Summary
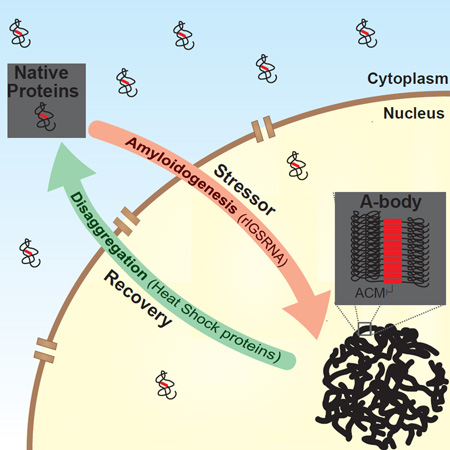
The amyloid state of protein organization is typically associated with debilitating human neuropathies and seldom observed in physiology. Here, we uncover a systemic program that leverages the amyloidogenic propensity of proteins to regulate cell adaptation to stressors. On stimulus, cells assemble the Amyloid-bodies (A-bodies), nuclear foci containing heterogeneous proteins with amyloid-like biophysical properties. A discrete peptidic sequence, termed the amyloid-converting motif (ACM), is capable of targeting proteins to the A-bodies by interacting with ribosomal intergenic noncoding RNA (rIGSRNA). The pathological β-amyloid peptide, involved in Alzheimer’s disease, displays ACM-like activity and undergoes stimuli-mediated amyloidogenesis in vivo. Upon signal termination, elements of the heat shock chaperone pathway disaggregate the A-bodies. Physiological amyloidogenesis enables cells to store large quantities of proteins and enter a dormant state in response to stressors. We suggest that cells have evolved a post-translational pathway that rapidly and reversibly converts native-fold proteins to an amyloid-like solid phase.
eTOC Paragraph
Audas et al. find that to adapt to stressors, heterogeneous proteins convert to an amyloid-like state and form sub-nuclear amyloid-bodies (A-bodies), which requires a specific protein motif and ribosomal intergenic lncRNA. Upon stimuli termination, Hsp chaperones disaggregate A-bodies. This pathway regulates reversible amyloidogenesis of the Alzheimer’s disease-associated pathological β-amyloid peptide.
Introduction
Eukaryotic cells are frequently exposed to adverse environmental stimuli including extreme temperatures, low oxygen availability and acidosis. Each stressor presents a unique risk to cellular sustainability, with the severity and duration dictating whether cells enact pro-survival or apoptotic responses. Numerous pathways have been identified to mitigate the effects of unfavorable growth conditions. High temperature causes protein denaturation (Pinto et al., 1991), activating the heat shock and unfolded protein responses. These pathways enhance the expression of chaperone proteins to alleviate the burden of misfolded proteins on the cell (Lindquist, 1986; Richter et al., 2010). The Hypoxia Inducible Factor family of transcription factors (Semenza and Wang, 1992; Wiesener et al., 1998) activate an array of genes that augment oxygen delivery and enhance glucose metabolism during periods of low [O2] (Semenza, 2012). Regardless of the stimulus, the goal of stress responsive pathways is to sustain viability during adverse environmental conditions, repair associated damage and restore cellular homeostasis.
Recently, noncoding RNAs (ncRNA) have emerged as important biological molecules in cellular adaptation to common stressors. Human Alu elements (Liu et al., 1995) and Satellite III transcripts (Jolly et al., 2004) are strongly induced in response to elevated temperatures. These ncRNAs reduce the burden on the protein folding machinery by impairing global transcription (Mariner et al., 2008) and mRNA maturation (Denegri et al., 2001), respectively. In response to DNA damage lincRNA-p21 represses apoptosis (Huarte et al., 2010), while Pint and TUG1 transcripts facilitate the epigenetic silencing of cell cycle factors (Khalil et al., 2009; Marin-Bejar et al., 2013), inhibiting proliferation until genomic integrity has been restored. The ribosomal intergenic spacer (IGS) appears to be a hub of long noncoding transcripts (Bierhoff et al., 2014; Zhao et al., 2016), notably producing an inducible class of ncRNA (rIGSRNA) that regulate cellular dynamics by capturing and immobilizing proteins in nuclear foci (Audas et al., 2012; Mekhail et al., 2005). This reversible process enables cells to regulate protein mobility (Lippincott-Schwartz and Patterson, 2003; Misteli, 2001) in response to stressors. Interestingly, fragments of the Huntington protein and the RNA-binding protein Rim4 display repressed mobility upon adopting an amyloid-like state in physiological settings (Berchowitz et al., 2015; Kayatekin et al., 2014).
Amyloids are a highly organized form of protein aggregation typically associated with human neuropathies, including Alzheimer’s, Parkinson’s and Huntington’s diseases. Under pathological settings, amyloids are believed to act in a dominant negative manner, converting native-fold species into irreversible β-sheet rich protein aggregates (Knowles et al., 2014). Physiological amyloids are quite uncommon compared to the native-fold, especially in higher eukaryotes. In mammals, functional amyloidogenesis has been associated with hormone storage (Maji et al., 2009), melanin production (Fowler et al., 2006), regulation of kinase activity (Li et al., 2012) and protein synthesis (Berchowitz et al. 2015). Yet, most proteins have an inherent amyloidogenic propensity and possess the capacity to adopt the amyloid-fold (Goldschmidt et al., 2010). Researchers have proposed the existence of kinetic and thermodynamic barriers (Baldwin et al., 2011; Knowles et al., 2014), as well as active suppressor programs to prevent the conversion of proteins to a toxic amyloid state (Dobson, 1999). This line of reasoning implies that the amyloidogenic propensity of proteins is essentially an undesirable byproduct of polypeptide assembly that cells must actively prevent. Still, it remains puzzling why cells have not evolved a comprehensive program that exploits the broad ability of proteins to assume the amyloid-fold.
In this report, we uncover the Amyloid-bodies (A-bodies), inducible and reversible subnuclear foci composed of an array of different proteins that adopt an amyloid-like immobile/insoluble state. Physiological amyloidogenesis of proteins is facilitated by the interaction between an amyloid-converting motif (ACM) and inducible rIGSRNA. Cells activate physiological amyloidogenesis to store large quantities of proteins and enter a state of dormancy in response to stress. These data challenge the concept that amyloids are an infrequent and mostly toxic protein-fold and introduce physiological amyloidogenesis as a cell-wide post-translational process.
Results
Uncovering the A-bodies: Nuclear protein foci with amyloid-like biophysical properties
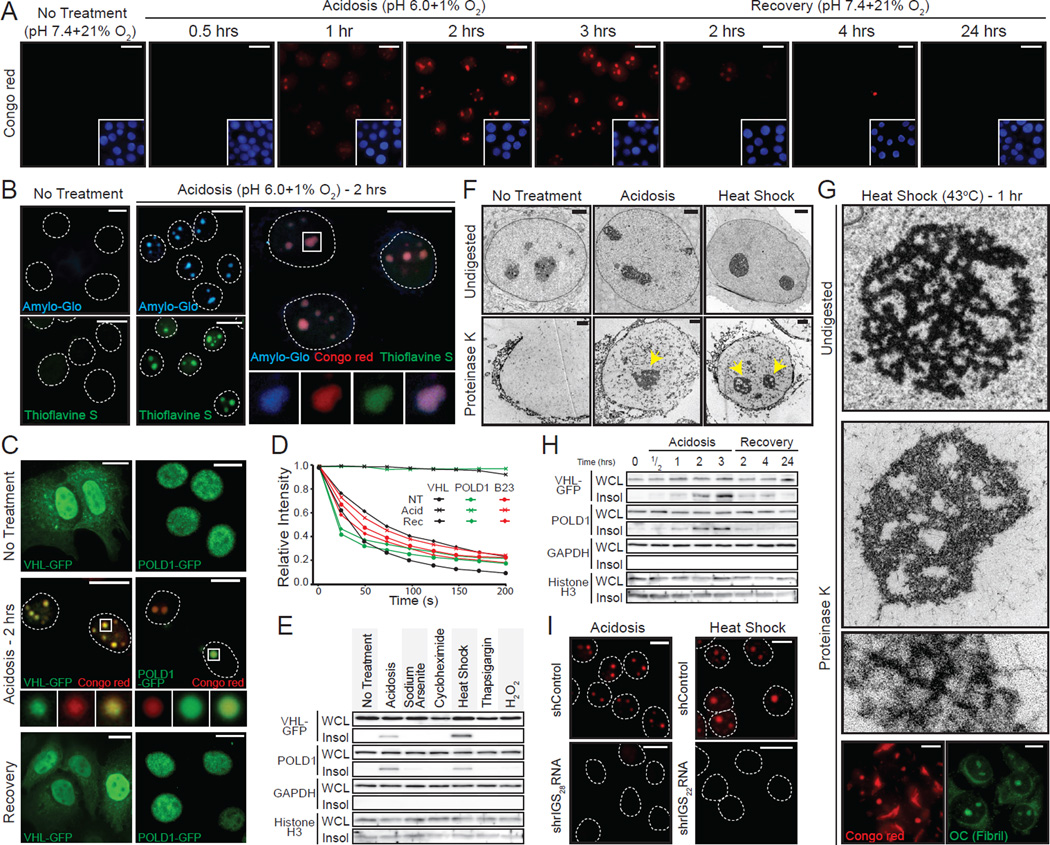
Highly mobile molecules establish functional networks by randomly diffusing in the cellular milieu in search of high affinity interactions (Lippincott-Schwartz and Patterson, 2003; Misteli, 2001). On stimulus, inducible rIGSRNA immobilize proteins in nuclear foci to regulate cellular dynamics (Audas et al., 2012). As protein immobilization is a property associated with cellular amyloids (Berchowitz et al., 2015; Kayatekin et al., 2014), we hypothesized that rIGSRNA participate in the conversion of mobile/soluble native-fold proteins to their immobile/insoluble amyloid-like counterparts. To test this hypothesis, we first stained cells with Congo red, the quintessential amyloidophilic dye. As expected, untreated cells lacked Congo red-positive foci, owing to the absence of detectable amyloid-like protein structures (Figure 1A). Cells exposed to stimuli that induce protein immobilization (Audas et al., 2012) (Figure S1A) displayed subnuclear foci with strong Congo red signatures, revealing the existence of widespread protein organization with amyloid features (Figure 1A and Figure S1B–D). Staining with additional dyes that recognize various biochemical features of amyloids, further highlighted the amyloidogenic properties of the nuclear foci in primary cultures and tumorigenic cells on stimuli (Figure 1B and Figure S1B–E) and in human tissues (Figure S1F). These amyloidogenic cellular bodies co-localized with VHL and POLD1, proteins that undergo stimulus-specific immobilization (Figure 1C, D and Figure S1A, B) (Audas et al., 2012) and insolubilization (Figure 1E). Stressors that do not target VHL to nuclear foci (sodium arsenite, cycloheximide, thapsigargin and H2O2) failed to generate insoluble structures that stained with amyloidophilic dyes (Figure 1E and Figure S1A). The Congo red-positive foci were resistant to proteinase K (Figure 1F, G), another amyloidogenic feature (McKinley et al., 1983), and the digestion revealed the presence of ~10nm fibers (Figure 1G and Figure S1G), structures that have been predicted for cellular amyloids/solid phase transition (Weber and Brangwynne, 2012). These fibers were recognized by the OC antibody (Figure 1G and Figure S1H), which targets the amyloid fibril conformation (Kayed et al., 2007). Stimuli termination caused the loss of nuclear amyloidogenic foci (Figure 1A and Figure S1C, D) and correlated with the release (Figure 1C) and re-solubilization (Figure 1H) of targeted proteins, suggesting that this form of amyloidogenesis is rapid and reversible. The appearance of Congo red-positive foci correlated with the expression of stimuli-specific rIGSRNA (e.g. rIGS22RNA induction by heat shock and rIGS28RNA induction by acidosis) (Figure S1I). Silencing of rIGSRNA impaired or delayed the formation of amyloidogenic foci (Figure 1I and Figure S1J), suggesting a role for these lncRNA in nuclear amyloidogenesis. Put together, these data identify the Amyloid-bodies (A-bodies): rIGSRNA-dependent nuclear foci of immobilized proteins with amyloid-like properties.
Figure 1. Uncovering the distinct biophysical properties of the cellular A-bodies.
(A) Physiological amyloidogenesis is rapid and reversible. MCF-7 cells exposed to extracellular acidosis and returned to standard growth conditions, for the indicated times, were stained with Congo red and Hoechst (blue inset). (B) Nuclear foci stain positively with amyloid-specific dyes. Untreated or acidotic MCF-7 cells were stained with Amylo-Glo (blue), Thioflavin S (green) and/or Congo red (red). Selected regions (white box) were expanded below with merged image included (far right panel). Dashed circles represent nuclei. (C) Established proteins are targeted to the A-bodies. MCF-7 cells expressing VHL-GFP or POLD1-GFP were grown under standard, hypoxic/acidotic conditions or recovered for 24 hours post-acidosis treatment. Acidotic cells were stained with Congo red. Selected regions (white box) were expanded (below). (D) A-body targets are reversibly immobilized. VHL-GFP, POLD1-GFP or GFP-B23 transfected MCF-7 cells were treated as above and bleached repeatedly for fluorescent loss in photobleaching. Quantification is presented as the mean relative intensity of at least 5 data sets. (E) Stimuli-specific insolubilization of A-body components. Insoluble proteins (Insol) were extracted from whole cell lysates (WCL) of untreated, acidotic (2 hrs), heat shocked (2 hrs), sodium arsenite (1 hr), cycloheximide (1hr), thapsigargin (8 hrs) or H2O2 (8 hrs) treated cells. A-body components; VHL-GFP and endogenous POLD1 or the GAPDH and Histone H3 control proteins were detected by western blot. (F) Stimuli-induced A-bodies are proteinase K resistant. Transmission electron microscopy (TEM) of untreated, heat shocked (1hr) and acidotic (1hr) MCF-7 cells were left undigested or exposed to proteinase K. Proteinase K-resistant nuclear bodies are indicated (yellow arrow). (G) Proteinase K-resistant fibrils possess amyloid-like properties. Heat shocked MCF-7 cells were left undigested or proteinase K-treated prior to TEM visualization. Proteinase K-resistant structures were stained with Congo red or the amyloid fibril conformation-specific antibody OC. (H) Protein insolubilization correlates with A-body assembly and disaggregation. MCF-7 cells were treated as above (A) and insoluble proteins were extracted from WCL. A-body targets/controls were detected as in (E). (I) Inhibition of rIGSRNA transcripts impairs amyloidogenesis. MCF-7 cells stably-expressing control or shRNA against rIGS28RNA or rIGS22RNA were grown in acidosis permissive media or exposed heat shock, respectively, prior to Congo red staining. Dashed circles represent nuclei. White scale bars represents 20mm. Black and white TEM scale box represent 1µM and 0.1mm. See also Figure S1.
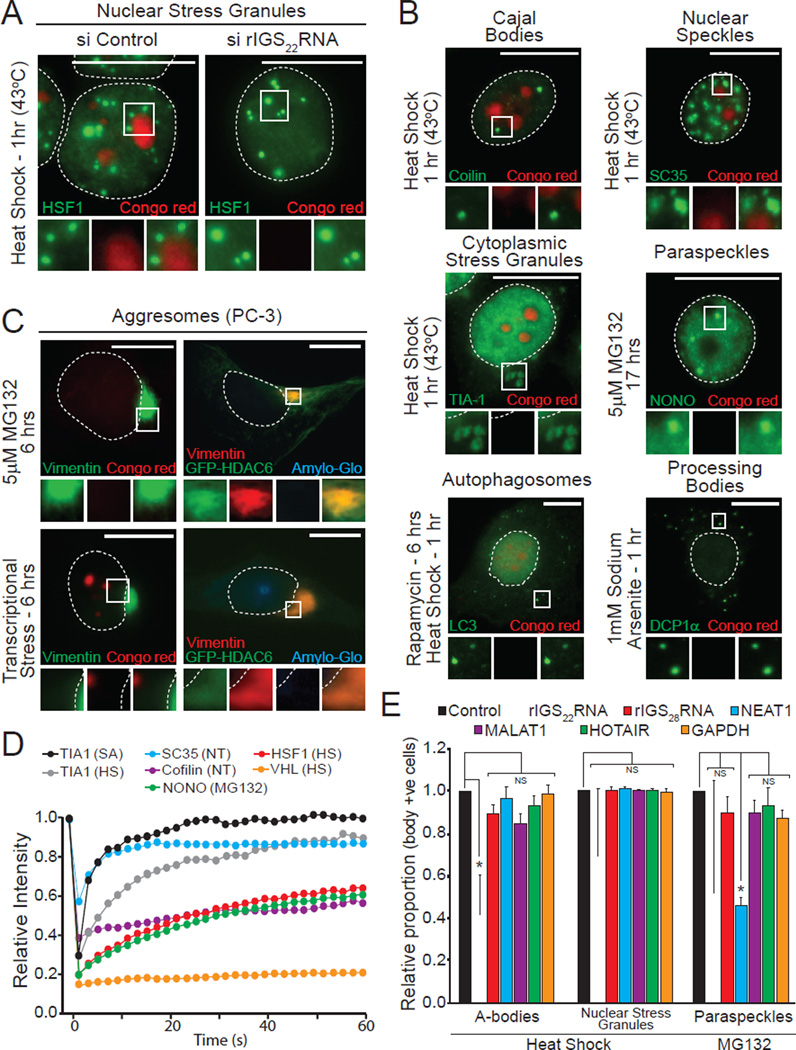
The unique biophysical properties of A-bodies was apparent when compared to known subcellular foci (nuclear and cytoplasmic stress granule, Cajal bodies and nuclear speckles, paraspeckles, PML bodies, autophagosomes, Processing-bodies (P-bodies) and aggresomes). A-bodies are spatially distinct from other domains (Figure 2A–C and Figure S2A–C), possess affinity for amyloidophilic dyes (Figure 1B and Figure S1B–E) and harbor proteins in an immobile/solid-phase organization (Figure 1D and Figure 2D). This is in stark contrast to other subcellular domains, which failed to stain with Congo red/Amylo-Glo (Figure 2A–C and Figure S2A–C), display mobile/liquid-phase properties (Figure 2D) and do not form fiber-like structures under physiological settings (Weber and Brangwynne, 2012). Like A-bodies, nuclear stress granules and paraspeckles are RNA-seeded domains. To confirm the specificity of the rIGSRNAs in the amyloidogenic process, we used siRNA to target several established lncRNAs (rIGS22RNA, rIGS28RNA, NEAT1, MALAT1 and HOTAIR) and assayed for foci formation (Figure S2D). Inhibition of rIGS22RNA delays heat shock-induced A-bodies, but has no effect on the genesis of other bodies under standard growth conditions (Figure 2A, E and Figure S2E). Impairing various lncRNA failed to disrupt the A-bodies, though NEAT1 depletion did reduce paraspeckle formation (Figure 2E), as previously observed (Sasaki et al., 2009). These results highlight the cellular and biophysical properties of A-bodies as rIGSRNA-induced endogenous protein foci with amyloidogenic characteristics.
Figure 2. A-bodies are unique rIGSRNA-seeded amyloidogenic structures.
(A) A-bodies are separate from nuclear stress granules. MCF-7 cells transfected with control or rIGS22RNA-specific siRNA were exposed to heat shock. HSF-1 was detected in Congo red stained cells. (B) Additional cellular bodies are not amyloidogenic. MCF-7 cells were treated as indicated. Foci-specific markers were detected (green) in Congo red (red) stained cells. (C) Aggresomes do not contain proteins in an amyloid-like conformation. Untransfected or HDAC6-GFP expressing PC3 cells were treated with stimuli that induce aggresome (5µM MG132) or aggresome and A-body (transcriptional stress: 8µM MG132 + 4µM Actinomycin D) formation. The aggresome marker vimentin was detected in Congo red or Amylo-Glo stained cells. (D) Other cellular domains possess mobile proteins. Fluorescence recovery after photobleaching was performed on MCF-7 cells expressing TIA1-GFP (sodium arsenite and heat shock – 1hr), SC35-GFP (no treatment), Coilin-GFP (no treatment), HSF1-GFP (heat shock – 1hr), NONO-GFP (5µM MG132 – 17hrs) and VHL-GFP (heat shock – 1hr). Quantified kinetics are presented as the mean relative intensity of at least 5 data sets. (E) A-body formation is mediated by rIGSRNA. PC3 cells transfected with siRNA targeting the indicated transcripts were exposed to heat shock (45 min) or 5µM MG132 (17 hrs) prior to detection of A-bodies (Congo red), nuclear stress granules (HSF1) or paraspeckles (NONO). The proportion of body positive cells was counted and compared to the control siRNA. Results are means and SEM (n=3). Significance was measured by Student’s t-test; *p < 0.01. Dashed circles represent nuclei. Selected regions (white box) were expanded below. White scale bars represents 20mm. See also Figure S2.
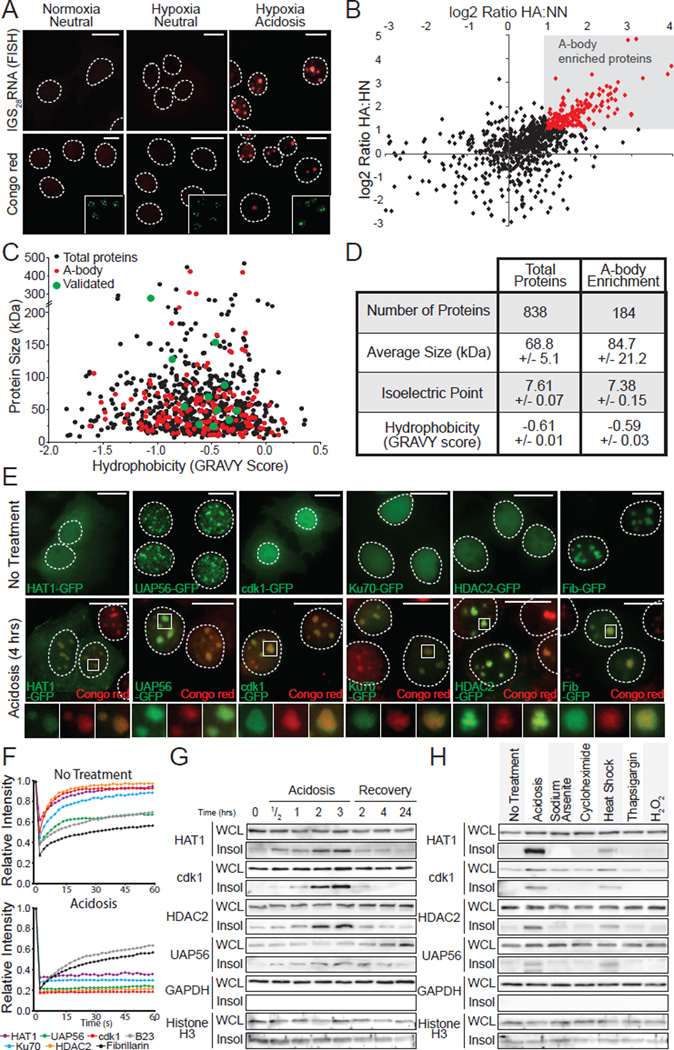
Next, we analyzed the proteomic composition of acidotic A-bodies (Figure 3A) using SILAC-MS analysis (Supp. Methods). This revealed a large influx (Figure 3B and Table S1) of heterogeneous proteins (Figure 3C, D and Figure S3A) into Congo red-positive nuclear foci, with >180 proteins over the 2 fold stringency threshold. We validated several of these molecules by western blotting (Figure S3B) and observed a re-localization of proteins from their original nuclear/cytoplasmic distribution to the A-bodies (Figure 3E and Figure S3C–F). This was accompanied by a loss of mobility (Figure 3F) and a reversible shift to the insoluble protein fraction (Figure 3G). Stressors that do not induce nuclear amyloidogenesis (Figure S1A) did not insolubilize SILAC-MS candidates (Figure 3H). Silencing of rIGSRNA prevented the shift of tested SILAC-MS candidate proteins to the A-bodies (Figure S3F). This indicates that the amyloidogenic properties of the A-bodies were dependent on the accumulation of SILAC-MS proteins, with a proteomic composition indistinguishable from the typical cellular distribution of proteins in term of size, hydrophobicity and isoelectric point (Figure 3D and Figure S3A). These data demonstrate that A-bodies contain a heterogeneous family of proteins with amyloid-like properties.
Figure 3. Heterogenic protein composition of the A-body.
(A) Acidosis induces the formation of the A-bodies. MCF-7 cells grown under normoxic-neutral (NN) and hypoxic-neutral (HN) as baseline controls, and hypoxic-acidotic (HA) conditions were stained with Congo red dye or fluorescence in situ hybridized with an anti-sense probe targeting rIGS28RNA. Nucleolar B23 (green) is inset. (B) Identification of the A-body residents. SILAC-MS analysis comparing proteins extracted from MCF-7 cells treated as above. Plot compares the log enrichment of HA:HN versus HA:NN. A-body-enriched (red) proteins fall in the upper right quadrant. (C) A-body components are biochemically similar to the total protein population. Scatter plot of protein size versus hydrophobicity. (D) A heterogeneous family of proteins are targeted to the A-bodies. The size, isoelectric point and hydrophobicity of total and amyloid-specific protein fractions is summarized. Values represent data set averages +/− SEM. (E) Localization of SILAC-MS candidates to the A-bodies. MCF-7 cells transfected with HAT1-GFP, UAP56-GFP, cdk1-GFP, Ku70-GFP, HDAC2-GFP and Fib-GFP were left untreated or exposed to extracellular acidosis prior to staining with Congo red. (F) Targets of the A-bodies are immobilized. Quantification of recovery after photobleaching kinetics for the proteins listed above (E) as the mean relative intensity of at least 5 data sets. (G) SILAC-MS candidates reversibly insolubilize in response to stimuli. MCF-7 cells were exposed to extracellular acidosis and returned to standard growth conditions, for the indicated times prior to harvesting WCL and insoluble proteins. HAT1, cdk1, HDAC2, UAP56, GAPDH and Histone H3 were detected by western blotting. (H) Stimuli-specific insolubilization of SILAC-MS candidates. WCL and insoluble proteins were extracted from untreated, acidotic (2 hrs), heat shocked (2 hrs), sodium arsenite (1 hr), cycloheximide (1hr), thapsigargin (8 hrs) or H2O2 (8 hrs) treated cells. A-body targets/controls were detected as in (E). Dashed circles represent nuclei. Selected regions (white box) were expanded below. White scale bars represents 20mm. See also Figure S3.
The amyloid-converting motif immobilizes proteins in the A-bodies
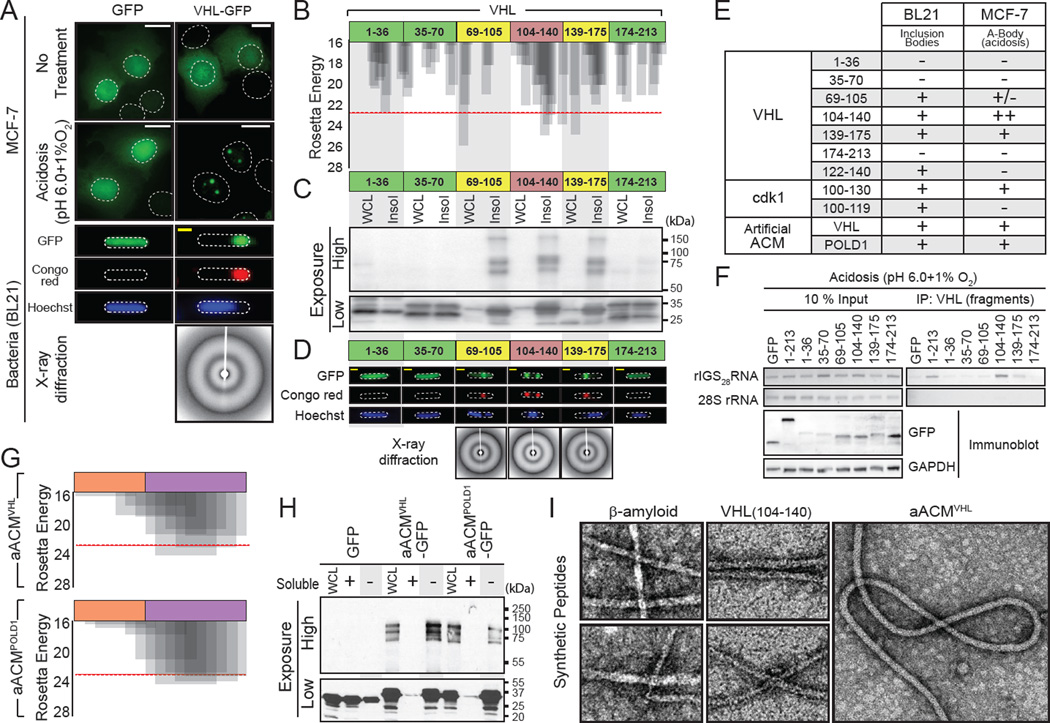
In principle, proteins that are immobilized in the A-bodies should possess amyloidogenic properties. We tested several immobile A-bodies constituents, including VHL, RNF8 (Audas et al., 2012) and the SILAC-MS identified cdk1 and UAP56 for their amyloid propensity using an established bacterial in vitro assay (Garcia-Fruitos et al., 2011; Wang et al., 2008). In this assay, proteins with amyloidogenic propensity are able to self-associate in amyloid-like structures likely due to their high concentration. Bacterially expressed A-body proteins formed Congo red-positive aggregates (Figure 4A and Figure S4A) containing crossed β-sheeted proteins, which generate the classic amyloid 4.7 and 10 Å ringed x-ray diffraction profile (Figure 4A). Conversely, mobile B23 and SILAC-MS-negatives cdk4 and Ran failed to form Congo red-positive structures under identical conditions (Figure S4A). Bioinformatics analysis of VHL (Figure 4B) identified several regions with amyloidogenic propensity based on a Rosetta energy score lower than the established −23 kcal/mol threshold (Goldschmidt et al., 2010; Thompson et al., 2006). There was excellent correlation between the predicted fibrillation propensity of the VHL fragments (Figure 4B) and their ability to insolubilize GFP in bacteria (Figure 4C; bottom panel), form SDS-boiling resistant multi-mers - another hallmark of amyloids - (Figure 4C; top panel), and Congo red-positive inclusion bodies (Figure 4D). This correlation was also maintained in mammalian cells, as fragments of VHL with low fibrillation propensity (Figure 4B) and poor amyloidogenic properties in bacteria (Figure 4C, D) failed to accumulate in insoluble A-bodies on stimulus (Figure 4E and Figure S4B, C). In contrast, the three VHL fragments with strong amyloidogenic properties in bacteria did accumulate in A-bodies on stimulus, albeit to different degrees (Figure 4E and Figure S4B). VHL fragment (104–140) was particularly efficient at targeting/immobilizing GFP within the A-bodies (Figure 4E and Figure S4B–D). RNA immunoprecipitation analysis revealed that this fragment assembles efficiently with endogenous rIGS28RNA during acidosis compared to other amyloidogenic or non-amyloidogenic regions (Figure 4F). A closer examination of VHL (104–140) revealed the presence of an arginine/histidine (R/H) cluster in close proximity to a highly amyloidogenic stretch of amino acids (Figure S4E, middle right panel). Bioinformatics analysis of the SILAC-MS candidate and A-body constituents cdk1, HAT1 and HDAC2 identified similar motifs (Figure S4E). As expected, within bacterial settings all tested fragments containing regions that cross the Rosetta energy threshold formed amyloid-like inclusion bodies (Figure 4F and Figure S4F). However, the presence of the R/H motif was essential for capture in the endogenous A-bodies in response to environmental stimuli (Figure 4F and Figure S4C, D, F, G). We named these bipartite domains amyloidogenic converting motifs (ACM). Based on these data, we generated artificial ACM sequences composed of peptidic fragments from VHL and POLDI to create molecules that harbor an R/H cluster flanking an amyloidogenic domain from the same proteins, termed aACMVHL and aACMPOLD1, respectively (Figure 4G). These artificial molecules displayed amyloidogenic properties in bacteria and were efficiently captured by the A-bodies (Figure 4E, H and Figure S4C, D, H). Synthetic peptides of VHL (104–140), or its artificial fragment, efficiently formed classic (Greenwald and Riek, 2010; Wang et al., 2008) ~10 nm fibrils in vitro, similar to β-amyloid, with the artificial molecule possessing considerably more steric flexibility than the other peptides (Figure 4I). Hence, the A-body targeted proteins examined in this study encode a bipartite domain capable of facilitating the conversion of proteins to an amyloid-like state.
Figure 4. Identification of the amyloid converting motif that targets proteins to A-bodies.
(A) VHL can obtain an amyloid-like conformation in bacteria. GFP or VHL-GFP expressing MCF-7 cells left untreated or exposed to acidosis or BL21 cells were stained with Congo red and Hoechst. X-ray diffraction was performed on BL21 bodies. (B) VHL contains fibril-forming peptidic regions. Results of ZipperDB analysis of full length VHL for fibrillation propensity. The Rosetta energy threshold of −23 kcal/mol is an indicator of fibril-positive regions. Truncated VHL fragments, used below, are indicated. (C) Amyloidogenic fragments of VHL insolubilize GFP and produce SDS-resistant multi-mers. Fragments of VHL (above) fused to GFP were expressed in BL21, prior to lysis and insoluble protein fractionation. Fusion proteins were detected with a GFP-specific antibody at low and high exposures to detect monomeric (low) and multi-meric (high) proteins. (D) Insoluble VHL fragments form bacterial inclusion bodies with an amyloid-like x-ray diffraction profile. BL21 expressing the VHL fragments (above) were fixed and stained with Congo red and Hoechst. Inclusion bodies were purified, where present, for x-ray diffraction. (E) Table summarizing inclusion body formation and targeting to the A-bodies (Figure 4D, S4B, F and H) for the indicated regions of VHL, cdk1 or POLD1 fused to GFP. (F) Regions of VHL can associate with rIGS28RNA. MCF-7 cells transfected with VHL or VHL mutants were exposed to acidosis for 1 hour prior to lysis and RNA immunoprecipitation. 28S rRNA and rIGS28RNA were detected by RT-PCR. Exogenous VHL fragments and GAPDH were detected by immunoblotting. (G) Generation of artificial ACM sequences. R/H-rich (orange) and amyloidogenic (purple) sequences derived from VHL (aACMVHL) and POLD1 (aACMPOLD1) were fused to generate artificial ACM motifs (sequence inset). The fibrillation propensity was calculated by ZipperDB. (H) Artificial ACMs are sufficient to create insoluble multi-mers. BL21 expressing GFP fused to the artificial ACMs (above) were lysed into soluble (+) and insoluble (−) fractions. (I) Amyloidogenic regions of VHL form 10nm amyloid-like fibrils. Peptides encoding VHL (104–140), aACMVHL and the classic pathological β-amyloid were synthesized and incubated for 1 week at 37°C. Fibrils were detected by TEM. White and yellow scale bars represent 20µm and 5µm, respectively. Dashed circles represent nuclei (MCF-7) or whole cell (BL21).TEM scale box represent 10nm. See also Figure S4.
The pathological β-amyloid displays ACM-like properties
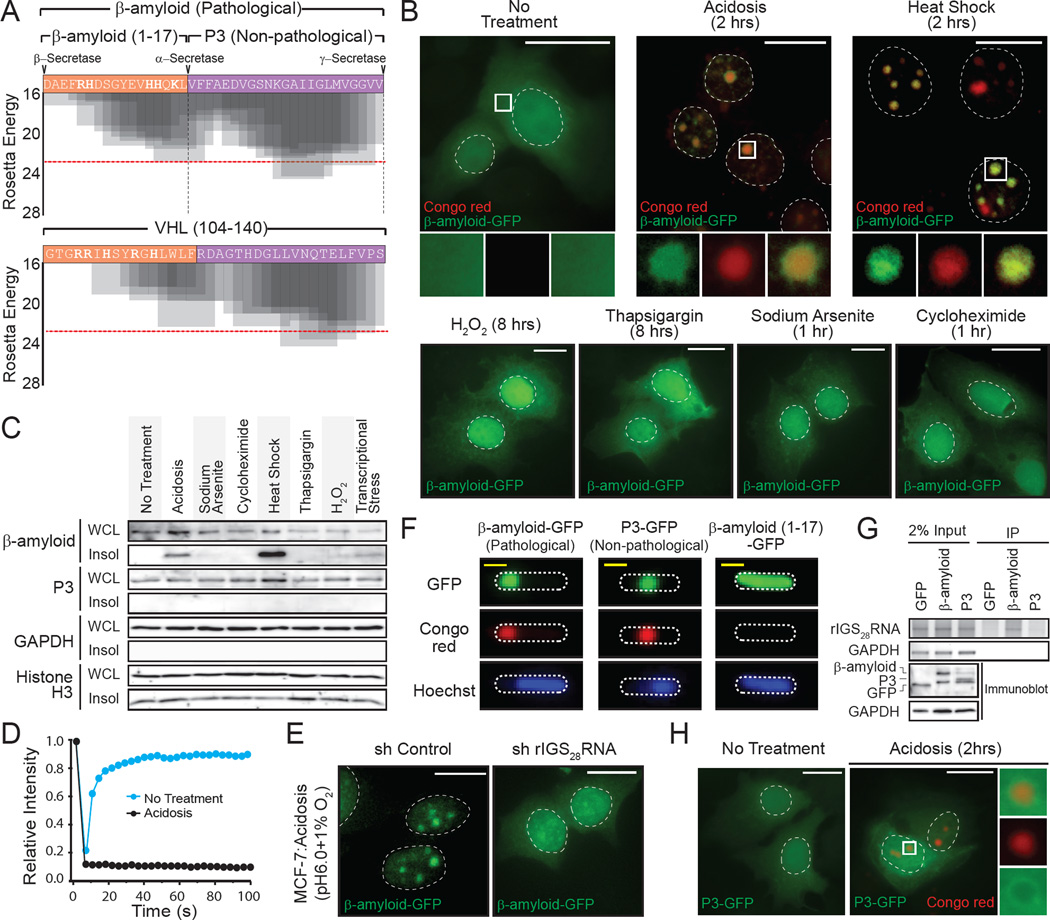
The identification of a physiological amyloidogenic program raises the obvious question as to its possible implication in disease-associated amyloid formation. Surprisingly, the prototypical pathological β-amyloid involved in Alzheimer’s disease harbors an R/H-rich cluster flanking a highly amyloidogenic domain analogous to VHL (100–140), and other ACM regions (Figure 5A and Figure S4E, S5A). Under standard growth conditions, the β-amyloid-GFP fusion protein has a diffuse and mobile cellular distribution, indistinguishable from GFP alone (Figure 5B–D). Environmental stimuli that activate physiological amyloidogenesis (Figure S1A) triggered the efficient capture, immobilization and insolubilization of β-amyloid within the A-bodies (Figure 5B–D and Figure S5B) in a rIGSRNA-dependent manner (Figure 5E). The amyloid precursor protein (APP) can be cleaved by β-secretase to produce the pathological β-amyloid or α-secretase to generate the non-pathological P3 fragment (Figure 5A and Figure S5A), lacking the R/H cluster necessary for mammalian cell amyloidogenesis seen in other ACMs (Figure S4F). P3 can form amyloid-like structures in bacteria (Figure 5F), however, it is unable to associate with rIGSRNA (Figure 5G) and failed to accumulate in the A-bodies in mammalian cells (Figure 5D, H and Figure S5B), similar to other regions of APP (Figure S5C) and ACM that lack the R/H cluster (Figure S4F, G). Hence, these data suggest that β-amyloid displays ACM-like properties and undergoes physiological amyloidogenesis in A-bodies in response to environmental stimuli.
Figure 5. The pathological β-amyloid peptide is a target of physiological amyloidogenesis.
(A) Amino acid sequence and Rosetta energy profiles for β-amyloid (1–40) and VHL (104–140). Secretase (α-, β- and γ-) cleavage sites are indicated. (B) Stress-specific targeting of β-amyloid to the A-bodies. β-amyloid-GFP expressing MCF-7 cells were left untreated or exposed to acidosis, heat shock, sodium arsenite, cycloheximide, thapsigargin or H2O2 and stained with Congo red. (C) Amyloidogenic stimuli insolubilize β-amyloid, not the non-pathological P3 peptide. Insoluble proteins were extracted from MCF-7 cells exposed to the stimuli (above). β-amyloid-GFP, P3-GFP, GAPDH and Histone H3 were detected by western blotting. (D) Immobilization of β-amyloid by acidosis. Quantification of fluorescence recovery after photobleaching kinetics for β-amyloid-GFP in untreated or acidotic MCF-7 cells. Mean relative intensity of at least 5 data sets. (E) rIGS28RNA is essential for the subnuclear targeting of β-amyloid. MCF-7 cells stably-expressing control or rIGS28RNA-specific shRNA were transfected with a plasmid encoding β-amyloid-GFP and grown under hypoxic conditions in acidosis-permissive media. (F) β-amyloid and P3 possess amyloidogenic propensity. β-amyloid-GFP, P3-GFP and β-amyloid (1–17)-GFP were expressed in BL21 cells prior to staining with Congo red and Hoechst. (G) The amino-terminus of β-amyloid is essential for rIGSRNA binding. RNA immunoprecipitation of acidified MCF-7 cells transfected with GFP, β-amyloid-GFP or P3-GFP. GAPDH mRNA and rIGS28RNA were detected by RT-PCR. Exogenous proteins and GAPDH were detected by immunoblotting. (H) P3 is not targeted to the A-bodies. P3-GFP-expressing MCF-7 cells were untreated or exposed to acidotic conditions, prior to Congo red staining. Dashed circles represent nuclei (MCF-7) or whole cell (BL21). Selected regions (white box) were expanded below. White scale bars represents 20µm. See also Figure S5.
Cellular amyloidogenesis is a reversible process
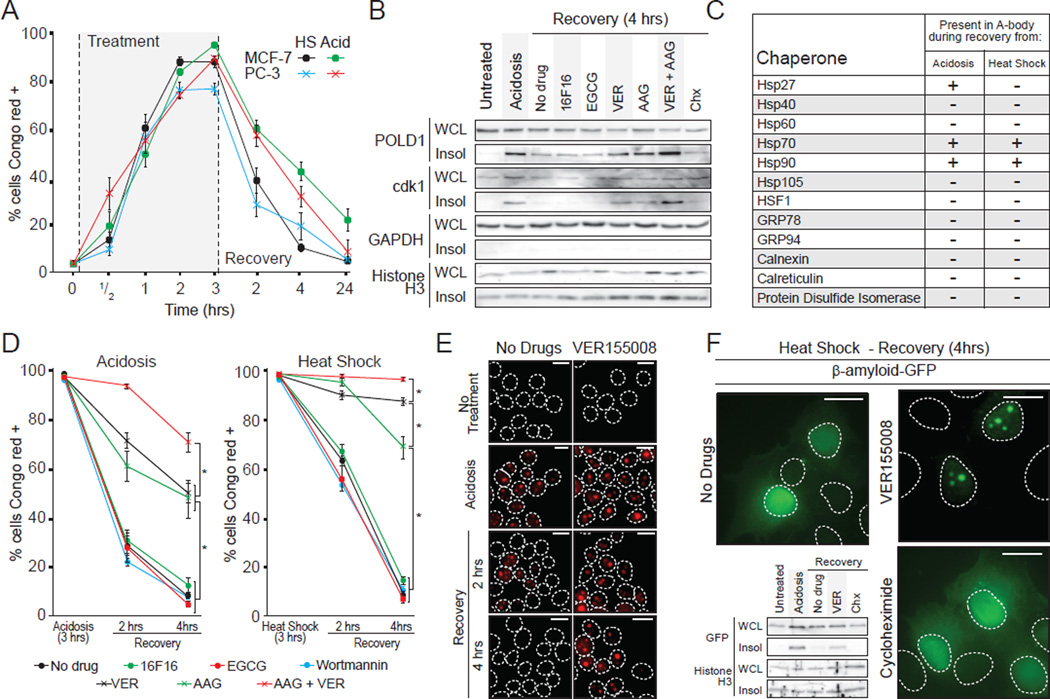
As seen in Figure 1A, the restoration of standard growth conditions results in a rapid dissipation of the A-bodies (Figure 1A, 6A), correlating with the release of proteins from the insoluble fraction (Figure 1H, 3G) and a downregulation of rIGSRNA levels (Figure S1I). We investigated refolding, rather than degradation, as a mechanism of A-bodies disassembly since the proteins return to their original localization and retain their steady state levels in the presence of cycloheximide, a protein synthesis inhibitor (Figure 6B and Figure S6A). Subcellular analysis of prominent chaperones revealed that members of the heat shock protein family (Hsp27, Hsp 70 and Hsp90) are associated with A-bodies during and after stimuli termination (Figure 6C and Figure S6B), suggesting a role for these molecules in amyloid disaggregation. To assay the effects of these chaperones, we used readily available drug inhibitors (Figure S6C). We preferred this approach to RNA interference or CRISPR technology, as depletion of the hsp members results in a loss of cellular viability during stress treatment and is therefore uninformative. Inhibition of Hsp70 and Hsp90, with VER155008 (VER) and 17-allylamino-17-demethoxygeldanamycin (AAG), respectively, after A-body formation significantly impaired the disaggregation of Congo red-positive foci upon signal termination (Figure 6B, D and E and Figure S6D). Inhibition of other chaperones, GRP78 (Epigallocatechin gallate [EGCG]), Protein Disulfide Isomerase (16F16) and the autophagosome (Wortmannin), failed to prevent this process (Figure 6B, D and Figure S6D). A combinatory treatment of VER and 17-AAG delayed recovery in an additive manner (Figure 6D), suggesting that both Hsp70 and Hsp90 are involved in disassembly. The pathological β-amyloid peptide was also released from the A-bodies in an Hsp70 activity-dependent manner (Figure 6F). This suggests that fibrillation of this neurotoxic molecule can be reversed by elements of the hsp pathway, returning to its pre-stress localization, even in the presence of cycloheximide (Figure 6F). While the hsp response is necessary for disassembly of the A-bodies, its inhibition alone is insufficient to induce the accumulation of cellular amyloid foci (Figure S6E). These data demonstrate the disaggregation of the A-bodies is regulated by elements of the heat shock machinery, which reversibly switch proteins from the amyloid- to the native-fold.
Figure 6. Heat shock chaperones regulate A-bodies disaggregation.
(A) Amyloidogenesis is rapid and reversible. MCF-7 and PC-3 cells exposed to acidosis or heat shock and allowed to recover (times indicated) were stained with Congo red. The proportion of cells containing Congo red-positive structures was assayed relative to Hoechst-positive nuclei. (B) Heat shock proteins mediate the solubilization of A-body components. Insoluble proteins were extracted from untreated, acidotic or recovering MCF-7 cells treated with the protein synthesis inhibitor cycloheximide (Chx) or PDI (16F16), GRP94 (EGCG), Hsp70 (VER155008) or Hsp90 (17-AAG) inhibitors. POLD1, cdk1, GAPDH and Histone H3 were detected by western blot. (C) Heat shock proteins are associated with the A-bodies during recovery. Table summarizing data in Figure S6B. (D) Heat shock proteins disaggregate the A-bodies. MCF-7 cells were exposed to acidotic (left panel) or heat shock (right panel) conditions for 3 hours, then returned to normal growth conditions for 2 or 4 hours in the presence 16F16, EGCG, VER, AAG or Wortmannin. The proportion of Congo red-positive cells was determined as above. (E) Congo red stained MCF-7 cells allowed to recover for 2 or 4 hours from a 3 hour acidosis or heat shock exposure in the presence or absence of VER155008. (F) Hsp70 activity enhances β-amyloid release during recovery. β-amyloid-GFP-expressing MCF-7 cells were allowed to recover for 4 hours from a 3 hour heat shock exposure in the presence or absence of VER or Chx. Western blots of insoluble fractionation for β-amyloid-GFP and Histone H3 are included (lower left). Results are means and SEM (n=4). Significance was measured by Student’s t-test; *p < 0.01. Dashed circles represent nuclei. White scale bars represents 20µm. See also Figure S6.
The rIGSRNA/A-bodies induce a state of cellular dormancy
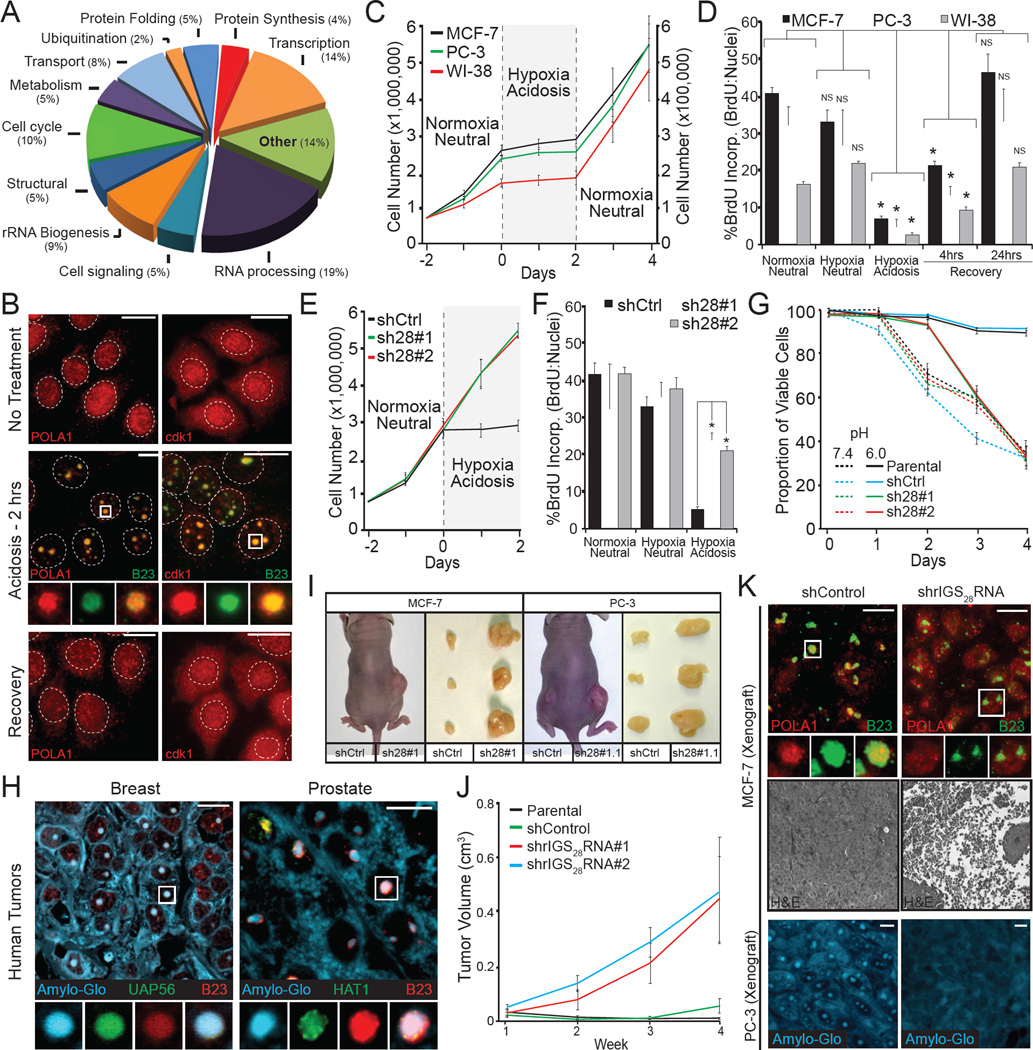
SILAC-MS analysis shown in Figure 3 revealed a large influx of proteins into the A-bodies with several constituents involved in cell cycle progression and DNA synthesis (Figure 7A, B and Figure S3E, F). As such, formation of the acidosis arrested proliferation (Figure 7C and Figure S7A) and DNA synthesis (Figure 7D) of cells in an A-body/rIGS28RNA-dependent manner (Figure 7E, F and Figure S7B–G). This enables cells to remain viable during prolonged periods of extracellular acidosis (Figure 7G), highlighting the non-toxic/protective nature of physiological amyloidogenesis. Restoration of neutral pH reverted these cells back to the untreated phenotype (Figure 7C, D), correlating with disaggregation of the A-bodies (Figure 1A). These hypoxic/acidotic conditions are prevalent within the tumor microenvironment and are believed to cause cancer cell dormancy (Giancotti, 2013; Sosa et al., 2014; Tannock and Rotin, 1989). We used this model system to assess for the presence and function of the A-bodies in an in vivo setting. Formalin fixed paraffin-embedded human tissues from human prostatic acinar and breast invasive duct carcinomas stained positive for the Amylo-Glo dye and the validated SILAC-MS proteins UAP56 and HAT1 (Figure 7H). To assess the effects of this amyloidogenic event we performed nude mouse xenograft assays with MCF-7 and PC-3 cells expressing control or rIGS28RNA-specific shRNA (Figure 1J and Figure S7C). MCF-7 cells only form minimal masses when injected into nude mice, without the addition of external growth factors (Benz et al., 1992). Consistent with our model, shRNA-mediated silencing of rIGS28RNA enabled MCF-7 cells to form large necrotic tumors four weeks post-injection (Figure 7I–K). PC-3 cells that generally form moderate sized masses also formed larger masses when rIGS28RNA was inhibited (Figure 7I and Figure S7H). Silencing of rIGS28RNA prevented capture of the validated SILAC-MS candidates POLA1, HAT1 and the formation of Congo red/Amylo-Glo positive nuclear foci in the core of these tumors (Figure 7K and Figure S7H, I). These data support the hypothesis that rIGS28RNA-mediated A-body formation force cells to enter a state of dormancy, maintaining viability during periods of extracellular stress.
Figure 7. rIGSRNA/A-bodies induce cellular dormancy.
(A) Functional classification of A-bodies constituents. SILAC-MS results were analyzed and grouped by function, percentages per group are indicated. (B) Proliferative factors are reversibly targeted to the A-bodies. Untreated, acidotic and recovered MCF-7 cells were stained for endogenous POLA1 and cdk1 (red) and the nucleolar marker B23 (green). Dashed circles represent nuclei. (C) Acidosis induces a reversible state of dormancy. 750,000 (MCF-7 and PC-3 – left axis) or 75,000 (WI-38 – right axis) cells were grown for 2 days, prior to the application of acidosis-permissive media and exposure to hypoxic (1% O2) conditions. Cells were returned to standard growth conditions two days post-acidification. Cells were counted daily with trypan blue staining to ensure viability. (D) DNA synthesis is reversibly inhibited by acidosis. MCF-7, PC-3 and WI-38 cells grown under the indicated conditions were incubated with BrdU prior to fixation. BrdU positive cells were counted relative to Hoechst stained nuclei. (E–F) Inhibition of rIGS28RNA restores proliferative capacity to acidotic cells. MCF-7 cells stably-expressing two independent shRNA against rIGS28RNA (sh28#1 and sh28#2) or a control sequence (shCtrl) were exposed to normoxic-neutral, hypoxic-neutral or hypoxic-acidosis and cell counts were performed each day (E) or incubated with BrdU for the incorporation assay described above (F). (G) Amyloidogenesis preserves cell viability during extracellular stress. MCF-7 cells described above were grown under hypoxic conditions in standard (pH7.4) or acidosis-permissive (pH6.0) low glucose media for the indicated times. Viability was calculated as propidium iodide-positive versus Hoechst-positive nuclei. (H) Human breast invasive duct carcinomas and prostatic acinar contain cellular amyloids. Paraffin-embedded prostate and breast tumors were stained with the Amylo-Glo, UAP56 or HAT1 and the nucleolar marker B23. (I–J) Inhibition of rIGS28RNA relieves tumor dormancy. Nude mouse xenograft assays with MCF-7 and PC-3 cells described above. Representative mice and excised tumors (I) are presented, with tumor volumes calculated weekly (J) (n=5). (K) Inhibition of rIGS28RNA prevents amyloidogenesis in situ, causing tumor necrosis. Paraffin-embedded MCF-7 andPC-3 tumor sections were stained for the SILAC-MS candidate POLA1 (red) and B23 (green), Amylo-Glo (blue) or hematoxylin/eosin. Results are means and SEM (n≥4) with significance measured by Student’s t-test; *p < 0.01. Selected regions (white box) were expanded below. White scale bars represents 20µm. See also Figure S7.
Discussion
This manuscript introduces A-bodies: rIGSRNA-seeded nuclear foci that containing proteins possessing biophysical properties associated with an amyloid-like state. Formation of the A-bodies is rapid, reversible and plays a role in the ability of cells to enter a dormant state as an adaptive response to severe environmental insults. Physiological amyloidogenesis represents a clever post-translational regulatory program, allowing for the prompt removal of a large family of heterogeneous proteins, without relying on complex covalent modifications or extensive protein degradation. In fact, A-body formation could be a physiological example of the liquid to solid phase transition of proteins, a process observed under in vitro settings (Kato et al., 2012) and attributed predominantly to pathological aggregates (Weber and Brangwynne, 2012). A-bodies are found in cells exposed to various stressors, the cores of tumors and normal human tissues, highlighting their ubiquitous nature. This challenges the widely held concept that amyloids are mostly aberrant/toxic aggregates and seldom observed in physiology compared to native-folded proteins. Based on these data, we suggest that the amyloid-fold should be considered, alongside the native-fold and unfolded, as a common protein organization in cell biology.
A-bodies join a group of established RNA-seeded cellular protein foci, including paraspeckles and nuclear stress granules (Chujo et al., 2016). Yet, they are currently unique in their biochemical properties as they include immobile proteins with amyloid-like properties. Other tested protein foci, such as stress granules and aggresomes, do not display amyloidogenic properties in a cellular context. rIGSRNA-impaired cells fail to produce A-bodies on stimuli but retain their ability to form other protein foci, highlighting the role of these lncRNA in amyloidogenic body formation. While the exact mechanisms remain unclear, we favor a model whereby rIGSRNA facilitate in vivo amyloidogenesis by operating as micro-concentrators of ACM-containing proteins. The ACM of proteins that we have studied so far harbor two distinct domains: an R/H rich sequence that flanks a highly amyloidogenic stretch of amino acids (Figure S5F). The amyloidogenic domains are independently capable of forming amyloids when overexpressed in bacteria, likely as a consequence of reaching a concentration threshold that initiates self-amyloidogenesis. In contrast, the amyloidogenic domains require the flanking R/H-rich sequences for RNA-mediated amyloidogenic conversion in mammalian cells. This implies that endogenous rIGSRNA enable the amyloidogenic domains to reach a sufficient concentration by interacting with the R/H residues, triggering the initial fibrillation event followed by polymerization of proteins, consistent with several models of amyloid formation. Precisely how rIGSRNA facilitates A-bodies formation and whether a combination of other endogenous or exogenous factors can also activate cellular amyloidogenesis remains to be studied.
A notable observation from this study is that β-amyloid shares striking similarities with the ACM of several A-body targets and undergoes physiological amyloidogenesis on stimuli. Unlike the non-pathological P3 peptide, which lacks the R/H rich motif (because of its naturally occurring cleavage by α-secretase), β-amyloid is efficiently captured in A-bodies. On signal termination, β-amyloid and other physiological amyloids can be reverted back to their soluble form by Hsp70/Hsp90, without undergoing degradation. This implies that the amyloid state is not a terminal/irreversible form of protein aggregation, even for pathological amyloid peptides. This cellular data is in good agreement with in vitro results showing a role for Hsp70 in disaggregation of β-amyloid and α-synuclein (Evans et al., 2006; Gao et al., 2015). While unproven, it is nonetheless tempting to speculate that pathological amyloidogenesis may be explained by dysregulation of the rIGSRNA-Hsp system. The data also suggest that β-amyloid/ACM activity is not unique to APP but present in many proteins that undergo amyloidogenesis in A-bodies.
Conceptually, these observations fundamentally change our understanding of amyloids from mysterious, toxic and irreversible protein aggregates to a common and reversible polypeptide-fold. We envision the existence of multiple different adaptive A-bodies, with proteins encoding variants of the ACM specific to rIGSRNA, and possibly other lncRNA and environmental cues. This work opens several avenues of investigation in protein folding, liquid to solid phase transition and cellular adaptation to stressors, while providing new conceptual insights in our efforts to resolve debilitating conditions associated with pathological amyloidogenesis, including Alzheimer’s disease and diabetes.
Experimental Procedures
Details of the plasmid design, peptides and antibodies can be found in Supplemental Information. Standard protocols were used for the following experiments and are described in the Supplemental Information: transmission electron microscopy, x-ray diffraction, SILAC-MS, RNA extractions, RT-PCR, RNA immunoprecipitation, western blotting, indirect immunofluorescence, cell proliferation and BrdU.
Cell Lines, Treatments and Stains
MCF-7, PC-3 and WI-38 cell lines were purchased from the ATCC and propagated in the suggested media. Stable MCF-7 and PC-3 cell lines were previously described(Audas et al., 2012). Hypoxia was induced by incubating cells in an H35 Hypoxystation at 37°C in a 1% O2, 5% CO2 and N2-balanced environment (HypOxygen). Acidosis was performed under hypoxic conditions by transferring cells to pH 6.0 media or growing cells in acidosis-permissive media as previously described (Audas et al., 2012). Heat shock was at 43°C for the indicated times. Sodium arsenite (125µM), H2O2 (300nM), thapsigargin (300nM), transcriptional stress (4µM Actinomycin D + 8µM MG132), rapamycin (5µM) and cycloheximide (25µg/ml) were treated for the indicated times. The Hsp70 (VER-155008: 40µM), Hsp90 (17-AAG: 5µM), PDI (16F16: 5µM) and GRP94 (Epigallocatechin gallate: 5µM) chaperone inhibitors and autophagosome inhibitor (Wortmannin: 5µM) were added 30 minutes before the 2 or 4 hour recoveries or for 3 hours in untreated cells. All recovered cells were grown in standard growth media at 21% O2 following stress treatment. Congo red (0.05%), Thioflavin S (0.002%), Amylo-Glo (1X), NIAD4 (10µM) and Methoxyl-X04 (20µM) were used to stain formaldehyde-fixed cells. Proteinase K (0.1µg/ml) treatment of methanol-fixed MCF-7 cells occurred at 25°C (1hr).
Insoluble Fractionation
Mammalian or bacterial cells were re-suspended in NP40 buffer (50µM Tris-HCl + 150µM NaCl + 1% NP40) and incubated at 25oC for 5min. Lysates were sonicated 2x for 10sec at 25% power (QSonica) and aliquots were taken as whole cell lysate fractions. Lysates were pelleted at 8000rpm for 10min. The pellet was washed 2x, prior to addition of NP40 buffer and one sonication for 10sec at 25% power to fully re-suspend the insoluble fraction.
Amyloid-Body Purification
Five 15cm plates per condition were washed in PBS and cells were scraped off of the surface. Osmotic buffer (10µM HEPES-pH 7.9, 10µM KCl, 1.2µM MgCl2, and 0.5µM DTT) was added and cells were incubated on ice for 5 minutes. Cells were dounced 10 times and pelleted. A sample of the supernatant was saved as the cytoplasmic fraction. Pellets were re-suspended in osmotic buffer, incubated on ice for 5 minutes, dounced 10 times and pelleted. Supernatant was discarded and a portion of the pellet was saved as the nuclear fraction. 1% TritonX-100 was used to re-suspend the pellet and slurry was sonicated twice for 10 seconds at 25% power (QSonica Sonicators). Solution was pelleted, the supernatant was discarded and the remaining material was re-suspended in 400µl 65% Percoll + 35% 1xNEH buffer (20µM HEPES-pH 7.4, 150µM NaCl, 0.2µM EDTA and 0.6% TritonX-100). Sample was layered with 400µl 55% Percoll + 45% 1xNEH buffer, 400µl 45% Percoll + 55% 1xNEH buffer and 400µl 35% Percoll + 65% 1xNEH buffer. Percoll gradients were spun at 1000 times gravity for 90 minutes and harvested into 3 400µl fractions. Fractions were analyzed by western blot for the nucleolar marker UBF1 and the established Amyloid-Body protein POLD1. These molecules were present predominantly at the 55–45% Percoll transition.
Stable isotope labeling by amino acids in cell culture–mass spectrometry
SILAC-labeling media was prepared using custom DMEM minus arginine and lysine (Athena Environmenal Sciences) supplemented with 5% dialyzed fetal calf serum (FCS) and 1% penicillin/streptomycin. MCF-7 cells were grown for 14 days in SILAC media containing 84 µg/µl L-arginine and 146 µg/µl L-lysine as follows: L-arginine and L-lysine (hypoxia-acidosis), L-arginine 13C6 and L-lysine 4,4,5,5-D4 (hypoxia-neutral) or L-arginine 13C6 15N4 and L-lysine 13C6 15N2 (normoxia-neutral) (Cambridge Isotope Labs). Normoxic-neutral cells were maintained at 21% O2 in neutral media (pH7.4), hypoxic-neutral cells were transferred to 1% O2 for 3 hours in pH7.4 media and hypoxic-acidotic cells were grown in pH 6.0 media at 1% O2 for 3 hours. Following treatment, nucleolar detention centers were isolated, described below, and total protein was extracted using 2% SDS and combined at a 1:1:1 ratio between the three treatments. Combined extracts were run on a precast TGX 4–12% gradient gel (BioRad), stained with Simply Blue Safestain (Invitrogen) and the entire lane was excised into 12 slices. Peptides were extracted from an in-gel tryptic digestion of each gel splice and analyzed by mass spectrometry as previously described (Andersen et al., 2005). Protein identification and quantitation were performed using the program Peaks Studio 7.0 (Thermo Scientific). Identification was set to a false discovery rate of 1% and proteins were considered to be enriched in the nucleoli of cells incubated under hypoxia-acidotic conditions if they possessed a minimum 2-fold hypoxia-acidosis:hypoxia-neutral and hypoxia-acidosis:normoxia-neutral ratio and significance (A) values. Protein sizes, grand average of hydrophobicity (GRAVY) and isoelectric point were calculated using the ProtParam program (SIB ExPASy Bioinformatics Resource Portal).
Bacterial Preparation
BL21 cultures were grown at 32°C (20hrs) prior to insoluble fractionation, inclusion body purification or staining. Microscopy was performed on fixed (4% formaldehyde 1hr) and permeabilized (0.5% Triton 10 min) cells stained with Congo red and Hoechst.
Fibrillation Propensity
Rosetta free energy scores for proteins and peptides was calculated using online ZipperDB software (UCLA) derived from published work (Goldschmidt et al., 2010).
Cell proliferation, BrdU incorporation and Congo red assays. 750,000 (10 cm plates) or 400,000 (3.5 cm plates) MCF-7 or PC-3 were plated for the cell proliferation and BrdU incorporation assay, respectively. 75,000 (6 cm plates) and 40,000 (3.5cm plates) WI-38 cells were also plated for the cell proliferation and BrdU assay, respectively. Media were changed at the indicated times to standard (pH 7.4) or acidosis-permissive (pH6.0) media, described previously (Audas et al., 2012; Mekhail et al., 2004), and grown at normoxic or hypoxic oxygen tensions for the indicated times. Post-acidification recovery was achieved by replacing acidosis-permissive media with standard media and a returning to normoxic conditions. Cell proliferation rates were determined by trypsinizing parallel plates and counting total cells for each time point for three independent repeats. Trypan blue was added to ensure variation was not due to excessive cell death. BrdU was incorporated for the last 20 minutes (MCF-7 and PC-3) or 60 minutes (WI-38) of each time point. Cells were fixed for 20 minutes at −20oC with an ethanol fixative (70% ethanol; 50µM glycine; pH 2.0), prior to BrdU staining according to manufacturer’s protocol (Roche). The ratio of BrdU stained nuclei versus Hoechst stained nuclei was assessed by fluorescence microscopy. Congo red assays to detect the proportion of A-body-positive cells was performed by staining untreated/treated cells with Congo red and Hoechst. Minimal exposure times were selected to eliminate background auto-fluorescence. The percentage of Congo red-positive cells was determined by counting the number of cells with Congo red-positive nucleoli and dividing by the total number of Hoecht-positive cells. The proportion of nuclear stress granule- and paraspeckle-positive cells was performed in the same manner on immunostained cells. At least three random fields were counted for three independent replicates.
Viability assay
Viability was calculated by growing the stable MCF-7 cell lines in standard or acidosis-permissive media for the indicated times. Each day parallel plates were stained for 5 minutes with 2µM propidium iodide and Hoechst 33342. Cells were immediately imaged by fluorescence microscopy. The cell death ratio was determined by counting the number of propidium iodide positive cells per field versus Hoechst stained nuclei. Experiments were performed in triplicate.
Tumor Xenografts
Animal experiments were performed in accordance with the University of Ottawa Animal Care Committee (CMM-181) policy. 107 exponentially growing cells suspended in 200 µL PBS were injected subcutaneously into the flanks of 6–8 week old CD-1 nude female mice (Charles River). Each mouse was injected with a control (left flank) and experimental (right flank) cell line. Tumor growth was recorded blind to the cell lines injected with calibers and animals were killed when the end point was reached or 5 weeks post-injection. Post-mortem, tumors were harvested, measured, fixed in 10% formalin, paraffin-embedded, sliced and stained.
Statistical analysis
Bar and line graphs represent the mean value from at least (n values indicated in figure legends) three independent replicates. Statistical analyses were performed with the error bars representing the standard error of the mean. p values were based on two-tailed Student’s t-test with the significance level indicated in the figure legend.
Supplementary Material
Highlights.
Nuclear amyloid bodies (A-bodies) form from systemic protein amyloidogenesis
An amyloid-converting motif and ribosomal intergenic lncRNA mediate amyloidogenesis
The heat shock chaperone pathway disaggregates A-bodies
Upon stimuli, cells activate physiological amyloidogenesis to enter a dormant state
Acknowledgments
We thank Y. Zhang, P.J. Salas, V. Shestopalov and I.S. Lossos for reagents, M.L. Bates at The Miami Project to Cure Paralysis for assistance with the TEM and M. Boulina at Analytical Imaging Core Facility of the Sylvester Comprehensive Cancer Center for assistance with the photobleaching experiments. Research in this publication was supported by grants from the National Institute of General Medical Sciences (R01GM115342) (SL) and National Cancer Institute (R01CA200676) (SL) of the National Institutes of Health, the Sylvester Comprehensive Cancer Center (SL, MLG), and the Canadian Institutes of Health Research (CHIR, SL). JJDH is a CIHR fellowship recipient.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
TEA performed the experiments, with assistance from DEA. MDJ (cell biology), JJDH and LTM (SILAC-MS and analysis), MK (cell growth and photobleaching), JP (flow cytometry), AM (x-ray diffraction), CB, ONK and MJ (data analysis), MW, TRH, SD (bacterial inclusion body preparation), provided technical assistance. TEA, MDJ, CG, LTM, MLG and SL conceived the experiments and analyzed the data. TEA and SL wrote the paper.
References
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Knowles TP, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL, et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc. 2011;133:14160–14163. doi: 10.1021/ja2017703. [DOI] [PubMed] [Google Scholar]
- Benz CC, Scott GK, Sarup JC, Johnson RM, Tripathy D, Coronado E, Shepard HM, Osborne CK. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24:85–95. doi: 10.1007/BF01961241. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell. 2014;54:675–682. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- Chujo T, Yamazaki T, Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta. 2016;1859:139–146. doi: 10.1016/j.bbagrm.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- Evans CG, Wisen S, Gestwicki JE. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1–42) aggregation in vitro. J Biol Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B. Human Hsp70 Disaggregase Reverses Parkinson’s-Linked alphα-synuclein Amyloid Fibrils. Mol Cell. 2015;59:781–793. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fruitos E, Sabate R, de Groot NS, Villaverde A, Ventura S. Biological role of bacterial inclusion bodies: a model for amyloid aggregation. FEBS J. 2011;278:2419–2427. doi: 10.1111/j.1742-4658.2011.08165.x. [DOI] [PubMed] [Google Scholar]
- Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci U S A. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald J, Riek R. Biology of amyloid: structure, function, and regulation. Structure. 2010;18:1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc’h C. Stress-induced transcription of satellite III repeats. J Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayatekin C, Matlack KE, Hesse WR, Guan Y, Chakrabortee S, Russ J, Wanker EE, Shah JV, Lindquist S. Prion-like proteins sequester and suppress the toxicity of huntingtin exon 1. Proc Natl Acad Sci U S A. 2014;111:12085–12090. doi: 10.1073/pnas.1412504111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, Margol L, Wu J, Breydo L, Thompson JL, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KP, Simon R, Schubert D, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, Huang L, Moreno I, Navarro A, Monzo M, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- McKinley MP, Bolton DC, Prusiner SB. A protease-resistant protein is a structural component of the scrapie prion. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol. 2004;6:642–647. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol. 2005;170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Pinto M, Morange M, Bensaude O. Denaturation of proteins during heat shock. In vivo recovery of solubility and activity of reporter enzymes. J Biol Chem. 1991;266:13941–13946. [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock IF, Rotin D. Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 1989;49:4373–4384. [PubMed] [Google Scholar]
- Thompson MJ, Sievers SA, Karanicolas J, Ivanova MI, Baker D, Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci U S A. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Maji SK, Sawaya MR, Eisenberg D, Riek R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008;6:e195. doi: 10.1371/journal.pbio.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Wiesener MS, Turley H, Allen WE, Willam C, Eckardt KU, Talks KL, Wood SM, Gatter KC, Harris AL, Pugh CW, et al. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1alpha. Blood. 1998;92:2260–2268. [PubMed] [Google Scholar]
- Zhao Z, Dammert MA, Grummt I, Bierhoff H. lncRNA-Induced Nucleosome Repositioning Reinforces Transcriptional Repression of rRNA Genes upon Hypotonic Stress. Cell Rep. 2016;14:1876–1882. doi: 10.1016/j.celrep.2016.01.073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.