Abstract
The fungal kingdom is the source of almost all industrial enzymes in use for lignocellulose bioprocessing. We developed a systems-level approach that integrates transcriptomic sequencing, proteomics, phenotype, and biochemical studies of relatively unexplored basal fungi. Anaerobic gut fungi isolated from herbivores produce a large array of biomass-degrading enzymes that synergistically degrade crude, untreated plant biomass and are competitive with optimized commercial preparations from Aspergillus and Trichoderma. Compared to these model platforms, gut fungal enzymes are unbiased in substrate preference due to a wealth of xylan-degrading enzymes. These enzymes are universally catabolite-repressed and are further regulated by a rich landscape of noncoding regulatory RNAs. Additionally, we identified several promising sequence-divergent enzyme candidates for lignocellulosic bioprocessing.
Lignocellulosic biomass from plant matter is an abundant, renewable starting material for biofuel and industrial chemical production (1, 2). Industrial-scale processes require fungal enzymes to convert biomass into fermentable sugars. However, to permit enzymatic degradation and sugar release (3), lignin must be removed from crude biomass via costly pretreatment processes (1). The need for multiple enzyme production processes increases this cost further, as genetically modified fungal platforms such as Trichoderma reesei and Aspergillus nidulans overproduce limited subsets of enzymes that are unable to independently digest even pretreated substrates completely to sugars (table S1) (4). Economical chemical production will require a versatile, unbiased platform to produce all of the enzymes needed to hydrolyze diverse lignocellulose feedstocks into fermentable sugars without pretreatment.
Microbes found in the digestive tract of large herbivores are attractive enzyme platforms for lignocellulose processing (5). Among these are Neocallimastigomycota (anaerobic gut fungi), the primary colonizers of biomass in ruminants and the earliest-branching nonparasitic fungi still living (6). Although Neocallimastigomycota account for ~8% of the gut microflora, they degrade up to 50% of the untreated biomass through invasive growth and enzyme secretion (7-9). Neocallimastigomycota contain a diverse repertoire of biomass-degrading enzymes (table S1) that degrade a range of feedstocks with equal efficiency (Fig. 1), making them rich untapped sources for previously unidentified lignocellulolytic enzymes. However, their strict anaerobic lifestyle, complex nutritional requirements, and culture recalcitrance have severely hindered early attempts at isolation, exploitation, and molecular characterization (10).
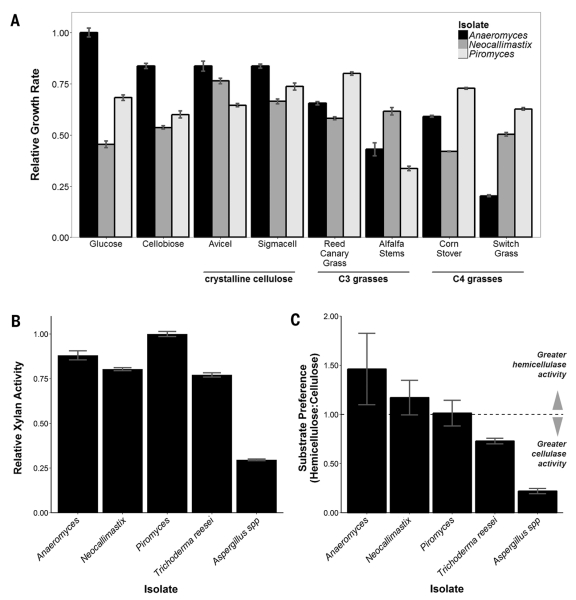
Fig. 1. Anaerobic fungi degrade crude biomass.
(A) Relative growth of gut fungal isolates on crystalline cellulose and crude C3 and C4 bioenergy crops (see table S3 for specific growth rates). (B) Relative xylan activity of cellulose-precipitated gut fungal secretions and commercial Trichoderma [Celluclast (Sigma-Aldrich)] and Aspergillus [Viscozyme (Sigma-Aldrich)]. (C) Relative hemicellulose:cellulose [xylan versus carboxymethylcellulose (CMC)] activity of cellulose-precipitated gut fungal secretions and commercial preparations. For all panels, data represent mean ± SEM (error bars) of more than three samples.
We isolated three previously uncharacterized cultures from the feces of different herbivorous mammals with varied diets. We used microscopy and ITS1 (internal transcribed spacer 1) sequencing (11) to verify that the isolates were distinct species, representing separate genera of Neocallimastigomycota (Anaeromyces robustus, Neocallimastix californiae, and Piromyces finnis). Each grew on C3 and/or C4 grasses at rates comparable to its growth on soluble substrates (Fig. 1A). Anaeromyces had a clear preference for glucose and grew more slowly on switch grass (~20% of the glucose growth rate). In contrast, the monocentric fungi, Piromyces and Neocallimastix, displayed limited substrate preference, with growth rates varying no more than 20% from the mean growth rate across all substrates. Similarly, these fungi had slight growth advantages on crude lignocellulose, growing up to 20% faster on reed canary grass (Phalaris arundinacea), an invasive species and bioenergy crop (12), when compared with glucose.
We collected and purified the biomass-degrading enzymes from fungal supernatants by exploiting the ability of many cellulases to bind to cellulose. We then tested the purified extracts for hydrolytic activity against several cellulosic substrates and analogs (fig. S1). Gut fungal secretions were active against all tested substrates, demonstrating cellulase, β-glucosidase, and hemicellulase activities comparable to those from engineered preparations of Trichoderma and Aspergillus. Neocallimastigomycota, and Piromyces in particular, displayed as much as a 300% increase in xylan-degradation activity when compared with commercial Aspergillus enzyme formulations (Fig. 1B). Gut fungi degrade cellulose at similar rates, demonstrating little preference for cellulose or hemicellulose (Fig. 1C), in agreement with their enzymatic distribution from genomic sequencing (table S1). This comprehensive array of biomass-degrading enzymes, and their inherent synergy, broadens the range of substrates that can be degraded effectively, making gut fungi better suited than later-diverging species to degrade diverse polymers found within crude plant biomass. More importantly, it is this synergy, and not enzyme diversity, that is responsible for the superior biomass-degradation abilities of Piromyces, making it an intriguing model system for further study.
Transcripts encoding biomass-degrading enzymes comprise ~2% of the gut fungal transcriptomes (data S1 to S3) containing diverse functions classified into distinct lignocellulolytic glycosyl hydrolase (GH) and other carbohydrate-active enzyme (CAZyme) domains (13) (Fig. 2A). The majority of these transcripts also encode non-catalytic dockerin domains thought to mediate self-assembly of an extracellular catalytic complex or cellulosome (Fig. 2, B and C) for synergistic degradation of lignocellulose (14). The hydrolytic capabilities of gut fungi on crude biomass are well explained by the functional expansions of many CAZyme families (table S1 and fig. S3). Neocallimastigomycota are rich in hemicellulases (notably GH10) and polysaccharide deacetylases, which allow gut fungi to remove hemicellulose and access the energy-rich cellulose core of plant biomass (15) in the absence of pretreatment. This process is greatly aided by pectin removal (16) with a number of polysaccharide lyases, carbohydrate esterases, and GH88s, allowing the anaerobic fungi to readily degrade an array of lignin-rich C3 and C4 bioenergy crops without pretreatment (Fig. 1A).
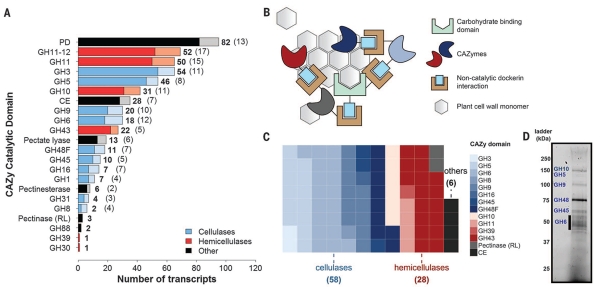
Fig. 2. Anaerobic fungi contain a wealth of biomass-degrading machinery.
(A) Distribution of cellulolytic carbohydrate-active enzyme (CAZy) transcripts and their regulatory antisense transcripts in Piromyces. CAZymes are shown in bold, whereas antisense transcripts are indicated in parentheses and plotted in a lighter shade. In the key, “Other” refers to pectinases and accessory enzymes that separate cellulose and hemicellulose from other cell wall constituents. PD, polysaccharide deacetylase (acetylxylan esterase); CE, carbohydrate esterase (excluding pectinesterases); RL, rhamnogalacturonate lyase. (B) Proposed model for an extracellular catalytic complex for cellulose degradation. (C) CAZyme composition of the putative extracellular complex. Each square represents a single enzyme that encodes a CAZyme fused to at least one dockerin domain. (D) Identity of predominant secreted gut fungal CAZymes in the cellulose-precipitated fraction. Bands were excised and mapped to the transcriptome by tandem MS (fig. S4).
Functional annotations of the transcriptome were validated within Piromyces, Anaeromyces, and Neocallimastix via a proteomic survey (data S5 to S7). Proteins secreted from Piromyces in the presence of reed canary grass were isolated by cellulose precipitation (Fig. 2D and fig. S4) and individually mapped by mass spectrometry (MS) (17) to more than 50 cellulolytic transcripts including 25 GH families enriched in or specific to the anaerobic fungal lineage (e.g., GH9, GH45, GH48, GH10, and GH11). Also present was the full complement of endoglucanases, exoglucanases, and β-glucosidases needed to fully depolymerize cellulose (GH5, GH6, GH9, GH45, and GH48) and hemicellulases (GH10 and GH11) (table S2), with many transcripts containing dockerin domains for extracellular fungal cellulosome formation.
A pervasive feature of gut fungal transcriptomes is long noncoding antisense transcripts (asRNA) (data S1 to S3). At least 11% of the Piromyces transcriptome is noncoding and complementary to putative targets involved in a range of catalytic and developmental pathways, including biomass degradation (Fig. 2A and fig. S2). asRNA is functionally enriched (hypergeometric test) in a number of gene ontology (GO) processes, such as the cellulose catabolic process (P = 0.02), ribosome biogenesis (P = 10−11), RNA-dependent DNA replication (P = 6 × 10−6), and amino acid transmembrane transport (P = 0.003) (data S4). These results infer a role for asRNA regulation in fungal cellulose catabolism and suggest that noncoding asRNAs may be as critical for function in early-branching Neocallimastigomycota as they are in higher fungal lineages (18-20).
To assess how the activities of biomass-degrading enzymes are coordinated, we grew Piromyces cultures on lignocellulose and perturbed the system with a small pulse of glucose to induce catabolite repression, after which we collected RNA samples until the glucose was consumed (Fig. 3A). Three hundred seventy-four transcripts showed more than a twofold change in expression (P ≤ 0.01), with a third of these transcripts containing cellulolytic domains (Fig. 3B). Among these regulated cellulolytic transcripts were all of the MS-validated proteins expressed under growth on reed canary grass (table S2), with the exception of GH45 and XylA. Transcripts associated with biomass degradation were almost exclusively repressed in response to glucose, as expected, and reflected activity trends from cellulose-isolated secretions (21). Expression levels of these transcripts returned to initial baselines once glucose was fully consumed (Fig. 3C and fig. S5). Cluster analysis revealed coordinated expression signatures of biomass degradation in the regulatory patterns of these transcripts (22).
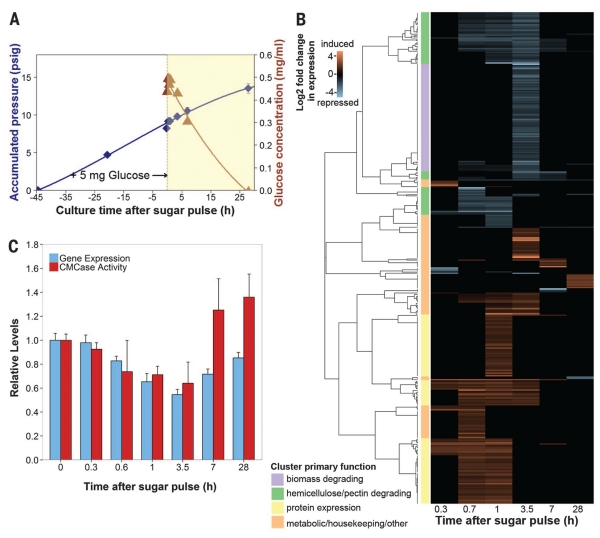
Fig. 3. Anaerobic fungal biomass-degrading machinery is catabolically repressed.
(A) Glucose consumption in fungal cultures. Exponential cultures of Piromyces were pulsed with 5 mg of glucose, and mRNA and secretome samples were collected during glucose depletion (yellow region). Blue diamonds, accumulated pressure; brown triangles, glucose concentration. Error bars indicate SEM. (B) Cluster analysis of genes strongly regulated by glucose. Transcript abundance data were compared to uninduced samples at time t = 0 to calculate the log2 fold change in expression.Transcripts with large, significant regulation are displayed (P ≤ 0.01, negative binomial distribution, ≥twofold change). Clusters were manually annotated based on the most common protein domains and/or BLAST (Basic Local Alignment Search Tool) hits. (C) Relative expression [fragments per kilobase of transcript per million mapped reads (FPKM)] of biomass-degrading enzymes (table S1) and their corresponding activity (cellulosome fraction) on CMC (21). Data represent the mean ± SEM (error bars) of at least two replicates.
Hierarchical cluster analysis revealed that glucose-regulated genes performing a common function grouped into 21 distinct clusters or regulons (Fig. 3B). Owing to the functional enrichment of these regulons, divergent transcripts of unknown function that co-regulate with biomass-degrading transcripts may be previously unidentified biomass-degrading enzymes for biotechnology. We identified 17 such candidates from Piromyces (table S4). Biomass-degrading regulons were either hemicellulose- and pectin-degrading and rapidly repressed within 40 min or contained a broad array of biomass-degrading enzymes that responded more slowly at 3.5 hours (Fig. 3B and data S8). The faster regulatory response of hemicellulases is conserved in higher fungi (23, 24) and thought to be an adaptation to lignocellulose structure. Hemicellulose and pectin surround cellulose; thus, cellulases act only after the hemicellulases and pectinases remove this outer coating. Given a common regulatory input, coordinating this expression leads to quicker regulation of hemicellulases and pectinases than of cellulases, in agreement with observation. Candidate mediators of this response include conserved orthologs of the fungal master carbon regulator (CreABC); xylose-sensitive transcription factors (Xlr-1 and XlnR); and other conserved cellulolytic activators such as ACE1-2, ClbR, and Clr1-2 (table S5). In contrast, up-regulated clusters contained an array of metabolic and housekeeping genes consistent with logarithmic growth, along with protein expression genes (such as those encoding chaperonins and ribosomal RNA processing proteins) that probably mediated the cellular response to the sugar pulse (data S8).
To better understand the regulation of key biomass-degrading enzymes, we analyzed expression as a function of substrate. Piromyces showed substantial remodeling of the transcriptome as the carbon source was varied (~10% of all transcripts), reflecting changes in the biomass-degrading machinery and internal processes of gut fungal cultures (fig. S6 and data S9). Among these transcripts were 194 of the differentially regulated transcripts from the glucose perturbation experiment described above. Overall, a twofold change in the expression of biomass-degrading enzymes occurred during the switch from glucose to complex reed canary grass. This trend was mirrored in the activity of cellulose-precipitated secretions (Fig. 4A). Discernible changes in the composition of the biomass-degradation machinery also accompanied variations in expression level (fig. S7).
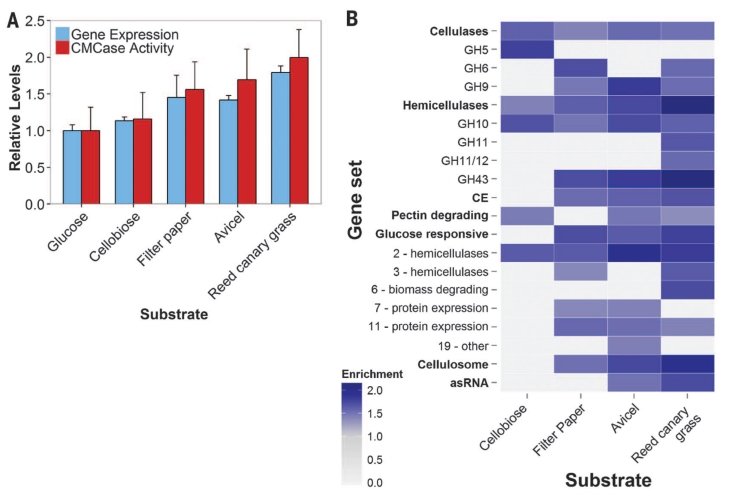
Fig. 4. Anaerobic fungi degrade complex substrates with increasingly diverse enzymes.
(A) Relative expression (FPKM) of biomass-degrading enzymes (table S1) and their activity (cellulosome fraction) on CMC. Data represent the mean ± SEM (error bars) of at least two replicates. (B) Normalized enrichment scores of positively enriched specified gene sets relative to growth on glucose. Gene sets containing genes that are expressed more highly in a given substrate are indicated (FDR ≤ 10%; Kolmogorov-Smirnov distribution). Enrichment scores are directly proportional to expression level. Gene sets shown in bold were analyzed in aggregate and in subsets (unbolded sets below). asRNA, antisense RNA that targets CAZy domains (Fig. 2A); Cellulosome, dockerin tagged transcripts. Numbers in the “Glucose responsive” subset indicate clusters.
Gene set enrichment analysis (25) of the transcriptomes confirmed that the number and functional diversity of CAZyme domains increased as a function of substrate complexity (Fig. 4B), with insoluble substrates [filter paper, Avicel (Sigma-Aldrich), and reed canary grass] inducing fungal cellulosomes for enhanced degradation. Nonhemicellulosic substrates (cellobiose, filter paper, and Avicel) up-regulated unneeded hemicellulases such as GH10, suggesting a common regulatory network for diverse enzymes. Nonetheless, the additional enzymes necessary to degrade crude reed canary grass are independently regulated. Our analyses also revealed shifts between enzyme types for similar reactions (e.g., GH5 to GH9 as a β-glucosidase) as a function of substrate, demonstrating a highly tailored catabolic response.
Among the gene sets we tested were clusters identified in the glucose perturbation experiment (Fig. 4B). Protein expression clusters (Fig. 3B) regulated by glucose were enriched on insoluble substrates, reaffirming their role in mediating expression of lignocellulolytic enzymes. Another regulon encoding diverse hemicellulases and a handful of cellulases (cluster 2: hemicellulases) was central to all growth phenotypes other than glucose. This enzyme prevalence, even on non-polymeric carbohydrates, suggests that enzymes play an integral role in the recognition of insoluble substrates (26): In the absence of glucose, these enzymes are expressed at low levels to partially solubilize available cellulosic materials that can be recognized to trigger a more specific catabolic response. Consistent with this hypothesis is the sixfold up-regulation (P ~ 0.02, negative binomial test) of the conserved transcription factor XlnR on reed canary grass and Avicel to better recognize solubilized sugars and induce fungal xylan degradation. This response is further regulated by asRNA targeting CAZyme domains, as evidenced by their functional enrichment on Avicel [P = 0.003, false discovery rate (FDR) = 0.03] and reed canary grass cultures (P ~ 0, FDR = 0.003). An independent analysis using a hypergeometric statistical test confirms that antisense transcripts targeting CAZyme domains (cellulose catabolic process GO annotation) are functionally enriched among the regulated transcripts (P ≈ 0.01) (data S10). Identities of the expressed asRNA, however, are substrate-specific to modify the catabolic response through a number of mechanisms (27) to conserve cellular resources (table S6).
Overall, our results show that anaerobic gut fungi tailor their hydrolytic response to lignocellulose, implying a coordination in catalysis between all expressed enzymes that may inform industrial hydrolysis strategies. The clear transcriptional signatures of these biomass-degrading enzymes provide a route to identify hundreds of sequence-divergent enzyme candidates with commercial potential from anaerobic microbial communities (28).
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Ngan, C. Daum, E. Lindquist, and K. Barry for supervising library construction, transcriptome sequencing and analysis, and project management; K. Lee and L. Choe for proteomics support; P. Weimer for lignocellulosic substrates; and the Broad Institute core facilities for sequencing and computational assistance. Sequence and cluster descriptions are included in data S1 to S3, S8, and S9 in the supplementary materials. Raw sequence data and transcriptomic profiles reported in this study are deposited under BioProject accession no. PRJNA 291757 (www.ncbi.nlm.nih.gov/bioproject/291757). Expression data are deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (29) and are accessible through GEO Series accession no. GSE64834 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64834). K.V.S., C.H.H., J.K.H., and M.A.O. are inventors on patent applications (UCSB 2014-075 and UCSB 2015-334), filed by The Reagents of the University of California, related to the production of anaerobic fungal enzymes. This work was supported by the Office of Science [Biological and Environmental Research (BER) program], DOE (grant DE-SC0010352); the U.S. Department of Agriculture (Award 2011-67017-20459); and the Institute for Collaborative Biotechnologies (grant W911NF-09-0001). A portion of this research was performed under the Joint Genome Institute (JGI)–Environmental Molecular Sciences Laboratory (EMSL) Collaborative Science Initiative and used resources at the DOE JGI and the EMSL, which are DOE Office of Science user facilities. Both facilities are sponsored by the Office of Science BER program and operated under contract nos. DE-AC02-05CH11231 (JGI) and DE-AC05-76RL01830 (EMSL). K.V.S., C.H.H., D.A.T., M.K.T., and M.A.O. planned the experiments. M.A.O., J.K.H., C.H.H., M.K.T., and K.V.S. isolated pure cultures of gut fungi. K.V.S., C.H.H., J.K.H., and M.A.O. performed growth and transcriptomic experiments. A.L. analyzed transcriptome sequencing. C.H.H., H.M.B., S.O.P., and A.T.W. performed proteomic analyses. S.P.G. performed enzyme characterization. K.V.S., D.B.-R., J.K.H., A.R., I.V.G., S.O.P., and S.P.G. facilitated bioinformatics analyses of the data sets. K.V.S., D.A.T., and M.A.O. wrote the manuscript.
Footnotes
www.sciencemag.org/content/351/6278/1192/suppl/DC1
Materials and Methods
Figs. S1 to S7
Tables S1 to S6
References (30–43)
Data S1 to S10
REFERENCES AND NOTES
- 1.Sanderson K. Nature. 2011;474:S12–S14. doi: 10.1038/474S012a. [DOI] [PubMed] [Google Scholar]
- 2.Dodds DR, Gross RA. Science. 2007;318:1250–1251. doi: 10.1126/science.1146356. [DOI] [PubMed] [Google Scholar]
- 3.Berlin A, et al. J. Biotechnol. 2006;125:198–209. doi: 10.1016/j.jbiotec.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Schülein M. Methods Enzymol. 1988;160:234–242. [Google Scholar]
- 5.Hess M, et al. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson MJ, Theodorou MK, Brookman JL. Microbiology. 2005;151:121–133. doi: 10.1099/mic.0.27353-0. [DOI] [PubMed] [Google Scholar]
- 7.Orpin CG, Gen J. Microbiol. 1977;99:107–117. doi: 10.1099/00221287-99-1-107. [DOI] [PubMed] [Google Scholar]
- 8.Theodorou MK, et al. Proc. Nutr. Soc. 1996;55:913–926. doi: 10.1079/pns19960088. [DOI] [PubMed] [Google Scholar]
- 9.Youssef NH, et al. Appl. Environ. Microbiol. 2013;79:4620–4634. doi: 10.1128/AEM.00821-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haitjema CH, Solomon KV, Henske JK, Theodorou MK, O’Malley MA. Biotechnol. Bioeng. 2014;111:1471–1482. doi: 10.1002/bit.25264. [DOI] [PubMed] [Google Scholar]
- 11.Tuckwell DS, Nicholson MJ, McSweeney CS, Theodorou MK, Brookman JL. Microbiology. 2005;151:1557–1567. doi: 10.1099/mic.0.27689-0. [DOI] [PubMed] [Google Scholar]
- 12.Lavergne S, Molofsky J. Crit. Rev. Plant Sci. 2004;23:415–429. [Google Scholar]
- 13.Lombard V, Ramulu H. Golaconda, Drula E, Coutinho PM, Henrissat B. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghothama S, et al. Nat. Struct. Mol. Biol. 2001;8:775–778. [Google Scholar]
- 15.Himmel ME, et al. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 16.Lionetti V, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:616–621. doi: 10.1073/pnas.0907549107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finehout EJ, Lee KH. Electrophoresis. 2003;24:3508–3516. doi: 10.1002/elps.200305615. [DOI] [PubMed] [Google Scholar]
- 18.Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK. Nature. 2003;421:948–952. doi: 10.1038/nature01427. [DOI] [PubMed] [Google Scholar]
- 19.Yassour M, et al. Genome Biol. 2010;11:R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donaldson ME, Saville BJ. Mol. Microbiol. 2012;85:405–417. doi: 10.1111/j.1365-2958.2012.08125.x. [DOI] [PubMed] [Google Scholar]
- 21.King BC, Donnelly MK, Bergstrom GC, Walker LP, Gibson DM. Biotechnol. Bioeng. 2009;102:1033–1044. doi: 10.1002/bit.22151. [DOI] [PubMed] [Google Scholar]
- 22.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakir U, Yavascaoglu S, Guvenc F, Ersayin A. Enzyme Microb. Technol. 2001;29:328–334. [Google Scholar]
- 24.Todero Ritter CE, Camassola M, Zampieri D, Silveira MM, Dillon AJP. Enzyme Res. 2013;2013:e240219. doi: 10.1155/2013/240219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glass NL, Schmoll M, Cate JHD, Coradetti S. Annu. Rev. Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 27.Pelechano V, Steinmetz LM. Nat. Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- 28.Solomon KV, Haitjema CH, Thompson DA, O’Malley MA. Curr. Opin. Biotechnol. 2014;28:103–110. doi: 10.1016/j.copbio.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Edgar R, Domrachev M, Lash AE. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.