Abstract
Our understanding of molecular motor function has been greatly improved by the development of imaging modalities, which enable real-time observation of their motion at the single-molecule level. Here, we describe the use of a new method, interferometric scattering microscopy, for the investigation of motor protein dynamics by attaching and tracking the motion of metallic nanoparticle labels as small as 20 nm diameter. Using myosin-5, kinesin-1, and dynein as examples, we describe the basic assays, labeling strategies, and principles of data analysis. Our approach is relevant not only for motor protein dynamics but also provides a general tool for single-particle tracking with high spatiotemporal precision, which overcomes the limitations of single-molecule fluorescence methods.
1. INTRODUCTION
Single-particle tracking (SPT) is a powerful tool for studying micro-and nanoscopic motions of cells, organelles, and molecules. Since the advent of video microscopy, there has been a continuous improvement in sensitivity, speed, precision, and spatial resolution, leading to our current ability to track single molecules on millisecond timescales and with nanometer precision. Early video microscopes used optical density, phase, or interference to provide contrast for visualizing large cargoes and natural tracks for motor proteins, such as mitochondria, and microtubules (MTs) (Allen, Metuzals, Tasaki, Brady, & Gilbert, 1982; Horio & Hotani, 1986; Howard, Hudspeth, & Vale, 1989; Vale et al., 1985). To detect motions of individual proteins, polymer microspheres (beads) were attached to individual motor molecules and imaged (Schnapp, Gelles, & Sheetz, 1988) or stabilized within an infrared optical trap (Rief et al., 2000; Svoboda, Schmidt, Schnapp, & Block, 1993). The average step size of kinesin-1, for instance, was determined to be 8 nm, matching the spacing of tubulin dimers along the MT axis (Svoboda et al., 1993). Optical trap experiments found that myosin-5a, which walks along actin filaments, exhibits ~36 nm steps (Mehta et al., 1999), the largest step size in the myosin superfamily.
In contrast to attaching beads, labeling single-protein molecules with fluorescent probes renders them clearly visible by fluorescence microscopy (Funatsu, Harada, Tokunaga, Saito, & Yanagida, 1995; Sakamoto, Amitani, Yokota, & Ando, 2000). The traditional spatial resolution limits in optical microscopy (including fluorescence) are related to diffraction of light, which results in the smallest objects producing an image whose size is equal to approximately half the wavelength of the light. This distance is much greater than the step size of molecular motors, such as the 8 nm steps of kinesin and the 5–40 nm steps, or power strokes, of myosin motors. Several investigators realized that when only one fluorescent object is present in the emitting volume, the center of the ~250 nm wide image of the single molecule, termed the point spread function, could be localized with very high precision (Thompson, Larson, & Webb, 2002; Yildiz et al., 2003). High-resolution tracking of myosin-5a and kinesin-1 (Forkey, Quinlan, Shaw, Corrie, & Goldman, 2003; Snyder, Sakamoto, Hammer, Sellers, & Selvin, 2004; Warshaw et al., 2005; Yildiz, Tomishige, Vale, & Selvin, 2004) found that these motors move in a “hand-over-hand” fashion, while recent studies on dynein showed comparatively erratic stepping behavior, demonstrating both “hand-over-hand” and “inchworm” mechanisms (DeWitt, Chang, Combs, & Yildiz, 2012; Qiu et al., 2012; Reck-Peterson et al., 2006).
As is apparent from the development of differential interference contrast and then fluorescence microscopy for SPT, continuous improvements to temporal and spatial resolution have led to deeper insights into the mechanisms of molecular machines responsible for cellular motion; many problems, however, remain unresolved. Outstanding questions in our understanding of molecular motor function include the details of myosin dynamics, while each step is being produced, how the two heads of kinesin coordinate their chemomechanical cycles to achieve processive stepping, and the determinants of dynein’s irregular stepping pattern. Here, we present detailed protocols for tracking different molecular motors by interferometric scattering microscopy (iSCAT), which enables orders of magnitude improved spatiotemporal resolution compared to single-molecule fluorescence and, thereby, provides a route to addressing questions such as those outlined earlier.
2. INTERFEROMETRIC SCATTERING MICROSCOPY
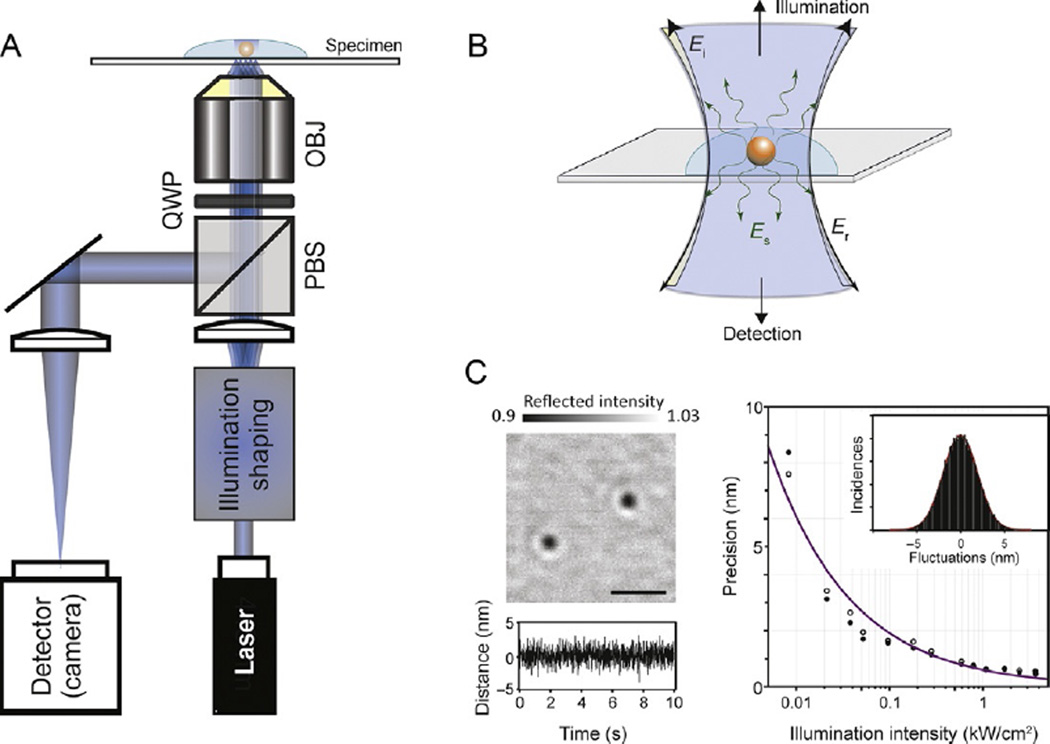
An iSCAT image results from the interference between a reference light, commonly provided by a weak reflection at the sample surface, and light scattered by an object in the medium. A typical iSCAT setup is based on an inverted microscope (Fig. 1A), where the sample is illuminated by a coherent light source passing through a beamsplitter, which enables any photons scattered or reflected by the sample (Fig. 1B) to be separated from the incident light beam and subsequently imaged onto a digital camera. As a result, the experimental setup is almost identical to those used for the various implementations of interference reflection microscopy (Curtis, 1964; Filler & Peuker, 2000), with the important distinction that a coherent light source is used, which enhances the interferometric contrast (Dulin, Barland, Hachair, & Pedaci, 2014).
Fig. 1.
Principles and performance of iSCAT for single-particle tracking. (A) Experimental setup. OBJ, microscope objective; PBS, polarizing beam splitter; QWP, quarter-wave plate. Illumination shaping refers to different schemes for illuminating the sample without generating excessive speckle such as slim field (Piliarik & Sandoghdar, 2014) or rapid beam scanning (Kukura et al., 2009). (B) Operating principle of iSCAT detection. Light reflected at the sample/glass interface is collected along with any scattered light from the object of interest. Ei, incident electric field; Er, reflected electric field; Es, scattered electric field. (C) Localization of 20 nm gold particles in the shot-noise-limited regime. Left: flat-field-corrected iSCAT image of 20 nm gold nanoparticles immobilized on a glass coverslip in water. Scale bar: 1 µm. The bottom trace shows the fluctuation in the distance between two nanoparticles as a function of time sampled at 1000 frames per second, caused solely by shot noise in the measurement. Right: localization precision as a function of illumination intensity for 20 nm gold nanoparticles. An increase in illumination intensity improves the localization precision for a given exposure time. Open and filled circles: localization precision in x and y, respectively. The line depicts the expected behavior for shot-noise-limited imaging.
The most critical aspect to iSCAT performance is the precise measurement and removal of any static imaging background, as it determines imaging sensitivity and precision. When controlled appropriately, iSCAT has been shown to detect and image single, unlabeled proteins (Ortega Arroyo et al., 2014; Piliarik & Sandoghdar, 2014) and to track 20 nm gold labels at up to 500 kHz frame rates with nm precision (Andrecka et al., 2015; Lin, Chang, & Hsieh, 2014; Mickolajczyk et al., 2015). Interferometric detection leads to a lesser drop in scattering contrast with decreasing particle size compared to dark-field microscopy, simplifying the detection of very weak scatterers. In comparison to fluorescence, iSCAT suffers from much poorer background rejection ability, but in exchange can completely remove the need for any fluorescent labeling, including labeling of actin filaments and microtubules, which themselves generate clear iSCAT signals (Andrecka, Arroyo, Lewis, Cross, & Kukura, 2016; Ortega Arroyo et al., 2014). As a result, the relevant single-molecule assays are in many instances easier to implement as they require fewer labeling steps. Most importantly, however, iSCAT provides access to experimental approaches and timescales that are inaccessible to single-molecule fluorescence-based studies, either due to the underlying photophysics or due to the difficulty associated with removing fluorescence background (Arroyo & Kukura, 2016; Ortega-Arroyo & Kukura, 2012).
3. METHODS AND PROTOCOLS
3.1 Flow Cell Preparation
In all experiments described later we used the same type of flow cell, which is common in studies of molecular motors with a variety of methods. Briefly, we clean a 50 × 25-mm, #1.5, coverslip (VWR) by washing with ultrapure water (MilliQ) followed by ethanol and again MilliQ water. After drying, passing the coverslip through a blue flame removes dust and moisture. Proper washing is important to remove any dirt adhered to the coverslip surface, which generates iSCAT signal. Importantly, however, iSCAT detection does not require special substrates, such as quartz glass, and the cleaning procedure is straightforward compared to single-molecule fluorescence measurements, where special care must be taken to minimize any residual fluorescence background. The final flow channel consists of the cleaned microscope slide and a 25 × 25-mm coverslip connected by double-sided sticky tape. The volume of the chamber is on the order of ~15–20 µL.
3.2 Preparation and Immobilization of Cytoskeletal Filaments
One of the strengths of iSCAT in the context of molecular motor studies is its ability to visualize cytoskeletal filaments such as actin or microtubules without the need for any labels. When illuminated with blue light (<500 nm), an actin filament generates ~1% iSCAT contrast, while a single microtubule produces ~8% contrast, comparable with a 20-nm gold nanoparticle (Andrecka et al., 2016; Ortega Arroyo et al., 2014). Complete immobilization of actin and microtubules on the glass surface is paramount to success when running single-molecule motor assays with high spatiotemporal precision. Poorly immobilized filaments will introduce additional fluctuations, which can reduce the ability to detect steps. In this section we describe how actin filaments and microtubules are obtained and attached to the glass surface.
3.2.1 Actin
Required materials
Globular (G) actin can be prepared as described (Pardee & Spudich, 1982) or purchased from Cytoskeleton
Polymerization buffer: 10 mM imidazole pH 7.3, 50 mM KCl, 1 mM MgCl2, 2mM DTT, 3 mM ATP, 1 mM EGTA
Motility buffer (MB): 20 mM MOPS pH 7.3, 5 mM MgCl2, 0.1 mM EGTA
Phosphate-buffered saline (PBS buffer) pH 7.4
Bovine serum albumin (BSA)
Poly(ethylene glycol)–poly-l-lysine branch copolymer (PEG–PLL) from Surface Solutions SuSoS, Switzerland
To polymerize and immobilize actin
Prepare 20 µM filamentous (F) actin in polymerization buffer and incubate on ice for 2 h.
Rinse the chamber (prepared as described in Section 3.1) with 1 mg/mL solution of PEG–PLL dissolved in PBS and incubate for 30 min.
Wash the chamber with MB before adding actin solution.
Dilute actin 20–50 times in MB, load into the chamber, and incubate for 2–5 min. Inspect the chamber under the iSCAT microscope and add more actin if necessary.
Wash the cell with MB and then block with BSA (2 mg/mL in MB) for 5 min.
NOTE: This procedure includes a minor modification of an established method (Dunn&Spudich, 2011). The polymer (PEG–PLL) is usually used to passivate the surface. In this protocol, however, PEG–PLL provides weak interactions with glass that immobilize actin. Additional blocking with BSA is required to avoid nonspecific binding of myosin and gold to the surface. Alternative methods, such as actin binding to nitrocellulose, are not recommended due to high iSCAT background of the nitrocellulose. Using biotinylated actin- and neutravidin-coated surfaces fails when gold nanoparticles are attached to myosin using the same moiety.
TIP: If actin binding is insufficiently strong, plasma cleaning the cover glass before assembling the flow cells improves the attachment.
3.2.2 Microtubules
Required materials
Free tubulin can be extracted from bovine or porcine brain as previously described (Ozeki et al., 2009) purchased directly from commercial sources (Cytoskeleton), or expressed in yeast cells (Johnson, Ayaz, Huddleston, & Rice, 2011)
Full-length Drosophila KHC R210A rigor mutant (see later)
BRB80 buffer: 80 mM PIPES pH to 6.8 with KOH, 1 mM EGTA, 1 mM MgCl2
Casein (Sigma Aldrich, Cat. No. C7078)
Taxol (Sigma Aldrich, Cat. No. T1912) dissolved in DMSO
To polymerize microtubules
Prepare a solution of 1.6 mg/mL free tubulin, 4 mM MgCl2, 1mM GTP, and 1:25 DMSO in BRB80 buffer, and incubate at 37°C for 30 min.
Stabilize by adding taxol to 10 µM.
Dilute to working concentration (~1:80) in BRB80 plus 10 µM taxol and 0.5 mg/mL casein.
Recent work has shown that a rigor form of kinesin-1 (full-length Drosophila KHCR210A) is highly effective in microtubule immobilization (Mickolajczyk et al., 2015). The tail domain of full-length kinesin binds strongly to casein-treated surfaces, while the head domains bind tightly to microtubules. Rigor kinesin binds microtubules more tightly than tubulin antibodies, improving microtubule immobilization. Cross-linking microtubules with glutaraldehyde is another effective method for immobilizing microtubules, but it greatly reduces the number of kinesin landing events likely due to damage to the microtubules (Turner, Chang, Fang, Cuomo, & Murphy, 1996).
To immobilize microtubules
Create a flow cell as described in Section 3.1.
Prepare casein solution (final 0.5 mg/mL in BRB80) and flow into the flow cell. Let sit for 5–10 min.
Dilute rigor mutant to 200 nM in casein solution plus 1 µM ATP, and add to the flow cell, washing out the previous solution. Incubate for 5 min to adsorb motors to the surface.
Wash any unbound rigor mutant out of the flow cell using casein solution.
Load diluted microtubules (see earlier) into the flow cell. Incubate for 5 min.
Wash excess microtubules out of the flow well using casein solution and observe microtubules under the iSCAT microscope.
NOTE: The rigor mutant requires at least 1 µMATP with equimolar MgCl2 at all times for proper function.
TIP: If there are multiple overlapping microtubules or clumps, then a further microtubule dilution is necessary. If microtubules appear to wobble, a higher concentration or a new preparation of rigor kinesin may be necessary.
3.3 Labeling Motor Proteins With Gold Nanoparticles
Attachment of nanoparticle labels to kinesin, myosin, and dynein motors has to date been achieved via biotin–streptavidin chemistry (Andrecka et al., 2015; Dunn & Spudich, 2007; Isojima, Iino, Niitani, Noji, & Tomishige, 2016; Mickolajczyk et al., 2015; Schneider, Glaser, Berndt, & Diez, 2013; Schneider, Korten, Walter, & Diez, 2015). One method of introducing biotin into kinesin is to create a cysteine-lite mutant, in which solvent-exposed cysteines are mutated out of the motor domain and a single cysteine is kept to take advantage of sulfhydryl chemistry for attaching a maleimide-functionalized label. This method has the advantage of direct, covalent attachment of the biotin and a minimal linker size, but has the disadvantage that cysteine-lite kinesin mutants exhibit markedly different mechanochemistry from wild type (Andreasson et al., 2015). For myosin-5a, a native or engineered cysteine residue in purified calmodulin can be labeled with a dye or gold nanoparticle and then exchanged for endogenous calmodulin in the myosin-5a (Dunn&Spudich, 2007; Sakamoto et al., 2000). However, in this method, it is difficult to control which of the six possible calmodulins is labeled. Since many existing constructs of kinesin and myosin possess an N- or C-terminal GFP moiety, another option is to use biotinylated anti-bodies against GFP (Andrecka et al., 2015; Schneider et al., 2013).
An alternative way of attaching a gold nanoparticle, which was used for the kinesin and myosin work here, is to fuse an Avi-Tag (GLNDIFEAQKIEWHE) to the motor and to biotinylate the protein with the biotin ligase BirA (Avidity) (Schatz, 1993). The constructs used to efficiently attach gold nanoparticles are described later.
3.3.1 Myosin
Mouse myosin-5a HMM was expressed in the presence of calmodulin and purified as described (Wang et al., 2000). The N-terminus was modified by the addition of a nucleotide sequence encoding an Avi-Tag peptide (Andrecka et al., 2015). Myosin was biotinylated at 4°C, overnight and dialyzed against 10 mM MOPS pH 7.3 buffer containing 0.1 mM EGTA, 1 mM DTT, and 100 mM KCl to remove excess of biotin. Before labeling, the myosin sample was diluted in MB containing 40 mM KCl, 5 mM DTT, 0.1 mg/mL BSA, and 5 µM calmodulin. Gold nanoparticles conjugated with streptavidin were purchased from BBI (UK) and directly mixed with biotinylated myosin-5a in a 4:1, gold particle to myosin molecule ratio, consistent with one or zero myosin molecules per gold particle and only one motor domain labeled per myosin dimer. The mixture was incubated on ice for at least 15 min (sample volume 50 µL, final concentration of myosin 300 pM). The myosin–gold conjugate sample was then diluted in the MB: 20 mM MOPS pH 7.3, 5 mM MgCl2, 0.1 mM EGTA supplemented with 20 mM KCl, 5 mM DTT, 0.1 mg/mL BSA, 1–1000 µM ATP and loaded into the flow cell prepared as described earlier.
3.3.2 Kinesin
Kinesin-1 (Drosophila KHC truncated at 560) was prepared with an N-terminal Avi-Tag and coexpressed with BirA in BL21(DE3) bacteria, which resulted in in vivo biotinylation of the motor (Mickolajczyk et al., 2015). Motors were purified by affinity chromatography using a C-terminal His-tag. The biotinylation percentage was kept low by waiting until 6 h after induction by isopropyl β-d-1-thiogalactopyranoside before adding biotin to avoid doubly biotinylating the homodimer motor. The extent of biotinylation was quantified using the colorimetric HABA assay (ThermoFisher, Cat No. 28005). Motors were diluted in imaging solution (0.5 mg/mL casein, 10 µM taxol, 20 mM glucose, 20 µg/mL glucose oxidase, 8 µg/mL catalase, 0.2 mg/mL BSA, 1:200 β-mercaptoethanol, 2 mM ATP-MgCl2 in BRB80) and mixed with streptavidin-coated 30 nm gold nanoparticles (Mickolajczyk et al., 2015).
NOTE: In order to ensure the presence of only one biotinylated motor per bead, a titration experiment was performed where the number of landing events per linear micron of microtubule is measured as a function of molar ratio of motors to nanoparticles (NPs). The assumption is that one motor is sufficient for a particle to bind to a microtubule and more motors on a bead do not increase the probability of binding. By Poisson statistics, this plot will be linear if there is truly one motor per NP (Block, Goldstein, & Schnapp, 1990), and will plateau when there are multiple motors per bead. Thus, any motor:gold ratio from the linear regime is acceptable for single-molecule assays. In our experiment we used the single-molecule landing rate which showed a linear relationship up to 6:1 motor-to-gold ratio, justifying the use of these molar ratios with the expectation of only one motor per nanoparticle. Kinesin concentrations are often obtained through absorbance at 280 nm; however, only a subset of total protein is active kinesin, and only a further subset is active and biotinylated, meaning that there is actually a superstoichiometric amount of gold in the linear regime. Each new batch of kinesin will have a different linear regime for the number of landing events per micron of microtubule.
3.3.3 Dynein
Tail-truncated (331 kDa) dynein mini-dimer (GST dimerized) from Saccharomyces cerevisiae (Reck-Peterson et al., 2006) was biotinylated in vivo at AAA5 and AAA6 by inserting biotinylation sites at A3666 and P4032 in the dynein ring (Kulman, Satake, & Harris, 2007). Dynein was diluted in dynein lysis buffer (30 mM HEPES pH 7.4, 2 mM Mg acetate, 1 mM EGTA, 10% glycerol, 1.25 mg/mL casein, 1 mM DTT, 20 µM taxol) and mixed with streptavidin-coated 30 nm gold nanoparticles in a 1:5, gold particle to dynein molecule ratio. The mixture was incubated on ice for at least 15 min. The dynein–gold conjugate sample was then diluted in the lysis buffer supplemented with oxygen scavenger system (300 µg/mL glucose oxidase (Sigma), 120 µg/mL catalase (Sigma), 0.4%, w/v glucose) and loaded into the flow cell.
3.4 Data Acquisition and Analysis
3.4.1 Image Operations: Flat-Fielding and Background Subtraction
The raw images from an iSCAT microscope contain static features resulting from nonuniform illumination, nonuniform pixel response, fixed-pattern noise, spurious back reflections, unwanted interference between back reflections, and imperfections of optical elements. These features are time invariant, do not change with lateral sample displacements, and can therefore be removed. We call the process of isolating and subsequently removing these signatures “flat-fielding” (Arroyo, Cole, & Kukura, 2016). First, the sample is translated laterally while recording a stack of 100 images. Second, a median image is generated by computing the median for each pixel across the stack of acquired images. Any subsequently recorded images are divided by this median image to account for illumination inhomogeneities.
TIP: The focus position of the sample must remain constant during acquisition of the flat-field stack of images, as some background signatures are focus dependent. In addition, the sample must be translated sufficiently quickly so that constant sample features are not imprinted onto the median image.
NOTE: Flat-fielding can only correct for truly static background features as the median image is acquired over several seconds.
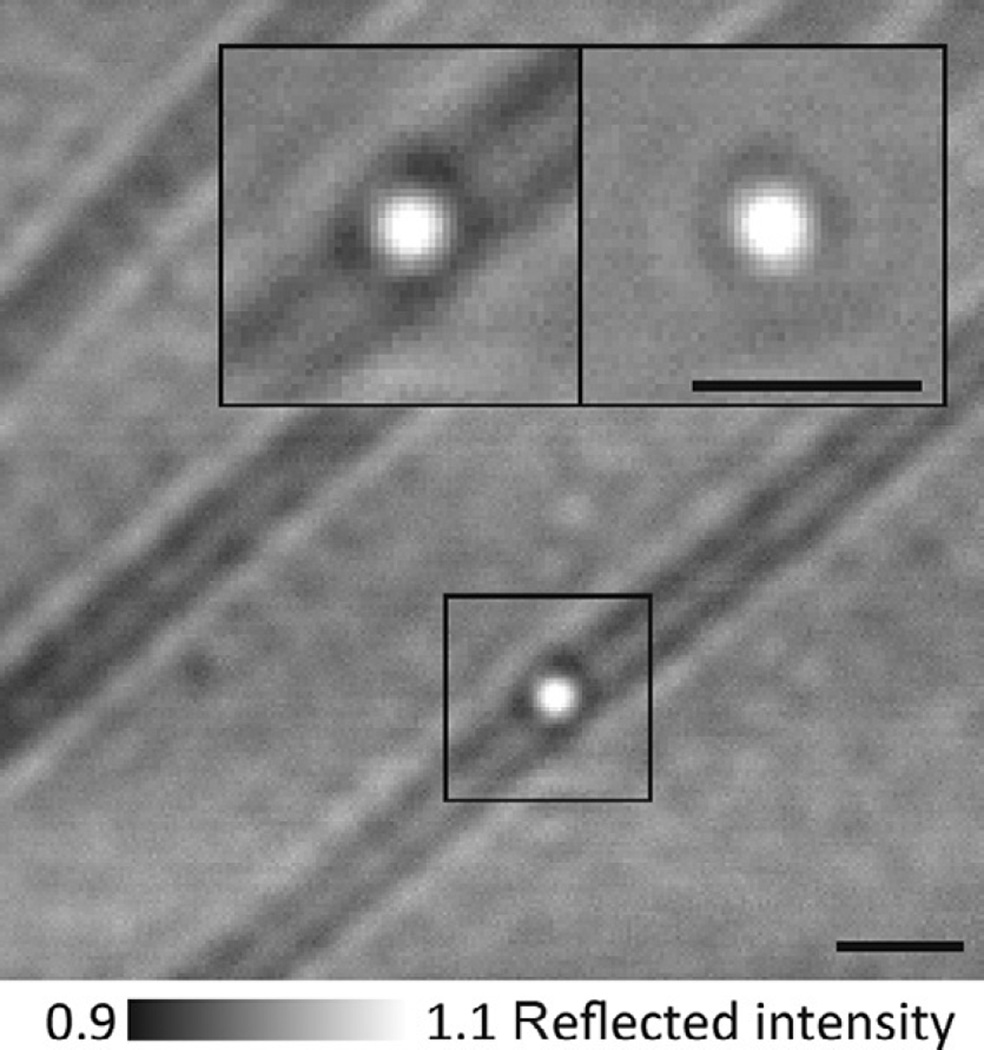
Despite the “flat-fielding” process, intrinsic sample features still contribute to the scattering background that limits the achievable signal-to-noise ratio (SNR). In the case of motor proteins, such background originates from actin filaments or microtubules, along with any inactive motors and other surface-bound objects. This scattering background can overwhelm the signal of interest, especially for microtubules, which produce ~8% iSCAT signal, comparable to 20 nm gold nanoparticles (Fig. 3). If the scattering background remains constant, however, it can be removed by background subtraction, as follows: from a recorded stack of images, a single median image is generated by averaging the entire stack (Figs. 2 and 3, insets). This median image contains only static features intrinsic to the sample (background). Subtracting this median image from each image in the acquired stack yields an image stack dominated by shot noise and the signal of the gold-labeled translocating motor.
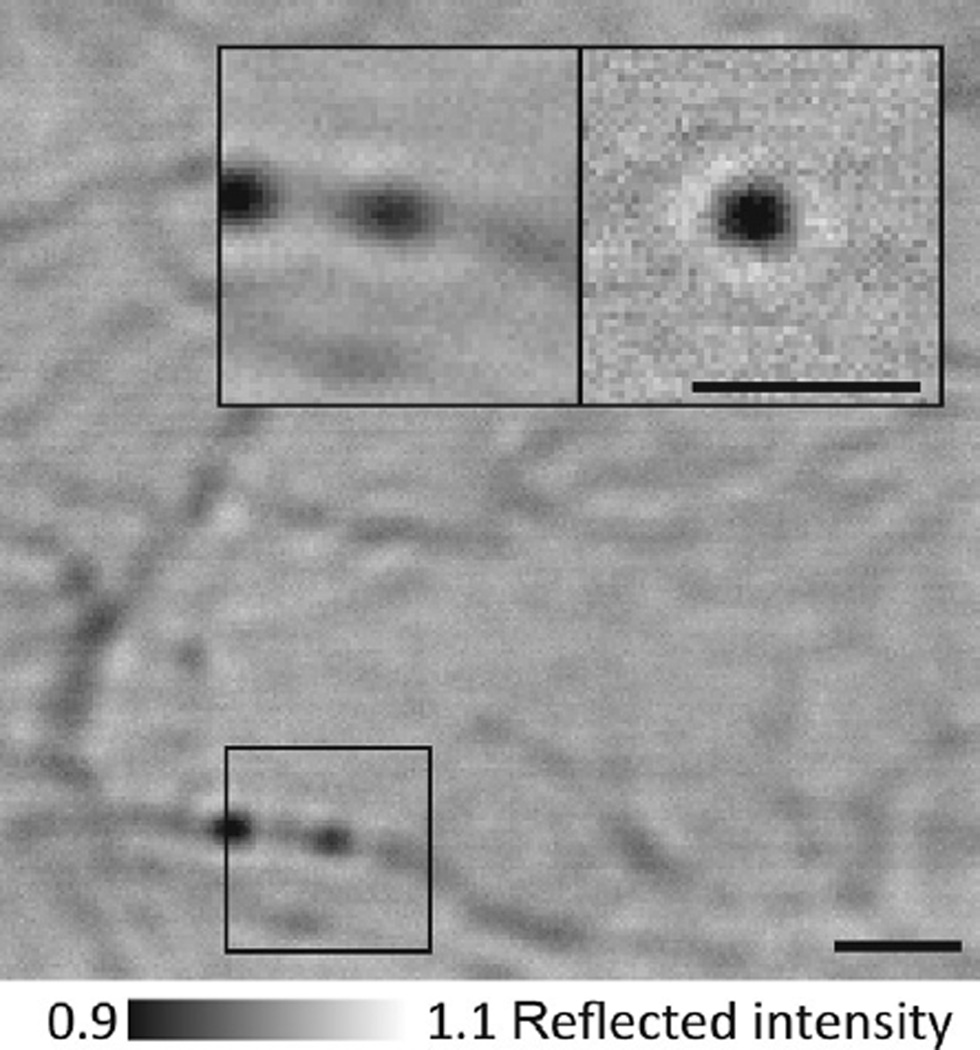
Fig. 3.
iSCAT imaging of gold-labeled dynein. 30 nm gold-labeled dynein bound to a microtubule. In this assay, a gold label produces a positive iSCAT contrast when placed into focus (bright) due to the increased distance between the label and the coverglass surface, in contrast to myosin-5a head-labeled gold, which invariably appeared dark (Fig. 2). Scale bars: 1 µm.
Fig. 2.
iSCAT assay for gold-labeled myosin-5a tracking. Flat-field-corrected image showing actin filaments and some 20 nm gold-labeled myosin-5a molecules bound to actin. Inset: zoom of the indicated region showing 20 nm gold-labeled myosin-5a bound to actin (left) and the same region after background subtraction, which removes all static features including the actin filament and any immobile particles (right). Scale bars: 1 µm.
NOTE: If a motor protein tends to pause or stops altogether, the median filter approach will fail unless static images are removed during the median image processing. The background image should only contain immobile filaments, therefore, it is necessary to select a substack of images before a particle binds to a filament or after it detaches. In this case a median (or average) image can be calculated and subtracted from the remaining stack to reveal a moving particle.
3.4.2 Fitting Diffraction-Limited Spots
From the processed images, diffraction-limited spots can be identified by a sequence of pattern recognition operations that output the location of a candidate feature to the nearest integer pixel value. First, each image is filtered using a Gaussian kernel with a size defined by the full width at half maximum of the diffraction-limited spots (~200 nm). This Gaussian blurring step artificially enhances the SNR of such features by smoothing out high-frequency noise caused by shot statistics. Next, a nonmaximum suppression algorithm identifies the local extrema within the optical resolution of the imaging system (Neubeck & Van Gool, 2006). In practice, selecting only pixel values exceeding twice the standard deviation of the entire image in the absence of outliers removes local maxima attributed to noise. Finally, candidate particles are segmented into regions of interest corresponding to approximately 1 µm2.
Subpixel localization information can be obtained from the candidate features, present in each segmented region of interest, by a nonlinear least square fit to an elliptical 2D Gaussian with a constant offset expressed by the following functional form:
where C represents the iSCAT contrast, xo and yo the center of mass location along each axis, sx and sy are proportional to the width of the Gaussian along each axis, and B a constant baseline. The nonlinear least square fit is performed by the iterative Levenberg–Marquardt algorithm, which requires an adequate initial guess for each of the adjustable parameters. This can be determined by a faster localization algorithm developed by Parthasarathy, termed radial symmetry centers (Neubeck & Van Gool, 2006). The advantage of this approach over iterative ones is that it is model free and requires a single iteration, which leads to an improvement in the execution time by two orders of magnitude, all while achieving similar performance to model-based techniques.
NOTE: The above fitting routine represents only one possible tracking algorithm. We also used open-source Fluorescent Image Evaluation Software for Tracking (FIESTA) to track gold-labeled kinesin (Ruhnow, Zwicker, & Diez, 2011).
3.4.3 Step-Finding Algorithms
Traditional step-finding algorithms analyze time vs distance traces in order to find time points where instantaneous and significant distance changes (steps) occur. In order to turn XY trajectories into time–distance traces that are amenable to step-finding analysis, the trajectory can simply be rotated such that the standard deviation in a given direction is minimized (Mickolajczyk et al., 2015). This rotation changes the coordinate system from XY axes to filament longitudinal and lateral axes and allows for more direct interpretation of how a motor is moving along the known structure of the filament.
Step-finding algorithms can broadly be split into two classes, model free and model dependent. Model-free algorithms make no assumptions about the data and simply perform significance testing to determine points where statistically significant changes in the data occur (Chen, Deffenbaugh, Anderson, & Hancock, 2014; Deffenbaugh, Chen, Anderson, & Hancock, 2015; Kalafut & Visscher, 2008). This class of algorithms is preferable when the goal of the study is to directly measure the size or number of steps present within a trace. A drawback of model-free algorithms is that they can suggest the existence of nonexisting step data if the significance level is set too low, and can miss steps if the significance level is set too high. The model-free detector algorithm, developed for counting the number of steps in GFP photobleaching data (Chen et al., 2014), has been applied with success to molecular motor data (Mickolajczyk et al., 2015). Model-dependent algorithms incorporate known information (a model) into the decision-making process for step finding (Kerssemakers et al., 2006; Milescu, Yildiz, Selvin, & Sachs, 2006a, 2006b; Mullner, Syed, Selvin, & Sigworth, 2010; Syed, Mullner, Selvin, & Sigworth, 2010). Example models might be the known lattice spacing of the filament or the known number of steps in a trace. Model-dependent algorithms are less-prone to over-fitting and are often the better choice when the goal of the study is to measure steps duration rather than step sizes. However, it is critical to justify the use of a model-dependent algorithm with prior experiments.
4. APPLICATIONS
4.1 Revealing the Motion of the Detached Head of Myosin-5a
For the movement ofmyosin-5a along actin, a body of biophysical work has led to a hand-over-hand stepping model in which the two heads alternate in leading, an average displacement of the striding head is 74 nm, and the detached head reaches the next binding site after Brownian rotation around a free joint at the head–head junction (Beausang, Shroder, Nelson, & Goldman, 2013; Dunn & Spudich, 2007; Forkey et al., 2003; Sakamoto, Webb, Forgacs, White, & Sellers, 2008; Veigel, Wang, Bartoo, Sellers, & Molloy, 2002; Warshaw et al., 2005; Yildiz et al., 2003). The last decade of research has been focused on understanding how unidirectional movement is achieved in the presence of random motion of the detached head (Karagiannis, Ishii, & Yanagida, 2014; Sellers & Veigel, 2006; Vale, 2003). Despite the variety of techniques with which myosin’s motion has been characterized, it has remained beyond reach to directly visualize individual head stepping in real time.
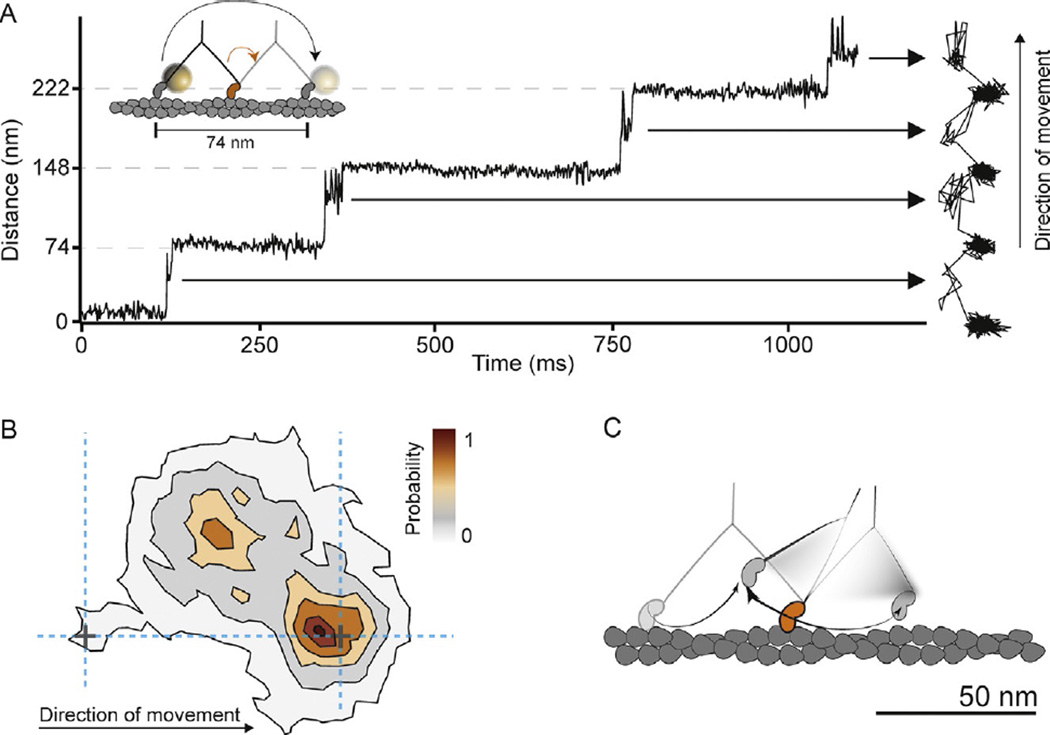
In order to resolve the detached head movement a 20-nm gold label was attached to the N-terminus of the myosin-5a head and we tracked its motion in an iSCAT microscope. According to the Brownian search model, we expected do detect periods of increased positional fluctuations of the labeled head between the 74 nm steps, which were previously interpreted as a signature of Brownian search (Dunn & Spudich, 2007). Surprisingly, instead of random motion of the bead between the two binding sites, we revealed a transient state with a center of mass just over half way between the two binding sites and offset by 40 nm perpendicular to the actin filament (Fig. 4A and B). The myosin head repeatedly moved back and forth between the transient state and the next actin-binding site (Andrecka et al., 2015). An important implication of our data is that the appearance of the transient state seems inconsistent with a purely rotational diffusion type of search. The position of the transient state, together with signatures of constrained diffusion, suggests that myosin side-steps along actin in a combined twisting and leaning motion, much more like a drawing compass rather than a freely jointed swivel (Fig. 4C).
Fig. 4.
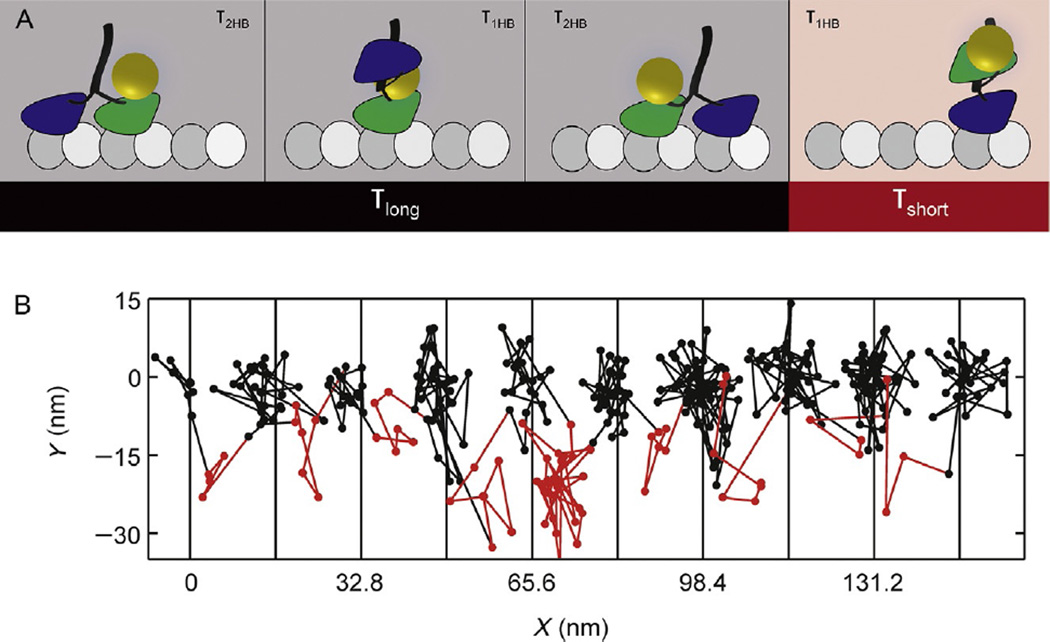
Myosin-5a head motion during processive stepping. (A) iSCAT tracking results: distance traveled as a function of time for a single-motor domain labeled with a 20-nm gold particle at its N-terminus and the corresponding 2D-trajectory, which is shown to the right of the time trace. The arrows indicate ~40 nm off-axis side position of the unbound motor domain (transient state). Inset: schematic of myosin-5a labeling and movement. Imaging speed: 1000 frames per second. (B) Contour map of a two-dimensional histogram with a 10 × 10-nm2 bin width obtained from the transient state of the unbound head x–y trajectories (number of steps=486). All contributing steps were aligned and those to the right of the filament when viewed in the direction of motion were mirrored. (C) The data lead to a hand-over-hand spinning model (shown schematically), in which each step proceeds via a single, spatially constrained transient state of the detached head (Andrecka et al., 2015).
4.2 Microtubule Motors
4.2.1 The Kinesin-1 Mechanochemical Cycle
The kinesin superfamily includes 45 genes in humans and is responsible for a huge diversity of tasks in the cell, including vesicle transport, microtubule dynamics control, and mitotic spindle organization (Hirokawa & Takemura, 2004). We recently applied iSCAT to solve long-standing questions regarding kinesin mechanochemistry by directly visualizing the dynamics of each step of the canonical kinesin-1 motor (Mickolajczyk et al., 2015). Attaching 30 nm gold nanoparticle to a single-motor domain of processive kinesin-1 dimers (Fig. 5A) enabled us to follow the motion of the motor over time. By identifying and quantifying the amount of time that kinesin-1 spent with one head bound vs two heads bound at various ATP concentrations, we determined the order and rates of mechanochemical transitions (Fig. 5B). We found the ATP-waiting state to be most similar to a two-heads-bound state, and the one-head-bound state duration was found to be ATP independent with a lifetime of about 8 ms. By using the slowly hydrolyzable ATP analog ATPγS, we further determined that hydrolysis precedes exit from the one-head-bound state, consistent with recent optical trapping studies (Milic, Andreasson, Hancock, & Block, 2014).
Fig. 5.
Example results of kinesin tracking with iSCAT. (A) Schematic showing detectable transitions in the two-step cycle of kinesin with only one motor domain tagged. The entire step (including both one- and two-head-bound durations) of the unlabeled head as well as the two-heads-bound duration of the labeled head precedes the first translocation of the bead (black), and the one-head-bound duration of the labeled head precedes the second translocation of the bead (red). (B) Example XY trajectory for kinesin-1 at saturating ATP. The amount of time that the motor spends on the microtubule and off the microtubule is easily determined. Black and red coloring corresponds to (A). Vertical lines show available microtubule-binding sites. Imaging speed: 1000 frames per second (Mickolajczyk et al., 2015).
4.2.2 Dynein
We also applied iSCAT tracking to the microtubule motor, cytoplasmic dynein from S. cerevisiae, brewer’s yeast. In contrast to kinesin-1, dynein walks toward the minus end of microtubules (Paschal & Vallee, 1987) and has an irregular stepping pattern, taking both forward and backward steps of variable size as well as moving laterally, switching between different protofilaments that make up the microtubule wall (Reck-Peterson et al., 2006). Dynein has been shown to use both a hand-over-hand stepping mechanism, where the leading head alternates with each step, and an inchworm-like mechanism where either the front or rear head steps without passing the other head (DeWitt et al., 2012; Qiu et al., 2012). iSCAT provides the opportunity to characterize in detail this unusual and highly variable stepping mechanism and the events that take place during each step.
Previous single-molecule fluorescence tracking studies of dynein with TIRF microscopy used low ATP concentrations (between 4 and 12 µM) to slow the stepping rate sufficiently to enable detection of individual steps (DeWitt et al., 2012; Qiu et al., 2012; Reck-Peterson et al., 2006). The high spatial and temporal resolution of iSCAT enabled us to track dynein at near saturating ATP concentrations (100 µM), which results in a mean velocity of 36.7 ± 20.4 nm/s.
As described in the protocol, we labeled yeast cytoplasmic dynein with 20–40 nm gold nanoparticles. As in earlier studies (DeWitt et al., 2012; Qiu et al., 2012), we observed a wide distribution of step sizes when we tracked the position of the particle on the dynein motor domain (Fig. 6). With the high spatial and temporal resolution of iSCAT, 16 and 24 nm steps were easily resolved as clusters of points in the lateral position of the tracked gold nanoparticle (Fig. 6), as well as smaller 8 nm steps. Yeast cytoplasmic dynein is highly processive (Gennerich, Carter, Reck-Peterson, & Vale, 2007), and we frequently observed runs >2 µm during the entire 40 s period of recording.
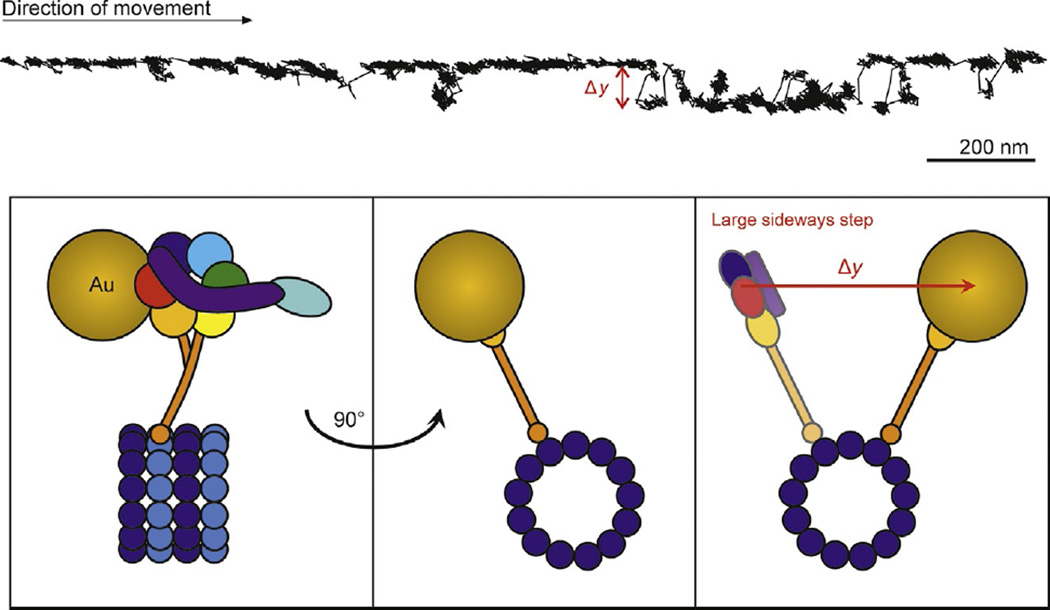
Fig. 6.
iSCAT tracking of dynein. Example x–y plot of cytoplasmic dynein position along a microtubule exhibiting side deviations from the track (Δy). The schematic presents one possible explanation of the observed trace where the dynein molecule switches microtubule protofilaments. Dynein was tracked at 200 frames per second using a 20-nm gold nanoparticle bound to AAA domains 5 and 6 (indicated by orange and red circles, respectively).
A prominent feature of many of the recordings was large off-axis motion, seen as changes perpendicular to the microtubule axis greater than 50 nm, followed by a return to the original axis of motion (Fig. 6). In some cases dynein appeared to transition between sequences of straight steps and off-axis steps. Off-axis steps were often correlated with substantial changes in iSCAT contrast, presumably due to z-axis motions with protofilament switching. Conversely, traces without off-axis stepping tended to have more constant contrast levels (not shown).
Our results indicate that during processive stepping along microtubules, dynein moves not only parallel to the microscope slide (x and y motions) but also closer and farther away from the slide (in z), consistent with changes to microtubule protofilament and azimuthal position around the microtubule. Further iSCAT studies should be valuable toward understanding the detailed motions of dynein’s puzzling transport mechanism.
5. OUTLOOK
We have shown how iSCAT coupled with nanoparticle labels can be used to produce nm-precise and highly time-resolved tracks of several processive motor proteins moving along their tracks in vitro. In addition to the capabilities presented here, there are numerous opportunities for both improving the capabilities of iSCAT for SPT and broadening the scope of iSCAT more generally.
In terms of improving the capabilities for SPT, there are three relatively straightforward avenues: (i) increasing localization precision for smaller labels; (ii) increasing imaging speed; and (iii) 3D tracking. Label size is important for two reasons. First, as imaging speed increases beyond the ms threshold, the observed motion will increasingly be affected by the drag or fluctuations of the bead. Thus, even though nominal localization precision may be on the order of a few nm, the positional fluctuations of the particle will be much larger, as the motions of the label itself are resolved. This effect is minimized for smaller labels, both because the positional fluctuations themselves become smaller and because the label moves more rapidly, causing any such fluctuations to average out until much higher imaging speeds are achieved. This distinction is also important when comparing dark field with interferometric scattering (Isojima et al., 2016), which have similar theoretical SNRs for the same incident power and scattering properties (Ortega-Arroyo & Kukura, 2012). Even though the nominal spatiotemporal precision can be very high (Isojima et al., 2016), large particle sizes require time averaging to remove localization noise caused by label motion, lowering the effective achievable localization precision and imaging speed. Thus, achieving true sub-ms and nm-level precision simultaneously will invariably require the use of labels with diameters of 20 nm or smaller.
Ideally, labels on the order of single fluorescent proteins would be desirable, with the smallest possible linker length. For instance, the high spatiotemporal precision for low incident power for 20 nm labels shown in Fig. 1C would translate into similar imaging performance with 5 nm silver labels and a moderate increase of illumination intensity. The major challenge associated with using labels with diameters on the order of 10 nm or less is purely one of convenience—they become difficult to detect outright due to the rough imaging background generated by microscope cover glass, however, in dynamic measurements where labeled motors bind to their tracks, move, and unbind, this background can be efficiently subtracted, paving the way for future studies with smaller labels and higher imaging speeds without any loss in localization precision. The ultimate limits are likely to be determined by light absorption and the resulting sample heating, although this could be alleviated by pulsed excitation, for example.
Beyond label size, extending localization from two to three dimensions would be valuable for many applications. There is precedent for using iSCAT for 3D tracking, although the precise calibration of the 3D response has generally been challenging and a clear methodology has yet to be developed (Andrecka et al., 2015; Goldfain, Garmann, Jin, Lahini, & Manoharan, 2016; Krishnan, Mojarad, Kukura, & Sandoghdar, 2010). The interferometric nature of the technique, however, lends itself well to very high localization precision along the optical axis on the length scale of a few tens of nm.
In many cases, it is highly advantageous to perform iSCAT and fluorescence experiments simultaneously (Andrecka et al., 2015; Kukura et al., 2009). The compatibility of iSCAT with single-molecule fluorescence imaging makes it possible to use well-established, fluorescent assays and combine them with the advantages of iSCAT imaging. Furthermore, multilabel experiments, where simultaneous tracking is performed, can be achieved with an iSCAT and fluorescence imaging modality (Andrecka et al., 2015). Such approaches could be of particular value for initial experiments on cell surfaces, which may be possible on cellular features that are particularly thin and produce little background scatter, such as filopodia, as opposed to cell bodies.
Finally, it is worth noting that there are ample opportunities for using iSCAT in a completely label-free manner. We initially demonstrated label-free tracking of myosin-5a (Ortega Arroyo et al., 2014), but such approaches could be of particular interest also for dynein tracking, which has a greater molecular weight and thereby produces a stronger iSCAT signal. The advantages here are that the center of mass of the protein can be tracked with very long observation times.
Acknowledgments
P.K. is supported by an ERC starting grant (NanoScope). J.A. was supported by a Marie Curie Fellowship (330215). K.J.M. and W.O.H. were supported by the NIH R01 GM076476. L.G.L. and Y.E.G. were supported by NIH Grants P01 GM087253 and R01 GM086352. L.G.L was supported by the NIH training grant T32- GM008275. J.R.S. and Y.T. were supported by funds from the NHLBI Intramural Research Program.
References
- Allen RD, Metuzals J, Tasaki I, Brady ST, Gilbert SP. Fast axonal-transport in squid giant-axon. Science. 1982;218(4577):1127–1129. doi: 10.1126/science.6183744. [DOI] [PubMed] [Google Scholar]
- Andreasson JOL, Milic B, Chen GY, Guydosh NR, Hancock WO, Block SM. Examining kinesin processivity within a general gating framework. eLife. 2015;4:e07403. doi: 10.7554/eLife.07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrecka J, Arroyo JO, Lewis K, Cross RA, Kukura P. Label-free imaging of microtubules with sub-nm precision using interferometric scattering microscopy. Biophysical Journal. 2016;110(1):214–217. doi: 10.1016/j.bpj.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrecka J, Ortega Arroyo J, Takagi Y, de Wit G, Fineberg A, MacKinnon L, Kukura P. Structural dynamics of myosin 5 during processive motion revealed by interferometric scattering microscopy. eLife. 2015;4:e05413. doi: 10.7554/eLife.05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JO, Cole D, Kukura P. Interferometric scattering microscopy and its combination with single-molecule fluorescence imaging. Nature Protocols. 2016;11(4):617–633. doi: 10.1038/nprot.2016.022. [DOI] [PubMed] [Google Scholar]
- Arroyo JO, Kukura P. Non-fluorescent schemes for single-molecule detection, imaging and spectroscopy. Nature Photonics. 2016;10(1):11–17. [Google Scholar]
- Beausang JF, Shroder DY, Nelson PC, Goldman YE. Tilting and wobble of myosin V by high-speed single-molecule polarized fluorescence microscopy. Biophysical Journal. 2013;104:1263–1273. doi: 10.1016/j.bpj.2013.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM, Goldstein LSB, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348(6299):348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Deffenbaugh NC, Anderson CT, Hancock WO. Molecular counting by photobleaching in protein complexes with many subunits: Best practices and application to the cellulose synthesis complex. Molecular Biology of the Cell. 2014;25(22):3630–3642. doi: 10.1091/mbc.E14-06-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis ASG. Mechanism of adhesion of cells to glass—Study by interference reflection microscopy. Journal of Cell Biology. 1964;20(2):199–215. doi: 10.1083/jcb.20.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffenbaugh NC, Chen YL, Anderson CT, Hancock WO. Novel step detection algorithms for photobleaching analysis of protein complexes with many subunits. Biophysical Journal. 2015;108(2):438a. [Google Scholar]
- DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335(6065):221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulin D, Barland S, Hachair X, Pedaci F. Efficient illumination for microsecond tracking microscopy. PLoS One. 2014;9(9):e107335. doi: 10.1371/journal.pone.0107335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AR, Spudich JA. Dynamics of the unbound head during myosin V processive translocation. Nature Structural & Molecular Biology. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- Dunn AR, Spudich JA. Single-molecule gold-nanoparticle tracking. Cold Spring Harbor Protocols. 2011;2011(12):1498–1506. doi: 10.1101/pdb.prot066977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filler TJ, Peuker ET. Reflection contrast microscopy (RCM): A forgotten technique? Journal of Pathology. 2000;190(5):635–638. doi: 10.1002/(SICI)1096-9896(200004)190:5<635::AID-PATH571>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Forkey JN, Quinlan ME, Shaw MA, Corrie JE, Goldman YE. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous-solution. Nature. 1995;374(6522):555–559. doi: 10.1038/374555a0. [DOI] [PubMed] [Google Scholar]
- Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131(5):952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfain AM, Garmann RF, Jin Y, Lahini Y, Manoharan VN. Dynamic measurements of the position, orientation, and DNA content of individual unlabeled bacteriophages. Journal of Physical Chemistry B. 2016;120(26):6130–6138. doi: 10.1021/acs.jpcb.6b02153. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Experimental Cell Research. 2004;301(1):50–59. doi: 10.1016/j.yexcr.2004.08.010. http://dx.doi.org/10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Horio T, Hotani H. Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature. 1986;321(6070):605–607. doi: 10.1038/321605a0. [DOI] [PubMed] [Google Scholar]
- Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342(6246):154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- Isojima H, Iino R, Niitani Y, Noji H, Tomishige M. Direct observation of intermediate states during the stepping motion of kinesin-1. Nature Chemical Biology. 2016;12(4):290–297. doi: 10.1038/nchembio.2028. [DOI] [PubMed] [Google Scholar]
- Johnson V, Ayaz P, Huddleston P, Rice LM. Design, overexpression, and purification of polymerization-blocked yeast alpha beta-tubulin mutants. Biochemistry. 2011;50(40):8636–8644. doi: 10.1021/bi2005174. [DOI] [PubMed] [Google Scholar]
- Kalafut B, Visscher K. An objective, model-independent method for detection of non-uniform steps in noisy signals. Computer Physics Communications. 2008;179(10):716–723. [Google Scholar]
- Karagiannis P, Ishii Y, Yanagida T. Molecular machines like myosin use randomness to behave predictably. Chemical Reviews. 2014;114:3318–3334. doi: 10.1021/cr400344n. [DOI] [PubMed] [Google Scholar]
- Kerssemakers JWJ, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442(7103):709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Krishnan M, Mojarad N, Kukura P, Sandoghdar V. Geometry-induced electrostatic trapping of nanometric objects in a fluid. Nature. 2010;467(7316):692–695. doi: 10.1038/nature09404. [DOI] [PubMed] [Google Scholar]
- Kukura P, Ewers H, Muller C, Renn A, Helenius A, Sandoghdar V. High-speed nanoscopic tracking of the position and orientation of a single virus. Nature Methods. 2009;6(12):923–927. doi: 10.1038/nmeth.1395. [DOI] [PubMed] [Google Scholar]
- Kulman JD, Satake M, Harris JE. A versatile system for site-specific enzymatic biotinylation and regulated expression of proteins in cultured mammalian cells. Protein Expression and Purification. 2007;52(2):320–328. doi: 10.1016/j.pep.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lin YH, Chang WL, Hsieh CL. Shot-noise limited localization of single 20 nm gold particles with nanometer spatial precision within microseconds. Optics Express. 2014;22:9159–9170. doi: 10.1364/OE.22.009159. [DOI] [PubMed] [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- Mickolajczyk KJ, Deffenbaugh NC, Arroyo JO, Andrecka J, Kukura P, Hancock WO. Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(52):E7186–E7193. doi: 10.1073/pnas.1517638112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milescu LS, Yildiz A, Selvin PR, Sachs F. Extracting dwell time sequences from processive molecular motor data. Biophysical Journal. 2006a;91(9):3135–3150. doi: 10.1529/biophysj.105.079517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milescu LS, Yildiz A, Selvin PR, Sachs F. Maximum likelihood estimation of molecular motor kinetics from staircase dwell-time sequences. Biophysical Journal. 2006b;91(4):1156–1168. doi: 10.1529/biophysj.105.079541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milic B, Andreasson JOL, Hancock WO, Block SM. Kinesin processivity is gated by phosphate release. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(39):14136–14140. doi: 10.1073/pnas.1410943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner FE, Syed S, Selvin PR, Sigworth FJ. Improved hidden Markov models for molecular motors, part 1 basic theory. Biophysical Journal. 2010;99(11):3684–3695. doi: 10.1016/j.bpj.2010.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubeck A, Van Gool L. Efficient non-maximum suppression. In Proceedings of the 18th international conference on pattern recognition. 2006;3:850–855. [Google Scholar]
- Ortega Arroyo J, Andrecka J, Spillane KM, Billington N, Takagi Y, Sellers JR, Kukura P. Label-free, all-optical detection, imaging, and tracking of a single protein. Nano Letters. 2014;14:2065–2070. doi: 10.1021/nl500234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Arroyo J, Kukura P. Interferometric scattering microscopy (iSCAT): New frontiers in ultrafast and ultrasensitive optical microscopy. Physical Chemistry Chemical Physics. 2012;14(45):15625–15636. doi: 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- Ozeki T, Verma V, Uppalapati M, Suzuki Y, Nakamura M, Catchmark JM, Hancock WO. Surface-bound casein modulates the adsorption and activity of kinesin on SiO2 surfaces. Biophysical Journal. 2009;96(8):3305–3318. doi: 10.1016/j.bpj.2008.12.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods in Cell Biology. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Vallee RB. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 1987;330(6144):181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- Piliarik M, Sandoghdar V. Direct optical sensing of single unlabelled proteins and super-resolution imaging of their binding sites. Nature Communications. 2014;5:4495. doi: 10.1038/ncomms5495. [DOI] [PubMed] [Google Scholar]
- Qiu W, Derr ND, Goodman BS, Villa E, Wu D, Shih W, Reck-Peterson SL. Dynein achieves processive motion using both stochastic and coordinated stepping. Nature Structural & Molecular Biology. 2012;19(2):193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126(2):335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief M, Rock RS, Mehta AD, Mooseker MS, Cheney RE, Spudich JA. Myosin-V stepping kinetics: A molecular model for processivity. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhnow F, Zwicker D, Diez S. Tracking single particles and elongated filaments with nanometer precision. Biophysical Journal. 2011;100(11):2820–2828. doi: 10.1016/j.bpj.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Amitani I, Yokota E, Ando T. Direct observation of processive movement by individual myosin V molecules. Biochemical and Biophysical Research Communications. 2000;272(2):586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Webb MR, Forgacs E, White HD, Sellers JR. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455(7209):128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate-specificity of a peptide-modifying enzyme—A 13 residue consensus peptide specifies biotinylation in Escherichia coli. Bio/Technology. 1993;11(10):1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Schnapp BJ, Gelles J, Sheetz MP. Nanometer-scale measurements using video light-microscopy. Cell Motility and the Cytoskeleton. 1988;10(1–2):47–53. doi: 10.1002/cm.970100109. [DOI] [PubMed] [Google Scholar]
- Schneider R, Glaser T, Berndt M, Diez S. Using a quartz paraboloid for versatile wide-field TIR microscopy with sub-nanometer localization accuracy. Optics Express. 2013;21(3):3523–3539. doi: 10.1364/OE.21.003523. [DOI] [PubMed] [Google Scholar]
- Schneider R, Korten T, Walter WJ, Diez S. Kinesin-1 motors can circumvent permanent roadblocks by side-shifting to neighboring protofilaments. Biophysical Journal. 2015;108(9):2249–2257. doi: 10.1016/j.bpj.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR, Veigel C. Walking with myosin V. Current Opinion in Cell Biology. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Snyder GE, Sakamoto T, Hammer JA, 3rd, Sellers JR, Selvin PR. Nanometer localization of single green fluorescent proteins: Evidence that myosin V walks hand-over-hand via telemark configuration. Biophysical Journal. 2004;87(3):1776–1783. doi: 10.1529/biophysj.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365(6448):721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- Syed S, Mullner FE, Selvin PR, Sigworth FJ. Improved hidden Markov models for molecular motors, part 2 extensions and application to experimental data. Biophysical Journal. 2010;99(11):3696–3703. doi: 10.1016/j.bpj.2010.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophysical Journal. 2002;82(5):2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D, Chang CY, Fang K, Cuomo P, Murphy D. Kinesin movement on glutaraldehyde-fixed microtubules. Analytical Biochemistry. 1996;242(1):20–25. doi: 10.1006/abio.1996.0422. [DOI] [PubMed] [Google Scholar]
- Vale RD. Myosin V motor proteins: Marching stepwise towards a mechanism. Journal of Cell Biology. 2003;163(3):445–450. doi: 10.1083/jcb.200308093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Schnapp BJ, Mitchison T, Steuer E, Reese TS, Sheetz MP. Different axoplasmic proteins generate movement in opposite directions along microtubules in vitro. Cell. 1985;43(3):623–632. doi: 10.1016/0092-8674(85)90234-x. [DOI] [PubMed] [Google Scholar]
- Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nature Cell Biology. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- Wang F, Chen L, Arcucci O, Harvey EV, Bowers B, Xu Y, Sellers JR. Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. Journal of Biological Chemistry. 2000;275:4329–4335. doi: 10.1074/jbc.275.6.4329. [DOI] [PubMed] [Google Scholar]
- Warshaw DM, Kennedy GG, Work SS, Krementsova EB, Beck S, Trybus KM. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophysical Journal. 2005;88:L30–L32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303(5658):676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]