Significance
Activation and deactivation of protein kinase A (PKA) are triggered by cAMP binding to and unbinding from two tandem domains of the regulatory subunit. Evidence indicates that both binding and unbinding of two cAMPs are ordered. Whereas sequential binding to the inactive holoenzyme complex can be attributed to masking of one binding site by the catalytic subunit, sequential unbinding from the regulatory subunit appears related to unidirectional allosteric communication between the two domains, although the mechanism for unidirectionality has been a mystery. Here, we present a solution through molecular dynamics simulations. One of the two cAMPs acts as a bridge between the domains and thereby gates interdomain communication. Directionality of allostery can facilitate PKA deactivation and may have broad functional importance.
Keywords: unidirectional allostery, cAMP, molecular dynamics, community analysis, NMR spectroscopy
Abstract
The holoenzyme complex of protein kinase A is in an inactive state; activation involves ordered cAMP binding to two tandem domains of the regulatory subunit and release of the catalytic subunit. Deactivation has been less studied, during which the two cAMPs unbind from the regulatory subunit to allow association of the catalytic subunit to reform the holoenzyme complex. Unbinding of the cAMPs appears ordered as indicated by a large difference in unbinding rates from the two sites, but the cause has remained elusive given the structural similarity of the two tandem domains. Even more intriguingly, NMR data show that allosteric communication between the two domains is unidirectional. Here, we present a mechanism for the unidirectionality, developed from extensive molecular dynamics simulations of the tandem domains in different cAMP-bound forms. Disparate responses to cAMP releases from the two sites (A and B) in conformational flexibility and chemical shift perturbation confirmed unidirectional allosteric communication. Community analysis revealed that the A-site cAMP, by forming across-domain interactions, bridges an essential pathway for interdomain communication. The pathway is impaired when this cAMP is removed but remains intact when only the B-site cAMP is removed. Specifically, removal of the A-site cAMP leads to the separation of the two domains, creating room for binding the catalytic subunit. Moreover, the A-site cAMP, by maintaining interdomain coupling, retards the unbinding of the B-site cAMP and stalls an unproductive pathway of cAMP release. Our work expands the perspective on allostery and implicates functional importance for the directionality of allostery.
Protein kinase A (PKA), also known as cAMP-dependent protein kinase, plays an important role in multiple cellular processes by catalyzing protein phosphorylation (1). Its dysregulation is involved in many diseases including cancer and inflammatory disorders (2). The holoenzyme complex, formed by two catalytic (C) subunits bound to a homodimer of regulatory (R) subunits, is inactive. Each regulatory subunit contains two tandem cAMP-binding domains (referred to as CBD-A and CBD-B) (3); in the holoenzyme complex, the A site is masked by the C subunit (4). Activation occurs when two cAMP molecules bind sequentially and cooperatively to the two sites on each R subunit (5). Possible structural changes in the activation process have been constructed from crystallographic studies (4). The first cAMP molecule binds to the B site and makes the A site accessible. Binding of the second cAMP molecule then leads to the release of the C subunit for catalyzing substrate phosphorylation. Deactivation, involving the unbinding of the two cAMP molecules from an R subunit and association of the R and C subunits to reform the holoenzyme complex, has been less studied, and a number of important questions remain open. In particular, kinetic studies have shown that the cAMP unbinding rate from the A site is ∼40-fold faster than from the B site (5), indicating that cAMP unbinding also occurs in a sequential manner (in the order opposite to cAMP binding). Although the masking of the A site by a C subunit provides a simple explanation for why the first cAMP molecule binds to the B site, it is not clear why unbinding should first occur from the A site, given the structural similarities between the two CBDs. Even more intriguingly, NMR studies have presented evidence that allosteric communication between the two cAMP-binding sites is unidirectional (6, 7), but the mechanism is uncertain. The present study aimed to address these questions on deactivation through extensive molecular-dynamics simulations.
In mammalian cells, the PKA R subunit is the primary receptor of cAMP and exists in four isoforms (RIα, RIβ, RIIα, and RIIβ). RIα is the most widely distributed and plays an essential role in maintaining the catalytic subunit under cAMP control (8). In the full-length R subunit, the C-terminal tandem CBD-A and CBD-B are preceded by an N-terminal dimerization/docking (D/D) domain and a flexible linker. The D/D domain is responsible for dimerization and docking to A-kinase anchoring proteins. The linker contains an inhibitory sequence that docks to the active-site cleft of the C subunit in the holoenzyme complex (4), thereby locking in the inactive state. The flexible linker has prevented crystallization of full-length RIα, but crystal structures of the tandem CBDs have been solved in a cAMP-bound form (3, 9–11) and in a C subunit-bound form (4, 12, 13).
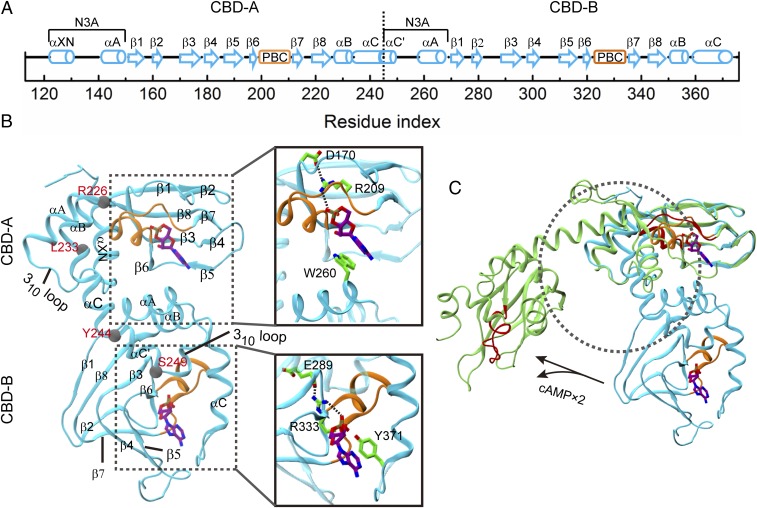
The structures of the tandem CBDs show significant differences between the cAMP-bound form and the C subunit-bound form, especially in the relative positioning of the two domains (Fig. 1). Each CBD comprises an eight-stranded β-barrel (β1–β8) and a sequentially noncontiguous helical subdomain (Fig. 1A). The latter contains a helix–turn–helix (known as N3A) motif preceding and two helices (αB and αC/αC′) following the β-barrel. In the cAMP-bound form, the αC/αC′ helix of CBD-A is bent between residues Y244 and E245 (Fig. 1B), which also serve to define the interdomain boundary. The αC′ portion of CBD-A, denoted as αC′:A (and analogously for other portions in a domain), also doubles as the first helix of N3A:B. The cAMP-binding site is located at the bottom of the β-barrel, with β4–β5 providing the base-binding region (BBR) and the helix-containing loop between β6 and β7 forming a phosphate-binding cassette (PBC). Within the PBC, a conserved arginine (R209 in PBC:A and R333 in PBC:B) forms a hydrogen bond with the phosphate group of cAMP (Fig. 1B, Inset). Mutation of this arginine to lysine significantly impaired cAMP binding at each site (5). The bound cAMP is capped by a π–π stack between the adenine ring and an aromatic residue (W260 at the A site and Y371 at the B site; Fig. 1B, Inset). Note that W260 resides on αA:B; its interaction with cAMP:A stabilizes the interdomain interface. The W260A mutation was apparently sufficient to disrupt the interdomain interface, as indicated by chemical shift changes in cAMP-bound CBD-A similar to those caused by the deletion of CBD-B (14). A main structural change from the cAMP-bound form to the C subunit-bound form occurs in αB–αC/αC′:A, which merge into a single long helix (now referred to as B/C), thereby separating the two CBDs and creating space for the C subunit (Fig. 1C). The C subunit interacts with the B/C helix and PBC:A, thereby masking the A site for cAMP (4). CBD-A is essential for forming the holoenzyme; after deletion of CBD-B, the R subunit retains high affinity for the C subunit (15).
Fig. 1.
Structures of PKA RIα tandem cAMP-binding domains. (A) Secondary structures. The boundary between the two domains is marked by a vertical dash. The helix-containing phosphate-binding cassette (PBC) in each domain is marked by a box in orange. (B) The crystal structure of the two-domain construct bound with two cAMPs (PDB ID code 1RGS). The PBCs are highlighted in orange. The B/C helix (residues R226–S249) linking the two domains is kinked at L233 and bent at Y244. (Inset) Important interactions of the cAMP at each site, including salt bridges involving the phosphate and π–π stacking involving the adenine ring. (C) Structural changes of the two tandem domains when the cAMPs are released and the catalytic subunit is bound. Two crystal structures (1RGS, cyan, and 2QCS, green) are superimposed on the β-barrel of CBD-A. The C subunit in the latter structure is omitted, but its approximate position is indicated by a circle in black dash. The 1RGS PBCs are shown in orange, whereas the 2QCS PBCs are shown in red.
The two CBDs are structurally similar but are well distinguished by cAMP-binding kinetics. The 40-fold difference in unbinding rates between the A and B sites was nearly unaffected by the deletion of the first 91 residues (containing the D/D domain) (5), suggesting that the cause for the different unbinding rates mainly resides in the two tandem CBDs of a single subunit. Moreover, the difference in unbinding rate reduced significantly when the other binding site was silenced by an arginine-to-lysine mutation. Apparently, occupation of one site affected cAMP unbinding at the other site, implicating allosteric communication between the two sites when the difference in unbinding rates was produced. Indeed, a prior study found that cAMP unbinding from the B site was retarded by occupation of the A site, but, curiously, the unbinding from the A site was not affected by occupation of the B site (16). Together these kinetic studies suggest that the two tandem CBDs possess unique allosteric properties during cAMP unbinding that potentially are important for the initiation of PKA deactivation.
Molecular insight into the allosteric communication between the two cAMP-binding sites in RIα N-terminal deletion constructs was provided by recent NMR and hydrogen/deuterium (H/D) exchange studies (6, 7). Gronenborn and coworkers carried out NMR titrations of cAMP and two site-selective analogs (6). Upon ligand binding to the A site alone, some CBD-B residues experienced chemical shift changes that corresponded to conformations intermediate between apo and cAMP-bound forms. In contrast, when the B site was occupied, CBD-A residues except for some on the B/C helix maintained their apo resonances. These contrasting responses were recognized by Gronenborn and coworkers as unidirectional allosteric communication.
A contemporaneous NMR and H/D exchange study (7) reinforced the notion of unidirectional allosteric communication. These authors compared chemical shifts and protection factors of the doubly bound wild-type construct and two single mutants, R209K and R333K, which lead to either cAMP release or much disrupted interactions of cAMPs with the binding sites (thereby partially mimicking cAMP release). The R209K mutation caused significant chemical shift changes in the β2–β3 loop in CBD-A, whereas the R333K mutation caused only minor changes in the corresponding region in CBD-B. A simple interpretation is that, with disruption of the A site, the CBD-A conformations changed, but with disruption of the B site, the CBD-B conformations remained in the cAMP-bound form. This interpretation was corroborated by the H/D exchange data. The R209K mutation induced global reductions in protection factors, both for CBD-A (β2–β3, BBR, PBC, and B/C helix) and for CBD-B (N3A, β2–β3, and αB–αC), consistent with conformational changes of CBD-A and parts of CBD-B toward the apo form. In contrast, loss of protection induced by the R333K mutation was largely limited to the B site (PBC:B and αB–αC:B). Evidently, cAMP binding induced stabilization of CBD-A propagated to CBD-B, but the reverse propagation did not occur. Interestingly, a crystal structure of the two tandem domains with cGMP bound only to the A site (10) superimposed well to the crystal structure of these domains with both sites occupied by cAMP (3): the Cα root-mean-square deviation (RMSD) calculated over both domains except for αB–αC:B is only 1.2 Å.
Computational studies, in particular molecular dynamics simulations, hold enormous potential to complement experimental studies such as NMR spectroscopy in elucidating allosteric mechanisms (17, 18). Previous simulation studies have revealed how unidirectional allosteric communication was achieved in Pin1 (19) and sortase A (20). Several simulation studies have already enriched our knowledge on the conformational properties of RIα, including the conformational space sampled by the tandem CBDs in the apo form (21, 22) and cAMP-induced conformational changes of the isolated CBD-A (23, 24). However, the mechanism of unidirectional allosteric communication between the two cAMP-binding sites has not been addressed and its potential role in PKA deactivation has yet to be explored.
Here, we present a mechanism of the unidirectional allosteric communication developed from extensive molecular dynamics simulations of the tandem CBDs in apo and different cAMP-bound forms. In the simulations, cAMP releases from the two sites elicited disparate responses in conformational flexibility and chemical shift perturbation, confirming unidirectional allosteric communication. Community analysis, which has achieved successes in the study of other allosteric proteins (19, 25–28), revealed that central to interdomain communication is a pathway between β-barrel:A and αA:B. The A-site cAMP, through the across-domain π–π stacking with W260, serves as a bridge between these two regions. Removal of cAMP from the A site leads to the separation of β-barrel:A from αA:B. In contrast, removal of cAMP from the B site keeps the pathway between β-barrel:A and αA:B intact. Correspondingly, the conformational ensemble of the A-site bound form largely overlaps with that of the doubly bound form, whereas the conformational ensemble of the B-site bound form, similar to that of the apo form, is much broader. It also becomes clear that the A-site cAMP, by maintaining interdomain coupling, retards the unbinding of the B-site cAMP, but not vice versa. The resulting faster cAMP unbinding from the A site allows the binding of the C subunit before cAMP unbinding from the B site, leading to efficient PKA deactivation.
Results
We carried out comparative molecular dynamics simulations of the RIα tandem CBDs in three different cAMP-bound forms, that is, both sites occupied (ABbound), only A site occupied (Abound), and only B site occupied (Bbound), as well as the apo form. The starting structure for ABbound was taken from Protein Data Bank (PDB) ID code 1RGS (3), containing RIα residues 113–376 and two cAMPs. For the other systems, one or both cAMPs were stripped. Abound and Bbound were used to model single cAMP release from the B site and the A site, respectively, whereas the apo form was to model complete cAMP release. The systems were simulated in conventional–molecular-dynamics (cMD) runs lasting 150 ns, and three replicate runs were performed for each system. The last 50 ns of each run was used for analysis on conformational flexibility, positional correlation, and community. In addition, we ran accelerated molecular dynamics (aMD) simulations, which have been shown to significantly broaden conformational sampling (26, 29–31). For each system, six replicate aMD runs were performed; each run was 20 ns long, and the last 10 ns was used for calculating chemical shifts and analyzing conformational changes. Last, three replicate cMD runs and six replicate aMD runs were carried out for the W260A mutant of ABbound to further assess the importance of the across-domain π–π stacking.
Nonreciprocal Responses in Conformational Flexibility to cAMP Releases from the Two Sites.
During the 150-ns cMD simulations, the β-barrel structures in both CBDs are well preserved in all of the four wild-type systems (SI Appendix, Fig. S1). Average RMSD values for β-barrel:A and β-barrel:B relative to their counterparts in 1RGS are all below 1.5 Å. The relative positioning of the two CBDs is also well maintained for the three systems with at least one site occupied. The average RMSD values calculated on the whole protein are 1.9 Å for ABbound, 2.5 Å for Abound, and 1.8 Å for Bbound. Deviations are pronounced in the termini, that is, N3A:A and αB–αC:B, and in the β4–β5 loop (part of BBR) of CBD-B. In one of the three replicate runs for Bbound, deviations are also prominent in N3A:B. In contrast, the apo form is much more mobile, with significant structural differences among the replicate runs and an average RMSD value at 4.2 Å. In one apo run, the B/C helix undergoes a large distortion and the whole CBD-B sways away from CBD-A (average RMSD for the whole protein at 6.7 Å), similar to observations from cMD simulations by Gullingsrud et al. (21).
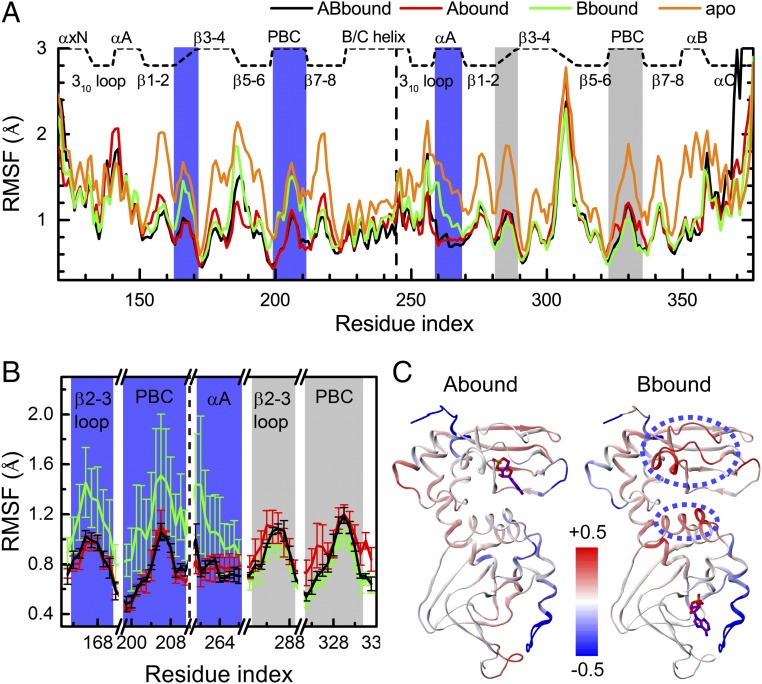
In the absence of dramatic conformational changes, conformational flexibility can provide a sensitive measure of allosteric effects (19). For each system, we calculated the average and standard deviation (SD) of the Cα root-mean-square fluctuations (RMSFs) for each residue over the three replicate runs. For ABbound, the protein core structure remains rigid, with the RMSF averages below 1 Å (Fig. 2A). Higher flexibility occurs only in N3A:A, αB–αC:B, and a few loops. Flexibility is pronounced in the β4–β5 loop of CBD-B, consistent with high B factors of this region in 1RGS (3) and with NMR relaxation data (14). The RMSF SDs for all ABbound residues are below 0.1 Å (Fig. 2B), indicating well-converged results. Hereafter, ABbound is used as the reference in describing the effects of cAMP release.
Fig. 2.
Changes in conformational flexibility upon cAMP release. (A) Cα RMSFs of four systems, each averaged over three replicate cMD runs. (B) RMSF SDs, shown as error bars, for five selected regions in the three cAMP-bound systems. (C) Changes in RMSFs of Abound and Bbound from those of ABbound, displayed on 1RGS as a color scale (red and blue corresponding to higher and lower flexibilities, respectively). For the Bbound form, regions showing significant increases in RMSF are highlighted by ovals in blue dash.
In Abound, CBD-A is as rigid as in ABbound, which is not unexpected because the A site is occupied by cAMP (Fig. 2 A and C, Left). However, CBD-B is also nearly as rigid as in ABbound, indicating as if, upon binding cAMP at the A site, the quenching of picosecond–nanosecond dynamics propagates from CBD-A to CBD-B. The RMSF SDs are also similar to those in ABbound (Fig. 2B). In contrast, in Bbound, the rigidity of the CBD-B core matches that in ABbound, but the RMSF averages are much higher for αA:B and the β2–β3 loop and PBC of CBD-A (Fig. 2 A and C, Right). The RMSF SDs illuminate the contrast in RMSF between CBD-A and CBD-B of Bbound (Fig. 2B). Whereas the SD values for the β2–β3 loop and PBC of CBD-B are as small as in ABbound, the SD values for the counterparts of CBD-A and for αA:B are approximately fivefold higher. The low RMSF SDs of CBD-B serve as an important internal control, and suggest that increased variability in conformational flexibility upon A-site cAMP release is an intrinsic property.
The difference in conformational flexibility is most easily explained by the strategic location of the A-site cAMP, at the interdomain interface to form an across-domain π–π stack (Fig. 1B, Inset). This interaction stabilizes the interface and is likely crucial for maintaining the rigidity of CBD-B in Abound. Upon removal of the A-site cAMP, αA:B loses the across-domain anchor and hence becomes more flexible. In addition, the A-site cAMP phosphate forms a salt bridge with R209 on PBC, which in turn forms a salt bridge with D170 on the β2–β3 loop (Fig. 1B, Inset). Removal of the A-site cAMP disrupts all these interactions and thus leads to increased RMSFs in the corresponding regions of CBD-A. Further removal of cAMP from the B site (as in the apo form) leads to the expected loss of rigidity for the entire protein (Fig. 2A).
Disparate Patterns in Interdomain Communication Between Abound and Bbound.
Community analysis can identify the pattern of motional coupling within a protein, using as input the residue–residue contact map and positional covariance matrix calculated on a cMD trajectory (25). A protein is partitioned into communities, whereby intracommunity contacts are dense but intercommunity contacts are sparse. The strength of intercommunity coupling is determined by the magnitudes of positional correlations within cross-community networks of contacting residues. For each system, community analysis was done separately on the three replicate runs.
First let us summarize the results on the positional covariance matrices, both to demonstrate the convergence of the replicate runs (27) and to highlight the motional correlations (or lack thereof) most responsible for the disparity in community structure among the four systems (see SI Appendix, Additional Results, for details). Among the three replicate runs for each system, the covariance matrices are similar to each other, with the average SD over all residue–residue pairs at only ∼0.1 (SI Appendix, Fig. S2). Importantly, the correlations of N3A:B with N3A:A and β-barrel:A do not differ significantly between ABbound and Abound (SI Appendix, Fig. S3). In contrast, relative to ABbound, in Bbound and apo the correlations of N3A:A with N3A:B increase, whereas the correlations of β-barrel:A with N3A:B decrease, both by 0.4–0.5, or approximately three times the average SDs.
In line with the convergence in the covariance matrices, the community structures, comprising the partitioning of communities and the interdomain couplings, are similar among the three replicate runs of each system, although differing in some details. Below, we present results for the first runs of the four systems, with emphasis on the features that are common among the replicate runs for each system (Fig. 3). Details on the other two runs for each system are given in SI Appendix, Additional Results and Fig. S4.
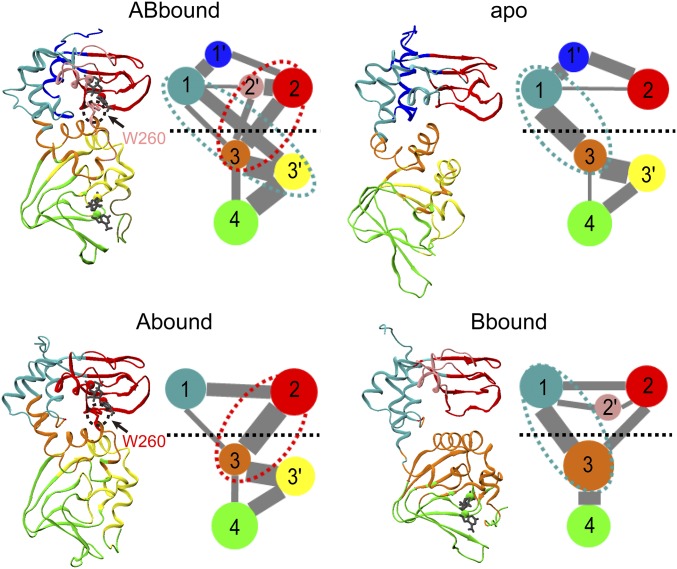
Fig. 3.
Community analysis of the four systems. For each system, the average cMD structure is shown in the left panel, with communities shown in different colors: 1, cyan; 1′, blue; 2, red; 2′, pink; 3, orange; 3′, yellow; and 4, green. In the right panel, communities are represented as colored circles, connected by lines with widths proportional to the cumulative intercommunity betweennesses. Communities with less than 10 residues are not included; neither are betweennesses that are smaller than 10% of the maximum. The horizontal dash separates the two domains. Ovals in cyan or red dash highlight the dominant pathways for interdomain communication.
For each of the four systems, the partitioning of the two tandem CBDs includes four major communities (numbered 1–4), divided first between the domains (1 and 2 versus 3 and 4), and then between the helical subdomain (1 or 3) and the β-barrel (2 or 4) in each domain. Overall, the partitioning is more conserved for the β-barrels than for the helical regions, giving rise to further separation of three minor communities (numbered 1′–3′). The latter comprise αXN:A, PBC:A, and PBC+αB–αC:B, respectively. All of the three minor communities occur in ABbound. In Abound, αC:A moves from community 1 to community 3, whereas 1′ and 2′ merge into communities 1 and 2, respectively. In Bbound, 1′ and 3′ merge into communities 1 and 3, respectively, and the latter also encroaches on the neighboring β-strands. In the apo form, αC′:A moves from community 3 to community 1, and 2′ merges into community 1. To simplify notation, below we combine a major community, for example, 2, and the corresponding minor community (i.e., 2′), if it exists, and denote the combination as 2/2′.
Although the four systems have similar community partitioning, they differ greatly in the intercommunity couplings, especially those across the two domains (highlighted by ovals in red or cyan dash in Fig. 3), implicating differences in interdomain allosteric communication. These differences could have been anticipated from the disparate interdomain correlations, with increased values for pairing N3A:A (community 1) with N3A:B (community 3/3′) but decreased values for pairing β-barrel:A (community 2/2′) with community 3/3′ in Bbound and apo relative to ABbound and Abound. In ABbound, communities 1 and 2/2′ couple to 3/3′ strongly and with nearly equal strengths, suggesting that two pathways, formed between either the helical subdomain or the β-barrel of CBD-A and the helical subdomain of CBD-B, are both effective in interdomain communication.
In Abound, the coupling between communities 1 and 3 is weakened, and the pathway between communities 2 and 3 becomes dominant in interdomain communication. The opposite is true in Bbound. The apo form appears as an extreme version of Bbound, where the coupling between communities 2 and 3 vanishes and the pathway between communities 1 and 3 becomes the only one for interdomain communication. The B/C helix links the two domains and largely accounts for the coupling between communities 1 and 3, thereby providing an intrinsic pathway preserved in all of the four systems for interdomain communication. On the other hand, the second pathway, connecting β-barrel:A and N3A:B, is prominent only when the A site is occupied by cAMP. The latter point is elaborated next.
As noted above, the A-site cAMP stabilizes the interdomain interface through a cross-domain π–π stack with W260. This interaction is so intimate that, in ABbound and Abound, W260, in contrast to the rest of αA:B, belongs to community 2/2′ instead of community 3 (Fig. 3). In this way, the A-site cAMP bridges the pathway between β-barrel:A and αA:B. Through this pathway, allosteric effects can be propagated from the A site to αA:B and PBC+αB–αC:B and finally to β-barrel:B. When the A-site cAMP is released, the domain interface is severely perturbed and the coupling between communities 2/2′ and 3 is significantly weakened. Consequently, allostery communication originating from the B site propagates to the B/C helix but not to β-barrel:A. This nonreciprocality in communication pathways explains why chemical shift changes induced by A-site cAMP binding spread to different regions of CBD-B, but those induced by B-site cAMP binding spill over to the B/C helix but not the rest of CBD-A (6).
Calculated Chemical Shift Perturbations Confirm Unidirectional Allostery.
To further validate our understanding on the interdomain allosteric communication against experimental observations (6, 7), we calculated backbone amide chemical shifts using the SPARTA+ program (32). We turned to aMD because this approach extends conformational sampling from the nanosecond timescale of cMD to beyond microseconds, which is necessary for predicting chemical shifts and other experimental observables (26, 29–31). The calculated chemical shift perturbations (CSPs) of Abound and Bbound relative to ABbound, averaged over six replicate aMD runs, are displayed in Fig. 4. Overall, Bbound shows much higher CSPs than Abound; Bbound has nearly twice as many residues with CSPs above mean plus 1 SD as Abound (35 versus 18). This difference is in qualitative agreement with the NMR data (6, 7), which indicate that, compared with CBD-B, removal of cAMP or disruption of its interactions with the binding site in CBD-A induces higher CSPs. It should be noted that, in ref. 7, CSPs were measured for the R209K and R333K mutants under excess cAMP. Still, because the mutations significantly disrupt the interactions of the ligand with the binding sites, the effectiveness of the ligand is compromised, and hence the mutations partially mimic cAMP removal. This caveat has to be kept in mind when we compare the calculated CSPs for Abound and Bbound with the experimental data for the R333K and R209K mutants, respectively. The CSP comparison is presented in detail in SI Appendix, Additional Results and Figs. S5 and S6, and can be summarized as showing similar patterns both for Abound and R333K and for Bbound and R209K. Below, we elaborate on the calculated CSPs and highlight pertinent experimental results from ref. 7.
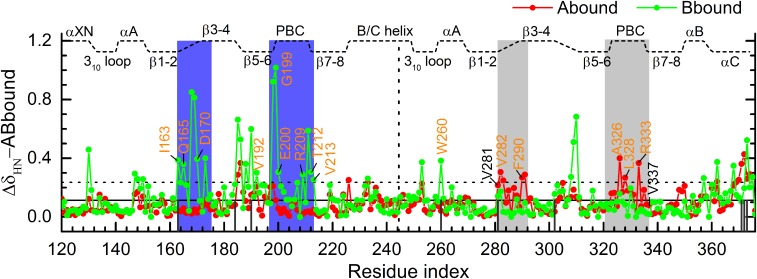
Fig. 4.
Backbone amide chemical shift perturbations (CSPs) of Abound (red) and Bbound (green) relative to ABbound. The mean CSP among the residues of both Abound and Bbound is indicated by the horizontal solid line; the value at 1 SD above the mean is indicated by the dashed line. Regions with significant calculated CSPs are highlighted by blue shading. Residues showing significant observed CSPs (7) are labeled in orange letters. Non-PBC residues within 4.5 Å of the cAMPs in 1RGS are indicated by black vertical lines.
For Abound, higher than mean plus 1 SD CSPs occur in three regions of CBD-B: β2–β3 loop, PBC, and αC C terminus. The latter two regions are in direct contact with the B-site cAMP in ABbound, whereas the β2–β3 loop interacts extensively with PBC, including a salt bridge between E289 and R333, which in turn forms a salt bridge with the cAMP phosphate (Fig. 1B, Inset). The residues that are identified here as showing significant CSPs largely overlap with those identified by Melacini and coworkers (7) for the R333K mutant, including V282 and F290 on the β2–β3 loop and A326, L328, and R333 on PBC (indicated by orange letters in Fig. 4). For Bbound, significant CSPs also occur in the A site and the neighboring β2–β3 loop. Again, the predicted residues largely overlap with those identified by Melacini and coworkers (7) for the R209K mutant. These include I163, Q165, and D170 on the β2–β3 loop; V192 on BBR; G199, E200, R209, T212, and V213 on PBC; and W260 on αA:B (indicated by orange letters in Fig. 4).
An important difference between the two singly bound forms is that regions of CBD-A in Bbound show more pronounced CSPs than the corresponding regions of CBD-B in Abound form, similar to what was found experimentally for the contrast between the R333K and R209K mutants (7). The contrast is most prominent in the β2–β3 loops and PBCs. In particular, I163 (at β2 C terminus) in Bbound has a much higher CSP than the corresponding residue, V281, in Abound; these two residues form van der Walls contacts with R209 and R333, respectively. V213 (at β7 N terminus) and its corresponding residue V337 behave likewise; these two residues form backbone hydrogen bonds with I163 and V281, respectively, between β2 and β7.
It is no coincidence that the regions of Bbound that show pronounced CSPs, including the β2–β3 loop and PBC of CBD-A, along with W260, are also the ones that exhibit high RMSFs (Fig. 2). Together, the two sets of results indicate that the chemical environments of these regions are significantly changed upon removing the A-site cAMP.
β-Barrel of CBD-A Experiences Separation from αA:B upon A-site cAMP Removal.
We now present details of the aMD conformations for the four systems. As expected, conformational sampling with aMD is more expansive than with cMD, with average RMSD values over six replicate aMD runs reaching 2.9, 3.4, 4.6, and 5.8 Å, respectively, for ABbound, Abound, Bbound, and the apo form. For both ABbound and Abound, average RMSDs are below about 4 Å for each of the six replicate runs. In contrast, for Bbound and the apo form, average RMSDs are above 4 Å for at least one-half of the six replicate runs and exceed 8 Å in some runs.
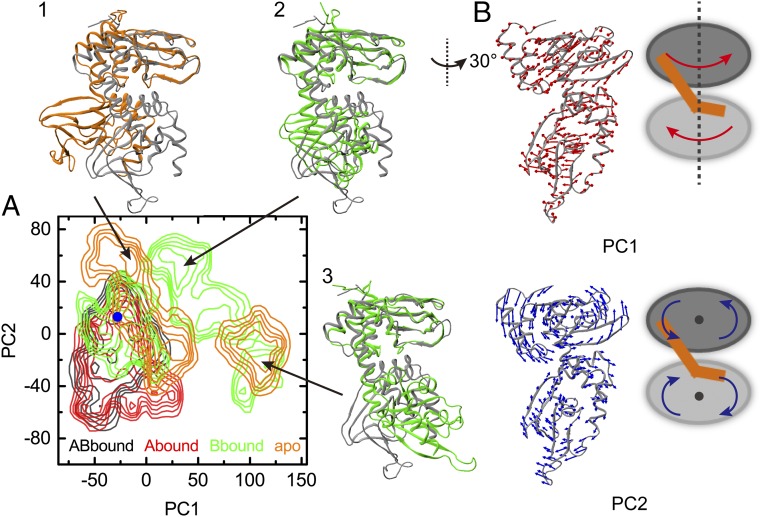
To better characterize the conformational differences among the four systems, we carried out principal component analysis on the snapshots collected from the 24 aMD runs. The resulting probability densities, translated to free-energy surfaces over the first two principal components (PC1 and PC2) according to the Boltzmann relation, are displayed in Fig. 5A. The free-energy basins of ABbound and Abound largely overlap and are centered near the position corresponding to the crystal structure 1RGS. The A-site cAMP is thus sufficient in maintaining the conformations of the whole protein close to those of the doubly bound form. Removal of this cAMP frees the protein to also sample other regions of conformational space, as in the cases for Bbound and the apo form.
Fig. 5.
Conformational ensembles of the four systems in aMD simulations. (A) Free-energy surfaces over the plane of the first two principal components (PC1 and PC2), contoured at 1.5 kcal/mol intervals. PC coordinates of 1RGS are indicated by a blue dot. Three snapshots (in green and orange) from the Bbound and apo simulations are displayed in superposition to 1RGS (in gray). (B) Motions along PC1 and PC2. Arrows on a structure near the origin of the PC plane represent the PC vectors, in a view rotated 30° from that for the snapshots in A; cartoons on the Right illustrate that PC1 can be described as interdomain twist and PC2 as gear grinding.
The motions along PC1 and PC2 are illustrated in Fig. 5B. PC1 represents an interdomain twist around the long principal axis; CBD-A rotates counterclockwise, whereas CBD-B rotates clockwise (top view). PC2 represents interdomain motions similar to the grinding of two gears; the two parallel axes go through the β7 residue A215 on the back of CBD-A and the β6 residue P318 on the front of CBD-B. Motions along PC1 and PC2 both achieve the effect of separating β-barrel:A from αA:B. This separation is illustrated in Fig. 5A by three snapshots from the Bbound and apo aMD runs. The extent of this separation is not as much as in the holoenzyme. It is possible that an initial separation such as suggested by our simulations merely provides an entry point for the C subunit. The full separation occurs only after the C subunit is fully situated into its binding site (Discussion).
As concluded from the cMD simulations, the A-site cAMP stabilizes the interactions between β-barrel:A and αA:B, and thereby bridges an interdomain allosteric pathway. Here, the aMD simulations further show that, upon removal of the A-site cAMP, even while the B-site cAMP is still bound, β-barrel:A and αA:B have the tendency to move away from each other and thereby break a pathway for interdomain communication. The aMD results thus accentuate the nonreciprocality in allosteric pathways between the two cAMP-binding sites.
W260A Mutation Mimics cAMP Release from the A Site.
As emphasized above, the π–π stack between the A-site cAMP and W260 is essential in bridging a pathway for interdomain allosteric communication. We thus expect that, if this interaction is eliminated, as by the W260A mutation, the allosteric pathway should be impaired. NMR experiments of Melacini and coworkers (14) have shown that the W260A mutation produces chemical shift changes in CBD-A regions, including β5–β6 and αC/αC′, that interface with CBD-B. This CSP pattern is similar to that from deleting CBD-B altogether. In addition, the global tumbling of the two domains becomes less coupled. Here, we carried out cMD and aMD simulations to assess how the W260A mutation on ABbound affects interdomain allosteric communication.
Similar to the cMD RMSF result of Bbound, the W260A mutation induces increases in flexibility for the β2–β3 loop and PBC of CBD-A and αA:B while not affecting the flexibility profile of the CBD-B core (Fig. 2A and SI Appendix, Fig. S7A). Again, the RMSF SDs tell the fuller story, which have values for the β2–β3 loop and PBC of CBD-B in the W260A mutant that are as small as those in ABbound, but are up to three times higher for the counterparts of CBD-A and for αA:B in W260A (SI Appendix, Fig. S7B). Much like cAMP removal from the A site (as in Bbound), the W260A mutation induces significant variability in CBD-A conformational flexibility.
Moreover, the aMD runs show that the W260A mutant, like Bbound, samples a wide range of relative positions and orientations between the two domains (SI Appendix, Fig. S7C). This loss in interdomain coupling is consistent with the NMR result of Melacini and coworkers (14) on global tumbling. The calculated chemical shifts, showing significant CSPs for β5–β6 and αC/αC′ in CBD-A (SI Appendix, Fig. S7D), also match the pattern of their NMR results (SI Appendix, Additional Results and Fig. S8). In short, the W260A mutant of ABbound behaves similarly to Bbound in conformational dynamics on both picosecond–nanosecond and longer timescales, and thus eliminating the cross-domain π–π stack largely mimics cAMP release from the A site.
Discussion
We have carried out extensive molecular dynamics simulations to develop a mechanism for the experimentally observed unidirectionality in allosteric communication between the two tandem cAMP-binding domains of the PKA R subunit (6, 7). The simulations confirm unidirectional allostery by showing disparate responses to removing cAMPs from the two domains, both in conformational flexibility on the picosecond–nanosecond timescales and in chemical shifts, which involve longer timescales. Our community analysis has revealed that the unidirectionality arises from nonreciprocal pathways for interdomain communication. Importantly, the pathway between the β-barrel of CBD-A and the αA helix of CBD-B is bridged by the A-site cAMP, via forming a π–π stack with W260. This pathway allows allosteric effects to be propagated from the A site to CBD-B. However, when cAMP is released from the A site, the resulting elimination of the cross-domain π–π stacking leads to significant destabilization of the interdomain interface. The two domains move away from each other and allosteric effects can no longer propagate from the B site to the β-barrel of CBD-A. Consequently, the conformational ensemble of the A-site bound form largely overlaps with that of the doubly bound form, whereas the conformational ensemble of the B-site bound form, similar to that of the apo form, is much broader.
We hope that our computational results will spur additional experimental work on PKA deactivation. In particular, the interdomain allosteric pathways identified here can potentially be verified by chemical shift covariance analyses (33, 34), which yield information on interresidue correlations and can probe allosteric effects mediated by both conformational changes and dynamic changes. Likewise, the predicted disparate effects of site-A and site-B disruptions on conformational ensemble can be tested by chemical shift projection analyses (35, 36).
It is worth remarking that the mechanism for unidirectional allostery in the PKA R subunit bears some resemblance to one developed for Pin1 (19). In the latter system, a substrate peptide can bind both to the catalytic site and to an exosite, on a WW domain (a small domain that folds into a three-stranded β-sheet and contains two conserved tryptophans) connected to the catalytic domain by a flexible linker. The exosite borders an interdomain cleft. Substrate binding to the exosite induces quenching of picosecond–nanosecond flexibility for catalytic-site loops, but substrate binding to the catalytic site has no effect on the flexibility of WW loops. The substrate bound to the exosite was also found to bridge an interdomain pathway and thereby gate the interdomain allosteric communication. Undoubtedly, there are other ways of achieving unidirectional allostery (20).
The mechanism developed here for unidirectional allostery in the two tandem domains of the PKA R subunit can provide explanations for several observations in kinetic studies of cAMP unbinding. There was evidence that cAMP occupation of the A site retarded cAMP unbinding from the B site, but occupation of the B site did not affect the unbinding rate from the A site (16). We now see that the A-site cAMP, by bridging the interdomain coupling, maintains the conformational ensemble and conformational dynamics of CBD-B to those of the bound form even as the B-site cAMP tries to unbind. Therefore, this unbinding is hindered. In contrast, as the A-site cAMP starts to unbind, the interdomain coupling weakens, and the allosteric communication between the two binding sites becomes compromised. Hence the unbinding from the A site takes place at effectively the same rate regardless of whether the B site is occupied. We further suggest that this nonreciprocal response in unbinding to occupation of the opposite site contributes to the 40-fold difference in the cAMP unbinding rates from the two sites (5).
The affinity of the PKA R subunit for cAMP may be so high (KD approximately in the low nanomolar range) that the decrease in cAMP level alone during deactivation may be insufficient to result in cAMP release. Recently, Anand and coworkers (37, 38) reported that phosphodiesterases (PDEs) can bind to and catalyze cAMP release from PKA RIα. The ligands are channeled from the tandem CBDs to the active sites of a PDE dimer and then hydrolyzed. However, the PDE dimer cannot simultaneously bind to the two tandem CBDs without significant changes in their relative orientation and position from those in the doubly bound structure.
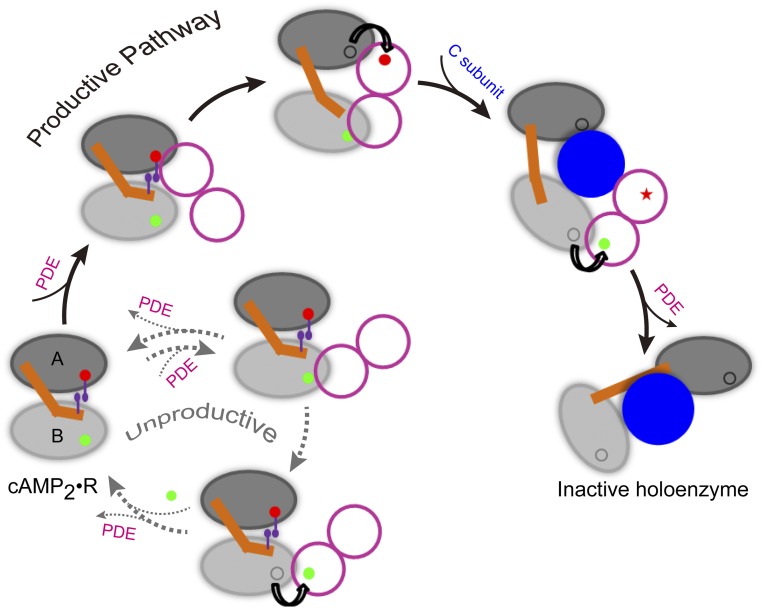
We propose that the unidirectional allostery in PKA RIα produces an elegant solution to the foregoing quandary of RIα:PDE binding and leads to efficient PKA deactivation (Fig. 6). A subunit in the PDE dimer may first contact either of the two CBDs. If the contact is formed with CBD-A (the “productive” pathway), the faster unbinding allows the A-site cAMP to be quickly channeled to the adjacent PDE active site. As suggested by our simulations of Bbound, the B/C helix is then expected to be distorted and the two domains to separate from each other. Consequently, the other subunit in the PDE dimer can now bind to CBD-B. In the meantime, the C subunit enters the space created between the two domains and displaces the PDE dimer as soon as the B-site cAMP completes channeling (37). Finally, the relative orientation and position of the two domains in the R subunit are further changed around the C subunit to form a stable complex.
Fig. 6.
A model of the PKA deactivation pathway. In the doubly bound form, the tandem cAMP-binding domains (labeled as “A” and “B”) of the R subunit are tightly coupled, in particular through the π–π stacking between the A-site cAMP and W260. The dimeric PDE first catalyzes the release of the A-site cAMP, generating the singly bound form in which interdomain coupling is weakened. After the PDE then extracts the B-site cAMP, the C subunit comes in to displace the PDE and forms a stable complex with the R subunit. Note that the unidirectional allostery in the R subunit stalls an unproductive pathway in which the PDE first extracts the B-site cAMP, both through retarding the release of the B-site cAMP and through disallowing the simultaneous binding of the two PDE subunits.
In contrast, if the initial contact of a PDE subunit is formed with CBD-B (the “unproductive” pathway), the retarded unbinding of the B-site cAMP (due to the occupation of the A site) means that, before ligand channeling, the PDE dimer has a high probability of dissociation. In the event that the B-site cAMP is channeled to the first PDE subunit, the interdomain interface would still be largely intact and the second PDE subunit would not be able to bind with CBD-A. The PDE dimer then dissociates, followed by quick refilling of the B site by cAMP from solution and regeneration of the doubly bound form. Hence the unidirectional allostery in PKA RIα forces the unproductive pathway to return quickly to the origin, giving the chance back to the productive pathway to proceed.
Although the full-length R subunit forms a homodimer via the N-terminal D/D domain, no evidence indicates that the two R subunits cooperate during cAMP release. Hence the two subunits may act independently according to the model in Fig. 6. After each R subunit forms a stable complex with a C subunit, the two R subunits may finally come together to produce the inactive holoenzyme tetramer. Further computational and experimental studies will be required to fill in more structural details for and validate the proposed model of PKA deactivation.
During their functional processes, proteins inevitably form higher complexes that contain multiple allosteric binding sites. The directionality of allosteric communication among these sites, either unidirectional or bidirectional, may help control the sequence of events and therefore influence the efficiency and fates of the functional processes. Given that the W260A mutation in PKA RIα breaks a pathway for interdomain communication, it can be easily imagined that some disease-associated mutations may exert their effects by disrupting allosteric pathways and altering the directionality of communication (39).
Materials and Methods
System Preparation.
The PKA RIα tandem cAMP-binding domains with two (ABbound), one at either the A site (Abound) or B site (Bbound), and no cAMP bound (apo) were studied. The initial structure of ABbound was the crystal structure of PKA RIα(113−376) bound with two cAMPs (PDB ID code 1RGS) (3). The initial structures of Abound, Bbound, and the apo form were obtained by removing cAMPs from the B site, A site, and both sites, respectively. For the W260A mutant, the mutation was modeled on 1RGS.
cMD Simulations.
All simulations were carried out with AMBER14 software package (40). The protein force field was AMBER99SB (41). Force field parameters for cAMP were taken from the general AMBER force field (gaff) and parm99 dataset (42), with atomic charges calculated by the R.E.D. server (upjv.q4md-forcefieldtools.org/RED/) while constraining the net charge to −1. Each system was solvated in a truncated octahedron box using TIP3P water (43) with at least 10 Å between the solute and the nearest box boundary. Na+ and Cl− ions were added to neutralize the net charge of the solute and generate a 150 mM salt concentration. The whole system was energy minimized in four steps: solvent minimization, protein backbone constrained, protein Cα atoms constrained, and full minimization. Subsequently, the system was heated from 0 to 300 K over 50 ps with protein backbone constrained and was equilibrated at 300 K with protein Cα atoms constrained for another 50 ps. Finally, the production run lasted 150 ns with a 2-fs time step. Langevin dynamics with a collision frequency of 2 ps and isotropic pressure coupling with a relaxation time of 2 ps were used to keep the system at constant temperature (300 K) and pressure (1 bar). The particle mesh Ewald sum method (44) was used for electrostatic interactions, with a direct space cutoff of 10 Å. Bonds involving hydrogens were constrained using the SHAKE algorithm (45). Three replicate cMD runs were carried out for each system. The last 50 ns of each run was used for analysis, with snapshots saved at every 4 ps.
aMD Simulations.
aMD, developed by McCammon and coworkers (29, 46), accelerates the exchange between low-energy conformational states by raising the energy minima to flatten the potential energy surface. A positive bias potential is added if the instantaneous potential falls below a predefined reference energy. Here, a dual-boost protocol (46) was followed, in which two bias terms were applied to the dihedral potential energy and the total potential energy, respectively. We took the parameters for determining the acceleration level from previous studies (26, 30). Six replicate aMD runs were performed for each system. Each run lasted 20 ns and the second 10 ns was used for analysis, with snapshots saved at every 4 ps. Following these previous studies, aMD snapshots were reweighted by the Boltzmann factors of the averages of the bias potential calculated over time blocks along the trajectory, to remove the effects of the bias introduced in the aMD simulations.
Analysis.
Cα RMSF calculation and principal component analysis were performed using the Ptraj module in AMBER. Each saved snapshot was first aligned to the starting structure using the protein backbone atoms. The average structure was then calculated and used as the reference for RMSF calculation. Community network analysis was performed using the NetworkView plug-in in VMD with default parameters (25). Chemical shifts of backbone H and N nuclei were calculated using SPARTA+ (32) on every other saved aMD snapshot. In calculating both the probability densities in the PC1–PC2 plane and the chemical shifts, aMD reweighting was accounted for. The HN chemical shift perturbation of each residue was reported according to the combination formula (7), where and were the differences of H and N chemical shifts from those of ABbound. All structure figures were prepared in VMD (47).
Supplementary Material
Acknowledgments
We thank Dr. Jian Dai for technical assistance and Dr. Giuseppe Melacini for kindly providing the chemical shift data. This work was supported by National Institutes of Health Grants GM058187 and GM118091.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610142113/-/DCSupplemental.
References
- 1.Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101(8):2381–2411. doi: 10.1021/cr000236l. [DOI] [PubMed] [Google Scholar]
- 2.Roskoski R., Jr A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol Res. 2015;100:1–23. doi: 10.1016/j.phrs.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Su Y, et al. Regulatory subunit of protein kinase A: Structure of deletion mutant with cAMP binding domains. Science. 1995;269(5225):807–813. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- 4.Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130(6):1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 5.Herberg FW, Taylor SS, Dostmann WRG. Active site mutations define the pathway for the cooperative activation of cAMP-dependent protein kinase. Biochemistry. 1996;35(9):2934–2942. doi: 10.1021/bi951647c. [DOI] [PubMed] [Google Scholar]
- 6.Byeon IJL, et al. Allosteric communication between cAMP binding sites in the RI subunit of protein kinase A revealed by NMR. J Biol Chem. 2010;285(18):14062–14070. doi: 10.1074/jbc.M110.106666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNicholl ET, Das R, SilDas S, Taylor SS, Melacini G. Communication between tandem cAMP binding domains in the regulatory subunit of protein kinase A-Ialpha as revealed by domain-silencing mutations. J Biol Chem. 2010;285(20):15523–15537. doi: 10.1074/jbc.M110.105783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amieux PS, McKnight GS. The essential role of RI alpha in the maintenance of regulated PKA activity. Ann N Y Acad Sci. 2002;968:75–95. doi: 10.1111/j.1749-6632.2002.tb04328.x. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Jones JM, Nguyen-Huu X, Ten Eyck LF, Taylor SS. Crystal structures of RIalpha subunit of cyclic adenosine 5′-monophosphate (cAMP)-dependent protein kinase complexed with (Rp)-adenosine 3′,5′-cyclic monophosphothioate and (Sp)-adenosine 3′,5′-cyclic monophosphothioate, the phosphothioate analogues of cAMP. Biochemistry. 2004;43(21):6620–6629. doi: 10.1021/bi0302503. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Brown S, Xuong N-H, Taylor SS. RIalpha subunit of PKA: A cAMP-free structure reveals a hydrophobic capping mechanism for docking cAMP into site B. Structure. 2004;12(6):1057–1065. doi: 10.1016/j.str.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Bruystens JGH, et al. PKA RIα homodimer structure reveals an intermolecular interface with implications for cooperative cAMP binding and Carney complex disease. Structure. 2014;22(1):59–69. doi: 10.1016/j.str.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boettcher AJ, et al. Realizing the allosteric potential of the tetrameric protein kinase A RIα holoenzyme. Structure. 2011;19(2):265–276. doi: 10.1016/j.str.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C, Xuong N-H, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIalpha) subunits of PKA. Science. 2005;307(5710):690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 14.Akimoto M, et al. Mapping the free energy landscape of PKA inhibition and activation: A double-conformational selection model for the tandem cAMP-binding domains of PKA RIα. PLoS Biol. 2015;13(11):e1002305. doi: 10.1371/journal.pbio.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringheim GE, Taylor SS. Dissecting the domain structure of the regulatory subunit of cAMP-dependent protein kinase I and elucidating the role of MgATP. J Biol Chem. 1990;265(9):4800–4808. [PubMed] [Google Scholar]
- 16.Døskeland SO, Ogreid D. Characterization of the interchain and intrachain interactions between the binding sites of the free regulatory moiety of protein kinase I. J Biol Chem. 1984;259(4):2291–2301. [PubMed] [Google Scholar]
- 17.Manley G, Rivalta I, Loria JP. Solution NMR and computational methods for understanding protein allostery. J Phys Chem B. 2013;117(11):3063–3073. doi: 10.1021/jp312576v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo J, Zhou H-X. Protein allostery and conformational dynamics. Chem Rev. 2016;116(11):6503–6515. doi: 10.1021/acs.chemrev.5b00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, Pang X, Zhou HX. Two pathways mediate interdomain allosteric regulation in pin1. Structure. 2015;23(1):237–247. doi: 10.1016/j.str.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang X, Zhou H-X. Disorder-to-order transition of an active-site loop mediates the allosteric activation of sortase A. Biophys J. 2015;109(8):1706–1715. doi: 10.1016/j.bpj.2015.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gullingsrud J, Kim C, Taylor SS, McCammon JA. Dynamic binding of PKA regulatory subunit RI α. Structure. 2006;14(1):141–149. doi: 10.1016/j.str.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 22.Dao KK, et al. The regulatory subunit of PKA-I remains partially structured and undergoes β-aggregation upon thermal denaturation. PLoS One. 2011;6(3):e17602. doi: 10.1371/journal.pone.0017602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vigil D, et al. A simple electrostatic switch important in the activation of type I protein kinase A by cyclic AMP. Protein Sci. 2006;15(1):113–121. doi: 10.1110/ps.051723606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malmstrom RD, Kornev AP, Taylor SS, Amaro RE. Allostery through the computational microscope: cAMP activation of a canonical signalling domain. Nat Commun. 2015;6:7588. doi: 10.1038/ncomms8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc Natl Acad Sci USA. 2009;106(16):6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasper PM, Fuglestad B, Komives EA, Markwick PRL, McCammon JA. Allosteric networks in thrombin distinguish procoagulant vs. anticoagulant activities. Proc Natl Acad Sci USA. 2012;109(52):21216–21222. doi: 10.1073/pnas.1218414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivalta I, et al. Allosteric pathways in imidazole glycerol phosphate synthase. Proc Natl Acad Sci USA. 2012;109(22):E1428–E1436. doi: 10.1073/pnas.1120536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao XQ, et al. Dynamic coupling and allosteric networks in the alpha subunit of heterotrimeric G proteins. J Biol Chem. 2016;291(9):4742–4753. doi: 10.1074/jbc.M115.702605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: A promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120(24):11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 30.Fuglestad B, et al. The dynamic structure of thrombin in solution. Biophys J. 2012;103(1):79–88. doi: 10.1016/j.bpj.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao Y, Nichols SE, McCammon JA. Free energy landscape of G-protein coupled receptors, explored by accelerated molecular dynamics. Phys Chem Chem Phys. 2014;16(14):6398–6406. doi: 10.1039/c3cp53962h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y, Bax A. SPARTA+: A modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48(1):13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc Natl Acad Sci USA. 2011;108(15):6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akimoto M, et al. Signaling through dynamic linkers as revealed by PKA. Proc Natl Acad Sci USA. 2013;110(35):14231–14236. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selvaratnam R, et al. The projection analysis of NMR chemical shifts reveals extended EPAC autoinhibition determinants. Biophys J. 2012;102(3):630–639. doi: 10.1016/j.bpj.2011.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selvaratnam R, Mazhab-Jafari MT, Das R, Melacini G. The auto-inhibitory role of the EPAC hinge helix as mapped by NMR. PLoS One. 2012;7(11):e48707. doi: 10.1371/journal.pone.0048707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moorthy BS, Gao Y, Anand GS. Phosphodiesterases catalyze hydrolysis of cAMP-bound to regulatory subunit of protein kinase A and mediate signal termination. Mol Cell Proteomics. 2011;10(2):M110.002295. doi: 10.1074/mcp.M110.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnamurthy S, et al. Active site coupling in PDE:PKA complexes promotes resetting of mammalian cAMP signaling. Biophys J. 2014;107(6):1426–1440. doi: 10.1016/j.bpj.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nussinov R, Tsai CJ. Allostery in disease and in drug discovery. Cell. 2013;153(2):293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 40.Case DA, et al. 2015. AMBER 2015 (University of California, San Francisco)
- 41.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Homeyer N, Horn AHC, Lanig H, Sticht H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: Phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model. 2006;12(3):281–289. doi: 10.1007/s00894-005-0028-4. [DOI] [PubMed] [Google Scholar]
- 43.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926. [Google Scholar]
- 44.Essmann U, et al. A smooth particle mesh ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 45.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 46.Hamelberg D, de Oliveira CAF, McCammon JA. Sampling of slow diffusive conformational transitions with accelerated molecular dynamics. J Chem Phys. 2007;127(15):155102–155109. doi: 10.1063/1.2789432. [DOI] [PubMed] [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.