ABSTRACT
Immunoglobulin G1 (IgG1) is the most abundant circulating human antibody and also the scaffold for many therapeutic monoclonal antibodies (mAbs). The destruction of IgG-coated targets by cell-mediated pathways begins with an interaction between the IgG Fc region and multiple varieties of membrane-bound Fc γ receptors (FcγRs) on the surface of leukocytes. This interaction requires the presence of an asparagine-linked (N-)glycan on the Fc, and variations in the N-glycan composition can affect the affinity of CD16A binding (an FcγR). Contemporary efforts to glycoengineer mAbs focus on increasing CD16A affinity, and thus treatment efficacy, but it is unclear how these changes affect affinity for the other FcγRs. Here, we measure binding of the extracellular Fc-binding domains for human CD16A and B, CD32A, B and C, and CD64 to 6 well-defined IgG1 Fc glycoforms that cover ∼85% of the pool of human IgG1 Fc glycoforms. Core α1–6 fucosylation showed the greatest changes with CD16B (8.5-fold decrease), CD16A (3.9-fold decrease) and CD32B/C (1.8-fold decrease), but did not affect binding to CD32A. Adding galactose to the non-reducing termini of the complex-type, biantennary glycan increased affinity for all CD16s and 32s tested by 1.7-fold. Sialylation did not change the affinity of core-fucosylated Fc, but increased the affinity of afucosylated Fc slightly by an average of 1.16-fold for all CD16s and CD32s tested. The effects of fucose and galactose modification are additive, suggesting the contributions of these residues to Fc γ receptor affinity are independent.
KEYWORDS: CD16, CD32, CD64, FcγRIII, FcγRII, FcγRI, N-glycosylation
Introduction
Immunoglobulin G (IgG) is a versatile molecule developed by the immune system to neutralize invading pathogens with high specificity, and it is now employed by clinicians to treat a wide range of diseases. Biologics are the fastest growing class of new prescription drugs driven largely by the phenomenal expansion of monoclonal antibody (mAb)-based therapies.1 Though some mAbs are able to affect disease simply by irreversibly binding the target epitope and blocking function (e.g., anti-tumor necrosis factor adalimumab), many mAbs recognize cell surface biomarkers and recruit circulating lymphoid and myeloid cells to destroy the diseased tissue. This latter process requires Fc γ receptors (FcγRs) that are expressed on the surface of recruited macrophages/monocytes, natural killer (NK) cells and neutrophils. The FcγRs bind mAb-coated tissues through the IgG crystallizable fragment (Fc) and kill the target by phagocytosis or cell-mediated cytotoxic processes.2
Human cells express 5 activating FcγRs (CD64, the “high affinity” FcγR, and the “low affinity” FcγRs CD32A, CD32C, CD16A, CD16B) and one “low affinity” inhibitory receptor (CD32B) (Fig. 1). The expression profile for these receptors is complex and discussed in detail elsewhere.3 A few features, however, should be noted. Macrophages express the entire complement of FcγRs; naïve NK cells express primarily CD16A and neutrophils primarily CD16B.4 Signaling through the low affinity receptors requires IgG oligomers, formed as a result of target opsonization.5 Even so, in vitro affinities of mAbs binding to the extracellular CD16A domains correlate with improved treatment outcome in treated patients.6,7 Thus, in vitro measurements of monovalent affinity are important metrics in mAb optimization.
Figure 1.
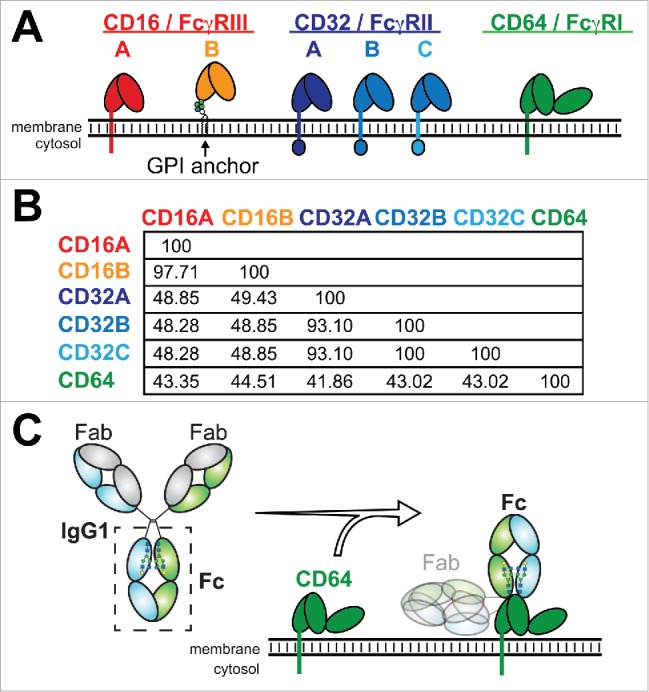
A comparison of the IgG-binding Fc γ receptors. (A) Domain organization of the Fc γ receptors and (B) a comparison of the percent sequence identity for the extracellular regions used in this study. (C) IgG1 binds these receptors through the crystallizable fragment (Fc) and recognizes antigens through the antigen-binding fragment (Fab).
IgG:FcγR interactions, and thus most mAb-mediated therapies, rely on the presence of an asparagine-linked (N)-glycan at residue Asn297 of the IgG Fc region.5,8 This complex carbohydrate is added to the newly synthesized immunoglobulin polypeptide chain during transport into the lumen of the endoplasmic reticulum (ER). The IgG N-glycan is remodeled by glycosyltransferases and glycosylhydrolases during transit through the ER and Golgi in a template-independent manner. The result is a distribution of IgG glycoforms that are primarily of a biantennary complex-type with ∼7–12 linked monosaccharide units and contain variable levels of fucose, galactose and N-acetylneuraminic acid.9 Proper N-glycan remodeling is required for efficient effector function, and the N-glycan composition is known to contribute to FcγR-mediated immune activation.10
The IgG1 Fc N-glycan forms an intramolecular carbohydrate/polypeptide interface that stabilizes the polypeptide loop containing Asn297, the site of N-glycan attachment.11 Truncating the N-glycan length or disrupting the carbohydrate/polypeptide interface reduces CD16A binding.12,13 Recent advances in expression technologies produce glycoengineered mAbs with reduced fucosylation to achieve tighter CD16A binding, stronger cell-mediated cytotoxicity and better patient outcomes.14,15
A number of studies have reported the effects of IgG carbohydrate composition on receptor-binding affinity (for examples, see ref. 13, 14, 16-21). These studies have used different FcγR ligands, including Fc and a wide range of mAbs; as a result, it is impossible to accurately compare different reports to quantify the effect of IgG N-glycan composition among different receptor fragments. Here, we measured the binding of 6 FcγRs to 6 IgG1 Fc glycoforms covering >85% of the glycan variability in the human IgG1 Fc pool22 to establish the relative contribution of each carbohydrate modification for each receptor. We were able to prepare and validate pure glycoforms, as well as quantify receptor binding in vitro with high precision. Our results provide clear insight into only the Fc:FcγR interaction component of FcγR-mediated cellular activation, and eliminates the influence of multiple variables present in cell activation studies, including immune complex size/construction, cell viability/health, FcγR density, and the effect of monovalent Fc:FcγR affinity in multivalent immune complex:cell surface interaction. We expect these results will be informative in the greater context of cell and immune system activation, and will be helpful to guide next-generation monoclonal antibodies with engineered Fc regions. Rather than full-length IgG, the Fc is used because full receptor binding properties of IgG are retained in the Fc portion, the IgG1 Fc is common to all IgG1-based mAbs, and removing the antigen-binding fragments likely does not affect Fc structure.23-25 The data reported herein revealed unanticipated differences among the FcγRs that will contribute to understanding of the effect of a single glycan modification on mAb binding to all FcγRs in the body.
Results
Fc γ receptor expression and homogenous IgG1 Fc preparation
Fc γ receptor extracellular soluble domains fused to an N-terminal green fluorescent protein (GFP) tag expressed from HEK293F cells at high glycoprotein yields (90–170 mg/L) and high purity for the CD16s and CD32s (Fig. 2). The extracellular domains of CD32B and CD32C are indistinguishable by protein sequence and are analyzed as a single construct (hereafter referred to as CD32B/C; Fig. 1.). An optimized CD64 construct without a GFP tag expressed at lower levels (4.3 mg/L) and appears as a smear on an SDS-PAGE with reduced Coomassie staining resulting from high N-glycosylation (7 sites; Fig. 2B). We estimate CD64 is 90–95% pure following purification. IgG1 Fc expression in the same cell line (with or without a reported inhibitor of fucosylation26-28) produced a limited range of glycoforms that were remodeled in vitro with a β-galactosidase and glycosyltransferases to produce nearly homogenous IgG1 Fc preparations29,30 that eluted in a single peak at ∼52 kDa from a size-exclusion chromatography column (Fig. 2C).
Figure 2.
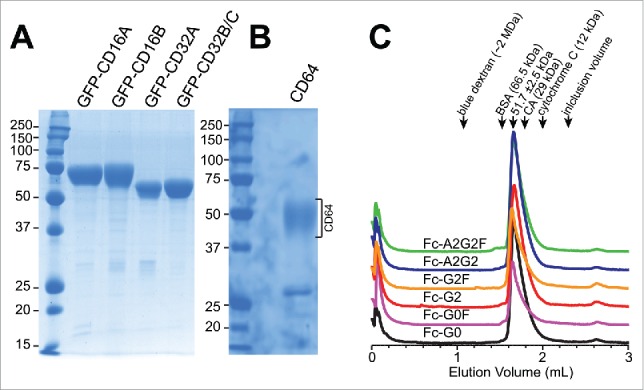
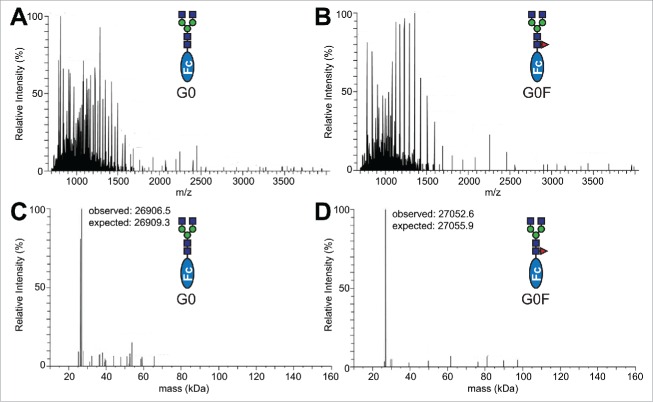
Analysis of Fc and receptor proteins. Reducing SDS-PAGE analysis of the purified Fc γ receptor fragments visualized with Coomassie brilliant blue staining. CD16A, 16B, 32A and 32B/C were expressed as N-terminal polyhistidine and GFP fusions (A). CD64 is expressed with a C-terminal polyhistidine tag (B). The receptors run as diffuse bands on a gel due to the high levels of N-glycosylation. CD16A has 5 N-glycan sites, CD16B: 6, CD32A: 2, CD32B/C: 3, CD64: 7. (C) All Fc materials eluted as a single peak from a S200 size-exclusion chromatography column at a retention time that indicated a molecular mass of ∼52 kDa. The elution volumes of several standard molecules are indicated with arrows at the top of the chromatogram.
Further analysis of the Fc glycoproteins by mass spectrometry (MS) confirmed their high homogeneity. An analysis of the PNGaseF-released N-glycans revealed little variation in the Fc N-glycan preparations (Fig. 3), particularly with respect to galactose and fucose modification. However, the sialylated forms were less homogenous. Though sialylation of IgG1 Fc is difficult to achieve,30 we consistently recovered at least 80% of the disialylated and core-fucosylated A2G2F form, but only 60% of the A2G2 form with the remainders being monosialylated. This level of conversion was consistent with repeated experiments and suggests differences in N-glycan accessibility to the α2–6 sialyltransferase (ST6Gal-I) due to the presence of a core α1–6-linked fucose residue. Analysis of the intact Fc glycoproteins by electrospray ionization-mass spectrometry (ESI-MS) confirmed the glycan remodeling and protein homogeneity. Representative spectra of the G0 and G0F forms are shown in Fig. 4. MS spectra of our Fc preparations show limited proteolysis of the C-terminal tail (removing LeuSerProGlyLys residues). These residues were not observed in structures of the Fc determined by X-ray crystallography.31-35 The G0 preparation revealed an additional, less abundant form that was consistent with proteolysis at the C-terminus that removes an additional Ser residue plus GluProLysSer residues at the N-terminus (Fig. 4C). These N-terminal residues occur well before the Cys residues that form the hinge disulfides in IgG1 Fc. They are not expected to affect receptor binding, and likewise do not appear in structures of the Fc determined by X-ray crystallography.
Figure 3.
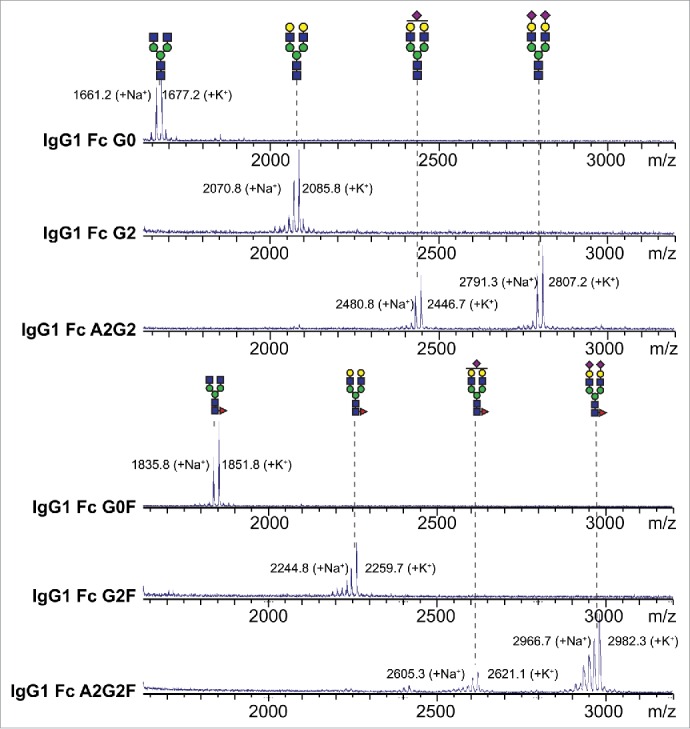
Mass spectrometric analysis of N-glycans following in vitro enzymatic remodeling on IgG1 Fc reveals a high level of homogeneity. N-glycans were cleaved from IgG1 Fc, purified, permethylated then analyzed by MALDI-MS. Cartoon diagrams show the potential N-glycan configuration (using the CFG convention; 59 http://glycomics.scripps.edu/CFGnomenclature.pdf); isobaric ions were not differentiated. Observed masses are indicated. Key: blue square: N-acetylglucosamine, green circle: mannose, yellow circle: galactose, red triangle: fucose, purple diamond: N-acetylneuraminic acid.
Figure 4.
Analysis of IgG1 Fc glycoforms by ESI-MS. Raw spectra for the intact G0 (A) and G0F (B) glycoforms. Deconvoluted spectra for G0 (C) and G0F (D) are also shown. Slightly lower-than-expected observed masses are likely due to incomplete reduction of the 7 Cys residues in this construct.
Binding analyses
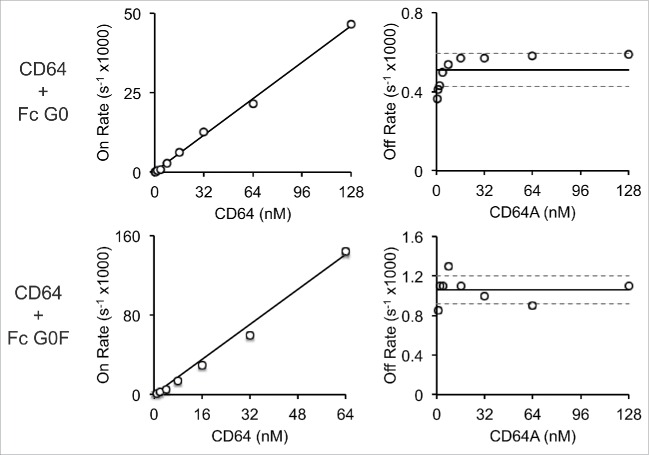
Surface plasmon resonance (SPR) provided kinetic and equilibrium binding measurements for the interaction between immobilized homogenous IgG1 Fc preparations and the Fcγ receptors. Sample binding interferograms and fits to equilibrium binding data for the IgG1 Fc G0 and G0F forms are shown in Figs. 5 and 6. The equilibrium dissociation constants measured for the IgG1 Fc G2F / CD16A interaction were similar to those measured previously by SPR with immobilized CD16A, by isothermal titration calorimetry, and by a bead binding assay.12 CD16 and CD32 proteins showed the clear establishment of binding equilibria at all concentrations analyzed and high quality fits of equilibrium binding isotherms (Table 1). The association of CD64 was much slower at the low concentrations tested, and many conditions failed to reach equilibrium within the timeframe of the experiment. As a result, we were unable to determine equilibrium binding constants for CD64 using equilibrium data, and instead analyzed the rates of association and dissociation to estimate binding affinity (Fig. 7). The CD64 kon and koff are compared to rates measured using CD16s and CD32s in Table 2.
Figure 5.
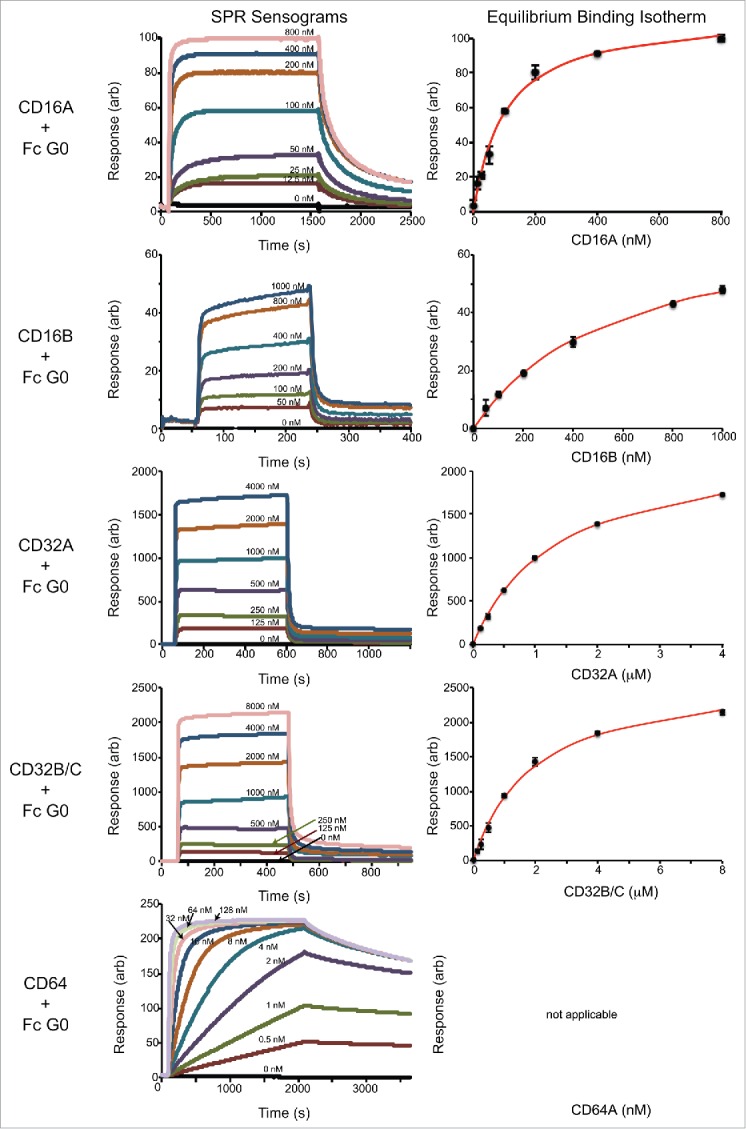
Representative SPR analysis of afucosylated IgG1 Fc (G0 form) binding to the Fc γ receptors. The left column shows the sensograms and the right column shows the analysis of maximum response values at binding equilibrium fitted with a binding isotherm.
Figure 6.
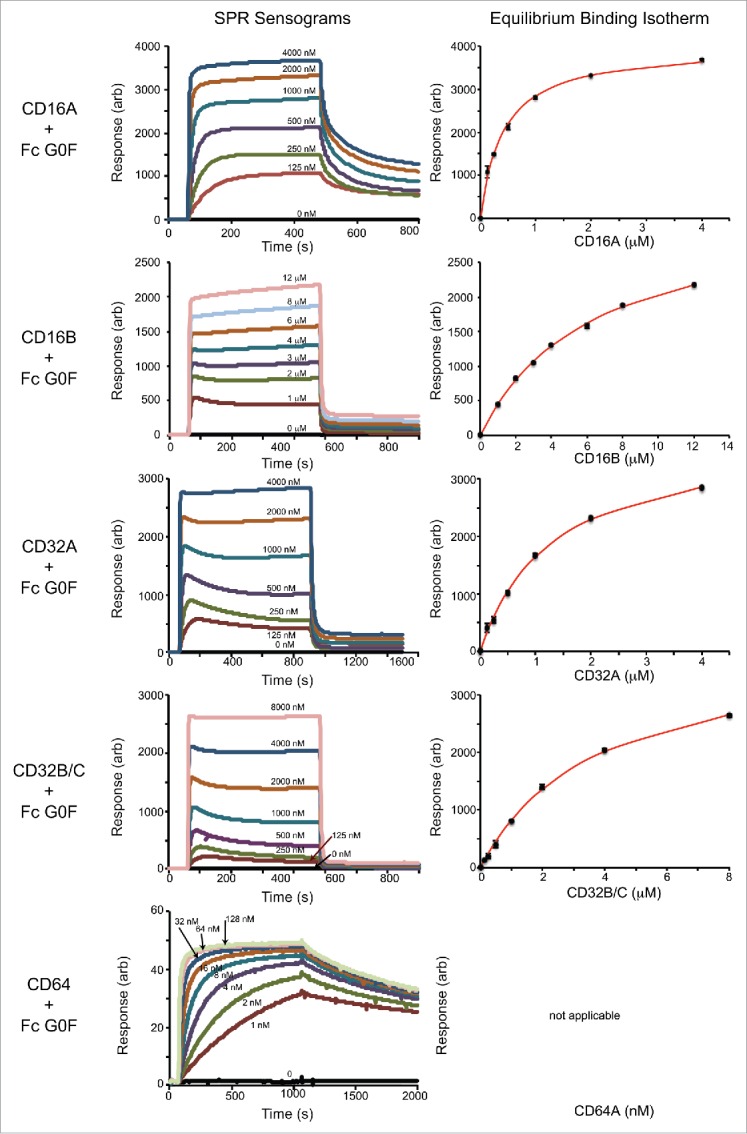
Representative SPR analysis of fucosylated IgG1 Fc (G0F form) binding to the Fc γ receptors. The left column shows the sensograms and the right column shows the analysis of maximum response values at binding equilibrium fitted with a binding isotherm.
Table 1.
Equilibrium dissociation constants for the Fc gamma receptors.
| afucosyl IgG1 Fc |
core fucosylated IgG1 Fc |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G0 |
G2 |
A2G2 |
G0F |
G2F |
A2G2F |
|||||||
| Receptor | KD (nM) | +/− err | KD (nM) | +/− err | KD (nM) | +/− err | KD (nM) | +/− err | KD (nM) | +/− err | KD (nM) | +/− err |
| CD16A+ | 101 | 12 | 64 | 8 | 51 | 8 | 409 | 32 | 208 | 14 | 220 | 3 |
| CD16B+ | 757 | 21 | 402 | 21 | 329 | 26 | 6250 | 300 | 3150 | 150 | 3090 | 130 |
| CD32A+ | 1370 | 70 | 785 | 26 | 765 | 31 | 1320 | 110 | 803 | 93 | 825 | 66 |
| CD32B+ | 1980 | 200 | 1540 | 160 | 1350 | 131 | 3740 | 260 | 2660 | 150 | 2470 | 1470 |
| CD64* | 1.41 | 0.09 | 1.69 | 0.32 | 1.1 | 0.20 | 0.48 | 0.16 | n.d. | n.d. | 0.63 | 0.71 |
- determined from fitting intensity data at equilibrium
- determined from fitting a kinetic model to the sensorgrams
n.d.- not determined
Figure 7.
Kinetic analysis of CD64 binding to immobilized IgG1 Fc (G0 and G0F forms). The left column shows a plot comparing observed association rates at each receptor concentration and the right column the observed dissociation rates at each receptor concentration measured from the SPR sensograms shown in Figs. 5 and 6. The kon was estimated by measuring the slope of a line that best fits the data in the right column and is reported in Table 2. The koff was determined by averaging the observed dissociation rates. Standard deviation of the mean is shown in the plots of koff rates as dashed lines.
Table 2.
Rate constants for Fc gamma receptor binding.
| afucosyl IgG1 Fc | core fucosylated IgG1 Fc | |||||||
|---|---|---|---|---|---|---|---|---|
| G0 |
G0F |
|||||||
| Receptor | kon (mM−1 s−1) | +/− err | koff (s−1) | +/− err | kon (mM−1 s−1) | +/− err | koff (s−1) | +/− err |
| CD16A | 420 | 11% | 0.00433 | 6% | 67.8 | 6% | 0.0138 | 6% |
| CD16B | n.d. | 0.27000 | 33% | n.d. | 0.2200 | 3% | ||
| CD32A | 110 | 17% | 0.11800 | 14% | 163 | 9% | 0.0780 | 22% |
| CD32B | n.d. | 0.16000 | 9% | n.d. | 0.2400 | 4% | ||
| CD64 | 360 | 1% | 0.00051 | 2% | 2200 | 3% | 0.0011 | 13% |
n.d.- not determined because the observed association rates were not linear with respect to IgG concentration
Receptor identity, not surprisingly, proved to be the largest contributor to observed binding affinity variability when using the predominant human IgG1 Fc glycoforms (G0F, G1F (not studied here) and G2F) with CD64 > CD16A > CD32A > CD32B/C > CD16B. These results are similar to those of previous studies that used full-length IgG1 and no explicit control of Fc glycosylation with CD64 (KD=15 nM) > CD32A (192 nM) > CD16A (500 nM) > CD16B (5 μM) > CD32B (8 μM).36,37 In addition to polypeptide sequence, we found IgG1 Fc N-glycan composition changes dramatically altered the binding of some, but not all, Fc γ receptors.
Fucosylation decreases CD16A,B and CD32B/C affinity but not CD32A
Core fucosylation of the IgG1 Fc N-glycan was shown to decrease the IgG1 Fc:CD16A affinity in vitro by 2.6–10 fold for IgG monomers16,17,38 and 19–50 fold for IgG dimers.14 We determined that Fc core fucosylation decreased the IgG1 Fc:CD16A affinity by 3.9 ± 0.6 fold after comparing the effect of fucose addition to each glycoform (e.g., G0 vs. G0F). A similar result was expected for CD16B; however, binding was reduced by 8.5 ± 0.8 fold. The effects on each Fc glycoform on CD16A and CD16B binding are shown in Fig. 8. This may be explained by the fact that CD16A and 16B have a conserved N-glycan (on Asn162) that was shown to stabilize the Fc:FcγR interaction when fucose is absent,33,34 although it is unclear why the effect of fucose is considerably greater for CD16B than 16A given the high degree of identity at the polypeptide level (Fig. 1B).
Figure 8.
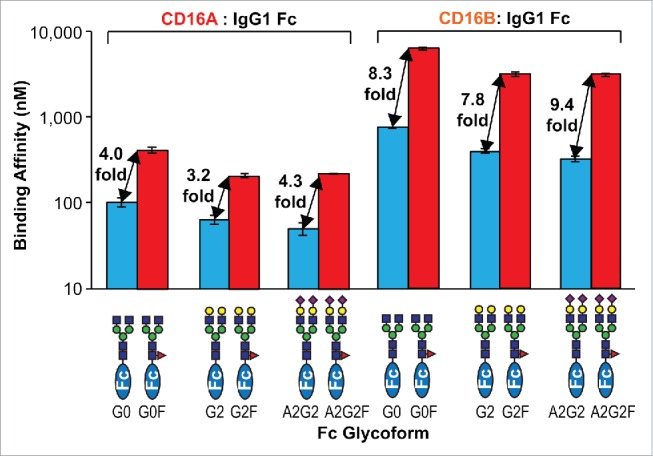
Relative effect of Fc core fucosylation on CD16 affinity. Errors of fit for the dissociation constants are shown.
Core fucosylation had no measureable effect on CD32A binding (1.02 ± 0.06-fold increase; Fig. 9), which is expected because CD32A does not contain a predicted N-glycan at the site homologous to Asn162 of CD16A and CD16B. The soluble CD32B/C construct also does not contain a homologous CD16A/B Asn162 N-glycan site; however, fucosylation reduced CD32B/C affinity by 1.8 ± 0.1 fold when removing the effects of galactosylation and sialylation. Fucosylation increased both kon and koff for CD64 binding, but had little measureable effect on affinity. The binding kinetics for CD16A, B CD32A and B/C were not affected in a similar manner and no clear connections between fucosylation and Fc γ receptor binding kinetics were found (data not shown), which agrees with previous reports.14,39 A comparison of the relative Fc γ receptor binding affinity to afucosylated Fc forms was CD64 > CD16A > CD16B > CD32A > CD32B/C.
Figure 9.
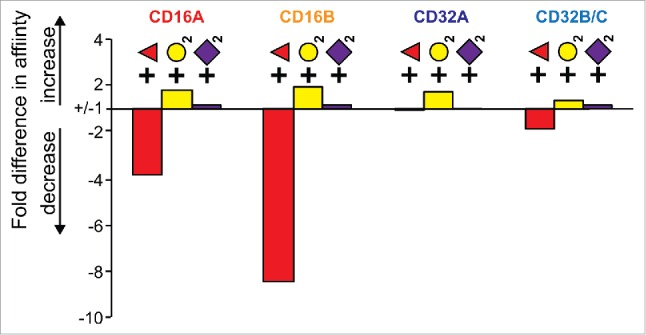
A comparison of the averaged relative effects of monosaccharide additions to the IgG1 Fc N-glycan on low affinity FcγR binding.
Fc stabilization through galactose modification
Yamaguchi et al. previously demonstrated that galactosylation of a fucosylated Fc increased CD16A affinity by 1.7 fold,13 close to our finding of 2.0 fold for G0F vs. G2F (Table 1). We found similar effects for CD16s and CD32s that increased affinity by an average of 1.7 ± 0.3 fold. There was no clear difference between the stabilization of fucosylated Fc versus afucosylated (data not shown). These data indicate galactosylation stabilizes the Fc in a general receptor binding conformation. This result is notable, considering the >15 Å distance between the galactose residues and the Fc γ receptor binding site on Fc, but can be explained by the allosteric effect of the N-glycan on receptor binding.12
N-glycan sialylation shows minimal effect
In previously published studies, IgG1 Fc sialylation was reported to have either a profound effect on CD16A binding40,41 or no measurable effect.19 In our experiments, N-acetylneuraminic acid addition (sialylation) to fucosylated Fc showed no measurable effect on binding to any CD16 or CD32 receptor (1.00 ± 0.06-fold increase). Sialylation of afucosylated Fc did show a slight increase in affinity (1.16 ± 0.11 fold). The lack of a sialylation effect is expected considering that the Fc receptors do not contact sugar residues on the non-reducing termini of the N-glycan32,42 and N-acetylneuraminic acid residues on the IgG1 Fc N-glycan do not interact with the Fc polypeptide directly.30,43
Discussion
The “low affinity” FcγRs responded differently to changes in IgG Fc N-glycan composition, with CD16B showing the greatest range followed by CD16A >> CD32B/C > CD32A (Fig. 9). Fucose addition accounted for the majority of the variation by reducing CD16A and B binding, but had a much smaller effect on CD32B/C and no measurable effect on CD32A. It is interesting to note that galactose addition consistently increased affinity for all low affinity receptors, indicating a mechanism that likely stabilizes the Fc itself in a receptor-independent manner. Furthermore, the effect of fucose and galactose addition was additive. Sialylation affected receptor binding to the least extent, with a statistically significant increase in affinity only noted for sialyation of afucosylated Fc binding to CD16B. There is growing evidence that the IgG1 Fc N-glycan composition changes in response to disease.44-48 Our data indicate variations in Fc N-glycan composition profiles alter relative FcγR binding affinities. Considering the tissue-specific expression profile for the FcγRs,3 changes to the IgG1 Fc N-glycan composition therefore have the potential to direct which leukocyte populations are affected and, as a result, direct the body's response to infection.
It was surprising that variation in the IgG Fc fucosylation caused a larger change to CD16B binding than observed for CD16A, considering the high degree of CD16A/B homology (Fig. 1) and the preservation in CD16B of an N-glycosylation site known to stabilize the afucosylated Fc:CD16A interaction.17,18,33,34 Although CD16A is often considered the primary target for the Fc of therapeutic mAbs,10,49,50 a growing body of evidence reveals a possible role for CD16B and neutrophil activation as an important mAb mechanism of action. Glycoengineered obinutuzumab, a CD20-targeting mAb that contains reduced fucose levels (plus a bisecting N-acetylglucosamine residue), has a 7-fold higher affinity to CD16B than the rituximab, which also targets CD20.51 Tighter CD16B binding correlated with neutrophil activation, and might explain the better suppression of cancer progression observed for chronic lymphocytic leukemia patients.49,52 These data implicate CD16B-targeting mAbs developed from afucosylated IgG1 with galactose-terminated N-glycans as a new direction for future mAbs designed to recruit neutrophils.
Multiple groups have speculated on the structural role of Fc N-glycan composition. Krapp et al. (2003) highlighted evidence for changes in the orientation of the Fc Cγ2 domains upon extending the N-glycan to a galactose-terminated form (G2F).53 These structures were still constrained by contacts within the crystal lattice, but offered a tangible hypothesis suggesting additions to the N-glycan non-reducing termini pushed the Cγ2 domains apart and provided an optimal Cγ2-Cγ2 distance for FcγR binding. This model is not supported by studies by our lab on molecules in solution at physiological pH that indicates the Fc N-glycan does not contribute much to the quaternary structure of the Fc, but rather stabilizes primarily the local secondary structure of a single polypeptide loop formed by residues including and adjacent to the site of N-glycan attachment (Asn297).11,12 Though the data we present here do not directly address the structural features of Fc, based on our previous reports we believe that galactosylation of the Fc N-glycan contributes to stabilization of the Asn297-containing C'E loop. The role of fucosylation has been addressed by multiple groups and is thought to specifically disrupt the Fc:CD16A interface.33,34
The data presented herein define the role of Fc N-glycan composition on FcγR binding. Our Fc constructs do not provide an opportunity to measure cell activation, although our results are quantitatively consistent with a recent report of Fc N-glycan composition on CD16A-expressing NK92 cell activation. Connell-Crowley and coworkers altered the N-glycan composition of 2 mAbs with glycosidase digestions, chromatography and antibody production techniques,54 and determined that a 1% decrease in fucosylation resulted in a 24% increase in antibody-dependent cell-mediated cytotoxicity (ADCC). Reducing galactose levels showed a decrease in ADCC, and neuraminidase treatment did not affect ADCC. These results perfectly mirror our results where fucosylation most dramatically affected CD16A binding, a small but significant effect was noted upon galactosylation, and no effect was observed with N-acetylneuraminic acid. We interpret this agreement between our results and those of Connell-Crowley and coworkers to indicate the strength of our experimental approach and the applicability of in vitro studies of the Fc:FcγR interaction to guide the next generation of IgG1-based antibodies through Fc-N-glycan optimization.
Materials and methods
Materials - All materials were purchased from Sigma-Aldrich unless otherwise noted.
Protein expression and purification - Human IgG1 Fc (residues 216–447) and CD16A (residues 19–193, V158 allotype) were expressed and purified as described. 12 The extracellular domains of receptors CD16B (residues 19–193), CD32A (residues 43–216, LR (H143) allotype) and CD32B (residues 43–216) with N-terminal His8 and GFP tags and a tobacco etch virus protease (TEV) digestion site were cloned into the pGen2 vector using the EcoRI and HindIII restriction sites.30, 55 As similar strategy to express CD64 failed to recover significant amounts of protein; as a result, CD64 (residues 16–292) was cloned with C-terminal TEV site and His8 tag between the NotI and HindIII sites of the pGen2 vector. Receptors were expressed via a transient transfection of HEK293F cells (Life Technologies) and purified using a Ni-NTA agarose (QIAGEN) column as described previously.12,55 Afucosylated Fc forms (G0, G2, A2G2) were expressed in the presence of 250 μM 2-deoxy-2-fluoro-L-fucose26 (Santa Cruz Biotech). IgG1 Fc glycovariants were analyzed with a 3.2/100 S200 column using an ÄKTA Pure FPLC (GE Healthcare). Fc samples (40 μL of 0.2 mg/mL) were injected and eluted at a flow rate of 0.075 ml/min with continuous monitoring at 280 nm. The column was equilibrated and eluted with a buffer containing 20 mM 3-morpholinopropane-1-sulfonic acid (MOPS), 100 mM sodium chloride, pH 7.2. The volume at which Fc eluted was compared to the elution volume of blue dextran, bovine serum albumin (BSA), carbonic anhydrase (CA) and cytochrome C.
Glycan remodeling - Fc glycovariants were prepared by previously described in vitro N-glycan remodeling methods.30,56 Approximately 5 mg of purified Fc from the HEK293F expression was treated with a β-1,4 galactosidase (New England Biolabs) to prepare the agalactosylated G0F forms. Samples were purified as previously described, and placed in a buffer containing 20 mM MOPS, 100 mM sodium chloride, 20 mM manganese chloride, pH 7.2. To prepare the G2F (digalactosylated) form, approximately 4 mg of Fc sample was treated with a β-1,4 galactosyltransferase (GalT) plus 10 mM UDP-galactose and incubated at 37°C for 24 hr. Equal amounts of GalT and UDP-galactose were added after 24 hr. Samples were purified and placed in a buffer containing 20 mM MOPS, 100 mM sodium chloride, and 20 mM manganese chloride, pH 7.2. The disialylated form (A2G2F), was prepared as previously described.30 Afucosylated forms (G0, G2 and A2G2) were prepared as described above except material was purified from HEK293F cells cultured in the presence of 250 µM 2-deoxy-2-fluoro-l-fucose. All samples were then placed in a buffer containing 20 mM MOPS, 100 mM sodium chloride, pH 7.2.
Analysis of remodeled glycans by Mass Spectrometry - Released N-glycans of all Fc glycovariants were analyzed as described previously56,57 using matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) on a Voyager-DE PRO (Applied Biosystems).
Analysis of Fc forms by Electrospray Ionization-Mass Spectrometry - Fc (0.1 mg/mL) in 100 μL of 20 mM MOPS, 100 mM sodium chloride, 50 mM dithiothreitol, pH 7.2 was denatured by heating at 95°C for 5 min. Samples were further analyzed by a liquid chromatography system coupled with ESI-MS (Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer; Thermo Scientific). Samples (10 μL) were injected at a flow rate of 0.1 mL/min onto a C4 reversed-phase column (5 μM, 30 mm × 1 mm, Restek) previously equilibrated with 95% solvent A (0.1% formic acid in water) and 5% solvent B (0.1% formic acid in acetonitrile) at 20°C. Samples were eluted using a linear gradient of 5–100% solvent B from 3 to 10 min followed by a 5 min elution with 100% solvent B before washing with 95% solvent A and 5% solvent B. The ESI-MS instrument was set to positive polarity with 60 eV in-source collision-induced dissociation (CID) with a mass scan range of 700–4000 m/z. Data were displayed and analyzed using Thermo Xcalibur Qual Browser (version 3.0.63). Average protein masses were deconvoluted using ProMass Deconvolution software (Thermo Scientific).
IgG1 Fc Immobilization - A Biacore T100 instrument was used to measure the binding affinities between Fc and its receptors. All SPR measurements were performed at 25°C by standard amine coupling procedures58 with Fc immobilized on a CM5 sensor chip (GE Life Sciences). The carboxymethyl dextran surface was activated by 1:1 mixture of 0.4 M 1-ethyl-3(3-dimethylaminopropyl)carbodiimide hydrochloride and 0.1 M N-hydroxy succinimide for 7 min at a flow rate of 5 µl/min. Fc between 1–50 µg/mL in 1 mL of 10 mM sodium acetate, pH 5.0 was coupled by injecting at a flow rate of 5 µl/min. Immobilization was completed by deactivating residual sites using 1 M ethanolamine, pH 8.5 for 7 min. Final immobilization response units were between 400–700 for 1 µg/mL and 5000–6000 for 50 µg/mL. Flow line 1 on all sensor chips was used as a blank with no immobilized Fc.
Binding analysis using surface plasmon resonance - All binding analyses were performed with binding buffer containing 20 mM MOPS, 100 mM sodium chloride, 1 µM BSA and 0.05% P20 surfactant (GE Life Sciences), pH 7.4. The CM5 chip surface was regenerated by a 100 mM glycine, pH 3.0 wash for 30 s to remove bound receptors. A minimum of one replication for each condition was collected on different days. Representative results are shown.
SPR data analysis - All the Biacore sensograms were processed using Biacore T100 Evaluation Software (Version 1.0). Sensograms were zeroed in the response unit axis and response of the blank injection was subtracted from the analyte injected flow cell responses to remove systematic artifacts. The equilibrium response units (RU) for different analyte concentrations [A] were fitted to obtain the dissociation constant (KD) by using Equation (1) with the maximum response unit (Rmax) obtained among the measured concentration range.
| (1) |
The observed rates of association and dissociation were obtained by fitting an exponential equation to the association or dissociation profiles observed in the SPR sensorgrams. kon was determined by measuring the slope of a line fitted to the observed association rates. koff was determined by averaging the observed dissociation rates. Presented data are representative of at least 2 independent experiments for each condition.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Joel Nott of the Iowa State University Protein Facility and Dan Falconer for assistance with the ESI-MS data collection and analysis as well as Aaron Marcella for assistance with the SEC.
Funding
This work was financially supported by the grant R01-GM115489 (NIGMS) from the National Institutes of Health (NIH), and by funds from the Roy J. Carver Department of Biochemistry, Biophysics & Molecular Biology at Iowa State University. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Allen LV, Popovich NG, Ansel HC. Ansel's pharmaceutical dosage forms and drug delivery systems. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins, 2011 [Google Scholar]
- 2.Stevenson GT. Three major uncertainties in the antibody therapy of cancer. Haematologica 2014; 99:1538-46; PMID:25271313; http://dx.doi.org/ 10.3324/haematol.2013.084640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nimmerjahn F, Gordan S, Lux A. FcgammaR dependent mechanisms of cytotoxic, agonistic, and neutralizing antibody activities. Trends Immunol 2015; 36:325-36; PMID:25981969; http://dx.doi.org/ 10.1016/j.it.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 4.Janeway C, Murphy KP, Travers P, Walport M. Janeway's immuno biology. New York: Garland Science, 2008 [Google Scholar]
- 5.Lux A, Yu X, Scanlan CN, Nimmerjahn F. Impact of immune complex size and glycosylation on IgG binding to human FcgammaRs. Journal Immunol 2013; 190:4315-23; PMID:23509345; http://dx.doi.org/22271896 10.4049/jimmunol.1200501 [DOI] [PubMed] [Google Scholar]
- 6.Persky DO, Dornan D, Goldman BH, Braziel RM, Fisher RI, Leblanc M, Maloney DG, Press OW, Miller TP, Rimsza LM. Fc gamma receptor 3a genotype predicts overall survival in follicular lymphoma patients treated on SWOG trials with combined monoclonal antibody plus chemotherapy but not chemotherapy alone. Haematologica 2012; 97:937-42; PMID:22271896; http://dx.doi.org/ 10.3324/haematol.2011.050419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Gordon M, Schultheis AM, Yang DY, Nagashima F, Azuma M, Chang HM, Borucka E, Lurje G, Sherrod AE, et al.. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol: Off J Am Soc Clin Oncol 2007; 25:3712-8; PMID:17704420; http://dx.doi.org/ 10.1200/JCO.2006.08.8021 [DOI] [PubMed] [Google Scholar]
- 8.Nose M, Wigzell H. Biological significance of carbohydrate chains on monoclonal antibodies. Proc Natl Acad Sci U S A 1983; 80:6632-6; PMID:6579549; http://dx.doi.org/ 10.1073/pnas.80.21.6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parekh RB, Dwek RA, Sutton BJ, Fernandes DL, Leung A, Stanworth D, Rademacher TW, Mizuochi T, Taniguchi T, Matsuta K, et al.. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature 1985; 316:452-7; PMID:3927174; http://dx.doi.org/ 10.1038/316452a0 [DOI] [PubMed] [Google Scholar]
- 10.Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A. A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol 2013; 6:1; PMID:23286345; http://dx.doi.org/ 10.1186/1756-8722-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subedi GP, Barb AW. The Structural Role of Antibody N-Glycosylation in Receptor Interactions. Structure 2015; 23:1573-83; PMID:26211613; http://dx.doi.org/ 10.1016/j.str.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subedi GP, Hanson QM, Barb AW. Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient FcgammaRIIIa binding. Structure 2014; 22:1478-88; PMID:25199692; http://dx.doi.org/ 10.1016/j.str.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi Y, Nishimura M, Nagano M, Yagi H, Sasakawa H, Uchida K, Shitara K, Kato K. Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochimica Et Biophysica Acta-General Subjects 2006; 1760:693-700; PMID:NOT_FOUND; http://dx.doi.org/ 10.1016/j.bbagen.2005.10.002 [DOI] [PubMed] [Google Scholar]
- 14.Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem 2002; 277:26733-40; PMID:11986321; http://dx.doi.org/ 10.1074/jbc.M202069200 [DOI] [PubMed] [Google Scholar]
- 15.Gabellier L, Cartron G. Obinutuzumab for relapsed or refractory indolent non-Hodgkin's lymphomas. Therapeutic Adv Hematol 2016; 7:85-93; PMID:27054024; http://dx.doi.org/ 10.1177/2040620715622613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanda Y, Yamada T, Mori K, Okazaki A, Inoue M, Kitajima-Miyama K, Kuni-Kamochi R, Nakano R, Yano K, Kakita S, et al.. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology 2007; 17:104-18; PMID:17012310; http://dx.doi.org/ 10.1093/glycob/cwl057 [DOI] [PubMed] [Google Scholar]
- 17.Shibata-Koyama M, Iida S, Okazaki A, Mori K, Kitajima-Miyama K, Saitou S, Kakita S, Kanda Y, Shitara K, Kato K, et al.. The N-linked oligosaccharide at Fc gamma RIIIa Asn-45: an inhibitory element for high Fc gamma RIIIa binding affinity to IgG glycoforms lacking core fucosylation. Glycobiology 2009; 19:126-34; PMID:18952826; http://dx.doi.org/ 10.1093/glycob/cwn110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrara C, Stuart F, Sondermann P, Brunker P, Umana P. The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms. J Biol Chem 2006; 281:5032-6; PMID:16330541; http://dx.doi.org/ 10.1074/jbc.M510171200 [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Baruah K, Harvey DJ, Vasiljevic S, Alonzi DS, Song BD, Higgins MK, Bowden TA, Scanlan CN, Crispin M. Engineering hydrophobic protein-carbohydrate interactions to fine-tune monoclonal antibodies. J Am Chem Soc 2013; 135:9723-32; PMID:23745692; http://dx.doi.org/ 10.1021/ja4014375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mimura Y, Sondermann P, Ghirlando R, Lund J, Young SP, Goodall M, Jefferis R. Role of oligosaccharide residues of IgG1-Fc in Fc gamma RIIb binding. J Biol Chem 2001; 276:45539-47; PMID:11567028; http://dx.doi.org/ 10.1074/jbc.M107478200 [DOI] [PubMed] [Google Scholar]
- 21.Hayes JM, Frostell A, Cosgrave EF, Struwe WB, Potter O, Davey GP, Karlsson R, Anneren C, Rudd PM. Fc gamma receptor glycosylation modulates the binding of IgG glycoforms: a requirement for stable antibody interactions. J Proteome Res 2014; 13:5471-85; PMID:25345863; http://dx.doi.org/ 10.1021/pr500414q [DOI] [PubMed] [Google Scholar]
- 22.Bakovic MP, Selman MH, Hoffmann M, Rudan I, Campbell H, Deelder AM, Lauc G, Wuhrer M. High-throughput IgG Fc N-glycosylation profiling by mass spectrometry of glycopeptides. J Proteome Res 2013; 12:821-31; PMID:23298168; http://dx.doi.org/ 10.1021/pr300887z [DOI] [PubMed] [Google Scholar]
- 23.Franklin EC. Structure and function of immunoglobulins. Acta endocrinologica Supplementum 1975; 194:77-95; PMID:47690 [DOI] [PubMed] [Google Scholar]
- 24.Nisonoff A, Wissler FC, Lipman LN, Woernley DL. Separation of univalent fragments from the bivalent rabbit antibody molecule by reduction of disulfide bonds. Arch Biochem Biophys 1960; 89:230-44; PMID:14427334; http://dx.doi.org/ 10.1016/0003-9861(60)90049-7 [DOI] [PubMed] [Google Scholar]
- 25.Porter RR. The hydrolysis of rabbit y-globulin and antibodies with crystalline papain. Biochem J 1959; 73:119-26; PMID:14434282; http://dx.doi.org/ 10.1042/bj0730119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okeley NM, Alley SC, Anderson ME, Boursalian TE, Burke PJ, Emmerton KM, Jeffrey SC, Klussman K, Law CL, Sussman D, et al.. Development of orally active inhibitors of protein and cellular fucosylation. Proc Natl Acad Sci U S A 2013; 110:5404-9; PMID:23493549; http://dx.doi.org/ 10.1073/pnas.1222263110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, Dell A, Haslam SM, Paulson JC. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol 2012; 8:661-8; PMID:22683610; http://dx.doi.org/ 10.1038/nchembio.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rich RL, Papalia GA, Flynn PJ, Furneisen J, Quinn J, Klein JS, Katsamba PS, Waddell MB, Scott M, Thompson J, et al.. A global benchmark study using affinity-based biosensors. Anal Biochem 2009; 386:194-216; PMID:19133223; http://dx.doi.org/ 10.1016/j.ab.2008.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barb AW, Prestegard JH. NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat Chem Biol 2011; 7:147-53; PMID:21258329; http://dx.doi.org/ 10.1038/nchembio.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barb AW, Meng L, Gao Z, Johnson RW, Moremen KW, Prestegard JH. NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry 2012; 51:4618-26; PMID:22574931; http://dx.doi.org/ 10.1021/bi300319q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry 1981; 20:2361-70; PMID:7236608; http://dx.doi.org/ 10.1021/bi00512a001 [DOI] [PubMed] [Google Scholar]
- 32.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature 2000; 406:267-73; PMID:10917521; http://dx.doi.org/ 10.1038/35018508 [DOI] [PubMed] [Google Scholar]
- 33.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, et al.. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci U S A 2011; 108:12669-74; PMID:21768335; http://dx.doi.org/ 10.1073/pnas.1108455108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima T, Yagi H, Takemoto E, Shibata-Koyama M, Isoda Y, Iida S, Masuda K, Satoh M, Kato K. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells: Devoted Mol Cell Mech 2011; 16:1071-80; PMID:NOT_FOUND; http://dx.doi.org/ 10.1111/j.1365-2443.2011.01552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank M, Walker RC, Lanzilotta WN, Prestegard JH, Barb AW. Immunoglobulin G1 Fc domain motions: implications for Fc engineering. J Mol Biol 2014; 426:1799-811; PMID:24522230; http://dx.doi.org/ 10.1016/j.jmb.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daëron M. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 2009; 113:3716-25; PMID:19018092; http://dx.doi.org/ 10.1182/blood-2008-09-179754 [DOI] [PubMed] [Google Scholar]
- 37.van der Poel CE, Spaapen RM, van de Winkel JG, Leusen JH. Functional characteristics of the high affinity IgG receptor, FcgammaRI. J Immunol 2011; 186:2699-704; PMID:NOT_FOUND; http://dx.doi.org/ 10.4049/jimmunol.1003526 [DOI] [PubMed] [Google Scholar]
- 38.Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, Shitara K. Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcgammaRIIIa. J Mol Biol 2004; 336:1239-49; PMID:15037082; http://dx.doi.org/ 10.1016/j.jmb.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 39.Derer S, Glorius P, Schlaeth M, Lohse S, Klausz K, Muchhal U, Desjarlais JR, Humpe A, Valerius T, Peipp M. Increasing FcgammaRIIa affinity of an FcgammaRIII-optimized anti-EGFR antibody restores neutrophil-mediated cytotoxicity. MAbs 2014; 6:409-21; PMID:24492248; http://dx.doi.org/ 10.4161/mabs.27457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anthony RM, Nimmerjahn F, Ashline DJ, Reinhold VN, Paulson JC, Ravetch JV. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science 2008; 320:373-6; PMID:18420934; http://dx.doi.org/ 10.1126/science.1154315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science 2006; 313:670-3; PMID:16888140; http://dx.doi.org/ 10.1126/science.1129594 [DOI] [PubMed] [Google Scholar]
- 42.Lu J, Chu J, Zou Z, Hamacher NB, Rixon MW, Sun PD. Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding. Proc Natl Acad Sci U S A 2015; 112:833-8; PMID:25561553; http://dx.doi.org/ 10.1073/pnas.1418812112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crispin M, Yu X, Bowden TA. Crystal structure of sialylated IgG Fc: implications for the mechanism of intravenous immunoglobulin therapy. Proc Natl Acad Sci U S A 2013; 110:E3544-6; PMID:23929778; http://dx.doi.org/ 10.1073/pnas.1310657110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauc G, Huffman JE, Pucic M, Zgaga L, Adamczyk B, Muzinic A, Novokmet M, Polašek O, Gornik O, Krištić J, et al.. Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet 2013; 9:e1003225; PMID:23382691; http://dx.doi.org/ 10.1371/journal.pgen.1003225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novokmet M, Lukic E, Vuckovic F, Ethuric Z, Keser T, Rajsl K, Remondini D, Castellani G, Gašparović H, Gornik O, et al.. Changes in IgG and total plasma protein glycomes in acute systemic inflammation. Sci Rep 2014; 4:4347; PMID:24614541; http://dx.doi.org/ 10.1038/srep04347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones MB, Nasirikenari M, Lugade AA, Thanavala Y, Lau JT. Anti-inflammatory IgG production requires functional P1 promoter in beta-galactoside alpha2,6-sialyltransferase 1 (ST6Gal-1) gene. J Biol Chem 2012; 287:15365-70; PMID:22427662; http://dx.doi.org/ 10.1074/jbc.M112.345710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trbojevic Akmacic I, Ventham NT, Theodoratou E, Vuckovic F, Kennedy NA, Kristic J, Nimmo ER, Kalla R, Drummond H, Štambuk J, et al.. Inflammatory bowel disease associates with proinflammatory potential of the immunoglobulin G glycome. Inflamm Bowel Dis 2015; 21:1237-47; PMID:25895110; http://dx.doi.org/ 10.1097/MIB.0000000000000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahan AE, Jennewein MF, Suscovich T, Dionne K, Tedesco J, Chung AW, Streeck H, Pau M, Schuitemaker H, Francis D, et al.. Antigen-specific antibody glycosylation is regulated via vaccination. PLoS Pathog 2016; 12:e1005456; PMID:26982805; http://dx.doi.org/ 10.1371/journal.ppat.1005456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dall'Ozzo S, Tartas S, Paintaud G, Cartron G, Colombat P, Bardos P, Watier H, Thibault G. Rituximab-dependent cytotoxicity by natural killer cells: influence of FCGR3A polymorphism on the concentration-effect relationship. Cancer Res 2004; 64:4664-9; PMID:15231679; http://dx.doi.org/ 10.1158/0008-5472.CAN-03-2862 [DOI] [PubMed] [Google Scholar]
- 50.Weiner GJ. Rituximab: mechanism of action. Semin Hematol 2010; 47:115-23; PMID:20350658; http://dx.doi.org/ 10.1053/j.seminhematol.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Golay J, Da Roit F, Bologna L, Ferrara C, Leusen JH, Rambaldi A, Klein C, Introna M. Glycoengineered CD20 antibody obinutuzumab activates neutrophils and mediates phagocytosis through CD16B more efficiently than rituximab. Blood 2013; 122:3482-91; PMID:24106207; http://dx.doi.org/ 10.1182/blood-2013-05-504043 [DOI] [PubMed] [Google Scholar]
- 52.Evans SS, Clemmons AB. Obinutuzumab: a novel anti-CD20 monoclonal antibody for chronic lymphocytic leukemia. J Adv Pract Oncol 2015; 6:370-4; PMID:26705497; http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4677809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol 2003; 325:979-89; PMID:12527303; http://dx.doi.org/ 10.1016/S0022-2836(02)01250-0 [DOI] [PubMed] [Google Scholar]
- 54.Brady LJ, Velayudhan J, Visone DB, Daugherty KC, Bartron JL, Coon M, Cornwall C, Hinckley PJ, Connell-Crowley L. The criticality of high-resolution N-linked carbohydrate assays and detailed characterization of antibody effector function in the context of biosimilar development. MAbs 2015; 7:562-70; PMID:25898160; http://dx.doi.org/ 10.1080/19420862.2015.1016692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subedi GP, Johnson RW, Moniz HA, Moremen KW, Barb A. High yield expression of recombinant human proteins with the transient transfection of HEK293 cells in suspension. J Vis Exp: JoVE 2015:e53568; PMID:26779721; http://dx.doi.org/ 10.3791/53568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barb AW, Brady EK, Prestegard JH. Branch-specific sialylation of IgG-Fc glycans by ST6Gal-I. Biochemistry 2009; 48:9705-7; PMID:19772356; http://dx.doi.org/ 10.1021/bi901430h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anumula KR, Taylor PB. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal Biochem 1992; 203:101-8; PMID:1524204; http://dx.doi.org/ 10.1016/0003-2697(92)90048-C [DOI] [PubMed] [Google Scholar]
- 58.Fischer MJ. Amine coupling through EDC/NHS: a practical approach. Methods Mol Biol 2010; 627:55-73; PMID:20217613; http://dx.doi.org/ 10.1007/978-1-60761-670-2_3 [DOI] [PubMed] [Google Scholar]
- 59.Varki A. Essentials of glycobiology. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press, 2009; PMID:2701005 [PubMed] [Google Scholar]