ABSTRACT
We identified active isoforms of the chimeric anti-GD2 antibody, ch14.18, a recombinant antibody produced in Chinese hamster ovary cells, which is already used in clinical trials.1,2,3 We separated the antibody by high resolution ion-exchange chromatography with linear pH gradient elution into acidic, main and basic charge variants on a preparative scale yielding enough material for an in-depth study of the sources and the effects of microheterogeneity. The binding affinity of the charge variants toward the antigen and various cell surface receptors was studied by Biacore. Effector functions were evaluated using cellular assays for antibody-dependent cell-mediated cytotoxicity and complement-dependent cytotoxicity. Basic charge variants showed increased binding to cell surface receptor FcγRIIIa, which plays a major role in regulating effector functions. Furthermore, increased binding of the basic fractions to the neonatal receptor was observed. As this receptor mediates the prolonged half-life of IgG in human serum, this data may well hint at an increased serum half-life of these basic variants compared to their more acidic counterparts. Different glycoform patterns, C-terminal lysine clipping and N-terminal pyroglutamate formation were identified as the main structural sources for the observed isoform pattern. Potential differences in structural stability between individual charge variant fractions by nano differential scanning calorimetry could not been detected. Our in-vitro data suggests that the connection between microheterogeneity and the biological activity of recombinant antibody therapeutics deserves more attention than commonly accepted.
KEYWORDS: Biosimilar, glycoforms, immunoglobulin, isoforms, linear pH gradient, Microheterogeneity, monoclonal antibody
Introduction
The exact reasons for why some antibodies show higher potency than others remains unclear. Lacking comprehensive information about the effects of product characteristics on a molecular level, there is an increased need to monitor the production process of antibody therapeutic products to ensure constant product quality.
An increased understanding of the structural and molecular basis of the efficacy of antibody therapeutics is of interest to the scientific, medical and bioprocess engineering communities, and will result in new approaches to develop more potent therapeutic products. Relevant information can be gained through the analysis of antibody variants, which can show quite profound differences in potency, as well as potential side effects, resulting from small structural modifications.4,5
Considering the large size of an IgG molecule (∼150 kDa) and the complexity of its structure, which consists of 4 subunits that are connected via disulfide bonds, it is not surprising that monoclonal antibodies contain multiple sites where protein modifications can occur. Such modifications can originate either during the production process or due to chemical reactions during product storage.6 Formulations of therapeutic monoclonal antibodies thus do not result in only one defined species of molecules, but rather in a large variety of so-called isoforms or protein variants that may differ in structure, biophysical characteristics, e.g., isoelectric point (pI), long-term stability, biological activity. The pattern of microheterogeneity created by these variants is considered to be of decisive importance for consistent product quality of monoclonal antibodies, and therefore must be monitored closely for changes during development and production.5,7
One of the most common, and possibly the best-studied, type of protein modification in antibody molecules is glycosylation. The highly conserved residue Asn 297 present in the CH2 domain of each heavy chain provides 2 potential N-glycosylation sites in each antibody molecule, resulting in a wide variety of glycosylation patterns. The glycan moieties predominately found at this site in human IgG are of the complex bi-antennary type, terminating in structures ranging from N-acetylglucosamine (GlcNAc) to galactose (Gal) and N-acetylneuraminic acid (Neu5Ac) molecules.8,9 Additionally, other O- and N-glycosylation sites can be present on the antibody molecule, further increasing the diversity of possible IgG glycosylation patterns.10,11 The presence of charged glycans containing sialic acid may be one of the most prominent reasons for the occurrence of different charge variants in IgG.
Other protein modifications that result in altered charge characteristics are C-terminal lysine processing, leading to the loss of up to one positive charge unit, deamidation of asparagine and glutamine, which introduces an additional negative charge unit, isomerization of aspartate to isoaspartate, which is slightly more acidic, and the cyclization of glutamic acid to pyroglutamate, which results in the loss of the positively charged primary amine.6
Furthermore, there are possible protein modifications that should formally not result in a change of the net charge of the molecule, including methionine oxidation12 and varying disulfide bond structures,13 even though they may affect other characteristics. The racemization of L-aspartic acid and L-iso-aspartic acid14 is another possible modification that should not introduce a change of the pI of the protein, but can result in conformational changes.
Recently, a method based on cation exchange chromatography combined with pH gradient elution, which allows the large-scale separation of IgG variants showing different surface charge characteristics, was reported.15,16 This method has now been applied to different production batches of ch14.18, a therapeutic mAb against neuroblastoma, which recently finished Phase 3 clinical studies, in order to obtain a closer insight into the origins and the biological effects of different charge variants present in a GMP-produced monoclonal antibody used in clinical settings.
We were able to gain new information about charge heterogeneity because we were able to separate closely related charge variants from each other in mg amounts, which allowed us to further characterize them in biological assays. For that purpose, the separated antibody variants were analyzed by isoelectric focusing, surface plasmon resonance (SPR)-based binding assays to GD2, FcγRIIIa and FcRn, and cellular assays to determine antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC).
Isoelectric focusing is the standard method for visualizing antibody charge variants and was therefore used to evaluate the results of the chromatographic experiments. SPR is a well-established platform for the label-free, real-time monitoring of interactions between biomolecules in vitro,17 and was therefore selected as an appropriate method for determining the strength of the interaction between the separated antibody charge variants to GD2 and the Fc receptors. The disialoganglioside GD2 is the molecular target of the mAb used for this study. It is a surface molecule, commonly expressed on many different kinds of tumors including neuroblastoma, melanoma and various sarcomas.18 The specificity of the antibody is a major feature of the molecule determining effectiveness and is therefore of great importance in evaluating differences between charge variants.
FcγRIIIa and FcRn, which are both cellular surface receptors binding the Fc domain of IgG, are involved in regulation of 2 major biological modes of action specific to antibodies.19 FcγRIIIa (CD16) is present on natural killer cells and macrophages.20 Binding of IgG to the receptor molecule is responsible for mediating ADCC21 and phagocytosis of tumor cells by macrophages.22 FcRn is expressed on monocytes and endothelial cells. Binding of FcRn to IgG is pH dependent, and it regulates the prolonged serum half-life of IgG by mediating the recycling of the antibody-receptor-complex from the early endosomes, which prevents degradation.23
To complement the data obtained in those in-vitro experiments, the separated antibody variants were also tested for their performance in cellular assays for ADCC and CDC.
When an antibody binds to the antigen GD2 on a target cell, the Fc domain interacts with its corresponding ligands (FcγRIIIa) on leukocytes, as well as with soluble blood proteins (C1q, C4 and C3) of the antibody-dependent complement activation pathway. The interaction with leukocytes triggers ADCC, a process in which target cells are lysed by leukocyte perforins that form pores in the membrane through which the cytoplasm leaks out of the cell. Granzymes induce programmed cell death in the target cell. The interaction with C1q induces CDC by forming a membrane attack complex, which disrupts the phospholipid bilayer leading to target cell lysis.24 For both types of effector functions, the extent of the target cell killing can be determined by labeling with a radioactive tracer, which is released from the cells when they disintegrate. The tracer, in our case 51Cr, can be detected in the cell supernatant after the immune reaction has taken place. Spontaneous release from labeled target cells, as well as maximum release by total cell lysis with a surfactant can be determined via control samples. Cell lysis by a test sample can then be calculated as a percentage value compared to the maximum lysis.25
Finally, we obtained insight into the structural basis of the variants that we separated and formed a structure-function relationship by analyzing the variants in a series of MS-based experiments, targeting the glycosylation pattern by glycan analysis and modifications on individual amino acids by peptide mapping. In addition, the samples were analyzed by nano differential scanning calorimetry (Nano DSC) to detect any possible differences in thermal stability of the separated variant fractions. For IgG, a Nano DSC experiment normally results in 2 peaks and the corresponding melting temperature (Tm) points, the first of which is associated with the unfolding of the CH2 domain and the antigen-binding fragment (Fab). The second one indicates the unfolding of the CH3 domain.26
To our knowledge, this is the first study to investigate the influence of the microheterogeneity pattern of a monoclonal antibody on various critical quality attributes, including target affinity, efficacy and stability, with a large scope. With this thorough data set, we were able to draw conclusions about the relationship of efficacy and overall performance of our antibody to its observed microheterogeneity pattern. Our data shows that antibody modifications can have a considerable influence on the in-vitro behavior of a therapeutic product. If the observed relationship can be confirmed for other mAb products, the results may aid future antibody and process design.
Results
Chromatographic separation, confirmation of identity and quality control of starting material
The method previously introduced and validated by Lingg et al.15,16 for the analytical separation of antibody charge variants using highly linear pH gradients was scaled up to preparative scale and applied to 3 different production batches of the ch14.18 a chimeric anti-GD2 monoclonal antibody intended for cancer immunotherapy27 (“Newton,” “Darwin,” “Curie”), which were manufactured at different stages during product development. Batch “Newton” contains the oldest material, produced in 2004, batch “Darwin” was the result of an engineering run in 2011 after introducing minor changes in the downstream process and batch “Curie” was produced in 2013 after the production process was transferred to a new contract manufacturer.
The chromatograms, as well as a comparison of the isoelectric focusing pattern of batches “Newton,” “Darwin” and “Curie,” (Figs. 1–4) show very similar isoform distributions. All three samples contain a main peak or gel band, flanked by a considerable number of acidic variants and a small amount of basic variants. This is a typical isoform distribution for IgG samples, given that many different kinds of protein modifications are possible, resulting in a shift of the pI value to either side of the main peak for a certain number of molecules.28 A clearly noticeable difference can be seen for batch “Newton,” which seems to contain less of the main fraction and larger amounts of both acidic and basic variants (Fig. 1). “Newton” is the oldest among the investigated batches and it was stored in a different formulation buffer, which may explain the increased number of variants. Environmental factors and storage conditions, such as temperature and the buffer system used, can have a profound effect on the microheterogeneity pattern of a protein solution,6 and different variants may have accumulated in the sample over the prolonged storage period. Another possible explanation is that the batches “Darwin” and “Curie” contain fewer variants due to changes in the downstream process.
Figure 1.
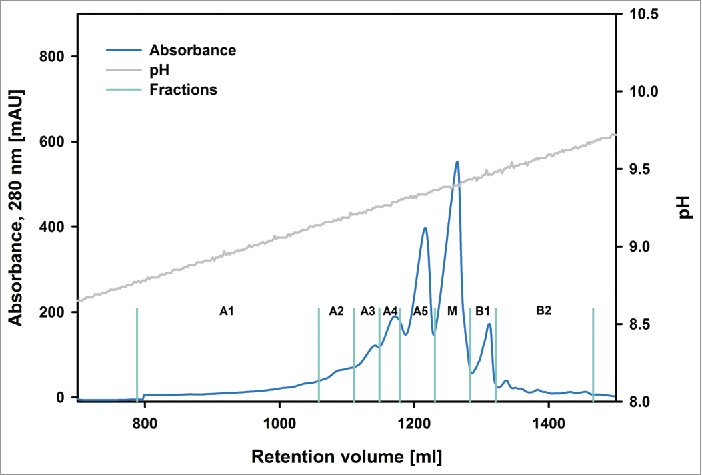
Preparative chromatogram of batch “Newton.” The blue trace indicates UV absorbance. A1-A5, M, B1-B2 denotes the collection of acidic, the main and basic variants, respectively. The fractionation boundaries are represented by turquoise lines. The gray trace shows the pH.
Figure 2.

Preparative chromatogram of batch “Darwin.” The yellow trace indicates UV absorbance. A1-A5, M, B1-B2 denotes the collection of acidic, the main and basic variants, respectively. The fractionation boundaries are represented by turquoise lines. The gray trace shows the pH.
Figure 3.

Preparative chromatogram of batch “Curie.” The red trace indicates UV absorbance. A1-A5, M, B1-B2 denotes the collection of acidic, the main and basic, respectively. The fractionation boundaries are represented by turquoise lines. The gray trace shows the pH.
Figure 4.

Isoelectric focusing of batches “Newton,” “Darwin” and “Curie” charge variants, positive control (PC) samples and starting material (SM). For batch “Curie” no positive control run was performed due to limited availability of the starting material. More acidic isoforms get enriched in the early eluting fractions, more basic ones are present in higher amounts in the later eluting fractions. No difference is detectable between the starting material and the positive control samples.
Despite the differences for batch “Newton” in comparison to the other batches, it was possible to fractionate the material eluting from the column in a similar fashion for all 3 batches. The chromatograms obtained for batches “Newton,” “Darwin” and “Curie” showed a qualitatively comparable shape, while exhibiting quantitative differences in the abundance of the variants (Figs. 1–3). Five acidic fractions (A1-A5), one main peak fraction (M) and 2 basic fractions (B1, B2) were identified and collected for each of the investigated antibodies. Isoelectric focusing (Fig. 4) showed that all those fractions still contained a mixture of different variants, corresponding to multiple bands visible on the gel. However, it was possible to enrich specific acidic and basic variants and obtain an almost pure main peak. Especially the fractions on both extremes of the pH spectrum contained a much larger amount of isoforms corresponding to bands that were hardly visible in the starting material. This high enrichment of specific variants provides the opportunity to collect enough material for extensive investigation using methods with high samples demand such as glycan analysis or peptide mapping.
To eliminate the possibility that these additional isoforms were created during the separation procedure, an additional preparative scale experiment was carried out in which the fractions were pooled together after fractionation, concentrated and analyzed for its microheterogeneity pattern via isoelectric focusing (IEF; positive control sample PC in Fig. 4). Compared to the starting material, no differences could be detected in the IEF profile of this control sample, which suggests that the ion-exchange chromatography (IEX) used is indeed appropriate for detection and separation of antibody charge variants without affecting the original isoform profile.
Analytical scale chromatography was performed to evaluate the quantitative differences in the microheterogeneity of the separated fraction. Fig. 5 shows an overlay of the analytical chromatograms of the “Newton” fractions compared to the starting material. Once again, a clear enrichment of individual peaks could be confirmed.
Figure 5.

Overlay of analytical chromatograms of batch “Newton” charge variants. A1-A5, M and B1-B2 shows the composition of the acidic, main and basic fractions, respectively. The dotted blue trace is the original material. The gray trace shows the % of buffer B.
For batch “Newton,” we collected enough of fraction B2 that it could be used as a starting material for a more refined separation step, applying a shallower pH gradient to achieve better isoform separation. Based on this procedure, the fractions B21-B25 were obtained (Fig. 6). This second separation step could not be done with the other batches because the B2 fraction did not contain enough material. An analytical scale pH gradient elution was performed from fractions B22-B25 (Fig. 7), clearly demonstrating that even those sub fractions are not pure charge variants. Fraction B21 contained too little protein to perform the analytical pH gradient method.
Figure 6.

Semi-preparative separation of the “Newton” B2 charge variant pool. The blue trace indicates UV absorbance, B21-B25 denotes the collection of variants of B2. The fractionation boundaries are represented by turquoise lines. The gray trace shows the pH.
Figure 7.
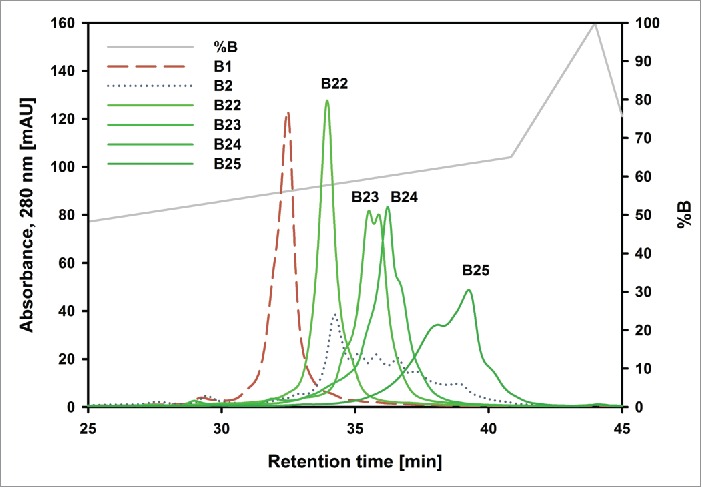
Overlay of analytical chromatograms of “Newton” basic charge variants. The dashed red trace is fraction B1. The dotted blue trace is fraction B2. B22-B25 are the further separated variants from B2. The gray trace shows the % of buffer B. Insufficient material from B21 made it impossible to analyze it.
In considering just 3 modifications responsible for charge variants, i.e., sialylation (0–4 sialic acids per mAb), pyroglutamate formation (0–4 affected N-termini per mAb) and lysine clipping (0–2 affected C-termini per mAb), the number of possible charge variants (213) is already in the high thousands. Even if only a fraction of those theoretically possible variants is actually present in a sample, it is not surprising that, in spite of high-resolution separation techniques, no pure isoform fractions could be observed.
To ensure that the starting material was free of proteins aggregates, which could compromise the results in later performed assays such as ADCC, size-exclusion chromatography coupled with multiple angle light scattering detection (SEC-MALS) was performed for all 3 batches (Fig. 8). Neither batch “Darwin” nor “Curie” contained any measurable amounts of aggregates or fragments. For batch “Newton” on the other hand, 3 different peaks were detected. The largest one, corresponding to the monomer, accounted for 95% of the sample, an earlier eluting aggregate peak was ∼1.5% and a later eluting fragment peak made up 3.5% of the total sample material. Given the fact that “Newton” is the oldest of the 3 batches and already out of specifications for clinical use, a certain amount of aggregation and fragmentation in this sample is not surprising. The detected levels are still in an acceptable range and should therefore not have a decisive influence on the biological assays. Typically, aggregation levels below 2% are not considered to be a cause for concern.5
Figure 8.
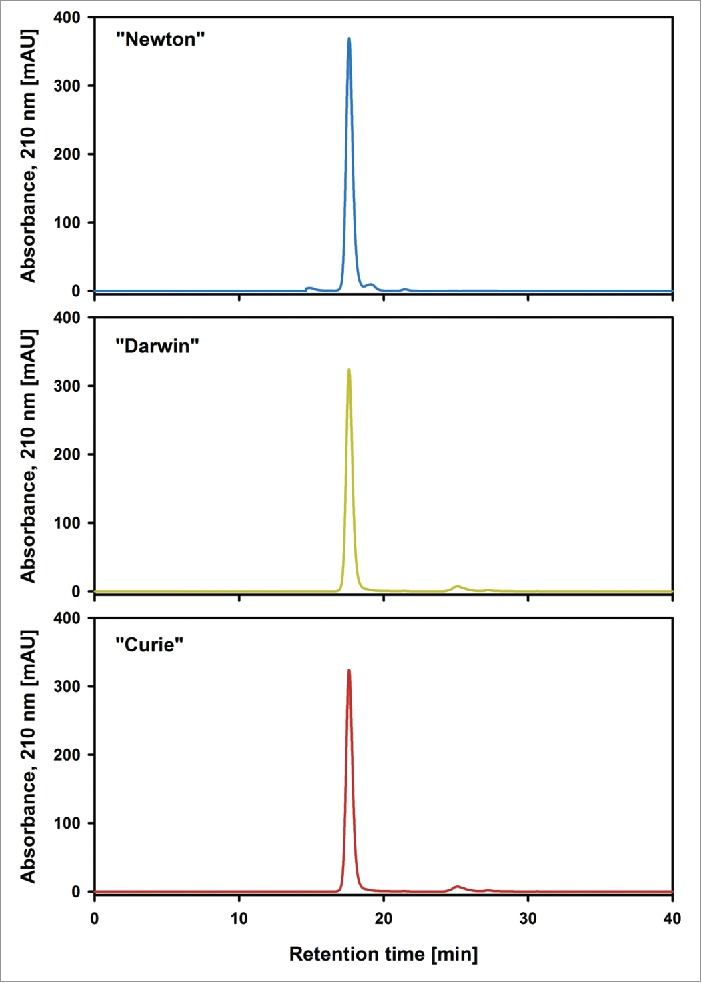
Size exclusion chromatogram of batches “Newton,” “Darwin” and “Curie” starting material. A significant amount of low and high molecular weight impurities are present in “Newton” starting material. No low and high molecular weight impurities were detected in “Darwin” and “Curie” starting material, but an additional peak at 25 minutes was present, which most likely stems from histidine from the sample buffer.
Influence of charge variants on antigen binding and effector functions
In vitro and cellular assays were performed to assess the biological efficacy of the individual charge variant fractions. Due to the high sample demand for some of these methods, not all analyses could be performed for all fractions.
SPR analysis for binding to GD2 (Fig. 9) showed a clear increase in target binding strength from the acidic toward the main peak fraction and the more basic charge variants for all investigated mAbs. A similar, though less pronounced, trend could be observed for the biding assay of the mAbs toward the Fc receptors, with main peak and basic variants showing better performance than acidic variants. (Figs. 10 and 11). In the FcγRIIIa assay, the only fraction that showed significantly higher binding strength than the reference in all 3 investigated batches was B2. For this fraction, a remarkably lower Kd, and therefore increased binding toward the FcγRIIIa receptor, was observed, with this effect being more pronounced in batch “Newton” than in the other 2 batches. A higher affinity for FcγRIIIa indicates an increased ADCC response, and therefore predicts a higher efficacy for the corresponding variants.
Figure 11.

Surface plasmon resonance FcRn binding data for batches “Newton” (blue), “Darwin” (yellow) and “Curie” (red). The main and basic fractions M, B1 and B2 show a lower Kd and therefore higher binding affinity toward the FcRn receptor than the more acidic variants.
Figure 9.

Surface plasmon resonance GD2 binding data for batches “Newton” (blue), “Darwin” (yellow) and “Curie” (red) reference material and charge variants. Main and basic fractions M, B1 and B2 show a lower Kd and therefore higher binding affinity to the antigen than the more acidic variants.
Figure 10.

Surface plasmon resonance FcγRIIIa binding data for batches “Newton” (blue), “Darwin” (yellow) and “Curie” (red). Fractions B2 and B22 from batch “Newton” show remarkably low Kd and therefore high binding affinity toward the FcγRIIIa receptor.
In the FcRn assay, the basic fractions again showed a slightly higher interaction strength toward the receptor than the more acidic fractions. This could be a first indication of a prolonged serum half-life of these fractions. However, it remains unclear whether the observed difference in binding strength is large enough to have a considerable influence on the in-vivo process. To confirm any such correlation, pharmacokinetic experiments must be conducted with the separated material, which exceeds the scope of this study.
The general trend of increased efficacy of the main and the basic variants compared to the acidic ones was confirmed by the biological assays for ADCC and CDC (Figs. 12 and 13). The main peak and the basic variant B1 showed a significantly better performance in both the ADCC and the CDC assay. The particularly strong binding of B2 toward FcγRIIIa, did not manifest itself as higher activity in the ADCC assay. Given that any kind of cellular assay will always be much more complex than a simple protein-protein interaction measurement, but also much more similar to the expected effect in vivo, we demonstrated that a simple characterization of antibody variants via receptor binding is not enough to estimate the efficacy of isoforms in vivo. The ADCC activity of an antibody is not solely defined by its affinity to the FcγRIIIa receptor, but by many other factors, e.g., structural modifications.
Figure 12.

ADCC assay for batches “Newton” (blue), “Darwin” (yellow) and “Curie” (red). For the main M and basic fraction B1, a lower antibody concentration is necessary to achieve 50% cell lysis.
Figure 13.

CDC assay for batches “Newton” (blue) and “Darwin” (yellow). For the main M and basic fraction B1, a lower antibody concentration is necessary to achieve 50% cell lysis.
Relationship of function and biochemical and biophysical properties
With a detailed set of data about the influence of microheterogeneity on antigen biding and effector functions available, we investigated the possible structural causes of the observed effects and their influence on structural integrity and stability of the product. Glycan analysis and peptide mapping were performed to address the first issue. As an indicator for overall structural stability, thermal unfolding was measured via Nano-DSC.
With respect to glycosylation, 4 groups of structures were previously reported to have significant impact on mAb efficacies and performance in cell based assays: sialylated structures, complex nonfucosylated structures, high mannose type structures and complex nongalactosylated structures.8 Glycan analysis was set up to investigate these 4 groups in detail. The first group of sialylated structures are a charged sugar moiety, and the presence of sialic acid influences the pI of a monoclonal antibody molecule. Furthermore, it is documented in the literature that monoclonal antibodies containing high amounts of this residue show a decreased performance in ADCC assays. 8 Complex type non-fucosylated structures were investigated because those are known to have a higher affinity toward the FcγRIIIa receptor.8,10,29 High mannose type glycan structures, on the other hand, are expected to show decreased CDC,30 and are therefore of interest for the performance of antibodies. The fourth group of target glycan structures in this study, the complex type non-galactosylated molecules, have been described to alter the affinity of an antibody molecule toward C1q, the initial component of the complement cascade and can therefore also be expected to affect the performance in CDC assays. Whether this affinity change occurs in a positive or negative manner remains unclear at this point, as both have been reported in the literature.8,31,32
Our glycan data (Fig. 14) shows a much higher amount of sialylated glycan structures in the acidic charge variant fractions compared to the main and basic ones (Fig. 14B), which suggests that sialic acid is indeed at least one of the modifications responsible for the observed microheterogeneity pattern. Furthermore, the obtained sialylation profile of the individual isoform fractions also correlates well with the ADCC data. Although sialylation might not be the only contributing factor, we demonstrated that, at least for our antibody, there is a strong link between antibody in-vitro efficacy and sialylation content.
Figure 14.
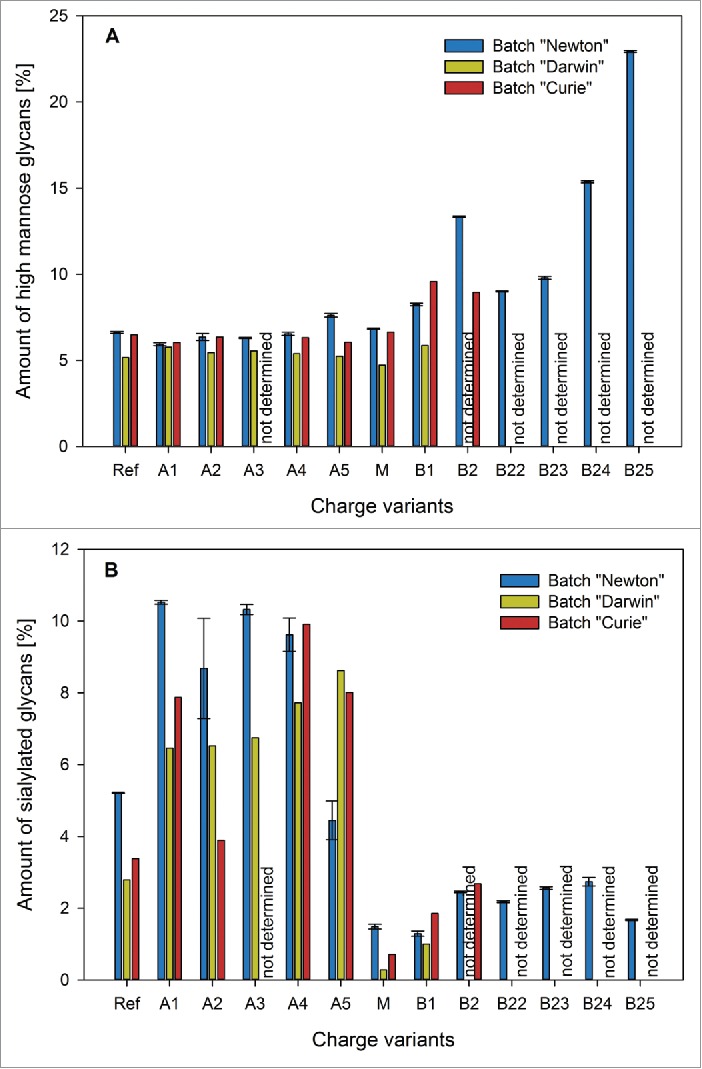
High mannose (A) and sialylated (B) glycans found in batches “Newton” (blue), “Darwin” (yellow) and “Curie” (red) charge variants. High mannose glycans are present in higher amounts only in fraction B2 of batch “Newton.” Sialylated glycans get enriched in the early eluting acidic fractions of all batches.
With respect to the complex type non-fucosylated glycans, only one fraction contained a considerably different amount of non-fucosylated N-glycans than the others (Figs. 14A and 15). This was B2 of batch “Newton,” which had also shown significantly stronger binding to the FcγRIIIa receptor. This confirms what was expected from the available literature. The same correlation could be observed for batch “Curie,” although to a lesser degree, as this batch B2 contains slightly lower amounts of non-fucosylated glycans than B1.
Figure 15.
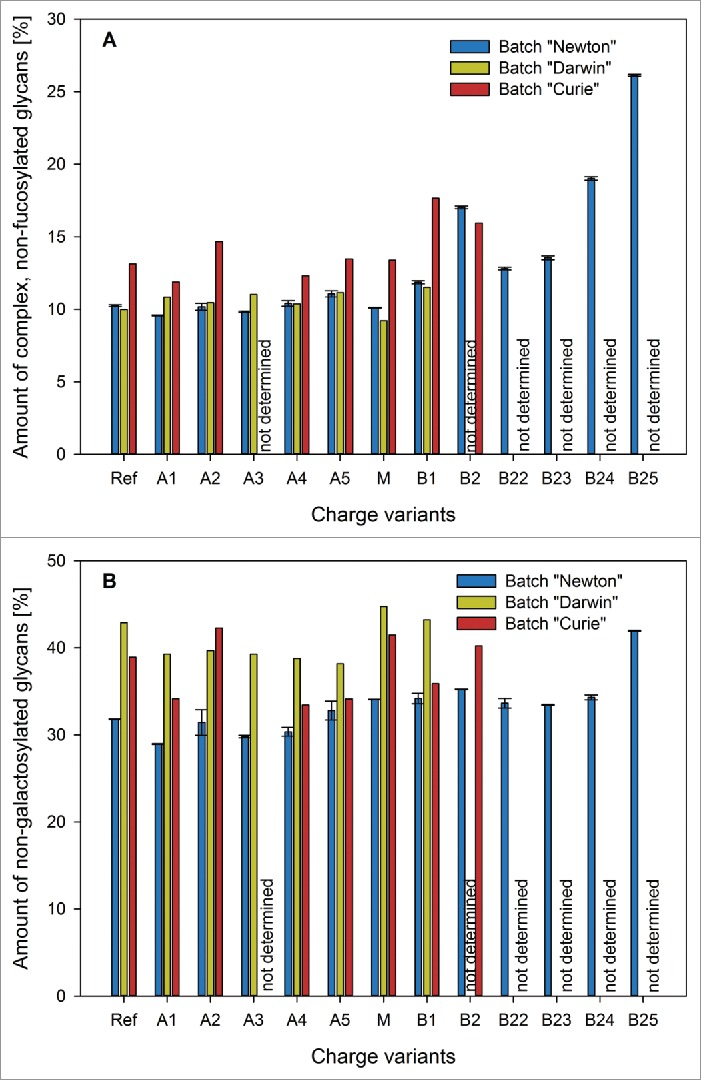
Complex non-fucosylated (A) and non-galactosylated (B) glycans found in batches “Newton” (blue), “Darwin” (yellow) and “Curie” (red) charge variants. Only fraction B2 of batch “Newton” contains considerably higher amounts of complex, non-fucosylated glycans than the other fractions of that batch. When it comes to non-galactosylated glycans, no characteristic differences could be observed between the fractions.
For the high-mannose type structures, the situation was similar (Fig. 14 A). Once again, the only fraction for which a noticeably different value could be measured was B2 of batch “Newton.” High-mannose type glycans therefore do not provide a satisfactory explanation for the observed behavior of our charge variants in the CDC assay. Though B2 of batch “Newton” does in fact show comparatively weak performance in the CDC assay, the general trend of an activity increase from the acidic variants toward M and B1 cannot be attributed to this group of glycan structures. Regarding the complex, non-galactosylated glycans, no characteristic differences were observed in the charge variant fractions (Fig. 15B). Thus, it remains doubtful if there really is a link between these types of sugar residues and the ability of an antibody to elicit an effective CDC response. Summarizing glycan data, our method is clearly able to enrich sialylated glycoforms in the early eluting fractions, and complex-non fucosylated and high-mannose-type structures in the later eluting ones.
Peptide map data (Fig. 16) was obtained focusing on 4 different types of modifications on amino acids located in or in close proximity to the complementarity-determining regions (CDRs), the binding sites for C1q and the Fc receptor or the C- and N-termini of the protein. With respect to the deamidation of asparagine residues, no significant differences could be observed between the charge variant fractions (data not shown).
Figure 16.

Peptide map data for batch “Schrödinger.” Increased levels of Oxidation at residue M34 (blue) were detected in fraction B2, N-terminal pyroglutamate (orange) is mainly present in the basic fractions B1 and B2, fraction B1 contains a considerably higher amount of molecules, where the C-terminal Lysine is still present (green).
In fraction B2, a remarkably high value for oxidation of methionine residue M34 in the CDR1 could be observed. Oxidation of methionine and tryptophan residues by itself should not have an effect on the charge heterogeneity of a given sample. The modifications may, however, result in conformational changes in the protein, bringing other charged residues to the surface of the molecule and thereby making them available for interaction in IEX chromatography. Oxidation reactions on residues located in the CDRs of the antibody could very well result in a different binding affinity toward its molecular target. For our fraction B2, the detected modification does not seem to result in a profound effect on the binding levels to GD2, which were only slightly lower than for the main fraction.
N-terminal pyroglutamate formation results in the change of the pKa values of the amine and carboxy group and a loss of a positive and a negative charge at neutral pH.6 At basic pH, only the loss of the negatively charged carboxyl group is relevant because the N-terminus gets deprotonated at a pH of around 7.7.33 Literature suggests that this type of modification has no substantial effect on antigen binding affinity, efficacy and safety of the antibody product.34 An increased amount of this modification was found mainly in fraction B1 and to a lesser extent in B2, suggesting an explanation for the observed behavior of those fractions in IEF and IEX. The high level of pyroglutamate in fraction A3 were surprising. We therefore tried to confirm this result by comparing it to peptide map data from stressed material from the same batch, which was also available to us. This material was stored at 40°C for 30 d and then separated with preparative pH gradient elution. The acidic variant A3 in the stressed sample did not show any elevated levels of pyroglutamate, which indicated that the value from the unstressed A3 sample is an outlier (data not shown).
Lysine clipping at the C-terminus results in the removal of one positive charge. Similar to N-terminal pyroglutamate formation, so far, no further effects on structure and biological functions of the molecule have been observed.34 Once again, this modification was found in significantly higher amounts in fractions B1 and seems to be contributing to the more basic characteristics of this variant fraction.
Overall, the peptide map data confirms once again that C-terminal Lysine clipping and N-terminal pyroglutamate formation are of decisive importance for the presence of basic isoforms in monoclonal antibodies.
What comes as a surprise is that our pH gradient elution method also seems to have a separation effect on molecules that show elevated oxidation levels at residue M34, since methionine sulfoxide does not have a charged side chain. It is likely that the oxidation of methionine changes the surrounding protein surface enough to influence binding to a cation-exchange stationary phase.
The lack of separation of mAbs with deamidated asparagine was likewise unexpected because the charged aspartate or iso-aspartate side chains should influence binding behavior. This may be due to the fact that we were only taking into account residues positioned in areas of the molecule involved in binding to Fc-receptors, antigen and C1q. It is still possible that the overall deamidation levels in the molecule have a decisive influence on the observed microheterogeneity pattern.
As it is easily, measureable thermal stability is often used as an indicator for overall structural integrity and long-term stability of proteins. In case of IgG molecules, a thermal unfolding experiment typically results in 2 or 3 peaks and their corresponding melting temperatures (Tm) are associated with the unfolding of the CH2 domain, the Fab, and the CH3 domain. In many cases, including ours, the first 2 peaks are not recognizable as 2 individual signals but are merged together into one bigger peak with a minor shoulder. The first thermal transition is usually taken into account as a reference number for stability determination because it occurs at lower temperature than the one for the CH3 domain. Concerning our variants fractions, little to no differences could be observed in between the separated charge variants (Fig. 17). If anything, the main peak and its neighboring fractions show a slightly higher Tm1 than the other variants, indicating a slightly better stability of the CH2 domain and the Fab in these fractions.
Figure 17.

Nano DSC data; (A) Transition temperature Tm1 corresponding to CH2 domain and Fab, (B) Transition temperature Tm2 corresponding to CH3 domain. No stability differences could be detected between the charge variant fractions
Discussion
With the pH gradient elution in cation-exchange chromatography, we could enrich sialylated glycoforms of antibody in the early eluting, acidic fractions, and complex non-fucosylated and high mannose type glycoforms in the later eluting, basic fractions. Our study can therefore be considered one of the first successful attempts at chromatographical separation of antibody glycoforms. This technique greatly facilitates research in the area of functional biology and may be of great use in cases when glycoengineering is not possible.
In our study, basic recombinant antibody charge variants separated by the pH gradient elution method exhibited more potent effector functions, which might correspond to better efficacy in vivo. These basic charge variants showed elevated levels of complex, non-fucosylated glycoforms, oxidation in methionine 34 and incomplete C-terminal lysine clipping. Only the absence of fucose in the N-glycosylation had an influence on effector functions. Non-fucosylated glycoforms showed elevated binding to FcγRIIIa, and therefore increased ADCC through increased recruitment of leukocytes. Since M34 is located in a CDR, it could potentially affect antigen binding and consequently ADCC and CDC, but this effect was not observed.
The separated charge variant fractions showed different binding to the FcRn receptor, which suggests that product microheterogeneity might have a noticeable effect on antibody half-life in serum. If the observed relation holds true in in-vivo studies, this may be used to optimize antibodies for increased half-life, reduced dosage and better overall performance.
No difference between the individual isoform fractions could be detected in terms of thermal stability, indicating that microheterogeneity does not greatly affect the overall structural stability of a recombinant antibody molecule. This opens possibilities for enhancing features such as better ADCC or CDC either during upstream or downstream processing without damaging the stability of the antibody.
Our data suggests that the connection between microheterogeneity and the efficacy of monoclonal antibody therapeutics is of more importance than commonly accepted. With the rise of biosimilar recombinant antibody development, and in context with quality by design approaches, the analysis and comparison of microheterogeneity patterns becomes even more relevant. In ensure equivalent biological characteristics, industry should emphasize microheterogeneity profiling. The ability to predict the effect of charge variant distribution might, however, obviate the need to closely match microheterogeneity of a biosimilar to the originator. The advent of new high-resolution separation and new tools in interaction analysis in recent years now allows the elucidation and quantification of differences in recombinant antibody isoforms. The work presented here lays the groundwork for future studies on this often overlooked phenomenon.
Material and methods
Materials and chemicals
All chemicals were of analytical grade, unless stated otherwise. 3-morpholino-2-hydroxypropanesulfonic acid (MOPSO) (M8389), 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) (H3375), N,N-bis(2-hydroxyethyl)glycine (bicine) (B3876), 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid (CAPSO) (C2278) and 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) (29337), 2-(N-morpholino)ethanesulfonic acid (MES) (M3671), NaH2PO4 (S8282), Tergitol (70% in water) (NP40S), acrylamido buffers (0.2 M in H2O with pKa 3.6 and >12, as well as 0.2 M in 1-propanol with pKa 8.5, 9.3, and 10.3) (01716, 01743, 01736, 01738, 01741) isopropanol (34959) and Coomassie® G250 (B0770) were purchased from Sigma–Aldrich. NaCl (1.06404.5000), Na2HPO4 (1.06586.2500), NaOH (1.06482.5000), urea (1.08487.5000), disodium dihydrogen ethylenediaminetetraacetate (EDTA, Titriplex III) (1.08421.1000), glycerol (1.04094.2500), paraffin oil (1.07174.2500) was purchased from Merck. GELbond-PAG-film (80-1129-36), Pharmalyte 3–10 (17-0456-01), CM 5 Sensor chips (BR-1000-12) and an amine coupling kit (BR-1000-50) for protein immobilization were purchased from GE Healthcare. Histidine, research grade (> 98.5% purity) (24820.03) was bought from Serva. Tween 20 (170–6606), 40% acrylamide-bis solution (37.5:1) (161–0148), ammonium persulfate (APS) (161–0700) and N,N,N',N'-tetramethylethylenediamine (TEMED) (161–0800) were obtained from Bio-Rad.
The recombinant monoclonal antibody CH14.18 was obtained from APEIRON biologics. It is a mouse-human chimeric IgG1 mAb, produced in Chinese hamster ovary (CHO) cells35 and directed against the disialoganglioside GD2, which is commonly expressed as a surface antigen on neuroblastoma and various other types of tumor cells, including melanoma and osteosarcoma.1-3
Preparative and analytical scale cation-exchange chromatography with linear pH gradient elution
All preparative separations were performed using an Äkta Explorer 100 (GE Healthcare) and a Dionex ProPac WCX-10 column, 22 × 250 mm (Thermo Fisher, SP5482)). The outlet was monitored at 280 nm. The flow rate was 10 ml/min (158 cm/h). The mobile phase used was a pH gradient buffer system as described in Lingg et al.16 and shown in Table 1.
Table 1.
The buffer system used for pH gradient elution.
| HEPES | bicine | CAPSO | CAPS | NaCl | pH | |
|---|---|---|---|---|---|---|
| Buffer A [mM] | 5.5 | 4.2 | 9.5 | 0.8 | 6.3 | 8.0 |
| Buffer B [mM] | 0.0 | 10.5 | 2.5 | 7.0 | 0.0 | 10.5 |
After loading 50 ml of mAb solution, with mAb concentrations between 3 and 4 mg/ml, to the equilibrated column (0% B) via a superloop, the method consisted of a 1 column volume (CV) wash step at 0% B, followed by a linear gradient to 70% B in 14 CV. Fractionation of the eluate was started at a concentration of 34% B into 7.5 ml fractions until the end of elution (70% B for mAb batch “Newton,” 60% B for the other batches).
The fractions were immediately neutralized with 10 × PBS, to avoid asparagine deamidation and pooled into a main fraction (M), acidic fractions named A1 – A5 and basic fractions B1 – B2. The fractions were concentrated and the buffer was exchanged with a Kvick Start Cassette, 50 kDa (GE Healthcare UFEST0005050ST) on a Labscale TFF System (Merck Millipore), using PBS as the ultrafiltration – diafiltration buffer. To further reduce the volume of the fractions, Amicon Ultra-15 Centrifugal Filter Units, 50 kDa (Merck Millipore UFC905096) were used.
In the case of the mAb batch “”Newton,” a significant amount of B2 fraction was purified (∼12 mg), which allowed further separation of this fraction into the B21 – B25 fractions. This additional step was performed on a semi-preparative Dionex ProPac WCX-10 column, 9 × 250 mm (Thermo Fisher Scientific 063474) on an Äkta Explorer 100 (GE Healthcare). Eight ml of B2 (1 mg/ml) were loaded onto the equilibrated (0% B) column via a super loop. The flow rate was 2.5 ml/min (236 cm/h). After a 2 CV wash step, an initial steep gradient to 25% B in 2.5 CV was followed by a shallower gradient to 70% B in 9 CV. Fractionation of the eluate was started at a concentration of 40% B into 1 ml fractions until end of elution. The fractions were, again, immediately neutralized with 10 × PBS, and pooled into the B21 – B25 fractions. Ultra- and diafiltration was performed with Amicon Ultra-15 Centrifugal Filter Units, 50 kDa (Merck Millipore UFC905096).
For analytical chromatography, an Agilent 1220 Infinity LC System equipped with a SIM sample-cooler (Scientific Instruments Manufacturer GmbH) was used. The column was a Dionex ProPac WCX-10, 4 × 250 mm (Thermo Fisher Scientific 054993). The outlet was monitored at 280 nm. The flow rate was 1 ml/min (477.5 cm/h). The buffers were the same as those used for the preparative experiments in Table 1. A gradient from 25% to 65% buffer B in 8 CV was used with an injection volume of 100 µl at a concentration of 1 mg/ml.
Isoelectric focusing
Immobilized pH gradient (IPG)-polyacrylamide-gels (size: 125 × 260 × 1 mm) were cast on a GELbond-PAG-film backing (GE Healthcare), polymerized, washed and dried following a procedure previously described by Westermaier et al.36 The desired pH gradient from 7.0 to 11.0 was obtained by graphic interpolation from a recipe previously published by Görg et al.37 Before use, the gel was cut into half, one half was used directly and rehydrated for 2 h in a solution containing 6 M urea, 2% Tergitol, 2% Pharmalyte 3–10, 10% glycerol and 16% isopropanol. The other one was stored at −20°C. The rehydrated gel was put onto the Multiphor instrument (GE Healthcare). The electrodes were positioned on the acidic and basic ends of the gel with wetted paper wicks between gel and electrode to ensure good contact. The samples were applied by cup loading under a covering layer of paraffin and electrophoresis was performed overnight (at 150 V for 1 h, followed by 300 V for 4 h, and 3500 V for 18 h). Afterwards, the gel was thoroughly rinsed with water and then stained with Coomassie G250.
Size exclusion chromatography coupled with multi-angle light scattering detection
IgG aggregation analysis was done using a previously described method. 38 Briefly, 45 – 80 µg of IgG was injected to an HPLC system (Shimadzu, Kyoto, Japan), which was connected in series with a TSK guard column SWXL, 6 × 40 mm, a TSK gel G3000 SWXL, 7.8 × 300 mm (both from Tosoh Corporation, 08543 08541), a UV-Vis detector (Shimadzu), followed by a MALS detector (Dawn 8 from Wyatt Technology Corporation). IgG aggregates, monomers, and fragments were separated by the SEC column, which was kept at 25°C, using an isocratic mobile phase of aqueous solution containing 0.2 M sodium phosphate (Merck Millipore 1063421000) and 0.1 M potassium sulfate at pH 6.0 (Merck Millipore 1051531000) at 0.5 ml/min flow rate. The molecular weight of each elution peak was estimated by ASTRA V 5.3.4.20 software (Wyatt Technology Corporation). Relative peak areas from the UV280 nm channel were used to calculate the percentage of IgG aggregates, monomers, and fragments. The SEC-MALS HPLC system was calibrated using monomeric bovine serum albumin. All IgG samples were analyzed in triplicates.
Surface plasmon resonance binding assay to GD2
For direct hydrophobic immobilization of ganglioside GD2, a CM5 sensor chip (GE healthcare) with 4 flow cells that had been reacted previously with ethanolamine using NHS/EDC chemistry and analyzed on a Biacore 2000 system (GE healthcare) was used. Therefore, GD2 was dissolved (1 mg/ml) in 50% ethanol, 50% methanol (v/v), further diluted 1/3 with HBS-N buffer (0.01 M HEPES, 0.15 M NaCl, pH 7.4) and injected (15 µl) at a flow rate of 5 µl/min over a single flow cell. Weakly bound GD2 was removed by 2 injections of 15 mM NaOH solution. For GD2 binding measurements, PBS, supplemented with 200 mM NaCl, pH 7.4, was used as running and dilution buffer for all antibody fractions, which were diluted to concentration levels of 24, 60, 120, 180 and 240 nM. Diluted antibody fractions were injected for 6 minutes at a flow rate of 20 µl/min over the GD2 immobilized and a control (blank) flow cell. GD2 binding levels were measured for each of the concentrations in steady state at the end of injection and were used to calculate the dissociation rate constant Kd by using a steady-state model. For these calculations, the maximum binding capacity value was calculated using the BiaEvaluation software (GE healthcare) using the Langmuir adsorption isotherm model fit. Blanked GD2 binding levels at the end of injection were used for calculation of the dissociation rate constant (Kd), by using the steady state model. The maximum binding response value (Rmax) used for all analysis was 988.5 RU, calculated by BiaEvaluation software using the 1:1 Langmuir absorption isotherm model fit. The final Kd value was calculated as an average of the Kd values at each concentration. Regeneration of the sensor chip surface was done by 2 injections of 15 mM NaOH solution for 15 seconds.
Surface plasmon resonance binding assay to FcγRIIIa and FcRn
Both receptors were immobilized on CM5 sensor chips (GE healthcare) using the amine coupling approach and analyzed on a Biacore 2000 system (GE healthcare). The running buffer for the FcγRIIIa binding assay was the standard Biacore HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, and 0.005% v/v surfactant P20) at pH 7.4 as recommended by the manufacturer of the instrument. For the FcRn binding assay, a similar MES buffer (0.01 M MES, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v surfactant P20) at pH 5.5 was used. All antibody fractions were diluted to concentration levels of 10, 25, 50, 75 and 100 nM before measurement. FcγRIIIa interaction was measured for each concentration level, FcRn interaction was only measured at concentrations of 25, 50 and 100 nM. The observed binding data was fitted to a protein-protein-interaction model to obtain rate and affinity constants. For FcγRIIIa, Kd values were calculated based on a Langmuir adsorption isotherm model. For FcRn, the 2:1 bivalent analyte model built into the BiaEvaluation Software was used. Regeneration was done by a one-minute injection of a 0.05% (w/v) solution of SDS in H2O for FcγRIIIa and a 50 mM Na2HPO4, 150 mM NaCl solution at pH 7.4 for FcRn.
ADCC and CDC assays
In our ADCC assays, we used GD2-positive M21 melanoma cells as target cells and labeled them by incubation in an isotonic labeling buffer containing Na251CrO4 (Perkin Elmer). After labeling, the cells were washed by centrifugation in fresh medium until no radioactivity was detected in the supernatant. As effector cells, a suspension of peripheral blood mononuclear cells was prepared by density gradient centrifugation of heparinized blood obtained by venipuncture of healthy human donors. These cells were used at an effector : target cell ratio of 30 : 1 in the assay. Dilutions of charge variant fractions were prepared in cell culture medium and used as test substance compared to the reference material from the original batches. The assays were conducted in 96-well plates. All samples were prepared in duplicates; supernatants were harvested and pipetted into micro tubes fitted with absorbent tissue. Said tubes were analyzed in a gamma counter and radioactivity values were used to calculate cell lysis of individual samples. All calculations and the 4-parameter sigmoidal dose-response fit with variable slope and a confidence interval of 95% for determination of the calibration function were performed using GraphPad Prism (GraphPad software) and Microsoft Excel Software.
In our CDC assays, we used GD2-positive M21 melanoma cells as target cells and labeled them by incubation in an isotonic labeling buffer containing Na251CrO4. After labeling, the cells were washed by centrifugation in fresh medium until no radioactivity was detected in the supernatant. As complement source, serum of healthy human donors was obtained by venipuncture and subsequent clotting of the blood sample. In the assay, a final dilution of 1:5 of serum in culture medium was used. Dilutions of charge variant fractions were prepared in cell culture medium and used as test substance compared to the reference material from the original batches. The assays were conducted in 96-well plates. All samples were prepared in duplicates; supernatants were harvested by pipetting into micro tubes fitted with absorbent tissue. Said tubes were analyzed in a gamma counter and radioactivity values were used to calculate cell lysis of individual samples. All calculations and the 4-parameter sigmoidal dose-response fit with variable slope and a confidence interval of 95% for determination of the calibration function were performed using GraphPad Prism and Microsoft Excel Software.
Glycan analysis
N-glycans were released directly from intact, purified glycoprotein samples, by PNGase F treatment and analyzed by mass spectrometry. Glycoprotein samples were desalted using a PD 10 column (GE Healthcare 17-0851-01) following manufacturer's protocol. Then, 100 µg IgG was mixed with 500 U of the PNGase F (New England Biolabs P0704 L) in the reaction buffer in a total volume of 100 µl and incubated at 37°C for 1 h. Such conditions will result in complete deglycosylation of IgG as indicated by SDS-PAGE-capillary gel electrophoresis (data not shown). The released glycans were then purified by HyperCarb porous graphitized carbon cartridge (Thermo Fisher Scientific 35003), and dried by CentriVap (Labconco).
The dried N-glycans were labeled with 2-aminobenzamide (2-AB) according to a published protocol. 39 The excess 2-AB was removed by passing the labeling mixture through a MiniTrap G-10 desalting column (GE Healthcare 28-9180-10) and the purified 2-AB-labeled glycans were then dried under vacuum. Before the analysis, the dried samples were reconstituted in 250 µl of solvent consisting of 70% (v/v) acetonitrile in water. The reconstituted 2-AB labeled glycan samples were analyzed by the UNIFI Biopharmaceutical platform (Waters Corporation). The entire platform consists of an UPLC-H class ultra-performance liquid chromatogram (UPLC) that is online-connected to a Xevo G2-S quadrupole-time of flight (QTOF) mass spectrometer, both under the control of UNIFI Biopharmaceutical scientific software platform (version 1.7). The UPLC-H class consists of a sample manager (kept at 10°C), a quaternary pump, a column oven (kept at 40°C) that houses a Waters BEH glycan column (2.1 mm ID × 150 mm length 186004742), and a fluorescence detector. Ten µl of the reconstituted 2-AB glycan sample were injected to the UPLC system. Glycans were separated on the hydrophilic interaction column using a binary solvent system. Solvent A was 50 mM ammonium formate (pH 4.4) and Solvent B is acetonitrile. The analytical run takes place in 16 min by ramping up Solvent A from 30% to 47%. The column was then regenerated with 80% Solvent A before re-equilibrated with 30% Solvent A for the next run. Glycan signal was detected at excitation wavelength of 330 nm and emission wavelength of 420 nm. Retention time for each chromatographic peak was converted to a specific glucose unit (GU) by fitting into a calibration curve established by a 2-AB-labeled dextran ladder (Waters Corporation 186006841). The GU value of each chromatographic peak was then used to search against an experimental database for N-glycans embedded in the UNIFI Biopharmaceutical platform. Primary assignment was done by alignment of observed and the expected GU values. In case of structural ambiguity, i.e., a GU value corresponding to more than one structure within the error tolerance (typically 0.1 GU), a decision was then made based on accurate mass confirmation (5 ppm error) by the online ESI-QTOF, exoglycosidase fingerprinting, and the knowledge of the N-glycan biosynthetic pathway in CHO cells. The online ESI-QTOF was operated under the following conditions: cone voltage: 80 kV; capillary voltage: 2.75 kV; source temperature: 120°C; desolvation gas flow: 800 l/h; desolvation temperature: 300°C. The QTOF was operated by scanning the mass range of 400 – 3,000 amu at the acquisition speed of 1 Hz. Mass accuracy was maintained by introducing a “lock spray” of Glu-fibrinopeptide (m/z = 785.8421).
Peptide map
For the peptide map analysis, samples were obtained from a newly produced antibody batch “Schrödinger,” which was separated into fractions A1–A5, M, B1 and B2 in the same fashion as described in section 4.2.
For the tryptic digest, 10 µg of each sample were diluted with PBS to a total volume of 10 µl. Then 36 µl of Buffer A (8 M guanidinium-HCl, 0.2 M His-HCl pH 6) were added. Reduction of cysteines was done using 6.4 µl of 0.02 mg/ml DTT and incubation at room temperature for 1 h. The sample was then diluted to 900 µl with buffer B (0.02 M His-HCl, pH 6), followed by the addition of 0.5 µg trypsin. Digestion was carried out at 37°C overnight. For MS-analysis the sample was acidified to 1% with formic acid.
For the double digest with Asp-N and Lys-C, 10 µg of each sample was diluted with PBS to a total volume of 10 µl. Then, 36 µl of Buffer A (8 M guanidinium-HCl, 0.4 M Tris-HCl pH 8.5) and 6.4 µl 0.02 mg/ml DTT were added and the samples were incubated at room temperature for 1 h. Cysteines were then alkylated by adding 1 µl iodoacetic acid (0.094 µg/ml) and incubating the mixture at room temperature in the dark for 30 minutes.
Following alkylation, a buffer exchange was done. The samples were diluted to 500 µl with buffer B (0.02 M His-HCl, pH 6) and applied to a Vivaspin column (cutoff 10 kDa VS0102). The sample was concentrated to a final volume of 25–30 µl with centrifugation. This procedure was repeated twice.
For digestion, 2.5 µl Lys-C and 1 µl Asp-N (both from stock solutions with 0.2 µg/µl were added and the samples were incubated at 37°C overnight. For MS analysis, the sample was acidified to 1% with formic acid.
Before the actual MS analysis, samples were desalted using a pre-column ACE C18, particle size 5 µm, pore size 100 Å, 2 cm length, 100 µm I.D.). Chromatographic separation was then performed using a 25 cm long C18 column particle size 3 µm, pore size 100 Å, 75 µm I.D.) with a linear gradient from 3–42% B in 90 minutes (B: 84% ACN in 0,1% FA) at a flow rate of 17 µl/min.
Samples were then analyzed online in an Orbitrap Velos mass spectrometer with a scan range from m/z 200–2000. The mass spectrometer was operating in the so called “data dependent” mode where after each full scan the 10 most intense signals are chosen automatically for MS/MS analysis. Here an inclusion list that contains the theoretical masses of the peptides of interest with and without modifications was used. This ensured that these peptides are detected even if they are present only in minute amounts.
Nano differential scanning calorimetry
Samples were diluted to a concentration of 0.5 mg/ml in PBS buffer at pH 6.94. Six hundred and 50 µl of this solution were loaded into the sample cell of a TA- Instruments Nano DSC instrument (model: 602000). The reference cell was filled with PBS buffer and a thermoscan from 25°C to 100°C with a scan rate of 0.7°C/min was performed. The obtained thermogram data was analyzed using the TA Instruments NanoAnalyse software.Between sample runs, the instrument was cleaned using a solution containing 0.5 M NaCl, 0.1 M acetic Acid and 1 mg/ml pepsin followed by flushing with water.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Toplab for providing the peptide map data, and Peter Satzer for critically reviewing the manuscript. This work was supported by the Austrian Research Promotion Agency (FFG). PZ, SW, and KMH were funded by the Strategic Positioning Fund (SPF) (“GlycoSing”) from the Biomedical Research Council (BMRC) of Agency for Science, Technology and Research (A*STAR), Singapore and A*STAR's Joint Council (JCO) Visiting Investigator Program (“HighGlycoART”).
References
- 1.Ladenstein R, Weixler S, Baykan B, Bleeke M, Kunert R, Katinger D, Pribill I, Glander P, Bauer S, Pistoia V, et al.. Ch14.18 antibody produced in CHO cells in relapsed or refractory Stage 4 neuroblastoma patients: a SIOPEN Phase 1 study. MAbs 2013; 5:801-9; PMID:23924804; http://dx.doi.org/ 10.4161/mabs.25215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desai AV, Fox E, Smith LM, Lim AP, Maris JM, Balis FM. Pharmacokinetics of the chimeric anti-GD2 antibody, ch14.18, in children with high-risk neuroblastoma. Cancer Chemother Pharmacol 2014; 74:1047-55; PMID:25212536; http://dx.doi.org/ 10.1007/s00280-014-2575-9 [DOI] [PubMed] [Google Scholar]
- 3.Siebert N, Eger C, Seidel D, Jüttner M, Zumpe M, Wegner D, Kietz S, Ehlert K, Veal GJ, Siegmund W, et al.. Pharmacokinetics and pharmacodynamics of ch14.18/CHO in relapsed/refractory high-risk neuroblastoma patients treated by long-term infusion in combination with IL-2. MAbs 2016; 8:604-16; PMID:26785755; http://dx.doi.org/ 10.1080/19420862.2015.1130196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brorson K, Jia AY. Therapeutic monoclonal antibodies and consistent ends: terminal heterogeneity, detection, and impact on quality. Curr Opin Biotechnol 2014; 30:140-6; PMID:25022603; http://dx.doi.org/ 10.1016/j.copbio.2014.06.012 [DOI] [PubMed] [Google Scholar]
- 5.van Beers MM, Bardor M. Minimizing immunogenicity of biopharmaceuticals by controlling critical quality attributes of proteins. Biotechnol J 2012; 7:1473-84; PMID:23027660; http://dx.doi.org/ 10.1002/biot.201200065 [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Gaza-Bulseco G, Faldu D, Chumsae C, Sun J. Heterogeneity of monoclonal antibodies. J Pharm Sci 2008; 97:2426-47; PMID:17828757; http://dx.doi.org/ 10.1002/jps.21180 [DOI] [PubMed] [Google Scholar]
- 7.Goetze AM, Schenauer MR, Flynn GC. Assessing monoclonal antibody product quality attribute criticality through clinical studies. mAbs 2010; 2:500-7; PMID:20671426; http://dx.doi.org/ 10.4161/mabs.2.5.12897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingg N, Zhang P, Song Z, Bardor M. The sweet tooth of biopharmaceuticals: importance of recombinant protein glycosylation analysis. Biotechnol J 2012; 7:1462-72; PMID:22829536; http://dx.doi.org/ 10.1002/biot.201200078 [DOI] [PubMed] [Google Scholar]
- 9.Zauner G, Selman MH, Bondt A, Rombouts Y, Blank D, Deelder AM, Wuhrer M. Glycoproteomic analysis of antibodies. Mol Cell Proteomics 2013; 12:856-65; PMID:23325769; http://dx.doi.org/ 10.1074/mcp.R112.026005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck A, Wagner-Rousset E, Bussat MC, Lokteff M, Klinguer-Hamour C, Haeuw JF, Goetsch L, Wurch T, Van Dorsselaer A, Corvaia N. Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr Pharm Biotechnol 2008; 9:482-501; PMID:19075687; http://dx.doi.org/ 10.2174/138920108786786411 [DOI] [PubMed] [Google Scholar]
- 11.Qian J, Liu T, Yang L, Daus A, Crowley R, Zhou Q. Structural characterization of N-linked oligosaccharides on monoclonal antibody cetuximab by the combination of orthogonal matrix-assisted laser desorption/ionization hybrid quadrupole-quadrupole time-of-flight tandem mass spectrometry and sequential enzymatic digestion. Anal Biochem 2007; 364:8-18; PMID:17362871; http://dx.doi.org/ 10.1016/j.ab.2007.01.023 [DOI] [PubMed] [Google Scholar]
- 12.Levine RL, Moskovitz J, Stadtman ER. Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life 2000; 50:301-7; PMID:11327324; http://dx.doi.org/ 10.1080/15216540051081056 [DOI] [PubMed] [Google Scholar]
- 13.Liu H, May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. MAbs 2012; 4:17-23; PMID:22327427; http://dx.doi.org/ 10.4161/mabs.4.1.18347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Yip H, Katta V. Identification of isomerization and racemization of aspartate in the Asp-Asp motifs of a therapeutic protein. Anal Biochem 2011; 410:234-43; PMID:21130067; http://dx.doi.org/ 10.1016/j.ab.2010.11.040 [DOI] [PubMed] [Google Scholar]
- 15.Lingg N, Tan E, Hintersteiner B, Bardor M, Jungbauer A. Highly linear pH gradients for analyzing monoclonal antibody charge heterogeneity in the alkaline range. J Chromatogr A 2013; 1319:65-71; PMID:24183595; http://dx.doi.org/ 10.1016/j.chroma.2013.10.028 [DOI] [PubMed] [Google Scholar]
- 16.Lingg N, Berndtsson M, Hintersteiner B, Schuster M, Bardor M, Jungbauer A. Highly linear pH gradients for analyzing monoclonal antibody charge heterogeneity in the alkaline range: Validation of the method parameters. J Chromatogr A 2014; 1373:124-30; PMID:25465369; http://dx.doi.org/ 10.1016/j.chroma.2014.11.021 [DOI] [PubMed] [Google Scholar]
- 17.Jason-Moller L, Murphy M, Bruno J. Overview of Biacore systems and their applications. Curr Protoc Protein Sci; Chapter 19:Unit 19.3 Wiley, 2006; PMID:18429302; http://dx.doi.org/ 10.1002/0471140864.ps1913s45 [DOI] [PubMed] [Google Scholar]
- 18.Ahmed M, Cheung NK. Engineering anti-GD2 monoclonal antibodies for cancer immunotherapy. FEBS Lett 2014; 588:288-97; PMID:24295643; http://dx.doi.org/ 10.1016/j.febslet.2013.11.030 [DOI] [PubMed] [Google Scholar]
- 19.Kapur R, Einarsdottir HK, Vidarsson G. IgG-effector functions: “the good, the bad and the ugly”. Immunol Lett 2014; 160:139-44; PMID:24495619; http://dx.doi.org/ 10.1016/j.imlet.2014.01.015 [DOI] [PubMed] [Google Scholar]
- 20.Gillis C, Gouel-Chéron A, Jönsson F, Bruhns P. Contribution of Human FcγRs to Disease with Evidence from Human Polymorphisms and Transgenic Animal Studies. Front Immunol 2014; 5:254; PMID:24910634; http://dx.doi.org/ 10.3389/fimmu.2014.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol 2014; 5:520; PMID:25368619; http://dx.doi.org/ 10.3389/fimmu.2014.00520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcγ receptors in dendritic cells and macrophages. Nat Rev Immunol 2014; 14:94-108; PMID:24445665; http://dx.doi.org/ 10.1038/nri3582 [DOI] [PubMed] [Google Scholar]
- 23.Sockolosky JT, Szoka FC. The neonatal Fc receptor, FcRn, as a target for drug delivery and therapy. Adv Drug Deliv Rev 2015; 91:109-24; PMID:25703189; http://dx.doi.org/ 10.1016/j.addr.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SY, Weiner G. Complement and cellular cytotoxicity in antibody therapy of cancer. Expert opinion on biological therapy 2008; 8:759-68; PMID:18476787; http://dx.doi.org/ 10.1517/14712598.8.6.759 [DOI] [PubMed] [Google Scholar]
- 25.Schuster M, Jost W, Mudde GC, Wiederkum S, Schwager C, Janzek E, Altmann F, Stadlmann J, Stemmer C, Gorr G. In vivo glyco-engineered antibody with improved lytic potential produced by an innovative non-mammalian expression system. Biotechnol J 2007; 2:700-8; PMID:17427997; http://dx.doi.org/ 10.1002/biot.200600255 [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Bulseco GG, Sun J. Effect of posttranslational modifications on the thermal stability of a recombinant monoclonal antibody. Immunol Lett 2006; 106:144-53; PMID:16831470; http://dx.doi.org/23386066 10.1016/j.imlet.2006.05.011 [DOI] [PubMed] [Google Scholar]
- 27.Parsons K, Bernhardt B, Strickland B. Targeted immunotherapy for high-risk neuroblastoma–the role of monoclonal antibodies. Ann Pharmacother 2013; 47:210-8; PMID:23386066; http://dx.doi.org/ 10.1345/aph.1R353 [DOI] [PubMed] [Google Scholar]
- 28.Du Y, Walsh A, Ehrick R, Xu W, May K, Liu H. Chromatographic analysis of the acidic and basic species of recombinant monoclonal antibodies. mAbs 2012; 4:578-85; PMID:22820257; http://dx.doi.org/ 10.4161/mabs.21328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang XR, Song A, Bergelson S, Arroll T, Parekh B, May K, Chung S, Strouse R, Mire-Sluis A, Schenerman M. Advances in the assessment and control of the effector functions of therapeutic antibodies. Nat Rev Drug Discov 2011; 10:101-11; PMID:21283105; http://dx.doi.org/ 10.1038/nrd3365 [DOI] [PubMed] [Google Scholar]
- 30.Kanda Y, Yamada T, Mori K, Okazaki A, Inoue M, Kitajima-Miyama K, Kuni-Kamochi R, Nakano R, Yano K, Kakita S, et al.. Comparison of biological activity among nonfucosylated therapeutic IgG1 antibodies with three different N-linked Fc oligosaccharides: the high-mannose, hybrid, and complex types. Glycobiology 2007; 17:104-18; PMID:17012310; http://dx.doi.org/ 10.1093/glycob/cwl057 [DOI] [PubMed] [Google Scholar]
- 31.Hodoniczky J, Zheng YZ, James DC. Control of recombinant monoclonal antibody effector functions by Fc N-glycan remodeling in vitro. Biotechnol Prog 2005; 21:1644-52; PMID:16321047; http://dx.doi.org/ 10.1021/bp050228w [DOI] [PubMed] [Google Scholar]
- 32.Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics 2010; 9:1716-28; PMID:20103567; http://dx.doi.org/ 10.1074/mcp.M900540-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci 2009; 18:247-51; PMID:19177368; http://dx.doi.org/ 10.1002/pro.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Ponniah G, Zhang HM, Nowak C, Neill A, Gonzalez-Lopez N, Patel R, Cheng G, Kita AZ, Andrien B. In vitro and in vivo modifications of recombinant and human IgG antibodies. MAbs 2014; 6:1145-54; PMID:25517300; http://dx.doi.org/ 10.4161/mabs.29883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Y, Fest S, Kunert R, Katinger H, Pistoia V, Michon J, Lewis G, Ladenstein R, Lode HN. Anti-neuroblastoma effect of ch14.18 antibody produced in CHO cells is mediated by NK-cells in mice. Mol Immunol 2005; 42:1311-9; PMID:15950727; http://dx.doi.org/ 10.1016/j.molimm.2004.12.018 [DOI] [PubMed] [Google Scholar]
- 36.Westermeier, R. Electrophoresis in Practice: A Guide to Methods and Applications of DNA and Protein Separations. 4th edn. Weinheim, Germany: Wiley; 2005; http://dx.doi.org/ 10.1002/3527603468 [DOI] [Google Scholar]
- 37.Gorg A, Obermaier C, Boguth G, Csordas A, Diaz JJ, Madjar JJ. Very alkaline immobilized pH gradients for two-dimensional electrophoresis of ribosomal and nuclear proteins. Electrophoresis 1997; 18:328-37; PMID:9150910; http://dx.doi.org/ 10.1002/elps.1150180306 [DOI] [PubMed] [Google Scholar]
- 38.Ho SC, Bardor M, Feng H, Mariati, Tong YW, Song Z, Yap MG, Yang Y. IRES-mediated Tricistronic vectors for enhancing generation of high monoclonal antibody expressing CHO cell lines. J Biotechnol 2012; 157:130-9; PMID:22024589; http://dx.doi.org/ 10.1016/j.jbiotec.2011.09.023 [DOI] [PubMed] [Google Scholar]
- 39.Shahrokh Z, Royle L, Saldova R, Bones J, Abrahams JL, Artemenko NV, Flatman S, Davies M, Baycroft A, Sehgal S, et al.. Erythropoietin produced in a human cell line (Dynepo) has significant differences in glycosylation compared with erythropoietins produced in CHO cell lines. Mol Pharm 2011; 8:286-96; PMID:21138277; http://dx.doi.org/ 10.1021/mp100353a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


