Significance
Resistance to antibiotics poses a serious threat in contemporary medicine. Avilamycin and evernimicin, polysaccharide antibiotics belonging to the orthosomycin family, possess inhibitory activity against multidrug-resistant pathogenic strains of Enterococci, Staphylococci, and other Streptococci gram-positive bacteria by paralyzing ribosomes function in protein biosynthesis. The crystal structures of the large ribosomal subunit from the eubacteria Deinococcus radiodurans in complex with avilamycin and evernimicin revealed their binding sites at the entrance to the A-site tRNA accommodating corridor, thus illuminating the mechanisms of their translation inhibition. Analysis of the binding interactions of these antibiotics depicted the features enabling their species discrimination (namely, selectivity) and elucidated the various mechanisms by which pathogens use single mutations to acquire resistance to those drugs.
Keywords: degradable antibiotics, ribosomes, resistance, species-specific antibiotics, Gram-positive
Abstract
Two structurally unique ribosomal antibiotics belonging to the orthosomycin family, avilamycin and evernimicin, possess activity against Enterococci, Staphylococci, and Streptococci, and other Gram-positive bacteria. Here, we describe the high-resolution crystal structures of the eubacterial large ribosomal subunit in complex with them. Their extended binding sites span the A-tRNA entrance corridor, thus inhibiting protein biosynthesis by blocking the binding site of the A-tRNA elbow, a mechanism not shared with other known antibiotics. Along with using the ribosomal components that bind and discriminate the A-tRNA—namely, ribosomal RNA (rRNA) helices H89, H91, and ribosomal proteins (rProtein) uL16—these structures revealed novel interactions with domain 2 of the CTC protein, a feature typical to various Gram-positive bacteria. Furthermore, analysis of these structures explained how single nucleotide mutations and methylations in helices H89 and H91 confer resistance to orthosomycins and revealed the sequence variations in 23S rRNA nucleotides alongside the difference in the lengths of the eukaryotic and prokaryotic α1 helix of protein uL16 that play a key role in the selectivity of those drugs. The accurate interpretation of the crystal structures that could be performed beyond that recently reported in cryo-EM models provide structural insights that may be useful for the design of novel pathogen-specific antibiotics, and for improving the potency of orthosomycins. Because both drugs are extensively metabolized in vivo, their environmental toxicity is very low, thus placing them at the frontline of drugs with reduced ecological hazards.
The emergence of multidrug-resistant pathogenic strains of Enterococci, Staphylococci, and Streptococci Gram-positive bacteria poses a serious threat to modern medicine (1–3). Several classes of antibiotics inhibit protein synthesis by targeting functional sites of the bacterial ribosome. Examples of antibiotic targets are the peptidyl transferase center (PTC), the nascent chain exit tunnel and the decoding center. Some of the ribosomal antibiotics bind in proximity to each other or share overlapping binding sites. Thus, single mutations can trigger cross-resistance to several antibiotics families.
The orthosomycins avilamycin (avi) and evernimicin (evn) discovered in the 1960s do not inhibit translation of in vivo or in vitro eukaryotic ribosomes (4), and therefore possess the selectivity required for clinical use of antibiotics (5). Currently, avi is used as growth promoter added to animal food (6), and evn was considered for clinical use for humans by the Schering-Plough Corporation (7, 8).
Avi and evn are produced by Streptomyces viridochromogenes Tü57 (S. virido Tü57) (9) and Micromonospora carbonaceae (M. carbo) (10), respectively. Both drugs possess activity against Gram-positive bacteria, including vancomycin-resistant enterococci (VRE), methicillin-resistant Staphylococcus aureus (MRSA), and penicillin-resistant pneumococci (6, 11); they also selectively inhibit protein translation in Gram-positive bacteria Bacillus brevis (12) and Halobacterium salinarum archaea (8, 13).
Avi and evn inhibit poly(U)-directed polyphenylalanine (Phe) synthesis (8, 12) but do not inhibit the single peptidyl transferase reaction of Phe–tRNA and puromycin (8, 13). Evn inhibits initiation factor 2 (IF2)-dependent formation of 70S initiation complex (70SIC) (7), thus indicating inhibition at translation initiation. Early as well as current studies show that avi largely inhibits tRNA binding (12). Evn shows sequence-dependent inhibition (14) and EF4 back translocation (4), indicating translation elongation inhibition. Avi and evn do not inhibit Gram-negative bacteria (12) or eukaryotic cells (4). Resistance to avi and evn in Gram-negative bacteria seems to arise from nonribosomal mechanisms, because avi and evn inhibits cell-free translation of E. coli ribosomes (4, 12).
Based on biochemical and genetic studies, the binding pockets of avi and evn were suggested to span from ribosomal protein (rProtein) uL16 to ribosomal RNA (rRNA) helices H89 and H91 of the large ribosomal subunit. Chemical footprinting showed protection of multiple nucleotides of H89 and H91 rRNA helices (8, 13). Furthermore, single mutations in several nucleotides of H89 and H91 confer resistance to avi and evn in Streptococcus pneumoniae (S. pneumonia), E. faecalis, and Halobacterium halobium (H. halobium) (8, 13, 15, 16). In addition, single nucleotide mutations in rplP gene coding for rProtein uL16 at Arg50, Ile51, and Arg55 (E. coli numbering is used throughout) were found to render resistance to avi and evn in S. aureus, S. pneumonia, E. faecium, and E. faecalis (6, 16–18). Also, methylation of nucleotides G2470, U2479, and G2535 results in resistance to avi and evn (19, 20).
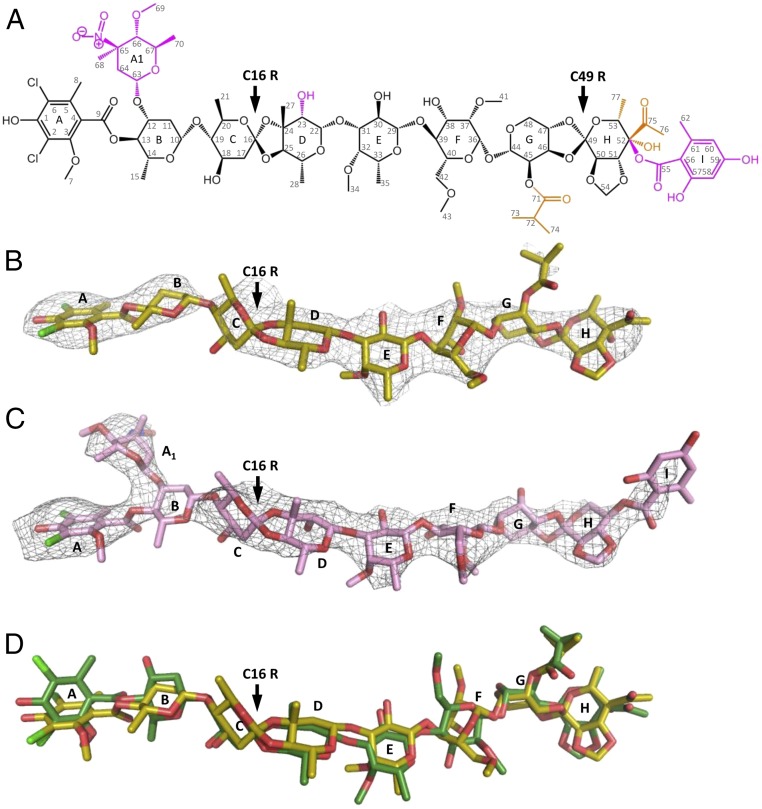
Orthosomycin drugs are composed of a common terminal dichloroisoeverninic acid (DIA) moiety and a heptasaccharide chain with two orthoester linkages at C16 and C49 (Fig. 1A). Avi and evn differ by the substitutes on their polysaccharide chain—notably, evn possess additional l-evernitrose and orsellinic acid residues. Several attempts were carried out to determine their structures by degradation (21), NMR (22), X-ray crystallography (23), and computational modeling (24). The evn structure has been fully determined by NMR and degradation studies (25). Avi C16 absolute configuration has not been determined before this study (11, 26); however, based on degradation studies, it was suggested to possess R chirality (27), similarly to evn.
Fig. 1.
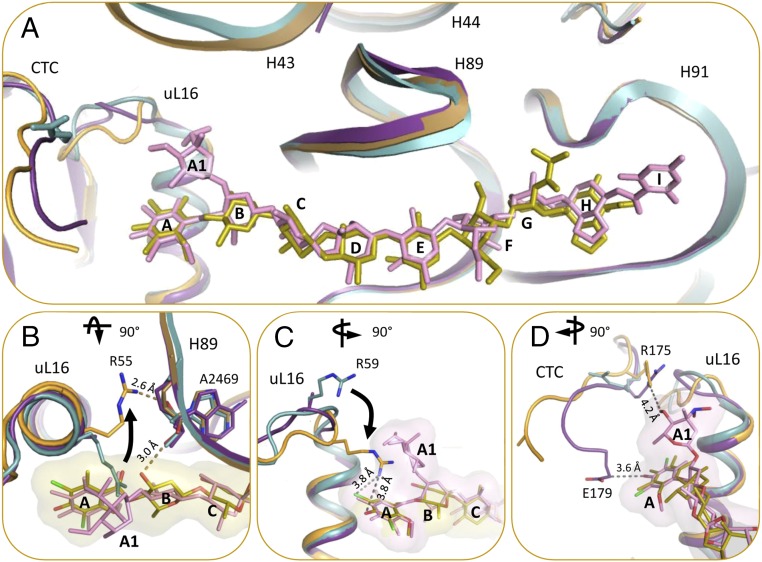
(A) The orthosomycin family consists of a common DIA residue (res A) and a heptasaccharide (residues B–H) shown in black, and two orthoester linkages at C16 and C49 pointed by an arrow. The heptasaccharide chain consisting of d-olivose (residues B and C), 2-deoxy-d-evalose (residue D), 4-O-methyl-d-fucose (residue E), 2,6-di-O-methyl-d-mannose (residue F), l-lyxose (residue G), and eurekanate (residue H). The additional chemical substitutes of avilamycin and evernimicin on the heptasaccharide chain are shown in orange and magenta, respectively. Evernimicin possess an additional l-evernitrose (res A1), orsellinic acid (res I), and a hydroxyl group on residue D. (B and C) Weighted 2Fo–Fc difference Fourier map contoured at 1σ of avilamycin (yellow) and evernimicin (pink) in complex with D50S. (D) Avilamycin conformation in D50S–avi complex (yellow) and free avi (green) superposed.
Here we present the high-resolution crystal structures of the large ribosomal subunit of Deinococcus radiodurans (D. radiodurans) in complex with avi and evn, as well as a high-resolution crystal structure of free avi, which shows that the absolute configuration of avi at position C16 is of R chirality. Our study demonstrates that avi and evn bind at a unique site of the ribosome that is not targeted by any other class of antibiotics. After submitting an abstract describing our results* and while our manuscript was in preparation, cryo-EM reconstructions of E. coli 70S (E70S) ribosomes in complexes with avi and evn were published (28). In this study (28), the low resolution and the quality of the EM maps did not enable a precise description of the interactions of avi and evn with the ribosome and consequently did not reveal the detailed mechanisms of resistance and selectivity. Conversely, our high-resolution crystal structures of (uniform at 3.35 and 3.58 A) provide findings regarding the binding, modes of action, resistance, and selectivity of avi and evn. Furthermore, because the EM studies were performed using a complex of ribosome from E. coli, which does not possess the domain 2 of CTC, the typical entity of many Gram-positive pathogens, the effect of this domain on avi and evn binding could not be assessed.
Results and Discussion
D50S–Avilamycin and D50S–Evernimicin Complex Crystal Structures and Free Avilamycin Conformation.
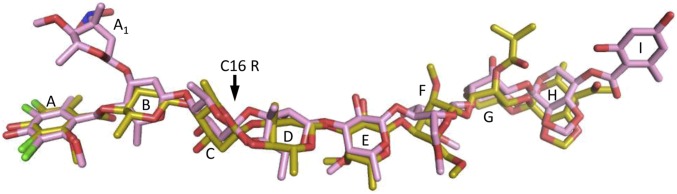
We determined the crystal structures of the 50S ribosomal subunit from D. radiodurans (D50S) in complex with either avi (D50S–avi) or evn (D50S–evn) at 3.35 and 3.58 Å resolution, respectively (Table 1). With the aim of determining the absolute configuration of avi, we studied the crystal structure of free avilamycin at 1.0 Å resolution (Table 1). The free avi crystal structure exhibits two copies of avi at an extended conformation in the asymmetric unit, with all sugars (res B-H) possessing chair conformation and R chirality of C16 in both copies, confirming previous crystallographic results of an avi fragment [BELJAD] (23) and NMR of all but C16 chirality assignments (11, 26), and in agreement with chemical degradation studies (27). According to this result, C16 of avi has the same chirality as evn C16 R (25), although not all other chiral centers of avi and evn share similar stereochemistry. Accordingly, we assigned R chirality to the C16 avi in the starting model of the drug and found that both avi and evn bind in an extended conformation to the large ribosomal subunit (Fig. 1 B and C and Fig. S1), similar to the free avi conformation (Fig. 1D).
Table 1.
Data collection and refinement statistics
| Statistics | D50S–avilamycin | D50S–evernimicin | Avilamycin-free |
| Crystal information | |||
| Space group | I222 | I222 | P21 |
| a, b, c, Å | 170.0, 412.6, 697.9 | 169.4, 407.4, 692.5 | 8.1, 34.5, 28.6 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 | 90, 97, 90 |
| Diffraction data statistics | |||
| Wavelength, Å | 0.873 | 1.033 | 0.710 |
| No. of crystals | 2 | 4 | 1 |
| <I>/<σI> | 6.5 (1.01) | 9.75 (1.43) | 20.7 (3.5) |
| Resolution, Å | 50–3.35 (3.41–3.35)* | 30–3.58 (3.64–3.58) | 20–1.00 (1.04–1.00) |
| Observed reflections | 1,115,549 (13,797) | 982,811 (13,017) | 25,930 |
| No. of unique reflections | 290,370 | 263,507(8,630) | 14,062 |
| Redundancy | 3.8 (3.3) | 3.7 (3.4) | 5.4 (3.6) |
| Completeness, % | 93.3 (89.2) | 94.4 (93.5) | 97.5 (99.0) |
| Refinement statistics | |||
| Rwork/Rfree | 0.215/0.259 | 0.206/0.249 | 0.1089†/0.1197‡ |
| RMSD bonds lengths, Å | 0.005 | 0.008 | — |
| RMSD angles, ° | 0.991 | 1.3 | — |
Values in parentheses are for highest-resolution shell.
R for all data.
R for data with I > 2σ(I).
Fig. S1.
Avilamycin (yellow) and evernimicin (pink) structures within their D50S complexes are superposed.
Binding Site of Orthosomycins.
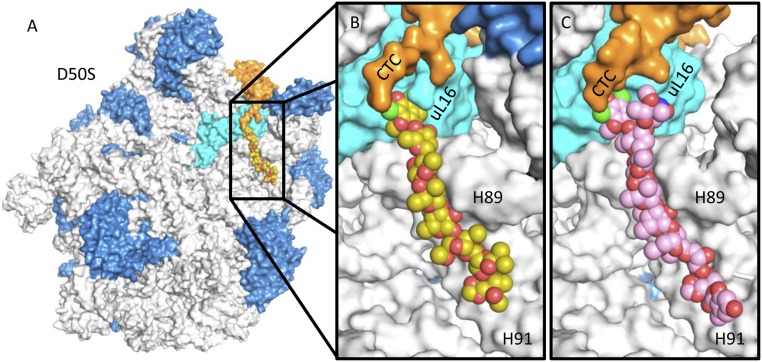
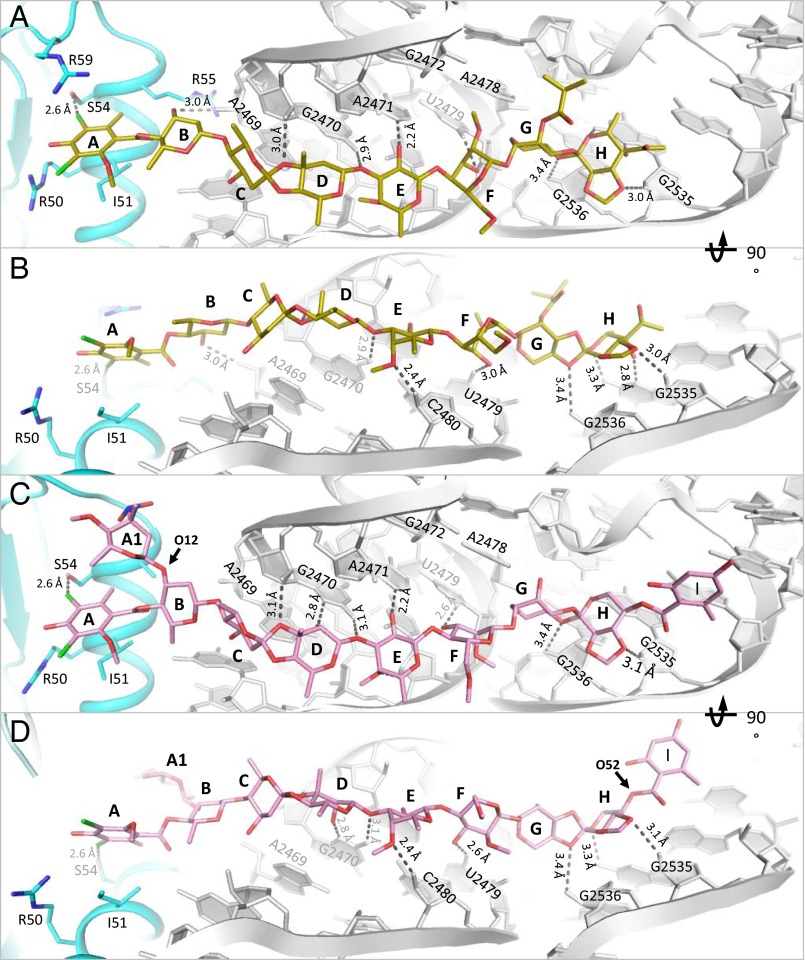
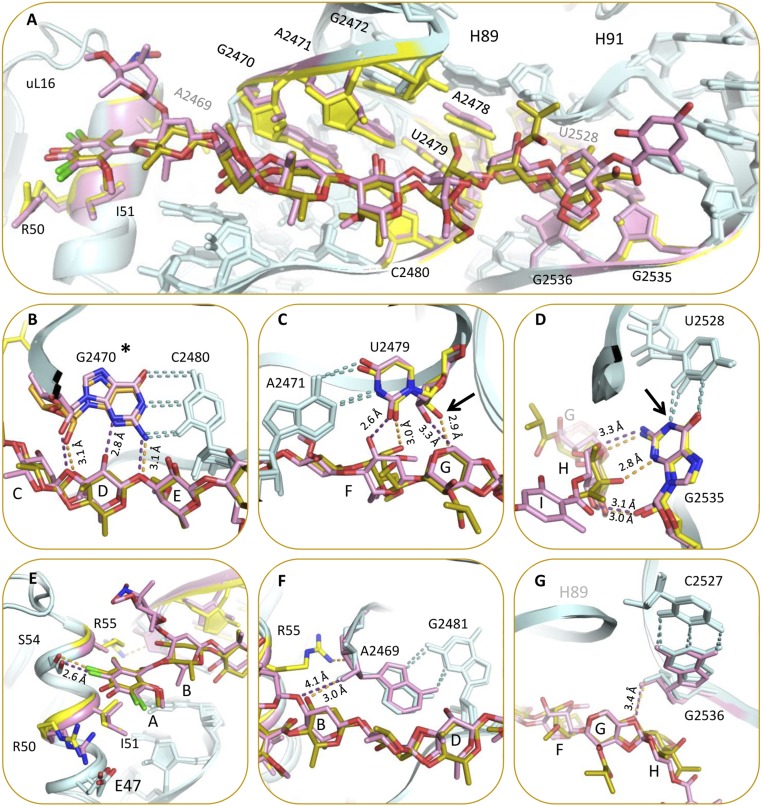
The structures of the complexes of avi and evn with D50S support and explain previous biochemical and genetic studies suggesting that the orthosomycins’ binding pocket spans rProtein uL16, H89, and H91 rRNA helices of the large ribosomal subunit. Both drugs bind with a full extended conformation to a cavity created by helix α1 of rProtein uL16, domain 2 of CTC, and the minor grooves of H89 and H91 of 23S rRNA (Fig. 2). The common heptasaccharide is in H-bond distance to the phosphate–ribose backbone of G2470, U2479, and C2480 from H89 and of A2530, G2535, and G2536 from H91; it also interacts with the nucleotide bases of G2470, A2471, A2478, and U2479 of H89 and G2535 of H91. The common DIA is found in a pocket created by rProtein uL16 side chains Arg50, Ile51, Ser54, Arg55, and Arg59. Ser54 is the closest residue to avi and evn and it creates hydrophilic interaction with the DIA (Fig. 3). The binding of both antibiotics displaces Arg55 away from the binding pocket (Fig. 4B) and draws Arg59 to the pocket (Fig. 4C).
Fig. 2.
(A) Binding site of the orthosomycins, represented by avi (yellow), in D50S, spanning uL16 (cyan), H89 and H91 (gray), and CTC (orange). The additional rRNA and rProteins are colored gray and blue, respectively. (B) Magnification of the binding pocket of avi (yellow) and (C) evn (pink) demonstrating the proximity of CTC domain 2 to the drug’s binding pocket.
Fig. 3.
The interactions of avi (yellow) and evn (pink) within their binding sites. H-bonds are colored gray. (A and B) Avi forms H-bonds with A2469, G2470, A2471, U2479, C2480, G2535, and G2536. (C and D) Evn forms H-bonds with G2470, A2471, A2479, C2480, G2535, G2536, and a hydrophilic bond with A2530 phosphate. Both avi and evn bond S54 of rProtein uL16 via hydrophilic interaction.
Fig. 4.
(A) The binding of avi and evn induce conformational changes in the rProtein uL16 loop, domain 2 of rProtein CTC, and 23S rRNA H43. D50S–avi (orange/yellow) and D50S–evn (purple/pink) complexes (orange) superposed on the apo D50S structure (PDB ID code 2ZJR) (teal). (B) D50S–avi superposed with the apo D50S (teal). R55 of rProtein uL16 is shifted away from avi compared with its apo conformation, and forms an H-bond with the A2469 backbone, which also binds to avi res B. (C) D50S–avi superposed with the apo D50S (teal). R59 of rProtein uL16 shifts toward avi upon binding, compared with its apo conformation. (D) R175 and E179 residues of CTC interact with evn (surface representation) res A1 and A, respectively. (B) 90° rotated to top view and (C and D) 90° rotated to side view, compared with A.
The main difference in the binding modes of avi and evn appears at the α1 helix of uL16 residue Arg59, which exhibits altered conformation owing to avi binding that shifts the entire loop Arg55–Arg63 towards the drug (Fig. 4C). This conformation encapsulates avi res A by creating an extra barrier within the binding pocket. In the D50S–evn complex, the conformation of the Arg59 loop is in an intermediate state between the D50S–avi complex and the apo ribosome structure, encapsulating both evn res A and l-evernitrose (res A1). Res B of the drugs interacts differently: avi O12 of res B interacts with A2469 backbone, whereas evn res O12 has an A1 substitute that interacts with Arg58 (Fig. 3C). Additionally, avi O52 of res H interacts with G2535 base, and evn C52 chirality is reversed with substituted orsellinic acid (res I) and O52 is facing away from G2535 (Fig. 3D).
In apo D50S structure, Arg58 of uL16 interacts with the backbone of C1075 of H43. The displacement of Arg58 upon the drugs’ binding alters the conformation of H43 that is located in the second shell around the antibiotics binding pocket (Fig. 4A). Benefiting from its inherent flexibility, domain 3 of protein CTC is displaced, and thus could not be traced. The displacement of Arg59 of uL16 in the D50S orthosomycin complexes toward the drug induces structural changes in domain 2 of CTC, so that in both complexes Arg175 of CTC is shifted to the position that is occupied by Arg59 of uL16 in the apo D50S structure (Fig. 4D). These structural rearrangements enable direct hydrophobic interactions between Arg175 and Glu179 of CTC domain 2 and evn res A and A1, respectively.
Translation Inhibition Mechanisms.
Inhibition of A-site tRNA binding.
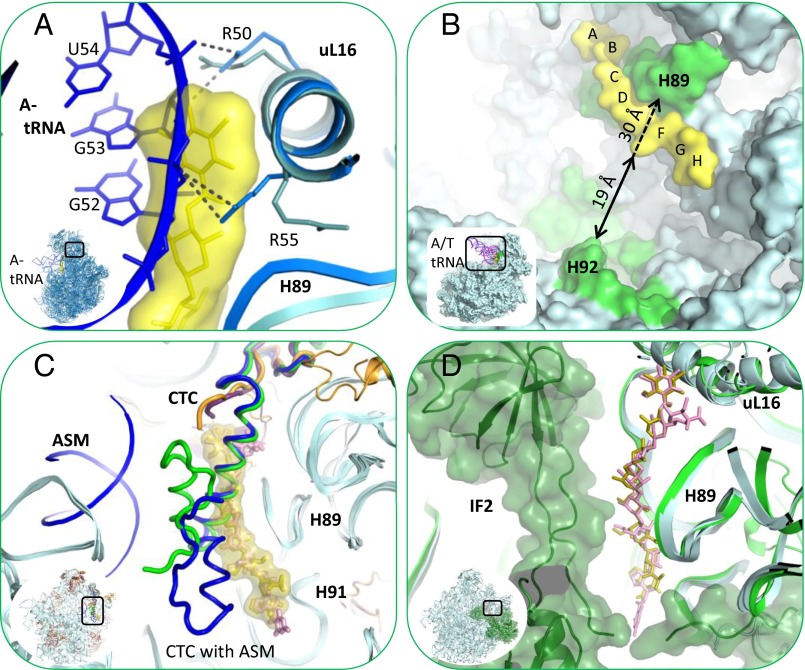
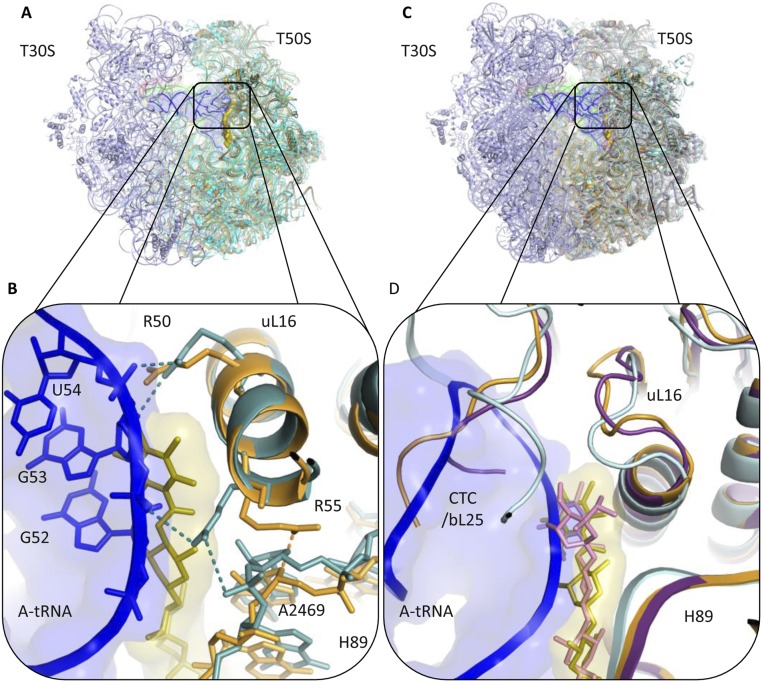
The elbow of A-site tRNA binds to uL16 and H89. A-tRNA G53 nucleotide interacts with uL16 residues Arg50 and Arg55 (29). In addition, G52 and A64 nucleotides of A-tRNA interact with A2469 and C2483 of H89, respectively (30). Superposition of the structure of 70S complex with three tRNAs from Thermus thermophilus (T70S) (PDB ID code 4VD5) on D50S–avi and D50S–evn complex structures reveals that the binding sites of both antibiotics and of A-tRNA overlap. A-tRNA cannot bind to the large ribosomal subunit, because A-tRNA elbow nucleotides G53 and G52 would clash with residues A and B of avi and evn (Fig. 5A and Fig. S2). In addition, whereas Arg50 of uL16 makes similar interactions with avi as it would with A-tRNA elbow, Arg55 shifts away from its A-tRNA binding conformation toward H89 and is stabilized by interactions with A2469 of H89. The access to nucleotide A2469 of H89 is blocked by avi and evn; thus, both drugs block the binding of A-tRNA by physically occupying its elbow binding site by using the Arg50, Arg55, and A2469 interactions designed for A-tRNA elbow binding. These findings support the evidence that avi largely prevents tRNA binding in poly(U)-directed poly-Phe synthesis (12).
Fig. 5.
Orthosomycins’ inhibition mechanism. Orthosomycin binding pocket overlaps A-tRNA, IF2, and domain 3 of rProtein CTC binding sites and the accommodation corridor. (A) Superposition of A-tRNA (blue) binding site at T70S (marine) crystal structure (PDB ID code 4VD5) with D50S–avi structure (teal, yellow). Avi is shown by surface representation. A-tRNA elbow U54 and G53 nucleotides backbone clash with avi residues A and B. Both avi and the A-tRNA elbow bind to R50 and R55 of rProtein uL16. The side chain of R55 is shifted toward H89 upon avi binding. (B) Accommodation corridor inhibition. A/T tRNA (pink) and A/A tRNA (blue) of E70S EM structure (PDB ID codes 1QZA and 1QZB) superposed on D50S–avi complex structure (teal, yellow). A/T tRNA is accommodated into A site, A/A tRNA, over H92 barrier. Zoom into the accommodation corridor (green), defined by H92 and H89. Avi and evn block the accessibility of accommodation corridor H89 rRNA nucleotides and narrows the accommodation corridor between H89 and H92, from 30 Å (dashed arrow) to 19 Å (black arrow). (C) rProtein CTC (bL25) location and conformation within apo D50S (green) and its complex with ASM (blue) crystal structures (PDB ID codes 1NKW and 1NJM) superposed on D50S–avi crystal structure (teal, yellow). Avi is shown with surface representation. The helix of both CTC conformations clash with avi res B, C, and D, all binding to H89 minor groove. (D) IF2 (dark green) binding site on E70S EM structure (PDB ID codes 3JCJ) superposed on the D50S–avi (teal, yellow) crystal structure. Zoom into IF2 interaction with H89, in proximity to avi and evn binding sites is shown.
Fig. S2.
A-tRNA binding site overlaps with the positions of avi and evn and CTC domain 2 in the D50S–avi and evn structures. (A) A-tRNA (blue) binding site at T70S (teal) crystal structure (PDB ID code 4VD5) superposed on D50S–avi structure (orange/yellow). Avi is shown with surface representation. Clash between A-tRNA and avi shown in black-framed zoom-in. (B) Magnification of A-tRNA elbow and avi interactions with rProtein uL16. Both avi and the A-tRNA interact with R50 and R55 of uL16. The backbone of A-tRNA nucleotides U54 and G53 clash with avi residues A and B. R55 side chain is shifted toward A2469 of H89 upon avi binding. (C) A-tRNA (blue) binding site at T70S (light cyan) crystal structure (PDB ID code 4V6F) superposed on D50S3 avi (orange/yellow) and D50S–evn structures (purple/pink). Avi is shown with surface representation. (D) Magnification of the clash between A-tRNA elbow and CTC second domain.
Accommodation corridor and inhibition of A-tRNA accommodation.
The accommodation corridor is the region involved in accommodating the A-tRNA into the ribosome (30), from the A/T partially bound state (31) to the A/A fully bound state (32). Dynamic simulations show that the accommodation step involves tRNA interactions with A2469–U2473 and G2481–C2483 nucleotides of H89 (33, 34). Avi and evn interact with G2470–G2472 residues of the accommodation corridor in H89 (Fig. 5B) and block their accessibility. Hence, we suggest that both drugs interfere with the A-tRNA accommodation by blocking the accessibility of H89 vital residues. Furthermore, drug binding narrows the corridor between H89 and of H92, which is involved in the accommodation (Fig. 5B).
CTC and inhibition of A-site binding regulation.
Domain 3 of rProtein CTC (D. radiodurans extended three domains homolog of rProtein bL25) interacts with uL16 and H89 (35, 36); it is connected to CTC domain 2 by a slim structural element, a flexible α-helix. CTC domain 3 serves as an A-site binding regulator, because it can either block A-site from binding tRNA or swing out and facilitate tRNA binding (36). The electron density maps of D5OS–avi and D5OS–evn show no density that could be assigned to CTC domain 3, and therefore could not be traced. We suggest that both avi and evn exclude this domain from binding to the large ribosomal subunit (at uL16 and H89), benefiting from its flexibility. Superposition of the CTC conformations in apo D50S and in the complex of D50S with acceptor stem mimic (ASM) (35, 36) on the structures of D50S–avi and D50S–evn complexes (Fig. 5C) indicated that D50S CTC domain 3 and the two drugs occupy H89 minor groove (A2469, G2470, A2471, C2480, and G2481). This overlap of binding sites suggests that avi and evn are interfering with CTC regulation of A-tRNA binding.
Domain 2 of CTC, or its homolog—namely, domain 2 of bL25 (37)—has an important role in tRNA accommodation (30) because it stabilizes the elbow of A-tRNA in the cognate complex. This domain of bL25 in T. thermophilus exists in many Gram-positive pathogens (Fig. S3). In the crystal structure of T70S in complex with the cognate A-tRNA, domain 2 of CTC adopts a conformation that alters the position of the Arg59 loop of uL16. These structural changes enable direct interactions between Arg59 of uL16 and the phosphate backbone of G53 in A-tRNA (30). Thus, binding of A-tRNA triggers conformational changes in CTC domain 2 and uL16 that stabilizes its interactions with the ribosome. However, binding of avi and evn induces structural rearrangements in uL16 that stabilize CTC domain 2 in a conformation that would clash with the bound A-tRNA (Fig. S2) and hence prevent its binding. This finding suggests that avi and evn may stabilize the bL25 second domain for further blockage of A-tRNA binding. In a cell-free translation assay, ribosomes from E. coli and S. aureus are inhibited in a similar level by evn (4) and ribosomes of E. coli and B. brevis are inhibited in a similar level by avi (12). Thus, both structures reveal that CTC domain 2 is involved in the inhibition mechanism, but thus far there is no evidence indicating that this involvement can be connected with the proposed linkage of this domain with elevated temperatures of thermophilic bacteria (38), although temperature rise is likely to be associated with bacterial infections.
Fig. S3.
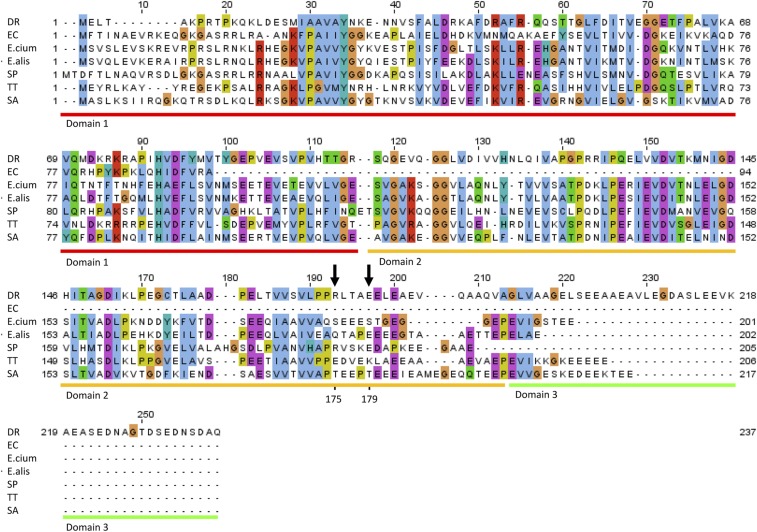
Sequence alignment of bL25 of D. radiodurans R1 (DR), E. coli K12 (EC), E. faecium V582 (E.cium), E. faecalis 29212 (E.alis), S. pneumoniae (SP), T. thermophilus HB8 (TT) and S. aureus NCTC 8325 (SA). Domains 1 (red), 2 (orange), and 3 (green) of DR bL25, or CTC, are underlined. R176 and E179 residues of DR CTC domain 2 that interact with evn are marked with black arrow. bL25 of EC is a one-domain protein, whereas other bacteria shown have two or three domains.
Inhibition of EF4 back translocation.
EF4 catalyzes back translocation of P-tRNA to the A-site (39, 40). Because A-site is blocked by avi and evn, the back translocation of P-tRNA will be physically blocked upon avi or evn binding, in accord with evn EF4 back translocation inhibition (7).
Inhibition of IF2-dependent translation initiation.
In prokaryotes, translation initiation involves the formation of the small ribosomal subunit (30S) initiation complex (30S IC), i.e., 30S; mRNA; fMet tRNA (tRNAfmet); and three initiation factors 1, 2, and 3 (IF1, IF2, and IF3). The latter leave upon 50S binding to the small subunit by surface complementarity, and upon intersubunit bridges formation, 70S elongation complex (70S EC) functionally active ribosome is formed. An intermediate 70S preinitiation complex (70S PIC) holds the three initiation factors, mRNA, and tRNAfmet (41). The 50S subunit binding to 30S IC, forming 70S PIC, triggers GTP hydrolysis by IF2; consequently, tRNAfmet is released from IF2 into the canonical P/P site of the 50S. Following this step, the initiation factors dissociate and the 70S initiation complex (70S IC) is formed (42).
IF2 domain C2 interacts with the CCA end of the initiator tRNA (43) and helps dock it in the initial P/I tRNA site at 50S of 70S PIC (44). Overlaying the recent 70SIC–IF2 complex (IF2–E70S) cryo-EM structure (45) with D50S–avi and D50S–evn complex structures shows that IF2 and the two drugs bind to H89, but from different directions (Fig. 5D). This close proximity may suggest that avi and evn can inhibit initiation complex formation by interfering with IF2 binding to 50S, thus inhibiting the formation of 70S PIC. This result is in accord with evn inhibition of IF2-dependent 70S IC formation in vitro (7) and evn inhibition of IF2-dependent peptide bond formation in vitro (8). Consequently, additional biochemical research is required to elucidate whether avi interferes with IF2-dependent 70S IC formation in a manner similar to evn.
Avi and evn Resistance Mechanisms.
Resistance to avi and evn acquired by 23S rRNA methylation.
The avi producer S. virido Tü57 possesses two enzymes and one transporter that protect the organism from its own product (19). These two enzymes are AdoMet-dependent rRNA methyltransferases aviRa and aviRb that methylate N1 of G2535 and O2′ of U2479 (46). Expressing aviRa and aviRb in Streptomyces lividans (S. livadas) confers resistance to avi with minimum inhibitory concentration (MIC) of 20 μg/mL and 250 μg/mL, respectively, compared with native S. livadas with MIC of <5 μg/mL (19). Mapping those methylation sites on the D50S–avi complex structure shows that both methylation targets are located at the avi binding site (Fig. 6 C and D). O2′ of U2479 does not form a hydrogen bond with avi but is located in a close proximity to its res G; this suggests that methylation of U2479 gives rise to resistance by clashing with res G and repulsing the drug. N1 of G2535 methylation hinders the G2535:U2528 wobble pair. This loss of wobble pair interaction can reshape the H91 loop, which can hinder avi’s binding; it is conceivable that these are the most crucial resistant mechanisms of avi with its target because they evolved in vivo by S. virido Tü57.
Fig. 6.
(A) Resistance to avi and evn acquired by a single mutation in 23S rRNA and uL16 rProtein. The mutations are located around the binding site or at its second shell. D50S–avi (teal/yellow) and D50S–evn (teal/pink) complex structures presented with mutation sites (yellow and pink, respectively). (B–D) Resistance to avi and evn is acquired by 23S rRNA methylation (same color scheme). Methylation sites (marked by arrow) at O2′ U2479 (C) and N1 G2535 (D). *G2470 methylation site is unknown (B). Magnification of res A binding pocket in uL16 α1 helix with amino acids R50, I51, and R55, which render resistance by mutation, are highlighted in yellow and pink for avi and evn, respectively (E). Purine:purine interaction of A2469:G2481. Distance to O12 is presented (F). Base pair between G2536:C2527, distance to O47 is presented (G). Distances (Å) are shown in light orange and purple, respectively.
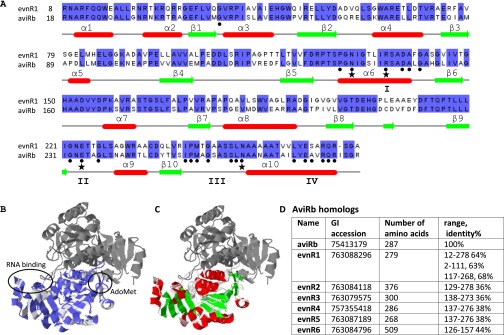
There are no reports to how the bacterium M. carbo protects itself from producing evn. A search for sequence homologs of aviRa and aviRb in recently sequenced M. carbo (taxonomy ID 47853) genome (47), yielded no aviRa homologs and six aviRb homologs (Fig. S4D). Though all six homologs share SpoU-methylase N-domain of aviRb (pfamoo588), only evnR1 (GI:763088296, 2–111, 63% identical amino acids) has a similar RNA binding N-domain of aviRb. Further comparison with aviRb reveals evnR1 possess the four sequence fingerprints of the SpoU MTase’s family, I–IV, as well as the four residues Asn139, Glu234, Asn262, and Arg145 that are important for catalysis of aviRb (48) (Fig. S4 A–C). Thus, we propose that M. carbo protects itself from evn by methylation of O2′ of U2479 rRNA via evnR1, AdoMet-dependent rRNA methyltransferases, similarly to S. virido.
Fig. S4.
Sequence alignment of evnR1 and aviRb superposed on aviRb structure. (A) The conserved nucleotides are highlighted in purple. (B) Secondary structure elements of aviRb are given for reference in red and green. (C) AviRb crystal structure (1 × 7O, 1 × 7P). The RNA-binding region of the N-terminal domain was derived from the structurally homologous ribosomal proteins L30 and L7Ae; it is marked in purple. The alignment is based on aviRb homologs, given in D.
A resistant strain of E. faecium, isolated from animal sources, has gained resistance to both avi and evn by methylation of G2470 by the EmtA methyltransferase enzyme (20), which is located on a plasmid-borne transposable element. The exact methylation site is not known yet, but the N1 position of the G2470 base is suggested (46). G2470 is located at the avi and evn binding sites (Fig. 6B). Methylation of the O2′, N1, N2, or N3 positions of G2470 will hinder hydrogen bonding to res D or E, respectively, and thus decrease avi and evn affinity and render resistance.
Resistance to avi and evn acquired by 23S rRNA single mutations.
Mutations in rRNA nucleotides of H89 and H91 of the binding site confer resistance to both avi and evn (8, 13, 15, 16). Eight of those mutations occur at nucleotides A2469, G2470, A2471, A2472, A2478, U2479, C2480, and A2535 that directly interact with avi and evn (Fig. 6A). Avi forms a hydrogen bond with A2469 ribose. Upon its binding, uL16 Arg55 swings to interact with A2469 backbone. The mutation A2469C will not directly compromise these bonds, but might change the conformation of A2469:G2481 purine–purine interaction, which may propagate to change the structure of the backbone hindering these interactions with the drug.
Both avi and evn form hydrogen bonds with N2 of G2470 as well as to the O2′ sugar of G2470 and C2480, where G2470:C2480 form a Watson–Crick (WC) base pair (Fig. 6B). The resistance mutation G2470U directly hampers these hydrogen bonds and generates a clash of O2 carbonyl with the drug in this tight binging pocket. Additionally, G2470U mutation directly hampers the G2470 N3 interaction with evn. Furthermore, G2470U and C2480U mutations will abolish G2470:C2480 base pairing interaction and may hinder the hydrogen bonds to O2′ of G2470 and C2480 of both drugs; both form hydrogen bonds to N3 of A2471 and O2 of U2479 where A2471:U2479 form a WC base pair (Fig. 6C). The mutation A2471C directly hampers these H-bonds, and may generate a clash of O2 carbonyl with the drug, and the mutation A2471G may create a clash with N2 of the amine. The mutation U2479C does not directly interfere with the hydrogen bond of O2 carbonyl to the drug. Importantly, the mutations A2471G, A2471C, and U2479C abolish the A2471:U2479 base pairing interactions and the stability provided for avi and evn binding by this anchor will be lost. A2478 is interacting with avi and evn residue G and G2472:A2478 form a purine–purine base pair. Thus, the mutations of G2472U or A2478C will generate a WC base pair, which might change the structure of the H89 backbone and hinder interactions with both drugs.
Both avi and evn form a hydrogen bond with N2 of G2535. The mutation G2535A will cause a loss of this hydrogen bond, explaining why G2535A mutation renders resistance to both drugs. U2528 forms a wobble pair with G2535, in which N2 of G2535 is free for interaction with both avi and evn. Mutations C2527A and U2528C in H91 confer resistance to both drugs, although they do not directly interact with avi and evn. C2527 and U2528 are base paired with G2536 and G2535, respectively, and are in second shell to the drug binding sites (Fig. 6D). The mutation U2528C will generate a WC base pair to G2535, which can shift G2535 further from the drugs and weaken the bond to N2 of G2535, as well as avi bond to N3 of G2535. Both drugs do not form hydrogen bonds with G2536 but the mutations G2536C and C2527A can change the conformation of G2536:C2527 base pair, which may change the structure of the H91 backbone, and propagate to hamper the binding to G2535. Thus, we suggest that although these nucleotides do not interact directly with avi and evn, their mutation induces a structural change to the conformation of H91, which propagates to the binding site.
Resistance to avi and evn acquired by uL16 single mutations.
Point mutations in the rplP gene resulting in substitutions in helix α1 of uL16 rProtein render resistance to both avi and evn in S. pneumonia, S. aureus, E. faecium, and E. faecalis (6, 8, 17, 18). The side chains of Arg50, Ile51, and Arg55 directly interact with avi (Fig. 6 A and E). Substitution mutations Ile51Ser and Ile51Thr add hydrophilic character, disrupting the Ile51 hydrophobic interactions and reducing the binding affinity. Arg50Cys, Arg50His, and Arg55His mutation substitutions conserve the positive environment required for binding A-tRNA, allowing ribosomal translation, but reduce contacts with avi and evn owing to a shorter side chain. Arg55His substitution mutation hinders the Arg55–A2469 interaction, which stabilizes the R55 at its swung out orientation.
Avi and evn Selectivity.
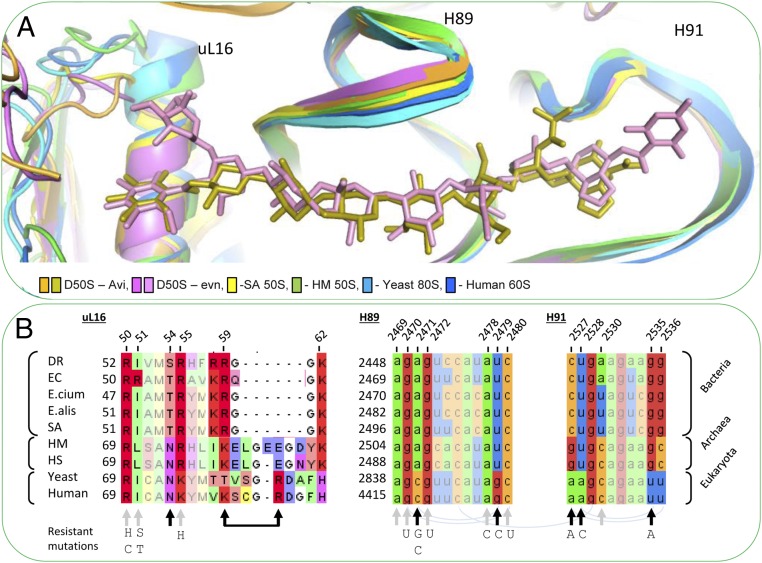
Avi and evn selectivity allows those compounds to inhibit protein translation in Gram-positive bacteria, but not to inhibit eukaryotic cells. Gram-negative bacteria are resistant to avi and evn, presumably due to their additional outer membrane of the Gram-negative bacteria, because both avi and evn inhibit cell-free translation of E. coli ribosomes (4, 12). Avi and evn also inhibit archaeal ribosomes from H. salinarum (or Halobacterium halobium) (8, 13). Ribosomes from eukaryotic cells of wheat germ are not inhibited by evn even at high concentrations (4). Examination of the structures of D50S–avi and D50–evn enables the rationalization of their selectivity mechanism by structural variations between prokaryotes and eukaryotes ribosomes. Though the overall structure of H89 and H91 is conserved among prokaryotes D. radiodurans and S. aureus (49), archaea Haloarcula marismortui (50) and eukaryotes Saccharomyces cerevisiae (S. cerevisiae) (51), and Homo sapiens (H. sapiens) (52), by corrective base pairing, the specific drug–rRNA interactions are hindered (Fig. 7 A and B). In addition, because the α1 helix of the uL16 structure is longer in eukaryotes (53, 54) (Fig. 7A), some of the specific drug–L16 interactions are hindered.
Fig. 7.
Orthosomycins’ selectivity. (A) Comparison among the conformations of H89, H91, and uL16 in 60S of H. sapiens (PDB ID code 3J3F; blue), 80S from the yeast S. cerevisiae (PDB ID code 3U5D; light blue), D50S–avi (orange-gold), D50S–evn (purple-pink), S. aureus 50S (PDB ID code 4WCE; yellow), and the archaeon H. marismortui 50S (PDB ID code 4HUB; green). The overall structure of H89 and H91 is conserved among bacteria, archaea, and eukaryotes. The rProtein uL16 in archaea and eukaryotes possesses a longer α1 helix. (B) Sequence alignment of uL16 (Left) of D. radiodurans R1 (DR), E. coli K12 (EC), E. faecium V582 (E.cium), E. faecalis 29212 (E.alis), S. aurous NCTC 8325 (SA), H. marismortui (Hmar), H. salinarum R1 (Hsal), S. cerevisiae 204508 (yeast), and H. sapiens 9606 (human), and 23S rRNA alignment of H89 (Middle) and H91 (Right) of the same organisms. Within the sequence alignments, paired bases (arch), avi and evn binding site (gray arrows), and variance in binding site (black arrows). Resistance-causing mutations are listed below sequence alignment.
uL16 rProtein homologs.
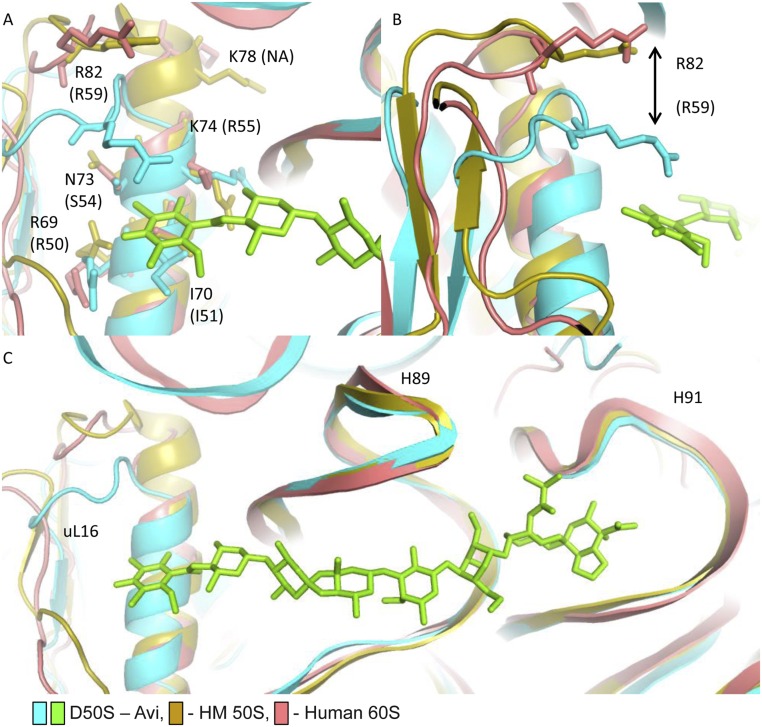
The human and archaeal homologs of uL16 possess a longer α1 helix than in the bacterial uL16 (Fig. 7A). Most uL16 key amino acids in avi and evn binding pocket (Arg50, ILe51, and Arg55) are highly conserved among bacteria, archaea, and humans (Fig. 7B), and possess a similar conformation (Fig. S5). ILe51Arg variation between Gram-negative and Gram-positive bacteria does not contribute to the Gram-negative resistance to evn, as was tested by mutation of Ile52Arg in susceptible S. pneumoniae (18). Ser54 in DR and Thr54 in E. coli, E. faecium, E. faecalis, and S. aureus are both susceptible to avi and evn. Also, Ser54 variation to Asn54 in archaea does not contribute to the selectivity (Fig. 7B), because they occur in susceptible ribosomes. Arg59 is located on a loop after α1 helix. Arg82 (human numbering, hArg82) in human and Glu82 in archaea (aGlu82) have the same orientation in the loop after α1 helix, and are the homologs of Arg59 in bacteria (Fig. S5). As a result of a longer α1 helix, hArg82 and aGlu82 are positioned further from the drug, compared with the bacterial Arg59. Thus, structural variation of α1 helix in humans compared with bacteria may still be in part contributing to the drug’s selectivity.
Fig. S5.
Orthosomycin selectivity. (A) The key residues of uL16 that interact with avi and evn res A (R50, I51, S54, and R55) and their equivalents in humans (R69, I70, N73, and K74) adopt similar conformations. K78 does not have a bacterial equivalent. (B) R82 of human uL16 is located further from avi compared with its bacterial equivalent, R59. Archaeal E82 possesses a similar conformation to the human R82. Although it appears that the structural variation of α1 helix length can hinder Arg82 ability to interact with res A of avi, it seems that this could not be the main contributor to selectivity, because it exists in susceptible archaeal ribosomes as well. However, because archaea possess a Glu82 at the position hosting Arg in human and bacteria, the archaeal susceptibility to avi and evn might arise from a “corrective” variation in this loop. (C) The superposition of the EM structure (PDB ID code 4UG0) of 60S from H. sapiens (pink) with the D50S–avi (teal/green) and H. marismortui (yellow) 50S crystal structure (PDB ID code 4HUB) demonstrates that the overall structure of H89 and H91 is conserved, and both human and archaeal uL16 possesses a longer α1 helix.
23S rRNA sequence variation.
The overall structure of H89 and H91 is conserved among eubacteria D. radiodurans, S. aureus (49), archaea H. marismortui (50), eukaryotes S. cerevisiae (51), and H. sapiens (52) (Fig. 7A). Most key rRNA nucleotides in avi and evn binding pocket (A2469, G2470, G2472, A2478, and C2480) are highly conserved between eubacteria and archaea (Fig. 7B). Avi and evn interact with five nucleotides that are not conserved between bacteria and humans (i.e., A2471C, U2479G, C2527A, U2528A, and G2535U; Fig. 7A). This sequence variation can eliminate up to four hydrogen bonds between avi and evn in its putative binding pocket in humans, compared with 10 H-bonds they form with D50S ribosome. A2471C variation can directly hinder the H-bond between N3 and res E of avi and evn. U2479G variation can exclude interactions between O2 and avi and evn res F. G2535U can hinder hydrogen bond between N3 and avi res H. In addition, altering G2535:U2528 wobble pair to A:U WC base pair can change the H91 loop conformation and prevent another H bonds between G2535 backbone O2′ and avi and evn res H. The C2527:G2536 base pair does not directly interact with the orthosomycins, and its variation to G:C in archaea do not contribute to the selectivity, because they occur in susceptible ribosomes (Fig. 7B). Evn interaction with the A2530 nucleotide backbone explains why its variation to A or U in bacteria and C in archaea do not contribute to selectivity, because they occur in susceptible ribosomes (Fig. 7B). The five variable nucleotides A2471, U2479, C2527, U2528, and G2535 seem to play a key role in the binding of avi and evn. Mutations of those nucleotides in bacteria render resistance to avi and evn (Fig. 7B). Thus, it is conceivable that the evolutionary distinction of those nucleotides hinders avi and evn binding and enables selectivity between bacterial and human ribosomes.
Comparison Between the Crystal Structures of D50S–Orthosomycins and the Cryo-EM Reconstructions of E70S–Orthosomycins Complexes.
After submitting a conference abstract† and while writing this manuscript, a study describing the cryo-EM single-particle reconstructions of E70S in complex with avi and evn was reported (28). These two independent studies show similar modes of action, namely, inhibition of A-tRNA accommodation. Moreover, a comparison of the avi and evn bound to D. radiodurans 50S subunit (this study) and to E. coli 70S ribosome (28) reveals that in both bacterial species the binding site of these antibiotic spans uL16, H89, and H91 in a similar way. However, there is a major difference between the two species: protein CTC second domain, or its homolog second domain of bL25, which participates in blocking A-tRNA accommodation by avi and evn, exists in many Gram-positive pathogens and in D50S but does not exist in the E. coli ribosome (Fig. S3). Thus, the D. radiodurans structures serve as a more suitable model for studying the orthosomycins inhibition mechanism against Gram-positive bacteria such as Enterococci (37). We suggest that the CTC domain 2 homolog participates in blocking A-tRNA accommodation and in binding avi and evn.
Our uniform higher-resolution structures provide a detailed description of the hydrogen bonds that are formed by avi and evn with the ribosome that is not found in the cryo-EM study. Based on this analysis, we explain the structural basis for the resistance mechanisms to avi and evn as well as to their mode of selectivity. We carefully examined the stereochemistry of all chiral centers of avi and evn and confirmed that all of them possess the correct chirality throughout all of the refinement cycles, benefiting from the higher uniform resolution of the crystal structures.
Summary.
By determining the crystal structures of the complexes of avi and evn with the large ribosomal subunit, we shed light on their binding sites and modes of action, as well as their selectivity and resistance mechanisms. Both drugs bind at a unique binding pocket spanning from the rProtein uL16 α1 helix and CTC domain 2 through H89– H91 rRNA helices. Protein translation inhibition is achieved by (i) blockage of an essential site in the large ribosomal subunit—namely, the A-tRNA site accommodation corridor, crucial for translation elongation; (ii) interaction with protein CTC, an A-tRNA regulatory ribosomal feature; and (iii) inhibition of IF2 binding by creating an additional barrier at the H89. Both the rRNA and rProtein uL16 mutations and methylations that give rise to resistance are part of their binding site, or are located in the second shell of it.
There are major structural differences between the prokaryotic helix α1 of rProtein uL16 and its eukaryotic homolog, the latter being longer, altering the shape of the orthosomycin binding pocket. In addition, sequence variations in rProtein uL16 and of H89 and H91 rRNA govern the avi and evn binding pocket character. These differences seem to account for avi and evn selectivity.
Many antibiotics are persistent in the environment, raising concerns of toxicity, resistance development, and other environmental risks (55). Avi is extensively metabolized in rats and pigs (56), at the orthoester C16 link. This unique feature of the orthosomycin family, and the Cl reduced derivatives, place this antibiotic family at the front line of antibiotics with reduced environmental hazards.
SI Text
Purification of D50S Ribosomes.
D50S Crystallization.
Crystals were grown by vapor diffusion in hanging drops. Ribosome solution containing 6.5 μM (180 A/mL) of D50S in 10 mM Hepes (pH 7.8) at 293 K, 15 mM MgCl2, and 75 mM NH4Cl crystallization buffer (H10M15N75) was mixed with 10 mM spermidine, 1% ethanol, and 0.5% 2-ethyl-1,3-hexanediol (EHD) precipitants. A 5-μL crystallization drop was hanged over 10% (vol/vol) ethanol and 5% (vol/vol) EHD in the H10M15N75 buffer, and left for 2 wk at 293 K. The crystals were kept in stabilizing solution containing H10M15N75 buffer with 20% (vol/vol) ethanol and 10% (vol/vol) EHD.
D50S Crystals Data Collection.
Crystals were soaked for 6 h in 50 μM avi or evn dissolved in stabilizing solution. Before the data collection, the crystals were transferred for 15 min to cryoprotecting solution containing H10M15N75 buffer with 10% (vol/vol) ethanol, 5% (vol/vol) EHD, 15% (vol/vol) MPD, and 50 μM avi or evn and flash-cooled in liquid nitrogen.
X-ray data of D50S–avi crystals were collected at 0.873 Å, under cryotemperature (100 K) at the microfocus beamline ID23-2, European Synchrotron Radiation Facility (ESRF). X-ray data of D50S–evn were collected at Beamline 19ID (1.033 Å) of the Structural Biology Center at the Advanced Photon Source at Argonne National Laboratory. We collected complete datasets from two D50S–avi crystals and four D50S–evn crystals. The data were indexed, integrated, and scaled using HKL2000 (58) and CCP4 (59). The crystals belong to orthorhombic space group I222.
Structures Solution, Model Building, and Refinement.
Each structure was solved by molecular replacement with Phaser as implemented in the PHENIX suite (60), using the coordinates of the native D50S (PDB ID code 2ZJR) (61) as a starting model. The initial solution was subjected to rigid-body refinement, followed by several cycles of positional and grouped B-factor refinement using CNS (62) and PHENIX (60), to yield an unbiased weighted difference Fourier Fo–Fc map. Clear difference electron densities showing avi and evn 2D structures (25, 26) were prepared using the ChemDraw suite (63). C16 R chirality of avi, previously unknown, was determined from the free avilamycin crystal structure. The 3D models of antibiotics were generated with ChemDraw3D (63), as a Mol2 file format (SYBYL). Geometrical restrains were generated via PRODRG server (64). The 3D models were fitted into positive unbiased weighted Fo–Fc density map using COOT (65) and refined in PHENIX (60). In addition to overall positional and geometrical parameters refinement, the occupancy and conformation of the eight rings were refined individually. Interactions of avi and evn with D50S were analyzed by LigPlot (66). Figures were generated using PyMOL (67) and Jalview (68).
Free avi Crystallization.
A total of 2.2 mg of avi were dissolved in 300 μL acetonitrile (ACN) and transferred to a small tube. Double distilled water (DDW) and ACN were added to 75% (vol/vol) ACN, and the tube was sealed with parafilm for 5 d. A small hole was pierced in the parafilm, and crystals with a thin-plate morphology appeared 2 d later. Crystals were fished out using paratone oil and flash-cooled in liquid nitrogen.
Free avi Crystals Data Collection and Processing.
A full X-ray data set of 1.0 Å resolution of free avi was collected under cryotemperature with λ = 0.710 Å at the beamline ID23-1, ESRF. Attempts to collect a dataset and solve it on a home source failed due to weak and low-resolution diffraction. The avi crystals gave a multiple pattern, which was successfully resolved at data processing. A monoclinic P21 space group was determined with XPREP from the SHELX software package (69) (Table 1). Data indexing, integration, and scaling of the data set were performed using CrysAlisPRO version 38.41 (70), and Friedel pairs were kept separate.
Free avi Structure Determination.
Avi structure was solved ab initio with a direct method using SHELXM. The initial solution was refined using SHELXT and further refined using SHELXL (SHELX software package). The disordered solvent contribution, which occupies ∼51% from the crystal structure, was eliminated by PLATON SQUEEZE tool (71). The solution contains two copies of avi in the asymmetric unit. The avi molecule is in extended conformation and all sugars (res B–H) possess chair conformation. The absolute configuration of avi at position C16 was determined to be R chirality for both copies. The avi molecules are organized inside the crystal are packed in a herringbone packing pattern.
Evn1 Identification.
AviRa and aviRb (gi|75413178, gi|75413179) were blasted against the genome of M. carbo (taxonomy ID 47853). Database searches were performed with the BLASTP 2.3.0 program (72) on the server of the National Center for Biotechnology Information.
Acknowledgments
We thank Shoshana Tel-Or for interest and experimental support; Dr. Harry Mark Greenblatt for IT help and discussions; Dr. Linda Shimon for skillful services; Prof. Micha Fridman for helpful discussions; and staff members at Beamlines ID23-1 and ID23-2 of the European Synchrotron Radiation Facility and ID19 of the Structural Biology Center at the Advanced Photon Source at Argonne National Laboratory for their assistance during data collection. Avilamycin A was kindly contributed by Prof. Andreas Bechthold. Funding was provided by the National Institutes of Health Grant GM34360; European Research Council Grant 322581; the Kimmelman Center for Macromolecular Assemblies; and the Adams Fellowship Program of the Israel Academy of Sciences and Humanities (M.K.). A.Y. holds the Martin S. and Helen Kimmel Professorial Chair at the Weizmann Institute of Science.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5JVG and 5JVH).
*Krupkin M, et al., The orthosomycins avilamycin and evernimicin block IF2 and A-tRNA binding to the large ribosomal subunit. In: Proceedings of the Ribosome Structure and Function EMBO Meeting, July 6–10, 2016, Strasbourg, France.
†Krupkin M, et al., The orthosomycins avilamycin and evernimicin block IF2 and A-tRNA binding to the large ribosomal subunit. In: Proceedings of the Ribosome Structure and Function EMBO Meeting, July 6–10, 2016, Strasbourg, France.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614297113/-/DCSupplemental.
References
- 1.Schaberg DR, Culver DH, Gaynes RP. Major trends in the microbial etiology of nosocomial infection. Am J Med. 1991;91(3B):72S–75S. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 2.Spera RV, Jr, Farber BF. Multidrug-resistant Enterococcus faecium. An untreatable nosocomial pathogen. Drugs. 1994;48(5):678–688. doi: 10.2165/00003495-199448050-00003. [DOI] [PubMed] [Google Scholar]
- 3.Swartz MN. Hospital-acquired infections: Diseases with increasingly limited therapies. Proc Natl Acad Sci USA. 1994;91(7):2420–2427. doi: 10.1073/pnas.91.7.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNicholas PM, et al. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 2000;44(5):1121–1126. doi: 10.1128/aac.44.5.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böttger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2(4):318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aarestrup FM, Jensen LB. Presence of variations in ribosomal protein L16 corresponding to susceptibility of enterococci to oligosaccharides (avilamycin and evernimicin) Antimicrob Agents Chemother. 2000;44(12):3425–3427. doi: 10.1128/aac.44.12.3425-3427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikolajka A, et al. Differential effects of thiopeptide and orthosomycin antibiotics on translational GTPases. Chem Biol. 2011;18(5):589–600. doi: 10.1016/j.chembiol.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belova L, Tenson T, Xiong L, McNicholas PM, Mankin AS. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc Natl Acad Sci USA. 2001;98(7):3726–3731. doi: 10.1073/pnas.071527498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buzzetti F, et al. Avilamycin. Experientia. 1968;24(4):320–323. German. doi: 10.1007/BF02140794. [DOI] [PubMed] [Google Scholar]
- 10.Wagman GH, Luedemann GM, Weinstein MJ. Fermentation and isolation of everninomicin. Antimicrob Agents Chemother (Bethesda) 1964;10:33–37. [PubMed] [Google Scholar]
- 11.Weitnauer G, et al. Novel avilamycin derivatives with improved polarity generated by targeted gene disruption. Chem Biol. 2004;11(10):1403–1411. doi: 10.1016/j.chembiol.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Wolf H. Avilamycin, an inhibitor of the 30 S ribosomal subunits function. FEBS Lett. 1973;36(2):181–186. doi: 10.1016/0014-5793(73)80364-3. [DOI] [PubMed] [Google Scholar]
- 13.Kofoed CB, Vester B. Interaction of avilamycin with ribosomes and resistance caused by mutations in 23S rRNA. Antimicrob Agents Chemother. 2002;46(11):3339–3342. doi: 10.1128/AAC.46.11.3339-3342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orelle C, et al. Protein synthesis by ribosomes with tethered subunits. Nature. 2015;524(7563):119–124. doi: 10.1038/nature14862. [DOI] [PubMed] [Google Scholar]
- 15.Adrian PV, et al. Evernimicin (SCH27899) inhibits a novel ribosome target site: Analysis of 23S ribosomal DNA mutants. Antimicrob Agents Chemother. 2000;44(11):3101–3106. doi: 10.1128/aac.44.11.3101-3106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarazaga M, Tenorio C, Del Campo R, Ruiz-Larrea F, Torres C. Mutations in ribosomal protein L16 and in 23S rRNA in Enterococcus strains for which evernimicin MICs differ. Antimicrob Agents Chemother. 2002;46(11):3657–3659. doi: 10.1128/AAC.46.11.3657-3659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNicholas PM, et al. Effects of mutations in ribosomal protein L16 on susceptibility and accumulation of evernimicin. Antimicrob Agents Chemother. 2001;45(1):79–83. doi: 10.1128/AAC.45.1.79-83.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adrian PV, et al. Mutations in ribosomal protein L16 conferring reduced susceptibility to evernimicin (SCH27899): Implications for mechanism of action. Antimicrob Agents Chemother. 2000;44(3):732–738. doi: 10.1128/aac.44.3.732-738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitnauer G, et al. An ATP-binding cassette transporter and two rRNA methyltransferases are involved in resistance to avilamycin in the producer organism Streptomyces viridochromogenes Tü57. Antimicrob Agents Chemother. 2001;45(3):690–695. doi: 10.1128/AAC.45.3.690-695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann PA, et al. EmtA, a rRNA methyltransferase conferring high-level evernimicin resistance. Mol Microbiol. 2001;41(6):1349–1356. doi: 10.1046/j.1365-2958.2001.02602.x. [DOI] [PubMed] [Google Scholar]
- 21.Wright DE. The orthosomycins, a new family of antibiotics. Tetrahedron. 1979;35(10):1207–1237. [Google Scholar]
- 22.Keller-Schierlein W, Heilman W, David Ollis W, Smith C. Stoffwechselprodukte von Mikroorganismen. 178. Mitteilung. Die Avilamycine A und C: Chemischer Abbau und spektroskopische Untersuchungen. Helv Chim Acta. 1979;62(1):7–20. German. [Google Scholar]
- 23.Kupfer E, Neupert-Laves K, Dobler M, Keller-Schierle W. Stoffwechselprodukte von Mikroorganismen. 210. Mitteilung. Über die Avilurekanosen A and C and weitere Abbauprodukte Avilamycine A and C. Helv Chim Acta. 1982;65(1):3–12. German. [Google Scholar]
- 24.Boll R, et al. The active conformation of avilamycin A is conferred by AviX12, a radical AdoMet enzyme. J Biol Chem. 2006;281(21):14756–14763. doi: 10.1074/jbc.M601508200. [DOI] [PubMed] [Google Scholar]
- 25.Ganguly AK, McCormick JL, Chan TM, Saksena AK, Das PR. Determination of the absolute stereochemistry at the C16 orthoester of everninomicin antibiotics: A novel acid-catalyzed isomerization of orthoesters. Tetrahedron Lett. 1997;38(46):7989–7992. [Google Scholar]
- 26.Mertz JL, et al. Isolation and structural identification of nine avilamycins. J Antibiot. 1986;39(7):877–887. doi: 10.7164/antibiotics.39.877. [DOI] [PubMed] [Google Scholar]
- 27.Beau JM, Jaurand G, Esnault J, Sinaÿ P. Synthesis of the disaccharide CD fragment found in everninomicin-C and -D, avalamycin-A and -C and curamycin-A: Stereochemistry at the spiro-ortholactone center. Tetrahedron Lett. 1987;28(10):1105–1108. [Google Scholar]
- 28.Arenz S, et al. Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome. Proc Natl Acad Sci USA. 2016;113(27):7527–32. doi: 10.1073/pnas.1604790113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16(5):528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17(9):1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 31.Valle M, et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10(11):899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 32.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 33.Whitford PC, et al. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA. 2010;16(6):1196–1204. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanbonmatsu KY, Joseph S, Tung C-S. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci USA. 2005;102(44):15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107(5):679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 36.Bashan A, et al. Ribosomal crystallography: Peptide bond formation and its inhibition. Biopolymers. 2003;70(1):19–41. doi: 10.1002/bip.10412. [DOI] [PubMed] [Google Scholar]
- 37.Gongadze GM, Korepanov AP, Korobeinikova AV, Garber MB. Bacterial 5S rRNA-binding proteins of the CTC family. Biochemistry (Mosc) 2008;73(13):1405–1417. doi: 10.1134/s0006297908130038. [DOI] [PubMed] [Google Scholar]
- 38.Bashan A, et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell. 2003;11(1):91–102. doi: 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 39.Qin Y, et al. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127(4):721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto H, et al. EF-G and EF4: Translocation and back-translocation on the bacterial ribosome. Nat Rev Microbiol. 2014;12(2):89–100. doi: 10.1038/nrmicro3176. [DOI] [PubMed] [Google Scholar]
- 41.Goyal A, Belardinelli R, Maracci C, Milón P, Rodnina MV. Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res. 2015;43(22):10700–10712. doi: 10.1093/nar/gkv869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461(7268):1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 43.Spurio R, et al. The C-terminal subdomain (IF2 C-2) contains the entire fMet-tRNA binding site of initiation factor IF2. J Biol Chem. 2000;275(4):2447–2454. doi: 10.1074/jbc.275.4.2447. [DOI] [PubMed] [Google Scholar]
- 44.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121(5):703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 45.Sprink T, et al. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci Adv. 2016;2(3):e1501502. doi: 10.1126/sciadv.1501502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treede I, et al. The avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA at the guanosine 2535 base and the uridine 2479 ribose. Mol Microbiol. 2003;49(2):309–318. doi: 10.1046/j.1365-2958.2003.03558.x. [DOI] [PubMed] [Google Scholar]
- 47.Jiang Y, Huang YH, Long ZE. De novo whole-genome sequence of micromonospora carbonacea jxnu-1 with broad-spectrum antimicrobial activity, isolated from soil samples. Genome Announc. 2015;3(2):e00174–15. doi: 10.1128/genomeA.00174-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosbacher TG, Bechthold A, Schulz GE. Structure and function of the antibiotic resistance-mediating methyltransferase AviRb from Streptomyces viridochromogenes. J Mol Biol. 2005;345(3):535–545. doi: 10.1016/j.jmb.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 49.Eyal Z, et al. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc Natl Acad Sci USA. 2015;112(43):E5805–14. doi: 10.1073/pnas.1517952112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gabdulkhakov A, Nikonov S, Garber M. Revisiting the Haloarcula marismortui 50S ribosomal subunit model. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 6):997–1004. doi: 10.1107/S0907444913004745. [DOI] [PubMed] [Google Scholar]
- 51.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 52.Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature. 2015;520(7549):640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 53.Harms J, et al. Protein structure: Experimental and theoretical aspects. FEBS Lett. 2002;525(1-3):176–178. doi: 10.1016/s0014-5793(02)02960-5. [DOI] [PubMed] [Google Scholar]
- 54.Nishimura M, et al. Solution structure of ribosomal protein L16 from Thermus thermophilus HB8. J Mol Biol. 2004;344(5):1369–1383. doi: 10.1016/j.jmb.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 55.Kumar RR, Lee JT, Cho JY. Fate, occurrence, and toxicity of veterinary antibiotics in environment. J Korean Soc Appl Biol Chem. 2012;55(6):701–709. [Google Scholar]
- 56.Magnussen JD, Dalidowicz JE, Thomson TD, Donoho AL. Tissue residues and metabolism of avilamycin in swine and rats. J Agric Food Chem. 1991;39(2):306–310. [Google Scholar]
- 57.McLellan TJ, et al. A systematic study of 50S ribosomal subunit purification enabling robust crystallization. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 12):1270–1282. doi: 10.1107/S0907444909039043. [DOI] [PubMed] [Google Scholar]
- 58.Otwinowski Z, Minor W. Denzo and Scalepack. In: Arnold E, Himmel DM, Rossmann MG, editors. International Tables for Crystallography. Volume F: Crystallography of Biological Macromolecules. Kluwer; Dordrecht: 2001. pp. 226–235. [Google Scholar]
- 59.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 61.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30(1):26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 62.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 63.PerkinElmer . ChemBioOffice, Ultra 13.0 Suite. PerkinElmer; Waltham, MA: 2012. [Google Scholar]
- 64.Schüttelkopf AW, van Aalten DMF. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 65.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 66.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 67.Schrodinger L. The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger; LLC, New York: 2010. [Google Scholar]
- 68.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 70. Rigaku (2016) CrysAlis(Pro), v38.41 (Rigaku, Tokyo)
- 71.Spek AL. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Crystallogr C Struct Chem. 2015;71(Pt 1):9–18. doi: 10.1107/S2053229614024929. [DOI] [PubMed] [Google Scholar]
- 72.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]