Significance
Embryonic stem cells (ESCs) maintain a low translation rate; therefore control of mRNA translation is critical for preserving their stemness. We identified a hitherto unstudied transcription factor, Yin-yang 2 (YY2), which is translationaly regulated and controls self-renewal and differentiation of mouse ESCs (mESCs). Although YY2 is essential for mESC self-renewal, increased YY2 expression directs differentiation of mESCs toward cardiovascular lineages. Examination of the Yy2 5′-UTR revealed a multilayer regulatory mechanism through which YY2 expression is dictated by the combined actions of the splicing regulator, Polypyrimidine tract-binding protein 1 (PTBP1), and the translation inhibitors, Eukaryotic initiation factor 4E-binding proteins (4E-BPs). YY2 directly controls the expression of several pluripotency and development-related genes. This study describes a synchronized network of alternative splicing and mRNA translation in controlling self-renewal and differentiation.
Keywords: mRNA translation, 4E-BPs, PTBP, embryonic stem cell, YY2
Abstract
Translational control of gene expression plays a key role during the early phases of embryonic development. Here we describe a transcriptional regulator of mouse embryonic stem cells (mESCs), Yin-yang 2 (YY2), that is controlled by the translation inhibitors, Eukaryotic initiation factor 4E-binding proteins (4E-BPs). YY2 plays a critical role in regulating mESC functions through control of key pluripotency factors, including Octamer-binding protein 4 (Oct4) and Estrogen-related receptor-β (Esrrb). Importantly, overexpression of YY2 directs the differentiation of mESCs into cardiovascular lineages. We show that the splicing regulator Polypyrimidine tract-binding protein 1 (PTBP1) promotes the retention of an intron in the 5′-UTR of Yy2 mRNA that confers sensitivity to 4E-BP–mediated translational suppression. Thus, we conclude that YY2 is a major regulator of mESC self-renewal and lineage commitment and document a multilayer regulatory mechanism that controls its expression.
Stringent control of mRNA translation is critical during early embryonic development, because relatively small changes in the expression of development-related genes can dramatically affect the self-renewal and differentiation of stem cells. In fact, a modest (twofold or less) increase or decrease in Octamer-binding protein 4 (OCT4) or Sex-determining region Y (SRY)-box 2 (SOX2) protein levels impairs ESC self-renewal and triggers differentiation (1, 2). mRNA translation, which is low in undifferentiated embryonic stem cells (ESCs) and multipotent somatic stem cells (e.g., hematopoietic stem cells and skin stem cells), increases significantly during differentiation (3–5). Importantly, genome-wide analysis of the transcriptome vs. proteome of ESCs during the early stages of differentiation demonstrated that protein levels correlate poorly with mRNA levels (Pearson’s R < 0.4), underscoring the importance of posttranscriptional regulation in ESC differentiation (6).
mRNA translation can be divided into three steps: initiation, elongation, and termination. Translational control has been documented most extensively at the initiation step, at which ribosomes are recruited to the mRNA by the concerted action of Eukaryotic translation initiation factors (eIFs) (7). Control of translation is exerted mainly by two key protein complexes: eIF4F (eIF4E–eIF4G–eIF4A) and the ternary complex (eIF2–GTP–Met-tRNAMeti) (7). The mammalian target of rapamycin complex 1 (mTORC1) controls the assembly of eIF4F through the phosphorylation of eIF4E-binding proteins (4E-BPs) (8, 9). The 4E-BPs consist of a family of small molecular weight (15–20 kDa) translational inhibitors (4E-BP1, -2, and -3 in mammals), that, when dephosphorylated, avidly bind eIF4E and block its association with eIF4G to form the eIF4F complex. Following phosphorylation by mTORC1, 4E-BPs dissociate from eIF4E, allowing the formation of the eIF4F complex and activation of translation (8, 10–12). 4E-BPs inhibit cap-dependent translation in embryonic and somatic stem cells (3, 4, 13, 14). Although eIF4E promotes cap-dependent translation of all cellular mRNAs, the translation of a subset of mRNAs, which generally contain a long and highly structured 5′-UTR, is strongly dependent on eIF4E (9, 15). These mRNAs are known as “eIF4E-sensitive” and encode proteins that control fundamental cellular processes such as cell proliferation and survival (16).
We showed that 4E-BPs are required for reprogramming mouse embryonic fibroblasts (MEFs) to induced pluripotent stem cells (iPSCs) (17). In the current study, we describe a tightly coordinated network in mESCs whereby the expression of the Yin-yang 2 (YY2) transcription factor is controlled by the splicing regulator Polypyrimidine tract-binding protein 1 (PTBP1) and the 4E-BP translation inhibitors. Our data reveal that stringent regulation of YY2 expression by this network is critical for mESC self-renewal and lineage commitment.
Results
Transcriptome and Translatome Profiling of WT and 4E-BP1/2–Null mESCs.
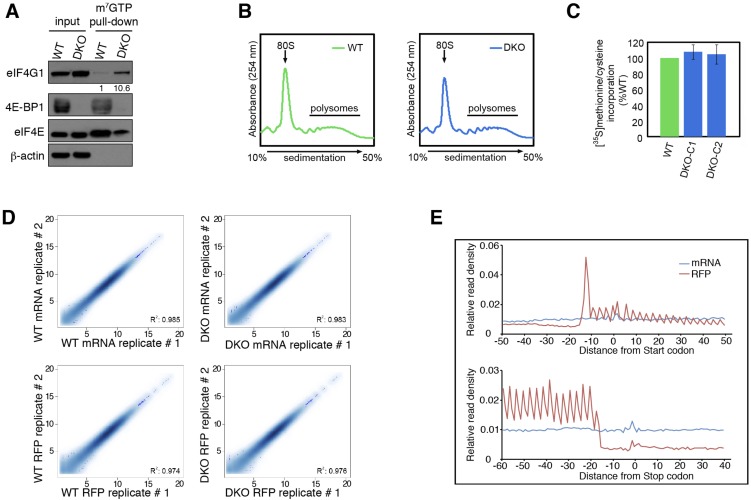
To investigate the role of 4E-BPs in mESCs, we first derived mESCs from WT and Eukaryotic translation initiation factor 4E-binding protein 1 (Eif4ebp1) and Eukaryotic translation initiation factor 4E-binding protein 2 (Eif4ebp2) double-knockout (DKO) mice and then examined the eIF4F complex using a 7-methyl-GTP (m7GTP)-agarose pull-down assay. The eIF4F amount was elevated, as demonstrated by increased (∼10.6-fold) eIF4G1 pull-down in DKO mESCs (Fig. S1A). However, polysome profiling (Fig. S1B) and [35S]methionine/cysteine-labeling assays (Fig. S1C) did not detect a substantial difference in global mRNA translation between WT and DKO mESCs. These data are consistent with previous findings that the lack of 4E-BPs affects the translation of a subset of mRNAs rather than affecting global translation (17, 18).
Fig. S1.
Translation control in WT and 4E-BP1/2–null mESCs. (A) Lysates from WT and Eif4ebp1 and Eif4ebp2 DKO mESCs were subjected to m7GTP pull-downs and analyzed for the indicated proteins. Numbers indicate the ratio of eIF4G1 in each pull-down to that in WT cells (B) Polysome profiles of WT and DKO mESCs treated with 100 µg/mL cycloheximide for 5 min. Absorbance light was set at 254 nm. (C) [35S] methionine/cysteine incorporation into newly synthesized proteins from WT and DKO mESCs grown in 10% (vol/vol) dialyzed FBS and pulsed for 30 min with [35S] methionine/cysteine. Data are mean ± SD (n = 3). (D) Correlation between replicates in mRNA-Seq and ribosome profiling datasets. R2 indicates the Pearson correlation. (E) Metagene analysis of randomly fragmented mRNAs and RFPs in mESCs. Normalized read counts are averaged across the entire transcriptome and aligned at the annotated start codons and stop codons.
To identify 4E-BP–sensitive mRNAs in mESCs, we performed ribosome profiling (19), which allows precise measurement of the translation of mRNAs on a genome-wide scale, by deep sequencing of ribosome-protected fragments (ribosome footprints; RFPs). We achieved a high degree of reproducibility between the replicates for mRNA sequencing (mRNA-Seq) and RFPs (R2 > 0.97, Fig. S1D). Metagene analysis confirmed the enrichment of RFP reads in coding sequences and the expected three-nucleotide periodicity (Fig. S1E). These analyses validated the quality of the mRNA and RFP libraries (Table S1). We used Babel analysis (20) to compute changes in the abundance of RFPs (Fig. 1A) independent of changes in the levels of their corresponding mRNAs (Fig. 1B). A significant enhancement in the translation efficiency of a small subset of mRNAs was detected in DKO mESCs [false discovery rate (FDR) < 0.1] (Fig. 1C and Dataset S1), as is consistent with the lack of global change in translation in the DKO mESCs (Fig. S1 B and C). Strikingly, mRNA-Seq data revealed down-regulation of mRNA levels for several pluripotency factors, such as PR domain-containing 14 (Prdm14), ES cell-expressed Ras (Eras), Estrogen-related receptor-β (Esrrb), and Nanog (−1.3, −1, −0.6, and −0.9, respectively; log2 DKO/WT) in DKO mESCs (Dataset S2). Possible reasons for this down-regulation are discussed below.
Table S1.
Number of uniquely mapped reads for each mRNA-Seq and RFP sample
| Sample | Uniquely mapped reads |
| WT_mRNA_1 | 34,574,032 |
| WT_mRNA_2 | 36,153,258 |
| WT_RFP_1 | 79,42,754 |
| WT_RFP_2 | 11,624,184 |
| DKO_mRNA_1 | 24,028,798 |
| DKO_mRNA_2 | 36,078,998 |
| DKO_RFP_1 | 8,946,514 |
| DKO_RFP_2 | 11,302,403 |
This table is related to Fig. 1.
Fig. 1.
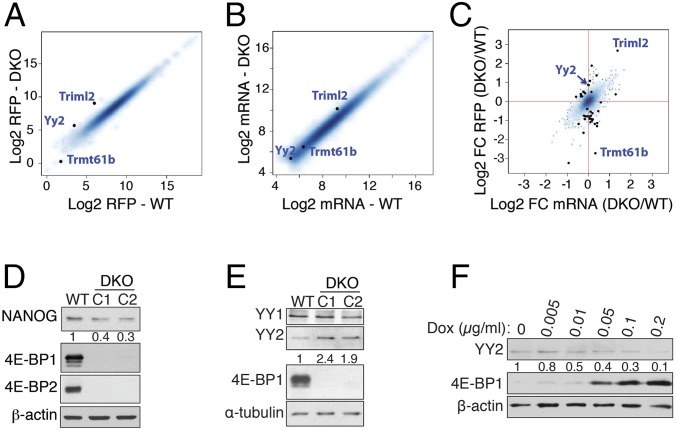
The lack of 4E-BPs deregulates the expression of pluripotency factors in mESCs. (A and B) The log2 abundance of RFPs (A) and mRNA (RNA-Seq) (B) of transcripts that were included in Babel analysis are plotted for WT and Eif4ebp1 and Eif4ebp2 DKO mESCs. (C) Babel analysis of transcripts with a significant change in RFPs independent of the corresponding change in mRNA abundance (black dots; FDR < 0.1). Triml2 and Trmt61b, respectively, are mRNAs with the highest and the lowest RFP ratios in DKO compared with WT mESCs; FC, fold change. (D) Western blot analysis of NANOG expression in a WT mESC and in two independent DKO mESC clones (C1 and C2). Numbers indicate the ratio of NANOG expression in each clone to that in the WT mESC followed by normalization with β-actin. (E) Western blot analysis of YY2 and YY1 expression in a WT mESC and two independent DKO mESC clones. Numbers indicate the ratio of YY2 expression in each clone compared with the WT mESC followed by normalization with α-tubulin. (F) DKO mESCs carrying the doxycycline-inducible 4E-BP1-4A mutant construct were treated with 0, 0.005, 0.01, 0.05, 0.1, or 0.2 μg/mL doxycycline for 24 h and were subjected to Western blot analysis. Numbers indicate the ratio of YY2 expression in each treatment compared with no doxycycline followed by normalization with β-actin.
Ablation of Eif4ebp1 and Eif4ebp2 Results in Reduced Expression of mESC Markers.
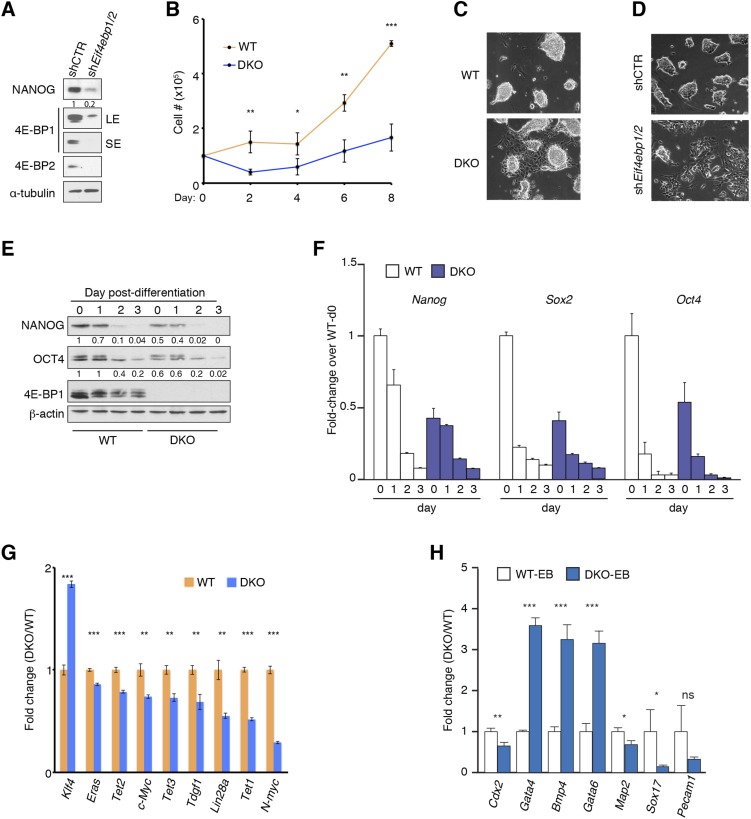
To validate the mRNA-Seq results, we examined the expression of pluripotency factors in undifferentiated WT and DKO mESCs, and also in WT and DKO mESCs after differentiation, by Western blot and quantitative RT-PCR (RT-qPCR). Although all DKO mESC lines maintained normal morphology under standard mESC culture conditions [in the presence of Leukemia inhibitory factor (LIF) and feeder layer (irradiated MEFs)], expression of the ESC marker NANOG was reduced by 60–70% (Fig. 1D). These changes did not result from unintended consequences of the knockout procedure, because RNAi-mediated depletion of 4E-BP1 and -2 (double knockdown; DKD) resulted in a strong (80%) reduction in NANOG (Fig. S2A). DKO mESCs proliferated more slowly than WT mESCs (Fig. S2B), and when cultured in the absence of feeder layer, they exhibited flattened morphology, which is indicative of cellular differentiation, whereas WT mESCs preserved their normal morphology (Fig. S2C). Similar morphological changes were observed in DKD ESCs (Fig. S2D). Notably, when cultured in the absence of LIF and a feeder layer, NANOG and OCT4 expression was strongly suppressed in DKO mESCs, whereas WT mESCs maintained a higher expression of these proteins (Fig. S2E). In addition to Nanog, mRNA levels of Oct4 and Sox2 were lower in DKO mESCs on day 0 of differentiation (Fig. S2F). Moreover, as determined by RT-qPCR, mRNA levels of several other ESC factors, such as Lin28a, Eras, and ten-eleven translocations (Tets) (Tet1, -2, and -3), were reduced in DKO mESCs (Fig. S2G). Therefore, 4E-BPs are required for the regulation of expression of pluripotency factors. Analysis of embryoid bodies (EBs) derived from WT and DKO ESCs revealed that the lack of 4E-BP1 and 4E-BP2 resulted in the differentiation of mESCs toward mesodermal and endodermal lineages, as indicated by significant up-regulation of Bone morphogenetic protein 4 (Bmp4), an early mesoderm marker, and Gata4 and Gata6, early endoderm markers (Fig. S2H). This up-regulation coincides with the down-regulation of the neuroectoderm markers Microtubule-associated protein 2 (Map2) and Sex-determining region Y (SRY)-box 17 (Sox17) mRNAs in DKO EBs.
Fig. S2.
Characterization of mESCs in the presence or absence of 4E-BP1/2. (A) Western blot analysis of mESCs transduced by lentivirus expressing shRNAs against 4E-BP1 and 4E-BP2 (shEif4ebp1/2) or control (shCTR). LE, long exposure; SE, short exposure. Numbers indicate the ratio of NANOG expression in each condition to that in control followed by normalization with α-tubulin. (B) Cell growth assay for WT and Eif4ebp1 and Eif4ebp2 DKO mESCs. Data are mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. (C) WT and DKO mESCs cultured in the absence of feeder layer. (D) Control and 4E-BP1 and DKD mESCs cultured in the absence of feeder layer. (E and F) Western blot (E) and RT-qPCR (F) analysis of expression of selected pluripotency factors in WT and DKO mESCs cultured in the absence of feeder layer and LIF for the indicated time. RT-qPCR values are normalized to β-actin. Data are mean ± SD (n = 3). Numbers in E indicate the ratio of NANOG or OCT4 expression in each condition to that at day 0 of differentiation of WT mESCs followed by normalization with β-actin. (G) RT-qPCR analysis of selected pluripotency markers in WT and DKO mESCs. Values are normalized to β-actin. Data are mean ± SD (n = 3). **P < 0.01, ***P < 0.001. (H) RT-qPCR analysis of WT and DKO EBs 2 wk postdifferentiation for expression of selected differentiation markers. Values are normalized to β-actin. Data are mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001; ns, nonsignificant.
The decrease in mRNA levels for several pluripotency factors, such as Nanog, Eras, and Esrrb in DKO mESCs compared with WT mESCs (Fig. 1D, Fig. S2 F and G, and Dataset S2) suggests that 4E-BP–dependent translational regulation of one or more factor(s) affects these changes in the transcriptome.
Stringent Control of YY2 Expression in mESCs.
One of the mRNAs exhibiting the most significant increase in translation efficiency in DKO mESCs versus WT cells is the Yy2 transcription factor (Babel P value = 0.0001) (Fig. 1C and Dataset S1). YY2 exhibits considerable sequence homology (56% identity) with the YY1 transcription factor (21). Like reduced expression 1 (Rex1) [Zinc finger protein 42 (Zfp42)], a well-known ESC marker (22, 23), YY2 is a retroposed copy of the YY1 gene, which evolved only in placental mammals (24). YY1 is a pleiotropic transcription factor that regulates diverse cellular processes and plays a critical role in early embryonic development, ESC biology, and reprogramming (25–27). Although the N-terminal domains of YY1 and YY2 differ significantly, the C-terminal DNA-binding domain of YY1 is highly conserved in Rex1 and YY2 (22), suggesting that YY2 may play an important role in the regulation of gene expression in mESCs. Although there is no significant change in the level of Yy2 mRNA in DKO mESCs compared with WT cells (Fig. S3A), YY2 protein, but not YY1, is elevated 1.9- to 2.4-fold in DKO mESCs (Fig. 1E). Importantly, expression of a phosphorylation-resistant 4E-BP1-4A mutant (28) in DKO mESCs reduced YY2 protein levels (Fig. 1F). These data demonstrate that translation of Yy2 mRNA is controlled by 4E-BPs.
Fig. S3.
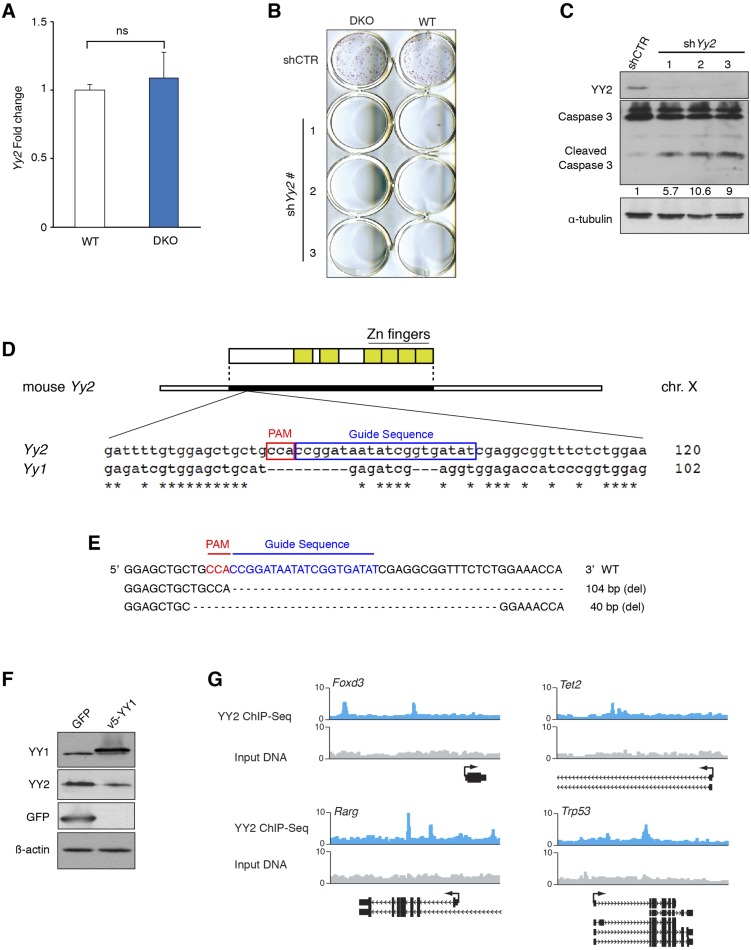
Role of YY2 in mESCs and blastocysts formation. (A) RT-qPCR analysis of Yy2 mRNA expression in WT and DKO mESCs. Values are normalized to β-actin. Data are mean ± SD (n = 3); ns, nonsignificant. (B) WT and DKO mESCs were transduced with three independent shRNAs against Yy2 or control shRNA. Resistant colonies were selected with puromycin (5 μg/mL) for 2 d and subjected to alkaline-phosphatase staining. (C) Western blot analysis of mESCs transduced with three independent shRNAs against Yy2 or control shRNA. Numbers indicate the ratio of cleaved caspase-3 in each condition to that in control after normalization with α-tubulin. (D) Schematic presentation of the CRISPR-Cas9 target site in the mouse Yy2 gene. The guide RNA was specifically designed to avoid regions with high homology to mouse Yy1. PAM, protospacer-adjacent motif. Numbers indicate the distance from the start codon. (E) The sequence of the Yy2 alleles in the WT and two mutant male blastocysts depicted in Fig. 2D. (F) Western blot analysis of mESCs overexpressing GFP or v5-YY1. (G) Graphical representation of selected YY2-binding peaks obtained from the UCSC browser. Twenty-kilobase windows are displayed.
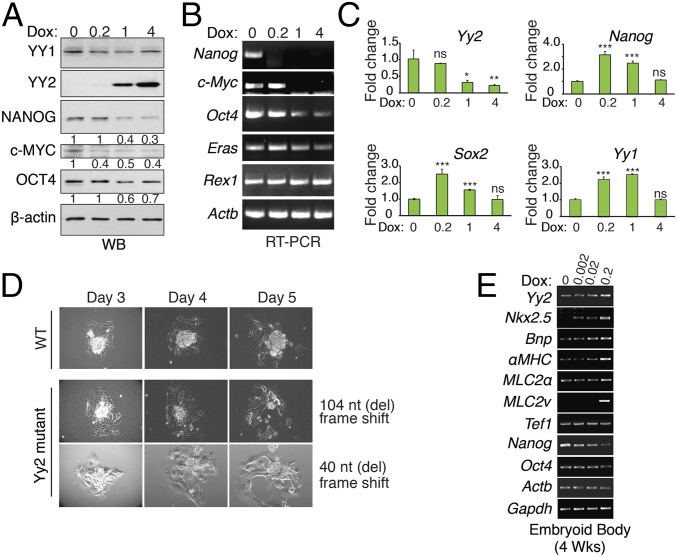
To study the functional consequence of YY2 up-regulation in mESCs, we generated an mESC line carrying a doxycycline-inducible YY2 construct (dox-YY2). Overexpression of YY2 caused a reduction in the expression of pluripotency factors Nanog, c-Myc, and Oct4 mRNAs (Fig. 2 A and B), indicating a negative role for YY2 in mESC self-renewal. However, constitutively expressed shRNA against Yy2 in mESCs caused depletion of mESCs in culture (Fig. S3B). This depletion coincided with increased levels (8.4- ± 2.4-fold) of the apoptosis marker, cleaved caspase-3, in Yy2-knockdown cells (Fig. S3C), suggesting that mESCs require a basal level of YY2 expression for survival. To study the impact of a moderate knockdown, we generated an mESC line carrying a doxycycline-inducible shRNA construct against Yy2 (shYy2). Although slight induction of shYy2 (0.2 μg/mL doxycycline) enhanced expression of the pluripotency factors Nanog and Sox2 (Fig. 2C), higher doses of doxycycline failed to do so (Fig. 2C), indicating a dose-sensitive effect of YY2 on the expression of mESC pluripotency factors. Importantly, the deleterious effect of complete depletion of YY2 was not limited to mESCs, because CRISPR/Cas9-mediated Yy2-knockout blastocysts were unable to maintain their inner cell mass, as demonstrated by blastocyst outgrowth assays (Fig. 2D and Fig. S3 D and E). A similar defective outgrowth has been described previously for Yy1−/− blastocysts (29), suggesting that the lack of YY2 may cause peri-implantation lethality, as described for Yy1−/− mice (26), and demonstrating that YY1 and YY2 fulfill nonredundant functions in blastocyst growth.
Fig. 2.
Stringent regulation of YY2 levels is critical for mESC survival and differentiation. (A and B) Western blot (A) and RT-PCR (B) analysis of WT mESCs carrying the doxycycline-inducible YY2 construct and treated with 0, 0.2, 1, or 4 μg/mL doxycycline for 24 h. Numbers indicate the ratio of the expression of the identified protein in each treatment compared with no doxycycline followed by normalization with β-actin. (C) RT-qPCR analysis of mESCs carrying doxycycline-inducible shRNA against Yy2 (shYy2) and treated with doxycycline (0, 0.2, 1, or 4 µg⋅mL−1⋅d−1) for 72 h. Values are normalized to β-actin. Data are mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001; ns, nonsignificant. (D) Blastocyst outgrowth assay in a WT embryo and in two independent CRISPR/CAS9-mediated Yy2-knockout embryos. Cas9 mRNA and sgRNAs targeting Yy2 were injected into zygotes. The blastocysts derived from injected embryos were subjected to the blastocyst outgrowth assay. The mutagenesis strategy and the sequence of mutant alleles are provided in Fig. S3 D and E. (E) RT-PCR analysis of EBs carrying the doxycycline-inducible YY2 construct and treated with 0, 0.002, 0.02, or 0.2 μg/mL doxycycline every other day for 4 wk.
Recent studies showing that YY1 directly regulates Nkx2.5 expression and promotes cardiogenesis uncovered a novel function for YY1 in cardiomyocyte differentiation and cardiac morphogenesis (30, 31). To examine the effect of YY2 on mESC differentiation toward cardiovascular lineages, EBs derived from dox-YY2 mESCs were exposed to doxycycline. One week after induction of YY2 expression, foci of beating cardiomyocytes began to appear in the plates with the highest level of YY2 induction (0.2 μg/mL doxycycline) (Movie S1), and the number of foci continued to increase during the following 2 weeks. No beating foci appeared in noninduced EBs up to the third week of differentiation. Consistently, expression of several cardiovascular-specific markers, such as Nkx2.5, Bone natriuretic peptide (Bnp), alpha-Myosin heavy chain (αMHC), Myosin light chain 2a (MLC2a), and MLC2v mRNAs was increased in the YY2-overexpressing EBs in a dose-dependent manner (Fig. 2E). These data suggest that YY1 and YY2 have overlapping functions in directing the differentiation of mESC toward cardiovascular lineages.
YY2 Binds to the Regulatory Regions of Key Genes for ESC Pluripotency and Differentiation.
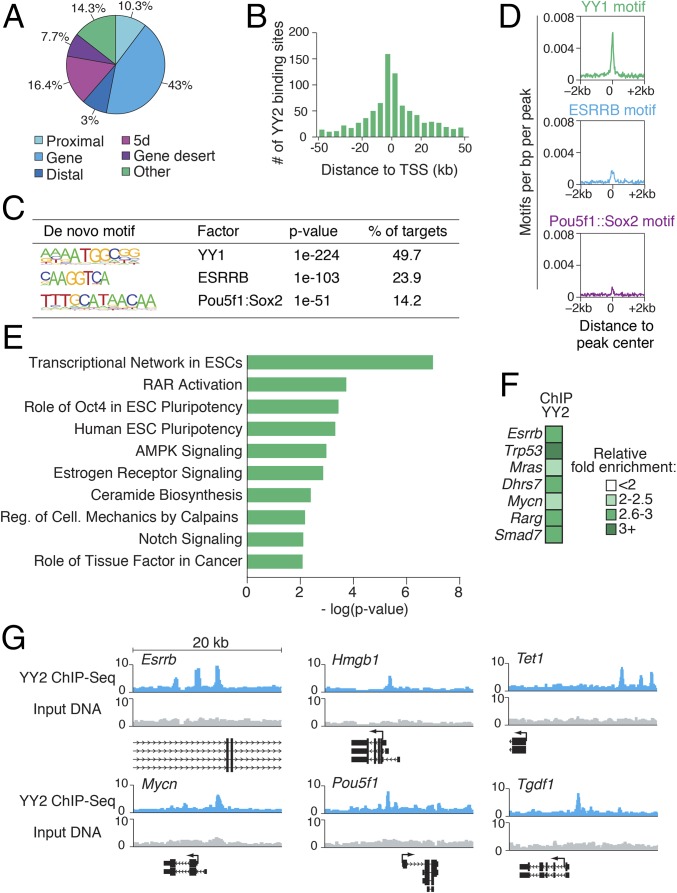
Genomic targets of YY1, but not YY2, in mESCs have been documented (25, 32). We determined the genome-wide binding sites of YY2 in mESCs by ChIP-Seq in WT mESC cells overexpressing YY2. Because of the high degree of similarity between the C-terminal domains of YY1 and YY2, we used a monoclonal antibody that specifically recognizes the N-terminal domain of YY2 (Fig. 2A and Fig. S3F; also see SI Materials and Methods). YY2-binding sites (Dataset S3) exhibit enrichment for the genomic loci of coding genes (exons or introns, indicated by gene, 43%) (Fig. 3A) and a preference for the promoter regions surrounding the transcription start sites (TSS) (Fig. 3B). Nearly half of the peaks contained the consensus YY1-binding motif (Fig. 3C), as is consistent with a similar sequence preference for YY1 and YY2 (21, 22). Motif distribution across binding peaks revealed enrichment for the known consensus YY1-binding motif directly at YY2 peak centers (Fig. 3D), indicating specific recognition of these binding sites by YY2.
Fig. 3.
YY2 controls the ESC transcriptional regulatory network and development-related genes. (A) Pie chart displaying the distribution of YY2 ChIP-Seq peaks across the genome based on the distance of the peaks from the nearest RefSeq gene: proximal, <2 kb upstream of the TSS; gene, exon or intron; distal, 2–10 kb upstream of the TSS; 5d, 10–100 kb upstream of the TSS; gene desert, >100 kb from a RefSeq gene; and other, anything not included in the above categories. (B) Histogram depicting the distance of YY2 ChIP-Seq peaks relative to the TSS of the nearest gene. (C) De novo motifs enriched in YY2-binding events. Enrichment P values and percentage of targets containing each motif are displayed, as generated by HOMER software. (D) Plots showing the average density of selected motifs in a window 2 kb from the YY2 peak center. (E) The most significantly enriched canonical pathways in genes associated with YY2 ChIP-Seq peaks, as identified by IPA. (F) Standard ChIP-qPCR validation of YY2-binding regions. Data are normalized to IgG. The Lmnb2 gene was used as a negative control. (G) Graphical representation of selected YY2-binding peaks, obtained from the University of California, Santa Cruz (UCSC) browser. Twenty-kilobase windows are displayed.
Pathway analysis of genes associated with YY2-binding peaks using the Ingenuity Pathway Analysis (IPA) program revealed significant enrichment for genes related to ESC pluripotency (Fig. 3E), such as Oct4, Teratocarcinoma-derived growth factor 1 (Tdgf1), Esrrb, and Forkhead box protein D3 (FoxD3). We also found enrichment for genes involved in the activation of the retinoic acid receptor (RAR)-signaling pathway. Previous studies showed that activation of the RAR pathway promotes differentiation of ESCs to cardiomyocytes, particularly MLC2v+ ventricular cardiomyocytes, and that the RAR-signaling pathway plays a critical role in cardiogenesis (33, 34). These findings indicate that activation of this pathway, along with other cardiovascular-related YY2 targets [e.g., Bnp, Mesoderm posterior protein 2 (Mesp2), and MKL (megakaryoblastic leukemia)/myocardin-like 1 (Mkl1)], is responsible for engendering the differentiation of mESCs toward the cardiovascular lineage by YY2 (Fig. 2E). We validated the ChIP-Seq results for a selected number of genes with ChIP-qPCR (Fig. 3 F and G and Fig. S3G).
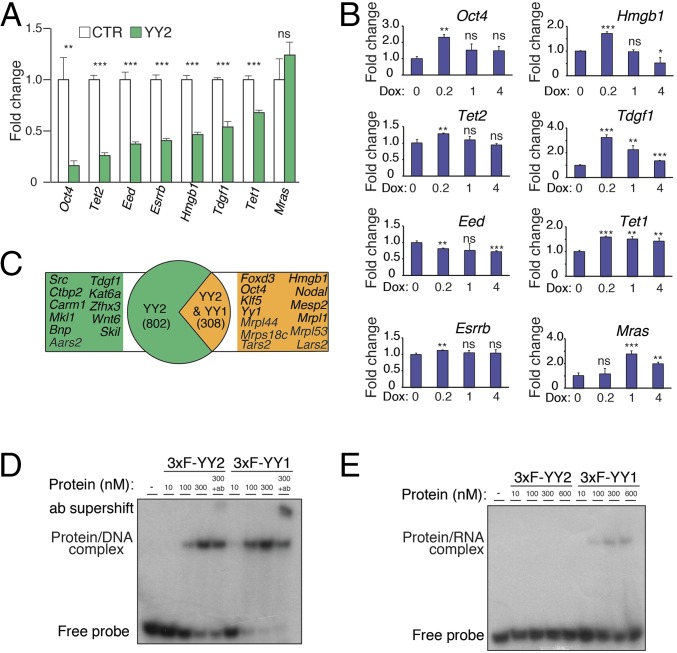
RT-qPCR analysis of a selected number of pluripotency factors in the ChIP-Seq dataset, such as Oct4, Esrrb, Tet1, and Tet2 mRNAs, revealed that YY2 overexpression suppresses their expression in mESCs (Fig. 4A). The ChIP-Seq analysis did not identify Nanog as a target of YY2. Thus, the decrease in Nanog mRNA expression in DKO mESCs (Dataset S2) or upon overexpression of YY2 (Fig. 2 A and B) is likely secondary to the down-regulation of other pluripotency factors. In agreement with our previous observation (Fig. 2C), doxycycline-inducible Yy2 knockdown in mESCs revealed a dose-sensitive regulation of its target genes (Fig. 4B).
Fig. 4.
The regulatory network and distinct mode of action of YY2 compared with YY1. (A) RT-qPCR analysis of selected YY2 targets in control and YY2-overexpressing mESCs. mESCs carrying the doxycycline-inducible YY2 construct were treated with 0 or 0.2 μg/mL doxycycline for 24 h. Values are normalized to β-actin. Data are mean ± SD (n = 3). **P < 0.01, ***P < 0.001; ns, nonsignificant. (B) RT-qPCR analysis of YY2 targets in mESCs carrying a doxycycline-inducible shYy2 and treated with doxycycline as described in Fig. 2C. Values are normalized to β-actin. Data are mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001; ns, nonsignificant. (C) Comparison of YY2 and YY1 ChIP-Seq targets in mESCs. Only peaks with at least a 1-nt overlap were considered as common targets. (D) EMSA with a radioactive-labeled dsDNA oligonucleotide probe derived from the promoter region of the mouse Arid1a gene (32) and purified recombinant mouse triple Flag-tagged (3xF)-YY1 and 3xF-YY2 proteins. The probes were incubated in the presence of increasing amounts of recombinant proteins and in the presence or absence of antibodies as indicated. (E) EMSA with a radioactively labeled single-stranded RNA oligonucleotide probe derived from the promoter region of the mouse Arid1a gene (32) and purified recombinant mouse 3xF-YY1 and 3xF-YY2 proteins.
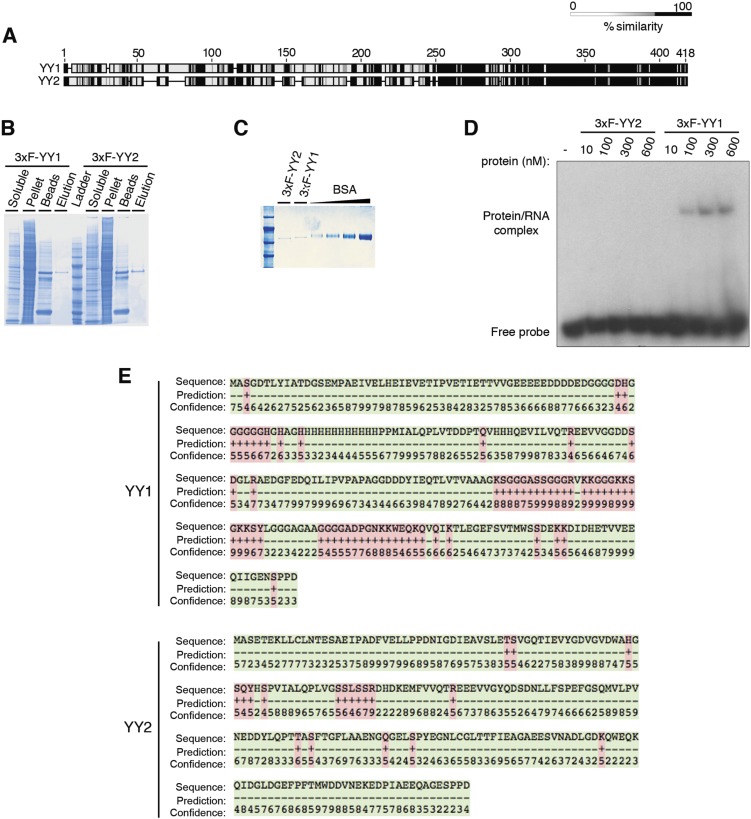
We analyzed the target genes identified by the YY2 ChIP-Seq assay relative to those of YY1 in mESCs (32) and found that 27.7% of YY2 targets are shared with YY1 (Fig. 4C and Dataset S3). Conversely, a large portion of the YY2 targets (∼72%) are not present among the YY1 targets (Fig. 4C), a finding that supports a previous study of genome-wide mRNA expression in HeLa cells demonstrating that YY1 and YY2 regulate some shared but mostly unique sets of genes (35). Although some pluripotency-related genes such as Tdgf1 are unique YY2 targets, Oct4, Krüppel-like factor 5 (Klf5), and Foxd3 are common targets of YY1 and YY2 (Fig. 4C). Notably, Yy1 is a target of both YY1 and YY2, and our data showed that YY2 has a dose-sensitive effect on YY1 expression (Fig. 2 A and C). The similar consensus-binding motifs of YY1 and YY2 are consistent with these factors exhibiting overlapping or competing effects on common target genes (21, 36–38). One plausible explanation for their distinct activities is the considerable divergence in their N-terminal domains (Fig. S4A). A recent report showed that the binding of YY1 to active promoters/enhancers in ESCs through its C-terminal DNA-binding domain is facilitated by concurrent binding of its N-terminal domain to RNA species transcribed from these regulatory elements (32). We hypothesized that the N-terminal domain of YY2 cannot interact with RNA. We examined this possibility by EMSA using purified mouse YY1 and YY2 proteins (Fig. S4 B and C) and DNA and RNA probes derived from the AT-rich interactive domain-containing protein A1 (ARID1A) promoter, which interact with YY1 (32). Notably, Arid1a is among the YY2 targets in our ChIP-Seq analysis (Dataset S3). Although YY2 interacts with the DNA probe with only slightly less efficiency than YY1 (Fig. 4D), only YY1 binds to the RNA probe (Fig. 4E), because no visible binding to the Arid1a promoter-derived RNA was detected for YY2, even after prolonged exposure (Fig. S4D). In agreement with these results, analysis of the N-terminal domains of mouse YY1 and YY2 proteins by BindN, an RNA-binding prediction server (39), identified two distinct RNA-binding motifs for YY1 that are not conserved in YY2 (Fig. S4E). Our data suggest a mechanism by which YY2 may act differently from YY1 and that their differential affinity for the promoter-derived RNAs underlies the opposing effects of YY1 and YY2 on certain shared promoters.
Fig. S4.
YY1, but not YY2, binds RNA through its N-terminal domain. (A) Pairwise alignment of mouse YY1 and YY2 protein sequences using Geneious software. The protein motifs are clustered according to their similarity. (B) Purification of 3xF-YY1 and 3xF-YY2 proteins from HEK293H cell lysate under nondenaturing conditions with anti-Flag M2 agarose beads and elution with 3xFlag-peptide. (C) Coomassie-stained 10% (wt/vol) SDS/PAGE gel of purified 3xF-YY1 and 3xF-YY2 proteins. (D) Prolonged exposure (3 wk) of the EMSA with the radioactively labeled single-stranded RNA oligonucleotide probe described in Fig. 4E. (E) RNA-binding prediction at the N-terminal domains of YY1 and YY2 using the BindN server (39). Predicted binding residues (pink) are labeled “+”; nonbinding residues (green) are labeled “−”. Numbers denote confidence from level 0 (lowest) to level 9 (highest).
A Retained 5′-UTR Intron Renders Yy2 mRNA Sensitive to 4E-BP–Dependent Translational Repression.
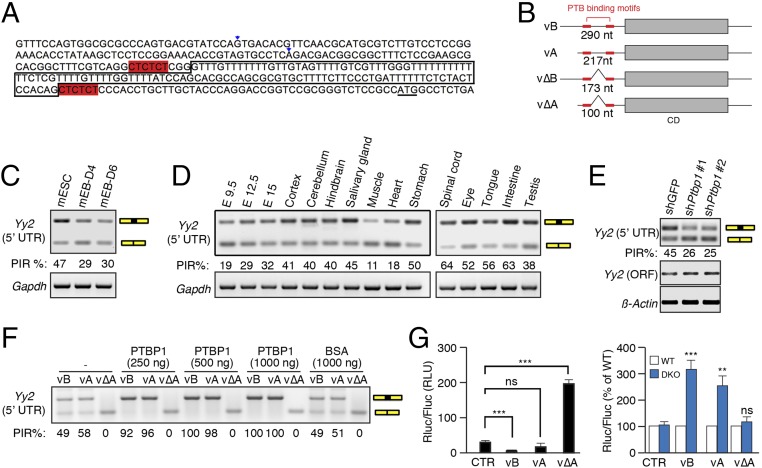
To search for eIF4E-sensitive elements in the 5′-UTR of Yy2 mRNA, we first used 5′-RACE to annotate the sequence in mESCs. In addition to the annotated mRNA sequence of the 217-nt 5′-UTR (RefSeq: NM_001098723.1), henceforth referred to as “variant A,” we uncovered two additional variants possessing 290-nt and 100-nt 5′-UTRs, respectively (Fig. 5 A and B). The 290-nt variant (designated “variant B”) is a 5′ extension of variant A; the 100-nt variant is a spliced version of variant A, lacking 117 nucleotides. The spliced region harbors all the features of a canonical intron, including a GU dinucleotide at the 5′ splice site (SS), an AG dinucleotide at the 3′ SS, and a polypyrimidine tract and putative branch site A nucleotide within 20 nt of the 3′ SS (Fig. 5A). Considering that the intronic sequence is shared between variants A and B, we termed their corresponding spliced variants ΔA and ΔB, respectively (Fig. 5B). Retroposed genes are generally intronless, because they are generated through the reverse transcription of mature mRNAs (40, 41). Therefore, the intron acquisition by Yy2 is most likely a recent evolutionary event that occurred after the retroposition of Yy2 from Yy1 in placental mammals.
Fig. 5.
Retention of the 5′-UTR intron renders Yy2 sensitive to 4E-BP–mediated translation suppression. (A) Sequence of the promoter region of the mouse Yy2 gene. The two alternative TSS are marked by arrowheads; the boxed sequence shows the retained intron; the two hexamers highlighted in red are the consensus PTBP-binding motifs; and the underlined ATG is the translation start codon for Yy2 mRNA. (B) A cartoon depicting the four variants of Yy2 5′-UTR. vB and vΔB represent a long variant with and without intron retention, respectively; vA and vΔA represent a short variant with and without intron retention, respectively. CD, coding DNA sequence. (C) RT-PCR using the primer pair Fw2 and Rv designed to recognize all four possible variants to estimate the splicing efficiency of the 5′-UTR intron in mESCs and mEBs on days 4 and 6 postdifferentiation. Gapdh mRNA was used as the control. (D) RT-PCR analysis of intron retention (IR) in the Yy2 5′-UTR in different mouse embryonic stages and adult tissues using the primers described in C. Gapdh mRNA was used as the control. (E) RT-PCR analysis of intron retention in the Yy2 5′-UTR using the primers described in C upon depletion of PTBP1 expression in mESCs by two independent shRNAs. Yy2-ORF primers amplifying a segment of the coding region of Yy2 transcript were used to demonstrate the change in overall expression of Yy2 mRNA. β-actin mRNA was used as the internal control. (F) RT-PCR amplification (primers Fw2 and Rv) of the in vitro splicing products of the A, B, and ΔA variants in WERI retinoblastoma cell extracts with different amounts of recombinant PTBP1 protein. Recombinant BSA was used as a negative control. (G) Luciferase reporter assay with Firefly (Fluc) and Renilla (Rluc) luciferase reporter mRNAs, as described in Fig. S5J. The in vitro-transcribed mRNAs were purified and transfected into WT and DKO mESCs. (Left) The normalized luciferase activity of each construct in WT mESCs. (Right) Comparison of the luciferase activity of each construct in DKO and WT mESCs. Fluc mRNA was cotransfected with Rluc mRNA as a transfection control. Data are mean ± SD (n = 3). **P < 0.01, ***P < 0.001; ns, nonsignificant. CTR, control; RLU, relative luminescence units.
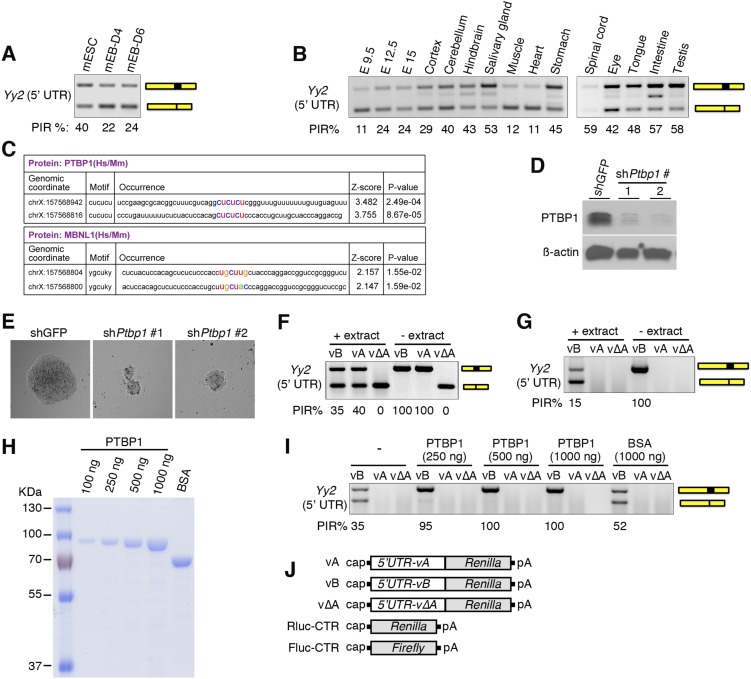
To measure intron retention in Yy2 mRNA during differentiation of mESCs by RT-PCR, we used two forward primers (Fw1 and Fw2) and a common reverse primer (Rv) to target the flanking exons (Table S2). Primers Fw1 and Rv detect only variants B and ΔB (297-bp and 180-bp PCR products, respectively), whereas Fw2 and Rv amplify a 209-bp PCR product for variants A and B and a 92-bp PCR product for variants ΔA and ΔB. We examined Yy2 intron retention events in mESCs and EBs at days 4 and 6 postdifferentiation and measured the degree of intron retention using the percent intron retention (PIR) as a metric (42). The percentage of nonspliced variants (A and B) is higher in mESCs than in EBs, demonstrating that differentiation coincided with a marked reduction in intron retention (Figs. 5C and Fig. S5A). Notably, this alternative splicing event is not restricted to mESCs, because various degrees of intron retention were detected at different embryonic stages and across different tissues (Fig. 5D and Fig. S5B), demonstrating developmental and tissue-specific regulation of Yy2 alternative splicing.
Table S2.
Primers used in this study
| Name | Sequence | Application |
| mYy2-5′UTR-Fw1 | GTTCAACGCATGCGTCTTGTCC | RT-PCR |
| mYy2-5′UTR-Fw2 | AAGCGCACGGCTTTCGTCAG | RT-PCR |
| mYy2-5′UTR-Rv | GTCTCAGAGGCCATGGCGGA | RT-PCR |
| mYy2-ORF-Fw | ACCAGCGTAGGCCAAACCATCGAAGTA | RT-PCR |
| mYy2-ORF-Rv | CGTCAAACCACAGAGATTCCCTTCATA | RT-PCR |
| mOct3/4-Fw | CTCGAACCACATCCTTCTCT | RT-PCR |
| mOct3/4-Rv | GGCGTTCTCTTTGGAAAGGTGTTC | RT-PCR |
| Nkx2.5-Fw | CAGTGGAGCTGGACAAAGCC | RT-PCR |
| Nkx2.5-Rv | TAGCGACGGTTCTGGAACCA | RT-PCR |
| Gata4-Fw | CTGTCATCTCACTATGGGCA | RT-PCR |
| Gata4-Rv | CCAAGTCCGAGCAGGAATTT | RT-PCR |
| Tef1-Fw | AAGACGTCAAGCCCTTTGTG | RT-PCR |
| Tef1-Rv | AAAGGAGCACACTTTGGTGG | RT-PCR |
| Tbx5-Fw | GGAGCCTGATTCCAAAGACA | RT-PCR |
| Tbx5-Rv | TTCAGCCACAGTTCACGTTC | RT-PCR |
| Bnp-Fw | ATGGATCTCCTGAAGGTGCT | RT-PCR |
| Bnp-Rv | TCTTGTGCCCAAAGCAGCTT | RT-PCR |
| MLC2v-Fw | GCCAAGAAGCGGATAGAAGG | RT-PCR |
| MLC2v-Rv | CTGTGGTTCAGGGCTCAGTC | RT-PCR |
| MLC2a-Fw | CAGACCTGAAGGAGACCT | RT-PCR |
| MLC2a-Rv | GTCAGCGTAAACAGTTGC | RT-PCR |
| aMHC-Fw | GGAAGAGTGAGCGGCCATCAAGG | RT-PCR |
| aMHC-Rv | CTGCTGGAGAGGTTATTCCTCG | RT-PCR |
| mKlf4-Fw1 | GACTATGCAGGCTGTGGCAA | RT-PCR |
| mKlf4-Rv1 | GGTAGTGCCTGGTCAGTTCA | RT-PCR |
| mGata6-Fw1 | AGAAATGCTGAGGGTGAGCC | RT-PCR |
| mGata6-Rv1 | ACAGAGCCACTGCTGTTACC | RT-PCR |
| mPecam1-Fw | GAAGTGTCCTCCCTTGAGCC | RT-PCR |
| mPecam1-Rv | TCGACCTTCCGGATCTCACT | RT-PCR |
| mLin28a-Fw | GTCTGCCAAGGGTCTGGAAT | RT-PCR |
| mLin28a-Rv | CTAGCCCACCGCAGTTGTAG | RT-PCR |
| m4EBP1-Fw | GCACATACCTCCTTGTGCCT | RT-PCR |
| m4EBP1-Rv | TCCCAGGTAACCCAGCCTAA | RT-PCR |
| m4EBP2-Fw | AGAGCAGCACAGGCTAAGAC | RT-PCR |
| m4EBP2-Rv | CAGATGTGGAAAATGGCCCG | RT-PCR |
| mGapdh-Fw | GTGTTCCTACCCCCAATGTGT | RT-PCR |
| mGapdh-Rv | ATTGTCATACCAGGAAATGAGCTT | RT-PCR |
| Cdx2-Fw | AGACAAATACCGGGTGGTGTA | RT-PCR |
| Cdx2-Rv | CCAGCTCACTTTTCCTCCTGA | RT-PCR |
| Bmp4-Fw | AGGAGGAGGAGGAAGAGCAG | RT-PCR |
| Bmp4-Rv | ACTGGTCCCTGGGATGTTCT | RT-PCR |
| mSox17-Fw | TGGAACCTCCAGTAAGCCAG | RT-PCR |
| mSox17-Rv | GACTTGCCTTGGGGAAAACT | RT-PCR |
| mRas-Fw | GCTTTGTTCCGTTCCGGGG | RT-PCR |
| mRas-Rv | TTAAGCCAGAGCCGGTGGTA | RT-PCR |
| Tdgf1-Fw | ACCTGAGGGTCCTACTCAACA | RT-PCR |
| Tdgf1-Rv | GCAGCCAAGATCTCCGTGTA | RT-PCR |
| Hmgb1-Fw | AGAGCGGAGAGAGTGAGGAG | RT-PCR |
| Hmgb1-Rv | CCGGCAAGTTTGCACAAAGA | RT-PCR |
| Esrrb-Fw | CCTTGCCCATGATCCAAGGT | RT-PCR |
| Esrrb-Rv | TTTGTCCAACCCTTGCTGGT | RT-PCR |
| Eed-Fw | GGGCGATTTGATTACAGCCAG | RT-PCR |
| Eed-Rv | TAGCCGCGCCACATTTATGA | RT-PCR |
| Tet1-Fw | CCTCACAGGCACAGGTTACA | RT-PCR |
| Tet1-Rv | ATTTGGGGCCATTTACTGGT | RT-PCR |
| Tet2-Fw | TGTTGTTGTCAGGGTGAGAATC | RT-PCR |
| Tet2-Rv | TCTTGCTTCTGGCAAACTTACA | RT-PCR |
| MycN-Fw | TGATCCTCAAGCGCTGTGTT | RT-PCR |
| MycN-RT-Rv | CTCAGGCTCTTCGCTTTTGC | RT-PCR |
| Myc-Fw | GCCGCCGCTGGGAAACTTTG | RT-PCR |
| Myc-Rv | GGCTGTCTGCGGGGTTTCCAAC | RT-PCR |
| Sox2-Fw | AGGAGTTGTCAAGGCAGAGAAGAGA | RT-PCR |
| Sox2-Rv | GCCGCCGCGATTGTTGTGATT | RT-PCR |
| Nanog-Fw | TGTCTTGCTCTTTCTGTGGG | RT-PCR |
| Nanog-Rv | TTGATGAGGCGTTCCCAGAAT | RT-PCR |
| Eras-Fw | ACTGCCCCTCATCAGACTGCTACT | RT-PCR |
| Eras-Rv | CACTGCCTTGTACTCGGGTAGCTG | RT-PCR |
| MAP2-Fw | CATCGCCAGCCTCGGAACAA | RT-PCR |
| MAP2-Rv | TCCACCAC CTGGCCTGTGAC | RT-PCR |
| TrP53-Fw | GCAGTGTGTGGTTCGAGAAG | ChIP-PCR |
| TrP53-Rv | ACTTATGCGAGGGTGGGAAA | ChIP-PCR |
| mRas-Fw | CTGGGAATTGTGCAGAAGGG | ChIP-PCR |
| mRas-Rv | CTTCTGGGATGTCTGGGTGT | ChIP-PCR |
| Dhrs7-Fw | AGCATTGATGGGTGAGGTGA | ChIP-PCR |
| Dhrs7-Rv | TGGCCCAGCAAAACAAACTT | ChIP-PCR |
| RarG-Fw | TCCCTCTCCTGTTCTTTCACA | ChIP-PCR |
| RarG-Rv | CCACTTATAGCCTTGGCAGC | ChIP-PCR |
| Smad7-Fw | TGTTTTCGGAGGGATGCTCA | ChIP-PCR |
| Smad7-Rv | GCGGTCATTGCTAACTCCTC | ChIP-PCR |
| YY2 Nt-F | CTCTCCCACCTGCTTGCTAC | CRISPR genotyping |
| YY2 Nt-R | GTCGGCTGGAGATAGTCGTC | CRISPR genotyping |
| SRY-F | TCTTAAACTCTGAAGAAGAGAC | Sex genotyping |
| SRY-R | GTCTTGCCTGTATGTGATGG | Sex genotyping |
| mYY2-ORF-Fw | GCTAGCATGGCCTCTGAGACA | RT-PCR-cloning |
| mYY2-ORF-Rv | GGATCCTTACTGGTCATTCTT | RT-PCR-cloning |
| mYY2-5′RACE-GSP-1 | GATTACGCCAAGCTTAATCAGCAGGAATCTCTGCGCTTTCTGTG | RACE |
| mYY2-5′RACE-GSP-2 | GATTACGCCAAGCTTCGATGGTTTGGCCTACGCTGGTTTC | RACE |
| KpnI-mYY2-ORF-Fw | TCAGTAGGTACCATGGCCTCTGAGACAGAGAAAC | RT-PCR cloning |
| BamHI-mYY2-ORF-Rv | TCAGTAGGATCCTTACTGGTCATTCTTGTTCTTAACATG | RT-PCR cloning |
| KpnI-mYY1-ORF-Fw | TCAGTAGGTACCATGGCCTCGGGCGACACCC | RT-PCR cloning |
| BamHI-mYY1-ORF-Rv | TCAGTAGGATCCTCACTGGTTGTTTTTGGCTTTAGCGTG | RT-PCR cloning |
Fig. S5.
PTBP1 regulates Yy2 5′-UTR intron retention. (A) RT-PCR analysis of mESCs and day 4 (D4) and day 6 (D6) mEBs using the primer pair designed to recognize the B and ΔB variants (Fw1 and Rv) to estimate the splicing efficiency of the Yy2 5′-UTR intron. (B) RT-PCR analysis (primers Fw1 and Rv) of intron retention in variant B of the Yy2 5′-UTR at different embryonic days (Left) and in different mouse tissues (Right). (C) Consensus PTBP1- or MBNL1-binding motifs in the 5′-UTR of mouse Yy2 mRNA, identified by the RBPmap web server. (D) Western blot analysis of PTBP1 expression in mESCs stably expressing two independent shRNAs against Ptbp1 or control shRNA (shGFP); this panel is related to Fig. 5E. (E) Representative images of the mESC colonies described in D. (F and G) RT-PCR analysis of the in vitro splicing products of the A, B, and ΔA variants in the presence (+) or absence (−) of the WERI retinoblastoma cell extract. Primers Fw2 and Rv were used in F to recognize all four possible variants, and primers Fw1 and Rv were used in G to recognize B and ΔB variants. (H) Coomassie-stained 10% (wt/vol) SDS/PAGE to detect the level of recombinant GST-PTBP1 protein used for the in vitro splicing assay in I and Fig. 5F. One microgram of BSA was loaded as control. (I) RT-PCR analysis (primers Fw1 and Rv) of the in vitro splicing products in WERI retinoblastoma cell extracts with different amounts of recombinant PTBP1 protein. Recombinant BSA was used as a negative control. (J) Schematic diagrams of the Renilla (Rluc) and Firefly (Fluc) luciferase reporter mRNA constructs used in G.
To identify the trans-acting factor(s) responsible for Yy2 alternative splicing, we used the RBPmap web server to predict consensus motifs for RNA-binding proteins (43). We identified two canonical PTBP recognition motifs (CUCUCU) flanking the Yy2 5′-UTR intron (Fig. 5 A and B and Fig. S5C). PTBP1 and PTBP2 (a neural- and testis-enriched paralog) are RNA-binding proteins implicated in several aspects of mRNA metabolism, including alternative splicing, stability, localization, and polyadenylation (44). PTBP1 is essential for embryonic growth before gastrulation, because Ptbp1−/− mESCs have severe proliferation defects (45, 46). To explore its role in Yy2 5′-UTR intron retention, PTBP1 was depleted in mESCs using shRNA (Fig. S5D). PTBP1 knockdown resulted in reduced Yy2 intron retention, demonstrating that it acts as an inhibitor of Yy2 splicing in mESCs (Fig. 5E). This knockdown was associated with reduced growth and smaller colonies of mESCs (Fig. S5E). To demonstrate that PTBP1 directly affects Yy2 5′-UTR splicing, we performed an in vitro splicing assay using a WERI retinoblastoma (WERI-Rb1) cell extract, purified recombinant PTBP1 protein, and the three Yy2 5′-UTR variants (A, B, and ΔA) transcribed in vitro. Incubation in the cell extract resulted in splicing of the introns from A and B variants but had no effect on the ΔA variant (Fig. S5 F and G). Importantly, the addition of recombinant PTBP1 protein (Fig. S5H) dramatically suppressed splicing and resulted in the complete retention of the intron in a dose-dependent manner (Fig. 5F and Fig. S5I).
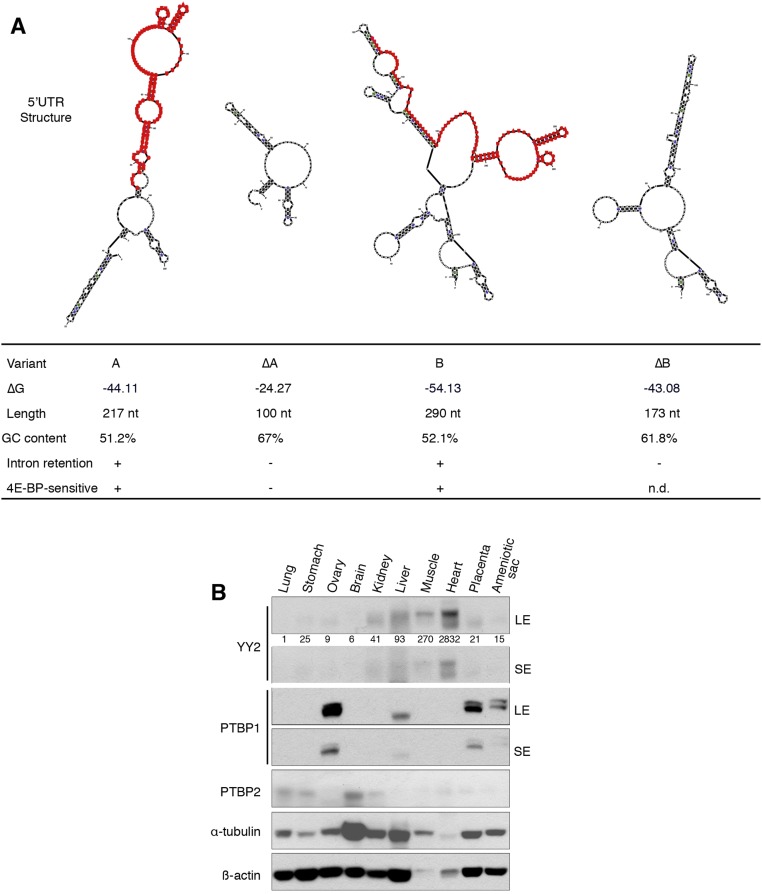
The length and complexity of the 5′-UTR play a critical role in mRNA translation, because mRNAs with long and structured 5′-UTRs are more sensitive to eIF4E activity (15, 47). The combined use of alternative TSS and alternative splicing determine the complexity of the Yy2 5′-UTR. To examine the effect of 5′-UTR variants on mRNA translation, we constructed luciferase reporters containing the 5′-UTRs of the A, B, and ΔA variants (Fig. S5J) and used the reporters to generate mRNAs that were transfected into WT and DKO mESCs. The B, and to a lesser extent A, variant mRNAs were poorly translated, whereas translation of the intron-less ΔA variant was markedly more efficient (more than sixfold higher than control) in WT mESCs (Fig. 5G, Left). These data demonstrate that the 5′-UTR containing the retained intron sequence inhibits translation. The translation of A and B variant mRNAs was significantly elevated in the DKO mESCs, whereas the ΔA variant remained insensitive to 4E-BPs levels (Fig. 5G, Right). These results demonstrate that intron retention in the Yy2 5′-UTR, in combination with the activity of 4E-BPs, determines the outcome of Yy2 mRNA translation. Intron retention adds an extra 117 nt to the Yy2 5′-UTR, increasing the complexity of its secondary structure (Fig. S6A) and rendering it sensitive to 4E-BP–mediated translation repression. As is consistent with the increased translation of the Yy2 spliced variant (Fig. 5G), the highest expression of YY2 protein was detected in the heart and muscle tissues (Fig. S6B), in which the lowest degree of Yy2 5′-UTR intron retention is observed (Fig. 5D and Fig. S5B).
Fig. S6.
Yy2 5′-UTR intron retention influences its secondary structure and expression in mouse tissues. (A) Effect of intron retention on the sequence and structure of Yy2 5′-UTR. The mfold web server (unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form) was used to predict the secondary mRNA structures and ΔG (Gibbs free energy). Nucleotides are numbered starting from the relevant TSS. The intronic sequence is highlighted in red. (B) Expression of YY2, PTBP1, and PTBP2 proteins in different tissues of a 4-wk-old female mouse. LE, long exposure; SE, short exposure. Numbers indicate the ratio of YY2 expression in each tissue compared with lung followed by normalization with α-tubulin.
This double-layered control mechanism, consisting of the retention of the Yy2 5′-UTR intron by PTBP1 and suppression of translation of the resulting mRNA variant by 4E-BPs, allows the modulation of Yy2 mRNA translation.
SI Materials and Methods
Antibodies, shRNAs, and Reagents.
The following antibodies and reagents were used: rabbit anti-PTBP1 (Cell Signaling; catalog no. 8776), rabbit anti-PTBP2 (Abcam; catalog no. ab154853), rabbit anti-eIF4G1 (Cell Signaling; catalog no. 2469), rabbit anti-YY1 (N-terminal) (Sigma; catalog no. SAB4200303), mouse anti-YY2 (Santa Cruz; catalog no. sc-377008), rabbit anti–4E-BP1 (Cell Signaling; catalog no. 9644), rabbit anti–4E-BP2 (Cell Signaling; catalog no. 2845), mouse anti-eIF4E (BD Biosciences; catalog no. 610270), mouse anti–α-tubulin (Santa Cruz; catalog no. sc-23948), mouse anti–β-actin (Sigma; catalog no. A5441), rabbit anti-NANOG (Bethyl Laboratories; catalog no. A300-397A), goat anti–OCT-3/4 (Santa Cruz; catalog no. sc-8628), goat anti-SOX2 (Y-17), rabbit anti-SOX2 (Cell Signaling; catalog no. 4195S), mouse anti–c-MYC (Santa Cruz; catalog no. sc-40), mouse anti-Flag (Sigma; catalog no. F3165), rabbit anti–Phospho-4E-BP1 Thr37/46 (Cell Signaling; catalog no. 2855), rabbit anti–Phospho-RPS6 (Ser240/244) (Cell Signaling; catalog no. 2215), mouse anti-ribosomal protein S6 (C-8), rabbit anti–caspase-3 (Cell Signaling; catalog no. 9665). The following shRNAs were obtained from the Sigma Mission library: anti-Ptbp1, shRNA no.1, TRCN0000295113; shRNA no. 2, TRCN0000295168; anti-Eif4ebp1 and Eif4ebp2, shRNAs TRCN0000075612 and TRCN0000075614, respectively. shRNAs against mouse Yy2 (V3LMM_474173, V3LMM_474175, and V3LMM_474177) were obtained from the Dharmacon.
Generation of the Inducible shRNA Construct.
The “all-in-one” system (64) for the inducible expression of shRNA against mouse Yy2 (shYy2) was generated by cloning the following oligos into the Tet-pLKO-puro lentiviral vector:
mYy2shRNA-T: CCGGGGCCAAACCATCGAAGTATATCTCGAGATATACTTCGATGGTTTGGCCTTTTT
mYy2shRNA-B: AATTAAAAAGGCCAAACCATCGAAGTATATCTCGAGATATACTTCGATGGTTTGGCC
Lentiviral particles generated by this vector were used to create stable doxycycline-inducible shRNA-expressing mESCs.
Lentivirus Production.
293FT cells (Invitrogen) were cultured in DMEM/10% (vol/vol) FBS medium containing 400 μg/mL neomycin (BioShop) according to the manufacturer’s instructions. Medium was replaced by antibiotic-free medium at least 8 h before transfection with Lipofectamine 2000. Ten micrograms of expression plasmid, 6.5 μg of psPAX2, and 3.5 μg of pMD2.G packaging plasmids were used to transfect 8 × 106 293FT cells in a 10-cm dish. Twenty-four hours after transfection, the medium was collected and replaced with fresh medium. Supernatant was collected for an additional 2 d, pooled, and subjected to centrifugation in a SW32-Ti rotor (Beckman Coulter) at 25,000 rpm for 1.5 h. The viral pellet was resuspended in DMEM and 10% (vol/vol) heat-inactivated FBS and was rotated overnight at 4 °C. The resulting concentrated virus solution was used to infect the cells directly in the presence of Polybrene (6 μg/mL) or was stored at −80 °C.
Cap Pull-Down Assay Using m7GTP-Sepharose.
m7GTP-Sepharose 4B (Jena Biosciences) was incubated with WT or DKO mESC lysate (500 μg) in buffer containing 50 mM Mops/KOH (pH 7.4), 100 mM KCl, 0.02% NaN3, and 0.5 mM EDTA for 30 min at 4 °C, washed five times (5 min each) with the same buffer, and eluted with 0.2 mM m7GTP for 15 min at 4 °C. Eluted proteins were subjected to SDS/PAGE followed by Western blotting using the indicated antibodies.
Analysis of Global Protein Synthesis.
Protein synthesis was measured by [35S]methionine/cysteine metabolic labeling. Briefly, mESCs were seeded in a 24-well plate the day before the experiment. Cells were incubated in methionine/cysteine-free DMEM supplemented with 10% (vol/vol) dialyzed FBS (GIBCO) and [35S]methionine/cysteine labeling mixture (10 µCi/mL) at 37 °C for 30 min, followed by lysis in Laemmli buffer. [35S]methionine/cysteine incorporation was determined by trichloroacetic acid (TCA) precipitation followed by scintillation counting.
Library Preparation and Sequencing.
Fragmented mRNAs and RFPs were dephosphorylated using T4 polynucleotide kinase (New England Biolabs). Denatured fragments were resuspended in 10 mM Tris (pH 7) and quantified using the Bio-Analyzer Small RNA assay (Agilent). A sample of 10 pmol of RNA was ligated to the 3′ adaptor with T4 RNA ligase 1 (New England Biolabs) for 2 h at 37 °C. Reverse transcription was carried out using the oNTI223 adapter (Illumina) and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturers’ instructions. Products were separated from the empty adaptor on a 10% (wt/vol) polyacrylamide Tris/borate/EDTA-urea (TBE-urea) gel and circularized by CircLigase (Epicentre). Ribosomal RNA (rRNA) amounts were reduced by subtractive hybridization using biotinylated rDNA complementary oligos (63). The mRNA and RFP libraries were amplified by PCR (10 cycles) using indexed primers and were quantified using the Agilent BioAnalyzer High-Sensitivity assay. DNA then was sequenced on the HiSeq 2000 platform with a read length of 50 nt (SR50), according to the manufacturer’s instructions, with sequencing primer oNTI202 (5CGACAGGTTCAGAGTTCTACAGTCCGACGATC).
Analysis of Ribosome-Profiling Data.
Reads were trimmed of known adaptor sequences using the FASTX toolkit. Clipped reads shorter than 24 nt were discarded, and the remaining reads were aligned against mouse rRNA, mitochondrial RNA, and tRNA. Then unaligned reads were mapped to the mouse genome (mm9) using bowtie2 (local mode) (bowtie-bio.sourceforge.net/bowtie2). Uniquely mapped reads with a Mapping Quality (MAPQ) score ≥10 were used for further analysis. mRNA translation efficiency within and between samples was quantified using the Babel analytical method (20). Briefly, a regression model-based approach was used to measure the negative binomial distribution to model the abundance of both mRNA and RFPs. A parametric bootstrap approach was used to test the null hypothesis that the RFP counts are similar to mRNA counts. This approach generated a high-confidence list of genes whose mRNA translation efficiency deviated significantly from the global transcript population. We used a cutoff of 40 counts for mRNAs to be included in this analysis, and the Benjamini–Hochberg method was used to adjust the P values for multiple comparisons. For metagene analysis of read distributions around start or stop codons, reads mapped to RefSeq transcripts were used. For a given region, only genes with at least 10 reads whose 5′ end was within the region were used. The 5′ end position of a read was used for plotting. The read number was normalized first within each gene and then across all genes. The final normalized read number is shown as read number per gene per nucleotide.
Analysis of Differential mRNA Expression.
Differential RNA expression was determined using the mapped mRNA counts and the DESeq method as described (65). The DESeq model counts data with negative binomial distribution and normalizes read counts based on library size. Genes were considered differentially expressed if they exhibited a fold change of ≥1.5 and FDR <0.05 (based on the Benjamini–Hochberg multiple testing adjustment).
5′-RACE Analysis.
5′-RACE was performed with the SMARTer RACE 5′/3′ kit (Clontech; catalog no. 634858). Briefly, 1 µg of total RNA extracted from mESCs was treated with DNase I (Fermentas), and cDNA was generated by the SMARTScribe reverse transcriptase (Clontech) according to the manufacturer’s instructions. The resultant cDNA was used for PCR amplification using the mouse Yy2 gene-specific forward primers (GSPs) (Table S2) together with a common universal reverse primer provided by the manufacturer. PCR products were resolved by agarose gel electrophoresis, and all visible bands were excised and digested by restriction enzymes followed by cloning into the PUC19 vector provided by the manufacturer and sequenced by Sanger sequencing.
Luciferase Reporter with in Vitro-Transcribed mRNAs.
PCR products encoding the specified 5′-UTR variants of Yy2 mRNA followed by the Renilla luciferase coding sequence were generated and inserted into the pBluescript II KS (+) vector (Stratagene) between the SacI and XhoI sites. Resultant plasmids (pBKS-Yy2-5′UTR-A, pBKS-Yy2-5′UTR-B, and pBKS-Yy2-5′UTR-ΔA) were used as templates for production of the luciferase reporter mRNAs by in vitro transcription using the MAXIscript T7 in vitro transcription kit (Ambion) according to the manufacturer’s protocol in the presence of the anti-reverse cap analog. A poly(A) tail was added using the Poly(A) Tailing kit (Ambion). WT and DKO mESCs were seeded in a 24-well plate and were cultured overnight. The next day cells were transfected with the specified Renilla luciferase mRNAs containing Yy2 5′-UTRs together with firefly luciferase mRNA as a transfection control, using Lipofectamine 2000 (Invitrogen). Cells were lysed the next day, and luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
RT-PCR Assays for Detection of Yy2 5′-UTR Intron Retention.
Poly(A) mRNAs from the indicated mouse tissues or cells were monitored for Yy2 intron retention by RT-PCR, as described previously (70). Briefly, 0.5 ng of poly(A)+ RNA was used for cDNA synthesis and amplification with the One-Step RT-PCR kit (Qiagen) according to the manufacturer’s recommendations. The number of amplification cycles was 24 for Gapdh and 32 for Yy2. Reaction products were resolved using a 2% (wt/vol) ethidium bromide agarose gel and were imaged using the Gel Doc XR System (Bio-Rad). Gel densitometry was used to calculate the PIR, measured as the amount of intron-retained isoform divided by the sum of spliced and retained isoforms.
In Vitro Splicing Assays.
In vitro splicing assays were performed as described (71). Briefly, in vitro splicing assays (20 µL) contained 1.5 mM ATP, 5 mM creatine phosphate, 5 mM DTT, 3 mM MgCl2, 2.6% (wt/vol) polyvinyl alcohol, 30 units of RiboLock RNase inhibitor (Fermentas), 10 ng of Yy2 RNA-splicing substrate, and 50–60 µg of Weri-Rb1 cell extract, in 12 µL of lysis buffer [20 mM Hepes (pH 7.9), 100 mM KCl, 0.2 mM EDTA, 10% (vol/vol) glycerol, 1 mM DTT] with or without recombinant proteins (PTBP1 or BSA). Transcripts used for in vitro splicing were prepared using Yy2 reporter constructs containing a T7 promoter. In vitro transcription was performed using the MEGAscript T7 transcription kit (Life Technologies) according to the manufacturer’s recommendations. The amounts of recombinant protein used were GST-PTBP1: 250, 500, or 1,000 ng, and BSA: 1,000 ng. Reactions were incubated for 1 h at 30 °C. RNA was recovered using TRI Reagent (Sigma Aldrich) and was resuspended in 10 µL of nuclease-free water. Intron retention was monitored using 2 µL of the recovered RNA in RT-PCR reactions, as described above in RT-PCR Assays for Detection of Yy2 5′-UTR Intron Retention.
ChIP and ChIP-Seq Assays.
Antibodies used for ChIP were anti-RNA Polymerase II (05-623; Millipore), anti-H3K4Me3 (07-473; Millipore), anti-YY2 (sc377008; Santa Cruz), and anti-mouse IgG (10400C; Thermo Fisher Scientific). Antibodies were prebound overnight at 4 °C to 20 µL of Dynabeads Protein A (10008D; Thermo Fisher Scientific) or 20 µL Dynabeads Protein G (10004D; Thermo Fisher Scientific), diluted in ChIP dilution buffer [1% Triton-X, 10 mM Tris⋅HCl (pH 8), 150 mM NaCl, 2 mM EDTA]. For each ChIP assay, chromatin prepared from ∼1 × 107 cells was diluted to 1 mL in ChIP dilution buffer and added to the antibody-bound beads. Chromatin was immunoprecipitated overnight with rotation at 4 °C. Beads were washed five times at 4 °C with wash buffer [50 mM Hepes-KOH (pH 7.5), 1 mM EDTA, 0.7% sodium deoxycholate, 1% Nonidet P-40, 0.5 M LiCl] and once with Tris-EDTA (TE) buffer [10 mM Tris⋅HCl (pH 8), 1 mM EDTA]. DNA was eluted in 200 µL of elution buffer [1% SDS, 50 mM Tris⋅HCl (pH 8), 10 mM EDTA], and cross-links were reversed overnight at 65 °C, followed by treatment with RNase A (19101; Qiagen) and Proteinase K (03115879001; Roche). DNA was purified with the Qiagen PCR Purification Kit (28106; Qiagen). Quantification of ChIP enrichment was carried out with SYBR Green-based qRT-PCR (04887352001; Roche) and the Roche LightCycler instrument. All primers used in this study are listed in Table S2.
Sequencing libraries of ChIP DNA and corresponding 10% (wt/wt) chromatin inputs were prepared at the McGill University and Génome Québec Innovation Centre, following quality assessment with the Agilent Bioanalyzer 2100. ChIP libraries were sequenced on the Illumina HiSeq 2000 sequencing system with 50-bp reads. Reads from biological replicates were combined.
Bioinformatics Analyses of the ChIP-Seq Results.
Sequence trimming and quality filtering were performed with Trimmomatic software v.0.32 (66), retaining reads with a phred33 score ≥30 and length ≥50 bp. Filtered sequences were aligned to the mm10 assembly with Burrows–Wheeler aligner v.0.7.10. Peaks were called with the model-based analysis of ChIP-Seq (MACS) algorithm v.2.0 (67) with P value 1 × 10−5 using input reads as the control. Peaks were annotated based on the nearest TSS with HOMER software v.4.7 (68). BedGraph files for ChIP-Seq visualization were performed with HOMER software and uploaded to the UCSC Genome Browser (69). For motif enrichment analysis, de novo motif finding was performed with HOMER software v.4.7 using the default parameters. Annotation of motif occurrences on ChIP-Seq peaks was performed in a 4-kb window from the center of the peak using HOMER software. For pathway enrichment analysis, gene lists were compiled using binding peaks occurring within 20 kb of the TSS of the nearest gene. Canonical pathway enrichment analysis was performed on these lists with IPA (Ingenuity Systems; www.ingenuity.com/). P values to identify significantly enriched biological pathways were calculated using Fisher’s exact test.
Expression and Purification of YY1 and YY2 Proteins.
Yy1 and Yy2 ORFs were amplified from the mESC cDNA library using the specific primers described in Table S2 and were cloned into the pCDNA5.A-3xF plasmid. HEK293H cells (a kind gift from Joseph Marcotrigiano, Department of Chemistry and Chemical Biology, Rutgers–New Brunswick, School of Arts and Sciences) were transfected with the indicated plasmid, and cells were collected 30 h posttransfection and resuspended in lysis buffer [25 mM Hepes-KOH (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40, 5% (vol/vol) glycerol, 15 µM ZnCl2, 0.5 mM DTT, 1 μg/mL RNase A/DNase I, and protease inhibitors] for 10 min on ice. After centrifugation, the Flag-tagged proteins were affinity purified using anti-Flag M2 agarose beads (Sigma) and were eluted with 5× bead volumes of 3xF-peptide (0.1 mg/mL). SYBR Gold and Coomassie staining were used to measure protein purification and ensure the absence of RNA/DNA impurities. Purified proteins were buffer exchanged to 50 mM Hepes-KOH (pH 7.5), 100 mM NaCl, 15 µM ZnCl2, 0.5 mM DTT buffer and concentrated to 1 mM using a 10-kDa Amicon centrifugal filter (Millipore). Aliquots in 10% (vol/vol) glycerol were frozen in liquid nitrogen at −80 °C for EMSA experiments.
EMSA.
The oligonucleotide DNA probe for Arid1a (adopted from ref. 32) was generated by annealing 30-nt single-stranded oligonucleotides (Fw: 5′-CTCTTCTCTCTTAAAATGGCTGCCTGTCTG-3′, Rv: 5′-CAGACAGGCAGCCATTTTAAGAGAGAAGAG-3′) to obtain 100 μM stock of dsDNA probe. Then 10 pmol were labeled with T4 polynucleotide kinase (PNK) (Thermo Scientific) and 20 mCi [γ-32P] ATP (Perkin-Elmer) for 30 min at 37 °C. PNK was inactivated at 68 °C for 10 min, and unincorporated label was removed by centrifugation through a spin column (TE-10; Clontech). The labeling incorporation was verified using 1 mL of labeled probe in scintillation liquid. Binding of YY1 and YY2 proteins to the DNA probe was performed according to the Gel-Shift Assay Kit protocol (Promega). For each gel shift reaction, 3,000 cpm of labeled probe was incubated with increasing concentrations (10, 100, 300 nM) of 3xF-YY2 or 3xF-YY1 proteins for 45 min on ice; then 1 mL of Flag antibody (Sigma) was added to one sample containing 300 nM protein, and all samples were incubated for an additional 60 min on ice. Free DNA–probe and DNA–protein complexes were separated on a 6% (wt/vol) DNA retardation gel (Novex; Thermo Scientific) at 250 V for 25 min in 0.5× TBE at 4 °C. Gels were vacuum-dried and exposed to an Amersham Hyperfilm MP (GE Healthcare Life Science) with a screen at −80 °C. The 30-nt single-stranded RNA probe for Arid1 (5′-rCrUrCrUrUrCrUrCrUrCrUrUrArArArArUrGrGrCrUrGrCrCrUrGrUrCrUrG-3′) (adopted from ref. 32) was labeled and purified in the same way as the DNA probe. EMSAs with the recombinant 3xF-YY2 or 3xF-YY1 proteins were performed in buffer containing 20 mM Hepes-KOH (pH 7.5), 100 mM KCl, 2 mM MgCl2, 15 µM ZnCl2, 0.5 mM DTT, 0.05% Nonidet P-40, 5% (vol/vol) glycerol, 0.1 mg/mL acetylated BSA (Sigma), and SUPERase-In RNase inhibitor (Life Technologies). Increasing amounts of 3xF-YY2 or 3xF-YY1 proteins (10, 100, 300, 600 nM) were incubated for 45 min on ice and applied to a 6% (wt/vol) retardation gel (Novex; Thermo Scientific) at 250 V for 25 min in 0.5× TBE at 4 °C. The gels were vacuum-dried and exposed to an Amersham Hyperfilm (GE Healthcare Life Science) with a screen at −80 °C.
PCR Genotyping.
Genomic DNA from the blastocysts was extracted and amplified using the primers listed in Table S2. PCR fragments then were subcloned into the pGEM-T Easy vector for Sanger sequencing. At least five clones per outgrowth were sequenced.
Discussion
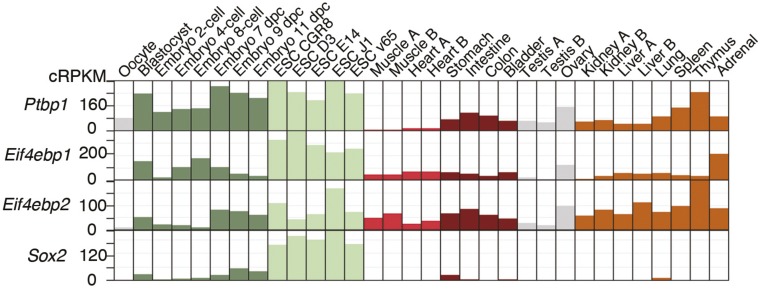
Many mammalian mRNAs contain heterogeneous 5′-UTRs (48). Length, structure, and sequence elements in the 5′-UTRs strongly impact translation (49, 50). We found that the combination of alternative TSS and splicing produces four Yy2 mRNA variants with different translation efficiencies. Thus, the relative level of each variant, in combination with 4E-BP activity, dictates the rate of YY2 protein synthesis. ESC differentiation, which is concomitant with the down-regulation of PTBP1 expression (51), triggers the splicing of the Yy2 5′-UTR intron. However, there is considerable variation in the extent of Yy2 5′-UTR intron retention among adult mouse tissues, with heart and skeletal muscle displaying the lowest rate of intron retention. Different degrees of intron retention among mouse tissues, most of which express very low levels of PTBP1 (Figs. S6B and S7 and Dataset S4), implies the existence of additional regulatory mechanisms that augment splicing of the Yy2 5′-UTR intron in heart and skeletal muscles. We also found consensus binding motifs for the MBNL1 splicing factor in the Yy2 5′-UTR (Fig. S5C). Muscleblind-like protein 1 (MBNL1) is highly expressed in cardiac and skeletal muscles (52) and is a known regulator of mRNA splicing in these tissues (53, 54). Intron retention has emerged as a widespread mechanism to regulate gene expression in different cell and tissue types as well as during stem cell differentiation (42, 55, 56). The finding that a retained intron at the 5′-UTR of Yy2 mRNA controls its translation underscores the intricate interplay between alternative splicing and translational control (57, 58).
Fig. S7.
Expression profiles of Ptbp1, Eif4ebp1, Eif4ebp2, and Sox2 in a cohort of embryonic and adult mouse samples. The references for the RNA-Seq studies used in this analysis are listed in Dataset S4.
We established Yy2 mRNA as a target of 4E-BP–dependent translation suppression in mESCs and demonstrated that a basal level of YY2 expression is essential for mESC survival. Similar to Yy1-knockout embryos, CRISPR-mediated Yy2-knockout embryos survived the preimplantation period, but the growth of their inner cell mass was impaired, as revealed by blastocyst outgrowth assays (Fig. 2D). These observations exclude the possibility of redundant functions for YY1 and YY2 in the early stages of embryonic development. The critical function of YY2 in mESCs is most likely mediated by its direct transcriptional regulation of ESC genes such as Oct4, Eras, Tet1, Tet2, and Tdgf1. We demonstrated that mESCs are highly sensitive to YY2 and that a modest manipulation of Yy2 levels significantly affects the expression of pluripotency factors, ESC self-renewal, and differentiation (Figs. 2 A–C and 4 A and B). Consistent with the observed phenotype in DKO mESCs, overexpression of YY2 in WT mESCs induces down-regulation of several pluripotency factors and promotes differentiation. Although in the current study we focused on the characterization of YY2, the ribosome-profiling experiment identified several other mRNAs, such as Triggering receptor expressed on myeloid cells 2 (Triml2), Sumo1, Amigo3, Lefty, and Nodal, that display 4E-BP sensitivity. The change in translation of these additional targets in DKO mESCs could contribute to the differentiation-prone phenotype of DKO mESCs and explain the differences that may be observed between DKO mESCs and mESCs overexpressing YY2.
We documented that increased expression of YY2 directs differentiation of mouse embryoid bodies (mEBs) toward the cardiovascular lineage. The importance of YY2 in cardiomyocytes is likely not limited to early differentiation, because the heart expresses a higher level of YY2 than other tissues of adult mouse (Fig. S6B). This function is likely mediated by transcriptional control of the RAR pathway and multiple YY2 targets, such as Bmp4, Nodal, Bnp, Mesp2, and Mkl1, as evident from the ChIP-Seq analysis. Recent studies showed binding of YY1 to the promoters of highly expressed ribosomal proteins and the nuclear encoded mitochondrial membrane, enzymes, and ribosomal proteins (35) and highlighted the importance of YY1 in cardiomyocyte differentiation and heart morphogenesis (30, 31). Notably, cardiac-specific ablation of Yy1 causes severe abnormalities in the heart, indicating that YY2 in the heart does not compensate for YY1 deletion (31). Comparison of YY2 and YY1 ChIP-Seq data (Fig. 4C and Dataset S3) demonstrated overlapping sets of nuclear encoded mitochondrial proteins, such as the aminoacyl tRNA synthetases Tars2, and Lars2, as well as mitochondrial ribosomal proteins Mrps18c, Mrpl1, Mrpl44, and Mrpl53 (Fig. 4C and Dataset S3). That YY2 and YY1 have both unique and overlapping target genes explains their convergent but nonredundant functions in development.
The reduced expression of several pluripotency markers (e.g., Oct4, Nanog, PRDM14, and Esrrb) in DKO mESCs and their greater tendency for differentiation (e.g., slower proliferation and a flattened morphology) resembles a “primed” state of pluripotency (59). Although the transcriptional regulatory network safeguarding the naive/primed state of pluripotency has been studied extensively, the role of posttranscriptional control mechanisms, particularly translational control, remained largely unexplored. The importance of posttranscriptional regulatory processes has been highlighted recently by the identification of the RNA-binding proteins Pum1 and Lin28 as regulators of naive/primed pluripotency (60, 61). Detailed analysis of mRNA translation therefore should provide valuable information about the role of translational control in these pluripotent states.
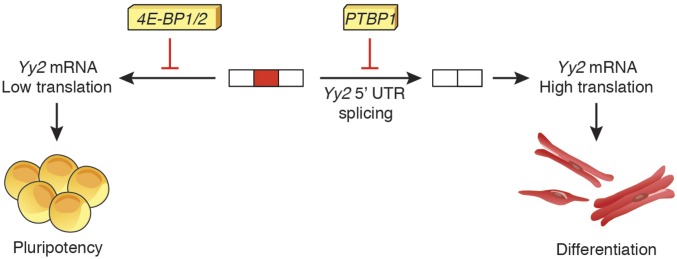
In summary, we have described a mechanism by which mESC self-renewal and lineage commitment are controlled via stringent regulation of YY2 expression at two stages: (i) by repressing the splicing of the Yy2 5′-UTR intron via PTBP1 and (ii) by suppressing the translation of the resulting mRNA variant by 4E-BP1 and -2. These two layers of control (Fig. 6) coalesce to limit the expression of YY2 protein to a low basal level in mESCs, thereby maintaining their self-renewal and pluripotency.
Fig. 6.
Regulation of YY2 expression by splicing and mRNA translational control. A proposed model depicting the regulation of YY2 expression at two different steps: (i) alternative splicing (via PTBP1) and (ii) mRNA translation (via 4E-BP1/2). A basal level of YY2 expression is required for maintenance of mESC self-renewal, but increased translation of Yy2 mRNA directs cardiovascular lineage commitment.
Materials and Methods
ESC Cell Culture and Differentiation.
Mouse ESCs were maintained in DMEM (Wisent Inc.), 1% nonessential amino acids (Gibco), 1% l-Glutamine (Wisent Inc.), 1% sodium pyruvate (100× stock from Invitrogen), 0.1 mM β-mercaptoethanol, 15% (vol/vol) FBS, 1,000 U mouse LIF/mL (ESGRO; Millipore), penicillin (50 μg/mL), and streptomycin (50 μg/mL) and were expanded on the feeder layer or gelatin. For mESC differentiation (62), 800–2,000 mESCs were cultured for 2 d in hanging drops containing differentiation medium [DMEM, 20% (vol/vol) FBS, 1% nonessential amino acids (Gibco), 1% l-Glutamine (Wisent Inc.), penicillin (50 μg/mL), and streptomycin (50 μg/mL)]. The resulting EBs were transferred to bacteriological dishes and cultured in suspension for 3 d. Then they were plated onto gelatin-coated tissue-culture plates for the rest of the differentiation process.
Single-Guide RNA Synthesis.
The DNA template for Yy2 single-guide RNA (sgRNA) was synthesized by PCR reactions using px330 (Addgene) as a template. Two primers were used: T7-Yy2-sgRNA forward (5′-TTAATACGACTCACTATAGGTTCGATGGTTTGGCCTACGCGTTTTAGAGCTAGAAATAGC-3′) and sgRNA reverse (5′-AAAAGCACCGACTCGGTGCC-3′). The PCR product was purified with the PCR DNA fragments extraction kit (Geneaid) and was used as a template for sgRNA synthesis with the T7 MAXIscript kit (Ambion). The synthesized sgRNA was EtOH-precipitated and dissolved in RNase-free water.
Cytoplasmic Microinjection, Embryo Culture, and Blastocyst Outgrowth.
The animal ethics committee of the McGill University approved all the animal procedures. Cas9 mRNA (50 ng/µL) (Sigma) and 50 ng/µL Yy2-sgRNA in 10 mM KCl were injected into CD1 zygotes in M2 medium (Zenith Biotech). Cytoplasmic injection was performed with a FemtoJet microinjector (Eppendorf) and a Cyto721 intracellular amplifier (World Precision Instruments) for the tickler’s oscillation to penetrate the zygote’s membrane. The injected zygotes were cultured for 4 d in KSOM drops covered with mineral oil (Zenith Biotech) in a 5% (vol/vol) CO2 incubator at 37 °C. The zona pellucidae of the developed blastocysts were removed with acid Tyrode’s solution (Millipore). The blastocysts were plated on gelatin-coated 24-well plates and were cultured 5–7 d in DMEM (Wisent) with ES-FBS (Wisent).
Cycloheximide Treatment and Hypotonic Cell Lysis.
Cells were pretreated with cycloheximide (100 µg/mL) (catalog no. CYC003; BioShop Canada) for 5 min and were lysed in hypotonic buffer [5 mM Tris⋅HCl (pH 7.5), 2.5 mM MgCl2, 1.5 mM KCl, 1× protease inhibitor mixture (EDTA-free), 100 µg/mL cycloheximide, 2 mM DTT, 200 U/mL RNaseIn, 0.5% Triton X-100, and 0.5% sodium deoxycholate] to isolate the polysomes with centrifugation (20,000 × g) at 4 °C for 5 min.
Polysome Fractionation.
Polysomes prepared as described in the previous paragraph (250 µg) were separated on a 10–50% (wt/vol) sucrose gradient by ultracentrifugation at 36,000 rpm for 2 h in an SW40 Ti rotor (Beckman Coulter) at 4 °C and were fractionated using an ISCO gradient fractionation system. OD at 254 nm was continuously recorded with a FOXO JR Fractionator (Teledyne ISCO).
Collection of RFPs.
The ribosome profiling assay was performed as described (63), with minor modifications. Briefly, 500 µg of the ribonucleoproteins (two biological replicates, prepared as described in the section Cycloheximide Treatment and Hypotonic Cell Lysis) were subjected to ribosome footprinting by RNase I treatment at 4 °C for 50 min with gentle mixing. Monosomes were pelleted by ultracentrifugation in a 34% (wt/vol) sucrose cushion at 70,000 rpm for 3 h (TLA 120.2, Beckman Coulter), and RNA fragments were extracted twice with acid phenol, once with chloroform, and were precipitated with isopropanol in the presence of NaOAc and GlycoBlue (Ambion). Purified RNA was resolved on a denaturing 15% (wt/vol) polyacrylamide urea gel, and the section corresponding to 28–32 nt containing the RFPs was excised, eluted, and precipitated by isopropanol.
Random RNA Fragmentation and mRNA-Seq.
Cytoplasmic RNA (150 µg) was used for mRNA-Seq analysis. Poly(A)+ mRNAs were purified using magnetic oligo-dT Dynabeads (Invitrogen) according to the manufacturer’s instructions. Purified RNA was eluted from the beads and mixed with an equal volume of 2× alkaline fragmentation solution (2 mM EDTA, 10 mM Na2CO3, 90 mM NaHCO3, pH 9.2) and incubated for 20 min at 95 °C. Fragmentation reactions were mixed with stop/precipitation solution [300 mM NaOAc (pH 5.5) and GlycoBlue], followed by isopropanol precipitation. Fragmented mRNA was size-selected on a denaturing 10% (wt/vol) polyacrylamide urea gel, and the area corresponding to 35–50 nt was excised, eluted, and precipitated with isopropanol.
Supplementary Material
Acknowledgments
We thank Martin Klar, Tommy Alain, Joshua Dunn, Jelena Popic, and Ola Larsson for discussions; Serge Gueroussov, Bushra Raj, and Vinagolu Rajasekhar for reagents; Joseph Marcotrigiano for the HEK293H cell line; and Annie Sylvestre and Annik Lafrance for animal care. This work was supported by Canadian Institutes of Health Research CIHR Grants MOP-142200 (to N.S.), MOP-125885 (to V.G.), MOP-115090 (to G.B.), MOP-111197 (to Y.Y.), and MOP-14609 (to B.J.B.). S.M.J. is the recipient of a CIHR Postdoctoral fellowship. S.T. is the recipient of a Richard and Edith Strauss fellowship. U.B. is the recipient of a Human Frontier Science Program Long Term Fellowship. T.G.-P. was supported by an EMBO (European Molecular Biology Organization) Fellowship and an Ontario Institute for Regenerative Medicine Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615540113/-/DCSupplemental.
References
- 1.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 2.Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26(4):903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 3.Sampath P, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2(5):448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 4.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509(7498):49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco S, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534(7607):335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462(7271):358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15(3):658–664. [PMC free article] [PubMed] [Google Scholar]
- 9.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: A critical nexus for cancer development. Cancer Res. 2015;75(2):250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunn GJ, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277(5322):99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 11.Hara K, et al. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272(42):26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 12.Pause A, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 13.Hartman NW, et al. mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Reports. 2013;5(2):433–444. doi: 10.1016/j.celrep.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 14.Signer RA, et al. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 2016;30(15):1698–1703. doi: 10.1101/gad.282756.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11(11):4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhat M, et al. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14(4):261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 17.Tahmasebi S, et al. Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell. 2014;14(5):606–616. doi: 10.1016/j.stem.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328(5982):1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olshen AB, et al. Assessing gene-level translational control from ribosome profiling. Bioinformatics. 2013;29(23):2995–3002. doi: 10.1093/bioinformatics/btt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen N, Zhang X, Olashaw N, Seto E. Molecular cloning and functional characterization of the transcription factor YY2. J Biol Chem. 2004;279(24):25927–25934. doi: 10.1074/jbc.M402525200. [DOI] [PubMed] [Google Scholar]
- 22.Kim JD, Faulk C, Kim J. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007;35(10):3442–3452. doi: 10.1093/nar/gkm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers MB, Hosler BA, Gudas LJ. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development. 1991;113(3):815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 24.Luo C, Lu X, Stubbs L, Kim J. Rapid evolution of a recently retroposed transcription factor YY2 in mammalian genomes. Genomics. 2006;87(3):348–355. doi: 10.1016/j.ygeno.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012;40(8):3403–3418. doi: 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donohoe ME, et al. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19(10):7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onder TT, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483(7391):598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25(1):43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 30.Gregoire S, et al. Essential and unexpected role of Yin Yang 1 to promote mesodermal cardiac differentiation. Circ Res. 2013;112(6):900–910. doi: 10.1161/CIRCRESAHA.113.259259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beketaev I, et al. Critical role of YY1 in cardiac morphogenesis. Dev Dyn. 2015;244(5):669–680. doi: 10.1002/dvdy.24263. [DOI] [PubMed] [Google Scholar]
- 32.Sigova AA, et al. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350(6263):978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wobus AM, et al. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J Mol Cell Cardiol. 1997;29(6):1525–1539. doi: 10.1006/jmcc.1997.0433. [DOI] [PubMed] [Google Scholar]
- 34.Pan J, Baker KM. Retinoic acid and the heart. Vitam Horm. 2007;75:257–283. doi: 10.1016/S0083-6729(06)75010-5. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, et al. Genome-wide analysis of YY2 versus YY1 target genes. Nucleic Acids Res. 2010;38(12):4011–4026. doi: 10.1093/nar/gkq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klar M. It is not necessarily YY1–the frequently forgotten Yin-Yang-2 transcription factor. Proc Natl Acad Sci USA. 2010;107(52):E190–author reply E191. doi: 10.1073/pnas.1011338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klar M, Bode J. Enhanceosome formation over the beta interferon promoter underlies a remote-control mechanism mediated by YY1 and YY2. Mol Cell Biol. 2005;25(22):10159–10170. doi: 10.1128/MCB.25.22.10159-10170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SH, et al. Yin-Yang 1 and Yin-Yang 2 exert opposing effects on the promoter activity of interleukin 4. Arch Pharm Res. 2016;39(4):547–554. doi: 10.1007/s12272-015-0622-7. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Brown SJ. BindN: A web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34(Web Server issue):W243–248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103(9):3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fablet M, Bueno M, Potrzebowski L, Kaessmann H. Evolutionary origin and functions of retrogene introns. Mol Biol Evol. 2009;26(9):2147–2156. doi: 10.1093/molbev/msp125. [DOI] [PubMed] [Google Scholar]
- 42.Braunschweig U, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24(11):1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paz I, Kosti I, Ares M, Jr, Cline M, Mandel-Gutfreund Y. RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014;42(Web Server issue):W361–W367. doi: 10.1093/nar/gku406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberstrass FC, et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309(5743):2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 45.Shibayama M, et al. Polypyrimidine tract-binding protein is essential for early mouse development and embryonic stem cell proliferation. FEBS J. 2009;276(22):6658–6668. doi: 10.1111/j.1742-4658.2009.07380.x. [DOI] [PubMed] [Google Scholar]
- 46.Suckale J, et al. PTBP1 is required for embryonic development before gastrulation. PLoS One. 2011;6(2):e16992. doi: 10.1371/journal.pone.0016992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colina R, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452(7185):323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 48.Melé M, et al. GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348(6235):660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. eLife. 2016;5:e10921. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Klerk E, t’Hoen PA. Alternative mRNA transcription, processing, and translation: Insights from RNA sequencing. Trends Genet. 2015;31(3):128–139. doi: 10.1016/j.tig.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Linares AJ, et al. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. eLife. 2015;4:e09268. doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19(17):4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kino Y, et al. MBNL and CELF proteins regulate alternative splicing of the skeletal muscle chloride channel CLCN1. Nucleic Acids Res. 2009;37(19):6477–6490. doi: 10.1093/nar/gkp681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han H, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498(7453):241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong JJ, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154(3):583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 56.Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26(11):1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sterne-Weiler T, et al. Frac-seq reveals isoform-specific recruitment to polyribosomes. Genome Res. 2013;23(10):1615–1623. doi: 10.1101/gr.148585.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maslon MM, Heras SR, Bellora N, Eyras E, Cáceres JF. The translational landscape of the splicing factor SRSF1 and its role in mitosis. eLife. 2014;3:e02028. doi: 10.7554/eLife.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4(6):487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 60.Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14(3):385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell. 2016;19(1):66–80. doi: 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 62.Wobus AM, Guan K, Yang HT, Boheler KR. Embryonic stem cells as a model to study cardiac, skeletal muscle, and vascular smooth muscle cell differentiation. Methods Mol Biol. 2002;185:127–156. doi: 10.1385/1-59259-241-4:127. [DOI] [PubMed] [Google Scholar]
- 63.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8):1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8(3):498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 65.Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 2010;11(9):R93. doi: 10.1186/gb-2010-11-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338(6114):1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 71.Gueroussov S, et al. An alternative splicing event amplifies evolutionary differences between vertebrates. Science. 2015;349(6250):868–873. doi: 10.1126/science.aaa8381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.