Significance
Despite the significant progress made in the last few years toward targeting phosphodiesterase-δ (PDEδ) for KRAS (Kirsten rat sarcoma isoform)-driven cancers, there is no structural information available on posttranslationally modified KRAS4b in complex with PDEδ. The KRAS4b–PDEδ structure reported here provides the structural details of the protein–protein interaction interface and the atomic details of the hypervariable region of KRAS4b. Structural comparison of the two crystal forms allowed identification of a 5-aa-long sequence motif in KRAS4b that could allow PDEδ to bind to both farnesylated and geranylgeranylated KRAS4b. Structural insights obtained from this study could be used to guide the development of improved and more specific inhibitors of the KRAS4b–PDEδ complex.
Keywords: prenylation, protein–protein interaction, KRAS4b, KRAS–PDEδ complex, phosphodiesterase-δ
Abstract
Farnesylation and carboxymethylation of KRAS4b (Kirsten rat sarcoma isoform 4b) are essential for its interaction with the plasma membrane where KRAS-mediated signaling events occur. Phosphodiesterase-δ (PDEδ) binds to KRAS4b and plays an important role in targeting it to cellular membranes. We solved structures of human farnesylated–methylated KRAS4b in complex with PDEδ in two different crystal forms. In these structures, the interaction is driven by the C-terminal amino acids together with the farnesylated and methylated C185 of KRAS4b that binds tightly in the central hydrophobic pocket present in PDEδ. In crystal form II, we see the full-length structure of farnesylated–methylated KRAS4b, including the hypervariable region. Crystal form I reveals structural details of farnesylated–methylated KRAS4b binding to PDEδ, and crystal form II suggests the potential binding mode of geranylgeranylated–methylated KRAS4b to PDEδ. We identified a 5-aa-long sequence motif (Lys-Ser-Lys-Thr-Lys) in KRAS4b that may enable PDEδ to bind both forms of prenylated KRAS4b. Structure and sequence analysis of various prenylated proteins that have been previously tested for binding to PDEδ provides a rationale for why some prenylated proteins, such as KRAS4a, RalA, RalB, and Rac1, do not bind to PDEδ. Comparison of all four available structures of PDEδ complexed with various prenylated proteins/peptides shows the presence of additional interactions due to a larger protein–protein interaction interface in KRAS4b–PDEδ complex. This interface might be exploited for designing an inhibitor with minimal off-target effects.
RAS mutations are found in one-third of all human cancers (1). Three RAS genes in humans encode four distinct but highly homologous RAS proteins: HRAS, NRAS, and the splice variants KRAS4a (Kirsten rat sarcoma isoform 4a) and KRAS4b (2). Among the three RAS isoforms, KRAS is the most commonly mutated gene in the RAS family and involved in 95% of pancreatic cancers and a high proportion of colorectal and lung cancers (3). RAS proteins function as molecular switches by alternating between inactive GDP-bound and active GTP-bound states (4–6). The active or inactive state of RAS proteins is regulated by guanine nucleotide exchange factors and GTPase-activating proteins (GAPs) (7–9). In the GTP-bound state, RAS proteins interact with a variety of effector proteins, such as Raf, PI3K, and RalGDS, leading to activation of several signaling cascades within the cell (10, 11). Oncogenic RAS mutations are predominantly found at amino acid positions G12, G13, and Q61 and impair intrinsic and GAP-mediated GTPase function (12).
RAS isoforms contain a highly conserved GTPase domain (G domain) and diverge substantially in the last 20 amino acids at the C-terminal end known as the hypervariable region (HVR) (10, 13). This region contains residues that undergo posttranslational modifications essential for targeting RAS proteins to the cytosolic leaflet of cellular membranes. All RAS proteins are farnesylated and carboxymethylated at the Cys residue present in the C-terminal CaaX motif, where a is an aliphatic amino acid and X is any amino acid. The terminal amino acid (X) determines the nature of the prenylated lipid chain. A leucine residue as X results in geranylgeranylation, and all other residues result in farnesylation (14). NRAS, HRAS, and KRAS4a are additionally modified by one or two palmitic acids on Cys residues located upstream of the CaaX motif. KRAS4b does not undergo palmitoylation. Instead, it has a polybasic region formed by a stretch of lysines that are believed to interact with negatively charged head groups of plasma membrane lipids (15). RAS proteins are further processed by RCE1 (RAS converting CaaX endopeptidase 1) (16), which removes the -aaX residues, followed by ICMT (isoprenylcysteine carboxyl methyltransferase) (17), which adds a methyl group on the carboxyl group of the C-terminal prenylated Cys. Almost two decades ago, RAS farnesyltransferase inhibitors (FTIs) were intensively explored in clinical trials, but because of alternative prenylation (geranylgeranylation) of KRAS and to a lesser extent, NRAS by geranylgeranyl transferases in the presence of FTIs, their high preclinical anticancer activity could not be translated into clinical efficacy (18, 19). Tumors driven by mutation of HRAS, which can only be farnesylated, are sensitive to FTIs, at least in preclinical models.
Phosphodiesterase-δ (PDEδ; also known as PDE6δ, PrBP/δ, and PDE6D) was initially identified as the noncatalytic δ-subunit of photoreceptor PDE6. It was later implicated in membrane release and localization of prenylated RAS and other prenylated proteins (20–23). PDEδ has been shown to sequester KRAS4b from the cytosol by binding the prenylated HVR, thus preventing it from binding to extensive endomembrane surfaces and thereby, enhancing its diffusion throughout the cell. KRAS4b is then released in perinuclear membranes by the release factors Arl2 and Arl3, where it binds the recycling endosome and is delivered to the plasma membrane via vesicular transport (24, 25). Binding of either GTP-bound Arl2/3 or prenylated RAS to PDEδ has been suggested to be mutually exclusive (26). Biochemical and cellular studies have shown that the interaction between PDEδ and KRAS4b is independent of its nucleotide (GTP/GDP) status (27).
The structure of PDEδ in a complex with fully modified Rheb (a member of the RAS subfamily) has been solved (26). Unlike KRAS4b, Rheb has a smaller HVR with a different amino acid composition and lacks the polybasic region. In this structure, the C-terminal farnesyl chain of Rheb is present in a hydrophobic pocket within the Ig-like β-sandwich fold of PDEδ, with three amino acids upstream of the prenylated cysteine interacting with PDEδ. Based on these studies, a high-throughput screen followed by a rational structure-based design approach resulted in the development of deltarasin as an inhibitor of the KRAS4b–PDEδ interaction (28). Deltarasin binds inside the hydrophobic pocket of PDEδ with low nanomolar affinity in vitro as well as in vivo. Cellular studies with adenopancreatic cancer cells showed that addition of 5 μM deltarasin blocks RAS signaling, reduces proliferation, and causes cell death in KRAS-dependent cells, whereas minimal effects were observed in KRAS-independent cells (28). Additionally, the administration of deltarasin to nude mice bearing s.c. xenografts of human Panc-Tu-I tumors caused a dose-dependent reduction of tumor growth. Overall, these results showed that PDEδ could be a potential target, and deltarasin and other small molecules represent a platform for finding better and more specific inhibitors of the KRAS4b–PDEδ complex.
Despite significant progress made in the last few years in identifying and targeting PDEδ for KRAS-driven cancers, there is no structural information available on fully processed KRAS4b in complex with PDEδ. In this paper, we describe structures of farnesylated–methylated KRAS4b (KRAS4b-FMe) in complex with PDEδ in two different crystal forms. Our structures provide atomic details of the HVR of KRAS4b in complex with PDEδ. Comparison of the two crystal forms describes not only how KRAS4b-FMe binds to PDEδ but also, how binding of prenylated KRAS4b and PDEδ is likely to be facilitated by the presence of a 5-aa-long sequence motif in the HVR of KRAS4b. The conserved sequence of this motif may account for the binding of PDEδ to both forms of prenylated KRAS4b. Finally, structure-based mutational studies suggest key roles played by various amino acids involved in the KRAS4b–PDEδ interaction.
Results
The Role of Farnesyl and Methyl Groups in Interactions Between KRAS4b and PDEδ.
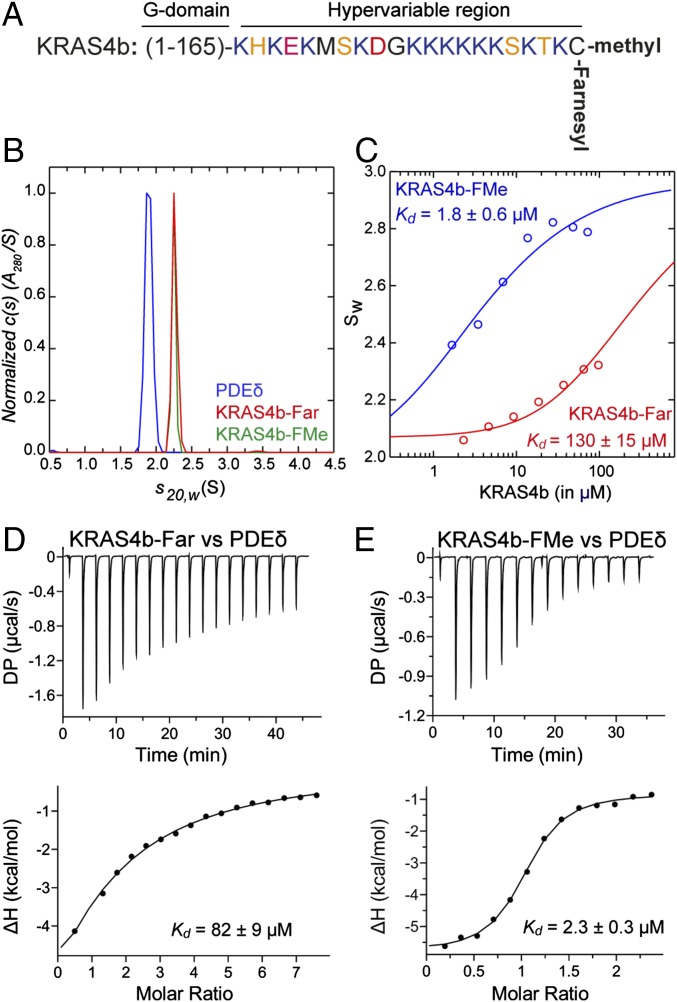
To understand the role of farnesylation and carboxymethylation of C185 of KRAS4b (Fig. 1A) in its interaction with PDEδ, we characterized the interaction of farnesylated KRAS4b (KRAS4b-Far) and KRAS4b-FMe with PDEδ using sedimentation velocity analytical ultracentrifugation (SV-AUC) and isothermal titration calorimetry (ITC). Sedimentation velocity studies on PDEδ, KRAS4b-FMe, and KRAS4b-Far showed the presence of a single species with sedimentation coefficients of 1.90 S, 2.25 S, and 2.24 S, respectively (Fig. 1B). The best fit molar masses of 18 ± 1 kDa for PDEδ, 22.9 ± 0.5 kDa for KRAS4b-FMe, and 22.9 ± 0.6 kDa for KRAS4b-Far show that these species are monomers in solution. To characterize the interaction of PDEδ with KRAS4b-FMe, solutions containing approximately equimolar amounts of these two species were studied at different concentrations. The c(s) profiles (sedimentation coefficient distributions) obtained indicate the formation of a high-affinity 1:1 complex (Fig. S1A). At the highest concentration, the predominant species observed has a sedimentation coefficient of 2.92 S and estimated molar mass of 36 ± 3 kDa. As the concentration was reduced, the relative proportion of complex decreased, and based on the c(s) profiles, it seems that the complex is in fast exchange with the free species (Fig. S1A). To determine the affinity of the interaction, a weighted average sedimentation coefficient obtained by integration of the c(s) distribution was used to construct an sw isotherm and fit to a model describing an A + B = AB interaction (Fig. 1C). The dissociation constant, Kd, was determined to be 1.8 ± 0.6 μM. Similar experiments characterizing the interaction of PDEδ with KRAS4b-Far led to c(s) profiles indicative of a much weaker interaction (Fig. S1B), with a significant proportion of uncomplexed material at the highest concentrations studied. The weak association was confirmed in an analysis of the sw isotherm, which returned an estimated dissociation constant Kd of 130 ± 15 μM (Fig. 1C). A similar trend was observed when we measured dissociation constants (Kd) of PDEδ with KRAS4b-Far and KRAS4b-FMe using ITC. ITC measurements showed a weak binding affinity with a Kd of 82 ± 9 μM between KRAS4b-Far and PDEδ (Fig. 1D), whereas the presence of both farnesyl and methyl groups on KRAS4b led to a relatively stronger binding with a Kd of 2.3 ± 0.3 μM (Fig. 1E). These results show that the presence of both the methyl and farnesyl modifications increases the affinity between KRAS4b and PDEδ close to two orders of magnitude compared with only KRAS4b-Far. Previous studies using a fluorescence–polarization (FP) assay showed a 10-fold higher binding affinity (Kd = 0.3 μM) between KRAS4b and PDEδ (29). This discrepancy is likely caused by the use of different techniques (analytical ultracentrifugation and ITC vs. FP), different assay conditions, and the use of semisynthetic and refolded KRAS4b vs. recombinant fully processed KRAS4b that we used for this study.
Fig. 1.
Carboxymethylation of farnesylated C185 is important for KRAS4b–PDEδ interaction. (A) Amino acid sequence of the HVR of KRAS4b showing the presence of a polylysine patch and farnesylated and carboxymethylated C185 at the C terminus. (B) Normalized absorbance c(s) profiles for PDEδ (15 μM; blue), KRAS4b-FMe (28 μM; red), and KRAS4b-Far (40 μM; green) in SV-AUC experiments show that these three proteins are monomers. Similar profiles were observed using the interference optical detection system. (C) Weighted absorbance sedimentation coefficient isotherms based on c(s) profiles for PDEδ and KRAS4b-FMe (blue) and PDEδ and KRAS4b-Far (red). Data were globally modeled in terms of an A + B = AB heteroassociation model, as described in the text, to yield the best fit shown. (D and E) ITC titration experiments to measure the dissociation constant (D) between KRAS4b-Far and PDEδ and (E) between KRAS4b-FMe and PDEδ. The DP is a measured differential power between the reference cell and the sample cell.
Fig. S1.
The association properties of PDEδ with KRAS4b-Far and KRAS4b-FMe measured using SV-AUC. (A) Absorbance c(s) profiles for approximately equimolar mixtures of PDEδ and KRAS4b-FMe at various concentrations. Based on a determination of the stock solution concentrations, the actual ratio is 1.08:1 KRAS4b-FMe:PDEδ. KRAS4b-FMe concentrations and cell path length are indicated. Similar profiles were observed using the interference optical detection system. (B) Absorbance c(s) profiles for approximately equimolar mixtures of PDEδ and KRAS4b-Far at various concentrations. Based on a determination of the stock solution concentrations, the actual ratio is 1.32:1 KRAS4b-Far:PDEδ. KRAS4b-Far concentrations and cell path length are indicated. Similar profiles were observed using the interference optical detection system.
Structures of KRAS4b-FMe in Complex with PDEδ in Two Crystal Forms.
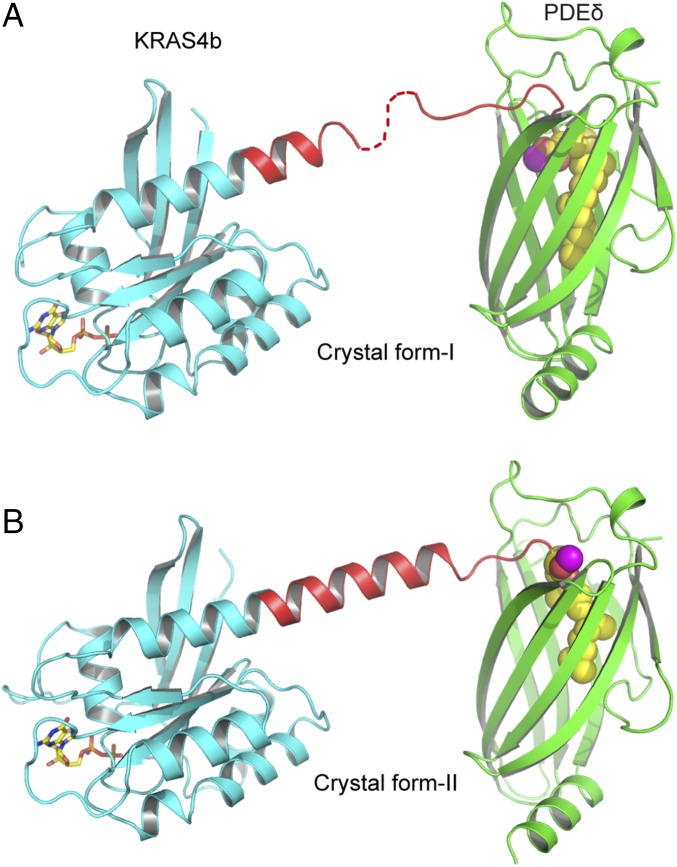
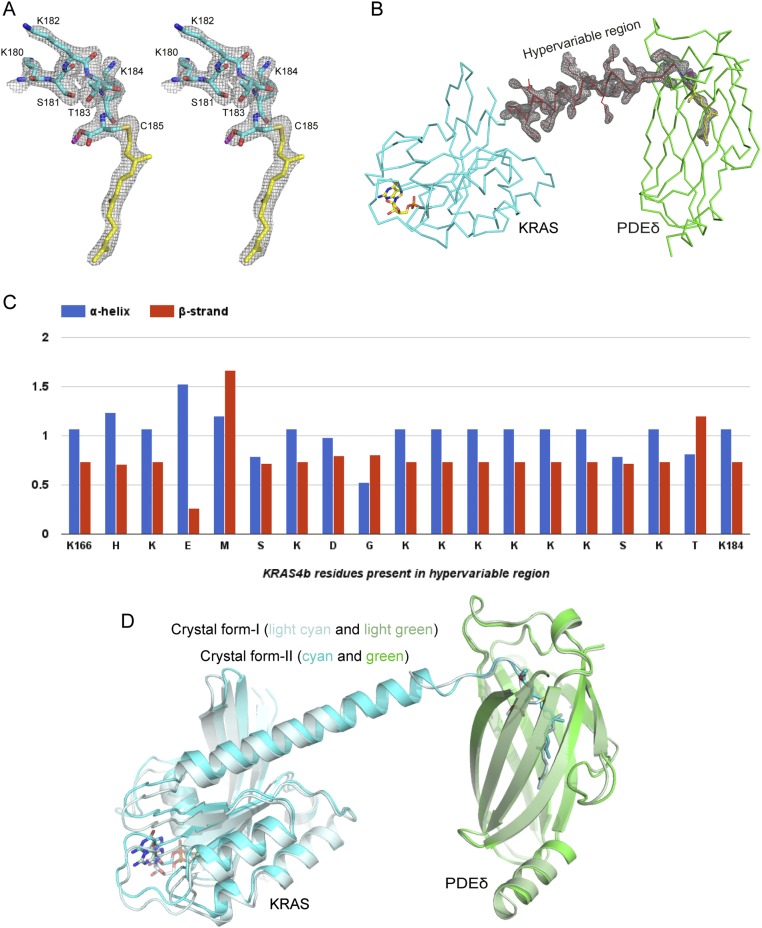
We attempted to crystallize KRAS4b-Far as well as KRAS4b-FMe in complex with PDEδ and were successful in crystallizing only the latter protein in complex with PDEδ in two different crystal forms referred to as crystal forms I and II (Fig. 2 and Table S1). In crystal form I, the asymmetric unit contains two molecules of KRAS4b–PDEδ complex, with PDEδ interacting with the last five residues located upstream of farnesylated–methylated C185 of KRAS4b. In this structure, we did not observe interpretable electron density for three to five amino acids (172–176 or 174–176) in the HVR in two KRAS4b–PDEδ complexes present in the asymmetric unit, indicating flexibility in these residues inside the crystal (Fig. 2A and Fig. S2A). In crystal form II, the asymmetric unit contained only one molecule of KRAS4b–PDEδ complex, where we see the electron density for the entire HVR of KRAS4b (Fig. S2B). In this crystal form, the C-terminal helix is extended and includes HVR residues up to K180, including six lysine residues (175–180) present in the polybasic region. Unlike crystal form I, in crystal form II, only three terminal residues of KRAS4b located upstream of farnesylated–methylated C185 were involved in complex formation. In crystal form II, most of the residues present in the HVR that were not involved in complex formation formed a long helical secondary structure (Fig. 2B). Secondary structure propensity analysis for residues present in HVR indicates that most of the residues have the tendency to form or favor α-helical structure (Fig. S2C).
Fig. 2.
Overall structure of KRAS4b–PDEδ complex in two different crystal forms. (A) Ribbon representation of GDP-bound KRAS4b-FMe in complex with PDEδ in crystal form I. (B) Ribbon representation of GDP-bound KRAS4b-FMe in complex with PDEδ in crystal form II. The PDEδ, GTPase domain, and HVR of KRAS4b are shown in green, cyan, and red, respectively. The farnesyl chain is shown as spheres and colored yellow. The carbon and oxygen atoms of carboxymethyl group are colored magenta and red, respectively. GDP is shown as a stick and colored yellow (carbon) and red (oxygen).
Table S1.
Crystallographic data collection and refinement statistics
| KRAS–PDEδ: Crystal form I | KRAS–PDEδ: Crystal form II | |
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 58.4, 70.7, 94.4 | 58.1, 70.0, 84.5 |
| α, β, γ (°) | 90.0, 92.4, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50–2.0 (2.1–2.0)* | 50–1.9 (2.0–1.9)* |
| Rmerge (%) | 5.0 (65.7) | 6.2 (82.7) |
| Average I/σ(I) | 21.6 (3.2) | 19.6 (3.3) |
| Completeness (%) | 99.3 (99.2) | 99.8 (99.0) |
| Redundancy | 6.2 (5.7) | 7.2 (7.0) |
| Refinement | ||
| Resolution (Å) | 50–2.0 | 50.0–1.9 |
| No. of reflections | 51,556 | 26,207 |
| Rwork/Rfree | 0.201/0.248 | 0.224/0.262 |
| No. of atoms | ||
| Protein | 5,172 | 2,688 |
| Ligand/ion | 94 | 55 |
| Water | 160 | 39 |
| B factors (Å2) | ||
| Protein | 49.1 | 37.6 |
| Ligand/ion | 47.1 | 43.8 |
| Water | 52.5 | 33.3 |
| rmsds | ||
| Bond lengths (Å) | 0.008 | 0.008 |
| Bond angles (°) | 1.012 | 1.297 |
Values in parentheses are for the highest-resolution shell. One crystal was used to collect each dataset.
Fig. S2.
Electron density corresponding to the HVR residues, superposition of two crystal forms, and secondary structure propensity analysis of the HVR residues. (A) 2Fo-Fc map contoured at 1.0σ for the HVR residues interacting with PDEδ and farnesylated–methylated C185 in the KRAS4b–PDEδ complex in crystal form I. (B) 2Fo-Fc map contoured at 1.0σ for the HVR residues and farnesylated–methylated C185 in the KRAS4b–PDEδ complex in crystal form II. (C) Secondary structure propensity for residues present in the HVR of KRAS4b. (D) Structural superposition of two crystal forms of KRAS4b–PDEδ complexes aligned using residues of PDEδ. KRAS4b and PDEδ in crystal form I are colored light cyan and light green, respectively and in crystal form II, are colored cyan and green, respectively. Farnesylated–methylated C185 and GDP are shown in stick representation.
In neither crystal form did we observe any specific interaction between the GTPase domain of KRAS4b and PDEδ (Fig. S2D), which provides a rationale for the broad specificity of PDEδ for prenylated proteins and also explains why the KRAS4b–PDEδ interaction is independent of the KRAS4b nucleotide (GDP/GTP) status. Although within the crystal, the GTPase domain of KRAS4b interacts with symmetry-related PDEδ molecules, these interactions are minimal and differ in the two crystal forms, implying that they are nonspecific and limited to crystal contact only. The overall structure and conformation of the switch I and II regions of KRAS4b resemble previously solved structures of KRAS4b–GDP complexes. In crystal form I, no electron density was observed for the switch I region.
Comparison of the interaction between KRAS4b and PDEδ in these two crystal forms suggests two different modes of binding between these two proteins (Fig. 2 and Fig. S2D). Unlike crystal form II, in crystal form I, HVR residues as well as farnesyl and methyl groups go deep inside the PDEδ hydrophobic pocket, presumably resulting in stronger interaction between these two proteins. We will first describe the stronger interaction observed between these two proteins in crystal form I and then, compare it with crystal form II.
Interaction of the HVR Residues of KRAS4b with PDEδ in Crystal Form I.
Residues present in the HVR.
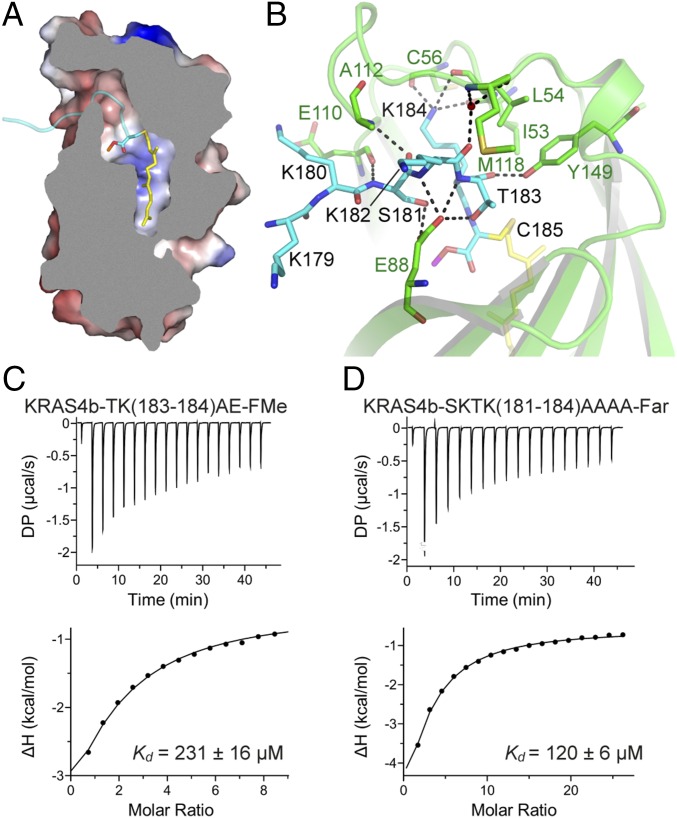
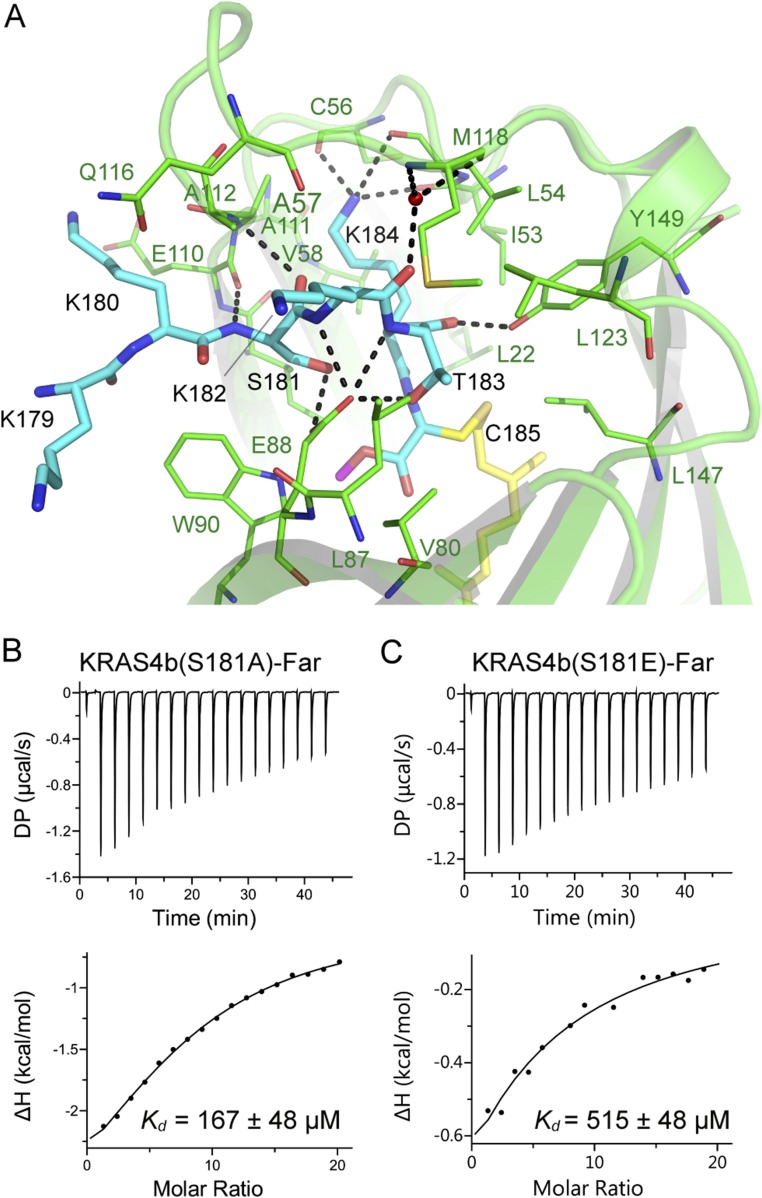
In crystal form I, KRAS4b interacts with PDEδ through the C-terminal amino acids S181, K182, T183, K184, and farnesylated–methylated C185 (Fig. 3 A and B). Residue K184 forms multiple hydrogen bonds with main chain atoms of PDEδ residues I53, L54, and C56 (Fig. 3B and Fig. S3A). Side chain oxygen and main chain oxygen/nitrogen atoms of T183 form hydrogen bonds with residues E88 and Y149 from PDEδ. The side chain of K182 points toward the surface and does not form any hydrogen bonds with PDEδ residues, but the main chain nitrogen atom of K182 forms a hydrogen bond with E88 from PDEδ. Residue S181 forms three hydrogen bonds with PDEδ via side chain oxygen atom with E88 and via main chain atoms with E110 and A112 from PDEδ. The HVR residue K180 present near the entry gate forms no hydrogen bonds with any PDEδ residues but does form relatively weak van der Waals and hydrophobic interactions with residues W90, E110, A111, and A112 of PDEδ (Fig. 3B and Fig. S3A).
Fig. 3.
Details of intermolecular protein–protein interactions in KRAS4b–PDEδ complex and impact of mutation of the HVR residues on KRAS4b–PDEδ interaction. (A) Sliced view of an electrostatic representation of PDEδ showing the region where the HVR residues (colored cyan) bind and a central hydrophobic pocket that accommodates farnesylated (colored yellow and shown in stick representation) and methylated C185 (colored cyan and shown in stick representation). (B) Intermolecular hydrogen bonding contacts involving residues from the HVR of KRAS4b and PDEδ. Residues from PDEδ and KRAS4b are shown in stick representation and colored green and cyan, respectively. The farnesyl and methyl groups are shown in stick representation and colored yellow and magenta, respectively. Dashed black lines indicate intermolecular hydrogen bonds. (C) ITC titration experiments to measure the dissociation constant between farnesylated–methylated double mutant (T183A-K184E) of KRAS4b and WT PDEδ. (D) ITC titration experiments to measure the dissociation constant between farnesylated quadruple mutant (S181A-K182A-T183A-K184A) of KRAS4b and WT PDEδ. This mutant could only be expressed and purified in farnesylated form in our engineered insect cell expression system. DP, differential power.
Fig. S3.
Details of intermolecular protein–protein interaction in KRAS4b–PDEδ complex and impact of mutation of the HVR residues on KRAS4b–PDEδ interaction. (A) Intermolecular hydrogen bonding contact involving residues from the HVR of KRAS4b and PDEδ. PDEδ residues involved in hydrophobic and van der Waals interactions with KRAS4b residues are also shown. Residues from PDEδ and KRAS4b are shown in stick representation and colored green and cyan, respectively. The farnesyl and methyl groups are shown in stick representation and colored yellow and magenta, respectively. Dashed black lines indicate intermolecular hydrogen bonds. (B) ITC measurement for binding of farnesylated S181A mutant of KRAS4b to WT PDEδ. (C) ITC measurement for binding of farnesylated S181E mutant of KRAS4b to WT PDEδ. DP, differential power.
We carried out structure-based mutational studies to examine the role of the HVR residues in the interaction between KRAS4b and PDEδ. A double mutant of KRAS4b, T183A and K184E, was expressed and purified in the farnesylated–methylated form and showed weaker binding by two orders of magnitude, indicating that the composition of residues present upstream of C185 also plays a key role in the interaction between KRAS4b and PDEδ (Fig. 3C). We also mutated S181 to alanine and glutamate and all four terminal residues from S181-K184 to alanine, but these three KRAS4b mutants could only be isolated in the farnesylated form (but not methylated) in our engineered insect cell expression system for reasons that remain to be determined. In these cases, we compared Kd values obtained on mutants with KRAS4b-Far. Farnesylated S181A and SKTK(181–184)AAAA showed slightly weaker binding to PDEδ compared with WT KRAS4b-Far, suggesting that smaller aliphatic residues are tolerated at these positions (Fig. 3D and Fig. S3B). Unlike farnesylated S181A, farnesylated S181E mutant showed much weaker binding with PDEδ, indicating that the presence of a larger and acidic amino acid at residue 181 affects KRAS4b binding to PDEδ (Fig. S3C). Overall, these studies showed that mutations of the HVR residues located upstream of C185 to small aliphatic amino acid, such as alanine, are tolerated but any drastic mutation to bulky and acidic amino acids, such as glutamate, severely affects the interaction between these two proteins.
Farnesylated and methylated C185.
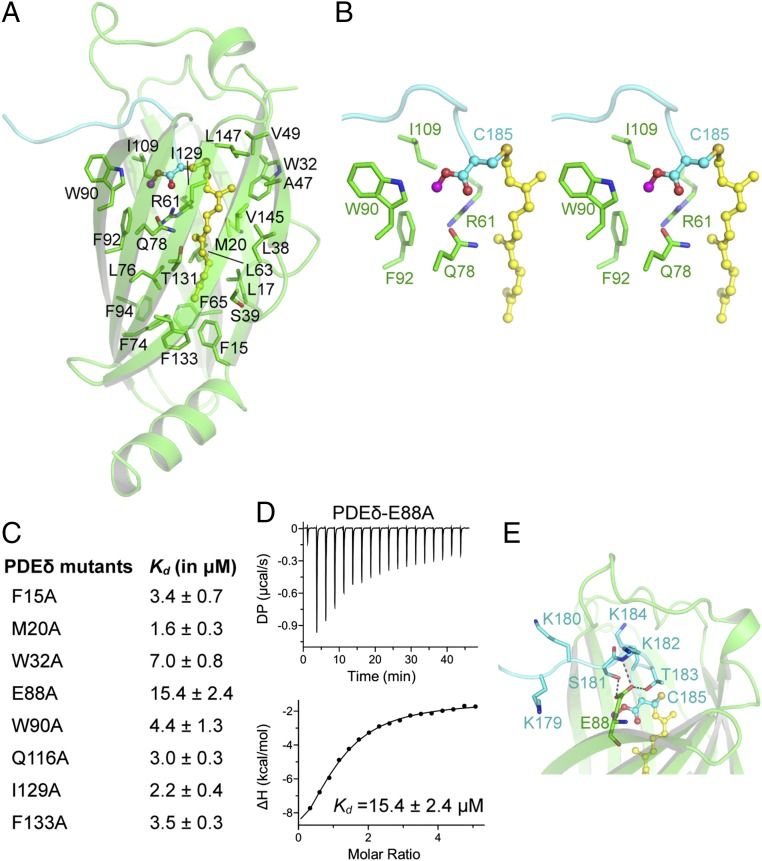
The farnesyl and methyl groups attached to C185 of KRAS4b bind in a hydrophobic pocket formed by ∼23 PDEδ residues, consisting mainly of Phe, Ile, Leu, Trp, Val, Met, and Ala (Fig. 4A). There are five phenylalanine residues, each present on individual β-strands that line the bottom of the hydrophobic pocket (Fig. 4A). Among these residues, F133 comes closest to the tail end of the farnesyl chain and forms hydrophobic interactions with terminal carbon atoms of the farnesyl chain. Residue W90 of PDEδ is located at the entry point of the pocket and expected to play a role in the entry of the HVR into the hydrophobic tunnel. Other aliphatic amino acids, such as Ile, Leu, Val, and Ala, of PDEδ position themselves around the aliphatic carbon atoms of the farnesyl chain to provide a hydrophobic environment when KRAS4b is trafficking inside the hydrophilic milieu of the cytoplasm (Fig. 4A). The carboxymethyl group of C185 binds at another pocket formed by PDEδ residues W90, F92, and I109. Residues R61 and Q78 of PDEδ are located close to this pocket and provide a polar environment for the carboxymethyl group of C185 (Fig. 4B).
Fig. 4.
Recognition of farnesylated and carboxymethylated C185 of KRAS4b by PDEδ and impact of mutation of PDEδ residues on KRAS4b–PDEδ interaction. (A) Nonpolar residues (colored green; stick representation) of PDEδ form a hydrophobic pocket that accommodates the farnesyl chain (colored yellow; ball and stick representation) and carboxymethyl group (magenta and cyan; ball and stick representation). (B) Stereo view of the PDEδ pocket formed by residues (colored green; stick) that accommodates the carboxymethyl group (colored magenta and cyan; ball and stick) of farnesylated C185 (colored yellow; ball and stick). (C) Dissociation constant of PDEδ mutants with KRAS4b-FMe measured using ITC titration experiments. (D) ITC titration experiments to measure the dissociation constant between PDEδ–E88A mutant and KRAS4b-FMe. (E) Intermolecular hydrogen bonds formed by PDEδ residue E88 (colored green; stick) with HVR residues (colored cyan; stick). DP, differential power.
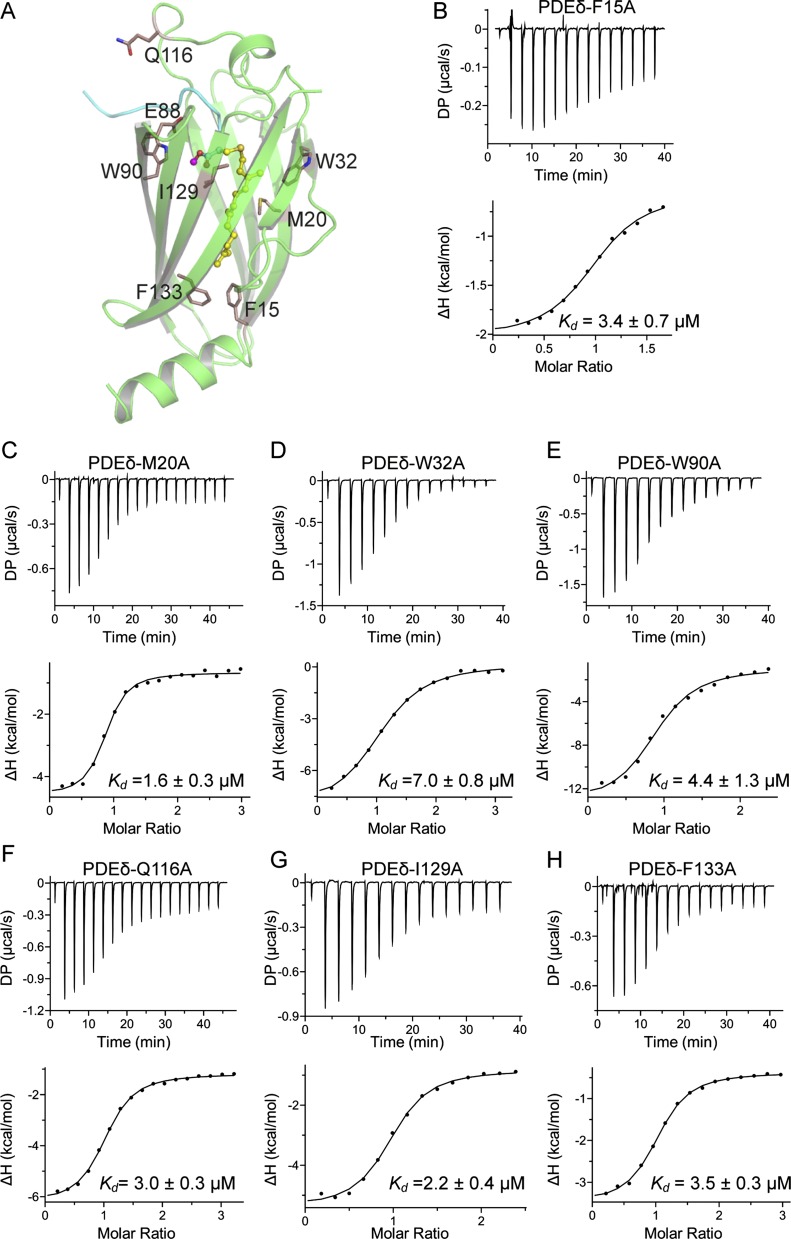
To understand the role of various PDEδ residues in the interaction with the farnesyl and methyl groups of KRAS4b, we selected eight different PDEδ residues based on the KRAS4b–PDEδ complex structure (F15, M20, W32, E88, W90, Q116, I129, and F133) and mutated those individually to alanine (Fig. 4C and Fig. S4A). We measured the dissociation constants (Kd) of these eight PDEδ mutants with KRAS4b-FMe using ITC. Surprisingly, most of these mutants could bind to KRAS4b-FMe with close to WT affinity, suggesting that single-amino acid substitutions of these amino acids into alanine do not substantially affect the interaction between KRAS4b and PDEδ (Fig. 4C and Fig. S4 B–H). Only the E88A mutation showed fivefold weaker binding to KRAS4b-FMe (Fig. 4D). Examination of the interactions mediated by E88 shows formation of multiple hydrogen bonds with side chain atoms of S181 and main chain atoms of T183, supporting the weaker binding for this point mutant (Fig. 4E).
Fig. S4.
Impact of mutations of PDEδ residues on KRAS4b–PDEδ interaction. (A) Position of eight PDEδ residues that were mutated in this study. ITC measurements showing dissociation constant of KRAS4b-FMe with PDEδ mutants (B) PDEδ-F15A, (C) PDEδ-M20A, (D) PDEδ-W32A, (E) PDEδ-W90A, (F) PDEδ-Q116A, (G) PDEδ-I129A, and (H) PDEδ-F133A. DP, differential power.
Comparison of Two Crystal Forms Suggests Two Possible Binding Modes Between Prenylated KRAS4b and PDEδ.
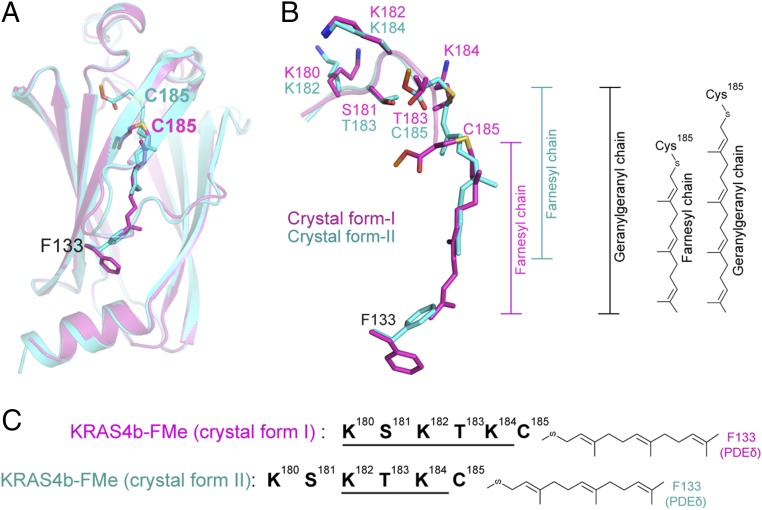
Structural superposition of the two crystal forms suggests two different binding modes between KRAS4b and PDEδ. Compared with crystal form I, the HVR residues in crystal form II are shifted upstream by two amino acids at the KRAS4b–PDEδ interface, and thus, only the last three amino acids of the HVR and prenylated C185 of KRAS4b interact with PDEδ (Fig. 5 A and B). Although it seems surprising at first that, even after shifting two amino acids, the HVR can still form a similar interaction with PDEδ residues, this can be explained by the fact that, in crystal form II, residues K182-T183-K184 occupy the same positions as those of residues K180-S181-K182 in crystal form I (Fig. 5 B and C). The methyl group of C185 in crystal form II occupies the same position as that of the side chain atoms of T183 in crystal form I. Consequently, in crystal form II, the farnesyl chain partially occupies the hydrophobic pocket of PDEδ, with the remainder of the empty space packed by conformational changes in the side chain atoms of F133 (Fig. 5B).
Fig. 5.
Structural comparison of two crystal forms of KRAS4b–PDEδ complex shows two possible modes of binding between KRAS4b and PDEδ. (A) Structural superposition of PDEδ residues from two crystal forms showing different positions of farnesyl and methyl groups attached to C185. Crystal forms I and II are colored magenta and cyan, respectively. (B) Enlarged view of structural superposition of HVR residues and farnesylated–methylated C185 from two crystal forms showing upstream shift of the HVR residues by two amino acids at the protein–protein interaction interface in crystal form II, resulting in the empty space in the central hydrophobic pocket of PDEδ. Positions of the farnesyl chains are shown. Structural superposition of the two crystal forms suggests that the empty space inside the hydrophobic pocket of PDEδ in crystal form II would fit an additional five carbons present on the geranylgeranyl chain. (C) Schematic diagram showing a 2-aa shift of the HVR residues in crystal form II. Because of the unique amino acid composition in the region, interaction interface formed by residues K182-T183-K184 residues in crystal form II mimics the interaction interface formed by residues K180-S181-K183 in crystal form I.
Structural superposition of two crystal forms indicates that the hydrophobic pocket of PDEδ in crystal form II can accommodate an additional five carbon atoms if F133 occupies the same rotamer conformation as seen in crystal form I (Figs. 5B and 6A). Previous studies have shown that, when farnesyl transferase inhibitors block farnesylation, KRAS4b undergoes alternative prenylation and is geranylgeranylated by geranylgeranyl transferase (19). The farnesyl chain is 15-carbon atoms long, whereas the geranylgeranyl chain has an additional five carbons attached to it. Structural superposition of the two crystal forms indicates there is space for these additional 5-carbon atoms in the hydrophobic pocket, thus indicating that the 20-carbon-long geranylgeranyl chain would also fit in this pocket (Figs. 5B and 6A).
Fig. 6.
KRAS4b–PDEδ structure in crystal form II mimics the binding of KRAS4b-GGMe to PDEδ and identification of a new sequence motif in the HVR of KRAS4b. (A) Schematic diagram showing the additional five carbon atoms present in the geranylgeranyl chain compared with the farnesyl chain. (B) Structural superposition of prenylated C185 and the HVR residues from KRAS4b-FMe–PDEδ structure in crystal form I (colored magenta) and modeled structure of KRAS4b-GGMe–PDEδ (colored green) using KRAS4b-FMe–PDEδ structure in crystal form II. Positions of farnesyl and geranylgeranyl chains are highlighted. (C) Structural superposition of geranylgeranylated–methylated pentapeptide of PDE6α′ in complex with PDEδ (colored light blue; PDB ID code 5ETF) and modeled structure of KRAS4b-GGMe in complex with PDEδ (colored green). Positions of geranylgeranyl chains in these two structures are similar. PDE6α′-GGMe, geranylgeranylated–methylated PDE6α′. (D) Sequence alignment of amino acid sequence of the HVR of KRAS4b from various organisms shows the presence of a unique 5-aa-long sequence motif that allows the binding of prenylated KRAS4b to PDEδ as seen in this study. The HVR residues proposed to maintain similar protein–protein interaction in the KRAS4b-FMe–PDEδ and the KRAS4b-GGMe–PDEδ complexes are highlighted by magenta- and green-colored lines above the sequence alignment.
Crystal Form II Mimics Geranylgeranylated–Methylated KRAS4b Binding to PDEδ.
Because crystal form II mimics a possible binding mode of geranylgeranylated–methylated KRAS4b (KRAS4b-GGMe) with PDEδ, we modeled the structure of KRAS4b-GGMe in complex with PDEδ using this structure (Fig. 6B). To allow the geranylgeranyl chain to bind in the hydrophobic pocket, we changed the rotamer conformation of side chain atoms of F133 to the conformation observed in crystal form I. Modeled structures showed an ideal fit for a geranylgeranyl chain in the hydrophobic pocket, with the last three HVR residues mimicking a similar interaction with PDEδ as observed in crystal form II (Fig. 6B). Recently, the structure of PDEδ in complex with geranylgeranylated–methylated peptide from PDE6α′ was reported (30). Comparison of this structure with our modeled structure of KRAS4b-GGMe in complex with PDEδ shows a similar position of the geranylgeranyl chain inside the hydrophobic pocket and similar HVR residues (KSKT in PDE6α′ vs. KSKTK in KRAS4b) upstream of the prenylated Cys, differing mainly in the position of the carboxymethyl group likely because of different penultimate residues in these two proteins (Fig. 6C).
Comparison Between KRAS4b–PDEδ and Rheb–PDEδ Structures.
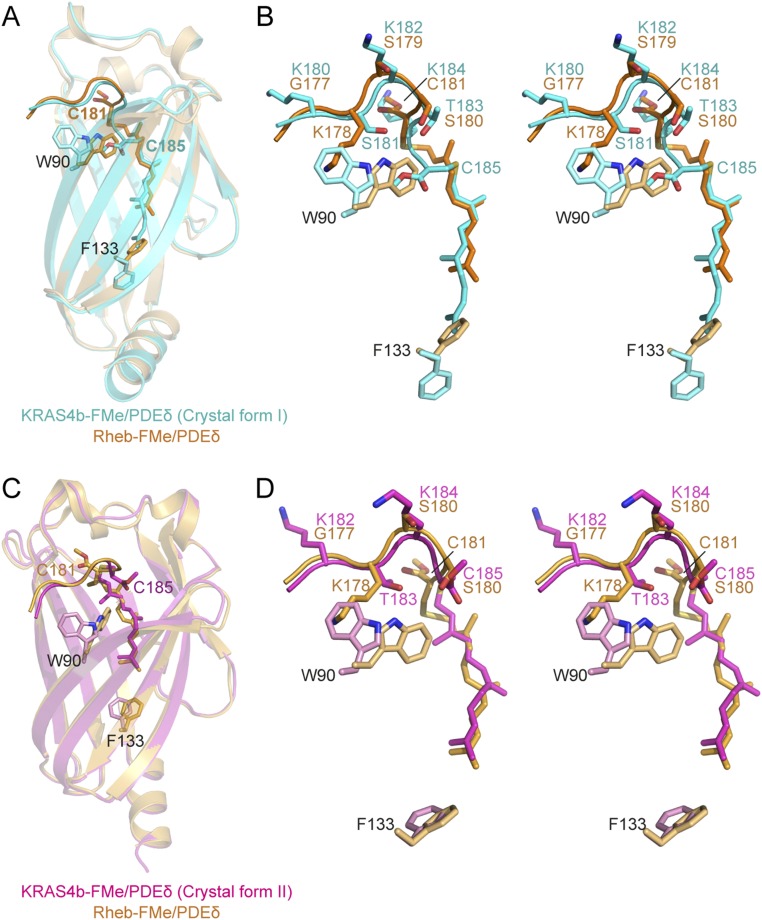
A structural comparison of KRAS4b–PDEδ and Rheb–PDEδ complexes revealed that neither KRAS4b nor Rheb interact with PDEδ via the GTPase domain (26), providing a rationale for the observed broad specificity of PDEδ to other members of the RAS superfamily. Along with this similarity, we observed a few significant differences between these two complexes. Unlike Rheb, the farnesyl and methyl groups in KRAS4b bind deeper inside the hydrophobic pocket of PDEδ in crystal form I (Fig. S5 A and B). In addition, the methyl groups present at the farnesylated cysteine in these two structures point in opposite directions with respect to the HVR. In the Rheb–PDEδ complex, the methyl group occupies the same position as that of K184 in the KRAS4b–PDEδ complex. Furthermore, in the Rheb–PDEδ complex, only three HVR residues and the prenylated cysteine interact with PDEδ (26), whereas in the KRAS4b–PDEδ complex (crystal form I), the last four residues and prenylated cysteine interact with PDEδ (Fig. S5B). Additional interactions in the case of KRAS4b are facilitated by large movements of side chain atoms of W90 at the entry point and F133 at the bottom of the hydrophobic pocket in PDEδ (Fig. S5B). The position of the farnesyl chain in crystal form II matches that of the farnesyl chain in the Rheb–PDEδ structure (Fig. S5 C and D). However, the position of the methyl group and interactions between the HVR and PDEδ residues differ in these two structures.
Fig. S5.
Structural comparison between KRAS4b–PDEδ and Rheb–PDEδ complexes. (A) Structural superposition of KRAS4b–PDEδ (crystal form I) and Rheb–PDEδ (PDB ID code 3T5I) aligned using PDEδ residues. KRAS4b–PDEδ and Rheb–PDEδ complexes are colored cyan and light orange, respectively. Two PDEδ residues, W90 and F133 (stick), undergo large conformational changes to facilitate the binding of farnesylated–methylated C185 deeper within the hydrophobic pocket of the KRAS4b–PDEδ complex (crystal form I). (B) Stereo view of superposition of KRAS4b–PDEδ (crystal form I) and Rheb–PDEδ complexes showing farnesylated–methylated cysteine, KRAS4b HVR, and PDEδ residues. The color-coding scheme is same as in A. (C) Structural superposition of KRAS4b–PDEδ (crystal form II) and Rheb–PDEδ (PDB ID code 3T5I) complexes aligned using PDEδ residues. KRAS4b–PDEδ and Rheb–PDEδ complexes are colored magenta and light orange, respectively. PDEδ residue W90 undergoes large conformation changes to facilitate binding of farnesylated–methylated C185 within the hydrophobic pocket in KRAS4b–PDEδ complex (crystal form II). (D) Stereo view of superposition of KRAS4b–PDEδ (crystal form II) and Rheb–PDEδ complexes showing farnesylated–methylated cysteine, KRAS4b HVR, and PDEδ residues. The color-coding scheme is same as in C. Rheb-FMe, farnesylated–methylated Rheb.
Identification of a Conserved Sequence Motif in KRAS4b and Its Role in Facilitating Binding of Prenylated KRAS4b to PDEδ.
The two different modes of binding observed in our crystal structures are possible because of the presence of a unique 5-aa-long sequence motif KSKTK present in KRAS4b (Fig. 6D). This motif maintains many KRAS4b–PDEδ interactions in the crystal form II, even when the HVR residues shift upstream by two amino acids at the protein–protein interaction interface. Sequence analysis of KRAS HVR residues in other organisms showed the presence of similar motifs that were compatible with binding of prenylated KRAS with PDEδ (Fig. 6D). This “prenylated KRAS recognition” motif contains large basic amino acids (Lys or Arg) at first, third, and fifth positions and smaller polar amino acids (Ser or Thr) at second and fourth positions. Interactions formed at the KRAS–PDEδ interface by the first three amino acids (first, second, and third) of this motif when KRAS is farnesylated would overlap with the last three amino acids (third, fourth, and fifth) present in this motif when KRAS is geranylgeranylated. Thus, conservation of this sequence motif in the KRAS4b sequences of many organisms indicates that PDEδ could bind to KRAS4b in both farnesylated and geranylgeranylated forms.
Structure-Based Sequence Analysis of PDEδ Binding Proteins.
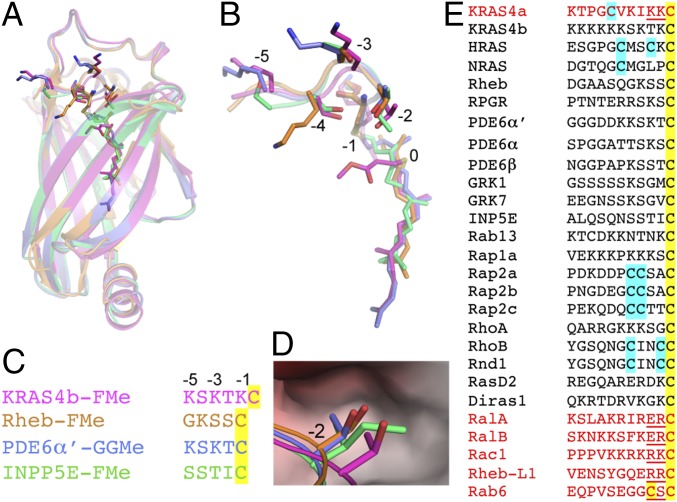
Previous studies have shown that a large repertoire of prenyl proteins, such as subunits of PDE6, GRK1, GRK7, Rab13, Rap1a, Rap2b, RhoA, RhoB, Rnd1, Rap2c, RasD2, and Reb-L1, bind to PDEδ (20, 22, 27, 31–33). Among RAS proteins, in addition to KRAS4b, NRAS and HRAS have also been shown to bind to PDEδ (20, 27). However, there have been some discrepancies in the literature on HRAS binding to PDEδ. Nancy et al. (27) showed that HRAS binds to PDEδ, but this observation could not be corroborated by later studies from two different groups (20, 31). Unlike many other prenylated members of RAS superfamily, KRAS4a, RalA, RalB, and Rac1 do not bind to PDEδ (27, 31, 34). To understand how PDEδ binds to a large number of prenylated proteins and why some prenylated proteins do not bind to PDEδ, we carried out structure-based sequence analysis using available structures of PDEδ in complex with prenylated proteins or peptides. Structural superposition of PDEδ in complex with the prenylated proteins KRAS4b and Rheb (26) and prenylated peptides from PDE6α′ (30) and INPP5E (35) showed that prenylated Cys occupies different positions in these complexes and PDEδ interacts with three or four amino acids located upstream of prenylated Cys (Fig. 7 A and B). Structure-based sequence alignment of residues interacting with PDEδ is shown in Fig. 7C, which shows that none of the residues located upstream of prenylated Cys in these structures are conserved. Structural superposition shows that, except Thr183, other residues located upstream of prenylated Cys in KRAS4b can be small or large and polar or charged amino acids. In the structural superposition, residue Thr183 in KRAS4b-FMe occupies a position corresponding to Ser, Thr, and Ile in Rheb, PDE6∝′, and INPP5E, respectively (−2 position in KRAS4b-FMe structure and −1 position in other structures). In the crystal form II, this position is occupied by carboxymethyl group of prenylated Cys. Examination of the protein–protein interface at −2 position corresponding to Thr183 in KRAS4b suggests that this position cannot accommodate large amino acids, like Lys, Arg, and Glu (Fig. 7D). Based on this rationale we propose that, for any prenylated protein to be able to bind to PDEδ, it should have small side chain containing residues at −1 and −2 positions.
Fig. 7.
Structural and sequence analysis of prenylated proteins that have been tested previously for binding to PDEδ. (A) Structural superposition of PDEδ in complex with KRAS4b-FMe (crystal form I), farnesylated–methylated Rheb (Rheb-FMe; PDB ID code 3T5G), geranylgeranylated–methylated PDE6α′ (PDE6α′-GGMe; PDB ID code 5ETF), and farnesylated–methylated INPP5e (INPP5E-FMe; PDB ID code 5F2U). (B) Enlarged view of prenylated protein/peptide shown in A. The residues located upstream of prenylated C185 are numbered from −1 (K184) to −5 (K180). (C) Structure-based sequence alignment of prenylated protein/peptide shown in A and B. (D) Enlarged view of the binding pocket formed by PDEδ residues, where Thr183 in KRAS4b and corresponding Ser, Thr, and Ile residues in Rheb, PDE6α′, and INPP5E localize at protein–protein interface. (E) Sequence alignment of various proteins that have been tested previously for binding to PDEδ. Prenylated and palmitoylated Cys are highlighted in yellow and cyan, respectively. Prenylated proteins that contain either large polar amino acids at −1 and −2 positions or a second prenylated Cys site (shown in red) would cause steric clash with PDEδ residues and are unlikely to form a protein–protein complex.
Sequence alignment of various proteins that have been previously tested for binding to PDEδ shows no conserved amino acids upstream of the prenylated Cys (Fig. 7E). Examination of residues present at −1 and −2 positions in this alignment shows the presence of large and charged amino acids, such as Lys, Arg, and Glu in case of KRAS4a, RalA, RalB, Rac1, and Rheb-L1, suggesting that the presence of these residues would prevent their ability to bind to PDEδ. Although Rheb-L1 has been previously shown to bind to PDEδ (20), our analysis indicates that the two arginines located upstream of prenylated Cys in Rheb-L1 will sterically clash with PDEδ at the protein–protein interface.
Discussion
The prenyl binding protein PDEδ has been shown to modulate signaling through RAS proteins by sustaining their dynamic distribution in cellular membranes (20, 23–25). Impairing KRAS4b localization at the plasma membrane by inhibiting PDEδ provides additional opportunities to target KRAS-driven cancer. Here, we present structures of the fully processed KRAS4b–PDEδ complex, which not only provide atomic details of the protein–protein interaction interface but also, reveal how PDEδ can bind to different forms of prenylated KRAS4b and why only some prenylated proteins bind to PDEδ. Our binding studies show that the carboxymethyl group present on farnesylated C185 plays a key role in the KRAS4b–PDEδ interaction (Fig. 1 B–E and Fig. S1). The weak binding observed in the absence of the methyl group is likely caused by the charge repulsion effect of the carboxylic acid group and the decreased hydrophobicity of farnesylated C185. Our results are in accordance with the previous findings showing that carboxymethylation of prenylated Rho peptide and catalytic subunits of PDE6 significantly increases their affinity for RhoGDI and PDEδ, respectively (33, 36). Carboxymethylation is the only step in the posttranslational modification of the CaaX motif that is potentially reversible (37). The importance of the methyl group in KRAS4b binding to PDEδ highlights the potential regulatory significance of this modification in trafficking KRAS4b to cellular membranes. Thus, our results suggest that KRAS4b molecules that are not fully processed (not farnesylated or methylated) will not bind to PDEδ and will not be delivered to the plasma membrane.
Although a large number of RAS structures have been solved during the preceding 25+ years of structural studies on RAS proteins, these studies did not provide any structural information on residues present in the HVR. The structure of the KRAS4b–PDEδ complex (crystal form II) described here provides atomic details of the entire HVR (165–185) (Fig. 2B and Fig. S2B) and likely represents a conformation of HVR residues in the cytoplasm when KRAS4b is bound to PDEδ. Additional studies are required to determine if some of the HVR residues could also form a similar α-helical conformation when KRAS4b is anchored to the membrane. Previous studies suggest that, in membrane-bound KRAS4b, the farnesyl and methyl groups are inserted inside the membrane bilayer, and the polybasic region present in the HVR interacts with acidic phospholipids, such as phosphatidylserine or phosphatidylinositol, that are preferentially located in the inner leaflet of the plasma membrane (38). Our mutational analysis of KRAS4b HVR residues showed that, other than the prenylated Cys (C185), the composition of the residues located upstream of C185 also plays a role in KRAS4b–PDEδ interactions. Although mutation to alanine of four residues (S181, K182, T183, and K184) preceding the prenylated C185 was tolerated, mutations of K184 and S181 to an acidic and larger residue like glutamate resulted in a significant loss of binding (Fig. 3C and Fig. S3C). Residue S181 is a part of the protein–protein interaction interface only in the crystal form I. The larger protein–protein interaction interface and role of S181 of KRAS4b in KRAS4b-FMe–PDEδ interaction indicate that the structure observed in crystal form I represents the biological interaction of KRAS4b-FMe with PDEδ.
Chandra et al. (20) showed that eight lysine residues present in the polybasic region of the KRAS4b are not necessary for binding to PDEδ and that the interaction becomes stronger when these lysine residues are mutated to glutamate. They also showed that diminishing the charge on the polybasic region by stimulating phosphorylation of S181 with bryostatin-1 enhances the ability of PDEδ to solubilize KRAS4b. Recent studies have also shown that KRAS suppresses Wnt/Ca2+ signaling pathway by direct binding with calmodulin and that the KRAS4b–calmodulin interaction is attenuated by phosphorylation of S181 by PKC (39, 40). Our structural data show that six lysine residues present between 175 and 180 do not interact with PDEδ and thus, are not likely to play any role in KRAS4b–PDEδ interaction. In contrast to the above-mentioned study, we saw that mutation of K184 (in the KRAS4b double-mutant T183A-K184E) to glutamate showed significant loss of binding affinity between KRAS4b and PDEδ. Also, the S181E mutant showed significant loss in the binding affinity compared with S181A. Analysis of the KRAS4b–PDEδ interaction interface suggests that phosphorylation of S181 is likely to decrease the binding affinity between KRAS4b and PDEδ because of steric hindrance and charge repulsion, because the side chain atoms of Glu88 of PDEδ form hydrogen bonds with the side chain atoms of KRAS4b S181.
Structure and sequence alignment of prenylated proteins that have been shown to bind to PDEδ shows that the residues involved at the protein–protein interface are not conserved and that prenylated cysteine can occupy multiple positions inside the central hydrophobic pocket of PDEδ. These observations are in accordance with the known broad specificity of PDEδ for prenylated proteins. The hydrophobic pocket in PDEδ is too small to accommodate another lipid chain. Thus, the prenylated proteins that undergo palmitoylation upstream of the prenylated Cys can bind to PDEδ only in the depalmitoylated state. For example, although Rab6 contains smaller amino acids upstream of the prenylated Cys, it cannot bind to PDEδ, because it becomes geranylgeranylated at the −3 position as well. Unlike palmitoylation, prenylation is not a reversible modification. Based on our structural and sequence analysis, we propose that prenylated proteins that contain a residue with a small side chain at −1 and −2 positions upstream of the prenylated Cys are likely to bind to PDEδ. This hypothesis explains why other prenylated members of the RAS superfamily, such as KRAS4a, RalA, RalB, and Rac1, do not bind to PDEδ. With this reasoning, one could predict if a given prenylated protein is likely to bind to PDEδ or not.
Unlike KRAS4b, the HVRs of KRAS4a, HRAS, and NRAS are mono- or dipalmitoylated at Cys residues located upstream of the C-terminal Cys. Palmitoylation of RAS proteins is readily reversible in vivo and plays a key role in their subcellular localization. Palmitoylation of HRAS/NRAS at the Golgi stabilizes their association with membranes. Depalmitoylation releases HRAS/NRAS into the cytoplasm, allowing their return to the Golgi for another round of palmitoylation. Sequence and structure analysis of KRAS4b and HRAS/NRAS indicates that PDEδ should be able to bind to depalmitoylated HRAS/NRAS, because it contains small side chain-containing residues upstream of prenylated Cys. As shown previously (20), it is possible that, in a steady state, the fraction of depalmitoylated HRAS is much less than that of depalmitoylated NRAS, because HRAS becomes palmitoylated at two positions in the HVR as opposed to just one position in NRAS. In accordance with the previous findings that, unlike KRAS4b, KRAS4a does not bind to PDEδ (31), our sequence and structural analysis suggests that the steric hindrance caused by two lysine residues located upstream of prenylated Cys is likely to interfere with interaction between depalmitoylated KRAS4a and PDEδ. NRAS has been shown to bind to PDEδ only in the farnesylated form and not in the geranylgeranylated form (20). Although NRAS contains small side chain-containing amino acids (Met-Gly-Leu-Pro) upstream of prenylated Cys, it is likely that the presence of a proline residue at −1 position results in limited flexibility, because proline and its preceding residue will have restricted φ–ψ torsion angles in the protein backbone. Limited flexibility of residues present at −1 and −2 positions could possibly affect the binding of geranylgeranylated NRAS to PDEδ.
Structural superposition of PDEδ in complex with prenylated proteins/peptides shows that the prenylated Cys of KRAS4b binds deeper inside the hydrophobic pocket of PDEδ, and thus, four additional KRAS4b residues (181–184), rather than three, are located inside the binding pocket of PDEδ. Comparison of the KRAS4b–PDEδ structure in crystal form I with the previously solved Rheb–PDEδ structure (26) shows that W90 and F133 residues in PDEδ undergo conformational changes to allow more extensive interaction between KRAS4b and PDEδ compared with that between Rheb and PDEδ (Fig. S5 A and B). This observation indicates that PDEδ interaction with KRAS4b could be more specific than other prenylated proteins and thus, despite PDEδ having broad specificity for prenylated proteins, it could serve as a potential target for KRAS-driven cancers. GTP-bound Arl2/Arl3 has been shown to disrupt RAS–PDEδ interaction at the membrane (26). Superposition of the KRAS4b–PDEδ and GTP-bound Arl2–PDEδ structures shows the possible formation of a ternary complex. However, based on an Rheb–PDEδ structure, it was suggested that binding of GTP-bound Arl2 to PDEδ forces the latter into a closed conformation by altering the conformation of two β-strands in PDEδ with the result that the RAS protein can no longer bind to PDEδ–Arl2 complex (26).
Recently, it was shown that PDEδ binds to geranylgeranylated–methylated PDE6α′ pentapeptide with an affinity (Kd = 2.3 ± 0.9 nM) ∼1,000 times higher than that of the KRAS4b-FMe (30). Sequence and structural similarity between our modeled structure of KRAS4b-GGMe–PDEδ and geranylgeranylated–methylated PDE6α′ pentapeptide in complex with PDEδ supports the potential binding of KRAS4b-GGMe by PDEδ (Fig. 6C). It is likely that the interaction formed by the additional five carbon atoms present in the geranylgeranyl chain within the hydrophobic pocket will result in a higher affinity between these two proteins.
Analysis of the amino acid sequence upstream of C185 showed the presence of a 5-aa sequence motif (K-S-K-T-K) that allows the formation of similar protein–protein interactions in both crystal forms of KRAS4b–PDEδ complex. The crucial role played by this motif in allowing prenylated KRAS4b binding to PDEδ is supported by the fact that this motif is highly conserved in KRAS4b across various organisms (Fig. 6D). Identification of this sequence motif strengthens our hypothesis that both the KRAS4b-FMe and KRAS4b–GGMe could bind to PDEδ by shifting the HVR residues by two amino acids toward the N terminus at KRAS4b–PDEδ interface, allowing PDEδ to target both forms of prenylated KRAS4b to cellular membranes. Based on our sequence and structural analyses, we suggest that crystal form I shows how KRAS4b-FMe binds to PDEδ, whereas crystal form II indicates how KRAS4b-GGMe could bind to PDEδ. It is important to note here that, until now, the only evidence that KRAS4b can be geranylgeranylated is in the presence of an FTI. No physiological condition has yet been found under which the KRAS4b modification is shifted to geranylgeranylation.
Despite the significant progress in characterizing the role of PDEδ in targeting RAS to the membrane and identifying nanomolar inhibitors that can break KRAS4b–PDEδ interactions, we need to overcome additional challenges before we can successfully target PDEδ for KRAS4b-driven cancers. Previous studies have shown that KRAS KO in mice is lethal (41, 42) but that PDEδ KO is not (43). This difference may be because PDEδ helps in the localization of KRAS4b, not of KRAS4a, and therefore, only affects one isoform of KRAS. Future studies need to focus on determining exactly how dependent RAS proteins are on PDEδ for proper localization to the membrane (20) and on PDEδ inhibitors that could target specific interactions between KRAS4b–HVR and PDEδ. The biggest challenge would be to design a specific inhibitor that can avoid any toxic or off-target effects of inhibiting PDEδ. One might also search for molecules that lock KRAS4b and PDEδ together so that they become resistant to dissociation in the presence of GTP-bound Arl2/Arl3 and thus, decrease population of active KRAS4b at the plasma membrane. A therapeutic strategy to alleviate, instead of eliminate, specific RAS proteins inside the cell might prove effective in targeting cancer, making PDEδ a target highly worthy of attention.
Materials and Methods
Cloning, Expression, and Purification.
KRAS4b-FMe (human) was cloned, expressed, and purified using the procedures described previously (44). Human PDEδ was cloned in pDest-566 vector with N-terminal His6-maltose binding protein tag and expressed in BL21 STAR (rne131) Escherichia coli strain. Highly purified untagged proteins were purified using a multistep purification process. SI Materials and Methods has a detailed methodology on cloning, expression, and purification.
Crystallization and Data Collection.
Before crystallization setup, purified KRAS4b-FMe and PDEδ proteins were mixed in a molar ratio of 1:1 and incubated on ice for 1 h. The complex crystals were grown with the sitting drop vapor diffusion method by mixing the protein–protein complex with an equal volume of reservoir solution containing 0.1 M citrate (pH 5.0) and 20% (wt/vol) PEG 6000 (crystal form I) and 0.1 M Hepes:NaOH (pH 7.5) and 2 M ammonium sulfate (crystal form II). Small crystals appeared in 2–3 d and grew to full size in 10 d. For data collection, crystals were flash frozen (100 K) in the above reservoir solutions supplemented with 30% (vol/vol) ethylene glycol as a cryoprotectant. Crystallographic datasets were integrated and scaled using the XDS (45). The high-resolution cutoffs of 2.0 and 1.9 Å were selected for crystal forms I and II, respectively, because reflections present in the high-resolution shell have average I/sigma(I) above three, multiplicity close to seven, and completeness close to 100%, despite having relatively high Rmerge values. Crystal form I belongs to the space group P21 and contains one copy of KRAS4b–PDEδ complex in the asymmetric unit. Crystal form II belonged to the space group P212121 and contains two copies of KRAS4b–PDEδ complex in the asymmetric unit. The crystal parameters and the data collection statistics are summarized in Table S1.
Structure Determination and Analysis.
Crystal structures of KRAS4b and PDEδ complex were solved by molecular replacement as implemented in the program PHASER (46) using the apo-structure of truncated KRAS4b [1–169; Protein Data Bank (PDB) ID code 4OBE] and protein atoms corresponding to PDEδ in the farnesylated–methylated Rheb peptide–PDEδ complex structure (PDB ID code 3T5G). Initial map showed interpretable electron density, in which both the proteins could be built manually. Additional model improvement was carried out with alternate rounds of model building via COOT (47) and refinement using Phenix.refine (48, 49) and Refmac (50), with rigid body, simulated annealing, positional, and translation–libration–screw (TLS) refinement at different stages. After a few rounds of refinement, inspection of the difference (Fo-Fc) map showed positive density for the farnesylated and methylated C185 and GDP. Placement of ligands were followed by identification of potential sites of solvent molecules by automatic water-picking algorithm in COOT (47). The positions of these automatically picked waters were checked manually during model building. The majority of the model has a clear and well-interpretable electron density map with the exception of switch 1 residues and three to five residues in HVR in crystal form I, which were omitted in the final model. Refinement statistics of both crystal forms are given in Table S1. Structural comparisons were performed using the SSM algorithm (51) implemented in COOT. Figures were prepared using the program PyMOL (52). All crystallographic and structural analysis software used in this study was compiled and maintained by the SBGrid consortium (53).
Binding Affinity Measurements by ITC.
ITC experiments were performed in a MicroCal PEAQ-ITC (Malvern). Both KRAS4b and PDEδ proteins were extensively dialyzed in 20 mM Hepes (pH 7.3), 300 mM NaCl, 5 mM MgCl2, and 1 mM TCEP [tris(2-carboxyethyl)phosphine]. Before titration, both proteins were centrifuged at 14,000 × g for 5 min to remove any debris and air bubbles. Protein concentration was measured using absorbance at 280 nm. Dialyzed KRAS4b and PDEδ proteins were loaded into the calorimeter cell and titration syringe, respectively. The calorimetric titrations were carried out at 25 °C using 15–18 injections of 2.4–2.6 μL each injected at 150- to 180-s intervals. A control experiment was performed by titrating PDEδ into a sample cell that contained buffer alone, with no significant heat release observed. Data analysis was performed based on a binding model containing “one set of sites” using a nonlinear least squares algorithm incorporated in the MicroCal PEAQ-ITC analysis software (Malvern).
SV-AUC.
To study the interaction of PDEδ with the different forms of KRAS4b, ∼1:1 stoichiometric mixtures of PDEδ:KRAS4b-FMe and PDEδ:KRAS4b-Far were studied at different concentrations ranging from ∼2–80 μM using SV-AUC technique. SI Materials and Methods has detailed information on sedimentation velocity experiments.
PDB ID Codes.
The atomic coordinates and structure factors for KRAS4b-FMe–PDEδ complex in crystal forms I and II have been submitted to the PDB and assigned ID codes 5TB5 and 5TAR, respectively.
SI Materials and Methods
Cloning, Expression, and Purification.
For cloning and expression of KRAS4b-FMe, the recently reported strategies were followed (44). To produce the desired KRAS4b mutants, a Gateway Entry clone of human KRAS4b with an upstream tobacco etch virus (TEV) protease cleavage site was mutated using QuikChange (Agilent). After sequence verification, these clones were subcloned into pDest-636, and baculovirus expression constructs were generated as for WT KRAS4b. KRAS4b was expressed in the modified baculovirus/insect expression system and purified as described previously (44), with the following modifications: the dialysis buffer before the SP Sepharose step contained 200 mM NaCl (effectively raising the NaCl concentration in the column load to 100 mM NaCl to reduce protein precipitation), and the SP Sepharose column was equilibrated with a buffer containing 100 mM NaCl. Note that this step leads to the separation of KRAS4b-Far from KRAS4b-FMe (44).
Full-length human PDEδ was cloned from a cDNA template using PCR. Initial amplification was carried out using primers 5′-GGCGAAAACCTGTACTTCCAAGGCTCAGCCAAGGACGAGCGGGCCAGGG and 5′-GGGGACAACTTTGTACAAGAAAGTTGATTAAACATAGAAAAGTCTCACTCTGGATG with Phusion polymerase (New England Biolabs) under standard conditions using a 30-s/kB extension time for five cycles; 200 nM adapter primer 5′-GGGGACAACTTTGTACAAAAAAGTTGGCGAAAACCTGTACTTCCAAGGC was then added, and amplification was continued for an additional 15 cycles to introduce a TEV protease cleavage site and Gateway recombination sites to the ends of the PCR product. PCR products were cleaned using the QIAQuick PCR Purification Kit (Qiagen). The final PCR products contain the genes of interest with an amino-terminal TEV protease site at the 5′ end and appropriate Gateway recombination sites on both ends. The cleaned PCR products were recombined into pDonr255 via a Gateway BP Recombination Reaction (Life Technologies) using the manufacturer’s protocols. The subsequent Entry clone was sequence-verified through the entire cloned region. The sequence-verified Entry clone was subcloned by Gateway LR Recombination (Life Technologies) into pDest-566 to produce an Escherichia coli expression clone for an N-terminal His6-maltose binding protein (MBP) fusion with PDEδ. Subsequent mutants of PDEδ were generated with QuikChange site-directed mutagenesis (Agilent) using the manufacturer’s protocols. Mutants were generated in the original Entry clone, sequence-verified, and then, subcloned into the same pDest-566 background as WT PDEδ. The BL21 STAR (rne131) E. coli strain containing the DE3 lysogen and rare tRNAs (pRare plasmid CmR) was transformed with the expression plasmid (His6-MBP-TEV-PDEδ, AmpR). The culture was grown at 37 °C; as the OD600 reached 6.0–8.0 (∼5 h), the culture was chilled to 16 °C, and isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM. The culture was grown overnight (∼17 additional hours), and the cells were collected by centrifugation. Clarified lysates were prepared, and the His6-MBP-TEV-PDEδ was purified by IMAC (immobilized metal affinity chromatography), essentially using the same protocol as described previously (44) with slight modifications (cells were resuspended with 10 mL buffer per 1,000 OD of cell paste). The pool created from the IMAC fractions was dialyzed against binding buffer and treated with His6-TEV protease [∼1:25 (wt/wt) TEV:fusion protein] for 60 min at room temperature and then, overnight at 4 °C. A second IMAC run was used to purify the target protein away from the contaminants of the protease digest. Fractions containing PDEδ were pooled and applied to a 26/60 Superdex 75 Gel Filtration Column (GE Healthcare) that was previously equilibrated in final buffer [20 mM Hepes, pH 7.3, 150 mM NaCl, 5 mM MgCl2, 1 mM TCEP [tris(2-carboxyethyl)phosphine]. Peak fractions were analyzed on SDS/PAGE, and fractions containing PDEδ protein were pooled and concentrated using Amicon Ultra Concentrator Units (Millipore).
SV-AUC.
Solutions of PDEδ, KRAS4b-FMe, and KRAS4b-Far were studied by sedimentation velocity in a buffer consisting of 20 mM Hepes (pH 7.3), 300 mM NaCl, 5 mM MgCl2, and 1 mM TCEP. To study the interaction of PDEδ with the different forms of KRAS4b, ∼1:1 stoichiometric mixtures of PDEδ:KRAS4b-FMe and PDEδ:KRAS4b-Far prepared by dilution of a stock preparation were studied at different concentrations ranging from ∼2 to 80 μM. Sedimentation velocity experiments were conducted at 50,000 rpm and 20 °C on a Beckman Coulter ProteomeLab XL-I Analytical Ultracentrifuge following standard protocols (54). Samples were loaded into cells with 12-mm (400-μL) or 3-mm (100-μL) double-sector charcoal-filled Epon centerpieces depending on their concentration, placed into an eight-hole An-50 Ti Rotor, and thermally equilibrated at zero speed. Absorbance and interference velocity scans were subsequently acquired at speed—absorbance data were collected in a continuous mode as single measurements at 280 nm using a radial spacing of 0.003 cm. Sedimentation data were temporally corrected (55) with REDATE 0.2.1 (biophysics.swmed.edu/MBR/software.html) and analyzed in SEDFIT 15.01b (https://sedfitsedphat.nibib.nih.gov) (56) in terms of a continuous c(s) distribution of sedimenting species using an uncorrected s range = 0–5 with a linear resolution of 100 and a maximum entropy regularization confidence interval of 0.68. In all cases, excellent fits were observed with rmsds ranging from 0.0028 to 0.0055 absorbance units and from 0.0026 to 0.0049 fringes. The partial specific volumes of PDEδ, KRAS4b-FMe, and KRAS4b-Far were calculated based on their amino acid composition using SEDNTERP (57) (sednterp.unh.edu/); the solution density and viscosity were also calculated in SEDNTERP based on the buffer composition. The absorbance extinction coefficient of PDEδ was calculated using the method of Pace et al. (58); interference signal increments for PDEδ, KRAS4b-FMe, and KRAS4b-Far were determined in SEDFIT 15.01b based on their composition (59). Sedimentation coefficients were corrected to standard conditions in water at 20 °C (s20,w); c(s) distributions for mixtures of PDEδ and KRAS4b were consistent with a reversible 1:1 association. To determine the association constants for the interaction of PDEδ with KRAS4b-FMe or KRAS4b-Far, weighted average sedimentation coefficients sw obtained from an integration of the c(s) distribution were analyzed in terms of a sedimentation isotherm accounting for mass balance:
where cA and cB represent the concentrations of PDEδ and KRAS4b, respectively; sA, sB, and sAB are the sedimentation coefficients of the free and complexed species; εA and εB are the absorption extinction coefficients or interference signal increments for the free species; and Ka is the association constant. The absorbance and interference isotherms for each interaction, containing the total concentrations of PDEδ and KRAS4b,
and sw were analyzed globally in SEDPHAT 12.1e (https://sedfitsedphat.nibib.nih.gov) (60) using an A + B = AB heteroassociation model to obtain Ka. Sedimentation coefficients for the free species were fixed to experimentally determined values. Errors in the best fit equilibrium constants were determined using the method of F statistics (61).
Acknowledgments
We thank members of the Protein Expression Laboratory (Frederick National Laboratory for Cancer Research) for their help in cloning, protein expression, and purification. We also thank the staff of ID-24-C/E beamline at the Advanced Photon Source, Argonne National Laboratory, for their help with data collection. We thank Drs. Monika Mehta, Jim Hartley, and David Heimbrook for critical reading of this manuscript. This project was funded in whole or in part with federal funds from National Cancer Institute, NIH Contract HHSN261200800001E. This work was funded in part by the Intramural Research Program of the NIH, the National Institute of Diabetes and Digestive and Kidney Diseases. Part of this work is based on research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from National Institutes of Health Grant P41 GM103403. The Pilatus 6M detector on 24-ID-C beamline is funded by NIH-Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the US Government.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5TB5 and 5TAR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615316113/-/DCSupplemental.
References
- 1.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 3.Forbes SA, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature. 1991;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 5.Wittinghofer A, Pai EF. The structure of Ras protein: A model for a universal molecular switch. Trends Biochem Sci. 1991;16(10):382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- 6.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81(1):153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 7.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10(12):842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93(1):269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 9.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129(5):865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1(1):2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinlan MP, Settleman J. Isoform-specific ras functions in development and cancer. Future Oncol. 2009;5(1):105–116. doi: 10.2217/14796694.5.1.105. [DOI] [PubMed] [Google Scholar]
- 13.Nussinov R, Tsai CJ, Chakrabarti M, Jang H. A new view of Ras isoforms in cancers. Cancer Res. 2016;76(1):18–23. doi: 10.1158/0008-5472.CAN-15-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid TS, Terry KL, Casey PJ, Beese LS. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J Mol Biol. 2004;343(2):417–433. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- 15.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63(1):133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 16.Manolaridis I, et al. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature. 2013;504(7479):301–305. doi: 10.1038/nature12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, et al. Mechanism of isoprenylcysteine carboxyl methylation from the crystal structure of the integral membrane methyltransferase ICMT. Mol Cell. 2011;44(6):997–1004. doi: 10.1016/j.molcel.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11(11):775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whyte DB, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272(22):14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 20.Chandra A, et al. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol. 2011;14(2):148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 21.Baehr W. Membrane protein transport in photoreceptors: The function of PDEδ: The Proctor lecture. Invest Ophthalmol Vis Sci. 2014;55(12):8653–8666. doi: 10.1167/iovs.14-16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, et al. Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J Biol Chem. 2004;279(1):407–413. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 23.Weise K, et al. Dissociation of the K-Ras4B/PDEδ complex upon contact with lipid membranes: Membrane delivery instead of extraction. J Am Chem Soc. 2012;134(28):11503–11510. doi: 10.1021/ja305518h. [DOI] [PubMed] [Google Scholar]
- 24.Schmick M, et al. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell. 2014;157(2):459–471. doi: 10.1016/j.cell.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 25.Schmick M, Kraemer A, Bastiaens PI. Ras moves to stay in place. Trends Cell Biol. 2015;25(4):190–197. doi: 10.1016/j.tcb.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Ismail SA, et al. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol. 2011;7(12):942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 27.Nancy V, Callebaut I, El Marjou A, de Gunzburg J. The delta subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. J Biol Chem. 2002;277(17):15076–15084. doi: 10.1074/jbc.M109983200. [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann G, et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013;497(7451):638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 29.Chen YX, et al. Synthesis of the Rheb and K-Ras4B GTPases. Angew Chem Int Ed Engl. 2010;49(35):6090–6095. doi: 10.1002/anie.201001884. [DOI] [PubMed] [Google Scholar]
- 30.Fansa EK, O’Reilly NJ, Ismail S, Wittinghofer A. The N- and C-terminal ends of RPGR can bind to PDE6δ. EMBO Rep. 2015;16(12):1583–1585. doi: 10.15252/embr.201541404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai FD, et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci USA. 2015;112(3):779–784. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karan S, Zhang H, Li S, Frederick JM, Baehr W. A model for transport of membrane-associated phototransduction polypeptides in rod and cone photoreceptor inner segments. Vision Res. 2008;48(3):442–452. doi: 10.1016/j.visres.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook TA, Ghomashchi F, Gelb MH, Florio SK, Beavo JA. Binding of the delta subunit to rod phosphodiesterase catalytic subunits requires methylated, prenylated C-termini of the catalytic subunits. Biochemistry. 2000;39(44):13516–13523. doi: 10.1021/bi001070l. [DOI] [PubMed] [Google Scholar]
- 34.Hanzal-Bayer M, Linari M, Wittinghofer A. Properties of the interaction of Arf-like protein 2 with PDEdelta. J Mol Biol. 2005;350(5):1074–1082. doi: 10.1016/j.jmb.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 35.Fansa EK, Kösling SK, Zent E, Wittinghofer A, Ismail S. PDE6δ-mediated sorting of INPP5E into the cilium is determined by cargo-carrier affinity. Nat Commun. 2016;7:11366. doi: 10.1038/ncomms11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mondal MS, Wang Z, Seeds AM, Rando RR. The specific binding of small molecule isoprenoids to rhoGDP dissociation inhibitor (rhoGDI) Biochemistry. 2000;39(2):406–412. doi: 10.1021/bi991856n. [DOI] [PubMed] [Google Scholar]
- 37.Chelsky D, Ruskin B, Koshland DE., Jr Methyl-esterified proteins in a mammalian cell line. Biochemistry. 1985;24(23):6651–6658. doi: 10.1021/bi00344a053. [DOI] [PubMed] [Google Scholar]
- 38.Roy MO, Leventis R, Silvius JR. Mutational and biochemical analysis of plasma membrane targeting mediated by the farnesylated, polybasic carboxy terminus of K-ras4B. Biochemistry. 2000;39(28):8298–8307. doi: 10.1021/bi000512q. [DOI] [PubMed] [Google Scholar]
- 39.Bivona TG, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21(4):481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Wang MT, et al. K-Ras Promotes Tumorigenicity through Suppression of Non-canonical Wnt Signaling. Cell. 2015;163(5):1237–1251. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 41.Johnson L, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11(19):2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plowman SJ, et al. While K-ras is essential for mouse development, expression of the K-ras 4A splice variant is dispensable. Mol Cell Biol. 2003;23(24):9245–9250. doi: 10.1128/MCB.23.24.9245-9250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, et al. Deletion of PrBP/delta impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc Natl Acad Sci USA. 2007;104(21):8857–8862. doi: 10.1073/pnas.0701681104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillette WK, et al. Farnesylated and methylated KRAS4b: High yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci Rep. 2015;5:15916. doi: 10.1038/srep15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 52.Schrodinger, LLC 2015. The PyMOL Molecular Graphics System, Version 1.8 (Schrodinger, LLC, New York)
- 53.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao H, Brautigam CA, Ghirlando R, Schuck P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr Protoc Protein Sci. 2013;20:20.12. doi: 10.1002/0471140864.ps2012s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao H, et al. Recorded scan times can limit the accuracy of sedimentation coefficients in analytical ultracentrifugation. Anal Biochem. 2013;437(1):104–108. doi: 10.1016/j.ab.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cole JL, Lary JW, P Moody T, Laue TM. Analytical ultracentrifugation: Sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4(11):2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao H, Brown PH, Schuck P. On the distribution of protein refractive index increments. Biophys J. 2011;100(9):2309–2317. doi: 10.1016/j.bpj.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao H, Schuck P. Combining biophysical methods for the analysis of protein complex stoichiometry and affinity in SEDPHAT. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 1):3–14. doi: 10.1107/S1399004714010372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson ML. Why, when, and how biochemists should use least squares. Anal Biochem. 1992;206(2):215–225. doi: 10.1016/0003-2697(92)90356-c. [DOI] [PubMed] [Google Scholar]