Summary
The anaphase-promoting complex or cyclosome (APC/C) is a ubiquitin ligase that polyubiquitinates specific substrates at precise times in the cell cycle, thereby triggering the events of late mitosis in a strict order. The robust substrate specificity of the APC/C prevents the potentially deleterious degradation of non-APC/C substrates, and also averts the cell cycle errors and genomic instability that could result from mistimed degradation of APC/C targets. The APC/C recognizes short linear sequence motifs, or degrons, on its substrates. The specific and timely modification and degradation of APC/C substrates is likely to be modulated by variations in degron sequence and context. We discuss the extensive affinity, specificity and selectivity determinants encoded in APC/C degrons, and we describe some of the extrinsic mechanisms that control APC/C-substrate recognition. As an archetype for protein motif-driven regulation of cell function, the APC/C-substrate interaction provides insights into the general properties of post-translational regulatory systems.
Introduction
The cell division cycle is a complex sequence of events, each of which must be initiated at the appropriate time. In the eukaryotic cell, these events are controlled by an oscillating regulatory system driven by changes in protein phosphorylation and ubiquitin-dependent protein degradation. Central to this system are the cyclin-dependent protein kinases (Cdks) and a ubiquitin-protein ligase called the anaphase-promoting complex or cyclosome (APC/C) (Morgan, 2007).
The APC/C, like other E3 ubiquitin ligases of the RING family, serves as a binding platform that brings together a specific substrate and an E2 co-enzyme, resulting in polyubiquitination and degradation of the substrate by the 26S proteasome. APC/C activity oscillates during the cell cycle, primarily due to changes in its association with an activator subunit, Cdc20 or Cdh1 (Pines, 2011; Primorac and Musacchio, 2013; Sivakumar and Gorbsky, 2015). The activator subunit serves as the primary site of APC/C-substrate interaction: binding pockets on its surface interact with short linear sequence motifs, called degrons, on the substrate (Glotzer et al., 1991; Pfleger and Kirschner, 2000; He et al., 2013; Primorac and Musacchio, 2013; Lu et al., 2014; Di Fiore et al., 2015). In early mitosis, rising Cdc20 levels and Cdk-mediated APC/C phosphorylation initiate the formation of active APC/CCdc20, which drives the destruction of the mitotic cyclins, the separase inhibitor securin, and other proteins, thereby triggering chromosome segregation in anaphase (Pines, 2011; Primorac and Musacchio, 2013; Sivakumar and Gorbsky, 2015). In early anaphase, Cdk inactivation leads to activation of Cdh1, and Cdc20 is degraded (Pines, 2011; Sivakumar and Gorbsky, 2015). APC/CCdh1 then governs the final stages of cell division and continues to function throughout G1 to suppress cyclin-Cdk activity. Deactivation of APC/CCdh1 at the end of G1 enables cyclin accumulation and progression into S phase (Pines, 2011).
The APC/C promotes the specific degradation of tens of substrates in a precise order (Min and Lindon, 2012). The APC/C modifies substrates with high specificity, recognizing specific degron-containing proteins in the crowded environment of the cell, and high selectivity, differentiating between APC/C targets to establish a hierarchy of substrates. The conserved ordering of substrate destruction suggests that degrading an APC/C target at the wrong time can have a profound effect on the fidelity of cell cycle progression. Substrate ordering is likely to depend on tight regulation of APC/C selectivity, but the mechanisms underlying this selectivity, and thus substrate ordering, are not clear. Recent advances have begun to clarify the determinants controlling the specific recruitment of APC/C substrates and the mechanisms that define the window of instability for these proteins. These findings suggest that multiple distinct mechanisms, both within a substrate and across all substrates, collaborate to trigger the degradation of the correct targets in the correct location at the correct time.
The general features of APC/C structure, function, and regulation have been well described in numerous review articles (Pines, 2011; Min and Lindon, 2012; Primorac and Musacchio, 2013; Chang and Barford, 2014; Sivakumar and Gorbsky, 2015). Here, we focus on APC/C substrates, with an emphasis on the mechanisms responsible for their specific and timely recognition by the APC/C. We describe some common misconceptions about degron motif composition, and we highlight the precise level of control that motif-binding systems can encode.
Substrate recognition by the APC/C
APC/C-mediated polyubiquitination governs the stability of over 100 distinct proteins across a range of eukaryotic species (Table 1; see a full list at http://slim.ucd.ie/apc/). As the APC/C promotes irreversible degradation of its targets, it is important that these targets are recognized with high specificity. Most APC/C substrates are recruited by interaction with the seven-blade β-propeller WD40 repeat domain in the C-terminal half of the APC/C activator subunit (He et al., 2013). This domain contains binding pockets that recognize APC/C degrons, of which there are three major types: the destruction box (D box) (Glotzer et al., 1991), the KEN box (Pfleger and Kirschner, 2000) and the ABBA motif (Burton et al., 2011; Lischetti et al., 2014; Lu et al., 2014; Di Fiore et al., 2015; Diaz-Martinez et al., 2015) (Figure 1). A fourth sequence, the CRY motif, also binds the WD40 domain (Alfieri et al., 2016; Yamaguchi et al., 2016), but only a single instance (in Cdc20) has been identified to date (Reis et al., 2006). Over two decades of biochemical studies have defined the sequence preferences for each of the major degrons, and recent structural studies of these degrons bound to the APC/C explain the preferred amino acids at each position (He et al., 2013; Chang et al., 2015; Tian et al., 2012)(Figures 1, 2).
Table 1.
Representative set of APC/C substrates. For a complete list of APC/C substrates and degrons see http://slim.ucd.ie/apc/.
| Activator | Degradation timing | Protein | Regulatory mechanism | Degrons |
|---|---|---|---|---|
| Budding yeast (Saccharomyces cerevisiae) | ||||
| Cdc20 | Pre-metaphase | S-phase cyclin 5 (Clb5) | Degron co-operativity | KEN-like box, D box, ABBA motif, Cks1 co-factor |
| Metaphase, Anaphase | M-phase cyclin 2 (Clb2) | KEN-like box, D box, Cks1 co-factor | ||
| Metaphase | Securin (Pds1) | Phosphorylation | KEN box, D box | |
| Metaphase | Cdc7 kinase regulatory subunit Dbf4 | Phosphorylation | D box | |
| Metaphase | Shugoshin (Sgo1) | D box | ||
| Metaphase | APC/C-Cdh1 modulator 1 (Acm1) | Degron hiding | D box | |
| Cdh1 | Anaphase | APC/C activator protein Cdc20 | KEN-like box, D box | |
| Anaphase | Polo-like protein kinase Cdc5 | KEN box, D boxes | ||
| Anaphase | Filament protein Fin1 | D box | ||
| Anaphase | Kinesin-like protein Kip1 | |||
| Cytokinesis | Contractile ring protein Iqg1 | |||
| Human (Homo sapiens) | ||||
| Cdc20 | Prometaphase | Cyclin A2 (CCNA2) | Degron co-operativity | KEN-like box, D box, ABBA motif, Cks1 co-factor |
| Prometaphase | Protein kinase Nek2 (NEK2) | Degron co-operativity | D box, KEN box, MR tail | |
| Metaphase | Mitotic cyclin B1 (CCNB1) | Degron co-operativity | KEN-like box, D box, Cks1 co-factor | |
| Metaphase | Kinesin-like protein Kif22 (KIF22) | KEN box | ||
| Metaphase | Securin (PTTG1) | Phosphorylation | KEN box, D box | |
| Metaphase | Shugoshin-like 1 (SGOL1) | KEN box, D box | ||
| Cdh1 | Anaphase | Mitotic cyclin B3 (CCNB3) | D box | |
| Anaphase | APC/C activator protein Cdc20 | KEN box | ||
| Anaphase | Kinesin-like protein Kifc1 (KIFC1) | Phosphorylation | D box | |
| Anaphase | Protein kinase Plk1 (PLK1) | D box | ||
| Mitotic exit/G1 | Actin-binding protein anillin (ANLN) | D box | ||
| Mitotic exit/G1 | Aurora kinase B (AURKB) | D box, KEN box | ||
| G1 | Origin-binding protein Cdc6 (CDC6) | Phosphorylation | KEN box, D box | |
| G1 | Geminin (GMNN) | D box | ||
| G1 | S-phase kinase-associated protein 2 (SKP2) | Phosphorylation | D box | |
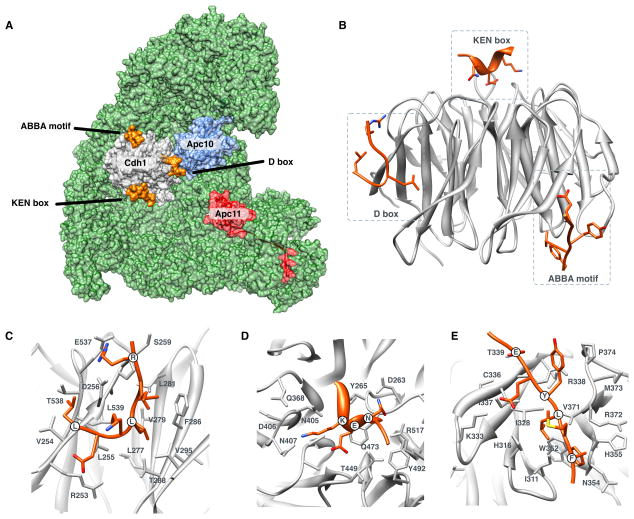
Figure 1. Structure and specificity of the WD40 domain of the APC/C activators.
(A) Structure of the Cdh1 activator subunit WD40 domain (gray) bound to APC/C (green) (He et al., 2013; Chang et al., 2015). The Apc10 subunit (blue), the Apc11 RING subunit (red), and the positions of the motif-binding pockets (with degrons in orange) are also shown. (B) The WD40 domain of Cdh1 with the three degron-binding pockets occupied by the D box, KEN box and ABBA motif. (C) The D box-binding pocket. (D) The KEN box-binding pocket. (E) The ABBA motif-binding pocket.
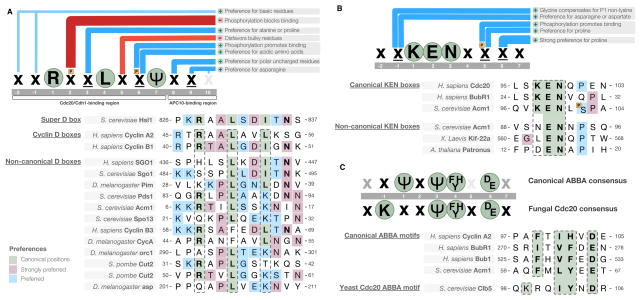
Figure 2. Sequence preferences of the APC/C degron-binding pockets.
(A) Preference of the D box-binding pocket and representative experimentally validated D box instances. “ψ” signifies a leucine, isoleucine or valine residue, “x” signifies any residue (unless certain residues are disfavored), underlined “x” signifies any residue but with strong preferences for particular residues based on information from characterized degrons, orange “P” signifies phosphorylation of the position, and residues in green circles are the consensus residues of the degron (those residues that are present in the majority of instances). Blue bars indicate preferences at a position. Red bars indicate disfavored residues at a position. (B) Preference of the KEN box-binding pocket and representative experimentally validated KEN box instances. (C) Preference of the ABBA motif-binding pocket, showing the divergent consensus of the fungal Cdc20 and representative experimentally validated ABBA motif instances (see also Figure 5 below). Few validated ABBA motifs are available, and thus the consensus may change in the future. Strong candidate motifs in BubR1 suggest that the canonical motif will allow a wider range of hydrophobic amino acids in place of the position +1 phenylalanine, and position 3 will allow proline. All degron instances used for the construction of the preferences in this figure can be found at http://slim.ucd.ie/apc/.
The D box-binding pocket is a composite binding site involving the activator WD40 domain and a second site on the Apc10/Doc1 subunit of the APC/C (Buschhorn et al., 2011; da Fonseca et al., 2011) (Figure 1A). The arginine and leucine of the consensus RxxL sequence contact an acidic patch and an aliphatic pocket on the activator surface, respectively (Figure 1C)(He et al., 2013). After the RxxL sequence, the D box adopts a tight turn that imposes strong constraints on the allowed amino acids in residues directly flanking the leucine, explaining the preference for proline and alanine in position +3 (the 3rd position of the motif) and for small residues in position +5 (Figure 2A). A hydrogen bond between an acidic position of the D box and an arginine on the activator surface explains the preference at position +6. A strong preference for hydrophobic residues at position +7 is explained by contacts with a non-polar surface of the activator. The remaining preferences for serine, threonine and asparagine at position +8, and the strong asparagine preference at position +9, reflect complementarity to the binding surface of the Apc10/Doc1 subunit (Chang et al., 2015).
The KEN box-binding pocket is situated on the top surface of the activator WD40 domain, in a depression at the center of the β-propeller (Chao et al., 2012; Tian et al., 2012; He et al., 2013)(Figure 1B, D). The bound KEN box peptide, named after its consensus sequence, forms an underwound helix that executes a tight turn in the pocket around the central glutamate residue. The charged residues of the consensus contact a highly conserved complementary binding pocket on the activator surface. The pocket also has a preference for aspartate or an asparagine in position −1 (Figure 2B). The preference for a C-terminal proline is a structural feature that breaks the helix and directs the exiting peptide away from the domain surface (Chao et al., 2012; He et al., 2013).
The ABBA motif-binding pocket recognizes peptides with a consensus of [ILVF]x[ILMVP][FHY]x[DE] (Figure 1E, 2C)(Di Fiore et al., 2015). The motif occupies a hydrophobic groove between blades 2 and 3 on the opposite surface of the WD40 domain from the KEN box-binding pocket (He et al., 2013)(Figure 1B). Position +1 and +3 are deeply buried, position +4 rests against blade 3 and position +6 reaches out of the pocket to contact the side of the WD40 repeat. The specificity of the budding yeast Cdc20 ABBA motif-binding pocket has diverged, losing the preference for the position +1 hydrophobic residue and instead preferring a position −1 basic residue (Lu et al., 2014). This change reflects a complementary change in the binding pocket, as discussed later in this article (see Figure 5 below).
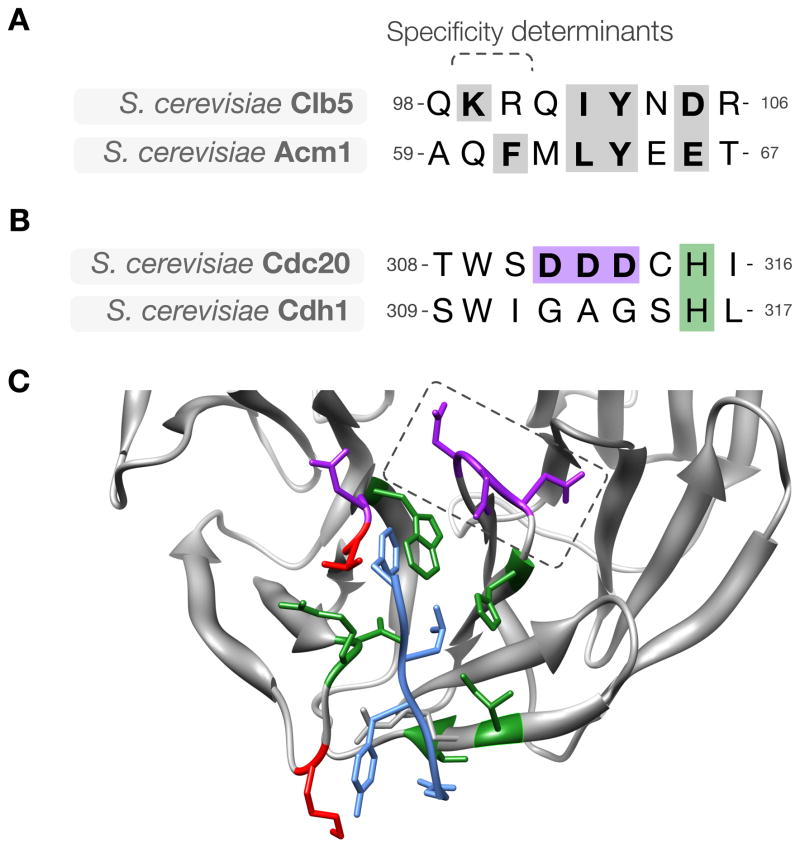
Figure 5. Divergence of ABBA motif binding specificity in evolution.
(A) Alignment of the ABBA motif in budding yeast Clb5 and Acm1, showing key conserved residues (gray). (B) The sequence of the activator region containing the specificity-determining loop in the ABBA motif-binding pocket. The loop is GAG in Cdh1 and DDD in Cdc20. (C) Modeled structure of budding yeast Cdc20 (gray) with the ABBA motif from Acm1 (blue). Pocket residues on Cdc20 are colored based on conservation between budding Cdh1 and Cdc20: green indicates residues conserved between Cdh1 and Cdc20, red indicates residues that are not conserved, and purple indicates Cdc20-specific acidic residues, including the specificity-determining acidic loop (dashed box).
Motif-binding pockets have the ability to bind a vast number of potential peptides with a broad range of binding affinities. Thus, the concept of a conserved consensus can be misleading and does not accurately describe the sequence preferences of the pocket (Figure 2). The core degron consensus, such as the RxxL of the D box, describes the residues with the highest complementarity to the core-binding pocket, and these residues are shared across the majority of peptides binding to the motif-binding pocket. However, degron-containing peptides are highly divergent outside these key residues, and this variability is likely to create major differences in specificity, affinity and selectivity of activator binding (Van Roey et al., 2014). Furthermore, many degrons vary from the consensus even for strongly preferred positions (Figure 2). Mutation of the canonical D box position 1 arginine is sufficient to stabilize many substrates (King et al., 1996), and yet the D boxes of shugoshin proteins (Karamysheva et al., 2009) and the D. melanogaster securin homolog Pimples (pim) (Leismann and Lehner, 2003) lack arginine at this position. Similarly, in budding yeast, securin (Pds1) (Hilioti et al., 2001) and Spo13 D boxes (Sullivan and Morgan, 2007), and the N-terminal D box of the Cdh1 inhibitor Acm1 (Enquist-Newman et al., 2008; He et al., 2013), have a lysine in the commonly observed +7 hydrophobic position. Structural information suggests that a position +7 lysine can contact the acidic residues of the position +1 binding site and position +7 non-polar binding surface, with their charged and aliphatic groups, respectively (He et al., 2013). Even the most conserved D box consensus position, the position +4 leucine, is variable. D. melanogaster cyclin A (Ramachandran et al., 2007) and H. sapiens cyclin B3 (Nguyen et al., 2002) both contain phenylalanine at this position. Several KEN box degrons also diverge from the classical consensus (Figure 2B). KEN boxes lacking the position +1 lysine have been identified, and asparagine and aspartate are also allowed (Cromer et al., 2013; He et al., 2013). In the Xenopus kinesin-like protein KIF22 (XKid) and several plant proteins, a glycine in the −1 position has been shown to compensate for the absence of a lysine in position +1 (Castro et al., 2003; Heyman et al., 2011; Cromer et al., 2012; He et al., 2013).
Divergence from degron consensus has led to some confusion, as novel classes of degrons were proposed for sequence motifs that are actually variations of D boxes and KEN boxes (Figure 2). These include the O box of Drosophila origin recognition complex subunit 1 (orc1) (Araki et al., 2005), Drosophila abnormal spindle (asp), and S. pombe securin (Cut2) (Funabiki et al., 1997; Araki et al., 2005; He et al., 2013), which are non-canonical D box degrons (Figure 2A). In budding yeast, several APC/C substrates with non-canonical degrons are stabilized by mutations in the D-box-binding pocket of the activator (Qin et al., 2016). Interestingly, most non-canonical degrons appear to compensate for the lack of consensus positions by using preferred residues in multiple other peptide positions that contact the activator surface (Figure 2) or are part of larger multi-degron interfaces (Funabiki et al., 1997; Hildebrandt and Hoyt, 2001; Hilioti et al., 2001; Heyman et al., 2011; Cromer et al., 2012; Cromer et al., 2013). For the remaining uncharacterized degrons (see http://slim.ucd.ie/apc/), it is possible that the substrate does not bind directly to the ligase; instead, uncharacterized stability-determining regions may promote scaffolding interactions with a degron-containing binding partner to indirectly recruit the APC/C.
In general, APC/C activators interact with degrons in substrate regions that are predicted to be both flexible and accessible (Guharoy et al., 2016). Thus, all high-confidence degrons (those backed by multiple complementary sources of validation) are found in the unstructured, intrinsically disordered regions of proteins. Several proposed APC/C degrons in the literature were later shown to be structurally buried; these include the D boxes of Ski-like protein (SKIL) and Aurora A and the KEN box of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3). These sites should be treated with caution without further investigation. Flexibility is highly important, as the peptides must adopt a strictly defined conformation when bound to the activator (He et al., 2013). Furthermore, the flexibility of the degron-containing polypeptide allows multiple lysines in nearby regions, or on an attached ubiquitin, to efficiently attack the E2-ubiquitin conjugate (Guharoy et al., 2016). The requirement for accessibility is self-explanatory, as the degron must be available for recognition by the APC/C. It is important to emphasize that most peptides matching a simplified APC/C degron consensus are unlikely to be recognized by the APC/C. It was suggested recently, for example, that ~70% of human proteins contain a D or KEN box-containing peptide (defined simply as RxxL or KEN)(Lu et al., 2015b), but the vast majority of these peptides are unlikely to be accessible in intrinsically disordered protein regions or co-localize with the APC/C. Most of the remaining peptides will not fit the complex sequence preferences of the degron-binding pockets.
Some APC/C substrates appear to contain multiple D box-like sequence motifs. In many cases, such as budding yeast Dbf4 (Lu et al., 2014) and human Sgo1 (Karamysheva et al., 2009), it is likely that only one of these motifs is a functional degron. Alternatively, multiple D boxes might provide fine-tuning of substrate affinity or specificity for different activators. These possibilities have not been explored systematically for any substrate.
Substrate ordering by the APC/C
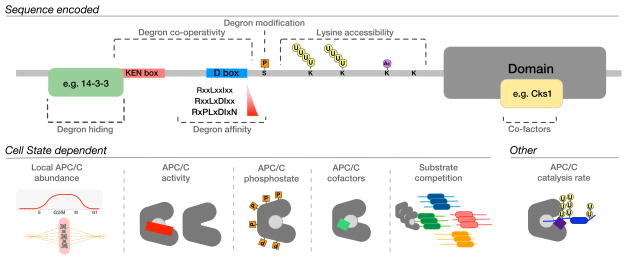
A remarkable feature of substrate recognition by the APC/C is the ability of a single holoenzyme, and two substrate recognition subunits, to modify a wide array of targets in a defined order (Table 1; see also http://slim.ucd.ie/apc/ for further discussion of the ordering of APC/C substrates). The ordering of substrate degradation is important for the fidelity of the cell cycle and is remarkably robust. It is now clear that no single mechanism controls ordering; indeed, substrate recruitment by the APC/C is controlled by a variety of cooperating mechanisms encoded by the degron-containing peptide, co-operativity between degrons, degron context, local activity of the APC/C, and competition among substrates (Figure 3).
Figure 3. General mechanisms of substrate recognition by the APC/C.
Intrinsic factors, encoded in the protein sequence, and extrinsic factors, encoded in the cell state, regulate substrate stability and are likely to determine the timing of substrate degradation.
Substrate binding affinity of the APC/C activators
Substrates typically bind with sufficient affinity to enable repeated ubiquitination during a single binding interaction (Carroll and Morgan, 2002; Rape et al., 2006). The binding affinity (or, more specifically, the dissociation rate) determines the residence time of a substrate on the APC/C, and, together with the enzyme catalytic rate, determines the number of ubiquitins attached per binding event (processivity). Several studies of other motif-binding domain families indicate that degeneracy of motif-binding pocket specificity permits peptides recognized by that pocket to exhibit a wide range of affinities (Roy et al., 2007; Hertz et al., 2016). In the case of the APC/C, the affinity of an individual degron has not been directly determined. The so-called “super D box” of budding yeast Hsl1 appears to be the most high-affinity D box tested to date, based on competition-based assays (Burton and Solomon, 2001; Frye et al., 2013). The D box of metazoan cyclin A appears to be a weak D box as, unlike the D box of cyclin B, it is not sufficient as a transplantable degradation signal (King et al., 1996; Klotzbucher et al., 1996). These observations can be rationalized by comparing the similarity of these motifs to the optimal D box consensus (Figure 2A). Drawing analogies to motifs with similar properties (e.g. the PP2AB56 LxxIxE docking motif, which physicochemically resembles the D box) (Hertz et al., 2016), it is likely that single APC/C degrons bind with dissociation constants (KD) in the low μM range.
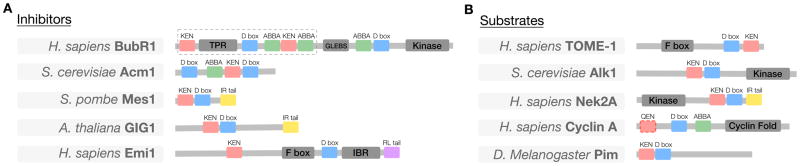
The presence of multiple degrons on the same substrate is likely to increase effective affinity to the low nM range (Bu et al., 1995). The structure of Acm1 bound to Cdh1 revealed that the D box-, KEN box- and ABBA motif-binding pockets of the activator WD40 domain can be occupied simultaneously (Chao et al., 2012; He et al., 2013). The enhanced avidity that results from multivalent binding is most easily observed in the pseudosubstrate inhibitors of the APC/C (Figure 4A). However, several APC/C substrates also illustrate the power of multivalent interactions (Figure 4B). These multivalent sites often appear to exist in close proximity, perhaps reflecting the spacing of the degron-binding pockets on the activator. Human cyclin A and yeast Clb5 each contain a D box, an ABBA motif, and a potential degenerate KEN box, and they also employ the Cdk-associated subunit Cks1 to provide additional binding affinity for the APC/C (Lu et al., 2014; Di Fiore et al., 2015). Another early mitotic target, human Nek2A, contains a KEN box and a D Box, and also uses a C-terminal MR tail motif to interact with a non-activator site on the APC/C (Hames et al., 2001; Sedgwick et al., 2013). These multiple interaction surfaces in early substrates allow them to compete effectively with the mitotic checkpoint complex (MCC), thereby allowing degradation in the presence of an active spindle assembly checkpoint (SAC) (Lu et al., 2014; Di Fiore et al., 2015). Even if the SAC is inactivated experimentally, these proteins are degraded earlier than the metaphase targets securin and M cyclins. Mutation of the ABBA motif in Clb5, together with mutations that block recruitment of Cks1, results in Clb5 destruction at the same time as securin destruction (Lu et al., 2014). Conversely, the addition of an ABBA motif to cyclin B1 can increase its rate of degradation, although cyclin B1ABBA cannot compete with the SAC (Di Fiore et al., 2015). These observations suggest that degron co-operativity contributes to substrate degradation ordering.
Figure 4. Modular architecture underlying the interaction of inhibitors and substrates with the APC/C.
(A) High-affinity pseudosubstrate inhibitors are commonly used to regulate enzymes. Numerous APC/C pseudosubstrate inhibitors have been identified, including: the MCC subunit Mad3/BubR1 (dashed box denotes the conserved region shared by all Mad3/BubR1-like proteins); the budding yeast Cdh1 inhibitor Acm1; the fission yeast protein Mes1; and the Arabidopsis proteins POLYCHOME (PYM) and GIGAS CELL1 (GIG1). The human APC/C regulator Emi1 also occupies the D box-binding pocket, although the major inhibitory mechanism is through regulation of E2 binding (Frye et al., 2013; Wang and Kirschner, 2013). Each of these APC/C pseudosubstrate inhibitors employs multiple distinct APC/C degrons to bind with high affinity to the APC/C activator, thereby competitively inhibiting the binding of APC/C substrates. (B) Representative examples of the modular architecture of likely multivalent APC/C substrates: Trigger of mitotic entry protein-1 (TOME-1), protein kinase haspin homolog Alk1, protein kinase Nek2, cyclin A and Pimples protein (Pim/securin). Details of the degron instances used in this figure can be found at http://slim.ucd.ie/apc/.
It is conceivable that high-affinity substrates act as competitive inhibitors of weaker binding substrates, thereby setting up the timing of substrate degradation (Kamenz et al., 2015; Lu et al., 2015a). The importance of competition to substrate ordering is largely untested. Early targets do appear to have multipartite APC/C degrons, suggesting that they are strong binders (Figure 4B). However, increasing the levels of Clb5, which should result in increased competition, does not result in delayed degradation of securin in budding yeast, suggesting that APC/CCdc20 activity is not limiting and that the ordering of Clb5 and securin destruction depends on intrinsic differences in their affinities, not on competition (Lu et al., 2015a). On the other hand, overexpression of the B-type cyclin Cdc13 delays the onset of securin (Cut2) destruction in the fission yeast S. pombe, arguing that these two substrates compete for limited APC/CCdc20 in this species (Kamenz et al., 2015).
Degrons and the rate of ubiquitination
The timing of APC/C substrate degradation is determined largely by the accumulation of sufficient ubiquitins to promote proteasomal targeting. The time taken to complete this process depends on the occupancy time of the substrate on the APC/C, and on the relative rates of substrate ubiquitination and deubiquitination. The catalytic rates of the E2s that work with the APC/C control the rates of polyubiquitin chain initiation and elongation (Rodrigo-Brenni and Morgan, 2007; Jin et al., 2008). These catalytic rates are often thought to be the same for different substrates. However, recent evidence suggests that activator binding dramatically increases the binding and catalytic rate of the E2, and degron binding to the activator is required for full E2 stimulation (Chang et al., 2014; Van Voorhis and Morgan, 2014). Different degrons can influence the rate of catalysis (Van Voorhis and Morgan, 2014; Lu et al., 2015a). Thus, specific degrons can modulate both the binding strength for the APC/C and the rate of ubiquitination, and both of these effects are likely to influence the timing of substrate degradation (Lu et al., 2015a).
Substrate specificity of APC/C activators
The two major activators of the APC/C are temporally regulated so that they occupy the APC/C at distinct times in the cell cycle. This sequential activation of two distinct APC/C isoforms led to the hypothesis that the ordering of substrate degradation is due in part to the different specificities of Cdc20 and Cdh1 (Visintin et al., 1997; Pfleger and Kirschner, 2000). The KEN- and D box-binding pockets of Cdc20 and Cdh1 are highly conserved and are expected to have very similar specificities for the core residues of substrate degrons. However, evolutionary refinement of activator binding surfaces outside the core of the degron-binding pocket could generate subtle specificity determinants that allow discrimination between different degron-containing peptides.
Little is known about the specificity determinants of the APC/C activators. APC/CCdc20 has a narrow substrate range in vivo (with only a handful of identified substrates), whereas APC/CCdh1 targets a much larger group of substrates that seems to include all substrates of APC/CCdc20. Cdc20 is required for the initiation of anaphase, and its essential function is to promote degradation of just two substrates: securin and cyclin (Thornton and Toczyski, 2003). Cdh1 is not essential for cell viability in any organism tested. Thus, Cdc20 interacts with the key substrates whose destruction is required for anaphase and mitotic exit, and Cdh1 tidies up afterwards, keeping Cdc20 targets and numerous other proteins at low levels, thereby stabilizing the G1 state. The major reason for the presence of two activators might not be their differing substrate specificities but rather their distinct regulation, as we describe later in this article. Nevertheless, activator specificity is responsible in part for substrate ordering: a small group of Cdc20-specific targets is degraded earlier than the numerous targets recognized only by Cdh1. The precise contribution of activator binding preferences to this specificity is poorly understood.
Degron regulation by post-translational modification
Post-translational modification of motif-containing regions is a widely used mechanism to integrate cell state information into regulatory decision-making (Van Roey et al., 2012). The ordered inactivation of cell cycle kinases by the APC/C provides the cell with information regarding the current stage of mitosis, and this temporal regulation of kinase activity can help order the timing of degradation of kinase targets. For example, Cdk1-mediated phosphorylation of a serine at position +2 of the D box stabilizes KIF1C and Dbf4, while Aurora kinase A-mediated phosphorylation at position +3 of the D box stabilizes geminin (Rape et al., 2006; Tsunematsu et al., 2013; Lu et al., 2014). Dephosphorylation of these sites promotes degradation of the protein. In other cases, degron phosphorylation can stimulate ubiquitination by the APC/C, as seen in the case of human securin, in which phosphorylation at position +6 of the D box enhances degradation (Hellmuth et al., 2014). The opposing outcomes of phosphorylation at different positions in the D box can be explained by the sequence preferences of D box degrons (Figure 2A): position +6 strongly prefers an acidic residue, and a phosphate at this position also fulfills this requirement and is therefore likely to increase the affinity of the D box for the activator; conversely, phosphorylation of position +2 inhibits binding. Phosphorylation can also modulate KEN box function. Cdk1-mediated phosphorylation near the KEN box of Cdc6 results in stabilization (Mailand and Diffley, 2005). Furthermore, the structure of the KEN box of Acm1 bound to Cdh1 included a phosphorylated serine residue that stabilized the kinked structure of the bound peptide (Hall et al., 2008; He et al., 2013). Substrate stability can also be influenced by phosphorylation of residues outside the degron, as seen in the destabilizing phosphorylation of Mcl-1, which promotes degradation (Harley et al., 2010), and the stabilizing phosphorylation of budding yeast securin (Holt et al., 2008; Lu et al., 2014). The mode of action in these cases is not known.
Regulation by motif hiding
Several targets of the APC/C are protected from degradation by association with proteins that block access of the APC/C to degron motifs. TPX2 protects Aurora A from APC/CCdh1 at the spindle, thereby linking activity, localization, and stability of Aurora A (Giubettini et al., 2011). HURP and NuSAP are protected from the APC/C through their association with importin subunit beta-1, an interaction relieved by the nuclear GTP-binding protein Ran (Song and Rape, 2010). Modification of substrates can also regulate degron access by driving the recruitment of a binding partner that interferes with activator binding. For example, phosphorylation of Acm1 by Cdk1 promotes recruitment of the 14-3-3 family members Bmh1 and Bmh2, thereby stabilizing Acm1 (Enquist-Newman et al., 2008; Hall et al., 2008; Ostapenko et al., 2008). Deactivation of Cdk1 and activation of the phosphatase Cdc14 are required to inactivate the phosphodependent 14-3-3 binding motif to allow Acm1 degradation (Hall et al., 2008). Similarly, NIPA is protected from degradation by a phosphodependent interaction with Skp1 (Klitzing et al., 2011).
Lysine accessibility
APC/C substrates are typically ubiquitinated at multiple lysines in disordered regions surrounding the activator-binding degron sequences (King et al., 1996; Ramachandran et al., 2007). No single ubiquitin-accepting lysine has been shown to be required for the degradation of any APC/C substrate, and multiple lysine mutations are required to stabilize a substrate. Nevertheless, lysine position is likely to be important: the degron-binding pockets of the activator are 20–40Å from the active site of the E2-ubiquitin conjugate, and ubiquitination is therefore likely to occur only at lysines located beyond this distance from degrons – about 5–10 residues in an unfolded polypeptide chain (Chang et al., 2015; Brown et al., 2016). The contribution of lysine density to substrate degradation is poorly understood, apart from the strict requirement for the presence of an accessible lysine for ubiquitin attachment. Furthermore, given the modular architecture and flexibility of disordered regions, acceptor lysines and degrons may not need to be in the same region of the protein. Sequence context of the ubiquitinated lysine might also contribute to the rate of substrate modification (Williamson et al., 2011; Min et al., 2013; Mattiroli and Sixma, 2014). Given the numerous enzymes that can modify lysines, blocking lysine accessibility through competitive modification of sites can be a powerful mechanism to control substrate degradation.
Activation state and local abundance of the APC/C
Several functionally distinct forms of inactive and active APC/C exist during the cell cycle, often simultaneously at distinct subcellular localizations (Sivakumar and Gorbsky, 2015). Inhibitory proteins and phosphorylation state modulate the activity of specific APC/C isoforms and thereby govern substrate ordering. In early mitosis, activation of the SAC leads to specific inhibition of APC/CCdc20 by the MCC, which contains multiple sequence motifs that block the degron-binding sites on Cdc20 (Alfieri et al., 2016; Yamaguchi et al., 2016). Some mammalian substrates, such as cyclin A, are degraded in the presence of the MCC, suggesting that resistance to the SAC determines the ordering of these early substrates. However, human cyclin A and securin are degraded in the correct order even in the absence of a functional SAC (Collin et al., 2013). Similarly, in budding yeast, the timing of Clb5 degradation, which begins before the SAC has been satisfied, still occurs earlier than securin degradation in mutants lacking the SAC (Lu et al., 2014). Other inhibitory proteins, such as Emi1 in metazoans and Acm1 in fungi, specifically inhibit Cdh1, thereby controlling the window of APC/CCdh1 activity (Frye et al., 2013; He et al., 2013). In budding yeast meiosis I, a third activator subunit called Ama1 associates with the APC/C (Pesin and Orr-Weaver, 2008). By unknown mechanisms, a core APC/C subunit, Mnd2, specifically inhibits ubiquitination of Pds1, Clb5 and Sgo1 by APC/CAma1, while permitting the degradation of Clb1, Clb4 and Clb5 (Oelschlaegel et al., 2005). Modification of the core APC/C also governs activity and might control substrate specificity. Phosphorylation by mitotic kinases is required for APC/CCdc20 activation in early mitosis (Rudner and Murray, 2000; Fujimitsu et al., 2016; Qiao et al., 2016; Zhang et al., 2016), and the large number of phosphorylation sites on multiple APC/C subunits raises the possibility that some modifications have an impact on activity toward specific substrates (Kraft et al., 2003). For example, dephosphorylation of the Cks1 binding site of the APC/C is likely to reduce the binding strength of the cyclins (Lu et al., 2014; Di Fiore et al., 2015).
The local concentration of active APC/C, and co-localization with its substrates, might regulate target selectivity (Arnold et al., 2015). Several studies have suggested that APC/CCdh1 activity is spatially restricted (Jaquenoud et al., 2002; Acquaviva et al., 2004; Arnold et al., 2015), and numerous APC/C substrates require nuclear localization for efficient degradation (Hu et al., 2011; Arnold et al., 2015). Export of Cdh1 from the nucleus might contribute to APC/C inactivation, and Cdh1 targets can be stabilized by forcing their cytoplasmic localization (Jaquenoud et al., 2002; Kraft et al., 2003). Different pools of a substrate can be degraded in a localization-dependent manner; for example, the majority of Aurora B is degraded at mitotic exit but a subpopulation at the cell cortex is still present in G1 (Floyd et al., 2013). In both human and Drosophila, spindle-associated cyclin B1 is destroyed while the remaining cyclin B1 is stable (Clute and Pines, 1999; Huang and Raff, 1999).
Evolution of the APC/C regulatory network
Origin and evolutionary tuning of the APC/C activators
The APC/C is present in most eukaryotic proteomes, and it has been suggested that it was present in the last eukaryotic common ancestor (Eme et al., 2011). It is likely that early in the evolution of the APC/C, it employed a single activator subunit, but this ancestral activator duplicated in early eukaryotes. All currently sequenced genomes contain at least two activators, suggesting that a strong adaptive pressure led to retention of early and late activators, despite their similar intrinsic specificities. This is likely related to the distinct regulatory mechanisms of the two activators: APC/CCdc20 activity is promoted by Cdk-dependent phosphorylation in early mitosis, whereas APC/CCdh1 activity is inhibited by phosphorylation but then stimulated when Cdks are inactivated in late mitosis and G1 (Primorac and Musacchio, 2013; Sivakumar and Gorbsky, 2015). This regulatory scheme allows APC/C activity to rise in mitosis to trigger anaphase, and then remain high, despite the loss of Cdk1 activity, throughout G1. The human proteome contains two activators (Cdc20 and Cdh1) and an activator homolog Cdc20B that appears to have lost the sequence features associated with APC/C-related functionality. Several species have acquired additional functional activators through gene duplication events. For example, Drosophila and budding yeast both express a meiosis-specific activator protein: Cortex (Cort) and Ama1, respectively (Pesin and Orr-Weaver, 2008). Plants seem to have taken activator duplication to an extreme: the Arabidopsis genome encodes nine activators (Capron et al., 2003).
The order of emergence of the D box-, KEN box- and ABBA motif-binding pockets has yet to be revealed. They appear to have evolved contemporaneously, as they are present in most known activators. In those cases where a binding pocket is missing, it is likely that the pocket has been lost. For example, it appears that the ABBA motif-binding site has been lost from Cdh1 in animal cells. The ABBA motifs of cyclin A, Bub1 and BubR1 do not bind human Cdh1, and there is no documented case of ABBA motif binding to a metazoan Cdh1 (Di Fiore et al., 2015). In agreement with this observation, several features of the ABBA-binding pocket have diverged significantly in human Cdh1 (and also in the yeast meiotic activator Ama1). A similar observation can be made for the KEN-binding pocket of C. elegans Cdc20, which has also diverged on a sequence level from other activators. Consistent with this divergence, the otherwise omnipresent KEN boxes of BubR1 are not present in C. elegans. No experimental information yet supports these observations, but they point to an interesting rewiring of APC/C regulatory mechanisms in C. elegans.
With the notable exception of C. elegans, the D box- and KEN box-binding pockets are well conserved throughout eukaryotic evolution and their specificity is nearly identical across all organisms. Subtle evolutionary changes in the specificity of the ABBA motif-binding pocket appear to be more common. For example, the specificity of Cdc20 and Cdh1 for the ABBA motif has diverged in the fungal lineage (Lu et al., 2014; Di Fiore et al., 2015). In S. cerevisiae, Cdc20 displays a preference for ABBA motifs with a lysine at position −1, whereas Cdh1 has a preference for a hydrophobic residue at position +1 (Lu et al., 2014; Di Fiore et al., 2015) (Figure 2C, 5). A lysine at position −1 is positioned to interact with an acidic patch in the yeast Cdc20 ABBA-binding pocket that is not present in Cdh1. Clb5, a Cdc20 target, has a complementary pair of basic residues at the beginning of its ABBA motif. This basic sequence is absent in the ABBA motif of the Cdh1 target Acm1, suggesting that this difference between Cdc20 and Cdh1 promotes specific Cdc20–Clb5 and Cdh1–Acm1 interactions, respectively. Indeed, replacing the acidic patch in Cdc20 with the Cdh1 residues inhibits Clb5 binding to Cdc20 (Lu et al., 2014).
Evolution of the APC/C substrate network
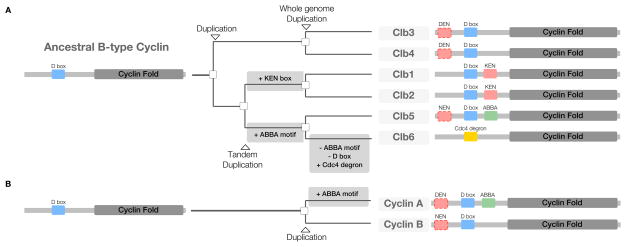
The vast majority of characterized APC/C degrons appear to have evolved ex nihilo – through addition of a functional degron to a previously non-functional region of protein sequence. Disordered regions generally tolerate high mutation rates that allow the rapid exploration of functional sequence space (Davey et al., 2015). The simplicity of APC/C degron sequences allows random mutations to continually create and destroy rudimentary degrons, resulting in selectable phenotypic diversity. Degron-containing alleles that result in more rapid or robust cell division will have large positive effects on an organism’s fitness and should spread through a population. Once established, functionally important degrons are protected from loss-of-function mutations by purifying selection (Davey et al., 2015). For example, the D boxes of cyclins are conserved across nearly all eukaryotes over a billion years of evolution. However, many other degrons are present in only a limited evolutionary window, as motifs are regularly gained and lost as regulatory networks are rewired (Davey et al., 2015). The ease of ex nihilo degron acquisition is highlighted by the evolution of the six budding yeast mitotic cyclins, known as Clbs, which evolved from a single ancestral B-type cyclin (Gunbin et al., 2011). Through the gain and loss of APC/C and SCF degrons, the Clbs are now degraded sequentially (Jackson et al., 2006; Lu et al., 2014) (Figure 6A). Interestingly, independent of the fungal Clb5 ABBA motif acquisition, cyclin A in the metazoan lineages gained an ABBA motif to promote the sequential degradation of cyclin A and cyclin B (Figure 6B). The separate acquisition of the ABBA motif by Cyclin A and Clb5 to order mitotic cyclin degradation is a beautiful example of convergent evolution.
Figure 6. Evolution of the N-terminal regulatory region of the B-type cyclins.
(A) Budding yeast have six B-type cyclins (Clb1–6), and their evolution can be traced through 3 duplication events to a single ancestor (Byrne and Wolfe, 2005; Gunbin et al., 2011). The common ancestor duplicated to produce Clb3/4 and Clb1/2/5/6, a tandem duplication led to the creation of Clb1/2 and Clb5/6 (Clb1/2 and Clb5/6 are still found adjacent to each other in the budding yeast genome), and finally a whole genome duplication produced the six Clbs seen today. The last common Clb ancestor, like all B-type cyclins, had a single D box and was likely destroyed in metaphase (Yamano et al., 1996; Gunbin et al., 2011). The Clb3/4 lineage retained this motif conformation. After the Clb1/2/5/6 duplication, Clb1/2 gained a KEN box and Clb5/6 gained an ABBA motif. Subsequently, after the Clb5/6 duplication, Clb6 lost both the D box and ABBA motif and gained a degron for a different ubiquitin ligase, SCFCdc4 (Amon et al., 1994; Hendrickson et al., 2001; Thornton and Toczyski, 2003; Jackson et al., 2006; Lu et al., 2014). Several of the Clbs also contain uncharacterized yet conserved non-canonical KEN boxes (DEN, NEN). The distinct motif content of the intrinsically disordered regulatory N-terminal regions of the Clb family is the major determinant of their ordered degradation. (B) Metazoan Cyclin A has acquired an ABBA motif independently of the fungal Clb5 ABBA motif acquisition.
Conclusions and future perspectives
The past decade has seen a rapid increase in the number of characterized protein motifs involved in cell regulation, but only recently have the general properties of the regulatory networks they support become apparent (Van Roey et al., 2012). It is now clear that all of the targets of a given motif-mediated regulatory network are not equal. A hierarchy of targets is created by intrinsic differences encoded in the sequence of the motif-containing peptide, encoded in the motif context, or modulated by crosstalk between regulatory networks acting at the motif. Although the vast majority of motif-centric networks remain uncharacterized (Van Roey et al., 2014), it is likely that the mechanisms described for the APC/C will be identified in all large regulatory networks. In recent years, the focus of APC/C studies has shifted from the search for degrons to uncovering the layers of regulation that govern degron recognition. Thus, the future of the APC/C field lies in teasing apart the cellular and biochemical mechanisms that determine the timing and localization of substrate degradation.
Acknowledgments
We apologize to the many colleagues whose work could not be cited due to space restrictions. We thank Jonathan Asfaha, Toby Gibson, Nairi Hartooni, Aino Järvelin, Arda Mizrak and Laura Rosen for fruitful discussions and critically reading the manuscript. NED is supported by an SFI Starting Investigator Research Grant (13/SIRG/2193), and DOM is supported by the National Institute of General Medical Sciences (R35GM118053).
Footnotes
Online resources
The accompanying website (http://slim.ucd.ie/apc/) contains summaries of degron structures and ordering mechanisms, a complete list of known APC/C substrates, and tools for searching protein sequences for potential APC/C degron motifs.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acquaviva C, Herzog F, Kraft C, Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat Cell Biol. 2004;6:892–898. doi: 10.1038/ncb1167. [DOI] [PubMed] [Google Scholar]
- Alfieri C, Chang L, Zhang Z, Yang J, Maslen S, Skehel M, Barford D. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature. 2016 doi: 10.1038/nature19083. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Araki M, Yu H, Asano M. A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo. Genes Dev. 2005;19:2458–2465. doi: 10.1101/gad.1361905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold L, Hockner S, Seufert W. Insights into the cellular mechanism of the yeast ubiquitin ligase APC/C-Cdh1 from the analysis of in vivo degrons. Mol Biol Cell. 2015;26:843–858. doi: 10.1091/mbc.E14-09-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NG, VanderLinden R, Watson ER, Weissmann F, Ordureau A, Wu KP, Zhang W, Yu S, Mercredi PY, Harrison JS, et al. Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C. Cell. 2016;165:1440–1453. doi: 10.1016/j.cell.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu JY, Shaw AS, Chan AC. Analysis of the interaction of ZAP-70 and syk protein-tyrosine kinases with the T-cell antigen receptor by plasmon resonance. Proc Natl Acad Sci USA. 1995;92:5106–5110. doi: 10.1073/pnas.92.11.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Xiong Y, Solomon MJ. Mechanisms of pseudosubstrate inhibition of the anaphase promoting complex by Acm1. EMBO J. 2011;30:1818–1829. doi: 10.1038/emboj.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhorn BA, Petzold G, Galova M, Dube P, Kraft C, Herzog F, Stark H, Peters JM. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron A, Okresz L, Genschik P. First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci. 2003;8:83–89. doi: 10.1016/S1360-1385(02)00028-6. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat Cell Biol. 2002;4:880–887. doi: 10.1038/ncb871. [DOI] [PubMed] [Google Scholar]
- Castro A, Vigneron S, Bernis C, Labbe JC, Lorca T. Xkid is degraded in a D-box, KEN-box, and A-box-independent pathway. Mol Cell Biol. 2003;23:4126–4138. doi: 10.1128/MCB.23.12.4126-4138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Barford D. Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Curr Opin Struct Biol. 2014;29:1–9. doi: 10.1016/j.sbi.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat Cell Biol. 2013;15:1378–1385. doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer L, Heyman J, Touati S, Harashima H, Araou E, Girard C, Horlow C, Wassmann K, Schnittger A, De Veylder L, et al. OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genet. 2012;8:e1002865. doi: 10.1371/journal.pgen.1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer L, Jolivet S, Horlow C, Chelysheva L, Heyman J, De Jaeger G, Koncz C, De Veylder L, Mercier R. Centromeric cohesion is protected twice at meiosis, by SHUGOSHINs at anaphase I and by PATRONUS at interkinesis. Curr Biol. 2013;23:2090–2099. doi: 10.1016/j.cub.2013.08.036. [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Cyert MS, Moses AM. Short linear motifs - ex nihilo evolution of protein regulation. Cell Comm Sig. 2015;13:43. doi: 10.1186/s12964-015-0120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Davey NE, Hagting A, Izawa D, Mansfeld J, Gibson TJ, Pines J. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev Cell. 2015;32:358–372. doi: 10.1016/j.devcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Martinez LA, Tian W, Li B, Warrington R, Jia L, Brautigam CA, Luo X, Yu H. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J Biol Chem. 2015;290:2431–2443. doi: 10.1074/jbc.M114.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L, Trilles A, Moreira D, Brochier-Armanet C. The phylogenomic analysis of the anaphase promoting complex and its targets points to complex and modern-like control of the cell cycle in the last common ancestor of eukaryotes. BMC Evol Biol. 2011;11:265. doi: 10.1186/1471-2148-11-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman M, Sullivan M, Morgan DO. Modulation of the mitotic regulatory network by APC-dependent destruction of the Cdh1 inhibitor Acm1. Mol Cell. 2008;30:437–446. doi: 10.1016/j.molcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S, Whiffin N, Gavilan MP, Kutscheidt S, De Luca M, Marcozzi C, Min M, Watkins J, Chung K, Fackler OT, et al. Spatiotemporal organization of Aurora-B by APC/CCdh1 after mitosis coordinates cell spreading through FHOD1. J Cell Sci. 2013;126:2845–2856. doi: 10.1242/jcs.123232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye JJ, Brown NG, Petzold G, Watson ER, Grace CR, Nourse A, Jarvis MA, Kriwacki RW, Peters JM, Stark H, et al. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat Struct Mol Biol. 2013;20:827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K, Grimaldi M, Yamano H. Cyclin-dependent kinase 1-dependent activation of APC/C ubiquitin ligase. Science. 2016;352:1121–1124. doi: 10.1126/science.aad3925. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Nagao K, Tanaka H, Yasuda H, Hunt T, Yanagida M. Fission yeast Cut2 required for anaphase has two destruction boxes. EMBO J. 1997;16:5977–5987. doi: 10.1093/emboj/16.19.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giubettini M, Asteriti IA, Scrofani J, De Luca M, Lindon C, Lavia P, Guarguaglini G. Control of Aurora-A stability through interaction with TPX2. J Cell Sci. 2011;124:113–122. doi: 10.1242/jcs.075457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Guharoy M, Bhowmick P, Tompa P. Design Principles Involving Protein Disorder Facilitate Specific Substrate Selection and Degradation by the Ubiquitin-Proteasome System. J Biol Chem. 2016;291:6723–6731. doi: 10.1074/jbc.R115.692665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunbin KV, Suslov VV, Turnaev II, Afonnikov DA, Kolchanov NA. Molecular evolution of cyclin proteins in animals and fungi. BMC Evol Biol. 2011;11:224. doi: 10.1186/1471-2148-11-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Jeong DE, Henderson JT, Choi E, Bremmer SC, Iliuk AB, Charbonneau H. Cdc28 and Cdc14 control stability of the anaphase-promoting complex inhibitor Acm1. J Biol Chem. 2008;283:10396–10407. doi: 10.1074/jbc.M710011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 2001;20:7117–7127. doi: 10.1093/emboj/20.24.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29:2407–2420. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Chao WC, Zhang Z, Yang J, Cronin N, Barford D. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol Cell. 2013;50:649–660. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth S, Bottger F, Pan C, Mann M, Stemmann O. PP2A delays APC/C-dependent degradation of separase-associated but not free securin. EMBO J. 2014;33:1134–1147. doi: 10.1002/embj.201488098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson C, Meyn MA, 3rd, Morabito L, Holloway SL. The KEN box regulates Clb2 proteolysis in G1 and at the metaphase-to-anaphase transition. Curr Biol. 2001;11:1781–1787. doi: 10.1016/s0960-9822(01)00564-4. [DOI] [PubMed] [Google Scholar]
- Hertz EP, Kruse T, Davey NE, Lopez-Mendez B, Sigurethsson JO, Montoya G, Olsen JV, Nilsson J. A Conserved Motif Provides Binding Specificity to the PP2A-B56 Phosphatase. Mol Cell. 2016;63:686–695. doi: 10.1016/j.molcel.2016.06.024. [DOI] [PubMed] [Google Scholar]
- Heyman J, Van den Daele H, De Wit K, Boudolf V, Berckmans B, Verkest A, Alvim Kamei CL, De Jaeger G, Koncz C, De Veylder L. Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell. 2011;23:4394–4410. doi: 10.1105/tpc.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol Biol Cell. 2001;12:3402–3416. doi: 10.1091/mbc.12.11.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilioti Z, Chung YS, Mochizuki Y, Hardy CF, Cohen-Fix O. The anaphase inhibitor Pds1 binds to the APC/C-associated protein Cdc20 in a destruction box-dependent manner. Curr Biol. 2001;11:1347–1352. doi: 10.1016/s0960-9822(01)00399-2. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Liu W, Wu G, Wan Y. Nuclear translocation of Skp2 facilitates its destruction in response to TGFbeta signaling. Cell Cycle. 2011;10:285–292. doi: 10.4161/cc.10.2.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Raff JW. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 1999;18:2184–2195. doi: 10.1093/emboj/18.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LP, Reed SI, Haase SB. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol Cell Biol. 2006;26:2456–2466. doi: 10.1128/MCB.26.6.2456-2466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M, van Drogen F, Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1) EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenz J, Mihaljev T, Kubis A, Legewie S, Hauf S. Robust Ordering of Anaphase Events by Adaptive Thresholds and Competing Degradation Pathways. Mol Cell. 2015;60:446–459. doi: 10.1016/j.molcel.2015.09.022. [DOI] [PubMed] [Google Scholar]
- Karamysheva Z, Diaz-Martinez LA, Crow SE, Li B, Yu H. Multiple anaphase-promoting complex/cyclosome degrons mediate the degradation of human Sgo1. J Biol Chem. 2009;284:1772–1780. doi: 10.1074/jbc.M807083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitzing C, Huss R, Illert AL, Froschl A, Wotzel S, Peschel C, Bassermann F, Duyster J. APC/C(Cdh1)-mediated degradation of the F-box protein NIPA is regulated by its association with Skp1. PLoS One. 2011;6:e28998. doi: 10.1371/journal.pone.0028998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzbucher A, Stewart E, Harrison D, Hunt T. The ‘destruction box’ of cyclin A allows B-type cyclins to be ubiquitinated, but not efficiently destroyed. EMBO J. 1996;15:3053–3064. [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leismann O, Lehner CF. Drosophila securin destruction involves a D-box and a KEN-box and promotes anaphase in parallel with Cyclin A degradation. J Cell Sci. 2003;116:2453–2460. doi: 10.1242/jcs.00411. [DOI] [PubMed] [Google Scholar]
- Lischetti T, Zhang G, Sedgwick GG, Bolanos-Garcia VM, Nilsson J. The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nat Comm. 2014;5:5563. doi: 10.1038/ncomms6563. [DOI] [PubMed] [Google Scholar]
- Lu D, Girard JR, Li W, Mizrak A, Morgan DO. Quantitative framework for ordered degradation of APC/C substrates. BMC Biol. 2015a;13:96. doi: 10.1186/s12915-015-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Hsiao JY, Davey NE, Van Voorhis VA, Foster SA, Tang C, Morgan DO. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J Cell Biol. 2014;207:23–39. doi: 10.1083/jcb.201402041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang W, Kirschner MW. Specificity of the anaphase-promoting complex: a single-molecule study. Science. 2015b;348:1248737. doi: 10.1126/science.1248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Sixma TK. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat Struct Mol Biol. 2014;21:308–316. doi: 10.1038/nsmb.2792. [DOI] [PubMed] [Google Scholar]
- Min M, Lindon C. Substrate targeting by the ubiquitin-proteasome system in mitosis. Semin Cell Dev Biol. 2012;23:482–491. doi: 10.1016/j.semcdb.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Min M, Mayor U, Lindon C. Ubiquitination site preferences in anaphase promoting complex/cyclosome (APC/C) substrates. Open Biol. 2013;3:130097. doi: 10.1098/rsob.130097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. The Cell Cycle: Principles of Control. London: New Science Press; 2007. [Google Scholar]
- Nguyen TB, Manova K, Capodieci P, Lindon C, Bottega S, Wang XY, Refik-Rogers J, Pines J, Wolgemuth DJ, Koff A. Characterization and expression of mammalian cyclin b3, a prepachytene meiotic cyclin. J Biol Chem. 2002;277:41960–41969. doi: 10.1074/jbc.M203951200. [DOI] [PubMed] [Google Scholar]
- Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell. 2005;120:773–788. doi: 10.1016/j.cell.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Ostapenko D, Burton JL, Wang R, Solomon MJ. Pseudosubstrate inhibition of the anaphase-promoting complex by Acm1: regulation by proteolysis and Cdc28 phosphorylation. Mol Cell Biol. 2008;28:4653–4664. doi: 10.1128/MCB.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. J Cell Biol. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao R, Weissmann F, Yamaguchi M, Brown NG, VanderLinden R, Imre R, Jarvis MA, Brunner MR, Davidson IF, Litos G, et al. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc Natl Acad Sci USA. 2016;113:E2570–E2578. doi: 10.1073/pnas.1604929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Guimaraes DS, Melesse M, Hall MC. Substrate Recognition by the Cdh1 Destruction Box Receptor is a General Requirement for APC/CCdh1-mediated Proteolysis. J Biol Chem. 2016;291:15564–15574. doi: 10.1074/jbc.M116.731190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Matzkies M, Dienemann A, Sprenger F. Cyclin A degradation employs preferentially used lysines and a cyclin box function other than Cdk1 binding. Cell Cycle. 2007;6:171–181. doi: 10.4161/cc.6.2.3716. [DOI] [PubMed] [Google Scholar]
- Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Reis A, Levasseur M, Chang HY, Elliott DJ, Jones KT. The CRY box: a second APCcdh1-dependent degron in mammalian cdc20. EMBO Rep. 2006;7:1040–1045. doi: 10.1038/sj.embor.7400772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Roy J, Li H, Hogan PG, Cyert MS. A conserved docking site modulates substrate affinity for calcineurin, signaling output, and in vivo function. Mol Cell. 2007;25:889–901. doi: 10.1016/j.molcel.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick GG, Hayward DG, Di Fiore B, Pardo M, Yu L, Pines J, Nilsson J. Mechanisms controlling the temporal degradation of Nek2A and Kif18A by the APC/C-Cdc20 complex. EMBO J. 2013;32:303–314. doi: 10.1038/emboj.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. A novel destruction sequence targets the meiotic regulator Spo13 for anaphase-promoting complex-dependent degradation in anaphase I. J Biol Chem. 2007;282:19710–19715. doi: 10.1074/jbc.M701507200. [DOI] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat Cell Biol. 2003;5:1090–1094. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- Tian W, Li B, Warrington R, Tomchick DR, Yu H, Luo X. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proc Natl Acad Sci USA. 2012;109:18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu T, Takihara Y, Ishimaru N, Pagano M, Takata T, Kudo Y. Aurora-A controls pre-replicative complex assembly and DNA replication by stabilizing geminin in mitosis. Nat Comm. 2013;4:1885. doi: 10.1038/ncomms2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr Opin Struct Biol. 2012;22:378–385. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Van Roey K, Uyar B, Weatheritt RJ, Dinkel H, Seiler M, Budd A, Gibson TJ, Davey NE. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem Rev. 2014;114:6733–6778. doi: 10.1021/cr400585q. [DOI] [PubMed] [Google Scholar]
- Van Voorhis VA, Morgan DO. Activation of the APC/C ubiquitin ligase by enhanced E2 efficiency. Curr Biol. 2014;24:1556–1562. doi: 10.1016/j.cub.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wang W, Kirschner MW. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat Cell Biol. 2013;15:797–806. doi: 10.1038/ncb2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A, Banerjee S, Zhu X, Philipp I, Iavarone AT, Rape M. Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Mol Cell. 2011;42:744–757. doi: 10.1016/j.molcel.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, VanderLinden R, Weissmann F, Qiao R, Dube P, Brown NG, Haselbach D, Zhang W, Sidhu SS, Peters JM, et al. Cryo-EM of Mitotic Checkpoint Complex-Bound APC/C Reveals Reciprocal and Conformational Regulation of Ubiquitin Ligation. Mol Cell. 2016;63:593–607. doi: 10.1016/j.molcel.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano H, Gannon J, Hunt T. The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J. 1996;15:5268–5279. [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chang L, Alfieri C, Zhang Z, Yang J, Maslen S, Skehel M, Barford D. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533:260–264. doi: 10.1038/nature17973. [DOI] [PMC free article] [PubMed] [Google Scholar]