SUMMARY
The cellular factor serine incorporator 5 (SERINC5) impairs HIV-1 infectivity but is antagonized by the viral Nef protein. We analyzed the anti-SERINC5 activity of Nef proteins across primate lentiviruses and examined whether SERINC5 represents a barrier to cross-species transmissions and/or within-species viral spread. HIV-1, HIV-2, and SIV Nefs counteract human, ape, monkey, and murine SERINC5 orthologs with similar potency. However, HIV-1 Nefs are more active against SERINC5 than HIV-2 Nefs, and chimpanzee SIV (SIVcpz) Nefs are more potent than those of their monkey precursors. Additionally, Nefs of HIV and most SIVs rely on the dileucine motif in the C-terminal loop for anti-SERINC5 activity, while the Nef from colobus SIV (SIVcol) evolved different inhibitory mechanisms. We also found a significant correlation between anti-SERINC5 potency and the SIV prevalence in the respective ape and monkey species. Thus, Nef-mediated SERINC5 antagonism may determine the ability of primate lentiviruses to spread within natural hosts.
In Brief
SERINC5 is a restriction factor that impairs the infectivity of retroviral particles. Heigele et al. show that the potency of SERINC5 antagonism by the accessory viral protein Nef correlates with the prevalence of corresponding SIVs in their respective wild-living hosts, suggesting that SERINC5 is a potential determinant of viral spread.
INTRODUCTION
The accessory protein Nef of human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) is required for efficient viral replication and substantially accelerates disease progression in vivo (Kestler et al., 1991; Deacon et al., 1995; Kirchhoff et al., 1995). Despite its small size of about 27–35 kDa, Nef performs a striking number of functions, including downmodulation of the CD4 receptor and MHC class I molecules from the cell surface and stimulation of viral replication in CD4+ T cells (Arhel and Kirchhoff, 2009; Pawlak and Dikeakos, 2015). Although these activities are conserved among primate lentiviral Nef proteins, important differences also exist. For example, the Nef proteins of HIV-2 and most SIVs downmodulate CD3 to suppress T cell activation, while the Nef proteins of HIV-1 and its vpu-containing simian precursors lost this function (Schindler et al., 2006). Moreover, most SIVs and the O group of HIV-1 use Nef to antagonize the restriction factor tetherin that inhibits the release of budding virions, while pandemic HIV-1 M strains use Vpu for the same function (Jia et al., 2009; Sauter et al., 2009; Zhang et al., 2009; Kluge et al., 2014). Thus, Nef acts throughout the replication cycle and promotes viral immune evasion and replication using a remarkable combination of diverse functions.
The ability of Nef to enhance the infectivity of HIV-1 particles was noted more than 20 years ago (Chowers et al., 1994; Miller et al., 1994), but the underlying mechanisms remained controversial. Nef-mediated downmodulation of CD4 increases the amount of Env glycoprotein in virus particles, and it was thus proposed that this enhances viral infectivity (Lama et al., 1999; Argañaraz et al., 2003). However, Nef also enhanced the infectivity of virus particles generated in CD4-negative producer cells, which was explained by reduced proteasomal degradation of virions (Qi and Aiken, 2007), increased cholesterol content of viral particles (Zheng et al., 2003), or enhanced transport of the viral pre-integration complex through the cortical actin network (Campbell et al., 2004). The effect of Nef on HIV-1 infectivity was also shown to depend on the envelope glycoprotein (Env) since VSV-G-pseudotyped particles were not affected (Aiken, 1997). Furthermore, Nef affects the infectivity of neutralization-sensitive CXCR4-tropic Env containing viruses more severely than those carrying neutralization-resistant CCR5-tropic Envs (Usami and Göttlinger, 2013). Another puzzling finding was that the glycosylated Gag protein (glycoGag) of murine leukemia virus (MLV) enhances HIV-1 infectivity in a manner very similar to Nef (Pizzato, 2010).
These seemingly discrepant results were recently reconciled by two elegant studies, which showed that Nef-mediated downmodulation of the multipass transmembrane proteins serine incorporator 3 and 5 (SERINC3 and SERINC5) from the cell surface prevents their incorporation into viral particles and thus increases virion infectivity (Rosa et al., 2015; Usami et al., 2015). Rosa et al. (2015) further demonstrated that the Nef proteins of SIVs infecting macaques and African green monkeys are capable of counteracting human SERINC5. Here, we examined the anti-SERINC5 activity of Nef proteins representing nearly the entire spectrum of primate lentiviruses to examine the role of this restriction factor in viral transmission within and between primate species. Our results show that Nef-mediated SERINC5 antagonism is largely species independent and suggest that the potency of this counteraction may determine, at least in part, the ability of primate lentiviruses to spread within their natural hosts.
RESULTS
SERINC5 Inhibits Primary HIV-1 and SIV Strains More Efficiently than SERINC3
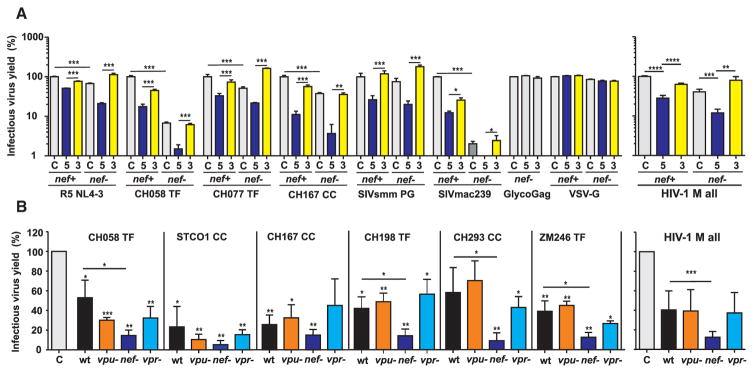
Although SERINC5 has been reported to impair HIV-1 infectivity to a greater extent than SERINC3 (Rosa et al., 2015; Usami et al., 2015), these data were mainly derived using the T cell line adapted HIV-1 NL4-3 molecular clone. We thus examined the ability of SERINC3 and SERINC5 to inhibit six HIV-1 and SIV strains containing either wild-type (WT) or defective nef genes. Two days post-transfection, infectious virus yield was determined by infection of TZM-bl cells. The susceptibility to SERINC5 and SERINC3 restriction varied among the different HIV-1 and SIV strains (Figures 1A and S1A). SERINC5 reduced the infectivity of WT HIV-1 strains by ~50% (NL4-3) to ~90% (CH167) and that of the nef-defective derivatives by ~80% to ~98%, while SERINC3 was generally poorly effective (Figure 1A, left). On average, transient SERINC5 expression reduced HIV-1 infectious titers by ~90% in the absence of Nef, while SERINC3 achieved only a 20% reduction (Figure 1A, right). Intact nef genes (Figure 1A) or coexpression of Nef in trans (Figure S1B) markedly counteracted SERINC5 inhibition. Intact nef genes also increased infectious virus yield in HEK239T cells in the absence of exogenous SERINC5 (Figure 1A), possibly because these cells express low levels of endogenous SERINC5 (Usami et al., 2015). Because SERINC3 generally reduced infectious HIV-1 yields only minimally, we focused on SERINC5 in subsequent experiments.
Figure 1. Role of Vpu, Vpr, and Nef in the Susceptibility of HIV-1 to SERINC5 and SERINC3 Inhibition.
(A) HEK293T cells were cotransfected with the indicated WT or nef-defective proviral HIV-1 or SIV constructs and SERINC5 or SERINC3 expression or control plasmid. Shown are the mean levels of infectious virus production by the respective IMCs in the presence of transient SERINC5 or SERINC3 expression (±SEM; n = 9) relative to those obtained in cells transfected with the control vector (100%). The results were derived from three independent experiments, each using triplicate infection of TZM-bl cells to determine infectious virus yield. CC, chronic HIV-1 strain; TF, transmitted/founder. p values represent reduction of infectious virus yield by SERINC expression or differences in susceptibility between WT and nef-defective HIV-1 IMCs. *p < 0.05; **p < 0.01; ***p < 0.001.
(B) Mean virus yield obtained for the six HIV-1 IMCs and vpu-, vpr-, or nef-defective derivatives thereof. The experiments were performed and the results obtained as described in the legend for (A).
See also Figure S1.
SERINC5 Is Counteracted by Nef, but Not by Vpu or Vpr
Nef shows functional redundancy with Vpu in modulating cellular receptors, such as CD4, CD1d, and tetraspanins (Haller et al., 2014), and the accessory protein Vpr has also been implicated in enhancing viral infectivity (Ueno et al., 2003). To determine the effect of all three accessory proteins on viral susceptibility to SERINC5 inhibition, we generated vpu-, vpr-, and nef-defective derivatives of HIV-1 infectious molecular clones (IMCs) representing two clade B and four clade C strains (Figure 1B). These included transmitted founder HIV-1 strains (CH058, CH198, ZM246), as well as viruses predominant during chronic infection (STCO, CH167, CH293) (Ochsenbauer et al., 2012). SERINC5 reduced the infectivity of all WT HIV-1 by 40%–75%, while nef-defective derivatives of these viruses exhibited an even greater reduction of 85%–93% (Figure 1B). In two cases (CH058, STCO1), vpu- and vpr-defective constructs were also slightly more susceptible to SERINC5 inhibition than the WT HIV-1 strains. However, these differences were not significant and most likely due to assay variation. Altogether, only lack of Nef, but not of Vpu or Vpr, significantly reduced infectious virus yield (Figure 1B, right). Thus, HIV-1 uses Nef, not Vpu or Vpr, to counteract SERINC5.
Antiviral Activity of SERINC5 and Counteraction by Nef Are Largely Species Independent
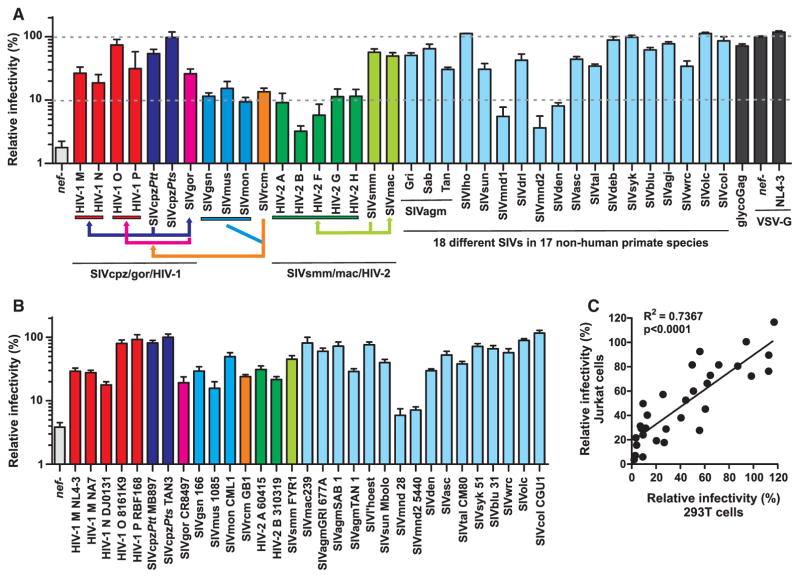
To determine the conservation of SERINC5 antagonism, we analyzed a set of 88 HIV-1 NL4-3 proviral constructs containing nef genes from all four groups of HIV-1 (M, N, O, and P), five groups of HIV-2 (A, B, F, G, and H), the simian precursors of both of these human viruses (SIVcpz, SIVgor, and SIVsmm), the descendants of SIVs that recombined to generate SIVcpz (SIVgsn/mus/mon and SIVrcm) (Bailes et al., 2003), and lentiviruses from 18 additional monkey species. The phylogenetic relationships between these highly divergent primate lentiviruses are shown in Figure S2A. Expression of most of these nef alleles has previously been reported (Schindler et al., 2006; Schmökel et al., 2011; Heigele et al., 2012; Kluge et al., 2014). SERINC5 coexpression reduced infectivity of the nef-defective control HIV-1 construct ~50-fold (Figures 2 and S2B). As expected (Aiken, 1997; Pizzato, 2010), MLV glycoGag and VSV-G pseudotyping abolished this inhibitory effect. With few exceptions (HIV-2 B, SIVmnd2), all HIV and SIV nef alleles displayed significant activity against human SERINC5 (Figure 2). Western blot analyses of AU1-tagged forms of Nefs confirmed that proteins lacking anti-SERINC5 activity were nonetheless efficiently expressed (Figure S2C). Notably, highly divergent nef alleles from various monkey SIVs were as active against human SERINC5 as nef genes obtained from viruses infecting humans or African great apes. Furthermore, nef genes derived from the ape (SIVcpz, SIVgor) and monkey (SIVsmm) precursors did not display higher activity against human SERINC5 than those obtained from SIVs that have not been found in humans.
Figure 2. Antagonism of Human SERINC5 by Primate Lentiviral Nef Proteins.
(A) Effect of various primate lentiviral Nefs on HIV-1 infectivity in the presence of human SERINC5. HEK293T cells were cotransfected with recombinant HIV-1 NL4-3 proviral constructs containing the indicated nef alleles (3 μg) and an empty control or SERINC5 expression vector (2.5 μg). For control, a glycoGag expression vector was cotransfected with the nef-defective (nef-) NL4-3 construct or the virions were produced in the presence of the VSV glycoprotein (VSV-G). Viral supernatants were obtained 2 days later and used to determine the quantity of infectious HIV-1 in the culture supernatants by infecting TZM-bl indicator cells. Shown are average values ± SD (n = 3) of infectious virion yield relative to those obtained in the absence of SERINC5 expression vector (100%). Results in (A) and (B) were derived from three independent experiments each using triplicate infection. The arrows indicate cross-species transmissions and the recombination event.
(B) Infectious virus yield in Jurkat T cells compared with a derivative thereof lacking SERINC5/3 expression (100%). Parental and SERINC5/3 knockout Jurkat T cells were transduced with HIV-1 NL4-3 proviral constructs containing the indicated nef alleles and infectious virus yield was determined by triplicate infection of TZM-bl cells 3 days later. SERINC5/3 expression and the presence of intact nef genes had no significant effect on p24 antigen production.
(C) Correlation between the infectious yield of HIV-1 nef recombinants in the presence of endogenous SERINC5/3 in Jurkat T cells and transiently expressed SERINC5 in HEK293T cells.
See also Figure S2.
To avoid overexpression artifacts due to the potent reduction of HIV-1 infectivity by SERINC5, we used the pBJ6 vector in our experiments, which expresses about 100-fold less SERINC5 than regular CMV promoter-driven expression plasmids (Rosa et al., 2015). Moreover, we also examined the infectious virus yield of 32 HIV-1 NL4-3 constructs containing a broad spectrum of primate lentiviral nef alleles in Jurkat T cells and a derivative thereof lacking SERINC5/3 expression. Endogenous SERINC5/3 reduced nef-defective infectious HIV-1 yield by 33-fold (Figure 2B) but had no significant effect on p24 antigen production (data not shown). Most primate lentiviral nef alleles enhanced infectious virus yield in the presence of endogenous SERINC5 expression (Figure 2B), and those that counteracted transiently expressed SERINC5 in HEK293T cells were also active against endogenously expressed SERINC5 in Jurkat T cells (Figure 2C).
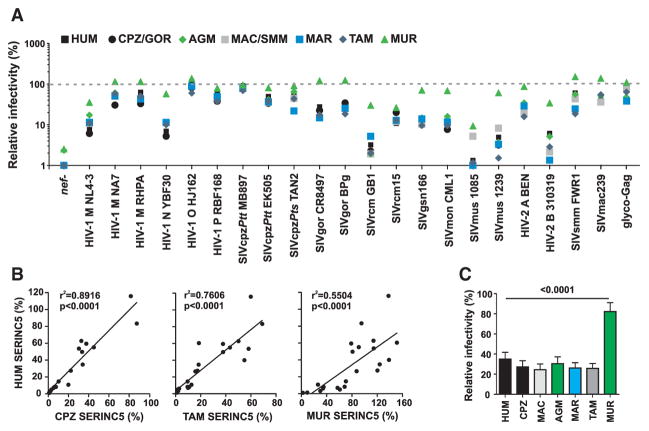
To determine whether the antiretroviral activity is evolutionarily conserved, we analyzed seven different SERINC5 orthologs, from humans (HUM), chimpanzees/gorillas (CPZ/GOR), rhesus macaques/sooty mangabeys (MAC/SMM), African green monkeys (AGM), common marmosets (MAR), tamarins (TAM) and mice (MUR). Consistent with their high degree of sequence conservation (Figure S3), all of them reduced the infectivity of the nef-defective HIV-1 control construct by about 2 orders of magnitude (Figure 3A). This was true for all primate derived SERINC5 proteins as well as the SERINC5 ortholog derived from mice. Moreover, 21 nef alleles from highly divergent HIV and SIV strains antagonized all of these SERINC5 orthologs, indicating that this counteraction is largely species independent. However, the efficiency with which the different nef alleles counteracted the various SERINC5 proteins differed considerably. For example, HIV-1 O, SIVcpz, SIVmac, and SIVsmm Nefs, as well as MLV glyco-Gag, restored viral infectivity almost completely, whereas SIVrcm, SIVmus, and HIV-2 B Nefs were much less potent (Figure 3A). Importantly, these varying efficiencies were observed for all SERINC5 proteins irrespective of the species origin. For example, the SIVrcm Nef protein was poorly active against human, ape, and three monkey-derived SERINC5 proteins. In contrast, SIVmac Nef counteracted not only its cognate, but also ape, human, and other monkey SERINC5s with great efficiency. In general, levels of infectious virus correlated well between different SERINC5 orthologs (Figure 3B), indicating that the antiretroviral activity of SERINC5 is conserved from mice to men and that the varying potency of the different Nefs in antagonizing monkey, ape, and human SERINC5 is largely species independent. Only murine SERINC5 was slightly less effective than primate SERINC5 at reducing viral infectivity and/or more susceptible to Nef-mediated counteraction (Figure 3C).
Figure 3. Antiretroviral Activity of SERINC5 and Nef-Mediated Counteraction Are Conserved and Species Independent.
(A) Inhibition of HIV-1 NL4-3 constructs containing the indicated nef alleles by SERINC5 orthologs from different primate species or mice. HEK293T cells were cotransfected with recombinant HIV-1 proviral vector and a control plasmid or vectors expressing SERINC5 from the indicated species. Viral supernatants were obtained and infectious HIV-1 production in the culture supernatants was determined as described in the legend to Figure 2. Each symbol represents the average infectious virus yield obtained in the presence of one SERINC5 ortholog relative to the vector control (100%). Values were derived from three to five experiments.
(B) Correlation between the infectious yield of HIV-1 nef recombinants in the presence of human SERINC5 with the yield obtained in the presence of CPZ, TAM, or MUR SERINC5. Values were derived from (A).
(C) Mean infectious virus yield (±SEM) obtained for the HIV-1 constructs expressing the 21 different nef alleles shown in (A) in the presence of the indicated SERINC5 orthologs.
See also Figure S3.
Antagonism of Human SERINC5 by HIV-1 and Its Primate Precursors
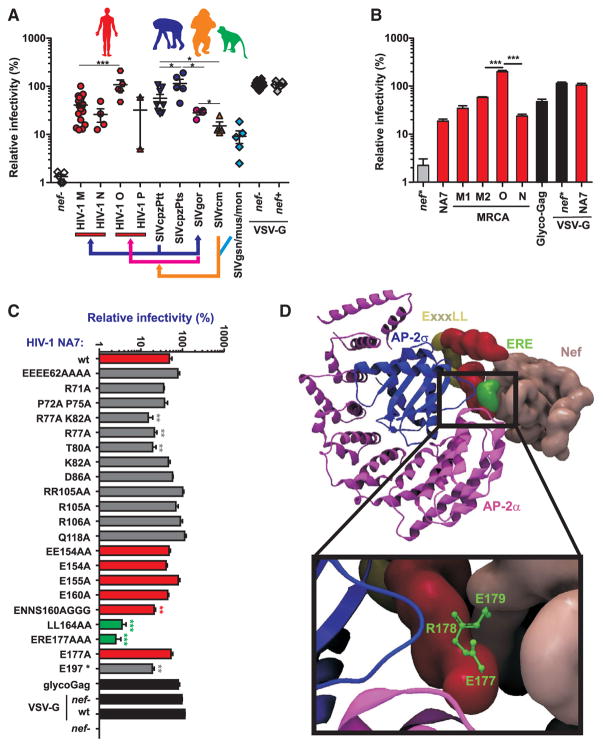
To determine whether the anti-SERINC5 activity of Nef may have changed after the cross-species transmissions that preceded the emergence of HIV-1, we analyzed a set of 57 nef alleles from the four groups of HIV-1 and their respective primate precursors. Two groups of HIV-1 (M and N) originated from SIVcpzPtt infecting central chimpanzees (Pan troglodytes troglodytes), whereas groups O and P originated from SIVgor found in western lowland gorillas (Gorilla gorilla gorilla) (Sharp and Hahn, 2011; D’arc et al., 2015). Eastern chimpanzees (P. t. schweinfurthii) are also infected with SIVcpz but have not transmitted their virus (SIVcpzPts) to humans. SIVcpz arose from a recombination of ancestors of today’s SIVgsn/mus/mon infecting Cercopithecus monkeys and SIVrcm from red-capped mangabeys (Bailes et al., 2003). We found that HIV-1 group M, N, and P Nefs do not differ significantly in their anti-SERINC5 activities (Figures 4A and S4A). HIV-1 group O Nefs were more active against human SERINC5 than Nef proteins from the other three HIV-1 groups (Figure 4A), although they exhibited a similar ability to downmodulate CD4 or MHC-I (Figure S4B). Functional analysis of the Nef proteins of previously inferred most recent common ancestors (MRCAs) of group M, N, and O viruses (Kluge et al., 2014) suggests that HIV-1 group O already possessed high anti-SERINC5 activity prior to its spread in the human population (Figure 4B). Human SERINC5 was potently counteracted by all SIVcpz Nef proteins (Figures 3 and 4A), indicating that this antiviral factor does not represent a barrier for cross-species transmission. Interestingly, however, SIVcpzPtt Nef proteins were significantly more active against human and ape SERINC5 than those of SIVrcm (Figures 3 and 4A), which gave rise to the SIVcpz nef gene (Schindler et al., 2006). Nefs derived from SIVgsn, SIVmus, and SIVmon whose common ancestor recombined with SIVrcm to become SIVcpz, also displayed poor activity against the human, ape and monkey orthologs of SERINC5 (Figures 3 and 4A). Thus, Nef gained anti-SERINC5 activity after transmission of SIVs from monkeys to chimpanzees.
Figure 4. SERINC5 Counteraction by nef Alleles from HIV-1 and Its Simian Precursors.
(A) Antagonism of human SERINC5 by nef alleles from HIV-1 and its simian precursors. Each symbol represents infectious virus yield in the presence of one individual HIV-1 or SIV nef allele analyzed relative to that obtained in the absence of SERINC5 expression vector (100%). Shown are mean values (±SEM) from at least three experiments. The arrows indicate cross-species transmissions and the recombination event.
(B) Infectious virus yield from HEK293T cells cotransfected with an HIV-1 NL4-3 ΔvpuΔnef construct and vectors expressing the indicated nef alleles in combination with a plasmid expressing SERINC5. Shown are mean values (±SEM) derived from triplicate infections of TZM-bl indicator cells relative to those obtained in the absence of SERINC5 expression vector (100%). The M1 and M2 group M MRCA Nefs differ by a single E-to-P substitution at their N terminus (Kluge et al., 2014).
(C) Activity of NA7 Nef mutants against SERINC5 inhibition. HEK293T cells were cotransfected with NL4-3 proviral constructs containing the indicated nef alleles (3 μg) and an empty control or SERINC5 expression vector (2.5 μg). Viral supernatants were obtained 2 days later and used to quantify infectious HIV-1 in the culture supernatants by infecting TZM-bl indicator cells. Shown are mean values (±SEM) derived from three experiments.
(D) Localization of amino acid residues involved in anti-SERINC5 activity in HIV-1 Nef complexed with the α and σ2 subunits of AP-2 (Ren et al., 2014). The ExxxLL (yellow) and ERE (green) motifs in the C loop (red) of Nef are critical for SERINC5 antagonism.
*p < 0.05; **p < 0.01; ***p < 0.001. See also Figure S4.
Determinants of Anti-SERINC5 Activity in HIV-1 M Nef
To define domains in HIV-1 Nef involved in SERINC5 antagonism, we examined 21 mutants of the highly active primary HIV-1 NA7 nef allele (Greenberg et al., 1997). Expression and functional analyses of these Nef proteins have previously been reported (Sauter et al., 2015). We found that mutation of R77A, T80A, and ENNS160AGGG as well as a premature stop at codon 197 significantly reduced Nef’s ability to counteract SERINC5 (Figure 4C). In contrast, mutations in the acidic region, the first two proline residues in the PxxP[3] domain, the RR PAK2 interaction site, and a putative β-COP binding site had no significant disruptive effect. Mutation of three charged C-terminal residues (ERE177AAA) in the C loop disrupted the effect of Nef on SERINC5 inhibition as severely as mutation of the ExxxLL motif (Figure 4C). These charged residues, which are critical for CD4 downmodulation and infectivity enhancement, but not MHC-I downmodulation (Sauter et al., 2015), are located in the C loop of Nef and directly interact with both AP-2 α and σ2 (Ren et al., 2014; Figure 4D). These results are in agreement with the previous finding that SERINC5 antagonism requires the interaction of Nef with AP-2 clathrin adaptor complexes (Rosa et al., 2015; Usami et al., 2015) but identify three additional amino acids in the C-terminal loop of HIV-1 Nef as key determinants of its ability to antagonize SERINC5.
Changes in O-Nef Residues that Confer Anti-tetherin Activity Enhance SERINC5 Antagonism
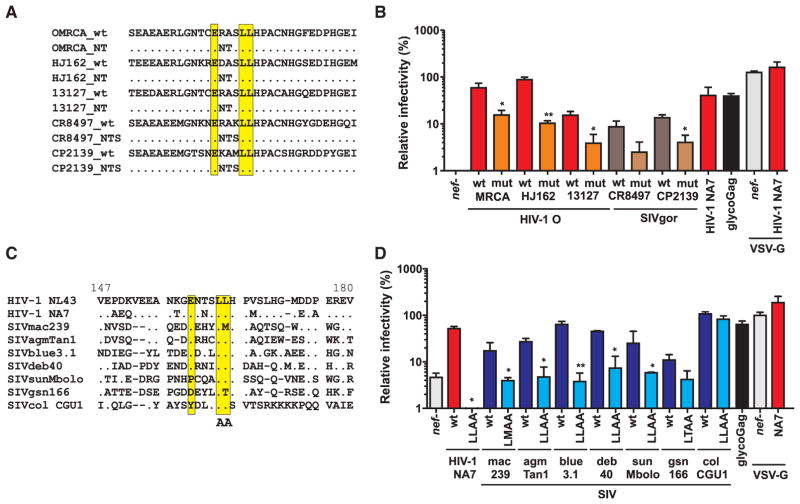
To dissect the mechanism(s) responsible for the higher anti-SERINC5 activity of group O Nefs (Figure 4A), we analyzed the impact of variable residues (x) in the otherwise highly conserved dileucine-based sorting signal (ExxxLL) in the C-terminal loop of Nef. Changes in these residues, which distinguish O-Nefs from those of M and N viruses, were critical for the acquisition of activity against human tetherin (Kluge et al., 2014). To determine their role in anti-SERINC5 activity, we compared the susceptibility of HIV-1 NL4-3 constructs expressing WT or NTS161–163 mutant HIV-1 O and SIVgor Nefs (Figure 5A) to this antiviral factor. Amino acid substitutions to NTS, a motif commonly found in group M Nefs, generally reduced the ability of Nef to antagonize SERINC5 (Figure 5B), although they did not significantly affect Nef expression levels or downmodulation of CD4 or MHC-I (Kluge et al., 2014). Thus, alterations in the variable residues of the ExxxLL motif of O-Nefs that conferred activity against human tetherin also increased their potency in SERINC5 antagonism.
Figure 5. Role of the ExxxLL Motif in Nef in SERINC5 Antagonism.
(A) Mutation of amino acid residues involved in anti-tetherin activity of HIV-1 group O and SIVgor Nefs. The dileucine motif involved in the interaction with AP complexes is highlighted and the mutations in the variable residues of the ExxxLL motif that impair tetherin antagonism are indicated.
(B) Effect of changes in the variable residues of the ExxxLL motif of Nef on SERINC5 antagonism. Values represent mean levels of infectious virus production (±SEM) in the presence of human SERINC5 relative to those obtained in the absence of this antiviral factor (100%).
(C) Alignment of the HIV-1 and SIV C-loop region of Nef. The NL4-3 C-loop sequence is shown on top for comparison. The [E/D]xxxLϕ motif and mutants analyzed in (B) are indicated. Dots indicate amino acid identity; dashes indicate gaps that were introduced to improve the alignment.
(D) Effect of primate lentiviral Nefs differing in the presence of the ExxxLL endocytosis motif on SERINC5 inhibition of infectious virus production. (B) and (D) show mean values ± SEM (n = 3) of infectious virion yield relative to those obtained in the absence of SERINC5 expression vector (100%).
*p < 0.05; **p < 0.01. See also Figure S5.
SIVcol Nef Counteracts SERINC5 Independently of an ExxxLL AP-2-Binding Motif
It has been shown that the ExxxLL motif in the C-terminal loop is critical for the ability of HIV-1 NL4-3 Nef to antagonize SERINC5 (Rosa et al., 2015). To determine whether this motif is required for anti-SERINC5 activity of other primate lentiviral Nef proteins, we mutated the leucine residues in the ExxxLL motifs of HIV-1 NA7, SIVmac, SIVagm, SIVblu, SIVdeb, SIVsun, SIVgsn, and SIVcol Nefs to alanines (Figure 5C). In most cases, these mutations disrupted the ability of Nef to counteract SERINC5 (Figure 5D) as well as to downmodulate CD4 (Figure S5A) but had no significant effect or even increased the potency of MHC-I downmodulation (Figure S5B). The single exception was the Nef protein of SIVcol from black-and-white colobus monkeys. This virus, which expresses a highly divergent Nef protein (Figure S2A) that contains a YxxxLL instead of the canonical ExxxLL motif, antagonized SERINC5 with high efficiency irrespective of the dileucine motif (Figure 5D). The SIVcol CGU1 Nef is known to have unusual functional properties because it increases virion infectivity and downmodulates CXCR4 with high efficacy but does not modulate CD4 and MHC-I (Figure S5; Wildum et al., 2006). We confirmed these properties for SIVcol Nefs from two additional colobus monkeys and showed that they are not just the result of species-specific differences in cellular receptors (data not shown). Thus, the ExxxLL sorting motif in the C loop that recruits AP-2 is critical for the anti-SERINC5 activity of most primate lentiviral Nef proteins, but not the divergent SIVcol Nef, which appears to have evolved different interactions to counteract this antiviral factor.
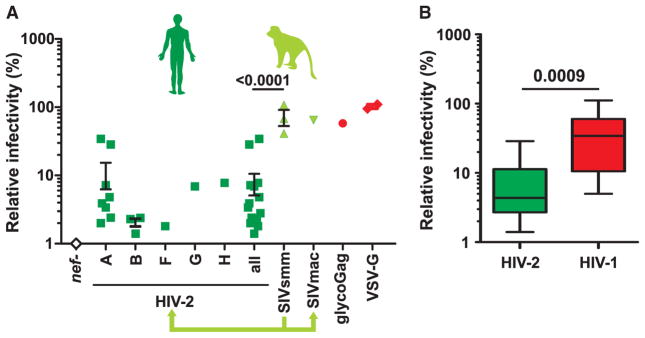
Most HIV-2 Nef Proteins Are Poor Antagonists of Human SERINC5
Our initial analyses of five HIV-2 nef alleles suggested that they might be less active against SERINC5 than those of HIV-1 and SIVsmm strains infecting sooty mangabeys (Figure 2), the original host of HIV-2. To follow up on this observation, we analyzed 14 different HIV-2 Nefs. Only two nef alleles (BEN, RH2-1-A8) from the A group of HIV-2, which accounts for the majority of HIV-2 infections (Campbell-Yesufu and Gandhi, 2011), showed relatively high activity against human SERINC5 (Figures 6A and S6A). Thus, a strikingly high percentage (86%) of HIV-2 Nefs displayed little activity against human SERINC5. Overall, HIV-2 Nefs were almost 10-fold less potent than SIVsmm Nefs (Figure 6A) and 5-fold less potent than HIV-1 Nefs (Figure 6B), although most of them efficiently downmodulated CD4 and MHC-I (Figure S6B).
Figure 6. SERINC5 Counteraction by nef Alleles from HIV-2 and Its Simian Precursors.
(A) Effect of nef alleles from HIV-2 and its simian SIVsmm counterparts on HIV-1 inhibition by human SERINC5. Each symbol represents the average infectious virus yield obtained for HIV-1 NL4-3 containing nef alleles from the indicated primate lentiviruses in the presence of human SERINC5 relative to the vector control (100%). Values were derived from at least three experiments and shown means ± SEM.
(B) Activities of nef alleles from HIV-2 and HIV-1. Shown are minimum and maximum values, 25% and 75% percentiles, and median values.
See also Figure S6.
Nef-Mediated Anti-SERINC5 Activity Correlates with SIV Prevalence Rates in the Wild
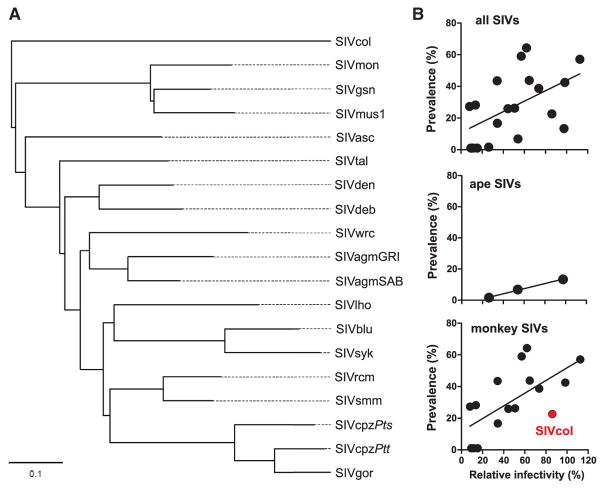
The higher anti-SERINC5 activity of HIV-1 nef alleles compared with those obtained from HIV-2 strains (Figure 6) suggested a potential role of this restriction factor in viral spread. To test this hypothesis, we examined whether the magnitude of Nef-mediated SERINC5 antagonism correlates with the prevalence of different SIVs in their respective natural hosts. A review of published SIV prevalence data identified 16 monkey species for which more than ten samples (average ~200) had previously been screened for SIV and for which functional nef alleles were also available (Table S1). Together with non-invasive screening results of wild-living chimpanzees and gorillas, infection rates and Nef-mediated SERINC5 antagonism data were available for 19 different SIV strains (Figure 7A).
Figure 7. Relationships between the Potency of Nef-Mediated SERINC5 Antagonism and SIV Prevalence Rates.
(A) Evolutionary relationships of the SIV strains included in the correlation analyses (B) based on Nef protein sequences (a maximum likelihood tree based on Pol protein sequences is shown in Figure S2).
(B) Relationships between the prevalence rates of SIVs infecting wild-living ape and monkey species and the average infectious yield of viruses expressing the corresponding Nef proteins in the presence of SERINC5 relative to the vector control (100%). Shown are results for 19 different SIV lineages for which functionally active nef alleles were available (top), for SIVcpzPts, SIVcpzPtt, and SIVgor infecting great apes (middle), and for SIVs infecting 16 African monkey species (bottom). The correlation analyses were performed using independent contrasts to correct for the underlying Nef phylogeny (A) and yielded significant values both for the 19 (top: R2 = 0.1872, p = 0.0377) and 16 (bottom: R2 = 0.2644, p = 0.0233) SIV panels. The SIVcol strain (indicated in red) expresses an unusual Nef protein that antagonizes SERINC5 but lacks other Nef functions. The numbers of nef alleles analyzed and number of apes and monkeys examined for SIV are shown in Table S1.
To determine whether there is an association between viral prevalence and the strength of Nef-mediated SERINC counteraction, we performed correlation analyses using phylogenetically independent contrasts (Felsenstein, 1985), which transforms comparative data to account for covariation of phenotypes due to an underlying shared ancestry. Using data from all 19 SIVs (Figure 7B), we found a significant correlation between the ability of a given SIV Nef to counteract SERINC5 and the prevalence of the corresponding virus in its natural host species (R2 = 0.1872, p = 0.0377) (Figure 7B, top). Thus, Nef-mediated anti-SERINC activity may explain about 19% of the variation in SIV prevalence among the different primate species. Because monkey and great ape-derived SIVs differ in their evolutionary history, with the ape SIVs representing more recent monkey SIV recombinants (Bailes et al., 2003), we also examined these two groups separately. Although SIV infection rates in apes are lower and more variable than those in most monkey species (Keele et al., 2006; Van Heuverswyn et al., 2007; Li et al., 2012; D’arc et al., 2015), SIVcpz is more prevalent in eastern than in central chimpanzees, and both SIVcpzPtt and SIVcpzPts are more common in chimpanzees than SIVgor is in western lowland gorillas (Figure 7B, middle; Table S1). Again, there was an association between prevalence rates of SIVcpzPts, SIVcpzPtt, and SIVgor and the potency of their respective Nef proteins in counteracting SERINC5 (Figure 7B, middle). However, the small number of ape SIVs studied precluded a meaningful statistical analysis. Interestingly, when the correlation analysis was restricted to the 16 monkey SIVs, the relationship between percent prevalence and viral infectivity was slightly more significant (R2 = 0.2644, p = 0.0233; Figure 7B, bottom), consistent with the longstanding relationship of monkey SIVs and their respective hosts. A similar level of significance was found when the Nef protein of SIVcol, which is an effective SERINC5 antagonist but lacks the otherwise conserved CD4 and MHC-I functions, was excluded (adjusted R2 = 0.2894, p = 0.0250).
DISCUSSION
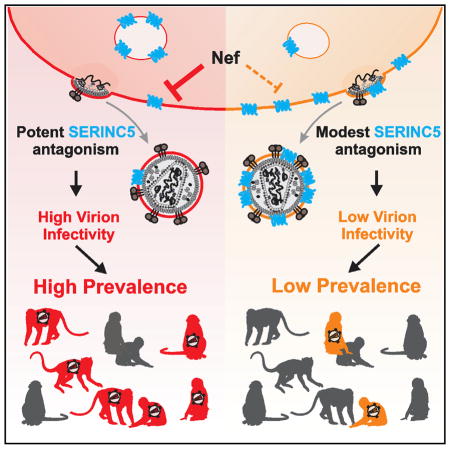
In this study, we examined the ability of primate lentiviral Nef proteins to counteract the antiviral restriction factor SERINC5. We show that this Nef function is highly conserved among all primate lentiviral lineages (Figure 2) and largely independent of the species origin of the SERINC5 ortholog (Figure 3). However, some interesting differences were also identified. First, all nef alleles from SIVs, whose ancestors recombined to generate SIVcpz (i.e., SIVgsn/mus/mon and SIVrcm), were poorly active against monkey, ape, and human SERINC5 orthologs (Figure 2). Because SIVcpz Nefs are generally highly effective antagonists of SERINC5, it seems clear that some mutations that increased anti-SERINC5 activity were selected for during viral adaptation to chimpanzees. Second, we found that HIV-1 Nefs are substantially more potent antagonists of SERINC5 than HIV-2 Nef proteins (Figure 4B). Because SIVsmm Nefs are also highly active against SERINC5, HIV-2 Nefs may have partly lost this function following the transfer to humans. Third, we found a significant, phylogenetically corrected correlation between the potency of Nef-mediated SERINC5 counteraction and the prevalence rates of the corresponding SIVs in great apes and African monkey species (Figure 7). Thus, high-level SERINC5 antagonism resulting in increased virion infectivity seems to affect the ability of primate lentiviruses to spread within their host species.
Previously identified restriction factors represent barriers to primate lentiviral cross-species transmission because they are counteracted (or evaded) in a species-specific manner (Pyndiah et al., 2015). One well-known example is tetherin antagonism (or lack thereof) by the four groups of HIV-1. Human tetherin contains a deletion conferring resistance to the Nef protein used by most SIVs to counteract this restriction factor (Jia et al., 2009; Sauter et al., 2009; Zhang et al., 2009). To counteract tetherin in humans, the M group of HIV-1 switched to Vpu (Sauter et al., 2009), while group O strains acquired the ability to target a different region in Nef (Kluge et al., 2014). Vpu proteins of rare group N HIV-1 strains are still evolving toward higher anti-tetherin activity (Sauter et al., 2012), and HIV-1 group P has not yet acquired an effective tetherin antagonist (Sauter et al., 2011; Yang et al., 2011). Thus, suboptimal viral adaptation following cross-species transmission may limit viral spread. In contrast to tetherin antagonism, however, SERINC5 counteraction is not host specific (Figure 3). Orthologs derived from monkeys, apes and humans exhibit very similar antiretroviral activity as well as susceptibility to Nef-mediated antagonism (Figure 3), suggesting that SERINC5 does not usually pose a barrier to cross-species transmission. Nonetheless, we identified substantial differences in the anti-SERINC5 activity of Nef proteins from different HIV/SIV lineages that are independent of the species origin of SERINC5 and may be the result of balancing different functions. Nef performs an enormous number of activities, and it is likely that the necessity to maintain multiple functions comes at a cost for some of them. For example, it has been shown that nef alleles obtained after AIDS progression are highly active in downregulation of CD4 and enhancement of HIV-1 infectivity but do not efficiently downmodulate MHC-I (Carl et al., 2001).
The finding of a significant correlation between the extent of Nef-mediated SERINC5 antagonism and the prevalence of the corresponding SIVs in their respective host species came as a surprise (Figure 7). SIV prevalence rates depend on many viral and non-viral factors, including population structure, transmission dynamics, host genetics, and immune responses. Although we analyzed more than 100 Nef proteins and included only those that were functionally active in at least one assay in addition to SERINC5 antagonism (Table S1), only single Nef proteins were available for analysis for some SIV strains. Moreover, the prevalence estimates in Table S1 can only serve as a very rough approximation, because the extent to which some primate species were sampled was very limited. Despite these limitations, the significant correlation, which was phylogenetically corrected, indicates that a considerable fraction (19%–26%) of the variation in SIV prevalence among the different primate species can potentially be explained by the strength of their Nef-mediated anti-SERINC activity. Notably, the prevalence rates of the four groups of HIV-1 did not correlate with the efficiency of SERINC5 antagonism, which is likely because HIV-1 is the result of very recent zoonotic transmissions. There are clear differences in the extent of adaptation of the four different HIV-1 groups to the human host (Sauter et al., 2010; Sharp and Hahn, 2011), and the spread of HIV-1 in humans is affected by many factors that do not apply to the spread of SIV in the wild. Thus, the HIV-1 data do not argue against a relationship between SIV prevalence and SERINC5 antagonism in wild-living primates.
Our finding that the majority of HIV-2 Nefs are less potent against SERINC5 than those of HIV-1 is in agreement with the previous observation that the latter are more effective in enhancing viral infectivity and replication (Münch et al., 2007). This is despite the fact that HIV-2 Nefs potently downmodulate CD3 and CD28 and are as effective as HIV-1 Nefs in modulating MHC-I (Münch et al., 2005; Schindler et al., 2006). Thus, the poor anti-SERINC5 activity of HIV-2 Nefs is not just the result of reduced expression levels or generally impaired function. In contrast, Nef proteins derived from SIVsmm (the direct precursor of HIV-2) are potent antagonists of human and simian SERINC5 proteins (Figure 3). SIVsmm Nefs are also highly active in other functions, such as modulation of CD4, CD3, CD28, or TCR-CD3 (Schmökel et al., 2009). Although HIV-2 Nef seems to have lost efficient anti-SERINC5 activity, it is possible that another HIV-2 protein may have acquired this function. One candidate is Env, which has been shown to confer SERINC5 resistance independently of Nef function in HIV-1 (Usami and Göttlinger, 2013).
Nef is one of the most variable proteins of primate lentiviruses, and some HIV and SIV Nefs show only 30% sequence identity at the amino acid level. Nonetheless, the dileucine-based sorting motif [E/D]xxxLϕ is conserved among the great majority of primate lentiviruses. Our data show that this motif is required for SERINC5 antagonism by HIV-1 and most SIVs but also identified an interesting exception. The SIVcol Nef is highly active against SERINC5 despite the absence of a canonical [E/D]xxxLϕ motif. This Nef protein is functionally and structurally different from all other SIV Nefs and lacks otherwise common activities, such as modulation of CD3, CD4, and MHC-I, although it is highly active in downmodulation of CXCR4. Thus, the SIVcol Nef may have evolved differently, and potent SERINC5 antagonism may represent an example of convergent evolution. We also identified three adjacent residues (ERE) in the C loop of HIV-1 Nef that are involved in AP-2 interaction and seem critical for SERINC5 antagonism (Figures 4C and 4D). Finally, we found that alterations in the variable residues of the ExxxLL motif that confer anti-tetherin activity to HIV-1 group O strains (Kluge et al., 2014) also significantly enhanced the efficiency of SERINC5 antagonism (Figure 5). Nef interacts with different adaptor proteins to modulate endocytosis as well as anterograde transport of cellular proteins (Roeth and Collins, 2006). Thus, further studies are required to determine, which Nef interactions besides AP-2 recruitment may be modulated by the alterations in the C-loop that affect Nef-mediated SERINC5 and tetherin antagonism.
In summary, we show that essentially all primate lentiviral Nef proteins antagonize SERINC5 in a largely species-independent manner but with different efficiencies. Nef seems to have acquired enhanced anti-SERINC5 activity after the cross-species transmission from monkeys to apes and from apes to humans. The significant correlation between the prevalence of SIVs and the ability of their Nefs to counteract SERINC5 supports a role of this restriction factor in the transmissibility of primate lentiviruses. It will be important to define the structural determinants in Nef involved in SERINC5 antagonism to selectively disrupt this activity and to determine its relevance for viral replication, pathogenesis, and transmission in vivo.
EXPERIMENTAL PROCEDURES
Expression Vectors
The pBJ6-SERINC5-HA as well as the pBJ5-MLV glycoGag 189-HA expression vectors have been described (Rosa et al., 2015). SERINC5 orthologs from other species were amplified from cDNA and compared with published sequences. For details, see Supplemental Experimental Procedures. Cloning of Nef mutants into the bi-cistronic CMV-based pCG expression vector coexpressing the GFP was essentially performed as described previously (Kluge et al., 2014). The pCG control vector expressing only eGFP (eGFP control) contains an NL4-3 nef gene with a mutation in the initiation codon and two premature stop codons at positions 3 and 40 of the reading frame. All PCR-derived inserts were sequenced to confirm their accuracy.
Proviral Constructs
Generation of HIV-1 (NL4-3-based) proviral constructs carrying functional nef genes followed by an IRES element and the eGFP gene has been described previously (Schindler et al., 2006). See Supplemental Experimental Procedures for details and generation of additional proviral HIV-1 and SIV constructs.
Cell Culture and Transfections
Cells were cultured and transfected or infected as described in Supplemental Experimental Procedures.
SERINC5/3 Knockout Jurkat Cells
SERINC5 and SERINC3 were knocked out sequentially in Jurkat Tag cells by targeting two different exons of each gene using CRISPR/Cas9 as described in Supplemental Experimental Procedures.
Virus Yield and Infectivity Assays
Virus yield and infectivity assay were conducted in principle as described (Münch et al., 2007; Zhang et al., 2009). For details, see Supplemental Experimental Procedures.
Virus Stocks and Transduction
HEK293T cells were transfected with HIV-1 (NL4-3) constructs coexpressing Nef and eGFP by the calcium phosphate method as described previously (Münch et al., 2007). Prestimulated CD4+ T cells were transduced as described in Supplemental Experimental Procedures.
Flow Cytometric Analysis
To determine the Nef-mediated down-modulation of CD4 and MHC-I from the surface of infected cells, CD4+ T cells were transduced with HIV-1 (NL4-3) constructs coexpressing Nef and eGFP. Three days post-transduction, receptor expression was examined by fluorescence-activated cell sorting analysis in GFP+ cells as described previously (Schindler et al., 2006). See Supplemental Experimental Procedures for details.
Western Blot
AU-1-tagged Nef proteins were generated and analyzed as described previously (Heigele et al., 2012). For details, see Supplemental Experimental Procedures.
Sequence Analyses and Accession Numbers
SERINC5 protein sequences were aligned using Multalign, followed by some manual editing. GenBank accession numbers for SERINC5 and Nef sequences utilized in this paper are provided in the Supplemental Information.
Structural Analysis of Nef
Protein structures are based on a complex of NL4-3 Nef with the α and σ2 subunits of AP-2 (Protein Data Bank accession number 4NEE) and were displayed with CLC Main Workbench 7.
Statistical Analysis
Statistical calculations and group comparisons were performed using a two-tailed Student’s t test implemented in the Prism package version 4.0 (Abacus Concepts). p values < 0.05 were considered to indicate statistical significance. Correlations were calculated with the linear regression module. The method of phylogenetically independent contrasts (Felsenstein, 1985), implemented in the R package Analysis of Phylogenetics and Evolution (Paradis et al., 2004), was used to assess the relationship of SIV prevalence rates to viral infectivity using the phylogenetic relationships of the corresponding Nef amino acid sequences to correct for the underlying shared ancestry. Because phylogenetically independent contrast values have an expected mean of zero, linear regressions were constrained to go through the origin (Garland et al., 1992).
Supplementary Material
Highlights.
SERINC5 antagonism is a fundamental property of primate lentiviral Nef proteins
Increases in Nef-mediated anti-SERINC5 activity preceded the emergence of HIV-1
HIV-1 Nefs are more potent SERINC5 antagonists than HIV-2 Nefs
The potency of SERINC5 antagonism correlates with SIV prevalence
Acknowledgments
We thank Daniela Krnavek for excellent technical assistance and Paul Sharp and Dominik Hotter for helpful comments and suggestions. TZM-bl cells were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. This work was supported by grants from Deutsche Forschungsgemeinschaft (DFG), European FP7 “HIT HIDDEN HIV” (305762), a Leibniz award of DFG, and an Advanced ERC investigator grant to F.K., as well as grants from the NIH (R37 AI 050529, R01 AI 114266, R01 AI 111789, P01 AI 045008) and the National Agency for Research on AIDS and Hepatitis (12325). M. Pizzato was supported by an FP7 Career Integration Grant (322130) and Caritro “Ricerca Biomedica” (2013.0248).
Footnotes
ACCESSION NUMBERS
The GenBank accession numbers for the SERINC5 and Nef sequence data reported in this paper are included in the Supplemental Information.
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2016.08.004.
AUTHOR CONTRIBUTIONS
A.H., D.K., and K.R. performed most experiments and analyzed the data. S.L., L.P., C.M.S., D.S., M. Peeters, and M. Pizzato provided reagents and expertise. G.H.L. performed phylogenetic and statistical analyses. A.H., B.H.H., and F.K. conceived and designed the experiments. F.K. and B.H.H. ensured rigorous analyses and wrote the manuscript.
References
- Aiken C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol. 1997;71:5871–5877. doi: 10.1128/jvi.71.8.5871-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argañaraz ER, Schindler M, Kirchhoff F, Cortes MJ, Lama J. Enhanced CD4 down-modulation by late stage HIV-1 nef alleles is associated with increased Env incorporation and viral replication. J Biol Chem. 2003;278:33912–33919. doi: 10.1074/jbc.M303679200. [DOI] [PubMed] [Google Scholar]
- Arhel NJ, Kirchhoff F. Implications of Nef: host cell interactions in viral persistence and progression to AIDS. Curr Top Microbiol Immunol. 2009;339:147–175. doi: 10.1007/978-3-642-02175-6_8. [DOI] [PubMed] [Google Scholar]
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Campbell EM, Nunez R, Hope TJ. Disruption of the actin cytoskeleton can complement the ability of Nef to enhance human immunodeficiency virus type 1 infectivity. J Virol. 2004;78:5745–5755. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell-Yesufu OT, Gandhi RT. Update on human immunodeficiency virus (HIV)-2 infection. Clin Infect Dis. 2011;52:780–787. doi: 10.1093/cid/ciq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl S, Greenough TC, Krumbiegel M, Greenberg M, Skowronski J, Sullivan JL, Kirchhoff F. Modulation of different human immunodeficiency virus type 1 Nef functions during progression to AIDS. J Virol. 2001;75:3657–3665. doi: 10.1128/JVI.75.8.3657-3665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’arc M, Ayouba A, Esteban A, Learn GH, Boué V, Liegeois F, Etienne L, Tagg N, Leendertz FH, Boesch C, et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci U S A. 2015;112:E1343–E1352. doi: 10.1073/pnas.1502022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Garland T, Jr, Harvey PH, Ives AR. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst Biol. 1992;41:18–32. [Google Scholar]
- Greenberg ME, Bronson S, Lock M, Neumann M, Pavlakis GN, Skowronski J. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J. 1997;16:6964–6976. doi: 10.1093/emboj/16.23.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller C, Müller B, Fritz JV, Lamas-Murua M, Stolp B, Pujol FM, Keppler OT, Fackler OT. HIV-1 Nef and Vpu are functionally redundant broad-spectrum modulators of cell surface receptors, including tetraspanins. J Virol. 2014;88:14241–14257. doi: 10.1128/JVI.02333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigele A, Schindler M, Gnanadurai CW, Leonard JA, Collins KL, Kirchhoff F. Down-modulation of CD8αβ is a fundamental activity of primate lentiviral Nef proteins. J Virol. 2012;86:36–48. doi: 10.1128/JVI.00717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 2009;5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestler HW, 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- Kluge SF, Mack K, Iyer SS, Pujol FM, Heigele A, Learn GH, Usmani SM, Sauter D, Joas S, Hotter D, et al. Nef proteins of epidemic HIV-1 group O strains antagonize human tetherin. Cell Host Microbe. 2014;16:639–650. doi: 10.1016/j.chom.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama J, Mangasarian A, Trono D. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol. 1999;9:622–631. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- Li Y, Ndjango JB, Learn GH, Ramirez MA, Keele BF, Bibollet-Ruche F, Liu W, Easlick JL, Decker JM, Rudicell RS, et al. Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J Virol. 2012;86:10776–10791. doi: 10.1128/JVI.01498-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Warmerdam MT, Gaston I, Greene WC, Feinberg MB. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch J, Schindler M, Wildum S, Rücker E, Bailer N, Knoop V, Novembre FJ, Kirchhoff F. Primary sooty mangabey simian immunodeficiency virus and human immunodeficiency virus type 2 nef alleles modulate cell surface expression of various human receptors and enhance viral infectivity and replication. J Virol. 2005;79:10547–10560. doi: 10.1128/JVI.79.16.10547-10560.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch J, Rajan D, Schindler M, Specht A, Rücker E, Novembre FJ, Nerrienet E, Müller-Trutwin MC, Peeters M, Hahn BH, Kirchhoff F. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J Virol. 2007;81:13852–13864. doi: 10.1128/JVI.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol. 2012;86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pawlak EN, Dikeakos JD. HIV-1 Nef: a master manipulator of the membrane trafficking machinery mediating immune evasion. Biochim Biophys Acta. 2015;1850:733–741. doi: 10.1016/j.bbagen.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Pizzato M. MLV glycosylated-Gag is an infectivity factor that rescues Nef-deficient HIV-1. Proc Natl Acad Sci U S A. 2010;107:9364–9369. doi: 10.1073/pnas.1001554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyndiah N, Telenti A, Rausell A. Evolutionary genomics and HIV restriction factors. Curr Opin HIV AIDS. 2015;10:79–83. doi: 10.1097/COH.0000000000000138. [DOI] [PubMed] [Google Scholar]
- Qi M, Aiken C. Selective restriction of Nef-defective human immunodeficiency virus type 1 by a proteasome-dependent mechanism. J Virol. 2007;81:1534–1536. doi: 10.1128/JVI.02099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Park SY, Bonifacino JS, Hurley JH. How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. eLife. 2014;3:e01754. doi: 10.7554/eLife.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeth JF, Collins KL. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol Mol Biol Rev. 2006;70:548–563. doi: 10.1128/MMBR.00042-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015;526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Münch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Specht A, Kirchhoff F. Tetherin: holding on and letting go. Cell. 2010;141:392–398. doi: 10.1016/j.cell.2010.04.022. [DOI] [PubMed] [Google Scholar]
- Sauter D, Hué S, Petit SJ, Plantier JC, Towers GJ, Kirchhoff F, Gupta RK. HIV-1 Group P is unable to antagonize human tetherin by Vpu, Env or Nef. Retrovirology. 2011;8:103. doi: 10.1186/1742-4690-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Unterweger D, Vogl M, Usmani SM, Heigele A, Kluge SF, Hermkes E, Moll M, Barker E, Peeters M, et al. Human tetherin exerts strong selection pressure on the HIV-1 group N Vpu protein. PLoS Pathog. 2012;8:e1003093. doi: 10.1371/journal.ppat.1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Hotter D, Van Driessche B, Stürzel CM, Kluge SF, Wildum S, Yu H, Baumann B, Wirth T, Plantier JC, et al. Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 2015;10:586–599. doi: 10.1016/j.celrep.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Münch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Müller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, et al. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell. 2006;125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Schmökel J, Li H, Bailes E, Schindler M, Silvestri G, Hahn BH, Apetrei C, Kirchhoff F. Conservation of Nef function across highly diverse lineages of SIVsmm. Retrovirology. 2009;6:36. doi: 10.1186/1742-4690-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmökel J, Sauter D, Schindler M, Leendertz FH, Bailes E, Dazza MC, Saragosti S, Bibollet-Ruche F, Peeters M, Hahn BH, Kirchhoff F. The presence of a vpu gene and the lack of Nef-mediated downmodulation of T cell receptor-CD3 are not always linked in primate lentiviruses. J Virol. 2011;85:742–752. doi: 10.1128/JVI.02087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno F, Shiota H, Miyaura M, Yoshida A, Sakurai A, Tatsuki J, Koyama AH, Akari H, Adachi A, Fujita M. Vpx and Vpr proteins of HIV-2 up-regulate the viral infectivity by a distinct mechanism in lymphocytic cells. Microbes Infect. 2003;5:387–395. doi: 10.1016/s1286-4579(03)00042-x. [DOI] [PubMed] [Google Scholar]
- Usami Y, Göttlinger H. HIV-1 Nef responsiveness is determined by Env variable regions involved in trimer association and correlates with neutralization sensitivity. Cell Rep. 2013;5:802–812. doi: 10.1016/j.celrep.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Wu Y, Göttlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn F, Li Y, Bailes E, Neel C, Lafay B, Keele BF, Shaw KS, Takehisa J, Kraus MH, Loul S, et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368:155–171. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Wildum S, Schindler M, Münch J, Kirchhoff F. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J Virol. 2006;80:8047–8059. doi: 10.1128/JVI.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Lopez LA, Exline CM, Haworth KG, Cannon PM. Lack of adaptation to human tetherin in HIV-1 group O and P. Retrovirology. 2011;8:78. doi: 10.1186/1742-4690-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YH, Plemenitas A, Fielding CJ, Peterlin BM. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc Natl Acad Sci U S A. 2003;100:8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.