ABSTRACT
Monoclonal antibodies are subjected to a wide variety of post-translational modifications (PTMs) that cause structural heterogeneity. Characterization and control of these modifications or quality attributes are critical to ensure antibody quality and to define any potential effects on the ultimate safety and potency of antibody therapeutics. The biopharmaceutical industry currently uses numerous tools to analyze these quality attributes individually, which requires substantial time and resources. Here, we report a simple and ultrafast bottom-up liquid chromatography-mass spectrometry (uLC-MS) method with 5 min tryptic digestion to simultaneously analyze multiple modifications, including oxidation, deamidation, isomerization, glycation, glycosylation, and N-terminal pyro-glutamate formation, which can occur during antibody production in mammalian cell culture, during purification and/or on storage. Compared to commonly used preparation procedures, this uLC-MS method eliminates assay artifacts of falsely-increased Met oxidation, Asp isomerization, and Asn deamidation, a problem associated with long digestion times in conventional LC-MS methods. This simple, low artifact multi-attribute uLC-MS method can be used to quickly and accurately analyze samples at any stage of antibody drug development, in particular for clone and media selection during cell culture development.
KEYWORDS: High throughput, monoclonal antibody, multi-attribute liquid chromatography-mass spectrometry method, peptide mapping, product quality attributes
Introduction
Monoclonal antibodies (mAbs) are the fastest growing sector within the pharmaceutical industry, and many mAb products have been approved for treating a wide variety of diseases including cancers, and immunological disorders.1-4 Most therapeutic mAbs are produced in mammalian cells where the cellular machinery can conduct complex post-translational modifications (PTMs), and they have many surface-exposed areas that are prone to various modifications during cell culture, purification, formulation, and storage.5-7 Modification events in vitro and particularly in vivo are the leading causes of structural heterogeneities. Common PTMs include glycosylation, deamidation, isomerization, and oxidation, incomplete disulfide bond formation, glycation, N-terminal glutamine cyclization, and C-terminal lysine processing.5,7-9
PTMs play important roles in modulating the physicochemical properties of proteins, changing enzymatic activity, and controlling protein interactions. In therapeutic antibodies, PTMs can affect biological function, efficient secretion, drug efficacy, and stability.6 The glycosylation that occurs on Asn297, which affects the effector functions and antibody stability, is of particular importance.10 The effects of oxidation and deamidation/isomerization greatly depends on the location of the affected residues. Modification of residues located in the complementarity-determining region (CDR) of an antibody can negatively affect binding to the target antigens, whereas those that occur in the constant domains of the Fc show few effects. Oxidation, specifically of methionine and tryptophan side chains, has been shown to result in conformational changes,11,12 to affect antibody binding to Fc receptors9,13 and antigens,14 and to alter mAb stability and half-life.12,15 Oxidation of the susceptible Met residues may also result in an increase in immunogenicity.16 Deamidation of Asn and isomerization of Asp in CDRs were linked to the loss of antibody binding affinity.17-20 Monitoring PTMs, in particular those considered as critical quality attributes that affect the safety or efficacy of the drug, such as glycosylation, deamidation/isomerization, and oxidation, is thus important for the quality control of antibody drugs during development, production, and storage.6 The processing control on critical quality attributes forms the basic principle of quality by design (QbD) in biopharmaceutical development.21
Deamidation of Asn on mAbs is normally analyzed by ion exchange chromatography and liquid chromatography-mass spectrometry (LC-MS),22 and isomerization of Asp is almost exclusively characterized by LC-MS. Oxidation of Met and Trp residues can be determined by hydrophobic interaction chromatography (HIC),23 reverse phase-HPLC coupled with Fabricator digestion,24 and mixed mode size-exclusion chromatography (SEC) using Sepax SEC-300MK25 and Waters BEH 200.26 However, the most common method to characterize site-specific oxidation, deamidation, and isomerization in proteins is bottom-up LC-MS, which is a very effective method for identifying and quantitating PTMs in protein molecules.27-30 Using the conventional LC-MS (cLC-MS) method, proteins are denatured in guanidine HCl or urea, reduced with dithiothreitol (DTT) or other reducing agents, which is then followed by alkylation of the peptides16,31 and/or buffer exchanged.31-33 The treated proteins are digested with trypsin or other proteases, and the peptides generated are separated by RP-HPLC for MS. The sample digestion step for the method is a time consuming process that could take up to 24 hours at 37°C, a step that could potentially induce artifacts34 such as asparagine deamidation,8,35-37 N-terminal glutamine cyclization,38 and methionine oxidation.39,40 In addition, the long preparation time hampers the high throughput analysis for a large number of samples in forced degradation studies and clone and media selection.
A multi-attribute method that is capable of detecting multiple types of modifications in mAbs is highly desired during the development and production of therapeutic antibodies. Because of the high detection sensitivity and selectivity of MS, bottom-up LC-MS is the ultimate technique for analyzing multiple PTMs simultaneously. A comprehensive method was recently described for simultaneous monitoring of mAb modifications and structural heterogeneity.32 Multi-attribute methods involve the generation of peptides with a protease such as trypsin and then detection of modifications in peptides by LC-MS. The masses of all tryptic peptides generated from a sample are acquired by LC-MS to create a sequence and PTM coverage library for the sample. Peptides with or without modifications can be automatically searched and extracted from the library for quantitation by software such as Pinpoint. In theory, any LC-MS method in conjugation with peptide generation should be able to generate the multi-attribute data. However, conventional sample preparation methods usually incorporate a time-consuming protease digestion procedure that can potentially introduce assay artifacts and hamper high throughput analysis.
We have developed an ultrafast multi-attribute liquid chromatography-mass spectrometry (uLC-MS) method with a 5 min trypsin digestion procedure that eliminates time-consuming alkylation, buffer exchange, and UV measurement steps present in the cLC-MS method.31,32 Incorporation of an ultrafast trypsin digestion step allows simultaneous monitoring of N-terminal pyro-glutamate, oxidation, deamidation, isomerization, glycation, and glycosylation in a single assay with minimal method artifacts. Because of the simplicity, our method enables high-throughput monitoring in the case of high sample loads, greatly reducing the labor cost of cLC-MS.
Results
Comparison of different digestion conditions
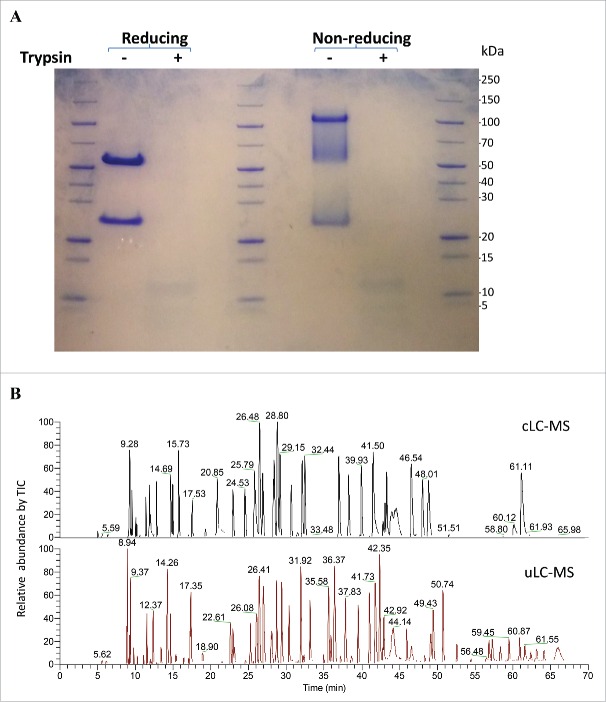
A cLC-MS method involves denaturation, reduction, and alkylation steps, as well as a labor-intensive buffer exchange step that makes it unsuitable for analysis of a large number of samples. To increase the throughput and minimize assay artifacts, we developed uLC-MS by using an easy and fast digestion strategy that was employed for fast glycan profiling and for mAb oxidation analysis.44,45 In this method, mAbs are first denatured and reduced in 4–8 M urea/10 mM DTT bicarbonate buffer at 70°C for 3 min, and then directly digested by trypsin at 37°C for 5 min. The alkylation and buffer exchange steps used in cLC-MS were eliminated. The completeness of antibody digestion by trypsin was analyzed by both reducing and non-reducing SDS-PAGE. All of intact mAb A was digested into peptides by trypsin within 5 min compared to undigested antibody (Fig. 1A). Fig. 1B shows a comparison of tryptic digestion of mAb A using the procedures in uLC-MS and cLC-MS methods on a C18-column. Differences in the peptide profile between these 2 methods were likely due to alkylation and different completeness of the digestion (Fig. 1B), which is also shown by the peak identification (supplemental Table S1). Almost all peptides detected by cLC-MS were also detected by uLC-MS. In addition, some miscleaved peptides were observed by uLC-MS. However, the cLC-MS method requires ∼6 h for the sample preparation to obtain peptides, while digestion in uLC-MS can be completed within 8 min (3 min for denaturation/reduction and 5 min for trypsin digestion). Importantly, uLC-MS also eliminates the buffer exchange step by spin columns and the potential sample loss in the columns.
Figure 1.
(A) SDS-PAGE analysis of mAb A digested by trypsin for 5 min under reducing or non-reducing conditions. Two μg of antibody or tryptic digests and MW standards were loaded. (B) Comparison of total ion counts of mAb A digests by trypsin using digestion procedures in conventional LC-MS and uLC-MS methods. Peptides were separated on a Zorbax C18 column.
Analysis of glycan profiles of mAb A
Glycosylation is a natural modification on therapeutic mAbs, and N-glycan profile can be a critical quality attribute for antibody drug development. To determine whether the peptide mixture of the fast digested antibody can be used to determine the glycan profiles, mAb A was subjected to trypsin digestion as described in the uLC-MS method section below. Peptides were separated on RP-HPLC, and determined by MS (Fig. 2). The total sequence coverage of peptides by uLC-MS was 96.5% with all the peptides identified (>0.07% total peak area), although some miscleaved peptides were observed (Table S1), while the total coverage for cLC-MS was 93.4% with identified peptides (>0.006% total peak area). The extracted ion chromatograms (EICs) of different glycopeptides of mAb A were determined. Multiple glycopeptides with different glycans,41 such as G0, G0F, G1F, G0F-GlcNac, and Man5, were detected. In addition, the glycan profile of mAb A determined by uLC-MS was highly similar to that obtained by the instant AB labeling/HILIC UPLC method, which is time-consuming and requires costly reagents (Fig. 2). Therefore, uLC-MS can be applied to screen the glycan profiles of antibodies with much less time and expense.
Figure 2.
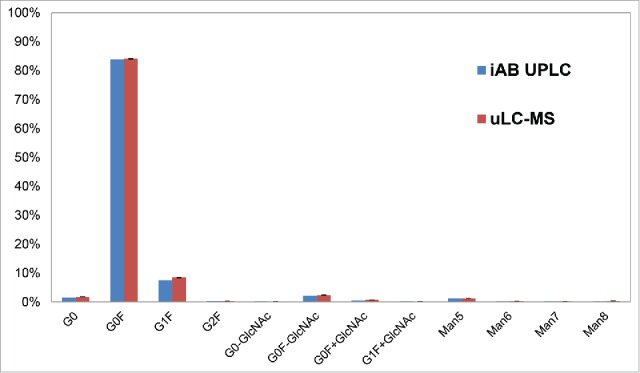
Glycan profiles determined by uLC-MS compared to iAB-HILIC-UPLC. EICs of each glycopeptide were performed by the software Pinpoint using the theoretical mass list of the glycopeptides in workbook. G0, G1, G0F, and G2F refer to different glycan forms associated with glycopeptides. G0, G1 and G2 represent glycans bearing zero, one or 2 galactose residues, respectively. G0F, G1F or G2F are glycan forms with fucose.41
Oxidation
Both cLC-MS and uLC-MS were used to assess the site-specific oxidation of mAb A following incubation at various temperatures for 12 months (Fig. 3). Both methods yielded almost identical % oxidation of Met253, Met429, and Met34 in the heavy chains (HC) at different temperatures. A significant increase of Met253 and Met429 oxidation was observed at 25°C at 12 months by both methods, whereas Met34 residues were not oxidized at 25°C.
Figure 3.
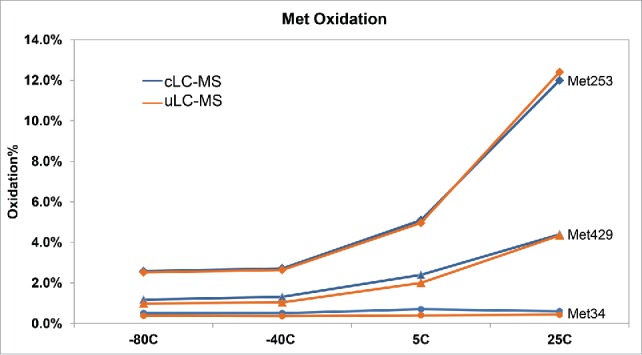
Detection of oxidation of mAb A at site-specific methionine residues (Met34, circles; Met429, triangles; Met253, diamonds) in the heavy chains by conventional LC-MS and uLC-MS following 12 month incubation at different temperatures.
Deamidation and isomerization
Antibodies are prone to isomerization at aspartate in DG motifs,42 and deamidation at asparagine residues, in particular in NG motifs.43 Following incubation at different temperatures for 12 months, mAb A was analyzed by both cLC-MS and uLC-MS. Results from both methods showed a significant increase in deamidation of asparagine residues at several positions at 25°C compared to that at −80, −40, and 5°C (Fig. 4A). In addition, the deamidation % at HC76-86 (Asn76, Gln81, and Asn83), Asn316, Asn277, and Asn385&Asn390 was significantly higher by cLC-MS than by uLC-MS for samples at different temperatures.
Figure 4.
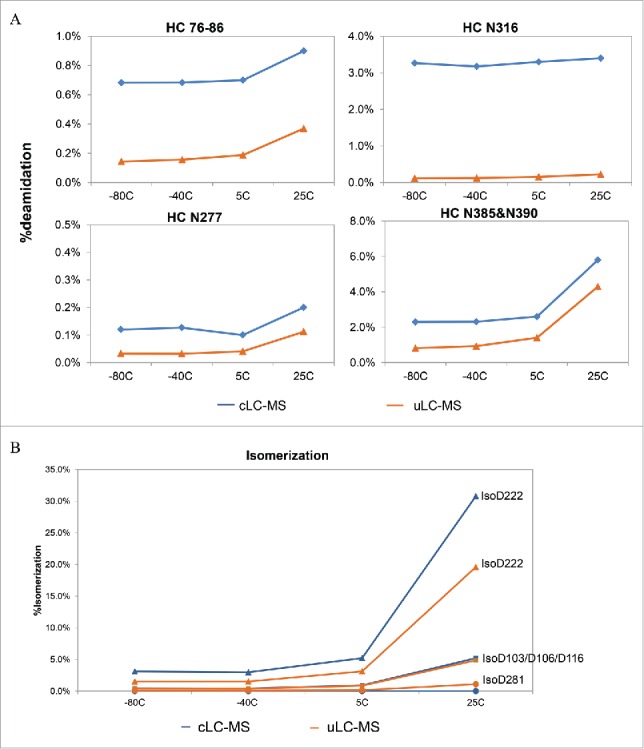
Deamidation (A) of Asn residues and isomerization (B) of Asp residues of mAb A at different temperatures determined by conventional LC-MS and uLC-MS following 12 month incubation at different temperatures. Sites of deamidation and isomerization are indicated.
Similarly, isomerization of several aspartate residues at positions 222, 103/106/116, and 281 in the HC also increased clearly at 25°C compared to that at −80, −40, and 5°C (Fig. 4B). cLC-MS and uLC-MS generated very similar results for Asp281 and Asp103/106/116. However, isomerization at Asp222 was significantly higher by cLC-MS than by uLC-MS.
Effects of digestion time
As described above, our results indicated higher levels of deamidation and isomerization at some sites of the antibody by cLC-MS compared to those by uLC-MS. The major differences might be artifacts resulting from the longer digestion time used in the LC-MS procedure. To confirm the effects of digestion time on detected oxidation and deamidation levels, mAb A was digested by trypsin using uLC-MS procedure in 20 mM NH4HCO3, pH 7.8 for both 5 min and 18 hours, and by our cLC-MS procedure in 20 mM Tris, pH 8.0 also for 18 hours, followed by analysis of Met oxidation, Asn deamidation, and Asp isomerization (Fig. 5).
Figure 5.
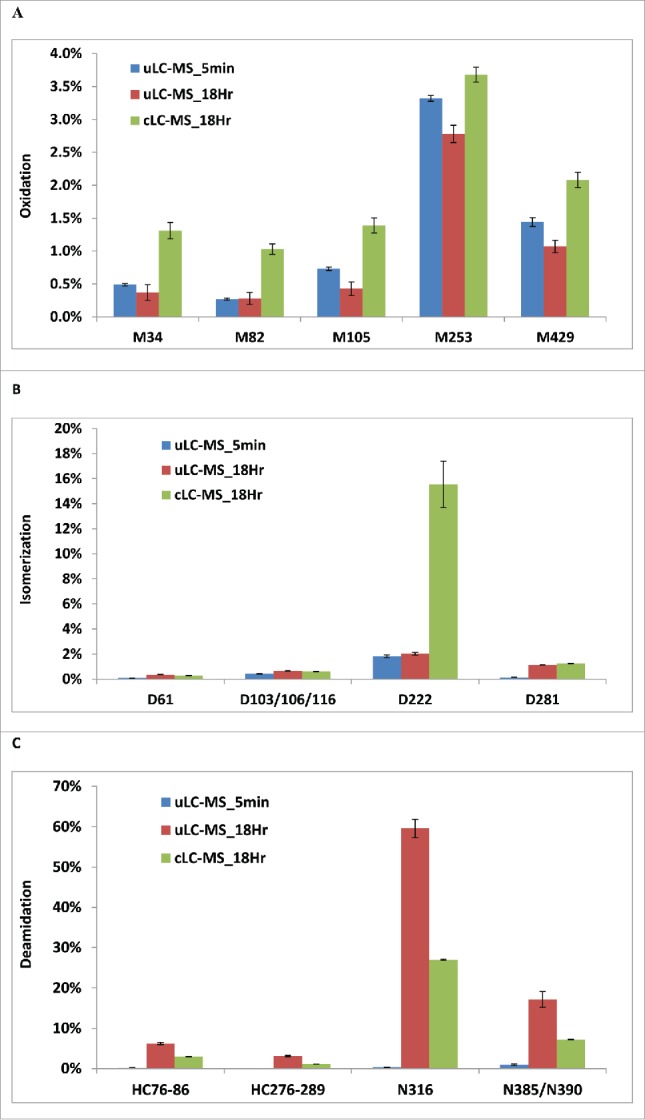
Impact of digestion time and digestion buffer on the detected levels of oxidation (A), isomerization (B), and deamidation (C). mAb A was digested by trypsin in NH4HCO3 using uLC-MS method for 5 min or 18 hours, or in Tris buffer using the conventional method for 18 hours ( n = 3 ).
Met oxidation at positions 34, 82, 105, 253, and 429 of HC showed similar levels using either 5 min or 18 hour digestion in uLC-MS. Therefore, the digestion time had little effect on detection of tested Met oxidation in mAb A by uLC-MS method. However, with the same 18-hour digestion, the cLC-MS method resulted in higher oxidation of detected Met residues than uLC-MS, indicating that buffer condition can induce higher Met oxidation (Fig. 5A). When Met34 oxidation from cLC-MS of mAb A incubated at different temperatures for 12 months with 4- or 18-hour digestion was compared (Table 1), the 18-hour digestion resulted in higher % of oxidized Met34 than the -hour digestion (Table 1), but had no effect on other oxidized Met residues (data not shown). Thus, the effect of the digestion time on Met oxidation is also site dependent.
Table 1.
Oxidation of Met34 of mAb A following 4- and 18-hour digestion in conventional LC-MS.
| Sample | 4 Hr digestion | 18 Hr digestion |
|---|---|---|
| −80°C | 0.5% | 0.9% |
| −40°C | 0.5% | 0.8% |
| 5°C | 0.7% | 1.9% |
| 25°C | 0.6% | 1.0% |
Note: samples were stored at indicated temperatures for 12 months.
Isomerization at Asp61 and Asp281 in HC was significantly increased by the longer digestion (18 hours) compared to 5 min digestion in uLC-MS, whereas isomerization at Asn103/106/116 and Asn222 was not affected. However, Asp222 by the 18-hour cLC-MS method demonstrated much higher % isomerization compared to uLC-MS with either 5 min or 18 hour digestion (16% vs 2%) (Fig. 5B). This reflects the buffer effect on Asp222 isomerization in solution.
Among others, the digestion time had the most impact on Asn deamidation. Long digestion (18 hours) in uLC-MS led to dramatic increases in detected deamidation in peptides HC76-86 and HC276-289, Asn316, and Asn385/390 (Fig. 5C). In particular, 60% of Asn316 was detected as deamidated by 18-hour digestion compared to only 0.5% by 5-min digestion in uLC-MS, reflecting a 120-fold artificial increase. Eighteen-hour digestion in the cLC-MS also resulted in higher detected deamidation, but not to the degree as by the 18-hour digestion in uLC-MS, suggesting the buffer effect on deamidation.
These data indicate that longer digestion could result in artifacts in detection of Met oxidation, Asn deamidation, and Asp isomerization. In addition, the buffer condition could also affect detection of all 3 of the modifications.
Analysis of forced degraded samples and real-time stability samples by uLC-MS
All results for our experiments suggest uLC-MS is able to reliably detect different modifications with very minimal artifacts. To test the feasibility of the method in the analysis of samples in forced degradation and real-time stability studies, 3 different mAbs were subjected to oxidant or light stress or exposure to elevated temperature and then tested. First, mAb B was subjected to treatment by 0.1% tert-butyl hydroperoxide (TBHP) and analyzed by uLC-MS (Table 2). TBHP is a methionine-specific oxidant,25 used to assess mAb oxidation.45 The location of methionine residues determines their sensitivity to TBHP. Up to 86% of Met105 in the HC was oxidized following 6 hours of TBHP treatment, whereas only 7.9% of Met252 was oxidized. Met358 and Met428 were little affected. As expected, TBHP did not induce deamidation or isomerization.
Table 2.
Analysis of PTMs in TBHP-treated mAb B at 25°C by uLC-MS.
| % modification by 0.1 % TBHP at 25C |
|||
|---|---|---|---|
| Modification | Initial | 1 Hour | 6 hours |
| M105 Oxidation | 1.4 | 36.4 | 85.5 |
| M252 Oxidation | 2.0 | 3.5 | 7.9 |
| M358 Oxidation | 0.2 | 0.4 | 0.7 |
| M428 Oxidation | 0.7 | 1.2 | 2.6 |
| N55 Deamidation | 0.5 | 0.5 | 0.6 |
| N384/N389 Deamidation | 0.3 | 0.3 | 0.4 |
To test antibody degradation after photo-irradiation, mAb C was exposed to different levels of light stress and analyzed by uLC-MS (Table 3). Two methionine sites (Met250 and M426) in the Fc domain were prone to oxidation by light stress. Up to 24% Met250 and 19% of Met426 were oxidized following the 1X UV light exposure, whereas 2 methionine residues in the CDR regions were relatively insensitive to light stress.
Table 3.
Analysis of PTMs in photo-irradiated mAb C at 25°C by uLC-MS.
| %oxidation |
|||
|---|---|---|---|
| Modification | Initial | 0.5X light | 1X light |
| M250 Oxidation | 1.7 | 14.3 | 23.9 |
| M356 Oxidation | 0.0 | 0.9 | 1.7 |
| M82 Oxidation | 0.2 | 0.3 | 0.3 |
| M105 Oxidation | 0.4 | 0.4 | 0.4 |
| M426 Oxidation | 0.8 | 11.6 | 19.2 |
To test antibody real-time stability, mAb A was incubated at 25°C for 0 to 12 months and then analyzed for various modifications by uLC-MS (Table 4). The N-terminal pyroglutamate significantly increased (by 11%) after a 12-month incubation. Met253 oxidation increased over time at 25 °C, up to 11.5% at 12 months. Only 4% of Met429 was oxidized at the same time points. Other Met residues were little changed. Asp222 and Asn326 displayed an increasing level of isomerization and deamidation over time, respectively. Up to 20.4% Asp222 isomerization, and 58.4% Asn326 deamidation was observed at 12 months. Asp and Asn at other locations showed little changes at 25 °C. Glycation at HC Lys327 and LC Lys96 was detected, but showed little changes over times at 25 °C.
Table 4.
Analysis of PTMs in real time stability samples of mAb A at 25 °C by uLC-MS.
| % modification at 25°C |
|||||
|---|---|---|---|---|---|
| Modification | 0 month | 1 month | 3 months | 6 months | 12 months |
| HC1-16pE | 80.2 | 81.8 | 84.7 | 87.8 | 91.3 |
| HC M82 Oxidation | 0.1 | 0.1 | 0.2 | 0.2 | 0.2 |
| HC M105 Oxidation | 0.3 | 0.3 | 0.3 | 0.3 | 0.4 |
| HC M253 Oxidation | 2.5 | 2.9 | 4.3 | 6.5 | 11.5 |
| HC M429 Oxidation | 1.0 | 1.1 | 1.6 | 2.3 | 4.0 |
| HC D103/D106/D116 IsoD | 0.4 | 0.6 | 1.4 | 2.5 | 4.8 |
| HC D222 IsoD | 2.0 | 3.2 | 6.7 | 11.2 | 20.4 |
| HC D281 IsoD | 0.1 | 0.2 | 0.3 | 0.5 | 1.0 |
| HC76-86 Deamidation | 0.2 | 0.2 | 0.3 | 0.4 | 0.5 |
| HC N316 Deamidation | 0.1 | 0.2 | 0.3 | 0.3 | 0.4 |
| HC N326 Deamidation | 4.5 | 10.4 | 21.2 | 39.4 | 58.4 |
| HC N385&N390 Deamidation | 0.9 | 1.2 | 1.6 | 2.1 | 3.2 |
| HC K327 Glycation | 0.4 | 0.4 | 0.4 | 0.4 | 0.4 |
| LC K96 Glycation | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 |
Our results thus indicate that uLC-MS is able to sensitively detect the change of various PTMs in forced degradation experiments and real-time stability studies of antibodies.
Linearity test
To demonstrate that uLC-MS is able to show a linear response to a modification, a series of mAb B samples with different amounts of oxidation were generated by mixing a control and antibody treated with 0.1% TBHP for 6 hours at different ratios. The oxidation at Met105 and Met252 was determined by uLC-MS. The measured percentage of oxidized Met105 and Met252 responded linearly to the expected % oxidation at these 2 Met sites (Fig. 6A and 6B). In addition, uLC-MS is also highly reproducible, as shown by Fig. 5.
Figure 6.
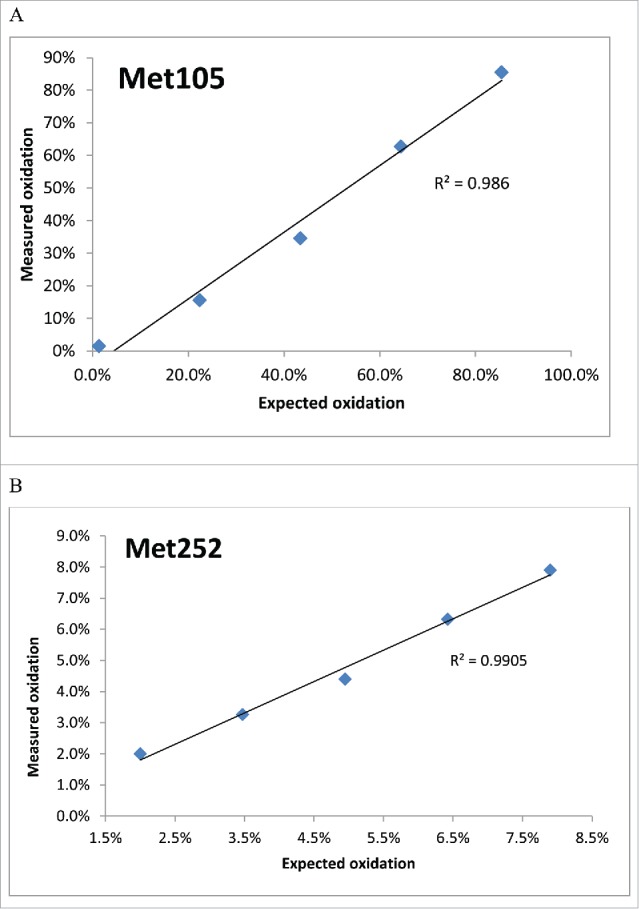
Linear response of the measured % of oxidized Met105 and Met252 in HC to the degree of oxidation in mAb B. The % oxidized peptides at Met105 (A) and Met252 (B) were integrated and plotted against the expected % of oxidation.
Discussion
MAbs harbor various PTMs that are introduced either during production in the expressing cells or during purification and storage. As PTMs can potentially affect the molecular structure and function of mAbs, it is vital to monitor and characterize them at different stages of production. A fast and simple method that enables analysis of various PTMs is needed for routine analysis of stability or forced degradation studies. Here, we describe an ultrafast and simple multi-attribute uLC-MS method that allows simultaneous monitoring of glycosylation, oxidation, deamidation, isomerization, glycation, and N-terminal pyroglutamate, or any modifications detectable by MS. This method adopted an ultrafast trypsin digestion protocol we developed for assays for glycan analysis44 and site-specific oxidation,45 and avoided the assay artifacts of oxidation, deamidation, and isomerization induced during the cLC-MS method.8,34-37,39
With uLC-MS available for comparison, assay artifacts for Asn deamidation and Asp isomerization at certain sites were clearly shown in the longer digestion procedure. For example, deamidation at Asn316 of mAb A was ∼120-fold overestimated by an 18-hour digestion in the cLC-MS compared to the 5-min digestion procedure (60% vs 0.5%). This result is very misleading because the deamidation was induced during the assay, and not originally present in the sample. The assay artifacts are normally unnoticed in routine LC-MS analysis during which the samples are digested from 1-2 hours up to 24 hours unless the shorter digestion protocol is available and is performed for comparison. Although different buffer conditions have been used to reduce deamidation/isomerization,37 the digestion step in our uLC-MS is only 5 min, and eliminates all artifacts associated with the cLC-MS. It is of interest that our data did not show that the digestion time had an effect on Met oxidation on mAb A by the uLC-MS method. However, the effect was observed for Met34 oxidation by the cLC-MS. The oxidation artifact could be specific to the antibody and oxidation site.
In addition, relative to cLC-MS, the sample preparation in uLC-MS is greatly simplified and eliminates the alkylation step, buffer exchange step, and UV concentration measurements. The alkylation is unnecessary in the assay because the digestion was so short, and the disulfide scrambling was inhibited at low pH when the digestion reaction was terminated by adding trifluoracetic acid (TFA) as we found no disulfide cross-linked peptides in any of the major peaks (>0.07% total peak area) (Table S1). In addition, our method is used to analyze PTMs but not disulfide linkages. Therefore, the major advantages of uLC-MS are the simple and faster procedure and avoidance of assay artifacts.
uLC-MS is able to analyze as many attributes of the antibody molecule as cLC-MS. Although modifications of many mAbs can be analyzed by HIC or mixed mode chromatography for oxidation and ion exchange chromatography for deamidation, the specificity of uLC-MS or cLC-MS is far superior, and particularly useful for mAbs that cannot be analyzed by the aforementioned assays.
uLC-MS has been successfully used to monitor antibody stability during forced degradation and real-time stability studies. Increasing oxidation of Met105 and Met252 in mAb B by TBHP treatment over time, and increasing Met253 oxidation, Asp222 isomerization, and Asn326 deamidation of mAb A in real-time stability at 25°C were all clearly demonstrated by this method. It can also be applied to screen a large number of samples during cell culture development, clone selection, and formulation studies.
As the digestion efficiency is antibody dependent, the digestion time and trypsin-to-antibody ratios in uLC-MS might need to be optimized for each case, although we found a 5 min digestion is sufficient for most antibodies tested thus far. Optimized uLC-MS is expected to be a useful tool for routine analytical needs in monitoring of antibody biochemical status. The method, however, should not be limited to monitoring only because it can be used in investigations, process development, and particular areas of product characterization that require quantitation of PTMs, although some miscleaved peptides might be present.
Materials and methods
Chemicals
Ammonium bicarbonate (cat # 09830-500G) was purchased from Fluka; urea (cat # 33247-250G), DTT, and iodoacetamide (IAM) were from Sigma-Aldrich. Sequencing grade modified trypsin (cat # V5111) was from Promega. Water (cat # W6-1) and acetonitrile (cat# A955-1), Tris buffer (1 M, pH 8.0), mass spectrometry grade water/0.1% formic acid, and acetonitrile/0.1% formic acid were purchased from Fisher Scientific. TFA (cat# 28904), guanidine hydrochloride (8 M), urea, Zeba spin desalting columns, 7 k MWCO (89883) were purchased from Thermo Scientific. TBHP, 70% solution (PN#180340050) was purchased from Acros Organics,
IgG monoclonal antibodies
All IgGs used in this study were expressed in Chinese hamster ovary cells and purified via a multiple-column purification process including Protein A chromatography. Antibodies were kept in their respective buffers for stability. The protein concentrations were determined by UV absorbance at 280 nm, and mAbs were provided at 25 mg/ml.
Forced degradation
Stressing of antibodies was performed according to our previous procedure.25
A. TBHP treatment
The antibodies were diluted to 5.0 mg/mL and incubated at 25°C for various lengths of time in an HPLC vial in the presence of 0.1% TBHP, and then injected for analysis.
B. Light stress
Each IgG under investigation (200 µL per exposure condition) was aliquoted into quartz cuvettes or HPLC glass vials. Light exposure was conducted in a photostability chamber (Caron model 6545–2) at 25°C. The levels selected ranged from 0X to 1X light exposure, where 1X = light exposure level equivalent to 1.2 million lux hours of white light and 200 watt hours/square meter of UV light based on ICH guidelines. After each designated exposure level was reached, the samples were covered and placed inside a dark box.
Trypsin digestion method in cLC-MS
In the cLC-MS method, the sample preparation was performed as described here, unless otherwise indicated. The antibody samples (100 μg of antibody) were denatured and reduced in a solution of 50 mM Tris at pH 8.0 with 6.0 M guanidine HCl and 20 mM DTT at 56°C for 30 min, followed by alkylation with 50 mM IAM for 30 min at room temperature in the dark. Then 11 mM DTT was added to quench the alkylation. The solution was buffer exchanged into 50 mM Tris pH 8.0 with 8 M urea using a Zeba spin column according to the manufacturer's instructions. Then 50 mM Tris pH 8.0 was added into the buffer-exchanged sample to dilute the 8 M urea to 2 M. Trypsin was added at a 1:20 enzyme: substrate ratio, and the mixture was incubated at 37°C for 4 hours. After digestion, 10% TFA was added at a 1:10 ratio to quench the digestion. The digested samples were stored at −80°C until analysis.
Ultrafast tryptic digestion in uLC-MS
The digestion was performed according to our previous procedure.44,45 Fifty μg of diluted mAb sample in 1 μl was thoroughly mixed with 20 μL of the reduction buffer (8 M urea in 20 mM ammonium bicarbonate and 10 mM DTT, and denatured and reduced at 70°C for 3 min in a water bath. Subsequently, the denatured and reduced sample was diluted 5X using 20 mM ammonium bicarbonate, and trypsin was added at a 1:20 enzyme: substrate ratio. The mixture was incubated at 37°C for 5 minutes. The digestion was quenched by mixing with 10% TFA at a 1:10 ratio. The digested samples were stored at −80°C until analysis.
The completeness of antibody digestion by trypsin was analyzed by reducing or non-reducing SDS-PAGE on a NuPAGE 4–12% Bis-Tris gel (Life Technologies, cat # NP0321BOX) according to the manufacturer's instructions. Two μg of intact antibody or antibody digest was loaded onto the gel, and separated at 200 volts for 35 minutes, and stained with Coomassie Bright Blue R-250. The MW standards (Thermo Scientific, Cat# 26630) were loaded as well.
LC-MS methods
An aliquot of 3 μg of the digest was injected onto a Zorbax C18 column (300-SB, 300 A pore size, 1.8 μm particle, 2.1 mm x 150 mm) at 50°C. Mobile phase A was 0.1% formic acid in water, and mobile B was 0.1% formic acid in acetonitrile. The gradient started at 1% B for 5 min, and then increased gradually to 10% in 1 minute. Next, the gradient was increased from 10% B to 35% B in 70 min or 90 min in cases when peak identification was performed. The column was then washed with 80% B for 5 minutes followed by equilibration at 1% B for 15 minutes.
The MS experiments were performed on a Q-Executive mass spectrometer (Thermo Fisher Scientific, San Jose, CA). For monitoring PTMs, the instrument was operated in MS1-only mode with AGC target of 1E6, maximum ion time of 200 ms with a scan range from m/z 300 to 1800. MS/MS was performed to confirm the peak identification. The instrument was operated in a data-dependent mode: the first survey MS (scan 1) from m/z 300 to 1800 with AGC target of 3E6 followed by 7 MS/MS scans. The MS/MS scan was set from m/z 200–2000 with AGC target of 5E5, maximum ion time of 250 ms. An m/z width of +/− 3 Da was employed to isolate the peptide precursor ions and 26% normalized collision energy was used to fragment the isolated peptides. Dynamic exclusion was utilized with exclusion duration of 8 s and a repeat count of 1.
Pepfinder
For characterization of modifications in different peptides, the raw data from the Q-Executive mass spectrometer were searched against the antibody database with different modifications using PepFinder version 1.0.10.11 (Thermo Scientific).
Pinpoint
For monitoring PTMs, amino acid sequences of peptides were manually entered into the Pinpoint software (version 1.2) (Thermo Scientific). Modifications, charge states, and isotopic distributions were also manually added to the peptides in the Pinpoint workbook. A tolerance of 5 ppm was used for the analysis in Pinpoint. EICs for desired peptides or ones with modifications of interest were generated for quantitation.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We want to thank Dr. Rich Rogers from Just for his assistance and discussion of this study, and Drs. Robin Ehrick and Kevin Turner for critical review of the manuscript.
References
- 1.Zitvogel L, Kroemer G. Cancer: Antibodies regulate antitumour immunity. Nature 2015; 7550:35-37; PMID:25924066; http://dx.doi.org/26325035 10.1038/nature14388 [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest 2015; 125:3384-91; PMID:26325035; http://dx.doi.org/ 10.1172/JCI80011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redman JM, Hill EM, AlDeghaither D, Weiner LM. Mechanisms of action of therapeutic antibodies for cancer. Mol Immunol 2015; 67:28-45; PMID:25911943; http://dx.doi.org/ 10.1016/j.molimm.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013; 39:1-10; PMID:23890059; http://dx.doi.org/ 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Gaza-Bulseco G, Faldu D, Chumsae C, Sun J. Heterogeneity of monoclonal antibodies. J Pharm Sci 2008; 97:2426-47; PMID:17828757; http://dx.doi.org/ 10.1002/jps.21180 [DOI] [PubMed] [Google Scholar]
- 6.Jenkins N, Murphy L, Tyther R. Post-translational modifications of recombinant proteins: significance for biopharmaceuticals. Mol Biotechnol 2008; 39:113-8; PMID:18327554; http://dx.doi.org/ 10.1007/s12033-008-9049-4 [DOI] [PubMed] [Google Scholar]
- 7.Manning MC, Chou DK, Murphy BM, Payne RW, Katayama DS. Stability of protein pharmaceuticals: an update. Pharm Res 2010; 27:544-75; PMID:20143256; http://dx.doi.org/ 10.1007/s11095-009-0045-6 [DOI] [PubMed] [Google Scholar]
- 8.Timm V, Gruber P, Wasiliu M, Lindhofer H, Chelius D. Identification and characterization of oxidation and deamidation sites in monoclonal rat/mouse hybrid antibodies. J Chromatogr B Analyt Technol Biomed Life Sci 2010; 878:777-84; PMID:20153988; http://dx.doi.org/ 10.1016/j.jchromb.2010.01.036 [DOI] [PubMed] [Google Scholar]
- 9.Bertolotti-Ciarlet A, Wang W, Lownes R, Pristatsky P, Fang Y, McKelvey T, Li Y, Drummond J, Prueksaritanont T, Vlasak J. Impact of methionine oxidation on the binding of human IgG1 to Fc Rn and Fc gamma receptors. Mol Immunol 2009; 46:1878-82; PMID:19269032; http://dx.doi.org/ 10.1016/j.molimm.2009.02.002 [DOI] [PubMed] [Google Scholar]
- 10.Hmiel LK, Brorson KA, Boyne MT 2nd. Post-translational structural modifications of immunoglobulin G and their effect on biological activity. Analytical and bioanalytical chemistry 2015; 407:79-94; PMID:25200070; http://dx.doi.org/ 10.1007/s00216-014-8108-x [DOI] [PubMed] [Google Scholar]
- 11.Liu D, Ren D, Huang H, Dankberg J, Rosenfeld R, Cocco MJ, Li L, Brems DN, Remmele RL Jr. Structure and stability changes of human IgG1 Fc as a consequence of methionine oxidation. Biochemistry 2008; 47:5088-100; PMID:18407665; http://dx.doi.org/ 10.1021/bi702238b [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Gaza-Bulseco G, Xiang T, Chumsae C. Structural effect of deglycosylation and methionine oxidation on a recombinant monoclonal antibody. Mol Immunol 2008; 45:701-8; PMID:17719636; http://dx.doi.org/ 10.1016/j.molimm.2007.07.012 [DOI] [PubMed] [Google Scholar]
- 13.Pan H, Chen K, Chu L, Kinderman F, Apostol I, Huang G. Methionine oxidation in human IgG2 Fc decreases binding affinities to protein A and FcRn. Protein Sci 2009; 18:424-33; PMID:19165723; http://dx.doi.org/ 10.1002/pro.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Z, Feng J, Lin HY, Mullapudi S, Bishop E, Tous GI, Casas-Finet J, Hakki F, Strouse R, Schenerman MA. Identification of a single tryptophan residue as critical for binding activity in a humanized monoclonal antibody against respiratory syncytial virus. Anal Chem 2007; 79:2797-805; PMID:17319649; http://dx.doi.org/ 10.1021/ac062311j [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Vlasak J, Li Y, Pristatsky P, Fang Y, Pittman T, Roman J, Wang Y, Prueksaritanont T, Ionescu R. Impact of methionine oxidation in human IgG1 Fc on serum half-life of monoclonal antibodies. Mol Immunol 2011; 48:860-6; PMID:21256596; http://dx.doi.org/ 10.1016/j.molimm.2010.12.009 [DOI] [PubMed] [Google Scholar]
- 16.Hermeling S, Crommelin DJ, Schellekens H, Jiskoot W. Structure-immunogenicity relationships of therapeutic proteins. Pharm Res 2004; 21:897-903; PMID:15212151; http://dx.doi.org/ 10.1023/B:PHAM.0000029275.41323.a6 [DOI] [PubMed] [Google Scholar]
- 17.Yan B, Steen S, Hambly D, Valliere-Douglass J, Vanden Bos T, Smallwood S, Yates Z, Arroll T, Han Y, Gadgil H, et al.. Succinimide formation at Asn 55 in the complementarity determining region of a recombinant monoclonal antibody IgG1 heavy chain. J Pharm Sci 2009; 98:3509-21; PMID:19475547; http://dx.doi.org/ 10.1002/jps.21655 [DOI] [PubMed] [Google Scholar]
- 18.Cacia J, Keck R, Presta LG, Frenz J. Isomerization of an aspartic acid residue in the complementarity-determining regions of a recombinant antibody to human IgE: identification and effect on binding affinity. Biochemistry 1996; 35:1897-903; PMID:8639672; http://dx.doi.org/ 10.1021/bi951526c [DOI] [PubMed] [Google Scholar]
- 19.Rehder DS, Chelius D, McAuley A, Dillon TM, Xiao G, Crouse-Zeineddini J, Vardanyan L, Perico N, Mukku V, Brems DN, et al.. Isomerization of a single aspartyl residue of anti-epidermal growth factor receptor immunoglobulin gamma2 antibody highlights the role avidity plays in antibody activity. Biochemistry 2008; 47:2518-30; PMID:18232715; http://dx.doi.org/ 10.1021/bi7018223 [DOI] [PubMed] [Google Scholar]
- 20.Wakankar AA, Borchardt RT, Eigenbrot C, Shia S, Wang YJ, Shire SJ, Liu JL. Aspartate isomerization in the complementarity-determining regions of two closely related monoclonal antibodies. Biochemistry 2007; 46:1534-44; PMID:17279618; http://dx.doi.org/ 10.1021/bi061500t [DOI] [PubMed] [Google Scholar]
- 21.Herwig C, Garcia-Aponte OF, Golabgir A, Rathore AS. Knowledge management in the QbD paradigm: manufacturing of biotech therapeutics. Trends Biotechnol 2015; 33:381-7; PMID:25980924; http://dx.doi.org/ 10.1016/j.tibtech.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 22.Huang L, Lu J, Wroblewski VJ, Beals JM, Riggin RM. In vivo deamidation characterization of monoclonal antibody by LC/MS/MS. Anal Chem 2005; 77:1432-9; PMID:15732928; http://dx.doi.org/ 10.1021/ac0494174 [DOI] [PubMed] [Google Scholar]
- 23.Haverick M, Mengisen S, Shameem M, Ambrogelly A. Separation of mAbs molecular variants by analytical hydrophobic interaction chromatography HPLC: overview and applications. mAbs 2014; 6:852-8; PMID:24751784; http://dx.doi.org/ 10.4161/mabs.28693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An Y, Zhang Y, Mueller HM, Shameem M, Chen X. A new tool for monoclonal antibody analysis: application of IdeS proteolysis in IgG domain-specific characterization. mAbs 2014; 6:879-93; PMID:24927271; http://dx.doi.org/ 10.4161/mabs.28762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavon JA, Li X, Chico S, Kishnani U, Soundararajan S, Cheung J, Li H, Richardson D, Shameem M, Yang X. Analysis of monoclonal antibody oxidation by simple mixed mode chromatography. J Chromatogr A 2016; 1431:154-65; PMID:26774436; http://dx.doi.org/ 10.1016/j.chroma.2015.12.068 [DOI] [PubMed] [Google Scholar]
- 26.Wong C, Strachan-Mills C, Burman S. Facile method of quantification for oxidized tryptophan degradants of monoclonal antibody by mixed mode ultra performance liquid chromatography. J Chromatogr A 2012; 1270:153-61; PMID:23182937; http://dx.doi.org/ 10.1016/j.chroma.2012.10.064 [DOI] [PubMed] [Google Scholar]
- 27.Mo J, Tymiak AA, Chen G. Structural mass spectrometry in biologics discovery: advances and future trends. Drug discovery today 2012; 17:1323-30; PMID:22819924; http://dx.doi.org/ 10.1016/j.drudis.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 28.Chen G, Warrack BM, Goodenough AK, Wei H, Wang-Iverson DB, Tymiak AA. Characterization of protein therapeutics by mass spectrometry: recent developments and future directions. Drug discovery today 2011; 16:58-64; PMID:21093608; http://dx.doi.org/ 10.1016/j.drudis.2010.11.003 [DOI] [PubMed] [Google Scholar]
- 29.Kaltashov IA, Bobst CE, Abzalimov RR, Wang G, Baykal B, Wang S. Advances and challenges in analytical characterization of biotechnology products: mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnol Adv 2012; 30:210-22; PMID:21619926; http://dx.doi.org/ 10.1016/j.biotechadv.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianferani S. Characterization of therapeutic antibodies and related products. Anal Chem 2013; 85:715-36; PMID:23134362; http://dx.doi.org/ 10.1021/ac3032355 [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Li H, Shameem M, Xu W. Development of a sample preparation method for monitoring correct disulfide linkages of monoclonal antibodies by liquid chromatography-mass spectrometry. Anal Biochem 2015; 495:21-8; PMID:26656925; http://dx.doi.org/ 10.1016/j.ab.2015.11.010 [DOI] [PubMed] [Google Scholar]
- 32.Rogers RS, Nightlinger NS, Livingston B, Campbell P, Bailey R, Balland A. Development of a quantitative mass spectrometry multi-attribute method for characterization, quality control testing and disposition of biologics. mAbs 2015; 7:881-90; PMID:26186204; http://dx.doi.org/ 10.1080/19420862.2015.1069454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Rossomando A, Wu SL, Karger BL. Comparability analysis of anti-CD20 commercial (rituximab) and RNAi-mediated fucosylated antibodies by two LC-MS approaches. mAbs 2013; 5:565-75; PMID:23751726; http://dx.doi.org/ 10.4161/mabs.24814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi H, Miyazaki M. Enzyme-immobilized reactors for rapid and efficient sample preparation in MS-based proteomic studies. Proteomics 2013; 13:457-66; PMID:23255229; http://dx.doi.org/ 10.1002/pmic.201200272 [DOI] [PubMed] [Google Scholar]
- 35.Krokhin OV, Antonovici M, Ens W, Wilkins JA, Standing KG. Deamidation of -Asn-Gly- sequences during sample preparation for proteomics: Consequences for MALDI and HPLC-MALDI analysis. Anal Chem 2006; 78:6645-50; PMID:16970346; http://dx.doi.org/ 10.1021/ac061017o [DOI] [PubMed] [Google Scholar]
- 36.Bongers J, Heimer EP, Lambros T, Pan YC, Campbell RM, Felix AM. Degradation of aspartic acid and asparagine residues in human growth hormone-releasing factor. Int J Pept Protein Res 1992; 39:364-74; PMID:1428526; http://dx.doi.org/ 10.1111/j.1399-3011.1992.tb01596.x [DOI] [PubMed] [Google Scholar]
- 37.Kori Y, Patel R, Neill A, Liu H. A conventional procedure to reduce Asn deamidation artifacts during trypsin peptide mapping. J Chromatogr B Analyt Technol Biomed Life Sci 2016; 1009–1010:107-13; PMID:26720699; http://dx.doi.org/ 10.1016/j.jchromb.2015.12.009 [DOI] [PubMed] [Google Scholar]
- 38.Dick LW Jr., Kim C, Qiu D, Cheng KC. Determination of the origin of the N-terminal pyro-glutamate variation in monoclonal antibodies using model peptides. Biotechnol Bioeng 2007; 97:544-53; PMID:17099914; http://dx.doi.org/ 10.1002/bit.21260 [DOI] [PubMed] [Google Scholar]
- 39.Liu H, Ponniah G, Neill A, Patel R, Andrien B. Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis. Anal Chem 2013; 85:11705-9; PMID:24200102; http://dx.doi.org/ 10.1021/ac403072w [DOI] [PubMed] [Google Scholar]
- 40.Zang L, Carlage T, Murphy D, Frenkel R, Bryngelson P, Madsen M, Lyubarskaya Y. Residual metals cause variability in methionine oxidation measurements in protein pharmaceuticals using LC-UV/MS peptide mapping. J Chromatogr B Analyt Technol Biomed Life Sci 2012; 895–896:71-6; PMID:22483985; http://dx.doi.org/ 10.1016/j.jchromb.2012.03.016 [DOI] [PubMed] [Google Scholar]
- 41.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nature reviews Drug discovery 2009; 8:226-34; PMID:19247305; http://dx.doi.org/ 10.1038/nrd2804 [DOI] [PubMed] [Google Scholar]
- 42.Wakankar AA, Borchardt RT, Eigenbrot C, Shia S, Wang YJ, Shire SJ, Liu JL. Aspartate isomerization in the complementarity-determining regions of two closely related monoclonal antibodies. Biochemistry 2007; 46:1534-44; PMID:17279618; http://dx.doi.org/ 10.1021/bi061500t [DOI] [PubMed] [Google Scholar]
- 43.Huang L, Lu J, Wroblewski VJ, Beals JM, Riggin RM. In vivo deamidation characterization of monoclonal antibody by LC/MS/MS. Anal Chem 2005; 77:1432-9; PMID:15732928; http://dx.doi.org/ 10.1021/ac0494174 [DOI] [PubMed] [Google Scholar]
- 44.Yang X, Kim SM, Ruzanski R, Chen Y, Moses S, Ling WL, Li X, Wang SC, Li H, Ambrogelly A, et al.. Ultrafast and high-throughput N-glycan analysis for monoclonal antibodies. mAbs 2016; 8:706-17; PMID:27082290; http://dx.doi.org/ 10.1080/19420862.2016.1156828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Xu W, Wang Y, Zhao J, Liu Y-H, Richardson D, Li H, Shameem M, Yang X. High throughput peptide mapping method for analysis of site specific monoclonal antibody oxidation. J chromatography A 2016; 1460:51-60; PMID:27432793; http://dx.doi.org/ 10.1016/j.chroma.2016.06.085 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.