How chilling leaches flavor from tomatoes
Vine tomatoes. Image courtesy of Wikimedia Commons/THOR.
Chilling fruits at temperatures below 12 °C hampers enzymes that help synthesize flavor-imparting volatile compounds, resulting in relatively fresh but insipid fruits. To uncover the genetic underpinnings of chilling-associated flavor loss, Bo Zhang et al. (pp. 12580–12585) stored heirloom and modern inbred varieties of red ripe tomatoes at 5 °C for 1, 3, or 7 days after which the fruits were transferred to 20 °C for 1 or 3 days. Measurement of volatile compounds revealed that 7 days of cold exposure reduced volatile levels by up to 65%; 3 days of recovery at the higher temperature failed to restore volatiles to normal levels. A panel of 76 consumers judged fruits stored at 20 °C after 7 days of chilling much less flavorful than fruits harvested a day before consumption. Though the fruits’ sugar and acid contents remained largely unaltered, chilling reduced the expression of genes implicated in volatile synthesis as well as the gene switch RIPENING INHIBITOR (RIN), the epigenetic action of which is tied to fruit ripening, among other switches. Analysis of rates of methylation of cytosine residues in the control regions of RIN-dependent genes suggested a link between chilling and DNA methylation of genes implicated in volatile synthesis and fruit maturation. According to the authors, the findings furnish a molecular view of the flavor-depleting effects of cold storage on fruits. — P.N.
Optimizing CRISPR/Cas9 for primary mouse cells
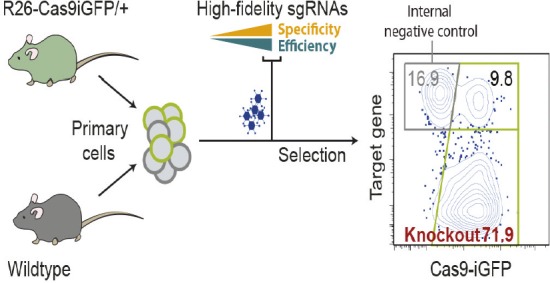
Efficient application of CRISPR/Cas9 to primary cells.
CRISPR/Cas9 technology has many applications in genome editing and genetic screening, but it remains to be optimized for use in primary cells. The efficiency of CRISPR/Cas9 in primary cells depends mainly on the levels of nuclear Cas9 and the effectiveness of the selected single guide RNA molecules (sgRNAs). By optimizing these two factors, Van Trung Chu et al. (pp. 12514–12519) developed a CRISPR/Cas9 system with high mutagenesis efficiency in primary mouse immune cells. The authors developed a sgRNA design tool called CrispRGold to select highly specific and efficient sgRNAs. The authors also generated a transgenic mouse line with high levels of Cas9 expression, and used the high-fidelity sgRNAs to mutagenize B cells, T cells, and macrophages isolated from the Cas9 transgenic mice. Using this approach, the authors achieved high-efficiency gene knockouts in the primary immune cells, with an average knockout efficiency of 80% in B cells. Further, the authors used the CRISPR/Cas9 system to perform a small-scale screen on primary B cells, and identified 22 genes needed for B-cell activation and proliferation and 8 genes required for plasma cell differentiation. The system could enable efficient mutagenesis and genetic screens in primary mouse immune cells and could be readily applied to other primary cells, according to the authors. — S.R.
RNA-mediated gene regulation across generations
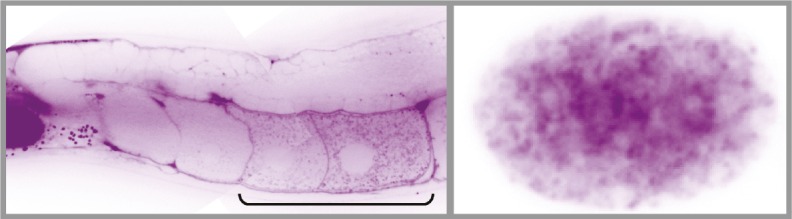
dsRNA (magenta) enters oocytes (bracket) from extracellular space in parent worms (Left) and reaches embryos (Right).
Studies using nematode worms, mice, and humans suggest that diet and stress can influence descendants’ metabolism and longevity. Genetic evidence indicates that double-stranded (ds) RNA molecules ingested by worms can silence sequence-matched genes in offspring through epigenetic mechanisms. By injecting fluorescent dsRNA into Caenorhabditis elegans worms and by feeding dsRNA-producing bacteria to the worms, Julia Marré et al. (pp. 12496–12501) found that extracellular dsRNA in parent worms was transported along with yolk to oocytes, where the molecules accumulated in concentrated clumps. Once transferred from oocytes to embryos, dsRNA spread between cells during embryonic development and silenced matching genes. Vertical transfer of dsRNA and cross-generational gene silencing occurred regardless of whether the target gene was present in the parent with extracellular dsRNA. Further, a receptor on the surface of oocytes called RME-2, which helps internalize yolk into oocytes, served as a conduit for dsRNA in nematode worms. Further experiments suggested that dsRNA molecules that originate in the circulation of parent worms can enter offspring cells without direct exposure to the cytoplasm of oocytes. According to the authors, the findings point to the tantalizing but unproven possibility that extracellular RNA molecules might serve as a vehicle for the transfer of experience-dependent gene regulatory information across generations. — P.N.
Improving DNA sequencing
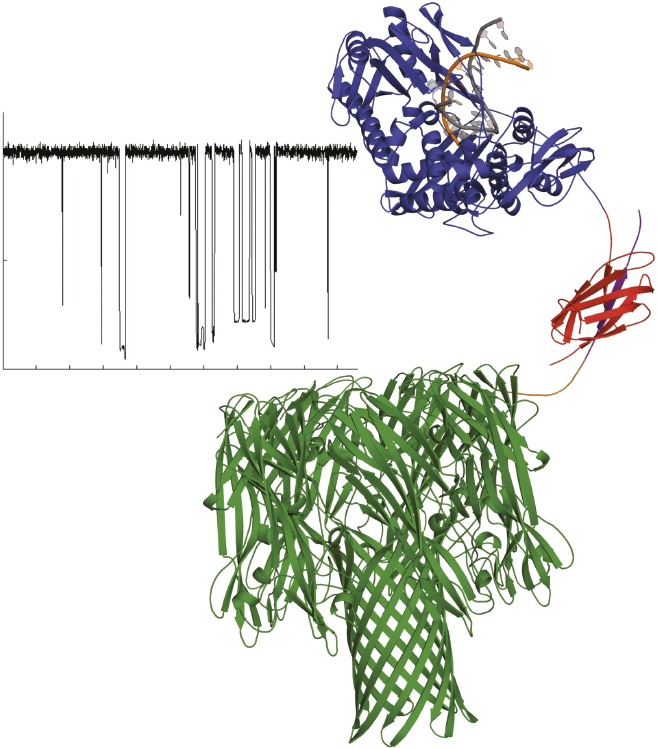
Model of DNA polymerase–nanopore complex and sequencing signal from electrode array.
The cost of genome sequencing limits its conventional use in clinics. Previous studies have found that nucleotides labeled with synthetic oligonucleotide tags can be detected in a nanopore and used in a synthesis-based sequencing platform, a common DNA sequencing method. The tags are removed during DNA synthesis and identified while they pass through a nanopore. This approach requires a protein construct that bridges sequencing to nanopore detection, as well as computational methods to differentiate the tag signals. P. Benjamin Stranges et al. (pp.E6749—E6756) designed a protein construct that couples a single ϕ29 DNA polymerase molecule to a seven-part protein nanopore complex called α-hemolysin. The authors embedded the polymerase–nanopore construct in a lipid bilayer on an electrode array. In the authors’ system, the polymerase feeds the nucleotide tags into the nanopore’s opening, and the electrode array measures resulting current changes in the nanopore. After measuring more than 200 tags for each of the four nucleotide bases, the authors found that a classification algorithm could distinguish between the different tags and background signals. This method identified the correct nucleotide by its tag 79–99% of the time, and identified a background event as a tag less than 1.2% of the time. According to the authors, this approach may provide the foundation for a low-cost, accurate, single-molecule, electronic DNA-sequencing platform. — L.C.



