Significance
The drug ataluren restores activity to otherwise nonfunctional nonsense alleles, a capability possibly reflecting the insertion of near-cognate aminoacyl tRNAs at premature termination codons during protein synthesis. Because nonsense alleles comprise a significant fraction of all alleles causing inherited disorders, drugs that promote such nonsense codon readthrough have broad therapeutic potential. However, the effectiveness of therapeutic nonsense suppression depends on the nature of the amino acids inserted at each of the three nonsense codons. Here we demonstrate that ataluren does indeed promote insertion of near-cognate tRNAs at nonsense codons, that the latter process yields functional proteins, and that specific codon:anticodon base pairings are critical to this process. These results should enable predictions of better clinical outcomes with therapeutic nonsense suppression.
Keywords: nonsense suppression, readthrough, Translarna, base mispairing
Abstract
A premature termination codon (PTC) in the ORF of an mRNA generally leads to production of a truncated polypeptide, accelerated degradation of the mRNA, and depression of overall mRNA expression. Accordingly, nonsense mutations cause some of the most severe forms of inherited disorders. The small-molecule drug ataluren promotes therapeutic nonsense suppression and has been thought to mediate the insertion of near-cognate tRNAs at PTCs. However, direct evidence for this activity has been lacking. Here, we expressed multiple nonsense mutation reporters in human cells and yeast and identified the amino acids inserted when a PTC occupies the ribosomal A site in control, ataluren-treated, and aminoglycoside-treated cells. We find that ataluren’s likely target is the ribosome and that it produces full-length protein by promoting insertion of near-cognate tRNAs at the site of the nonsense codon without apparent effects on transcription, mRNA processing, mRNA stability, or protein stability. The resulting readthrough proteins retain function and contain amino acid replacements similar to those derived from endogenous readthrough, namely Gln, Lys, or Tyr at UAA or UAG PTCs and Trp, Arg, or Cys at UGA PTCs. These insertion biases arise primarily from mRNA:tRNA mispairing at codon positions 1 and 3 and reflect, in part, the preferred use of certain nonstandard base pairs, e.g., U-G. Ataluren’s retention of similar specificity of near-cognate tRNA insertion as occurs endogenously has important implications for its general use in therapeutic nonsense suppression.
Nonsense mutations are responsible for ∼10–15% of the single-base-pair mutations that cause human disease, with some disease genes having considerably higher nonsense mutation frequencies (1). Patients with genetic disorders attributable to nonsense mutations tend to have more serious ramifications than those with missense mutations, presumably because of marked reductions in specific gene expression. Because all diseases caused by nonsense mutations share the same key gene expression problems, namely premature translation termination and accelerated mRNA decay, therapeutic approaches to these problems are being investigated, with the understanding that a single drug has the potential to treat a large number of different disorders (1, 2). Normally, the in-frame UAG, UAA, and UGA codons that arise in mRNA from nonsense mutations serve as signals for translation termination, but this role can be functionally overridden by alterations in the translation machinery or by the presence of certain small molecules that compromise the fidelity of translation termination (1, 2). Such nonsense suppression, or nonsense codon readthrough, by a ribosome engaged in peptide elongation has been predominantly observed in the context of premature termination codons (PTCs) and is thought to reflect ribosomal A site acceptance of a near-cognate tRNA and subsequent incorporation of an amino acid into the nascent polypeptide at the position of the PTC (1, 2).
The small-molecule drug ataluren (3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]-benzoic acid; also known as Translarna or PTC124) is being evaluated both preclinically and clinically for its ability to promote therapeutic nonsense suppression (1, 3). To date, ataluren has been shown to restore function to more than 20 different disease-specific or reporter nonsense alleles in systems ranging in complexity from in vitro translation to cell culture to mouse models and human patients (1, 3–14). In the latter, Translarna clinical trials have demonstrated restoration of full-length functional protein in Duchenne muscular dystrophy and cystic fibrosis nonsense mutation patients (10, 11, 13, 15, 16). However, a lack of activity has been reported for some cell systems (17, 18).
Here, we have investigated the mechanistic basis of PTC readthrough induced by ataluren in human and yeast cells and compared it to the activity of two other readthrough-promoting compounds, the aminoglycosides G418 and gentamicin. Using multiple reporters and a physiologically relevant CFTR (cystic fibrosis transmembrane conductance regulator) nonsense allele, we have determined the nature of the translation events at PTCs when cells are treated with these drugs. Our study provides comprehensive characterization of the products of PTC readthrough and elucidates the nature of the nonstandard codon:anticodon base pairings that are favored under physiological conditions in the cell. These proof-of-concept experiments establish unambiguously that these compounds promote insertion of near-cognate tRNAs at PTCs and provide insight into the likelihood of functionality for the products of therapeutic nonsense suppression.
Results
Ataluren Promotes Nonsense Suppression in Mammalian Cells and Yeast.
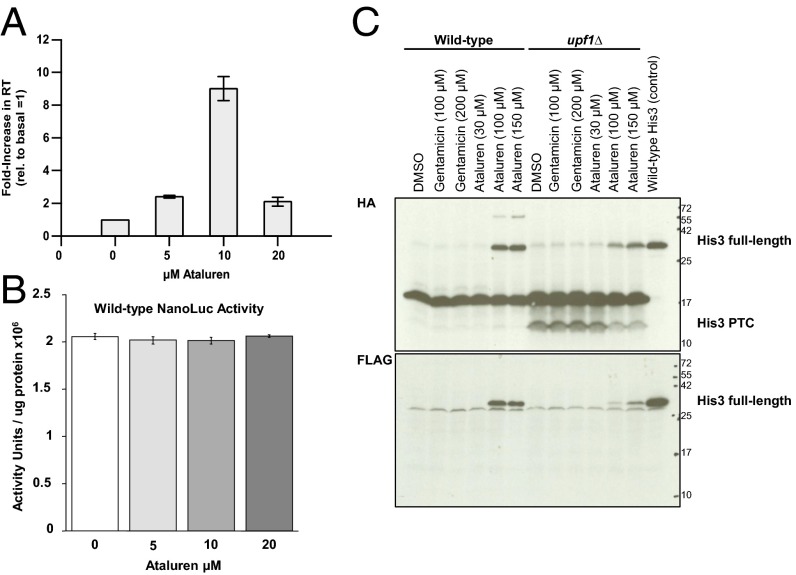
Six different PTC reporter genes, including some with multiple alleles, were used to monitor different aspects of nonsense codon readthrough in human cells and yeast (SI Appendix, Fig. S1). PTC-containing transcripts arising from selected reporters are substrates for nonsense-mediated mRNA decay (SI Appendix, Fig. S2). Ataluren treatment of mammalian 293H cells stably transfected with wild-type (WT) or PTC(W12X)-containing NanoLuc (19) reporters promoted a bell-shaped dose–response, with a peak ninefold increase in luciferase activity derived from the PTC reporter at 10 µM (Fig. 1). This result was attributable to readthrough of the reporter mRNA’s premature UGA codon, and not protein stabilization (18), because ataluren treatment of 293H cells stably expressing WT NanoLuc had no effect on NanoLuc activity (Fig. 1B). In SI Appendix, Fig. S3 A and B demonstrate that ataluren (and G418) also promote readthrough of different p53 UGA alleles in Calu6 and HDQ cells and of a UGA PTC in an H2B-GFP reporter (6) expressed in 293H cells.
Fig. 1.
Ataluren treatment of mammalian or yeast cells increases PTC readthrough. Dose–response of ataluren treatment in 293H cells stably transfected with (A) PTC(UGAW12X) or (B) wild-type (WT) NanoLuc reporters. The 293H cells stably expressing the NanoLuc reporters were treated with ataluren at the indicated concentrations. The data are expressed as the mean ± SD of the NanoLuc activity units normalized to total protein. (C) Western analyses of yeast cells showing readthrough products expressed from HA-HIS3(UAA100)-SF reporters after ataluren or gentamicin treatment. Upper and Lower blots, respectively, were probed for the HA (N-terminal) and FLAG (C-terminal) epitopes. The predominant band at ∼17 kDa in the HA blot is an HA artifact detected only in cells expressing HIS3(UAA100)-SF, but not HIS3(WT)-SF.
Yeast WT [PSI−] cells devoid of the prion form of Sup35 (to ensure that the readthrough observed was solely attributable to drug treatment) showed ataluren-dependent increases in readthrough from the HA-HIS3(UAA100)-SF or HA-LUC(UGA20)-SF reporter mRNAs without concomitant changes in mRNA levels albeit at higher concentrations than used for mammalian cells, a difference probably attributable to the yeast cell membrane (Fig. 1C; SI Appendix, Figs. S2 A and B and S4). This indicated that ataluren treatment directly affected reporter protein abundance, not alterations in transcription, mRNA processing, or mRNA stability. In cycloheximide chase assays, ataluren treatment also did not change the half-life of the full-length His3 readthrough protein, indicating that the drug affects PTC readthrough per se (SI Appendix, Fig. S5). Consistent with this conclusion, the prematurely terminated His3 polypeptide fragment that is detectable only by its N-terminal tag in upf1Δ cells (20) decreases in abundance as synthesis of the full-length His3 product increases (Fig. 1C, Top; see “His3 PTC” in DMSO and ataluren lanes in upf1Δ cells).
Near-Cognate tRNAs Facilitate Ataluren-Mediated Readthrough.
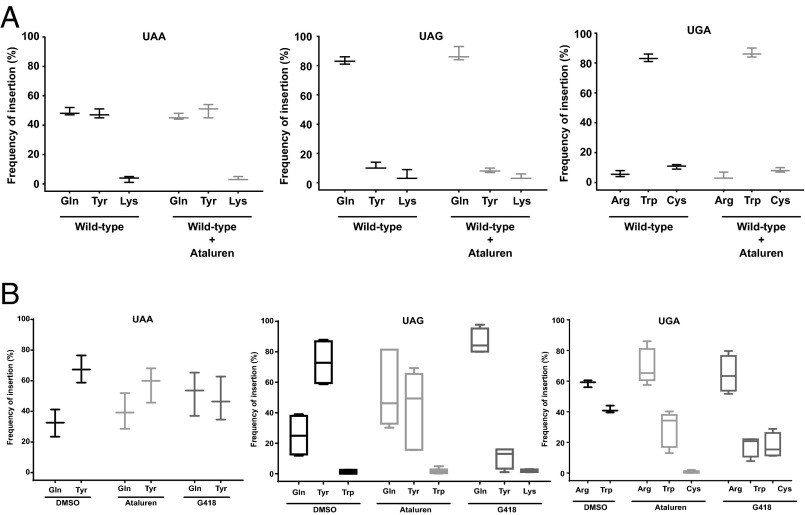
We identified the amino acids inserted at PTCs in drug-treated cells using a firefly luciferase (LUC) reporter system that yields highly purified readthrough products and well-resolved peptides suitable for mass spectrometry and amino acid sequence determination (SI Appendix, Fig. S1C) (21). It has been suggested that ataluren is an inhibitor of firefly luciferase and that its reported nonsense suppression activity simply reflects stabilization of the luciferase enzyme (18, 22). We consider this claim to be spurious because (i) ataluren-mediated readthrough has been observed in numerous other systems (Fig. 1 and SI Appendix, Fig. S3) (1–14, 23); (ii) the effects on luciferase activity could not be independently replicated by others (24) or by us (SI Appendix, Fig. S3C); (iii) ataluren’s putative luciferase inhibitory activity depends on a specific enzyme substrate (25); and (iv) most of the hypothetical inhibitory molecule, PTC124-AMP (22), is rapidly converted to the active readthrough molecule PTC124 (ataluren) under conditions of in vitro translation (SI Appendix, Fig. S6). Accordingly, three of the HA-LUC(PTC20)-SF reporters were separately expressed in yeast WT [PSI−] cells. Strep-Tactin purification of the respective readthrough products synthesized in the absence or presence of ataluren was monitored by gel electrophoresis to resolve full-length proteins (SI Appendix, Fig. S7). Following silver staining of the gel, the band corresponding to full-length luciferase protein was excised and subjected to endo-LysC digestion and LC-MS/MS analysis, and amino acid insertion frequencies were determined as described previously (21). Analysis of the readthrough products purified from untreated cells showed the incorporation of Gln, Lys, and Tyr at UAA and UAG and insertion of Trp, Arg, and Cys at UGA, all of which were similar to results reported previously for endogenous background readthrough (Fig. 2A) (21, 26). Readthrough products purified from cells treated with ataluren showed that Gln, Lys, and Tyr were also inserted when UAA was the PTC (Fig. 2A). Three independent experiments revealed that the frequencies of insertion of Tyr and Gln were comparable (51.5 ± 2% and 46.4 ± 2%, respectively) and that Lys was inserted at a much lower frequency (2 ± 1.7%) (Fig. 2A). Readthrough from UAG resulted in the insertion of the same sets of amino acids, but with different frequencies. Gln insertion predominated, with a frequency of 88 ± 3.6%, whereas Tyr (8.6 ± 2%) and Lys (3.3 ± 1.5%) had somewhat similar insertion frequencies (Fig. 2A). With UGA as the PTC, ataluren treatment resulted in the incorporation of Trp, Arg, and Cys, with Trp insertions occurring at a much higher frequency (86.3 ± 3%) than those of Arg (4.6 ± 2%) or Cys (9 ± 2%) (Fig. 2A). A comparison of the endogenous readthrough events with those mediated by ataluren indicates that there are no major differences in the respective amino acid insertion frequencies in yeast cells.
Fig. 2.
Ataluren-mediated decoding of nonsense codons. Comparison of amino acid insertion at PTCs during termination readthrough of HA-LUC(PTC20)-SF reporters after ataluren treatment in yeast and 293H cells. Epitope-tagged luciferase was purified from the respective cells and subjected to mass spectrometry analyses. Box and whisker graphs depict the amino acids inserted for each nonsense codon. Horizontal lines are the means and vertical lines are the SD. (A) Data from WT [PSI−] yeast cells treated with 30 µM ataluren (n = 3). (B) Data from 293H cells treated with 30 µM ataluren. Boxes represent the range of at least four determinations. Where there are no boxes, three determinations were done. Data used to generate graphs in B are in SI Appendix, Table S6.
In contrast to the results obtained when readthrough was promoted by ataluren, treatment of [PSI−] yeast cells with 7.5 µM G418 showed a significantly different amino acid insertion profile for two (UAG and UGA) of the three PTCs (SI Appendix, Table S1). With UAA as the PTC, the average of two independent experiments showed that the amino acid insertion pattern was similar to all other readthrough-inducing conditions with relatively comparable insertion frequencies of Gln (46.1%) and Tyr (51.8%) and a low insertion rate for Lys (1.9%) (Fig. 2A; SI Appendix, Table S1). With UGA as the PTC, insertion of the same amino acids (Trp, Arg, and Cys) was observed, but, compared with either endogenous readthrough events or ataluren treatment, significantly higher levels of Cys insertion (27.2%) were observed (Fig. 2A; SI Appendix, Table S1). Surprisingly, the amino acid insertion pattern at UAG was atypical after G418 treatment, with identification of 12 different amino acids at the PTC, some of which were inserted at very low levels (SI Appendix, Table S1). Lys was the predominant insertion at UAG with a frequency of 90% (SI Appendix, Table S1). Gln and Tyr, the other two amino acids that were normally inserted with much higher frequencies under all other conditions, were detected after G418 treatment, but with much lower frequencies of 4.7% and 1.5%, respectively (SI Appendix, Table S1). Treatment of cells with a higher concentration of G418 (15 µM) resulted in insertion of various amino acids at all three PTCs (SI Appendix, Table S2).
We next tested whether ataluren promoted insertion of similar sets of near-cognate tRNAs when HA-LUC(PTC20)-SF alleles were expressed in human cells. As expected, 293H cells stably expressing HA-LUC(PTC20)-SF alleles manifested increases in luciferase protein and activity after incubating the cells with ataluren or G418, and for all combinations other than UAA and ataluren these increases were clearly dependent upon the presence of a PTC in the luciferase allele (SI Appendix, Fig. S3 C and D). Luciferase readthrough products were purified and subjected to LC-MS/MS analyses as described above. Endogenous readthrough products purified from cells treated with 0.02% DMSO showed incorporation of two amino acids when UAA was the PTC: Gln (32.4 ± 8.9%) and Tyr (67.5 ± 8.9%) (Fig. 2B). When UAG was the PTC, Tyr (73.1 + 15%), Gln (25.2 ± 13.7%), and Trp (1.3 ± 1.4%) were inserted (Fig. 2B). Surprisingly, the decoding of UAG by Trp-tRNA is indicative of mispairing at the second position of the nonsense codon (SI Appendix, Table S3), a phenomenon not seen with yeast reporters or with the other two nonsense codons in these cells. Analysis of UGA endogenous readthrough products revealed insertion of Trp (41.4 ± 2.3%) and Arg (58.6 ± 2.3%) (Fig. 2B).
Characterization of readthrough products from 293H cells treated with ataluren (30 µM) showed insertion of Tyr (57.9 ± 11.3%) and Gln (39.9 ± 11.7%) at UAA; insertion of Tyr (43.9 ± 20.9%), Gln (53.2 ± 20.3%), and Trp (1.8 ± 1.9%) at UAG; and insertion of Arg (69.7 ± 11.3%), Trp (28.8 ± 11.4%), and Cys (0.7 ± 0.7%) at UGA (Fig. 2B). For UAA and UAG, the ataluren-induced readthrough products were similar to the endogenous readthrough products in identity and relative insertion frequencies for the respective amino acids. UGA readthrough products from ataluren-treated cells showed a low but consistent frequency of Cys insertion that was not observed in the endogenous readthrough products. Treatment of human cells with another small-molecule readthrough effector, G418 (150 µM), resulted in the insertion of Tyr (47.9 ± 14.1%) and Gln (52 ± 14.2%) at UAA; Gln (86.5 ± 8.3%), Tyr (10.8 ± 7.0%), and Lys (2.0 ± 0.8%) at UAG; and Arg (64.5 ± 11.8%), Trp (17.9+6.8%), and Cys (17.7 ± 8.0%) at UGA (Fig. 2B). Thus, G418 treatment yielded a pattern and frequency of amino acid insertion that differed significantly from endogenous readthrough, with the greatest difference at the UAG PTC, followed by UGA, and then UAA. Comparisons to endogenous readthrough indicated that G418 altered PTC decoding in both yeast and human cells and in part may reflect G418’s interference with the decoding process (1, 27). This was found to be true for another aminoglycoside readthrough effector, gentamicin, in both yeast (21) and human cells (SI Appendix, Table S4).
Readthrough Products Are Functional.
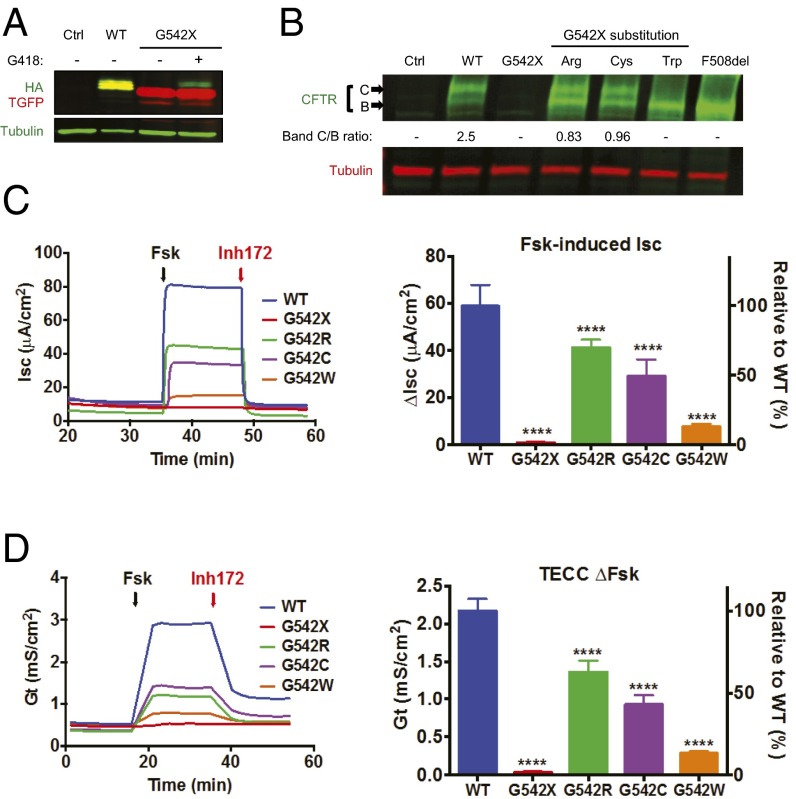
We used three approaches to determine whether the readthrough products synthesized after ataluren or aminoglycoside treatment result in functional proteins. First, the growth of yeast cells harboring the HA-HIS3(UAA100)-SF allele was monitored when media lacking histidine was or was not supplemented with readthrough-promoting drugs. In SI Appendix, Fig. S8A shows that cell growth was restored after ataluren (or gentamicin) treatment, demonstrating that the amino acids inserted at the HA-HIS3(UAA100)-SF PTC result in functional readthrough products. Second, we assessed the functionality of ataluren-induced luciferase readthrough products by replacing the WT glycine residue at position 20 with the six different amino acids identified in the yeast-based mass spectrometry analyses of Fig. 2A. These comparisons demonstrated comparable activity from all position 20 substitutions (SI Appendix, Fig. S8B), indicating that, at least with respect to luciferase position 20, enzymatic activity is unlikely to be affected by ataluren- or G418-mediated readthrough. Finally, we determined whether protein produced via readthrough therapy from a disease-causing nonsense allele could also retain function. Recognizing that G542X is the most frequent CFTR nonsense mutation (28), we used that allele to test this possibility. CFTR is a large, hydrophobic protein of generally low abundance, the expression of which is normally limited to epithelial cells. Because previous studies have shown that the local sequence context around a PTC acts as a primary determinant of readthrough efficiency (29, 30), we inserted the CFTR-G542X mutation (TGA) and its local sequence context (three codons on either side) into a construct amenable to protein purification from HEK293 cells and mass spectrometry. This construct contained TGFP, the CFTR-G542X mutation and context, and C-terminal HA and 6× His tags (SI Appendix, Fig. S1D). After PTC suppression, the full-length protein will contain TGFP, along with the HA and His tags, whereas the prematurely terminated protein will contain TGFP only. We observed an increase of full-length protein production after G418 (150 µM) treatment of HEK293 cells harboring this reporter (Fig. 3A) and then identified the amino acids inserted at the PTC of the TGFP-CFTR-542X mRNA with G418 treatment. Consistent with the data obtained from LUC reporters in yeast and 293H cells, the same three amino acids (Arg, Cys, and Trp) were identified in this UGA suppression reporter system, but with different frequencies (SI Appendix, Fig. S9 and Table S5).
Fig. 3.
Characterization of the amino acid insertions at human CFTR-G542X. (A) Western blot showing an increased amount of full-length protein (containing both TGFP and HA tag) (yellow-green bands) compared to the TGFP protein lacking the HA tag (red bands) after G418 treatment in HEK293 cells transiently transfected with the G542X construct. (B) Western blot showing the level of CFTR maturation observed in HEK293 cells transiently transfected with CFTR G542 variants. “Band B” is the ER form of CFTR, whereas “Band C” is the mature form. CFTR-F508del is a mutant form of CFTR that is retained in the ER and consequently only produces Band B. (C) Representative tracings (Left) and quantitation results (Right) of short-circuit current (Isc) measurements of CFTR chloride channel activity in FRT epithelial cell monolayers stably expressing the CFTR G542 variant constructs. (D) Representative tracings (Left) and quantitation (Right) of Conductance (Gt) measurements of CFTR chloride channel activity in FRT monolayers stably expressing the CFTR G542 variant constructs.
We then determined if these amino acid substitutions affected CFTR function. Site-directed mutagenesis was used to change the position 542 Gly codon to Arg, Cys, or Trp codons in a full-length cDNA. Transient transfections were then used to introduce each construct into HEK293 cells, and the core glycosylated (band B) and mature (band C) forms of CFTR were visualized by Western blotting (Fig. 3B). We found that the cell lines expressing the G542R and G542C forms of CFTR exhibited less maturation of band B to band C than WT CFTR, whereas the G542W cell line did not exhibit any band C. These results suggest that these three CFTR variants at position G542 either cause a partial defect in maturation of the endoplasmic reticulum (ER) form of CFTR to the mature cell-surface form or reduce the stability of the mature form of CFTR at the cell surface (Fig. 3B). To address these potential defects at the functional level, we constructed stable cell lines by transfecting the WT CFTR, CFTR-G542X, G542R, G542C, or G542W constructs into Fischer rat thyroid (FRT) cells, a cell type for which the conditions required for cell growth, PTC readthrough, and electrophysiological assays were compatible with G418 treatment. Using these stable FRT cell lines grown to confluence on filters, we performed short-circuit current (Isc) and transepithelial chloride conductance (Gt) assays to measure CFTR function. Forskolin was used to activate CFTR, and the CFTR inhibitor CFTRInh-172 was used to confirm the specificity of CFTR function. To correct for any differences in CFTR expression, these measurements were then normalized to the CFTR mRNA level measured in each stable cell line by qPCR analysis. As expected, all three G542 substitutions displayed lower CFTR activity than WT (Fig. 3 C and D). G542R and G542C showed ∼40–70% of WT Isc, which correlates well with less band C (Fig. 3B). We measured a corrected CFTR activity of only ∼10% for G542W, consistent with little or no band C observed in the Western blot. Taken together, these results demonstrate that CFTR variants resulting from readthrough retain variable but significant functionality.
Rules of Near-Cognate tRNA Insertion.
With the exception of some aspects of G418-mediated nonsense suppression treatment, which may be atypical (see above), a comparison of the amino acids inserted at PTCs under different readthrough-inducing conditions, as well as with different reporters, shows considerable similarity in yeast and mammalian cells. In both organisms, Gln, Lys, and Tyr are inserted at UAA and UAG, and Trp, Arg, and Cys are inserted at UGA (Fig. 2; SI Appendix, Tables S1, S2, S4, and S6) (21, 26). Our analyses of readthrough products indicate that such biases arise, in part, because some nonstandard codon:anticodon base pairs are preferred over others (SI Appendix, Table S3). For example, in all instances where individual tRNA choices can be identified, position 1 U-G mispairing appears to be preferred over U-U (Fig. 2; SI Appendix, Tables S1, S2, S4, and S6) (21). Furthermore, the lack of Gly insertion at UGA and Gln insertion at UAA or UAG (with the exception of G418 treatment) suggests that U-C mispairing at position 1 of the codon is not readily accommodated. Given that the decoding center of the ribosome is geometrically constrained (31, 32), the structure of the base pairs might be key determinants of mispairing. Alternatively, the relative abundance of tRNAs may govern insertion frequencies. To assess whether favored geometry of certain base pairs or tRNA abundance is the key parameter for this bias, we examined tRNA utilization in UGA readthrough. When UGA is the PTC, yeast cells show preferential insertion of Trp whereas 293H cells prefer insertion of Arg (Fig. 2 A and B). Arg insertion at UGA can be mediated by two near-cognate tRNAs: tRNA-Arg-UCG (using U-G mispairing at position 1 of the codon) and tRNA-Arg-UCU (using U-U mispairing at position 1 of the codon) (SI Appendix, Table S3). In yeast, Arg insertion at UGA most likely occurs by mispairing of tRNA-Arg-UCU, as tRNA-Arg-UCG expression has not been observed in these cells (33). On the other hand, tRNA-Arg-UCG is expressed in mammalian cells (33), and their insertion of Arg could be mediated via either of the Arg near-cognate tRNAs. We introduced and overexpressed a recombinant tRNA-Arg-UCG in WT [PSI−] yeast cells and monitored the insertion of Arg in the readthrough products. As a control, we overexpressed the endogenous tRNA-Arg-UCU. Northern analyses of total RNA isolated from the cells overexpressing the corresponding tRNA-Arg plasmids show considerable and comparable overexpression of the respective tRNA-Arg species (SI Appendix, Fig. S10A). A comparison of the luciferase activity from these strains showed that cells expressing tRNA-Arg-UCG also showed increased luciferase activity (SI Appendix, Fig. S10B), indicating that the expression of this specific tRNA correlated with enhanced readthrough efficiency. Moreover, expression of the recombinant tRNA-Arg-UCG shifted the bias toward Arg insertion at a frequency of 89.3 ± 1.5%, in contrast to a frequency of 6.6 ± 3% in WT cells (SI Appendix, Fig. S10C). In comparison, overexpression of the endogenous tRNA-Arg-UCU in yeast cells increased the frequency of Arg insertion to 30.6 ± 4.7% (SI Appendix, Fig. S10C). These observations are consistent with the notion that U-G codon:anticodon mispairing is favored over U-U mispairing in position 1.
We also tested whether a U-C codon:anticodon mispairing event can be introduced at position 1 by overexpressing the corresponding near-cognate tRNA, tRNA-Glu-UUC. Even though strains overexpressing tRNA-Glu-UUC showed increased levels of the tRNA species compared with WT (SI Appendix, Fig. S10D), the readthrough efficiency, as monitored by luciferase activity, was not altered (SI Appendix, Fig. S10E), and the readthrough products from those cells showed insertion of the same three amino acids; i.e., Glu insertion was still not observed (SI Appendix, Fig. S10F). Taken together, these data suggest that some nonstandard base pairs are favored over others for near-cognate tRNA insertion at PTCs and that the geometry of the base pairs is the most likely determinant of this bias.
The Ribosome as a Likely Target of Ataluren.
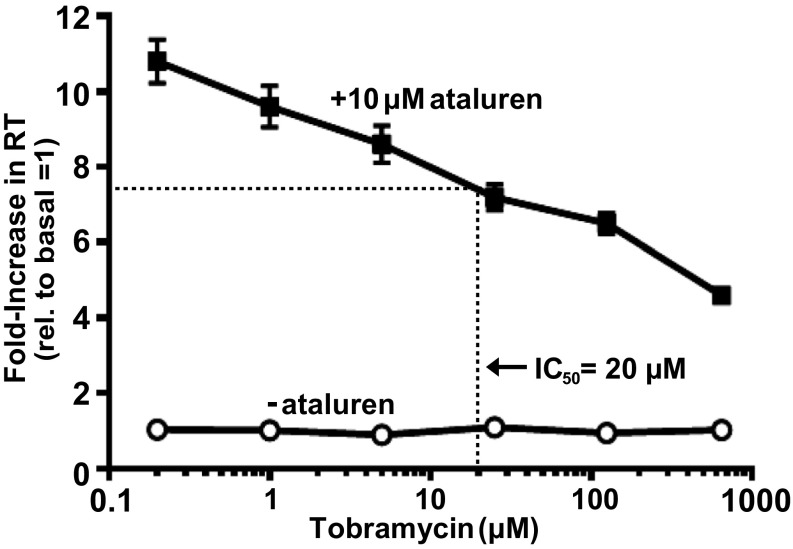
In a recent clinical trial (12), cystic fibrosis patients with nonsense mutations failed to show a significant response to ataluren if they were concurrently being treated with inhaled tobramycin, an aminoglycoside with strong affinity for the ribosomal A site (34). This observation suggested that tobramycin might be an inhibitor of ataluren’s nonsense suppression activity, a possibility considered in the experiments of Fig. 4. In HEK293 cells, stably expressing the W12X NanoLuc reporter treatment with a wide range of tobramycin concentrations failed to elicit any detectable readthrough activity of the drug (Fig. 4). However, in cells manifesting substantial ataluren-mediated readthrough of the PTC-containing NanoLuc mRNA, addition of tobramycin showed strong, dose-dependent reduction of ataluren’s readthrough activity (Fig. 4), consistent with previous work showing that aminoglycosides reduce the ability of ataluren to promote readthrough but nonaminoglycoside antibiotics have no effect (12). Quantitation of this effect indicated that tobramycin inhibition of ataluren’s activity had an IC50 of 20 µM (Fig. 4). These results not only provide insight into the results of the aforementioned clinical trial, but also suggest that ataluren, like tobramycin, might also bind to the ribosome and thus affect near-cognate tRNA selection during readthrough.
Fig. 4.
Tobramycin is a potent inhibitor of ataluren-mediated readthrough. The effect of tobramycin on NanoLuc activity in HEK293 cells expressing a UGA (W12X) NanoLuc reporter with either no prior treatment or following treatment with 10 µM ataluren for 48 hours. Initial stimulation of readthrough by ataluren in the absence of tobramycin was 15-fold above background.
Discussion
Therapeutic nonsense suppression is a potentially powerful approach to the treatment of the large number of genetic disorders caused by nonsense mutations, and several small-molecule drugs are being investigated for their capability to engender such therapy (1, 2). Multiple experimental approaches with cell lines, patient cells, animal models, and genetic disorder patients have demonstrated that the drug ataluren can restore expression to genes and mRNAs otherwise inactive because of the presence of premature nonsense codons (1, 3–14, 23). Collectively, such experiments have strongly suggested that ataluren promotes nonsense suppression, i.e., the insertion of near-cognate tRNAs at PTCs, but direct evidence for this activity has been lacking. Here, we determined the amino acid sequences of full-length LUC and chimeric CFTR proteins accumulating in control and readthrough effector-treated yeast and human cells, an approach that leads us to conclude that ataluren does indeed stimulate the incorporation of near-cognate tRNAs at PTCs. Notably, our analysis of the products of ataluren-induced PTC readthrough demonstrates that a distinct subset of amino acids is inserted during nonsense suppression of reporter and disease-related mRNAs in human cells. These results, the observation that the amino acid insertions occurring as a consequence of ataluren treatment are similar but not identical to those occurring endogenously (Fig. 2), and the finding that tobramycin specifically antagonizes the restoration of NanoLuc activity from a PTC allele by ataluren (Fig. 4) negate other hypotheses that sought to explain ataluren’s mechanism of action (17, 18).
Ataluren’s enhancement of near-cognate tRNA insertion favors a subset of tRNAs, generally leading to incorporation of Gln, Lys, and Tyr at UAA and UAG codons and of Trp, Arg, and Cys at UGA codons. The weak readthrough activity seen here with the HA-LUC(UAA20)-SF allele is atypical of that seen with UAA PTCs in other mRNAs (3, 4, 14) and most likely reflects the influence of codon context on readthrough (29). Ataluren’s influence over the extent of specific tRNA selection implies that this drug’s target could be the ribosome, a conclusion consistent with the drug’s markedly diminished efficacy in the presence of tobramycin (Fig. 4), an aminoglycoside known to bind to the ribosome’s A site (34). The apparent tRNA selection bias also suggests that both endogenous and ataluren-stimulated near-cognate tRNA mispairing are not random processes and that factors contributing to the apparent preferences are conserved. One component of this selection process is a set of favored mispairings, primarily at codon position 1, where U-G mispairing is preferred over U-U and where U-C mispairing is strongly disfavored (Fig. 2; SI Appendix, Table S3) (21). As noted previously (21), U-G mispairing at position 1 is likely to be favored because of its ability to act as a geometric mimic of a U-A base pair (32). It is also plausible that the stability of nonstandard base pairs in position 1 is enhanced by modification of neighboring tRNA anticodon bases, e.g., thiolation of the wobble position uridine (35). At position 3, multiple nonstandard mispairings are tolerated including A-C, G-G, A-G, and possibly others (Fig. 2; SI Appendix, Table S3) (21). Another determinant of tRNA insertion choice is the extent to which the tRNA is expressed, if at all. As shown in the experiments in SI Appendix, Fig. S10C, yeast cells prefer Trp insertion over Arg insertion at UGA, largely because they lack tRNA-Arg-UCG, a tRNA that allows U-G mispairing. When tRNA-Arg-UCG was provided, it both stimulated readthrough and led to a marked increase in the relative frequency of Arg insertion at the PTC, unlike tRNA-Arg-UCU overexpression (SI Appendix, Fig. S10C) (36).
An important observation from these experiments is that the readthrough products of ataluren- or aminoglycoside-mediated nonsense suppression tested here are at least partially functional and, in many cases, fully functional. Earlier experiments with reporter genes, cultured cells, mouse models, and clinical trials of human DMD and CF nonsense mutation patients indicated that readthrough products are functional (1, 3–13, 23, 37–39), but could not address the question of specific amino acid substitutions at specific codon positions. Although our studies have addressed substitutions in only a limited number of positions in luciferase, His3, and CFTR, it is nevertheless reassuring with respect to the potential of therapeutic nonsense suppression that all of the substitutions analyzed here retained some degree of function. It will be of interest to determine whether the observed activities can be increased by parallel approaches, e.g., antagonists of mRNA decay (2), specific protein correctors or potentiators [as have been successfully used for CFTR (39)], or enhancers of the expression of certain tRNAs.
The efficacy of therapeutic nonsense suppression will also depend on the relative toxicity of the drug used to promote specific PTC readthrough. Ataluren has been shown to exhibit a favorable safety profile (27), and we speculate that at least two aspects of its activity support this attribute. First, as discussed above, ataluren stimulates PTC insertion of a set of near-cognate tRNAs that closely resemble those inserted endogenously at much lower levels. As a consequence, the full-length proteins that accumulate in ataluren-treated cells are unlikely to have antigenic differences from the repertoire of readthrough proteins normally accumulating in a patient, albeit at low levels. This low-level expression of full-length proteins by endogenous readthrough of nonsense-containing mRNAs may be sufficient to allow the polypeptides that accumulate during ataluren-stimulated readthrough to be recognized as “self” by the immune system, thus minimizing the potential for an autoimmune reaction. Second, at least compared with G418, ataluren generally has modest readthrough activity, showing only readthrough of premature termination events, but lacking readthrough activity on the considerably more efficient process of normal termination (3, 40). Ataluren’s failure to promote detectable readthrough of normal termination codons undoubtedly minimizes the possible accumulation of C-terminally extended proteins and their likely inhibitory activity and immunogenicity. In this regard, the much higher readthrough activity of G418, as well as its deviation from the pattern of endogenous insertion of near-cognate tRNAs, may contribute to that drug’s known toxicity (2). These results increase our understanding of readthrough therapies and how they might be tailored to specific disease alleles.
Materials and Methods
Detailed methods, given in SI Appendix, SI Materials and Methods, are summarized here.
Reporter and Expression Plasmids.
LUC- and HIS3-based reporters were used to analyze readthrough in yeast, and NanoLuc-, H2B-GFP-, p53-, LUC-, and TGFP-based reporters were used for the same purpose in human cells. For yeast, the construction and use of epitope-tagged LUC-PTC20 reporters has been described previously (21); the HIS3-PTC100 reporter used comparable epitope tags. In human cells, (i) using oligonucleotides DB4084 and DB4085 (SI Appendix, Table S7), UGA was introduced at codon 12 of the NanoLuc ORF expressed from pFN[Nluc/CMV/Neo] (Promega); (ii) the H2B-GFP reporter was constructed as described by Lentini et al. (6); (iii) UGA PTCs at positions 196 or 213 of the p53 ORF were introduced using site-directed mutagenesis, and the respective constructs were expressed from pcDNA 3.1 Hygro(+); (iv) LUC-PTC20 constructs used in yeast were modified to include TPI1 intron 6 at nucleotide 239 and expressed from pSELECT Zeo; (v) CFTR-G542X (UGA) and CFTR-WT, along with codons 539–541 and 543–545, were fused to TurboGFP (TGFP) along with HA and 6× His tags and expressed from pcDNA 3.1 Zeo(+); and (vi) CFTR-G542R, G542C, and G542W were created by site-directed mutagenesis of a WT CFTR cDNA and expressed from pcDNA 3.1 Zeo(+).
RNA and Protein Analyses.
Procedures for RNA analysis, protein purification, and luciferase assays were as described previously (3, 19, 21, 38) or in SI Appendix. Western analyses and protein purification largely used antibodies or affinity resins targeted to the HA, FLAG, StrepII, T-GFP, and 6× His epitope tags expressed from the respective reporter constructs.
Amino Acid Sequence Analysis of Luciferase and GFP-CFTR Readthrough Products.
Luciferase readthrough proteins were purified by epitope tag affinity chromatography and gel electrophoresis, subjected to proteolytic digestion, and analyzed by LC-MS/MS as described previously (21) and in SI Appendix. Raw data files were subjected to database searches, using SwissProt indices for Saccharomyces cerevisiae and human polypeptides. Comparable analyses based on CFTR used SEQUEST searches and Scaffold Q+.
Supplementary Material
Acknowledgments
We are indebted to Elizabeth Grayhack and Christina Brule for generously providing tRNA expression plasmids and Shirley Yeh and Angela Esteves for technical support. This work was supported by National Institutes of Health Grants R37 GM27757 (to A.J.), R21 NS090928 (to D.M.B.), R21NS090928 (to K.M.K.), and P30 DK072482 (to S.M.R.); by the Cystic Fibrosis Foundation (D.M.B. and S.M.R.); and by the University of Alabama Institutional Core Support Program from National Cancer Institute Grant P30CA13148-38 (to J.M.)
Footnotes
Conflict of interest statement: B.R., W.J.F., Y.T., J.Z., B.J., J.D., C.R.T., X.X., and E.M.W. are employees of PTC Therapeutics Inc. (PTCT). A.J. is a cofounder, director, and consultant for PTCT, and D.M.B. is a consultant for PTCT. S.M.R. receives grant support from PTCT to conduct clinical trials for the treatment of cystic fibrosis.
This article is a PNAS Direct Submission.
See Commentary on page 12353.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605336113/-/DCSupplemental.
References
- 1.Peltz SW, Morsy M, Welch EM, Jacobson A. Ataluren as an agent for therapeutic nonsense suppression. Annu Rev Med. 2013;64:407–425. doi: 10.1146/annurev-med-120611-144851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welch EM, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447(7140):87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 4.Li M, Andersson-Lendahl M, Sejersen T, Arner A. Muscle dysfunction and structural defects of dystrophin-null sapje mutant zebrafish larvae are rescued by ataluren treatment. FASEB J. 2014;28(4):1593–1599. doi: 10.1096/fj.13-240044. [DOI] [PubMed] [Google Scholar]
- 5.Gregory-Evans CY, et al. Postnatal manipulation of Pax6 dosage reverses congenital tissue malformation defects. J Clin Invest. 2014;124(1):111–116. doi: 10.1172/JCI70462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lentini L, et al. Toward a rationale for the PTC124 (Ataluren) promoted readthrough of premature stop codons: A computational approach and GFP-reporter cell-based assay. Mol Pharm. 2014;11(3):653–664. doi: 10.1021/mp400230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake KM, Dunmore BJ, McNelly LN, Morrell NW, Aldred MA. Correction of nonsense BMPR2 and SMAD9 mutations by ataluren in pulmonary arterial hypertension. Am J Respir Cell Mol Biol. 2013;49(3):403–409. doi: 10.1165/rcmb.2013-0100OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldmann T, Overlack N, Wolfrum U, Nagel-Wolfrum K. PTC124-mediated translational readthrough of a nonsense mutation causing Usher syndrome type 1C. Hum Gene Ther. 2011;22(5):537–547. doi: 10.1089/hum.2010.067. [DOI] [PubMed] [Google Scholar]
- 9.Sánchez-Alcudia R, Pérez B, Ugarte M, Desviat LR. Feasibility of nonsense mutation readthrough as a novel therapeutical approach in propionic acidemia. Hum Mutat. 2012;33(6):973–980. doi: 10.1002/humu.22047. [DOI] [PubMed] [Google Scholar]
- 10.Haas M, et al. European Medicines Agency review of ataluren for the treatment of ambulant patients aged 5 years and older with Duchenne muscular dystrophy resulting from a nonsense mutation in the dystrophin gene. Neuromuscul Disord. 2015;25(1):5–13. doi: 10.1016/j.nmd.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Bushby K, et al. PTC124-GD-007-DMD STUDY GROUP Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve. 2014;50(4):477–487. doi: 10.1002/mus.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerem E, et al. Cystic Fibrosis Ataluren Study Group Ataluren for the treatment of nonsense-mutation cystic fibrosis: A randomised, double-blind, placebo-controlled phase 3 trial. Lancet Respir Med. 2014;2(7):539–547. doi: 10.1016/S2213-2600(14)70100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkel RS, et al. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS One. 2013;8(12):e81302. doi: 10.1371/journal.pone.0081302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moosajee M, et al. June 21, 2016. Functional rescue of REP1 following treatment with PTC124 and novel derivative PTC-414 in human choroideremia fibroblasts and the nonsense-mediated zebrafish model. Hum Mol Genet, pii: ddw184.
- 15.Sermet-Gaudelus I, et al. Ataluren (PTC124) induces cystic fibrosis transmembrane conductance regulator protein expression and activity in children with nonsense mutation cystic fibrosis. Am J Respir Crit Care Med. 2010;182(10):1262–1272. doi: 10.1164/rccm.201001-0137OC. [DOI] [PubMed] [Google Scholar]
- 16.Kerem E, et al. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: A prospective phase II trial. Lancet. 2008;372(9640):719–727. doi: 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- 17.McElroy SP, et al. A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays. PLoS Biol. 2013;11(6):e1001593. doi: 10.1371/journal.pbio.1001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auld DS, Thorne N, Maguire WF, Inglese J. Mechanism of PTC124 activity in cell-based luciferase assays of nonsense codon suppression. Proc Natl Acad Sci USA. 2009;106(9):3585–3590. doi: 10.1073/pnas.0813345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall MP, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7(11):1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroha K, Tatematsu T, Inada T. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 2009;10(11):1265–1271. doi: 10.1038/embor.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy B, Leszyk JD, Mangus DA, Jacobson A. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc Natl Acad Sci USA. 2015;112(10):3038–3043. doi: 10.1073/pnas.1424127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auld DS, et al. Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proc Natl Acad Sci USA. 2010;107(11):4878–4883. doi: 10.1073/pnas.0909141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Jiang Q, Takahagi S, Shao C, Uitto J. Premature termination codon read-through in the ABCC6 gene: Potential treatment for pseudoxanthoma elasticum. J Invest Dermatol. 2013;133(12):2672–2677. doi: 10.1038/jid.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green L, Goff SP. Translational readthrough-promoting drugs enhance pseudoknot-mediated suppression of the stop codon at the Moloney murine leukemia virus gag–pol junction. J Gen Virol. 2015;96(11):3411–3421. doi: 10.1099/jgv.0.000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peltz SW, et al. 2009. Nonsense suppression activity of PTC124 (ataluren). Proc Natl Acad Sci USA 106(25):E64; author reply E65.
- 26.Blanchet S, Cornu D, Argentini M, Namy O. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42(15):10061–10072. doi: 10.1093/nar/gku663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirawat S, et al. Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers. J Clin Pharmacol. 2007;47(4):430–444. doi: 10.1177/0091270006297140. [DOI] [PubMed] [Google Scholar]
- 28.Bobadilla JL, Macek M, Jr, Fine JP, Farrell PM. Cystic fibrosis: A worldwide analysis of CFTR mutations: Correlation with incidence data and application to screening. Hum Mutat. 2002;19(6):575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 29.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6(7):1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keeling KM, Bedwell DM. Clinically relevant aminoglycosides can suppress disease-associated premature stop mutations in the IDUA and P53 cDNAs in a mammalian translation system. J Mol Med (Berl) 2002;80(6):367–376. doi: 10.1007/s00109-001-0317-z. [DOI] [PubMed] [Google Scholar]
- 31.Westhof E, Yusupov M, Yusupova G. Recognition of Watson-Crick base pairs: Constraints and limits due to geometric selection and tautomerism. F1000Prime Rep. 2014;6:19. doi: 10.12703/P6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. New structural insights into the decoding mechanism: Translation infidelity via a G·U pair with Watson-Crick geometry. FEBS Lett. 2013;587(13):1848–1857. doi: 10.1016/j.febslet.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141(2):344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 34.Salian S, et al. Structure-activity relationships among the kanamycin aminoglycosides: Role of ring I hydroxyl and amino groups. Antimicrob Agents Chemother. 2012;56(12):6104–6108. doi: 10.1128/AAC.01326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar RK, Davis DR. Synthesis and studies on the effect of 2-thiouridine and 4-thiouridine on sugar conformation and RNA duplex stability. Nucleic Acids Res. 1997;25(6):1272–1280. doi: 10.1093/nar/25.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beznosková P, Gunišová S, Valášek LS. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA. 2016;22(3):456–466. doi: 10.1261/rna.054452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bedwell DM, et al. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med. 1997;3(11):1280–1284. doi: 10.1038/nm1197-1280. [DOI] [PubMed] [Google Scholar]
- 38.Du M, et al. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc Natl Acad Sci USA. 2008;105(6):2064–2069. doi: 10.1073/pnas.0711795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue X, et al. Synthetic aminoglycosides efficiently suppress cystic fibrosis transmembrane conductance regulator nonsense mutations and are enhanced by ivacaftor. Am J Respir Cell Mol Biol. 2014;50(4):805–816. doi: 10.1165/rcmb.2013-0282OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amrani N, et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432(7013):112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.