Abstract
A genome-wide methylation study was conducted among a sample of 114 infants (M age = 13.2 mo.; SD = 1.08) of low-income urban women with (n=73) and without (n=41) Major Depressive Disorder (MDD). The Illumina HumanMethylation450 BeadChip array with the GenomeStudio Methylation Module and Illumina Custom model were used to conduct differential methylation analyses. Using the 5.0 × 10−7 p-value, 2119 loci were found to be significantly different between infants of depressed and non-depressed mothers. Infants of depressed mothers had greater methylation at low methylation sites (0–29%) compared to infants of non-depressed mothers. At high levels of methylation (70–100%) the infants of depressed mothers were predominantly hypomethylated. The mean difference in methylation between the infants of depressed and infants of non-depressed mothers was 5.23%. Disease by biomarker analyses were also conducted using GeneGo MetaCore Software. Results indicated significant cancer-related differences in biomarker networks such as prostatic neoplasms, ovarian and breast neoplasms, and colonic neoplasms. Results of a process networks analysis indicated significant differences in process networks associated with neuronal development and central nervous system functioning, as well as cardiac development between infants of depressed and non-depressed mothers. These findings indicate that early in development, infants of mothers with MDD evince epigenetic differences relative to infants of well mothers that suggest risk for later adverse health outcomes.
Introduction
As has been widely documented in the scientific literature, children of mothers with depression are at risk for a range of negative developmental sequelae over the course of development (Cicchetti & Schneider-Rosen, 1986; Cicchetti & Toth, 1998; Goodman & Gotlib, 1999). Infants of mothers with depression also exhibit problems with developmental attainments (Cicchetti & Aber, 1986; Field, 2008). Included among these negative effects of maternal depression on infant outcome are homeostatic and physiological dysregulation, affect differentiation and emotional responsivity, emotion dysregulation, insecure and disorganized attachment, hemispheric activation asymmetries, and self-other differentiation (Cicchetti & Toth, 1995, 1998; Cicchetti, Rogosch, Toth, & Spagnola, 1997; Cole, Luby & Sullivan, 2008; Davidson & Fox, 1982; Radke-Yarrow, Cummings, Kuczynski & Chapman, 1985).
The developmental cascade of negative developmental outcomes is attributed to interactive, experiential aspects of the mother-child relationship (e.g., maternal insensitivity, inappropriate responsivity, greater blunted, depressed, angry affect, etc.). (Cole et al., 2008; Cummings & Cicchetti, 1990). Thus, it is thought that infants of depressed mothers go through atypical, “not good enough,” rearing experiences and do not receive the appropriate level of relational “nutrients” to foster adaptive development.
The same scenario holds for infants who are maltreated. Stress mechanisms have been invoked as the “cause” of epigenetic modification in these children. Moreover, the infant of a depressed mother also may experience “stress,” but of a rather different kind. In addition to stress, there may be other aspects of the infant of a depressed mother’s relational experience that may be related to epigenetic modification, relative to infants with “good enough” rearing experiences.
We hypothesize that epigenetic modifications occur that are a consequence of rearing by a depressed mother. If there is no appreciable modification, then developmental differences in offspring may be more experientially based and not influenced by epigenetics. However, if there are differences, then consideration of the role of epigenetics in the developmental process and trajectories of these infants may be an important area of inquiry. A first step, then, is to determine if there is evidence for epigenetic differences.
This is the first study, or at least one of the very first studies, conducted with infants from low-SES backgrounds who are the offspring of mothers with, or without, major depressive disorder (MDD). Thus, this investigation contains both the risks associated with poverty and those associated with having a mother with MDD.
We hypothesize that infant offspring of mothers with MDD will evince methylation differences relative to infants of well mothers.
Compared to infants of well mothers, infants with MDD will demonstrate greater adverse physical and mental health risk based on patterns of differential methylation associated with disorder outcomes.
Method
Participants
Participants included 114 infants (M age = 13.2 mo.; SD = 1.08) of low-income urban women. The sample was drawn from a larger RCT investigation evaluating preventive interventions for mothers with Major Depressive Disorder (MDD) (Toth, Rogosch, Manly, & Cicchetti, 2006) and was comprised of infants of mothers with MDD (n = 73) and a comparison group of infants of mothers without depression (n = 41). More mothers and children were recruited for the depressed group for random assignment to intervention arms. Data from the current investigation were drawn from baseline assessments prior to randomization. Mothers provided informed consent for participation prior to the initiation of data collection, and the research was conducted in accord with the Institutional Review Board approval.
Recruitment
All mothers in the depressed group met criteria for MDD. We recruited a community sample of non-treatment-seeking women from primary care clinics serving low-income women and from Women, Infant and Children (WIC) clinics. To be eligible, women needed to reside at or below the federal poverty level. Seventy-eight percent of the sample was below the U.S. Department of Health and Human Services definition of poverty level, and 96% met WIC criteria (185% of the poverty level). A project recruitment coordinator initially screened women with the Center for Epidemiologic Studies-Depression Scale (CES-D; Radloff, 1977), and those scoring above 16 were targeted for further assessments to determine eligibility for inclusion. Women who subsequently scored 19 or higher on the Beck Depression Inventory II (BDI-II; Beck, Steer, & Brown, 1996), and who met MDD diagnostic criteria based on the operational criteria on the Diagnostic Interview Schedule (DIS-IV; Robins, Cottler, Bucholz, & Compton, 1995) were eligible to participate. For all but 6.3% of the women, the onset of their first major depressive episode had been over one year ago, thus preceding the infant’s birth. Accordingly, the current sample was not comprised of women with depression restricted to the post-partum period, but rather was of longer standing duration. At baseline, consistent with inclusion criteria, all mothers in the depressed group had BDI-II scores above 19 and also met criteria for a current MDD diagnosis. Women meeting diagnostic criteria for lifetime bipolar disorder or for any lifetime psychotic spectrum disorder were excluded. Women with Mood Disorder Due to a General Medical Condition and Substance-Induced Mood Disorder also were excluded, as were women with any current alcohol or substance abuse disorder, as defined by DSM-IV criteria. Women with other co-morbid disorders were not excluded.
Demographically comparable low-income mothers of infants in the nondepressed group also were recruited by the project recruitment coordinator at primary care clinics and WIC offices. These mothers also were screened for depressive symptoms with the CES-D, and those scoring below the clinical cutoff were invited for further screening. Baseline assessments with the BDI-II and DIS-IV were used to exclude mothers with BDI-II scores greater than 12 or with histories of MDD or other DSM-IV diagnoses.
Sample Characteristics
Infants in the depressed and nondepressed mother groups were comparable on a range of demographic variables. See Table 1. No significant differences were observed between groups for child age, t (112) = .25, p = .80, child gender, χ2 (1) = .20, p = .66, child race, χ2 (1) = .05, p = .83, or child ethnicity, χ2 (1) = .63, p = .43. Similarly, to characterize maternal and family demographics, the depressed and nondepressed groups were comparable in terms of maternal age, t (112) = .25, p = .80, marital status of never married, χ2 (1) = 2.28, p = .13, years of education, t (112) = 1.59, p = .12, low SES status based on the Hollingshead scale, χ2 (1) = 1.76, p = .19, and current receipt of public assistance, χ2 (1) = .17, p = .68. Mothers in the depressed and nondepressed groups differed substantially on BDI-II scores, t (102.3) = 22.72, p < .001, consistent with respective group recruitment criteria.
Table 1.
Demographic Characteristics
| Depressed Group M(SD) or % |
Nondepressed Group M (SD) or % |
|
|---|---|---|
| Child Age (mo.) | 13.27 (1.09) | 13.22 (.96) |
| Gender (% female) | 49.3 % | 53.2% |
| Child Race (% African-American) | 58.9% | 61.0% |
| Child Ethnicity (% Latino) | 28.8% | 22.0% |
| Maternal Age | 24.85 (5.04) | 24.63 (5.06) |
| Marital Status (% Never Married) | 80.8% | 68.3% |
| Years of Education | 11.68 (1.70) | 12.24 (1.97) |
| Current Public Assistance | 98.6% | 97.6% |
| BDI-II score | 31.23 (8.89) | 4.63 (3.48) |
Note: All contrasts were nonsignificant for demographic variables; BDI-II scores: p < .001.
Procedures
As part of the larger investigation, mothers and their infants participated in baseline assessments during laboratory- and home-based research sessions. All assessments were conducted by trained research assistants who were unaware of group condition or study hypotheses. During laboratory sessions, DNA samples were obtained from infants, as described below.
Maternal Measures
The Center for Epidemiologic Studies-Depression (CES-D; Radloff, 1977). The CES-D is a frequently used, well-validated 20-item scale to screen for depression. Scores > 16 predict a high likelihood of MDD.
Diagnostic Interview Schedule-IV (DIS-IV; Robins et al, 1995)
The DIS-IV is a structured interview designed to assess diagnostic criteria for Axis I disorders, as well as for antisocial personality disorder, as outlined in the Diagnostic and Statistical Manual of Mental Disorders (4th edition; American Psychiatric Association, 1994). The DIS-IV ascertains diagnoses present in the past year, the past six months, and those that are current or remitted. The DIS has been shown to be reliable and valid for use in psychiatric epidemiological field studies (Robins, Helzer, Croughan, & Ratcliff, 1981; Robins, Helzer, Ratcliff, & Seyfried, 1982). Robins et al. (1981) compared DSM diagnoses made using the DIS to those made by psychiatrists and reported mean κ = .69, sensitivity of 75%, and specificity of 94%. Given the forced choice structured format of the DIS, interviewers do not need to be trained clinicians. All interviewers were trained to criterion reliability in the administration of the DIS and computer-generated diagnoses were utilized.
Beck Depression Inventory-II (BDI-II; Beck, Steer, & Brown, 1996)
The BDI-II is the most widely used self-report instrument for measuring the severity of depression. It includes 21-questions in a multiple-choice format and scores of 17 or above indicate levels of depression with clinical significance. Previous studies report that the BDI-II demonstrates good internal consistency (coefficient alpha of .91) and validity (Dozois, Dobson & Ahnberg 1998; Storch, Roberti, & Roth, 2004). In the current study, the average internal consistency of the BDI-II based on the three assessments was α = .94.
DNA Sampling and Methylation
Buccal swab samples were collected from infants during baseline laboratory sessions using the BuccalAmp™ DNA Extraction Kit (Epicentre, Item#BQ0901). After collection, the swabs were processed into the provided QuickExtract solution and stored at −80°C. The entire sample of approximately 450 ul was purified using a chloroform extraction in 1.5 ml MaXtract High Density tubes (Qiagen, Item# 129046). The aqueous layer was recovered and DNA precipitated by adding 15 ul 3M Sodium Acetate and 1 ml 100% Ethanol and incubating at −20°C for 1 hr. The sample was then pelleted by centrifugation in an Allegra 25R (Beckman Coulter, Inc.) at 12,000 × g for 20 min. The samples were then washed in 80% Ethanol 3 times, dried and resuspended in 50 ul 1× TE.
The diluted DNA samples were submitted to the BioMedical Genomics Center (BMGC) at the University of Minnesota for quality analysis in preparation for testing of whole genome methylation analysis using the HumanMethylation450 BeadChip (Illumina). The samples were assayed for quality by determining the concentration by Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Item #P7589) and Real Time PCR (TaqMan) quantification of human DNA concentration. DNA samples were included based on the previously determined criteria of greater than 40% of the total DNA in the sample to be human (Real Time PCR divided by PicoGreen) and a minimum of 40 ng human DNA.
For samples that passed quality control testing, 40 ng of each sample, as well as 3 study samples at 60 ng, 40 ng (replicated) and 20 ng, were subjected to bisulfite conversion using the EZ-96 DNA Methylation Kit (Zymo Research, D5003) which converts unmethylated Cytosine bases to Uracils. This method utilizes the methyl group attached to a Cytosine as a protecting group to deamination and subsequent conversion to a Uracil. After bisulfite conversion, the total amount of DNA was increased by methylation specific amplification (MSA) using a whole genome amplification process which copies the converted Uracils to Thymine bases. The DNA was then enzymatically fragmented in an end-point fragmentation process.
Microarray processing and analysis of the Illumina Infinium HumanMethylation450 BeadChip then proceeded at U. of Minnesota’s BMGC. This BeadChip covers over 485,000 individual sites with single nucleotide resolution of CpG sites both inside and outside CpG islands and greater than 90% of the content in common with the HumanMethylation27 BeachChip. The HumanMethylation450 BeadChip offers comprehensive genome-wide coverage including 99% of RefSeq genes with high quality by using more than 600 negative controls. Bisulfite converted samples were then hybridized to these BeadChips followed by washing and staining per protocols prescribed by Illumina. The microarray bead chips were then imaged using a HiScan SQ system.
The fluorescence data was subsequently analyzed using the Methylation Module v1.9.0 of the GenomeStudio software package v2011.1 (Illumina). All data were background corrected and negative control normalized producing average beta values. This average beta value represents the relative quantity of methylation at an individual site ranging from 0 to 1 (unmethylated to completely methylated). Tests that produced different results from technical replicates of the three control DNA samples at 60 ng, 40 ng and 20 ng Human DNA were identified as poor and removed from subsequent analyses. This was accomplished by using differential methylation analysis of replicate sample average beta and the loci with a |DiffScore| > 13, which is equivalent to a p<0.01 as determined by comparing each sample individually at 40 ng to the 20, 40 and 60 ng quantities. These suspect loci, N=75,512, and those tests with p-values of greater than 0.01, N=4612, were excluded (N=77018, 15.9%). Beta values were analyzed using principle component analysis (PCA) in Partek Genomics Suite, Partek Inc. Review of the data distribution identified 8 samples as outliers which were subsequently removed from further analysis.
Methylation Analysis Approach
Differential methylation analyses were conducted for infants of depressed and non-depressed mothers. These analyses were performed after subtracting background noise and normalizing to array controls using GenomeStudio, Methylation Module and the Illumina Custom Model. The resulting measure of this calculation set is delta beta, which represents the amount of change to the average beta at a site, or relative percent methylation change between defined groups. Positive delta beta values indicate an elevation in relative methylation and a negative value indicates a reduction. Because the data set includes both male and female infants, the X and Y chromosome data were removed from subsequent analyses. To correct for multiple testing, the significance threshold to determine differential methylation was set to 5.0 X 10−7, which is consistent with prior research (i.e. Yang et al., 2013) and recommendations by Raykan and colleagues (2011). To investigate the association maternal depression with known disease biomarkers, the delta beta values of differentially methylated loci (p<5.0 × 10−7) were then analyzed using GeneGo MetaCore Software (Thomas-Reuters, MetaCore Version 6.23).
Results
Differential methylation analysis
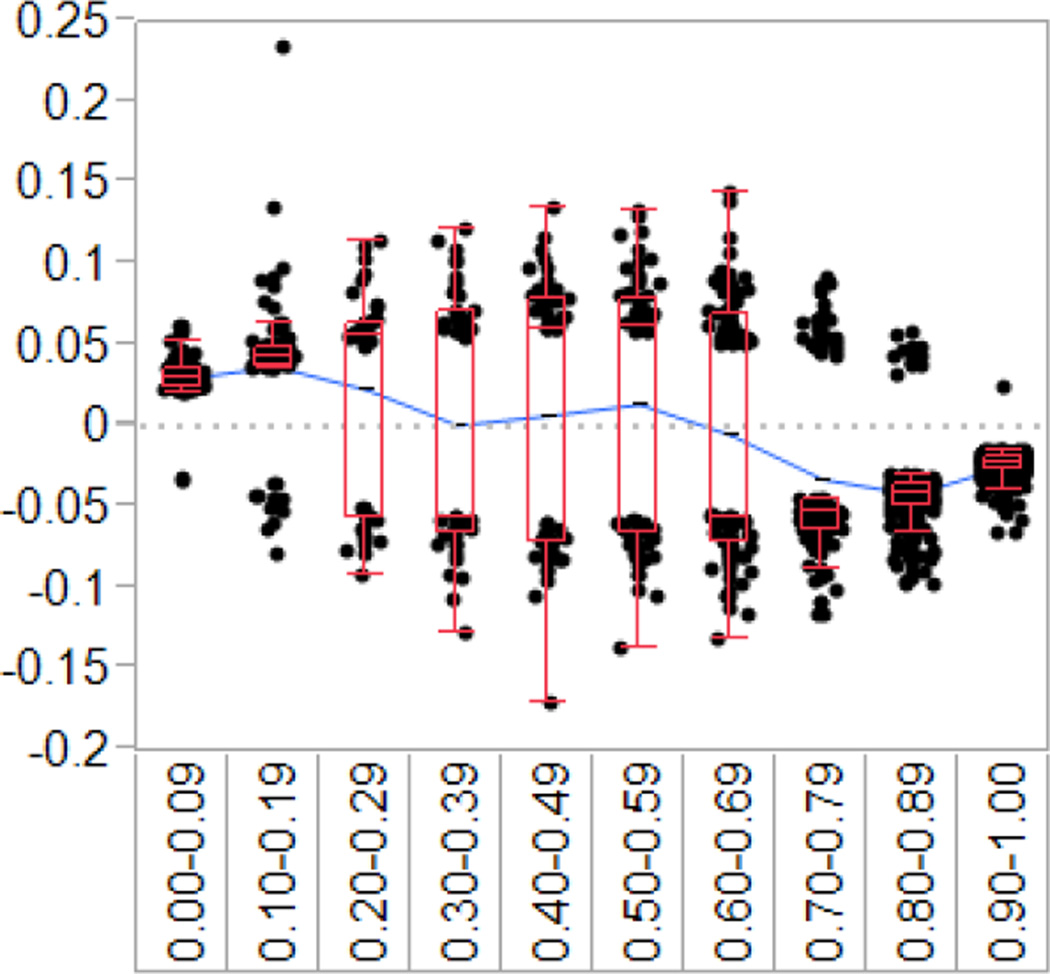
Using the 5.0 × 10−7 p-value, 2119 loci were found to be significantly different between infants of depressed and non-depressed mothers. The delta beta information was binned to 10% increments based on the expected amount of percent methylation from the infants of non-depressed mother group, or expected average beta. Results indicated a pattern of greater methylation from 0–20% and less methylation from 70–100%. The data are also heavily weighted to the 70–100% end on the continuum. (see Figure 1 for graphical representation). Data were further classified at low methylation (0–29%), medium methylation (30–69%), and high methylation (70–100%). Table 2 illustrates that infants of depressed mothers had greater methylation at low methylation sites (0–29%) compared to infants of non-depressed mothers. At high levels of methylation (70–100%) the infants of depressed mothers were predominantly hypomethylated. The mean difference in methylation between the infants of depressed and infants of non-depressed mothers was 5.23% with a range of 1–68% for the differentially methylated (p<5.0 × 10−7).
Figure 1.
Difference in methylation of the infants of depressed mothers compared to the infants of non-depressed mothers
Table 2.
Genome-wide methylation differences among infants of depressed mothers compared to expected methylation (infants of non-depressed mothers)
| Expected Methylation Range |
N, Depressed Greater |
N, Depressed Less |
Total Loci Differentially Methylated |
|---|---|---|---|
| 0.00–0.29 | 232 | 29 | 260 |
| 0.30–0.69 | 144 | 135 | 279 |
| 0.70–1.00 | 51 | 1528 | 1579 |
| Total | 427 | 1692 | 2119 |
Disease by biomarker analysis
Loci that evidenced differential methylation between infants of depressed and infants of non-depressed mothers were uploaded into the GeneGo MetaCore to examine the association of infants of maternal depression with known disease biomarkers. First, a disease biomarker network analysis was conducted to examine differences between infants of depressed mothers and infants of non-depressed mothers in multiple disease components. Table 3 lists the number of network objects associated with each disease biomarker network and the number of objects that were differentially methylated for maltreated and nonmaltreated groups. The significance values for differential methylation rates are lower than the adjusted false discovery rate (FDR) p-values, indicating significant depressed/nondepressed group differences. Results indicated significant cancer-related differences in biomarker networks such as prostatic neoplasms, ovarian and breast neoplasms, and colonic neoplasms. Next, a disease by biomarker analysis was conducted to identify a broader range of individual components of potential diseases. Significant differences between infants of depressed and infants of non-depressed mothers were found for mental disorders, immune system diseases, respiratory tract diseases, central nervous system diseases, neurodegenerative diseases, and cardiovascular diseases (see Table 4). Finally, a process networks analysis was conducted to determine differences in general biological processes among infants of depressed and non-depressed mothers. Results indicated significant differences in process networks associated with neuronal development and central nervous system functioning, cardiac development, as well as others (see Table 5).
Table 3.
MetaCore analysis of disease biomarker network differences between infants of depressed and infants of non-depressed mothers
| Networks | p-value | False Discovery Rate- adjusted p-value |
# sig. different network objects |
Total network objects |
|---|---|---|---|---|
| Prostatic Neoplasms_Regulation of progression through cell cycle | 9.0E-03 | 2.4E-01 | 14 | 70 |
| Ovarian Neoplasms (core network 2) | 9.1E-03 | 2.4E-01 | 18 | 99 |
| Breast neoplasm_Transcription | 9.8E-03 | 2.4E-01 | 11 | 50 |
| Prostatic Neoplasms_Cell proliferation | 1.4E-02 | 2.4E-01 | 9 | 39 |
| Breast neoplasm_Transcription regulation | 1.4E-02 | 2.4E-01 | 24 | 150 |
| Breast neoplasm_Cell-cell signaling | 2.1E-02 | 3.0E-01 | 17 | 100 |
| Colonic Neoplasms_Cell cycle | 3.6E-02 | 4.0E-01 | 11 | 60 |
| Breast neoplasm_p53 | 3.7E-02 | 4.0E-01 | 10 | 53 |
Table 4.
MetaCore analysis of diseases by biomarker differences among infants of depressed and infants of non-depressed mothers
| Diseases | p-value | False Discovery Rate-Adjusted p-value |
# sig. different network objects |
Total network objects |
|---|---|---|---|---|
| Mental Disorders | 3.6E-14 | 2.1E-11 | 195 | 1610 |
| Immune System Diseases | 1.1E-13 | 4.2E-11 | 574 | 6257 |
| Respiratory Tract Diseases | 8.1E-11 | 4.4E-09 | 1401 | 18398 |
| Central Nervous System Diseases | 1.1E-10 | 5.5E-09 | 305 | 3060 |
| Neurodegenerative Diseases | 4.1E-07 | 6.7E-06 | 206 | 2087 |
| Cardiovascular Diseases | 5.0E-07 | 8.2E-06 | 324 | 3565 |
Table 5.
MetaCore analysis of process network differences among infants of depressed and infants of non-depressed mothers
| Networks | p-value | False Discovery Rate- adjusted p-value |
# sig. different network objects |
Total network objects |
|---|---|---|---|---|
| Neuronal development | ||||
| Development_Neurogenesis_Synaptogenesis | 2.6E-05 | 4.1E-03 | 33 | 180 |
| Development_Neurogenesis_Axonal guidance | 2.7E-03 | 2.1E-01 | 33 | 230 |
| Central nervous system functioning | ||||
| Development_Neuromuscular junction | 2.9E-02 | 2.6E-01 | 20 | 147 |
| Neurophysiological process_GABAergic neurotransmission | 2.9E-02 | 2.6E-01 | 19 | 138 |
| Neurophysiological process_Transmission of nerve impulse | 4.5E-02 | 2.8E-01 | 26 | 212 |
| Cardiac development | ||||
| Cardiac development_FGF_ErbB signaling | 2.1E-02 | 2.6E-01 | 18 | 124 |
| Cardiac development_Role of NADPH oxidase and ROS | 2.2E-02 | 2.6E-01 | 19 | 134 |
| Cardiac development_BMP_TGF_beta_signaling | 2.4E-02 | 2.6E-01 | 17 | 117 |
| General transcription/transduction | ||||
| Signal transduction_NOTCH signaling | 7.3E-03 | 2.1E-01 | 32 | 236 |
| Signal transduction_ESR1-membrane pathway | 1.1E-02 | 2.1E-01 | 15 | 91 |
| Signal transduction_ERBB-family signaling | 1.2E-02 | 2.1E-01 | 13 | 75 |
| Signal transduction_ESR1-nuclear pathway | 1.2E-02 | 2.1E-01 | 29 | 216 |
| Signal transduction_WNT signaling | 1.9E-02 | 2.6E-01 | 24 | 177 |
| Transcription_mRNA processing | 3.6E-02 | 2.8E-01 | 21 | 160 |
| Signal Transduction_TGF-beta, GDF and Activin signaling | 4.4E-02 | 2.8E-01 | 20 | 154 |
| Other | ||||
| Cell adhesion_Synaptic contact | 4.6E-03 | 2.1E-01 | 27 | 184 |
| Cell adhesion_Cell junctions | 6.5E-03 | 2.1E-01 | 24 | 162 |
| Cytoskeleton_Regulation of cytoskeleton rearrangement | 8.2E-03 | 2.1E-01 | 26 | 183 |
| Reproduction_Gonadotropin regulation | 2.3E-02 | 2.6E-01 | 26 | 199 |
| Transport_Calcium transport | 2.6E-02 | 2.6E-01 | 25 | 192 |
| Apoptosis_Anti-apoptosis mediated by external signals via NF- kB |
3.0E-02 | 2.6E-01 | 16 | 111 |
| Reproduction_FSH-beta signaling pathway | 3.6E-02 | 2.8E-01 | 21 | 160 |
| Cell adhesion_Cadherins | 3.9E-02 | 2.8E-01 | 23 | 180 |
| Cytoskeleton_Cytoplasmic microtubules | 4.0E-02 | 2.8E-01 | 16 | 115 |
| Cell cycle_G0-G1 | 4.1E-02 | 2.8E-01 | 11 | 71 |
| Reproduction_Male sex differentiation | 4.8E-02 | 2.9E-01 | 29 | 243 |
| Cell adhesion_Attractive and repulsive receptors | 4.9E-02 | 2.9E-01 | 22 | 175 |
| Cell adhesion_Amyloid proteins | 5.1E-02 | 2.9E-01 | 24 | 195 |
Discussion
To our knowledge, this is the first study to examine methylation differences between infants of mothers with MDD and infants of well mothers in a low income sample. Moreover, we obtained DNA samples and tested for methylation in the infants at approximately one year of age, very early in development. Mothers in the depressed group met diagnostic criteria for current MDD rather than elevated depressive symptoms alone.
We found that there were significant differences in methylation between infants of mothers with MDD and infants of well mothers across the epigenome. Infants of depressed mothers tended to have higher levels of methylation at CpG sites where infants of well mothers evinced low levels of methylation, and lower levels of methylation at CpG sites where infants of well mothers evidenced higher levels of methylation (see Table 2 and Figure 1). Specifically, between 0 and 70% methylation, the infants of mothers with MDD showed a generalized increase in methylation, whereas for loci with 70 to 100% methylation in the well mothers group, the infants of depressed mothers had decreased methylation. This suggests that there are genes that should be turned “on” that are turned “off” and genes that should be turned “off” that are turned “on”.
The pattern of methylation differences was related to differential physical and mental health risk for infants of mothers with MDD and infants of well mothers. Infants of depressed mothers evinced differential methylation in genes associated with disease biomarkers for mental disorders, as well as a range of physical diseases, biomarker network effects on different forms of cancer, and process network risks for neuronal development, CNS functioning, cardiac development, transcription and transduction signaling, among others. Thus, current epigenetic differences observed in infants of mothers with MDD suggest future liabilities for future health outcomes.
In future research, further probing of the nature of epigenetic transmission should be conducted. Subsequent research will need to investigate the extent to which these methylation differences are experienced-based, involving the rearing experiences with a depressed mother in the first year, or initiated in utero (Braithwaite, Kundakovic, Ramchandani, Murphy, & Champagne, 2015; Oberlander, Weinberg, Papsdorf, Grunau, Misri, & Devlin, 2008), as over 90% of the mothers with MDD were depressed prior to the infant’s birth. In addition, methylation differences may have been inherited epigenetically from the mothers with depression. Differentiating the source of these methylation differences will be important.
Longitudinal research would be valuable to investigate epigenetic changes over time and determine how they are related to the development of physical and mental health outcomes. It will be important to investigate the potential for additional methylation variation to occur as these infants develop, particularly if mothers continue to be depressed over time. This will contribute to developing a better understanding of the role of epigenetics in the risk associated with having a depressed mother. Linking methylation differences to the cascade of developmental vulnerabilities observed in the offspring of depressed mothers, (e.g., emotion regulation, attachment organization, self development, and neurocognitive abilities) will be important. Longitudinal mediational analysis will be useful in establishing whether methylation may be portrayed as a veridical mechanism of the risk of maternal depression on various developmental processes, and subsequent mental and physical health outcomes.
If DNA methylation changes are driving human disease and health problems, it may be possible to reverse (demethylate) maladaptive DNA methylation marks with pharmacological or behavioral interventions (Szyf & Bick, 2013). As the pathways that lead from specific to experiences to phenotypic DNA methylation changes become increasingly understood, this should enable prevention scientists to design interventions to decrease disease and increase health and wellness (Szfy & Bick, 2013). Through early intervention with depressed mothers and their young children, research will be able to ascertain whether the observed patterns of methylation may be altered, whether normalization of methylation relative to comparisons may occur, and role of methylation in adaptive intervention outcomes.
Acknowledgments
This research was supported by grants from the Jacobs Foundation to Dante Cicchetti and The National Institute of Mental Health (R01 MH67792) to Dante Cicchetti and Sheree L. Toth.
References
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4th. Washington, DC: Author; 1994. [Google Scholar]
- Beck AT, Steer RA, Brown GK. Manual for the Beck Depression Inventory – II. San Antonio, TX: Psychological Corporation; 1996. [Google Scholar]
- Braithwaite EC, Kundakovic M, Ramchandani PG, Murphy SE, Champagne FA. Maternal prenatal depressive symptoms predict infant NR3C1 1F and BDNF IV DNA methylation. Epigenetics. 2015;10(5):408–417. doi: 10.1080/15592294.2015.1039221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti D, Aber JL. Early precursors to later depression: An organizational perspective. In: Lipsitt L, Rovee-Collier C, editors. Advances in infancy. Vol. 4. Norwood, NJ: Ablex; 1986. pp. 87–137. [Google Scholar]
- Cicchetti D, Rogosch FA, Toth SL, Spagnola M. Affect, cognition, and the emergence of self-knowledge in the toddler offspring of depressed mothers. Journal of Experimental Child Psychology. 1997;67:338–362. doi: 10.1006/jecp.1997.2412. [DOI] [PubMed] [Google Scholar]
- Cicchetti D, Schneider-Rosen K. An organizational approach to childhood depression. In: Rutter M, Izard C, Read P, editors. Depression in young people, clinical and developmental perspectives. New York: Guilford; 1986. pp. 71–134. [Google Scholar]
- Cicchetti D, Toth SL. Developmental psychopathology and disorders of affect. In: Cicchetti D, Cohen DJ, editors. Developmental psychopathology: Risk, disorder, and adaptation. Vol. 2. New York: Wiley; 1995. pp. 369–420. [Google Scholar]
- Cicchetti D, Toth SL. The development of depression in children and adolescents. American Psychologist. 1998;53:221–241. doi: 10.1037//0003-066x.53.2.221. [DOI] [PubMed] [Google Scholar]
- Cole PM, Luby J, Sullivan MW. Emotions and the Development of Childhood Depression: Bridging the Gap. Child Development Perspectives. 2008;2(3):141–148. doi: 10.1111/j.1750-8606.2008.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings EM, Cicchetti D. Toward a transactional model of relations between attachment and depression. In: Greenberg MT, Cicchetti D, Cummings EM, editors. Attachment in the preschool years. Chicago: University of Chicago Press; 1990. pp. 339–372. [Google Scholar]
- Davidson RJ, Fox NA. Asymmetrical brain activity discriminates between positive versus affective stimuli in human infants. Science. 1982;218:1235–1237. doi: 10.1126/science.7146906. [DOI] [PubMed] [Google Scholar]
- Dozois DJ, Dobson KS, Ahnberg JL. A psychometric evaluation of the Beck Depression Inventory-II. Psychological Assessment. 1998;10:83–89. [Google Scholar]
- Field T, Diego M. Maternal depression effects of infant frontal EEG asymmetry. International Journal of Neuroscience. 2008;118:1081–1108. doi: 10.1080/00207450701769067. [DOI] [PubMed] [Google Scholar]
- Goodman SH, Gotlib IH. Risk for psychopathology in the children of depressed mothers: A developmental model for understanding mechanisms of transmission. Psychological Review. 1999;106:458–490. doi: 10.1037/0033-295x.106.3.458. [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- Radke-Yarrow M, Cummings EM, Kuczynski L, Chapman M. Patterns of attachment in two-and-three-year-olds in normal families and families with parental depression. Child Development. 1985;56:884–893. [PubMed] [Google Scholar]
- Radloff L. The CES-D Scale: A self-report depression scale for research in the general population. Applied Psychological Measurement. 1977;1:385–401. [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-Wide Association Studies for common human diseases. Nature Reviews. Genetics. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins L, Cottler L, Bucholz K, Compton W. Diagnostic interview schedule for DSM-IV. Washington University: St. Louis; 1995. [Google Scholar]
- Robins L, Helzer J, Croughan J, Ratcliff K. The NIMH Diagnostic Interview Schedule: Its history, characteristics, and validity. Archives of General Psychiatry. 1981;38:381–389. doi: 10.1001/archpsyc.1981.01780290015001. [DOI] [PubMed] [Google Scholar]
- Robins LW, Helzer JE, Ratcliff KS, Seyfried W. Validity of the diagnostic interview schedule, version II: DSM-III diagnoses. Psychological Medicine. 1982;12:855–865. doi: 10.1017/s0033291700049151. [DOI] [PubMed] [Google Scholar]
- Storch EA, Roberti JW, Roth DA. Factor structure, concurrent validity, and internal consistency of the Beck Depression Inventory- Second Edition in a sample of college students. Depression and Anxiety. 2004;19:187–189. doi: 10.1002/da.20002. [DOI] [PubMed] [Google Scholar]
- Szyf MBJ. DNA methylation: a mechanism for embedding early life experiences in the genome. Child Development. 2013;84(1):49–57. doi: 10.1111/j.1467-8624.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth SL, Rogosch FA, Manly JT, Cicchetti D. The efficacy of toddler-parent psychotherapy to reorganize attachment in the young offspring of mothers with major depressive disorder. Journal of Consulting & Clinical Psychology. 2006;74(6):1006–1016. doi: 10.1037/0022-006X.74.6.1006. [DOI] [PubMed] [Google Scholar]
- Yang BZY, Zhang H, Ge W, Weder N, Douglas-Palumberi H, Perepletchikova F, Kaufman J. American Journal of Preventive Medicine. 2013;44:101–107. doi: 10.1016/j.amepre.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]