
| Q64268 | Serpind1 | Heparin cofactor 2 | 0.74 | 0.70 | 0.72 | 0.24 | 0.31 | 0.27 |
| Q68FP1 | Gsn | Gelsolin | 0.56 | 0.58 | 0.57 | 0.31 | 0.52 | 0.41 |
| Q00918 | Ltbp1 | Latent-transforming growth factor | 0.44 | 0.50 | 0.47 | 0.44 | 0.47 | 0.46 |
| P23680 | Apcs | Serum amyloid P-component | 0.73 | 0.69 | 0.71 | 0.22 | 0.21 | 0.22 |
| Q8R2H5 | Gpld1 | Phosphatidylinositol | 0.53 | 0.69 | 0.61 | 0.30 | 0.31 | 0.31 |
| P31211 | Serpina6 | Corticosteroid-binding globulin | 0.54 | 0.50 | 0.52 | 0.36 | 0.36 | 0.36 |
| Q9QUH3 | Apoa5 | Apolipoprotein A-V | 0.53 | 0.53 | 0.53 | 0.39 | 0.28 | 0.33 |
| P48199 | Crp | C-reactive protein | 0.72 | 0.72 | 0.72 | 0.10 | 0.19 | 0.14 |
| O70535 | Lifr | Leukemia inhibitory factor receptor | 0.33 | 0.40 | 0.36 | 0.46 | 0.52 | 0.49 |
| P55797 | Apoc4 | Apolipoprotein C-IV | 0.33 | 0.53 | 0.43 | 0.43 | 0.41 | 0.42 |
| P18292 | F2 | Prothrombin | 0.65 | 0.64 | 0.65 | 0.13 | 0.25 | 0.19 |
| P07092 | Serpine2 | Glia-derived nexin | 0.52 | 0.37 | 0.44 | 0.42 | 0.14 | 0.28 |
| P19939 | Apoc1 | Apolipoprotein C-I | 0.34 | 0.35 | 0.34 | 0.32 | 0.34 | 0.33 |
| P07808 | Npy | Pro-neuropeptide Y | 0.63 | 0.29 | 0.46 | 0.13 | 0.24 | 0.19 |
| O08770 | Gp5 | Platelet glycoprotein V | 0.37 | 0.56 | 0.46 | 0.17 | 0.14 | 0.15 |
| P55159 | Pon1 | Serum paraoxonase/arylesterase 1 | 0.27 | 0.19 | 0.23 | 0.31 | 0.24 | 0.28 |
| P06759 | Apoc3 | Apolipoprotein C-III | 0.41 | 0.28 | 0.35 | 0.13 | 0.11 | 0.12 |
| P04638 | Apoa2 | Apolipoprotein A-II | 0.11 | 0.13 | 0.12 | 0.25 | 0.27 | 0.26 |
M, model group; C, control group. 114/113 and 116/115, two biological replicates; Run 1 and Run 2, two technical replicates.
Figure 4.
Data mining of the set of differentially expressed proteins in YDH syndrome rats. (a) Biological process. (b) Cellular component. (c) Molecular function. (d) The interacted network of proteins was analyzed by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) software.
Table 3.
KEGG pathways analysis of the differentially expressed proteins categorized by organismal systems.
| Pathway | Number of matched proteins | Organismal systems | Pathway map |
|---|---|---|---|
| Complement and coagulation cascades | 9 | Immune system | rno04610 |
| Antigen processing and presentation | 7 | Immune system | rno04612 |
| Leukocyte transendothelial migration | 5 | Immune system | rno04670 |
| Fc gamma R-mediated phagocytosis | 4 | Immune system | rno04666 |
| NOD-like receptor signaling pathway | 2 | Immune system | rno04621 |
| B cell receptor signaling pathway | 1 | Immune system | rno04662 |
| Natural killer cell mediated cytotoxicity | 1 | Immune system | rno04650 |
| Intestinal immune network for IgA production | 1 | Immune system | rno04672 |
| Chemokine signaling pathway | 1 | Immune system | rno04062 |
| Fc epsilon RI signaling pathway | 1 | Immune system | rno04664 |
| Hematopoietic cell lineage | 1 | Immune system | rno04640 |
| Neurotrophin signaling pathway | 8 | Nervous system | rno04722 |
| Long-term potentiation | 1 | Nervous system | rno04720 |
| Vasopressin-regulated water reabsorption | 2 | Excretory system | rno04962 |
| Endocrine | 1 | Excretory system | rno04961 |
| Circadian rhythm - mammal | 1 | Environmental adaptation | rno04710 |
| Insulin signaling pathway | 3 | Endocrine system | rno04910 |
| Progesterone-mediated oocyte maturation | 1 | Endocrine system | rno04914 |
| Renin-angiotensin system | 1 | Endocrine system | rno04614 |
| PPAR signaling pathway | 2 | Endocrine system | rno03320 |
| Mineral absorption | 1 | Digestive system | rno04978 |
| Pancreatic secretion | 2 | Digestive system | rno04972 |
| Axon guidance | 1 | Development | rno04360 |
| Cardiac muscle contraction | 4 | Circulatory system | rno04260 |
Table 4.
KEGG pathways analysis of the differentially expressed proteins categorized by metabolism.
| KEGG categories | Number of matched proteins | Metabolism | Pathway map |
|---|---|---|---|
| Glycolysis/gluconeogenesis | 4 | Carbohydrate metabolism | rno00010 |
| Pyruvate metabolism | 2 | Carbohydrate metabolism | rno00620 |
| Starch and sucrose metabolism | 1 | Carbohydrate metabolism | rno00500 |
| Fructose and mannose metabolism | 1 | Carbohydrate metabolism | rno00051 |
| Propanoate metabolism | 1 | Carbohydrate metabolism | rno00640 |
| Pentose phosphate pathway | 1 | Carbohydrate metabolism | rno00030 |
| Glycosylphosphatidylinositol- (GP-I) anchor biosynthesis | 1 | Glycan biosynthesis and metabolism | rno00563 |
| Arginine and proline metabolism | 1 | Amino acid metabolism | rno00330 |
| Drug metabolism, cytochrome P450 | 1 | Xenobiotics biodegradation and metabolism | rno00982 |
| Folate biosynthesis | 1 | Metabolism of cofactors and vitamins | rno00790 |
| Glutathione metabolism | 1 | Metabolism of other amino acids | rno00480 |
| Purine metabolism | 2 | Nucleotide metabolism | rno00230 |
| Metabolism of xenobiotics by cytochrome P450 | 1 | Xenobiotics biodegradation and metabolism | rno00980 |
| Cysteine and methionine metabolism | 1 | Amino acid metabolism | rno00270 |
| Valine, leucine, and isoleucine biosynthesis | 1 | Amino acid metabolism | rno00290 |
3.3. Validation of Proteins by ELISA
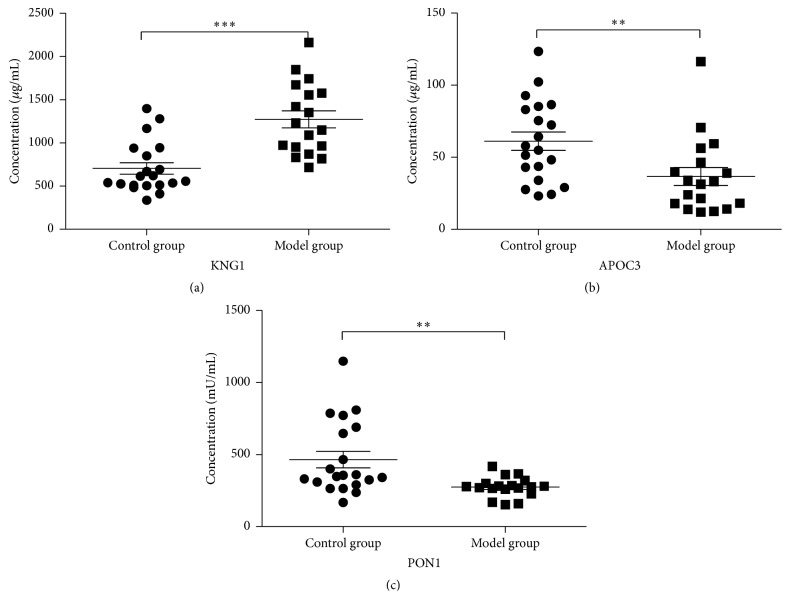
According to the results of the bioinformatics analysis, several abnormally expressed proteins (KNG1, APOC3, and PON1) were validated by ELISA. The results showed that the serum levels of KNG1 in YDH syndrome rats (1271.80 ± 413.65 μg/mL) were significantly higher (p < 0.0001) than that in normal rats (702.89 ± 296.43 μg/mL) (Figure 5(a)). The serum levels of APOC3 in YDH syndrome rats and normal rats were 36.60 ± 26.43 μg/mL and 61.04 ± 28.32 μg/mL, respectively (Figure 5(b)), and the serum levels of PON1 in YDH syndrome rats and normal rats were 274.76 ± 69.23 mU/mL and 465.73 ± 254.91 mU/mL, respectively (Figure 5(c)). These data indicated that the levels of APOC3 (p < 0.005) and PON1 (p < 0.001) were significantly decreased in YDH syndrome rats. The results were in line with the iTRAQ data.
Figure 5.
Validation of KNG, APOC3, and PON1 in serum. Levels of these proteins were measured by ELISA in serum of the control group (n = 20) and the model group (n = 18). Median values are shown by a horizontal line. p values were calculated with the Mann–Whitney U test. ∗∗∗ p < 0.001; ∗∗ p < 0.01.
3.4. ROC Analysis
The data of serum protein levels (18 YDH syndrome rats and 20 normal rats) were subjected to forward stepwise multivariate regression analysis. The results indicated that KNG1, APOC3, and PON1 were all included in the diagnostic model as follows:
| (1) |
Odds ratios for KNG1, APOC3, and PON1 in the model were 1.006, 0.991, and 0.986, respectively. The area under the ROC curve (AUC) in the diagnostic model was 0.950 (95% CI, 0.826–0.994, p < 0.0001, cutoff value was 0.3154). It was much higher than the AUC of KNG1 (0.886, 95% Cl, 0.741–0.966, p < 0.0001), APOC3 (0.771, 95% Cl, 0.606–0.891, p = 0.0006), and PON1 (0.767, 95% Cl, 0.601–0.888, p = 0.0009) alone (Figure 6). The sensitivity and specificity of KNG1, APOC3, and PON1 for detection of YDH syndrome were 100.0% and 70.0%, 72.2% and 75.0%, and 83.3% and 70.0%, respectively. The sensitivity and specificity of the diagnostic model (KNG1, APOC3, and PON1 combination) were 100.0% and 85.0%. In addition, the positive values and the negative predictive values for YDH syndrome diagnosis were also improved by the combination of KNG1, APOC3, and PON1 (Table 5).
Figure 6.
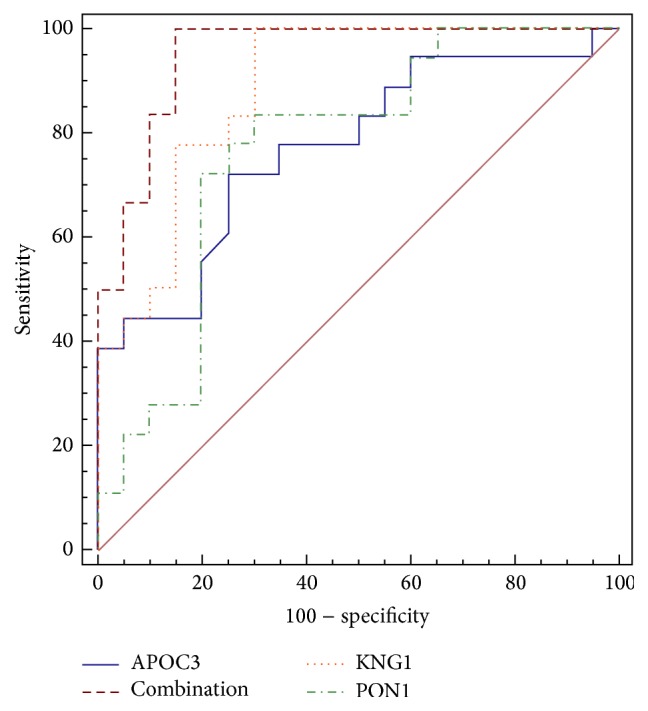
The receiver operation characteristics (ROC) curve analyses. ROC curve analyses of the serum protein KNG1, APOC3, and PON1 as well as the combination of the three proteins to discriminate YDH syndrome from controls.
Table 5.
Diagnostic value for YDH syndrome detection of the individual markers and KNG1, APOC3, and PON1 combination.
| Protein | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | AUC | 95% CI | p value |
|---|---|---|---|---|---|---|---|
| KNG1 | 100.0 | 70.0 | 75.0 | 100.0 | 0.886 | 0.741–0.966 | p < 0.0001 |
| APOC3 | 72.0 | 75.0 | 72.2 | 75.0 | 0.771 | 0.606–0.891 | p = 0.0006 |
| PON1 | 83.3 | 70.0 | 71.4 | 82.4 | 0.767 | 0.601–0.888 | p = 0.0009 |
| KNG1 + APOC3 + PON1 | 100.0 | 85.0 | 85.7 | 100.0 | 0.950 | 0.826–0.994 | p < 0.0001 |
AUC, area under the curve; PPV, positive predictive values; NPV, negative predictive values; 95% CI, 95% confidence interval.
4. Discussion
YDH syndrome is common in TCM practice and has been widely studied in recent decades. Many former studies have observed the abnormalities of immune function in YDH syndrome individuals. Wang found decline in immune function with a decrease of immunological substances in YDH constitution [17]. In addition, TNF-α, IL-1β, and IL-6 levels have been found to be elevated, while IL-2 levels have been found to be decreased in YDH syndrome individuals [18, 19]. Liu demonstrated decreased CD3+ and CD4+ levels in the peripheral blood of YDH syndrome individuals [20], indicating that the immune function is declined in YDH syndrome. Notably, we found that the weight of the thymus, one of the most important immune organs, was significantly decreased in YDH syndrome rats, suggesting reduced immune function in YDH syndrome. In iTRAQ-2DLC-MS/MS experiment, it was interesting to find that a majority of differentially expressed proteins were involved in immune response and metabolic processes. KEGG pathway analysis revealed that the immune response-associated proteins were mainly clustered at complement and coagulation cascades. To our knowledge, the deficiency of complement significantly reduces IL-1β levels [21, 22], and TNF-α contributes to coagulation and complement activation in virus-induced fulminant hepatitis [23], accounting for the upregulation of IL-1β and TNF-α levels in YDH syndrome individuals. Based on these findings, we speculated that the abnormally expressed proteins involved in complement and coagulation cascades disrupted the signaling pathways related to immune response. Accordingly, immune function in YDH syndrome was disturbed.
Previous studies have revealed that the activities of energy-yielding and energy-consuming reactions were enhanced in YDH syndrome rats [24], which was in line with our results of proteomic experiment. By comparing the differentially expressed serum proteins between YDH syndrome rats and normal rats, we found that approximately half of these proteins were associated with the metabolic processes. KEGG pathway analysis demonstrated that proteins participating in carbohydrate metabolism pathway, especially glycolysis and gluconeogenesis, accounted for a great proportion of the metabolism-associated proteins, which was consistent with previous studies revealing the lower levels of glycogen in YDH syndrome rats [24] and the hyperactivity of glycolysis in epithelial tissue with YDH syndrome [25]. Our results revealed that the enhanced energy metabolism in YDH syndrome attributed mostly to glucose metabolism, which might account for the weight loss in YDH syndrome individuals and animal models.
In the present study, we found that the level of KNG1 was significantly increased in the serum of YDH rats (p < 0.0001). KNG1, a precursor protein of vasoactive kinin [26], is known to participate in inflammation, coagulation, and innate immunity [27]. KNG1 has been found to be significantly elevated in uterine proliferative lesions rats [28] and the mRNA expression of the KNG1 gene has been shown to be upregulated in mouse lung tumors [29]. High-molecular-weight kininogen (HK) is cleaved by kallikrein to release bradykinin (BK) and HKa. HKa induces mononuclear cells to release cytokines (TNF- α, IL-1, and IL-6) and chemokines (IL-8 and MCP-1) [30]. Interestingly, it has been demonstrated previously that the levels of TNF-α, IL-1β, and IL-6 were higher in YDH syndrome individuals, suggesting that KNG1 is highly associated with the immune abnormalities in YDH syndrome.
PON1 is an important antioxidant enzyme against oxidative stress [31], which protects low-density lipoprotein (LDL) against oxidative processes [32] and prevents the formation of oxidized- (Ox-) LDL [33]. Ox-LDL facilitates the proliferation of endothelial cells and the accumulation of macrophages in arterial wall [34] and has been shown to be associated with high oxidative stress [35, 36]. So, PON1 may reduce the oxidative stress in the body. In this study, we found that the serum level of PON1 declined considerably in YDH syndrome rats (p = 0.0052), which was consistent with the PON1 alterations in cardiovascular disorders, cancer, and acute influenza infection [37, 38]. YDH syndrome, induced by emotional stress or long-term intake of spicy and hot food, is characterized by mucosal lesions. Meanwhile, oxidative stress plays an important role in the pathogenesis of mucosal lesions [39]. Therefore, the low levels of PON1 may indirectly increase oxidative stress in YDH individuals, subsequently contributing to the mucosal lesions.
APOC3 is closely correlated to lipid metabolism. It is an inhibitor of hepatic lipase and lipoprotein lipase [40], which interferes with lipolysis. Maeda et al. found that the plasma triglyceride levels in mice lacking gene APOC3 reduced to approximately 70% of normal level [41]. Another study indicated that the overexpression of APOC3 can lead to hypertriglyceridemia in vivo [42]. In the present study, obvious weight loss was observed in YDH syndrome rats, and the rate of weight gain was consistently lower than normal rats. Serum APOC3 levels in YDH syndrome rats decreased significantly (p = 0.0046). So, we suggested that rats with lower APOC3 have higher activities of hepatic lipase and lipoprotein lipase and undergo more adipose tissue lipolysis compared with normal rats.
5. Conclusion
In this study, we found that YDH syndrome is highly associated with the disturbance in immune response and metabolic processes. Further proteomic analysis revealed that YDH syndrome was mainly attributed to the disruption in complement and coagulation pathway and the glucose metabolism pathway. Besides, we revealed that the increased serum KNG1 levels and the decreased serum APOC3 and PON1 levels were closely related to YDH syndrome. KNG1 may regulate cytokines and chemokines release during the abnormal immune responses in YDH syndrome, while the low levels of PON1 and APOC3 may increase oxidative stress and lipolysis in YDH syndrome, respectively. The results indicated that KNG1, APOC3, and PON1 may act as potential biomarkers for the diagnosis of YDH syndrome. To accelerate the process of applying our findings in clinical practice, further study to evaluate the effectiveness in a relatively large sample size in YDH syndrome patients is needed. This study provides a novel method to diagnose YDH syndrome and may have significant implications for revealing the molecular mechanism of YDH syndrome.
Supplementary Material
The quality control reports of iTRAQ-2D LC-MS/MS experiments
Acknowledgments
This work was supported by grants from National Basic Research Program of China (no. 2014CB543002), National Natural Science Foundation of China (no. 81573709, no. 81273882), and National Special Sci-Tech Projects (no. 2012ZX10005001-006).
Competing Interests
The authors have declared no conflict of interests.
References
- 1.Wang Q., Zhu Y. B. Epidemiological investigation of constitutional types of Chinese medicine in general population: base on 21,948 epidemiological investigation data of nine provinces in China. China Journal of Traditional Chinese Medicine and Pharmacy. 2009;24(1):7–12. [Google Scholar]
- 2.Yu H. J., Wang Y., Yang L. P. Epidemiological investigation of Yin deficiency and Yang deficiency constitutions in TCM of Henan University. Liaoning Journal of Traditional Chinese Medicine. 2013;40(4):619–621. [Google Scholar]
- 3.Wrotnowski C. The future of plasma proteins—compounds represent both greatest good and worst danger. Genetic Engineering News. 1998;18(11):p. 14. [Google Scholar]
- 4.Mangé A., Dimitrakopoulos L., Soosaipillai A., Coopman P., Diamandis E. P., Solassol J. An integrated cell line-based discovery strategy identified follistatin and kallikrein 6 as serum biomarker candidates of breast carcinoma. Journal of Proteomics. 2016;142:114–121. doi: 10.1016/j.jprot.2016.04.050. [DOI] [PubMed] [Google Scholar]
- 5.Sun L., Hu S., Yu L., et al. Serum haptoglobin as a novel molecular biomarker predicting colorectal cancer hepatic metastasis. International Journal of Cancer. 2016;138(11):2724–2731. doi: 10.1002/ijc.29993. [DOI] [PubMed] [Google Scholar]
- 6.Jiang T.-T., Wang C., Wei L.-L., et al. Serum protein gamma-glutamyl hydrolase, Ig gamma-3 chain C region, and haptoglobin are associated with the syndromes of pulmonary tuberculosis in traditional Chinese medicine. BMC Complementary and Alternative Medicine. 2015;15(1, article 243) doi: 10.1186/s12906-015-0686-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y. S., Fan Y. L., Zhang Y. P., Jiang X. L., Zou S. J., Wang L. F. Development of animal model of heat syndrome due to insufficiency of yin fluids. China Journal of Basic Medicine in Traditional Chinese Medicine. 2001;7(9):23–25. [Google Scholar]
- 8.Yang C. P., Xue C. M. The establisment and the influence on the antioxidant effect of yin-deficiency mice model induced by pungent Chinese herbs. Journal of Sichuan of Traditional Chinese Medicine. 2004;22(10):14–15. [Google Scholar]
- 9.Diz A. P., Truebano M., Skibinski D. O. F. The consequences of sample pooling in proteomics: An Empirical Study. Electrophoresis. 2009;30(17):2967–2975. doi: 10.1002/elps.200900210. [DOI] [PubMed] [Google Scholar]
- 10.Cao W., Zhou Y., Li Y., et al. ITRAQ-based proteomic analysis of combination therapy with taurine, epigallocatechin gallate, and genistein on carbon tetrachloride-induced liver fibrosis in rats. Toxicology Letters. 2015;232(1):233–245. doi: 10.1016/j.toxlet.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y.-S., Cao R., Jin H., et al. Altered protein expression in serum from endometrial hyperplasia and carcinoma patients. Journal of Hematology and Oncology. 2011;4, article 15 doi: 10.1186/1756-8722-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin G.-Z., Li Y., Cong W.-M., et al. ITRAQ-2DLC-ESI-MS/MS based identification of a new set of immunohistochemical biomarkers for classification of dysplastic nodules and small hepatocellular carcinoma. Journal of Proteome Research. 2011;10(8):3418–3428. doi: 10.1021/pr200482t. [DOI] [PubMed] [Google Scholar]
- 13.Han D., Moon S., Kim H., et al. Detection of differential proteomes associated with the development of type 2 diabetes in the Zucker rat model using the iTRAQ technique. Journal of Proteome Research. 2011;10(2):564–577. doi: 10.1021/pr100759a. [DOI] [PubMed] [Google Scholar]
- 14.Oberg A. L., Mahoney D. W., Eckel-Passow J. E., et al. Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. Journal of Proteome Research. 2008;7(1):225–233. doi: 10.1021/pr700734f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juang Y.-M., Lai B.-H., Chien H.-J., Ho M., Cheng T.-J., Lai C.-C. Changes in protein expression in rat bronchoalveolar lavage fluid after exposure to zinc oxide nanoparticles: an iTRAQ proteomic approach. Rapid Communications in Mass Spectrometry. 2014;28(8):974–980. doi: 10.1002/rcm.6866. [DOI] [PubMed] [Google Scholar]
- 16.Xu D.-D., Deng D.-F., Li X., et al. Discovery and identification of serum potential biomarkers for pulmonary tuberculosis using iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics. 2014;14(2-3):322–331. doi: 10.1002/pmic.201300383. [DOI] [PubMed] [Google Scholar]
- 17.Wang J. M. The application of doctrine of Yin and Yang in the immune response. Journal of Zhejiang College of Traditional Chinese Medicine. 1979;5(1):4–5. [Google Scholar]
- 18.Wang Q., Ren X.-J., Yao S.-L., Wu H.-D. Clinical observation on the endocrinal and immune functions in subjects with yin-deficiency constitution. Chinese Journal of Integrative Medicine. 2010;16(1):28–32. doi: 10.1007/s11655-010-0028-9. [DOI] [PubMed] [Google Scholar]
- 19.Yan H. F., Ma J. L., Zhu H. H., et al. Yin deficiency syndrome vexing heat in chest, palms and soles was related to TNF-α IL-1β IL-6 clinical observation. Chinese Archives of Traditional Chinese Medicine. 2008;26(2):293–295. [Google Scholar]
- 20.Liu Y. M. Comparative study on T-lymphocyte subsets in the patients of excess-heat syndrome and Deficiency-heat syndrome. Journal of Traditional Chinese Medicine. 2005;46(4):289–290. [Google Scholar]
- 21.Ning C., Li Y.-Y., Wang Y., et al. Complement activation promotes colitis-associated carcinogenesis through activating intestinal IL-1β/IL-17A axis. Mucosal Immunology. 2015;8(6):1275–1284. doi: 10.1038/mi.2015.18. [DOI] [PubMed] [Google Scholar]
- 22.An L.-L., Mehta P., Xu L., et al. Complement C5a potentiates uric acid crystal-induced IL-1β production. European Journal of Immunology. 2014;44(12):3669–3679. doi: 10.1002/eji.201444560. [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Tan Y., Zhang J., et al. C5aR, TNF-α, and FGL2 contribute to coagulation and complement activation in virus-induced fulminant hepatitis. Journal of Hepatology. 2015;62(2):354–362. doi: 10.1016/j.jhep.2014.08.050. [DOI] [PubMed] [Google Scholar]
- 24.Han B., Wang S., Li L., Wang Y., Zhao H. Gene expression profiling of rat livers with Yin-deficiency-heat syndrome. Journal of Traditional Chinese Medicine. 2013;33(3):378–383. doi: 10.1016/s0254-6272(13)60182-x. [DOI] [PubMed] [Google Scholar]
- 25.Zhao P., Xingyu Z., Yongshen F. The relationship between excessive internal heat and intestinal microflora from the aspects of energy metabolism and mucosal immunity. Journal of Zhejiang Chinese Medicine University. 2015;39(8):591–594. [Google Scholar]
- 26.Kitagawa H., Kitamura N., Hayashida H., Miyata T., Nakanishi S. Differing expression patterns and evolution of the rat kininogen gene family. Journal of Biological Chemistry. 1987;262(5):2190–2198. [PubMed] [Google Scholar]
- 27.Long A. T., Kenne E., Jung R., Fuchs T. A., Renné T. Contact system revisited: an interface between inflammation, coagulation, and innate immunity. Journal of Thrombosis and Haemostasis. 2016;14(3):427–437. doi: 10.1111/jth.13235. [DOI] [PubMed] [Google Scholar]
- 28.Kakehashi A., Tago Y., Yoshida M., et al. Hormonally active doses of isoflavone aglycones promote mammary and endometrial carcinogenesis and alter the molecular tumor environment in Donryu rats. Toxicological Sciences. 2012;126(1):39–51. doi: 10.1093/toxsci/kfs016. [DOI] [PubMed] [Google Scholar]
- 29.Alyaqoub F. S., Liu Y., Tao L., Steele V. E., Lubet R. A., Pereira M. A. Modulation by bexarotene of mRNA expression of genes in mouse lung tumors. Molecular Carcinogenesis. 2008;47(3):165–171. doi: 10.1002/mc.20383. [DOI] [PubMed] [Google Scholar]
- 30.Khan M. M., Bradford H. N., Isordia-Salas I., et al. High-molecular-weight kininogen fragments stimulate the secretion of cytokines and chemokines through uPAR, Mac-1, and gC1qR in monocytes. Arteriosclerosis, Thrombosis, and Vascular Biology. 2006;26(10):2260–2266. doi: 10.1161/01.atv.0000240290.70852.c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowalska K., Socha E., Milnerowicz H. Review: the role of paraoxonase in cardiovascular diseases. Annals of Clinical and Laboratory Science. 2015;45(2):226–233. [PubMed] [Google Scholar]
- 32.La Du B. N., Aviram M., Billecke S., et al. On the physiological role(s) of the paraoxonases. Chemico-Biological Interactions. 1999;119-120:379–388. doi: 10.1016/s0009-2797(99)00049-6. [DOI] [PubMed] [Google Scholar]
- 33.Rosenblat M., Gaidukov L., Khersonsky O., et al. The catalytic histidine dyad of high density lipoprotein-associated serum paraoxonase-1 (PON1) is essential for PON1-mediated inhibition of low density lipoprotein oxidation and stimulation of macrophage cholesterol efflux. Journal of Biological Chemistry. 2006;281(11):7657–7665. doi: 10.1074/jbc.M512595200. [DOI] [PubMed] [Google Scholar]
- 34.Kita T., Kume N., Yokode M., et al. Oxidized-LDL and atherosclerosis-role of LOX-1. Annals of the New York Academy of Sciences. 2000;902:95–102. doi: 10.1111/j.1749-6632.2000.tb06304.x. [DOI] [PubMed] [Google Scholar]
- 35.Steinberg D., Witztum J. L. Lipoproteins and atherogenesis—current concepts. Journal of the American Medical Association. 1990;264(23):3047–3052. doi: 10.1001/jama.264.23.3047. [DOI] [PubMed] [Google Scholar]
- 36.Permpongpaiboon T., Nagila A., Pidetcha P., Tuangmungsakulchai K., Tantrarongroj S., Porntadavity S. Decreased paraoxonase 1 activity and increased oxidative stress in low lead-exposed workers. Human and Experimental Toxicology. 2011;30(9):1196–1203. doi: 10.1177/0960327110388536. [DOI] [PubMed] [Google Scholar]
- 37.Goswami B., Tayal D., Gupta N., Mallika V. Paraoxonase: a multifaceted biomolecule. Clinica Chimica Acta. 2009;410(1-2):1–12. doi: 10.1016/j.cca.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 38.Van Lenten B. J., Wagner A. C., Nayak D. P., Hama S., Navab M., Fogelman A. M. High-density lipoprotein loses its anti-inflammatory properties during acute influenza A infection. Circulation. 2001;103(18):2283–2288. doi: 10.1161/01.CIR.103.18.2283. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharyya A., Chattopadhyay R., Mitra S., Crowe S. E. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiological Reviews. 2014;94(2):329–354. doi: 10.1152/physrev.00040.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larsson M., Vorrsjo E., Talmud P., Lookene A., Olivecrona G. Apolipoproteins C-I and C-III inhibit lipoprotein lipase activity by displacement of the enzyme from lipid droplets. Journal of Biological Chemistry. 2013;288(47):33997–34008. doi: 10.1074/jbc.M113.495366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maeda N., Li H., Lee D., Oliver P., Quarfordt S. H., Osada J. Targeted disruption of the apolipoprotein C-III gene in mice results in hypotriglyceridemia and protection from postprandial hypertriglyceridemia. Journal of Biological Chemistry. 1994;269(38):23610–23616. [PubMed] [Google Scholar]
- 42.Ito Y., Azrolan N., O'Connell A., Walsh A., Breslow J. L. Hypertriglyceridemia as a result of human Apo CIII gene expression in transgenic mice. Science. 1990;249(4970):790–793. doi: 10.1126/science.2167514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The quality control reports of iTRAQ-2D LC-MS/MS experiments