Significance
Substrate rigidity has important roles for physiological processes, such as stem cell differentiation and cell growth. Although substrate rigidity clearly modulates gene expression, the mechanism of rigidity control of gene expression remains unknown. Our work reveals that four-and-a-half LIM domains (domain discovered in the proteins, Lin11, Isl-1, and Mec-3) 2 moves from adhesion sites to the nucleus on soft substrates through focal adhesion kinase activity and up-regulates p21 gene expression. Thus, we show a molecular pathway for inhibiting cell growth on soft substrates.
Keywords: mechanotransduction, four-and-a-half LIM domains 2, focal adhesion kinase, substrate rigidity, gene expression
Abstract
Substrate rigidity affects many physiological processes through mechanochemical signals from focal adhesion (FA) complexes that subsequently modulate gene expression. We find that shuttling of the LIM domain (domain discovered in the proteins, Lin11, Isl-1, and Mec-3) protein four-and-a-half LIM domains 2 (FHL2) between FAs and the nucleus depends on matrix mechanics. In particular, on soft surfaces or after the loss of force, FHL2 moves from FAs into the nucleus and concentrates at RNA polymerase (Pol) II sites, where it acts as a transcriptional cofactor, causing an increase in p21 gene expression that will inhibit growth on soft surfaces. At the molecular level, shuttling requires a specific tyrosine in FHL2, as well as phosphorylation by active FA kinase (FAK). Thus, we suggest that FHL2 phosphorylation by FAK is a critical, mechanically dependent step in signaling from soft matrices to the nucleus to inhibit cell proliferation by increasing p21 expression.
The mechanics of extracellular matrix affect cell growth and differentiation and represent an important aspect of cancer progression. For example, the invasion of premalignant mammary epithelium is promoted by increased tissue stiffness through collagen cross-linking and integrin clustering (1), whereas a decrease in substrate stiffness results in TGF-β1–induced apoptosis instead of the epithelial-mechenchymal transition (2). The events at the molecular level that lead from the initial mechanosensing to the eventual changes in cell growth or differentiation are not well understood, however.
Matrix rigidity and external forces are critical mechanical signals in cell differentiation and growth pathways, (3, 4). The mechanosensing of matrix rigidity occurs at cell substrate focal adhesion (FA) sites through actomyosin contraction of the matrix to a constant length (5–7). Previous studies have suggested that FA sites are composed of more than 150 components and have a 3D architecture (8, 9). Importantly, force is required for FA stabilization through recruitment of FA components (10, 11). Furthermore, many FA components bind to FAs in a force-dependent manner (12). At a finer level, sarcomere-like contractile units displace the matrix by a constant length of approximately 60 nm through stepwise contractions, and if the force exceeds a certain threshold, then a rigid signal is generated. In contrast, if the surface is soft, then the adhesion disassembles (13). This rigidity response transduces mechanical forces into chemical signals, such as tyrosine phosphorylation and the activation of small G proteins (14, 15). Similarly, forces applied to cells on soft surfaces through substrate stretch can activate cell growth (4). These signals cause short-term responses in the reinforcement of adhesions and the further spreading of the cells. Long-term responses to mechanotransduction signals involve changes in gene expression (16, 17).
An important finding linking extracellular matrix stiffness and gene expression is that stiffness of extracellular environment correlates with the amount of Lamin-A in the nucleus (18). Furthermore, actin dynamics mediates the translocation of MRTF-A, a mechanosensitive transcriptional factor, to the nucleus through Lamin A/C (19). The mechanism by which the signal is transmitted from an adhesion on a rigid or soft matrix to the nucleus to alter gene expression remains largely unknown, however. Both direct mechanical transduction through cytoskeleton–nuclear links and chemical signals have been postulated as the critical signals in mechanotransduction (20, 21).
LIM domain (domain discovered in the proteins, Lin11, Isl-1, and Mec-3) proteins have been implicated as potential mechanotransducers owing to their ability to move between FAs and the nucleus (22). Proteomic studies have shown that some LIM domain proteins localize at FAs in a myosin II contraction-dependent manner (12, 23). One example is zyxin, an LIM domain protein that localizes to adhesion sites in a force-dependent manner during actin filament assembly (24, 25). The movement of zyxin to the nucleus has been found to depend on stretch-dependent hormone activation of a cytoplasmic kinase in endothelial cells, however (26). Although many LIM domain proteins have roles as transcriptional cofactors, to date none has been found to move to the nucleus as a result of direct mechanotransduction (22).
Four-and-a-half LIM domains 2 (FHL2) is an LIM domain protein involved in cancer progression or suppression as a transcriptional coactivator and a scaffold protein in cell adhesions (27, 28). In spread cells, FHL2 localizes to adhesions and also binds to a number of FA proteins, including FA kinase (FAK) and various integrins (29, 30). Moreover, a previous study reported that Rho signaling mediates nuclear localization of FHL2 in NIH 3T3 cells under serum-starved conditions (31). FHL2 is overexpressed in some highly metastatic cell lines and has dramatic effects on cell migration (32–34). Thus, FHL2 appears to have a number of signaling roles mediating communication between the cytoskeleton and the nucleus, but the molecular steps remain unclear.
Here we report that FHL2 moves to the nucleus in a substrate rigidity-dependent manner. Specifically, soft substrates and myosin inhibition cause FHL2 to concentrate in the nucleus. At a molecular level, tyrosine phosphorylation by FAK is required for FHL2 concentration, and mutation of a specific tyrosine residue to phenylalanine in FHL2 blocks its movement to the nucleus. Once in the nucleus, FHL2 binds to active sites of transcription, as indicated by active RNA polymerase (Pol) II, and this correlates with increased p21 gene expression. FHL2 appears to be a chemical transducer of mechanical signals from soft matrix adhesions to the nucleus for the regulation of gene expression.
Results
FHL2 Localization Depends on Substrate Rigidity.
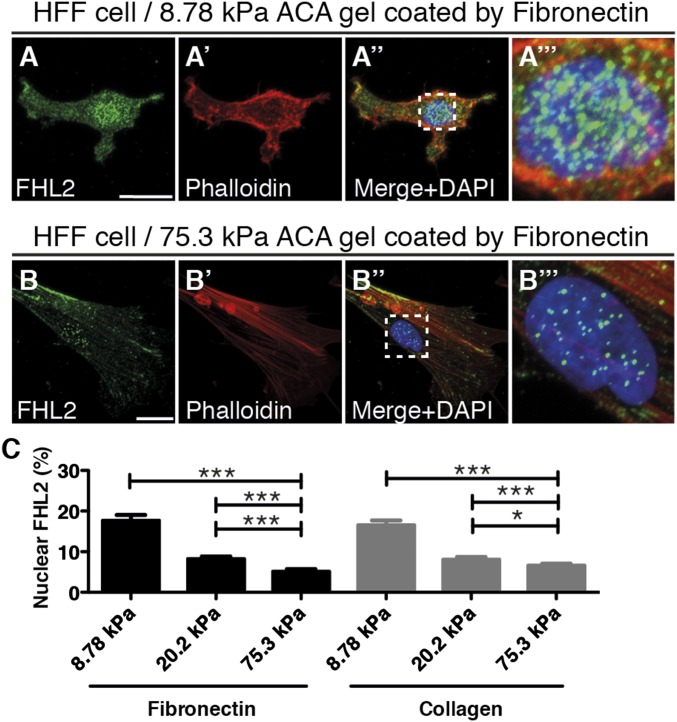
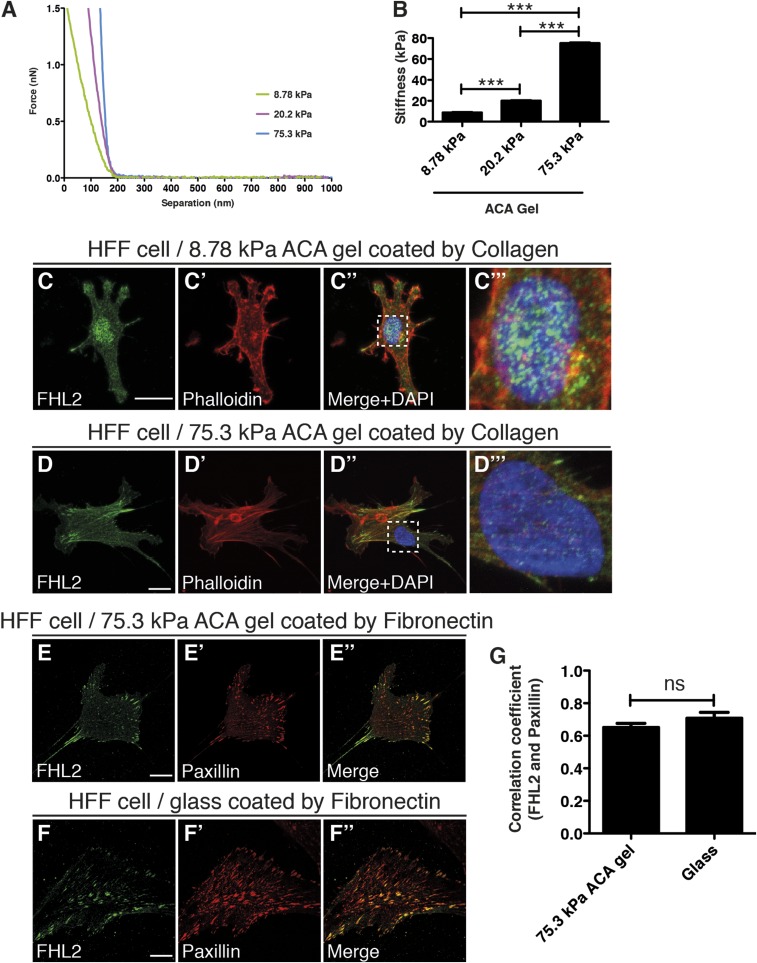
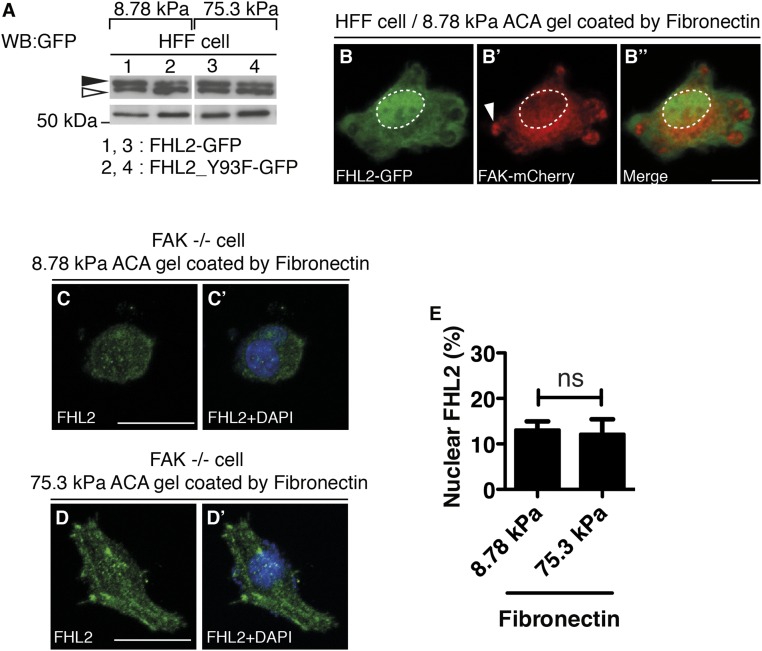
To determine whether substrate rigidity affects FHL2 transport to the nucleus, we examined FHL2 localization in cells on substrates with different rigidities (Fig. 1 and Fig. S1 C and D′′). We altered the rigidity of the substrate by varying the concentrations of acrylamide and N,N′–methylenebisacrylamide (BIS) (7). Treatment with N-acryloyl-6-aminocaproic acid (ACA) was used to covalently attach fibronectin or collagen to the polyacrylamide (35, 36). The stiffness of each gel was characterized by atomic force microscopy (AFM) (Fig. S1 A and B). Human foreskin fibroblasts (HFFs) on 75.3-kPa ACA gel (hard substrate) formed FAs with stress fibers and FHL2 localized to the FAs (Fig. 1 B–B′′ and Fig. S1 D–D′′ and E–E′′). Pearson correlation coefficient analysis quantified the extent of correlation between FHL2 intensity and the intensity of paxillin, another LIM domain protein at adhesion sites in HFF cells on glass or 75.3-kPa ACA gel (Fig. S1 E–G) (37). The correlation coefficient in HFF cells was 0.71 on glass, compared with 0.65 on 75.3-kPa ACA gel (Fig. S1G). In contrast, HFF cells on 8.78-kPa ACA gel (soft substrate) did not spread or form stress fibers (Fig. 1 A–A′′ and Fig. S1 C–C′′). FHL2 was concentrated in the nucleus of HFF cells on the 8.78-kPa ACA gel compared with HFF cells on the 75.3-kPa ACA gel (Fig. 1 A′′′, B′′′, and C and Fig. S1 C′′′ and D′′′). Thus, soft substrates stimulate FHL2 transport to the nucleus.
Fig. 1.
FHL2 localization depends on substrate rigidity. Confocal immunofluorescence images of FHL2, actin, and nuclear staining in HFF cells cultured on ACA gels of different rigidities for 17 h. FHL2 localization was labeled by the FHL2 antibody (green), actin filaments were stained with Alexa Fluor 594 phalloidin (red), and the nucleus was counterstained with DAPI (blue). (Scale bars: 20 μm.) (A–A′′) HFF cells on an 8.78-kPa ACA gel coated with fibronectin. (A′′′) Zoomed-in view of the dotted box in A′′, showing nuclear localization of FHL2 and actin. (B–B′′) HFF cell on a 75.3-kPa ACA gel coated with fibronectin. (B′′′) Zoomed-in view of the dotted box in B′′, showing nuclear localization of FHL2 and actin. (C) Graph showing the intensity ratio of FHL2 immunofluorescence staining between the nucleus and whole cell area (nucleus/whole cell area), fibronectin- or collagen-coated gels of varying rigidity. All images are shown as projected images from adhesion sections to nuclear sections. n > 30. Error bars represent SEM. *P < 0.05; ***P < 0.0001.
Fig. S1.
Characterization of ACA gel stiffness and FHL2 localization in HFF cells on ACA gel. (A) Representative force curve for the characterization of ACA gel stiffness. (B) Graph showing the stiffness of each gel. (C–C′′) FHL2 and phalloidin staining of HFF cells on the 8.78-kPa ACA gel coated with collagen. (C′′′) The box in C′′, magnified. (D–D′′) FHL2 and phalloidin staining of HFF cells on the 75.3-kPa ACA gel coated with collagen. (D′′′) The box in D′′, magnified. (E–E′′) FHL2 and paxillin staining of HFF cells on the 75.3-kPa ACA gel coated with fibronectin. (F–F′′) FHL2 and paxillin staining of HFF cells on glass coated with fibronectin. (G) Graph showing the correlation coefficient between the intensity of FHL2 staining and the intensity of paxillin staining on a 75.3-kPa ACA gel or glass coated with fibronectin.
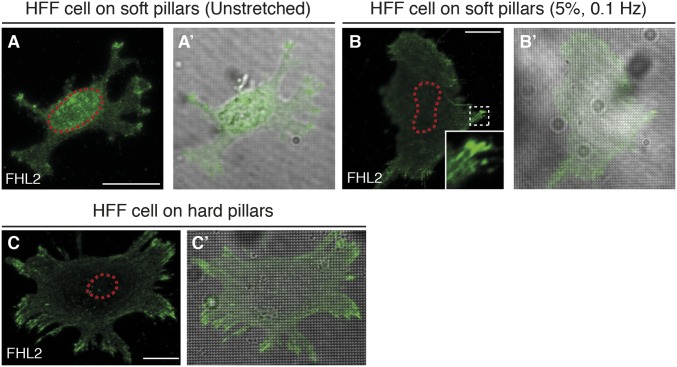
Previously our group developed a cyclic stretching system with soft pillars that mimics cycles of cell stretching and relaxation in soft tissues (4). We tested whether FHL2 localization to the nucleus on soft pillars would be affected by cyclic stretching. As expected, FHL2 localized to the nucleus on soft pillars that were not stretched, but after stretching, the cell spread area increased and FHL2 bound to FAs (Fig. S2 A–C′). Thus, cycles of cell stretching and relaxation of soft surfaces can cause FHL2 to bind to adhesions.
Fig. S2.
FHL2 localization in HFF cells with cyclic stretching and relaxation on soft pillars. Confocal immunofluorescence images of FHL2 (green) in HFF cells on the soft or hard pillars (diameter of pillar top, 500 nm; distance between each pillar, 1 μm; height, 750 nm for hard pillars and 2 μm for soft pillars). (Scale bars: 20 μm.) (A and A′) FHL2 localization in HFF cells on soft pillars. (B and B′) FHL2 localization in HFF cells on soft pillars after cyclic stretching (5%, 0.1 Hz, 2 h). (C and C′) FHL2 localization in HFF cells on hard pillars. All images are projected images from adhesion sections to nuclear sections.
Myosin II Activity and Actin Polymerization Are Involved in FHL2 Transport to the Nucleus.
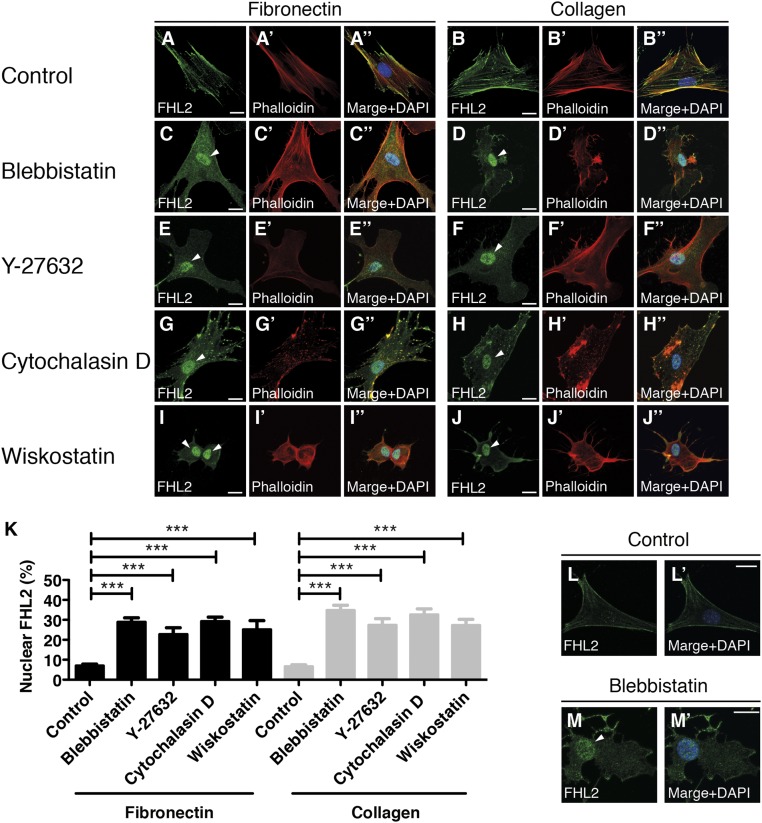
Because cells generate less force on soft substrates, we speculated that FHL2 movement from FAs to the nucleus depends on myosin II contraction. In HFF cells on fibronectin- or collagen-coated glass, FHL2 localized to FAs and stress fibers (Fig. S3 A–B′′); however, inhibition of myosin II activity with either blebbistatin (38) or the Rho-associated kinase (ROCK) inhibitor Y-27632 (39) caused a dramatic increase in FHL2 localization to the nucleus (Fig. S3 C–F′′ and K). Correspondingly, localization of FHL2 at FAs diminished after treatment with these inhibitors (Fig. S3 C–F′′). To check whether FHL2 localization is affected by actin dynamics, we inhibited actin polymerization with cytochalasin D or the N-WASP inhibitor wiskostatin. Under these conditions, FHL2 also accumulated in the nucleus (Fig. S3 G–K). Western blot analysis of FHL2 protein levels from the nuclear fraction of lysed cells verified that FHL2 localized to the nucleus (Fig. S4A). Both blebbistatin and cytochalasin D treatment caused an increase in the amount of FHL2 protein in the nuclear fraction (Fig. S4A). Taken together, the foregoing findings indicate that the reduction of contractile force by either inhibition of myosin II activity or actin depolymerization induces transport of FHL2 to the nucleus.
Fig. S3.
Inhibition of either myosin II activity or actin polymerization induces FHL2 localization to the nucleus. Confocal immunofluorescence images of FHL2 (green), nucleus (blue), and actin (red) in HFF cells treated with various inhibitors. (Scale bars: 20 μm.) Arrowheads indicate nuclear accumulation of FHL2. (A–B′′) HFF cells treated with DMSO or water (control) on fibronectin- or collagen-coated glass. (C–D′′) HFF cells treated with blebbistatin (50 μM, 1 h) on fibronectin- or collagen-coated glass. (E–F′′) HFF cells treated with Y27632 (10 μM, 0.5 h) on fibronectin- or collagen-coated glass. (G–H′′) HFF cells treated with cytochalasin D (2 μM, 0.5 h) on fibronectin- or collagen-coated glass. (I–J′′) HFF cells treated with wiskostatin (2 μM, 0.5 h) on fibronectin- or collagen-coated glass. (K) Graph showing the intensity ratio of FHL2 between nucleus and whole cell after each drug treatment. (L and L′) NIH 3T3 cells treated with DMSO (control) on fibronectin-coated glass. (M and M′) NIH 3T3 cells treated with blebbistatin on fibronectin-coated glass. All images are projected images from adhesion sections to nuclear sections. n > 25. Error bars represent SEM. ***P < 0.0001.
Fig. S4.
Inhibition of either myosin II activity or actin polymerization increases FHL2 protein levels in the nucleus. (A) Western blot analysis for FHL2 with samples isolated from cytoplasmic or nuclear fraction of HFF cells treated with DMSO (control), blebbistatin (50 μM, 1 h), or cytochalasin D (2 μM, 0.5 h). (B, Top) SDS/PAGE with polyacrylamide containing Mn2+ and Phos-tag. Black arrow, phosphorylated FHL2; white arrow, unphosphorylated FHL2-GFP. (B, Bottom) SDS/PAGE with polyacrylamide, Western blot analysis for FHL2-GFP. Lane 1, cytoplasmic fraction in control cell; lane 2, nuclear fraction in control cell; lane 3, cytoplasmic fraction in the cell with blebbistatin treatment, lane 4, nuclear fraction in the cell with blebbistatin treatment; lane 5, cytoplasmic fraction in the cell with blebbistatin washout; lane 6, nuclear fraction in the cell with blebbistatin washout. HFF cells were treated with DMSO (control) or blebbistatin (50 μM, 1 h) or 1 h after washout of blebbistatin (washout). The cytoplasmic marker was α-tubulin; the nuclear marker was Lamin B1.
FHL2 Transport to the Nucleus Depends on Force but Not on Adhesion Disassembly.
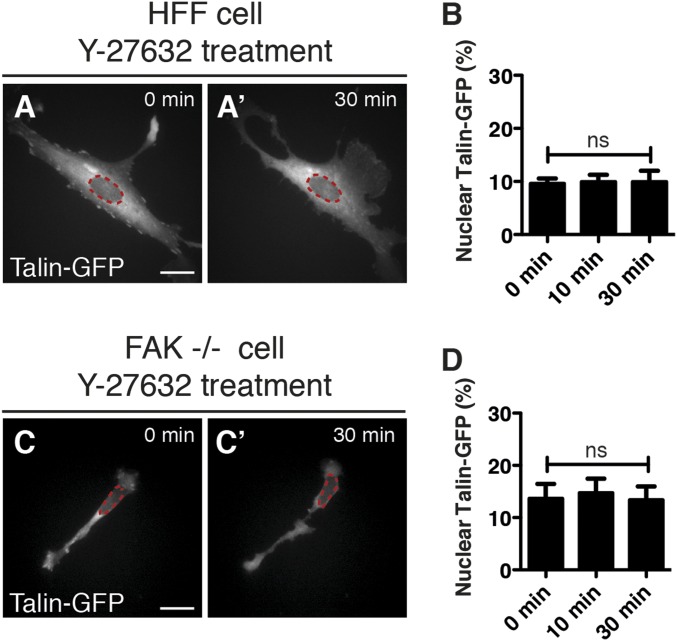
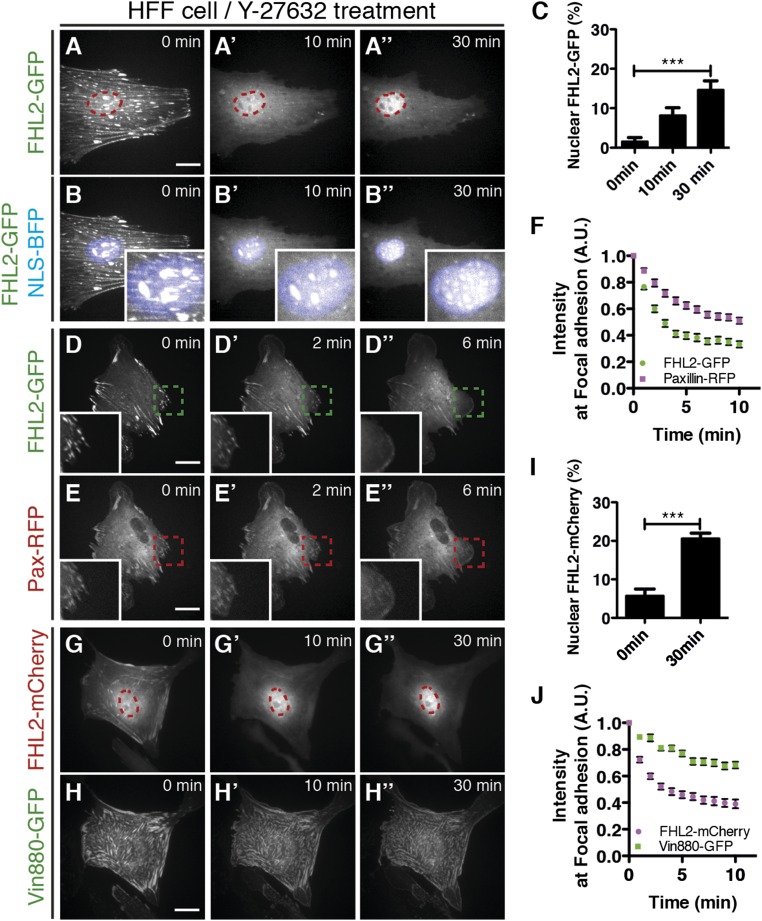
To examine FHL2 protein dynamics after inhibition of myosin II activity, we followed a GFP chimera with human FHL2. After expression in HFF cells, the transport of FHL2-GFP protein to the nucleus was quantified after Y-27632 treatment with custom software using a nuclear marker, BFP-NLS. As a negative control to determine background intensity in the nucleus with this optical technique, we measured the apparent nuclear accumulation of a Talin-GFP chimera (Fig. S5 A–D). In the spread cells, FHL2-GFP localized to FAs and stress fibers, but after Y-27632 addition, fluorescence from the adhesions decreased and fluorescence in the nuclei increased (Fig. 2 A–A′′ and B–B′′). At 30 min after Y-27632 treatment, FHL2-GFP localized to puncta in the nucleus, but not in the perinuclear region (magnified images in Fig. 2B′′). Western blot analysis showed increased FHL2 protein levels in the nuclear fraction after blebbistatin treatment (Fig. S4A). These data are consistent with the imaging data.
Fig. S5.
Talin-GFP dynamics after Y-27632 treatment in HFF and FAK−/− cells. The magenta circle indicates NLS-BFP (nuclear marker). (A–A′′) Talin-GFP dynamics in HFF cells after Y-27632 treatment. (B) Graph showing relative intensity of Talin-GFP at the nucleus in HFF cells after Y-27632 treatment. (C–C′′) Talin-GFP dynamics in FAK−/− cells after Y-27632 treatment. (D) Graph showing the relative intensity of Talin-GFP at the nucleus in FAK−/− cells after Y-27632 treatment (n > 5).
Fig. 2.
Dynamics of FHL2 transport to the nucleus. Live confocal imaging of several proteins fused with fluorescence proteins in HFF cells. (Scale bars: 20 μm.) The magenta circle indicates NLS-BFP (nuclear marker). (A–A′′) FHL2-GFP dynamics in HFF cells after Y-27632 treatment. (B–B′′) FHL2-GFP dynamics with NLS-BFP (nuclear marker) in HFF cells after Y-27632 treatment. (Insets) Magnified nucleus at each time point. (C) Graph showing the relative intensity of FHL2-GFP at the nucleus after Y-27632 treatment. (D–D′′) FHL2-GFP dynamics at FAs in HFF cells after Y-27632 treatment. The dotted green box indicates the magnified region shown in the Inset. (E–E′′) Paxillin-RFP dynamics at FAs in HFF cells after Y-27632 treatment. The dotted magenta box indicates the magnified region shown in the Inset. (F) Graph showing the relative intensity of FHL2-GFP and paxillin-RFP at each FA after Y-27632 treatment (>40 adhesions at the edge in five cells). (G–G′′) FHL2-mCherry dynamics at FAs in HFF cells with Vin880-GFP expression after Y-27632 treatment. (H–H′′) Vin880-GFP dynamics at FAs in HFF cells after Y-27632 treatment. (I) Graph showing the relative intensity of FHL2-mCherry with Vin880-GFP after Y-27632 treatment at the nucleus. (J) Graph showing the relative intensity of FHL2-mCherry with Vin880-GFP after Y-27632 treatment at each FA (>30 adhesions at the edge in five cells). All images are projected images from adhesion sections to nuclear sections. n > 10. Error bars represent SEM. ***P < 0.0001.
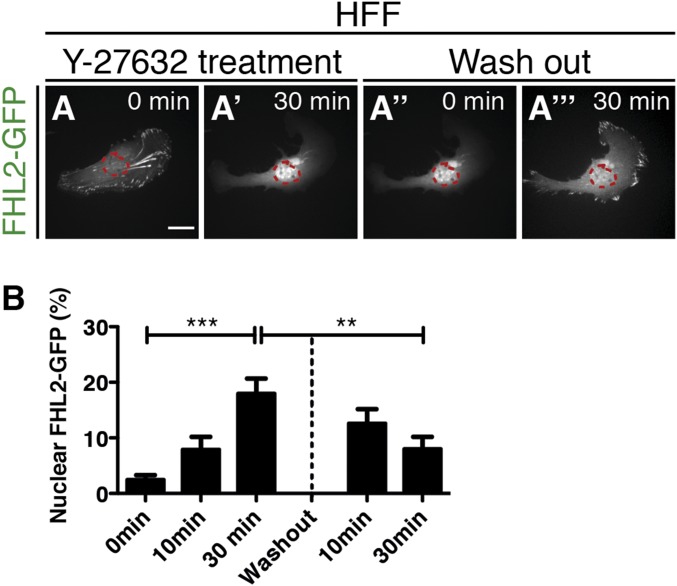
As perhaps expected, the loss of force caused a dramatic increase in the cytoplasmic concentration of FHL2 released from adhesions that preceded nuclear accumulation (Fig. 2C). In contrast, washing out of Y-27632 induced the shuttling of FHL2 from the nucleus to adhesions (Fig. S6 A and B). Thus, shuttling of FHL2 between adhesions and nucleus is rapidly reversible.
Fig. S6.
FHL2 dynamics after Y-27632 washout. Live confocal imaging of several proteins fused with fluorescence proteins in HFF cells. (Scale bars: 20 μm.) The magenta circle indicates NLS-BFP (nuclear marker). (A–A′′) FHL2-GFP dynamics in HFF cells after Y-27632 treatment. (A′′ and A′′′) FHL2-GFP dynamics after Y-27632 washout. (B) Graph showing the relative intensity of FHL2-GFP at the nucleus after Y-27632 treatment and washout.
Because inhibition of myosin II contraction caused general disassembly of FAs, we tested whether loss of myosin II contraction causes the release of other adhesion components simultaneously with FHL2. The dynamics of FHL2 release from FA was compared with the release of paxillin. After ROCK inhibition with Y27632 treatment, FHL2 release from FAs was quicker than paxillin release (Fig. 2 D–F). Thus, FHL2 release appears to be mediated by changes in myosin II-mediated contractility rather than by the general disassembly of FAs.
To further test whether FA disassembly is required for FHL2 transport to the nucleus, we overexpressed the Vinculin head and neck domain (vin880), which stabilizes adhesions even after the inhibition of myosin contractility (40). After FA stabilization by overexpression of vin880-GFP, inhibition of myosin II was still able to induce FHL2 transport to the nucleus (Fig. 2 G–I). Under these conditions, FHL2-mCherry intensity in adhesions decreased, whereas vin880-GFP intensity remained constant (Fig. 2J). Thus, transport of FHL2 to the nucleus is coupled to the loss of myosin II contractility, not to FA disassembly.
FA Kinase Is Involved in FHL2 Shuttling Between FA and Nucleus.
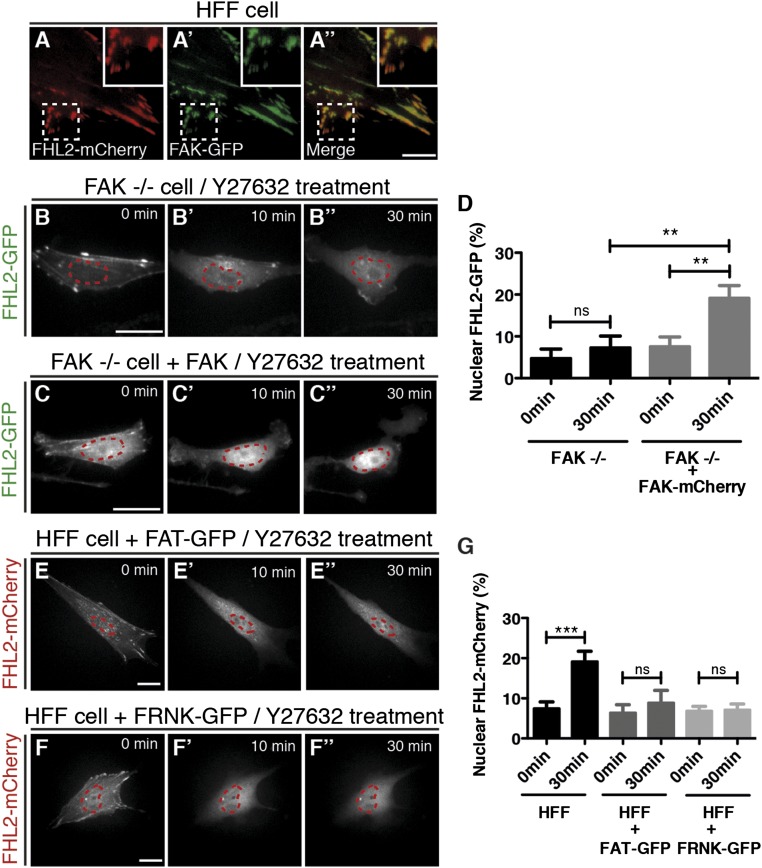
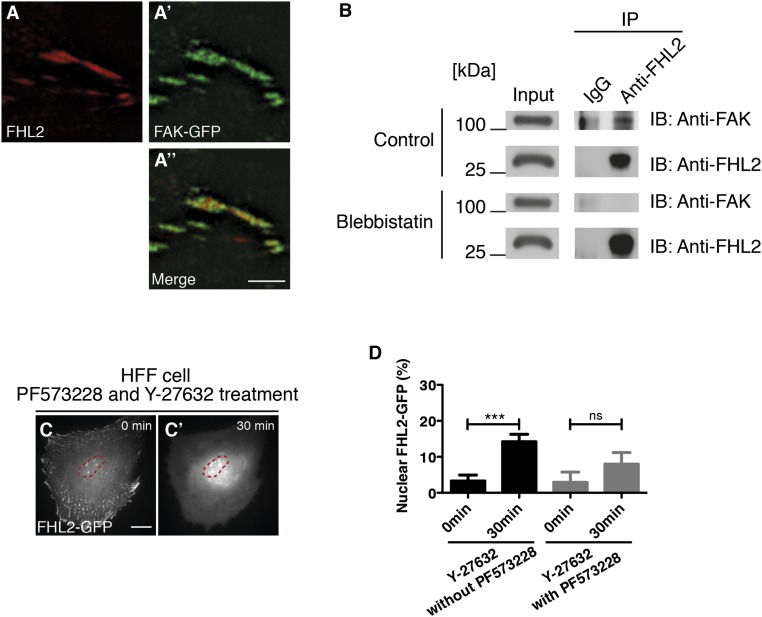
FA kinase (FAK) is a nonreceptor tyrosine kinase located at the FA (41) that is involved in collagen rigidity sensing (42). A previous study found that FHL2 interacts with FAK (29). Furthermore, because FAK autophosphorylation depends on myosin contraction (43, 44), we hypothesized that FAK is involved in contractility-dependent nuclear localization of FHL2. Our initial examination of colocalization of FAK and FHL2 in HFF cells revealed that FAK-mCherry and FHL2-GFP partially colocalized in HFF cells (Fig. 3 A–A′′). In addition, partial colocalization of FHL2 and FAK-GFP was observed using superresolution microscopy (Fig. S7 A–A′′). Moreover, as expected, the biochemical interaction between FHL2 and FAK in HFF cells was detected by co-immunoprecipitation (IP) (Fig. S7B).
Fig. 3.
FAK is involved in FHL2 shuttling from FAs to the nucleus. (A–A′′) TIRF image of FHL2-mCherry (red) and FAK-GFP (green) in HFF cells. The dotted white box indicates the magnified region shown in the Inset. (B–B′′) FHL2-GFP dynamics in an FAK KO cell (FAK−/−) after Y-27632 treatment. (C–C′′) FHL2-GFP dynamics in an FAK KO cell (FAK −/−) expressing FAK-mCherry after Y-27632 treatment. (D) Graph showing the intensity ratio of FHL2-GFP between the nucleus and whole cell area after Y-27632 treatment in FAK−/− cells, with and without expression of FAK-mCherry. (E–E′′) FHL2-mCherry dynamics in HFF cells with FAT-GFP expression after Y-27632 treatment. (F–F′′) FHL2-mCherry dynamics in HFF cells with FRNK-GFP expression after Y-27632 treatment. (G) Graph showing the intensity ratio of FHL2-mCherry between the nucleus and whole cell before and after Y-27632 treatment in HFF cells with either FAT-GFP or FRNK-GFP expression. The magenta circle indicates NLS-BFP (nuclear marker). (Scale bars: 20 μm.) All images are shown as projected images from adhesion section to nuclear section. n > 15. Error bars represent SEM. **P < 0.001; ***P < 0.0001.
Fig. S7.
FHL2 interacts with FAK in HFF cells. (A–A′′) Images obtained with a Nikon N-SIM superresolution microscope. (A) Superresolution image of FHL2 localization at the adhesion in HFF cells. (A′) Superresolution image of FAK-GFP localization at the adhesion in HFF cells. (A′′) Merged image of A and A′. (B) Western blot analysis for FAK with the cell lysate from HFF cells after IP with FHL2 antibody with or without blebbistatin. (C and C′) FHL2-GFP dynamics after PF573228 (10 μM, 7 h) and Y-27632 treatment. (D) Graph showing the relative intensity of FHL2-GFP at the nucleus in HFF cells after PF573228 and Y-27632 treatment. n > 20. Error bars represent SEM. ***P < 0.0001. The magenta circle indicates NLS-BFP (nuclear marker).
To further test whether FHL2 transport to the nucleus after the addition of Y-27632 is dependent on FAK, we measured the movement of FHL2 to the nucleus in FAK knockout (KO) cells (FAK−/− cells). FHL2 still localized to the adhesions in FAK−/− cells, but the addition of Y-27632 did not cause nuclear concentration (Fig. 3 B–B′′ and D). Instead, FHL2 left the adhesions and accumulated in cytoplasm after the addition of Y-27632 (Fig. 3 B–B′′ and D). In contrast, in FAK−/− cells expressing FAK-mCherry, FHL2 moved to the nucleus after Y-27632 treatment (Fig. 3 C–C′′ and D). Because FAK was required for FHL2 nuclear localization in a force-dependent manner, we speculated the soft substrates could not induce FHL2 nuclear localization in FAK−/− cells. To test this hypothesis, we checked FHL2 localization in FAK−/− cells on soft or stiff substrates. FHL2 localized to adhesions in FAK−/− cells on rigid substrates, but not on soft substrates (Fig. S8 C–D′); however, FHL2 did not accumulate in the nucleus on soft substrates (Fig. S8 C and E). Thus, FAK is necessary for the movement of FHL2 to the nucleus on soft substrates as well as in the absence of force.
Fig. S8.
FHL2 nuclear localization driven by substrate rigidity is dependent on FAK. (A) SDS/PAGE with polyacrylamide containing Mn2+ and Phos-tag (Top). SDS/PAGE without Mn2+ and Phos-tag (Bottom). Western blots for FHL2-GFP or FHL2_Y93F-GFP from each sample isolated from HFF cells on the soft substrate or the hard substrate. Lane 1, FHL2-GFP from HFF cells on soft substrate; lane 2, FHL2_Y93F-GFP from HFF cells on soft substrate; lane 3, FHL2-GFP from HFF cells on hard substrate; lane 4, FHL2_Y93F-GFP from HFF cells on hard substrate. Black arrowhead, phosphorylated FHL2 protein; white arrowhead, FHL2 protein. (B) FHL2-GFP localization in an HFF cell on soft substrate. (B′) FAK-mCherry localization in an HFF cell on soft substrate. (B′′) Merged image of B and B′. (C) FHL2 localization in an FAK−/− cell on 8.78-kPa ACA gel coated with fibronectin. (C′) Merged image of C and DAPI. (D) FHL2 localization in an FAK −/− cell on 75.3-kPa ACA gel coated with fibronectin. (D′) Merged image of D and DAPI. (E) Graph showing the intensity ratio of FHL2 immunofluorescence staining between the nucleus and whole cell area (nucleus/whole cell area) on fibronectin-coated gels of varying rigidity. n > 20. Error bars represent SEM. All images are projected images from adhesion sections to nuclear sections.
FAK contains three specific domains: the FERM, kinase, and FRNK domains (consisting of a Pro-rich region and FAT) (41, 45). Normally, overexpression of FAT or the FRNK domain acts as a dominant-negative form by releasing FAK from adhesions (46, 47). We found that after FRNK-GFP or FAT-GFP overexpression in HFF cells, FHL2 was still bound to FAs and released from adhesions on the addition of Y-27632, but accumulation of FHL2 in the nucleus was blocked (Fig. 3 E–F′′ and G), consistent with our results with FAK−/− cells.
FAK Activity Is Required for FHL2 Accumulation in the Nucleus.
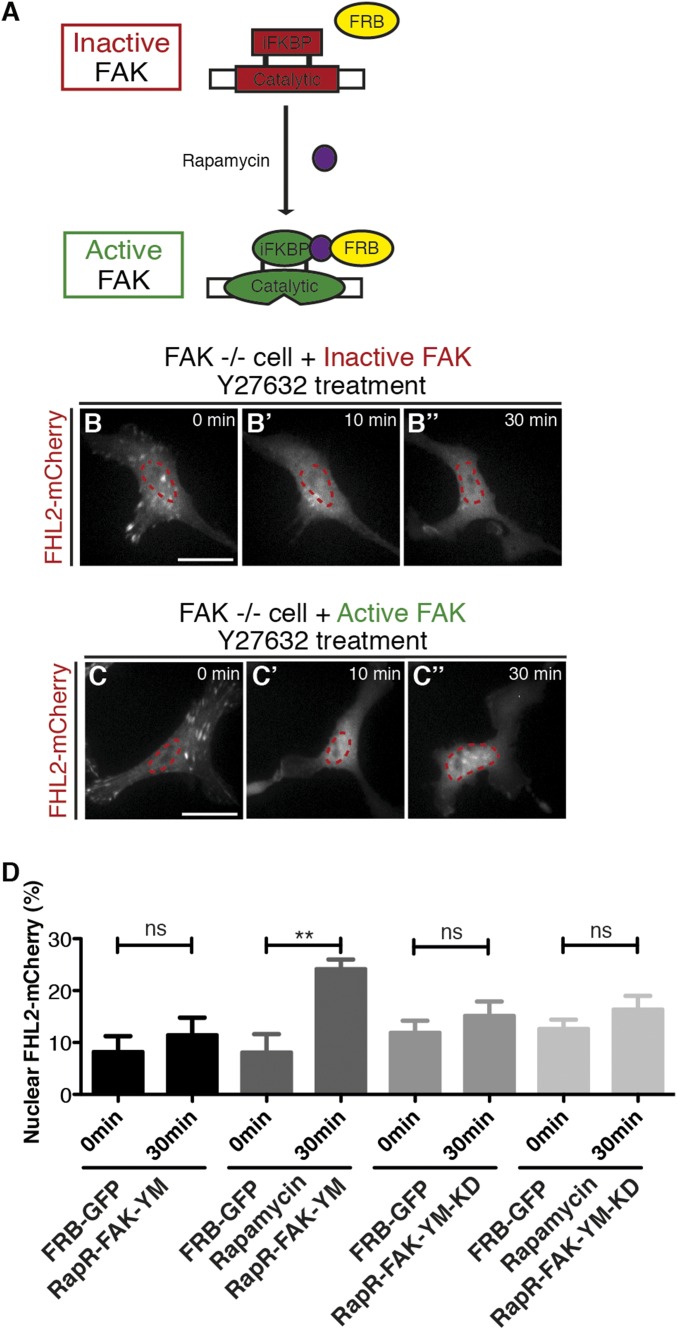
Although we found that FAK is involved in shuttling of FHL2 between FAs and the nucleus, the molecular mechanism was not elucidated. It is possible that, as a kinase, FAK-mediated phosphorylation of FHL2 plays a key role in nuclear concentration. In support of this hypothesis, inhibition of FAK activity by PF573228 partially inhibited FHL2 movement to the nucleus after Y-27632 treatment (Fig. S7 C and D). Using an engineered allosteric activation construct, we performed a series of rescue experiments in FAK−/− cells to confirm the importance of kinase activity. The RapR-FAK-YM construct was activated by FRB expression and rapamycin treatment (Fig. 4A) (48). In FAK−/− cells transfected with the RapR-FAK-YM construct, FRB-GFP expression and rapamycin treatment restored FHL2-mCherry transport to the nucleus after the addition of Y-27632 (Fig. 4 C–C′′ and D). When rapamycin was omitted, FHL2 was not transported to the nucleus (Fig. 4 B–B′′ and D). In addition, transfection with the kinase-dead construct RapR-FAK-YM-KD did not rescue FHL2 transport to the nucleus after Y-27632 treatment in FAK −/− cells with FRB-GFP and rapamycin (Fig. 4D). Thus, active FAK kinase is needed to induce a nuclear concentration of FHL2 after myosin inhibition.
Fig. 4.
Kinase activity in FAK is responsible for FHL2 transport to the nucleus. (A) Schematic image illustrating the allosteric activation of RapR-FAK by rapamycin treatment (47). (B–B′′) FHL2-mCherry dynamics in FAK−/− cells expressing RapR-FAK-YM and FRB-GFP expression (inactive FAK) after Y-27632 treatment. (C–C′′) FHL2-mCherry dynamics in FAK−/− cells with RapR-FAK-YM and FRB-GFP expression and rapamycin treatment (active FAK) after Y-27632 treatment. (D) Graph showing the intensity ratio of FHL2-mCherry in FAK−/− cells between nuclei and whole cells using the RapR-FAK system before and after Y-27632 treatment. The magenta circle indicates NLS-BFP (nuclear marker). (Scale bars: 20 μm.) All images are projected images from adhesion sections to nuclear sections. n > 15. Error bars represent SEM. **P < 0.001.
A Critical Tyrosine for FHL2 Concentration in the Nucleus.
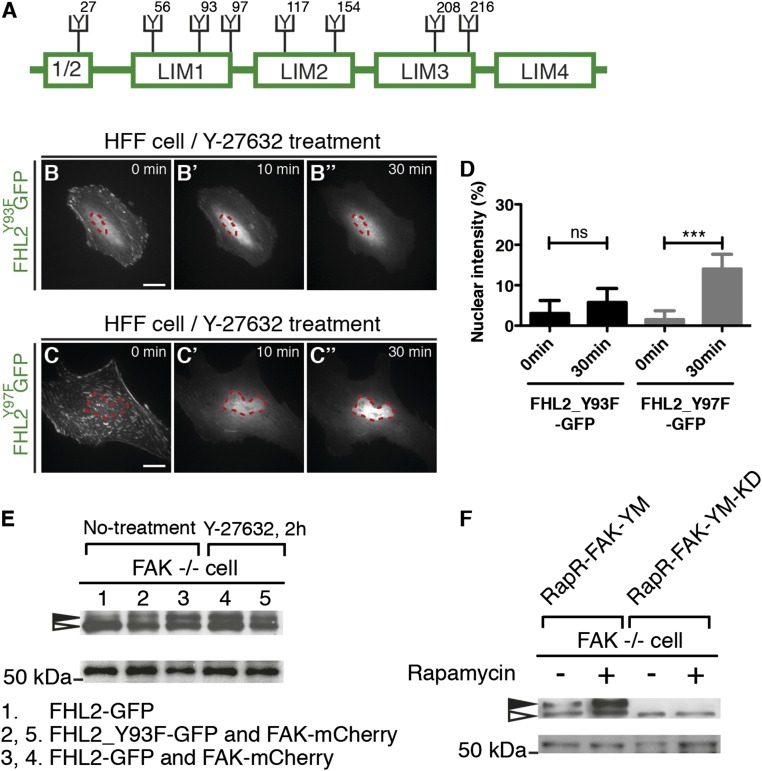
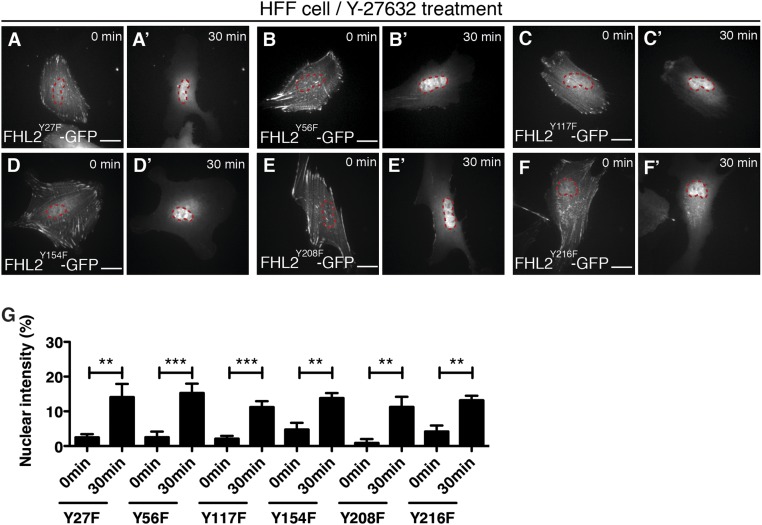
The FHL2 protein contains eight tyrosines that could be substrates of tyrosine kinases (Fig. 5A). We hypothesized that if FHL2 phosphorylation by FAK is important for FHL2 transport and concentration in the nucleus, then an alteration of one or more of those tyrosines would block FHL2 movement to the nucleus. Therefore, we mutated each tyrosine in FHL2 to a phenylalanine and observed FHL2 accumulation in the nucleus after the addition of Y-27632. One mutation of FHL2, Y93F, failed to accumulate in the nucleus after Y-27632 treatment, similar to what was seen in FAK−/− cells (Fig. 5 B–B′′ and D). In contrast, the Y97F mutation of FHL2 and the other six tyrosine-to-phenylalanine mutations were still able to accumulate in the nucleus (Fig. 5 C–C′′ and D and Fig. S9 A–G). Thus, we suggest that FAK-mediated phosphorylation of the Y93 residue of FHL2 after Y-27632 treatment is important for FHL2 concentration in the nucleus.
Fig. 5.
The tyrosine-93 residue in FHL2 is responsible for FHL2 nuclear transport. (A) Schematic image showing the location of each tyrosine residue in FHL2. (B–B′′) The dynamics of FHL2-GFP with tyrosine-93 mutation (Y93F) in HFF cells after Y-27632 treatment. (C–C′′) The dynamics of FHL2-GFP with tyrosine-97 mutation (Y97F) in HFF cells after Y-27632 treatment. (D) Graph showing the intensity ratio of FHL2_Y93F-GFP or FHL2_Y97F-GFP between nuclei and whole cells after Y-27632 treatment. (Scale bars: 20 μm.) (E, Top) SDS/PAGE with polyacrylamide containing Mn2+ and Phos-tag. Black arrow, phosphorylated FHL2; white arrow, unphosphorylated FHL2. (E, Bottom) SDS/PAGE with polyacrylamide. Western blot analysis for FHL2-GFP or FHL2_Y93F-GFP from each sample isolated from FAK−/− cells and FAK−/− cells rescued by FAK-mCherry. Lane 1, FHL2-GFP from FAK−/− cells; lane 2, FHL2_Y93F_GFP from FAK−/− cells rescued by FAK-mCherry; lane 3, FHL2-GFP from FAK−/− cells rescued by FAK-mCherry; lane 4, FHL2-GFP from FAK−/− cells rescued by FAK-mCherry with Y-27632, lane 5; FHL2_Y93F-GFP from FAK −/− cell rescued by FAK-mCherry with Y-27632. (F, Top) SDS/PAGE with polyacrylamide containing Mn2+ and Phos-tag. Black arrow, phosphorylated FHL2; white arrow, unphosphorylated FHL2. (F, Bottom) SDS/PAGE with polyacrylamide. Western blot analysis for FHL2-GFP from each samples isolated from FAK−/− cells with RapR-FAK-YM or RapR-FAK-YM-KD and myc-FRB expression, with and without rapamycin treatment. (F, Left) FHL2-GFP from FAK−/− cells with RapR-FAK-YM and myc-FRB expression, without and with rapamycin treatment. (F, Right) FHL2-GFP from FAK−/− cells with RapR-FAK-YM-KD and myc-FRB expression, without and with rapamycin treatment. (Scale bars: 20 μm.) All images are projected images from adhesion sections to nuclear sections. n > 10. Error bars represent SEM. ***P < 0.0001.
Fig. S9.
FHL2 nuclear localization with mutations of tyrosine residues in FHL2. (A–F′) Dynamics of FHL2-GFP with mutations at each tyrosine after Y-27632 treatment. The magenta circle indicates NLS-BFP (nuclear marker). (G) Graph showing the relative intensity of FHL2-GFP with mutation at each tyrosine after Y-27632 treatment. All images are projected images from adhesion sections to nuclear sections. n > 10. Error bars represent SEM. **P < 0.001; ***P < 0.0001.
The question remained of whether FHL2 phosphorylation is dependent on FAK activity. The Phos-tag system separates phosphorylated proteins in SDS/PAGE (49) and also separates multiple phosphorylated forms of FHL2. In FAK−/− cells, phosphorylation of FHL2-GFP was reduced, and phosphorylation was rescued by FAK-mCherry expression in FAK−/− cells (Fig. 5E). In contrast, the Y93F mutation of FHL2 was only weakly phosphorylated by FAK-mCherry expression in FAK−/− cells (Fig. 5E). Furthermore, transfection with the RapR-FAK-YM and FRB-myc constructs followed by rapamycin treatment restored FHL2 phosphorylation; however, phosphorylation of FHL2 was not rescued by transfection with the kinase-dead construct RapR-FAK-KD in FAK−/− cells with FRB-myc and rapamycin (Fig. 5F). Thus, the Y93 residue is the critical FAK phosphorylation site in FHL2, and an active FAK is needed to phosphorylate FHL2.
Given that the Y93F mutation inhibited FHL2 nuclear accumulation after Y-27632 treatment, we checked whether Y-27632 treatment increased FHL2 phosphorylation. In FAK−/− cells rescued by FAK expression, Y-27632 treatment increased control FHL2 but not Y93F mutant phosphorylation (Fig. 5E). Furthermore, HFF cells on a soft substrate exhibited increased FHL2 phosphorylation (Fig. S8A). As shown in Fig. S4A, blebbistatin treatment induced an increase in nuclear FHL2 protein (Fig. S4A). We speculated that the nuclear FHL2 protein is more phosphorylated than the cytoplasmic FHL2. To test this hypothesis, we measured the amount of phosphorylated FHL2 in the nuclear fraction with or without blebbistatin treatment. We found that phosphorylated FHL2 indeed was increased in the nuclear fraction in HFF cells after blebbistatin treatment (Fig. S4B). Thus, the cellular response to the loss of force or a soft substrate was to increase FHL2 phosphorylation, which caused it to concentrate in the nucleus.
FHL2 Localizes to Active RNA Pol II Sites in the Nucleus.
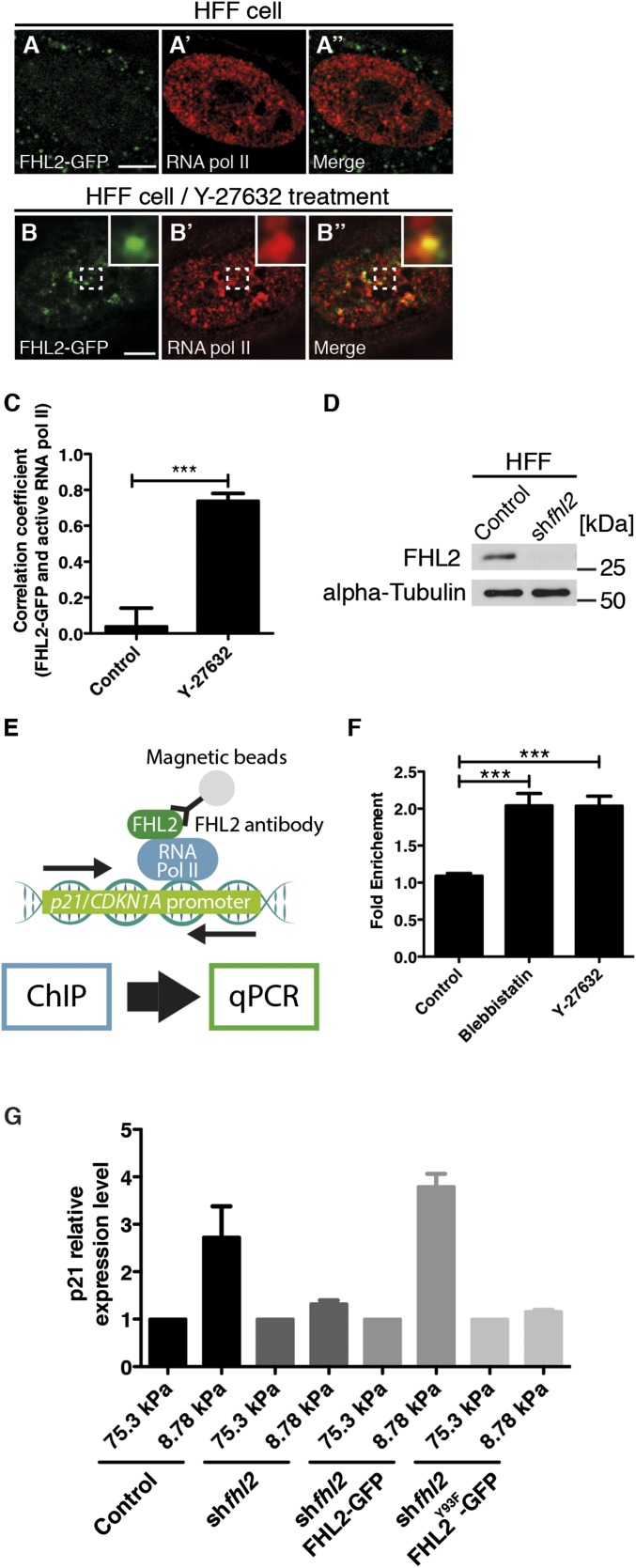
Importantly, FHL2 functions as a transcriptional cofactor in the nucleus (28). Consistent with that role, FHL2-GFP showed punctate localization in the nucleus after Y-27632 treatment (Fig. 6 A and B–B′′). To check whether FHL2 localizes to active sites of transcription after inhibition of myosin II activity, we examined the colocalization of FHL2-GFP and active RNA Pol II using superresolution microscopy. Active RNA Pol II is a marker for sites of initiation of transcription (50). After Y27632 or blebbistatin treatment, we observed a marked increase in FHL2-GFP puncta, many of which colocalized with active RNA Pol II (Fig. 6 B–B′′). We performed Pearson correlation coefficient analysis quantified the extent of correlation of FHL2-GFP intensity with RNA Pol II intensity after Y-27632 treatment, and found a correlation coefficient of 0.73 (Fig. 6 B–B′′ and C) (37). Thus, inhibition of myosin II activity causes FHL2 localization to sites of activated gene expression.
Fig. 6.
Nuclear FHL2 colocalizes with RNA Pol II and promotes p21 gene expression. The images were obtained using a Nikon N-SIM superresolution microscope. (A) FHL2-GFP localization at the nucleus in an HFF cell. (A′) RNA polymerase II staining at the nucleus in an HFF cell. (A′′) Merged image of A and A′. (B) FHL2-GFP localization at the nucleus in an HFF cell after Y-27632 treatment. (B′) RNA polymerase II staining at the nucleus in an HFF cell after Y-27632 treatment. (B′′) Merged image of A and A′. In B and B′′, the dotted white box indicates the magnified region shown in the Inset. (C) Graph showing the Pearson’s correlation coefficient of images between FHL2-GFP and RNA polymerase II staining at the nucleus in HFF cells with and without Y-27632 treatment. (D) Western blots showing the efficiency of FHL2 gene suppression in HFF cells infected with control or FHL2 shRNA-expressing retrovirus. (E) Schematic illustrating quantitative real-time PCR after the ChIP assay. (F) Graph showing normalized fold enrichment of DNA in the p21 gene promoter region after the ChIP assay. (G) Graph showing relative gene expression level of p21 gene in HFF cell with various conditions. The p21 expression level in 75.3-kPa ACA acrylamide gel served as a reference. (Scale bars: 5 μm.) All images are projected images from adhesion sections to nuclear sections. n > 15. Error bars represent SEM. ***P < 0.0001.
FHL2 Nuclear Localization with Loss of Force Induces p21 Gene Expression.
Previous studies have shown that soft surfaces inhibit cell proliferation (4, 51). In a possibly related finding, p21 inhibits cell proliferation through inhibition of cyclin protein gene expression (52). Specifically, FHL2 regulates p21 gene expression in breast cancer cells through an interaction with the p21 gene promoter (53, 54). We first checked whether less force induces a stronger interaction between FHL2 and the p21 gene promoter through chromatin IP (ChIP) assays. The FHL2 protein–DNA complex was pulled down using an FHL2-specific antibody or normal IgG antibody, after which the p21 gene promoter level was quantified by quantitative real-time PCR (Fig. 6E). Blebbistatin and Y-27632 treatment induced a significant increase in the amount of FHL2 protein bound to the p21 gene promoter (Fig. 6F). We next used quantitative real-time PCR to check whether FHL2 concentration in the nucleus on soft substrates causes changes in p21 gene expression, and found a significant induction of p21 gene expression on soft surfaces (Fig. 6G). After knockdown of FHL2 expression in HFF cells, there was no increase in p21 expression on soft surfaces compared with rigid surfaces (Fig. 6 D and G). The normal increase in p21 expression level on soft surfaces was restored in knocked-down cells by overexpression of FHL2-GFP, but not by overexpression of the Y93F mutant FHL2-GFP (Fig. 6G). These results indicate that FHL2 concentration in the nucleus on soft surfaces is necessary for p21 gene expression. Thus, we suggest that FHL2 regulates p21 gene expression in a force-dependent manner by binding to the p21 promoter.
Discussion
We have found that the well-documented inhibition of cell growth on soft surfaces correlates with the increased expression of p21 as a result of FHL2 movement to the nucleus (Fig. 7). At a molecular level, the phosphorylation of Y93 in FHL2 by FAK is increased on soft surfaces, which causes FHL2 to concentrate at nuclear sites of active pol II transcription, including the promoter sequence of p21, which in turn increases p21 expression.
Fig. 7.
Schematic of a working model of FHL2 shuttling depending on substrate rigidity. The presence of a rigid substrate keeps FHL2 at the FA through mechanical tension. In contrast, a soft substrate induces nuclear localization of FHL2, through the release of phosphorylated FHL2 by FAK from FAs into the cytoplasm. This step requires phosphorylation of a specific tyrosine residue (Y93) in FHL2. Free FHL2 accumulates in the nucleus, where it colocalizes with active RNA Pol II to modulate p21 gene expression.
FHL2 moves in a force- and rigidity-dependent manner from adhesions to the nucleus. This movement is seen in several different cell types (i.e., HFF, 3T3, and mouse embryonic fibroblasts), and several alterations in contractility can affect shuttling, including myosin contractility, FAK activity, and a specific tyrosine residue in FHL2. In FAK−/− cells, FHL2 localizes to adhesions and leaves the adhesions after ROCK inhibition by Y-27632, but there is no significant movement to the nucleus. Furthermore, overexpression of the FAT and FRNK domains of FAK blocks the increase in FHL2 nuclear concentration even in the presence of normal FAK. Because inhibition of FAK kinase activity, kinase-dead FAK, and mutation of Y93F all inhibit movement of FHL2 to the nucleus, it appears that the kinase activity of FAK is important for this shuttling. Furthermore, biochemical experiments show that phosphorylation of Y93 in FHL2 is dependent on FAK activity. Finally, we show that this pathway induces FHL2 colocalization with active RNA Pol II and promotes FHL2-dependent p21 expression.
Several mechanical signaling pathways have been shown to use tyrosine phosphorylation, including the activation of p130Cas in force sensing (55) and the phosphorylation of many adhesion components (56). Tyrosine kinases such as FAK are involved in the sensing of matrix rigidity, and it is logical to propose that FAK will phosphorylate FHL2 on soft substrates and on the loss of contractile force (7). As we have shown, FHL2 phosphorylation and nuclear localization depend on FAK activity (Fig. 4 B–D and Fig. 5F); however, inhibition of FAK activity by the small molecule inhibitor PF573228 only partially inhibited FHL2 movement to the nucleus (Fig. S7 C and D). These data suggest that the FAK inhibition may be incomplete, meaning that even a low level of activity could result in the phosphorylation of significant levels of FHL2 because of its concentration in an FAK complex.
The movement of FHL2 to the nucleus is dissociated from the disassembly of adhesions, because FHL2 can still move to the nucleus when adhesions are stabilized by vinculin head domain expression (Fig. 2 G–J), and it does not concentrate in nuclei after adhesion disassembly in the absence of FAK activity. The release of FHL2 from adhesions occurs faster than that of paxillin in control cells (Fig. 2 D–E′′ and F). Surprisingly, both the presence of FAT domains and the loss of FAK activity keep FHL2 in the cytoplasm. Furthermore, phosphorylation of FHL2 is reduced in the FHL2 mutant Y93F and the expression of a kinase-dead FAK, indicating that the specific phosphorylation of Y93 is critical for nuclear transport.
Recent studies have shown that FAK activity as measured by autophosphorylation at Y397 is increased with myosin contractility (43, 44) and decreased on inhibition of myosin. These findings raise the question of how FHL2 is phosphorylated after the inhibition of contractility, where there is reduced FAK activity. One possibility is that FAK inactivation on myosin inhibition activates another kinase that phosphorylates FHL2. Alternatively, an FAK complex with FHL2 and a GRB7 family protein (e.g., Grb14) may be force-sensitive, such that the complex assembles under force and can phosphorylate FHL2, but releases the phosphorylated form only on relaxation. Overexpression of the FAT domain (either as the FAT domain alone or in FRNK) can then compete with endogenous FAK in the adhesion and prevent phosphorylation from occurring (Fig. 3 E–G).
The mechanism of how FHL2 is transported and concentrates within the nucleus is not fully understood. We tried two inhibitors of the importin complex, importazole and ivermectin, but they had no effect on the nuclear concentration of FHL2. Furthermore, FHL2 does not have a classical nuclear localization signal. Similarly, STAT3 moves to the nucleus on tyrosine phosphorylation, which provides a precedence for concentration in the nucleus by this mechanism (57). Thus, we suggest that nuclear binding sites for phosphorylated FHL2 can cause it to be concentrated at punctate sites in the nucleus, as observed on superresolution microscopy. On the other hand, a previous study showed that stimulation of Rho signaling by the bioactive lipid lysophosphatidic acid (LPA) mediates FHL2 nuclear localization (31). To determine whether this pathway was somehow related, we checked FHL2 nuclear localization in NIH 3T3 cells and HFF cells both with and without serum starvation on noncoated glass. As with HFF cells, blebbistatin induced FHL2 nuclear localization in 3T3 cells with serum (Fig. S3 L–M′). In addition, as reported previously, LPA treatment induced FHL2 nuclear localization in NIH 3T3 cells after serum starvation, but the same effect was not seen in HFF cells. In contrast, LPA treatment was able to induce nuclear localization of FHL2 containing the Y93F mutation in serum-starved NIH 3T3 cells. These results indicate that there are two distinct pathways for FHL2 movement to the nucleus, and that the transport mechanism may be through passive diffusion, because FHL2 is small enough to diffuse through nuclear pores.
The mechanism of how FHL2 is recruited to adhesion sites remains elusive. After washout of Y-27632, FHL2-GFP localization was restored at adhesion sites (Fig. S6). Related to this, blebbistatin treatment inhibited the interaction between FAK and FHL2 (Fig. S7B). These results support the idea that the tension might be sufficient to recruit FHL2 to the adhesion sites similar to other FA molecules (12). On the other hand, our data indicate that phosphorylated FHL2 protein does not go out from the nucleus completely after washout of Y-27632 (Fig. S6). These data are consistent with the results of biochemical experiments (Fig. S4B). The foregoing results further support the hypothesis that FHL2 freely diffuses into the nucleus and, once inside, binds to nuclear sites, thereby concentrating it in the nucleus; however, once FAs are restored, dephosphorylation of FHL2 in the nucleus will release it from nuclear sites, and FHL2 will diffuse out of the nucleus to bind to FAs.
There is considerable interest in the mechanisms that control the movement of LIM domain proteins to the nucleus. Movement of the LIM domain protein Hic-5 to the nucleus is also related to FAK activity, but the stimulus for nuclear uptake correlates with increases in reactive oxygen species (58). Once in the nucleus, LIM domain proteins are thought to alter gene expression profiles (22), and FHL2 is responsible for changes in the expression of many different genes (28). LIM domain proteins are hypothesized to play a role in mechanotransduction, but this report more specifically shows that changes in substrate rigidity cause the uptake of a LIM domain protein into the nucleus. Mass spectrometry screens of FA proteins after inhibition of myosin contraction support the hypothesis that the binding of the LIM domain proteins to adhesions is force-dependent (12, 23). Many adhesion proteins exhibit force dependence because of the general disassembly of adhesions on the inhibition of contractility, however. Hic-5 has been shown to move from adhesions to stress fibers, but not to the nucleus, on mechanical stretching (59). The fact that FHL2 can move to the nucleus in response to decreases in contractility or rigidity indicates that those physiological stimuli may cause long-term changes in cell function through FHL2-dependent changes in gene expression.
Interestingly, we found that FHL2 colocalizes with activated RNA Pol II sites, suggesting an important link between matrix rigidity and global transcription in such physiological processes as cell differentiation and cell proliferation (60, 61). Previous studies showed that RNA Pol II subunits promote centromere transcription and RNAi-dependent heterochromatin assembly (62, 63). Following these studies, we checked whether FHL2 localized at centromeres, but found no FHL2 colocalization with the centromeric protein CENP-A (64) (Fig. S10 C–C′′). Thus, we suggest that the nuclear localization of FHL2 with RNA pol II is not at centromeres, but rather at sites of transcription.
Fig. S10.
FHL2 nuclear localization at the active RNA polymerase II site in HFF cells on soft substrate. The images were obtained with a Nikon N-SIM superresolution microscope. (A) FHL2-GFP localization at the nucleus in an HFF cell on 8.78-kPa ACA gel coated with fibronectin. (A′) Active RNA polymerase II staining at the nucleus in an HFF cell on 8.78-kPa ACA gel coated with fibronectin. (A′′) Merged image of A and A′. (B) FHL2-GFP localization at the nucleus in an HFF cell on 75.3-kPa ACA gel coated with fibronectin. (B′) RNA polymerase II staining at the nucleus in an HFF cell on 75.3-kPa ACA gel coated with fibronectin. (B′′) Merged image of A and A′. (C) FHL2-GFP localization at the nucleus in an HFF cell on glass coated with fibronectin. (C′) CENP-A staining at the nucleus in an HFF cell on glass coated with fibronectin. (C′′) Merged image of C and C′. (D) Graph showing the Pearson correlation coefficient between FHL2-GFP and active RNA polymerase II staining at the nucleus in HFF cells on 8.78-kPa and 75.3-kPa ACA gels coated with fibronectin. (E) Graph showing the Pearson correlation coefficient between FHL2-GFP and CENP-A staining at the nucleus in HFF cells on fibronectin-coated glass. n > 20. Error bars represent SEM. ***P < 0.0001.
Previous studies have identified FHL2 as a positive regulator of p21 gene expression (53, 54) and found that p21 negatively regulates cell proliferation through inhibition of cyclin proteins (52). Thus, we suggest that soft surfaces will cause growth inhibition by activating movement of FHL2 to the nucleus to increase p21 gene expression. The prominent role of FHL2 in cancer metastasis indicates that it has an important role in overriding mechanical signals that would otherwise inhibit tumor growth and metastasis (27, 34).
Materials and Methods
Cell Culture and Transfections.
HFF cells (American Type Culture Collection) and FAK−/− mouse fibroblast cells (14) were cultured in DMEM (Gibco) with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco). Transfections were performed with the Neon Transfection System (Life Technologies). The Vin880-GFP construct was a kind gift from Christoph Ballestrem, The University of Manchester, Manchester, United Kingdom. pmCherry-C1-FAK-HA (Addgene plasmid 35039) was a gift from Anna Huttenlocher, University of Wisconsin–Madison, Madison, WI. pGFP-FAT (Addgene plasmid 50517) was a gift from Kenneth Yamada, National Institutes of Health, Bethesda. myc-Rapr-FAK-YM, pEGFP-RapR-FAK-YM-KD, and pEGFP-FRB (Addgene plasmids 25927, 25929, and 25919) were gifts from Klus Hahn, University of North Carolina, Chapel Hill, NC. EBFP2-Nucleus-7 (Addgene plasmid 55249) was a gift from Michael Davidson, Florida State University, Tallahassee, FL. Paxillin-RFP and FRNK-GFP have been described previously (44, 65).
ACA Acrylamide Gel Experiment.
The ratios of acrylamide and BIS in the gels with various stiffness were as follows: 8.78 kPa: acrylamide, 2.81%, BIS, 0.120%; 20.2 kPa: acrylamide, 5.50%, BIS, 0.230%; 75.3 kPa: acrylamide, 10.7%, BIS, 0.495%. The same concentration of ACA was used for all gels (100 mM). All gels were functionalized with 0.01 mg/mL type I collagen (C3867; Sigma-Aldrich) or 0.01 mg/mL fibronectin (F0635; Sigma-Aldrich) in HEPES buffer (0.5 M HEPES, pH 9.0). All cells were cultured on the ACA acrylamide gels for 17 h.
AFM Measurements of Acrylamide Gel Stiffness.
Rigidity modulus measurements were performed using a Dimension Icon atomic force microscope (Bruker). The acrylamide gels were prepared on coverglasses and immersed in PBS for force measurements. The probe consisted of a 4.5-µm-diameter polystyrene bead attached to a cantilever. The spring constant of the cantilever was determined by the thermal tune method (66) and was typically in the range of 0.0404 N/m. Force volume measurements were carried out at 1 Hz, in five reference areas of 16 × 16 µm2 for each sample. Young’s modulus values were calculated for each recorded curve using Nanoscope Analysis 1.7 software (Bruker), which uses a Hertz contact model for spherical indenters (tip radius, 2.25 mm; Poisson ratio, 0.5) fitted to the indentation curves (67).
Inhibitors.
Blebbistatin treatment was performed as described previously (68). Y-27632, cytochalasin D, and PF573228 were purchased from Sigma-Aldrich. Wiskostatin was purchased from Tocris Bioscience.
Immunostaining and Microscopic Analysis of Cells.
Cells were fixed by 4% formaldehyde/PBS for 30 min. After fixation, cells were washed with PBS and then treated with 5% FBS/PBS for 30 min. Primary and secondary antibodies were diluted with Can Get Signal Immunostain Solution A (TOYOBO). The primary antibodies used for immunostaining were rabbit anti-FHL2 (HPA006028, 1:700; Sigma-Aldrich), rabbit anti-transcriptionally active CTD phosphorylated RNA Pol II (ab5131, 1:500; Abcam), and CENP-A rabbit mAb (C51A7, 1:500; Cell Signaling Technology). Secondary antibodies used for visualization were Alexa Fluor 488 goat anti-rabbit IgG (H+L) and Alexa Fluor 594 donkey anti-rabbit IgG (H+L) (Invitrogen). Stained cells were mounted in Vectashield mounting medium with DAPI/PBS (1:200; Vector Laboratories).
Plasmids.
To generate FHL2-GFP and FHL2-mCherry constructs, human FHL2 cDNA in pENTR223 vector from the DNASU Plasmid Repository (Biodesign Institute/Arizona State University) was recombined into pDest-eGFP-N1 or pDest-mCherry-N1 (Addgene 31796 and 31907) by LR recombination using Gateway cloning vectors with LR clonase (Invitrogen).
Each tyrosine mutation was generated using the Agilent QuikChange Lightning Site-Directed Mutagenesis Kit using the following primers: tyrosine-27: forward, 5′-GAGGAGAGCCCCTTCTGCGTGGTGTGCTTT-3′; reverse, 5′-AAAGCACACCACGCAGAAGGGGCTCTCCTC-3′; tyrosine-56: forward, 5′-AAGGACTTGTCTTTCAAGGACCGGCACTGG-3′; reverse, 5′-CCAGTGCCGGTCCTTGAAAGACAAGTCCTT-3′; tyrosine-93: forward, 5′-TGTACAGACTGCTTTTCCAACGAGTACTCA-3′; reverse, 5′-TGAGTACTCGTTGGAAAAGCAGTCTGTACA-3′; tyrosine-97: forward, 5′-TATTCCAACGAGTTCTCATCCAAGTGCCAG-3′; reverse, 5′- CTGGCACTTGGATGAGAACTCGTTGGAATA-3′; tyrosine-117: forward, 5′-CGCAAGATGGAGTTCAAGGGCAGCAGCTGG-3′; reverse, 5′-CCAGCTGCTGCCCTTGAACTCCATCTTGCG-3′; tyrosine-154: forward, 5′-TGTGTGCCCTGCTTTGAGAAACAACATGCC-3′; reverse, 5′-GGCATGTTGTTTCTCAAAGCAGGGCACACA-3′; tyrosine-208: forward, 5′-GATGACTTTGCCTTCTGCCTGAACTGCTTC-3′; reverse, 5′-GAAGCAGTTCAGGCAGAAGGCAAAGTCATC-3′; tyrosine-216: forward, 5′-TTCTGTGACTTGTTTGCCAAGAAGTGTGCT-3′; reverse, 5′-AGCACACTTCTTGGCAAACAAGTCACAGAA-3′.
For generation of retroviruses shRNAs against FHL2 gene, the DNA fragment including FHL2 gene target sequence 5′-CCGGGAGACTTTCTTCTAGTGCTTTCTCGAGAAAGCACTAGAAGAAAGTCTCTTTTT-3′ was cloned into a pSUPER retro.puro vector (Oligoengine).
Microscopy and Data Analysis.
The fixed cells were examined using an A1R-Si (Nikon) or iLAS TIRF (Olympus) microscopy system. Superresolution images were obtained using a Nikon N-SIM system with a CFI Apo TIRF 100× oil objective lens (NA 1.49, immersion oil type NF, ND 1.515) and an EMCCD camera (DU897; Andor Technology). Imaging was carried out in the 3D SIM mode, and each image was reconstructed from three directions and five phases (a total of 15 images) using NIS-elements Ar (Nikon). The parameters for the image reconstruction of SIM data were as follows: illumination modulation contrast, 1; high-resolution noise suppression, 0.5. Out of Focus blur suppression: 0.05. Resolution under these conditions was estimated to be ∼120 nm. Live cell images were taken following previous protocols (68), but using a 60× water lens (UPLSAPO 60×, NA 1.20, WD 0.28 mm) instead of a 100× oil immersion lens. Image analysis was processed with ImageJ or MATLAB (MathWorks). The intensity ratio of talin-GFP or talin-mCherry between the nucleus and whole cell in HFF or FAK−/−cells was subtracted as the background intensity ratio of GFP or mCherry.
Detection of Protein in Nuclear and Cytoplasmic Fractions.
Cells were solubilized with ice-cold extraction buffer A (10 mM HEPES-Na pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.4% Nonidet P-40, 1 mM DTT, and Protease Inhibitor Mixture; Sigma-Aldrich). Lysates were incubated on ice for 5 min and then centrifuged at 9,100 × g for 15 min. The supernatants were removed as the cytoplasmic fraction. The remaining pellets were solubilized with extraction buffer B (20 mM HEPES-Na pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, and Protease Inhibitor Mixture; Sigma-Aldrich). Lysates were vortexed for 30 min at 4 °C and then centrifuged at 20,400 × g for 20 min. The supernatants were used for the nuclear fraction. The primary antibodies for Western blot analysis were rabbit anti-FHL2 (HPA006028, 1:100; Sigma-Aldrich), rabbit anti-Lamin B1 (ab16048, 1:500; Abcam), and mouse anti–α-tubulin (T6199, 1:2,000; Sigma-Aldrich). Secondary antibodies used for detection were goat anti-mouse IgG (H+L)-HRP conjugate and goat anti-rabbit IgG (H+L)-HRP conjugate (Bio-Rad).
Detection of Phosphorylated Protein Using Phos-tag SDS/PAGE.
Cells were solubilized with ice-cold extraction RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10 mM EDTA, 1% Triton 100, 10 mM NaF, 1 mM Na3VO4, 1% SDS, 25 mM β-glycerol phosphatase, and Protease Inhibitor Mixture; Sigma-Aldrich). SDS/PAGE with polyacrylamide-bound Mn2+ Phos-tag was described previously (49). For Western blot analysis, mouse anti-GFP (sc-9996, 1:200; Santa Cruz Biotechnology) was the primary antibody and goat anti-mouse IgG (H+L)-HRP conjugate (Bio-Rad) was the secondary antibody for detection.
ChIP Analysis.
The procedure for the purification of DNA with antibody pulldown was as described previously with modifications (69). Mouse monoclonal anti-FHL2 antibody (5 μg; MBL) and normal mouse IgG (sc-2025; Santa Cruz Biotechnology) were used for IP analysis. The primers for DNA amplification of the p21 gene promoter were described previously (53).
IP Analysis.
IP was performed using rabbit anti-FHL2 (HPA006028, 5 μg; Sigma-Aldrich) and normal rabbit IgG (sc-2027; Santa Cruz Biotechnology). FAK antibody (c-20, 1:500; Santa Cruz Biotechnology) was used for immunoblotting.
Quantitative Real-Time PCR.
Total RNA was extracted using the E.Z.N.A Total RNA Kit I (Omega Bio-tek). cDNA was prepared using the Tetro cDNA Synthesis Kit (Bioline). Quantitative real-time PCR was performed using SsoFast EvaGreen Supermix with the CFX96 Real-Time System (Bio-Rad). The following primers were used: human cyclophilin A: forward, 5′-CTCGAATAAGTTTGACTTGTGTTT-3′; reverse, 5′-CTAGGCATGGGAGGGAACA-3′; human p21: forward, 5′-GAGGCCGGGATGAGTTGGGAGGAG-3′; reverse, 5′-CAGCCGGCGTTTGGAGTGGTAGAA-3′ (70).
Statistical Analysis.
Data were analyzed using the unpaired t test.
Acknowledgments
We thank Dr. Keiko Kawauchi, Dr. Cheng-Han Yu, Dr. Shota Yamauchi, Dr. Hiroaki Hirata, Dr. Yukako N Motegi, Dr. Ichiro Harada, Dr. Prasuna Rao, and Ms. Yidan Cui for useful discussions; Dr. Chwee Teck Lim and Dr. Brenda Nai Mui Hoon for assistance with the AFM experiments; and Chun Xi Wong and Dr. Andrew Ming-shang Wong for scientific illustration and clarification. This work was supported by the National Research Foundation of Singapore and the Ministry of Education of Singapore through the Mechanobiology Institute, National University of Singapore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608210113/-/DCSupplemental.
References
- 1.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leight JL, Wozniak MA, Chen S, Lynch ML, Chen CS. Matrix rigidity regulates a switch between TGF-β1–induced apoptosis and epithelial–mesenchymal transition. Mol Biol Cell. 2012;23(5):781–791. doi: 10.1091/mbc.E11-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Cui Y, et al. Cyclic stretching of soft substrates induces spreading and growth. Nat Commun. 2015;6:6333. doi: 10.1038/ncomms7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saez A, Buguin A, Silberzan P, Ladoux B. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys J. 2005;89(6):L52–L54. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghassemi S, et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc Natl Acad Sci USA. 2012;109(14):5328–5333. doi: 10.1073/pnas.1119886109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelham RJ, Jr, Wang Yl. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9(8):858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468(7323):580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balaban NQ, et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3(5):466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 11.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo JC, Han X, Hsiao CT, Yates JR, 3rd, Waterman CM. Analysis of the myosin-II–responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13(4):383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfenson H, et al. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat Cell Biol. 2016;18(1):33–42. doi: 10.1038/ncb3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang G, Huang AH, Cai Y, Tanase M, Sheetz MP. Rigidity sensing at the leading edge through alphavbeta3 integrins and RPTPalpha. Biophys J. 2006;90(5):1804–1809. doi: 10.1529/biophysj.105.072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prager-Khoutorsky M, et al. Fibroblast polarization is a matrix rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13(12):1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 16.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 17.Iskratsch T, Wolfenson H, Sheetz MP. Appreciating force and shape: The rise of mechanotransduction in cell biology. Nat Rev Mol Cell Biol. 2014;15(12):825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swift J, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341(6149):1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497(7450):507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shivashankar GV. Mechanosignaling to the cell nucleus and gene regulation. Annu Rev Biophys. 2011;40:361–378. doi: 10.1146/annurev-biophys-042910-155319. [DOI] [PubMed] [Google Scholar]
- 21.Aggarwal V, Dickinson RB, Lele TP. Concentration sensing by the moving nucleus in cell fate determination: A computational analysis. PLoS One. 2016;11(2):e0149213. doi: 10.1371/journal.pone.0149213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadrmas JL, Beckerle MC. The LIM domain: From the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5(11):920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 23.Schiller HB, Friedel CC, Boulegue C, Fässler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12(3):259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirata H, Tatsumi H, Sokabe M. Mechanical forces facilitate actin polymerization at focal adhesions in a zyxin-dependent manner. J Cell Sci. 2008;121(Pt 17):2795–2804. doi: 10.1242/jcs.030320. [DOI] [PubMed] [Google Scholar]
- 25.Uemura A, Nguyen TN, Steele AN, Yamada S. The LIM domain of zyxin is sufficient for force-induced accumulation of zyxin during cell migration. Biophys J. 2011;101(5):1069–1075. doi: 10.1016/j.bpj.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suresh Babu S, et al. Mechanism of stretch-induced activation of the mechanotransducer zyxin in vascular cells. Sci Signal. 2012;5(254):ra91. doi: 10.1126/scisignal.2003173. [DOI] [PubMed] [Google Scholar]
- 27.Cao CY, Mok SW, Cheng VW, Tsui SK. The FHL2 regulation in the transcriptional circuitry of human cancers. Gene. 2015;572(1):1–7. doi: 10.1016/j.gene.2015.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleiber K, Strebhardt K, Martin BT. The biological relevance of FHL2 in tumour cells and its role as a putative cancer target. Anticancer Res. 2007;27(1A):55–61. [PubMed] [Google Scholar]
- 29.Gabriel B, et al. Focal adhesion kinase interacts with the transcriptional coactivator FHL2 and both are overexpressed in epithelial ovarian cancer. Anticancer Res. 2004;24(2B):921–927. [PubMed] [Google Scholar]
- 30.Samson T, et al. The LIM-only proteins FHL2 and FHL3 interact with alpha- and beta-subunits of the muscle alpha7beta1 integrin receptor. J Biol Chem. 2004;279(27):28641–28652. doi: 10.1074/jbc.M312894200. [DOI] [PubMed] [Google Scholar]
- 31.Müller JM, et al. The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J. 2002;21(4):736–748. doi: 10.1093/emboj/21.4.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebrahimian T, et al. Inhibition of four-and-a-half LIM domain protein-2 increases survival, migratory capacity, and paracrine function of human early outgrowth cells through activation of the sphingosine kinase-1 pathway: Implications for endothelial regeneration. Circ Res. 2014;114(1):114–123. doi: 10.1161/CIRCRESAHA.113.301954. [DOI] [PubMed] [Google Scholar]
- 33.Verset L, et al. Epithelial expression of FHL2 is negatively associated with metastasis-free and overall survival in colorectal cancer. Br J Cancer. 2013;109(1):114–120. doi: 10.1038/bjc.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, et al. Four-and-a-half LIM protein 2 promotes invasive potential and epithelial-mesenchymal transition in colon cancer. Carcinogenesis. 2010;31(7):1220–1229. doi: 10.1093/carcin/bgq094. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu T, et al. Dual inhibition of Src and GSK3 maintains mouse embryonic stem cells, whose differentiation is mechanically regulated by Src signaling. Stem Cells. 2012;30(7):1394–1404. doi: 10.1002/stem.1119. [DOI] [PubMed] [Google Scholar]
- 36.Yip AK, et al. Cellular response to substrate rigidity is governed by either stress or strain. Biophys J. 2013;104(1):19–29. doi: 10.1016/j.bpj.2012.11.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300(4):C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299(5613):1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 39.Uehata M, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389(6654):990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 40.Carisey A, et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol. 2013;23(4):271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: In command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6(1):56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 42.Wang HB, Dembo M, Hanks SK, Wang Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci USA. 2001;98(20):11295–11300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore SW, Zhang X, Lynch CD, Sheetz MP. Netrin-1 attracts axons through FAK-dependent mechanotransduction. J Neurosci. 2012;32(34):11574–11585. doi: 10.1523/JNEUROSCI.0999-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Moore SW, Iskratsch T, Sheetz MP. N-WASP–directed actin polymerization activates Cas phosphorylation and lamellipodium spreading. J Cell Sci. 2014;127(Pt 7):1394–1405. doi: 10.1242/jcs.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi I, Vuori K, Liddington RC. The focal adhesion targeting (FAT) region of focal adhesion kinase is a four-helix bundle that binds paxillin. Nat Struct Biol. 2002;9(2):101–106. doi: 10.1038/nsb755. [DOI] [PubMed] [Google Scholar]
- 46.Richardson A, Parsons T. A mechanism for regulation of the adhesion-associated proteintyrosine kinase pp125FAK. Nature. 1996;380(6574):538–540. doi: 10.1038/380538a0. [DOI] [PubMed] [Google Scholar]
- 47.Gervais FG, Thornberry NA, Ruffolo SC, Nicholson DW, Roy S. Caspases cleave focal adhesion kinase during apoptosis to generate a FRNK-like polypeptide. J Biol Chem. 1998;273(27):17102–17108. doi: 10.1074/jbc.273.27.17102. [DOI] [PubMed] [Google Scholar]
- 48.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM. Engineered allosteric activation of kinases in living cells. Nat Biotechnol. 2010;28(7):743–747. doi: 10.1038/nbt.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5(4):749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Maharana S, Wang MD, Shivashankar GV. Super-resolution microscopy reveals decondensed chromatin structure at transcription sites. Sci Rep. 2014;4:4477. doi: 10.1038/srep04477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh YT, et al. Matrix stiffness regulates endothelial cell proliferation through septin 9. PLoS One. 2012;7(10):e46889. doi: 10.1371/journal.pone.0046889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong Y, et al. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366(6456):701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 53.Lee SW, Kim EJ, Um SJ. FHL2 mediates p53-induced transcriptional activation through a direct association with HIPK2. Biochem Biophys Res Commun. 2006;339(4):1056–1062. doi: 10.1016/j.bbrc.2005.11.116. [DOI] [PubMed] [Google Scholar]
- 54.Martin BT, et al. FHL2 regulates cell cycle-dependent and doxorubicin-induced p21Cip1/Waf1 expression in breast cancer cells. Cell Cycle. 2007;6(14):1779–1788. doi: 10.4161/cc.6.14.4448. [DOI] [PubMed] [Google Scholar]
- 55.Sawada Y, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127(5):1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev Cell. 2004;7(5):709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 57.Vogt M, et al. The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J Cell Sci. 2011;124(Pt 6):900–909. doi: 10.1242/jcs.072520. [DOI] [PubMed] [Google Scholar]
- 58.Shibanuma M, Mori K, Kim-Kaneyama JR, Nose K. Involvement of FAK and PTP-PEST in the regulation of redox-sensitive nuclear-cytoplasmic shuttling of a LIM protein, Hic-5. Antioxid Redox Signal. 2005;7(3-4):335–347. doi: 10.1089/ars.2005.7.335. [DOI] [PubMed] [Google Scholar]
- 59.Kim-Kaneyama JR, et al. Uni-axial stretching regulates intracellular localization of Hic-5 expressed in smooth-muscle cells in vivo. J Cell Sci. 2005;118(Pt 5):937–949. doi: 10.1242/jcs.01683. [DOI] [PubMed] [Google Scholar]
- 60.Johannessen M, Møller S, Hansen T, Moens U, Van Ghelue M. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci. 2006;63(3):268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Labalette C, et al. The LIM-only protein FHL2 regulates cyclin D1 expression and cell proliferation. J Biol Chem. 2008;283(22):15201–15208. doi: 10.1074/jbc.M800708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Djupedal I, et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19(19):2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato H, et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309(5733):467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 64.Stellfox ME, Bailey AO, Foltz DR. Putting CENP-A in its place. Cell Mol Life Sci. 2013;70(3):387–406. doi: 10.1007/s00018-012-1048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iskratsch T, et al. FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev Cell. 2013;27(5):545–559. doi: 10.1016/j.devcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev Sci Instrum. 1993;64:1868–1873. [Google Scholar]
- 67.Sneddon IN. The relation between load and penetration in the axisymmetric Boussinesq problem for a punch of arbitrary profile. Int J Eng Sci. 1965;3:47–57. [Google Scholar]
- 68.Yu CH, Law JB, Suryana M, Low HY, Sheetz MP. Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proc Natl Acad Sci USA. 2011;108(51):20585–20590. doi: 10.1073/pnas.1109485108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang G, Xu Y, Chen X, Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2007;26(4):594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]