Significance
We show that the changes in DNA methylation that occur in F1 hybrids of Arabidopsis are mostly dependent on the presence of 24-nt siRNAs at the locus. The methylation change at a locus results in the two alleles becoming similar to each other in methylation pattern. The methylation changes occur through the processes of trans-chromosomal methylation and trans-chromosomal demethylation. These altered methylation states can be inherited in the F2 generation and can be associated with changes in levels of gene activity, which may contribute to the phenotypic heterogeneity in the F2.
Keywords: heterosis, gene expression, hybrid vigor, DNA methylation, RNA-dependent DNA methylation
Abstract
Hybrid Arabidopsis plants undergo epigenetic reprogramming producing decreased levels of 24-nt siRNAs and altered patterns of DNA methylation that can affect gene expression. Driving the changes in methylation are the processes trans-chromosomal methylation (TCM) and trans-chromosomal demethylation (TCdM). In TCM/TCdM the methylation state of one allele is altered to resemble the other allele. We show that Pol IV-dependent sRNAs are required to establish TCM events. The changes in DNA methylation and the associated changes in sRNA levels in the F1 hybrid can be maintained in subsequent generations and affect hundreds of regions in the F2 epigenome. The inheritance of these altered epigenetic states varies in F2 individuals, resulting in individuals with genetically identical loci displaying different epigenetic states and gene expression profiles. The change in methylation at these regions is associated with the presence of sRNAs. Loci without any sRNA activity can have altered methylation states, suggesting that a sRNA-independent mechanism may also contribute to the altered methylation state of the F1 and F2 generations.
Hybrid vigor, heterosis, is important in agriculture due to the increased performance of hybrid plants over parental lines. The increase in performance is attributable to changes in gene expression (1, 2) but how these changes result in the increased performance of the hybrid is still not known. The level of hybrid vigor is believed to correlate with the genetic distance between parental lines; however, hybrid vigor can occur in progeny derived from crosses of genetically similar parents. An example is found in intraspecific crosses between Arabidopsis thaliana accessions. In crosses between C24 and Landsberg erecta (Ler), up to a 100% increase in plant biomass and seed yield is observed (3). The different epigenomes of the two parental accessions could provide the diversity required to initiate gene expression changes and vigor-related characteristics. A number of laboratories have investigated whether there are epigenetic changes in Arabidopsis hybrids, whether they affect gene expression, and whether they contribute to the hybrid vigor phenotype (1, 3–6).
We have previously established that F1 hybrids between Arabidopsis accessions undergo some epigenetic reprogramming. We found that the abundance of 24-nt siRNAs is reduced in F1 hybrids and that a second epigenetic system, DNA methylation (mC), is also altered in the hybrid genome (1, 3, 5). The altered mC patterns in the hybrids involve the processes trans-chromosomal methylation (TCM) and trans-chromosomal demethylation (TCdM), where the mC state of one parental allele is altered to resemble the other allele (reviewed in ref. 7). Due to the presence of siRNAs at regions that undergo TCM/TCdM, we have suggested that siRNAs are the initiating molecules that establish these TCM/TCdM events (reviewed in ref. 8). In the hybrid nucleus, the complement of 24-nt siRNAs can interact with homologous sequences from either parent.
The TCM and TCdM events do not always occur in the early cell divisions of the developing embryo (9). A TCM event overlapping At3g43340 and At3g43350 results in a decrease in expression of both genes and is stably maintained through successive generations where it continues to correlate with altered levels of gene expression (9, 10). The TCM characteristics defined at At3g43340/50 may not be representative of how all TCM events are established in the C24/Ler F1 hybrid system. A more comprehensive approach is required to assess the impact of TCM/TCdM across the entire C24/Ler F1 hybrid genome. In this paper, we determine when the TCM/TCdM events occur during development and if they are inherited into the F2. We examine what impact these events have on gene expression and the importance of sRNAs in initiating the TCM/TCdM events. Understanding of the key mechanisms underlying these altered epigenetic states is essential for determining what impact they may have on the hybrid vigor phenotype.
Results
TCM/TCdM Events Influence mC States in Subsequent Generations.
TCM-mediated mC patterns at the At3g43340/50 locus are inherited into the F2 generation (9). We examined the ability of altered mC states to be retained in subsequent generations on a genome-wide scale by profiling parental, F1, and F2 individuals via whole genome bisulphite sequencing (11, 12). There are 17,382 mCG, 3,149 mCHG, and 8,270 mCHH differentially methylated regions (DMRs) (H represents A, C, or T) between the two parental accessions that could potentially undergo TCM/TCdM in the F1 and be tracked into the F2 generation (Table 1 and SI Appendix, SI Materials and Methods).
Table 1.
Differentially methylated regions
| Context | Methylation level above parent (TCM) in F2 | Methylation level below parent (TCdM) in F2 | Differentially methylated regions |
| CG | 301 | 241 | 17,382 |
| CHG | 111 | 339 | 3,149 |
| CHH | 247 | 4,717 | 8,270 |
Individual F2 plants were genotyped and chromosomal segments defined as being homozygous C24, homozygous Ler, or heterozygous (SI Appendix, Table S1 and Fig. S1A). mC levels of homozygous regions of the F2 were compared with the mC level of the parent of origin for that segment. If the segment in the F2 had mC levels different from the parent of origin, it was assumed to have inherited a TCM/TCdM-mediated mC pattern from the F1 hybrid (SI Appendix, Fig. S1B and SI Materials and Methods). In the F2 individuals, there were 542 mCG, 450 mCHG, and 4,964 mCHH DMRs that differed in mC levels from the parent of origin (Table 1 and SI Appendix, SI Materials and Methods). We classified these windows as TCM- or TCdM-inherited windows. In the F2, mCG-inherited windows contained 11% more TCM events than TCdM events, whereas mCHG- and mCHH-inherited windows are predominantly TCdM (Table 1).
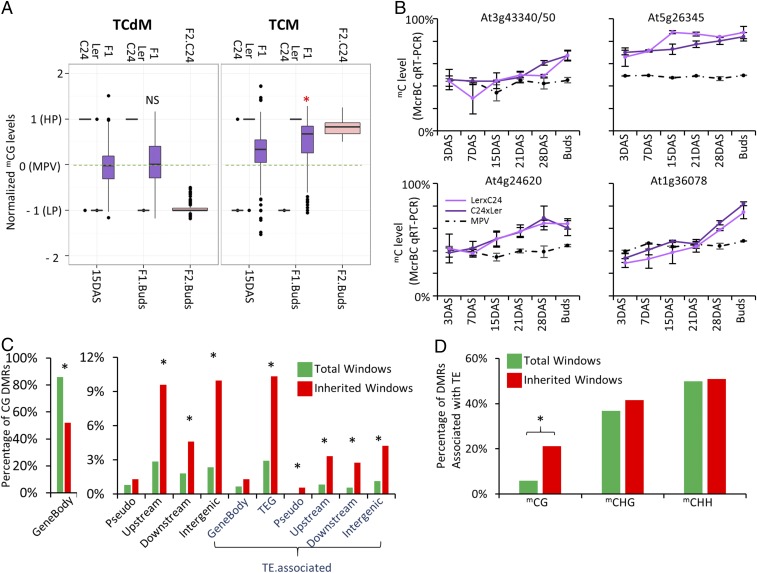
We found that the TCM event at At3g43340/50 had not occurred at 15 days after sowing (DAS) but was present in the immature inflorescence of F1 hybrids (9). We suggested that TCM events may occur due to a build-up of an inducing molecule that triggers a TCM event (reviewed in refs. 8 and 13). To test this on a genome-wide scale, we compared mC levels of the F2-inherited windows at two time points in the F1 hybrid, 15 DAS and immature inflorescence (buds; Fig. 1A). The mCG level at these regions increased from the earlier time point (15 DAS) to the later time point (immature inflorescence), which had levels similar to the F2 (Fig. 1A). Similar observations applied to TCM mCHG- and mCHH-inherited windows (SI Appendix, Fig. S2). Only TCdM mCHH-inherited windows showed clear signs of TCdM at both time points in the F1 hybrid (SI Appendix, Fig. S2). The TCM results were confirmed by analyzing mC levels at four regions in the F1 hybrid over several time points of development (3, 7, 15, 21, and 28 DAS and immature inflorescence). All four regions displayed a build-up of mC over the lifecycle of the F1 hybrid (Fig. 1B). The timing of the TCM event differed between loci, with some attaining mC levels consistent with TCM late in the F1 lifecycle (At3g43340 and At1g36078), whereas at At5g26345, TCM-mediated mC levels occurred earlier than the 3 DAS time point (Fig. 1B).
Fig. 1.
Inheritance of TCM/TCdM-mediated mC states. (A) The mC levels of F2-inherited windows in two time points of F1 hybrid development. F2.C24, inherited windows homozygous for C24 in the F2 individuals (see SI Appendix, Fig. S2 for F2.Ler); 15 DAS, mC levels in F1 15 d whole seedlings; F1.buds, mC levels in F1 hybrid immature inflorescence; and F2.buds, mC levels in F2 immature inflorescence. Dotted green line represents MPV. The * above F1.buds represents a statistical difference in normalized mC levels between F1 15 DAS and F1 buds (P ≤ 0.01; ANOVA test). NS, normalized mC levels between F1 15 DAS and F1 buds that are not statistically significant. (B) McrBC qRT-PCR was used to analyze mC levels at several time points of the F1 hybrid life cycle. (C) Total windows (green) and inherited windows (red) were mapped to different genomic features for mCG. Features were split into those overlapping TEs (TE.associated) and features not associated with TEs. Features mapped to include intragenic (GeneBody), TE genes (TEG), pseudogenes (Pseudo), 1 kb upstream, 1 kb downstream, and intergenic. (D) Association of total DMRs and inherited windows with TEs. χ2 test, *P ≤ 0.01.
mCG-Inherited Windows Are Enriched in Transposable Elements.
To determine whether there were genomic features more prone to the inheritance of altered mC states, we identified where in the genome the inherited windows were located. mCG-inherited windows were underrepresented in the gene body, but enriched in upstream, downstream, transposable element (TE) genes, and intergenic regions (Fig. 1C). There was a 3.5-fold enrichment for mCG-inherited windows in TEs; Fig. 1D), whereas mCHG- and mCHH-inherited windows showed no enrichment for any particular genomic feature (SI Appendix, Fig. S3). TEs are normally heavily methylated in Arabidopsis accessions (5, 14) and a large proportion of the DMRs, both total and inherited, were associated with TEs (Fig. 1D). Our data suggest that TEs that are differentially methylated between parents are susceptible to TCM/TCdM with the resulting methylated state capable of being retained into the F2 generation.
Altered mC States Vary in Their Ability To Be Maintained in the F2 Generation.
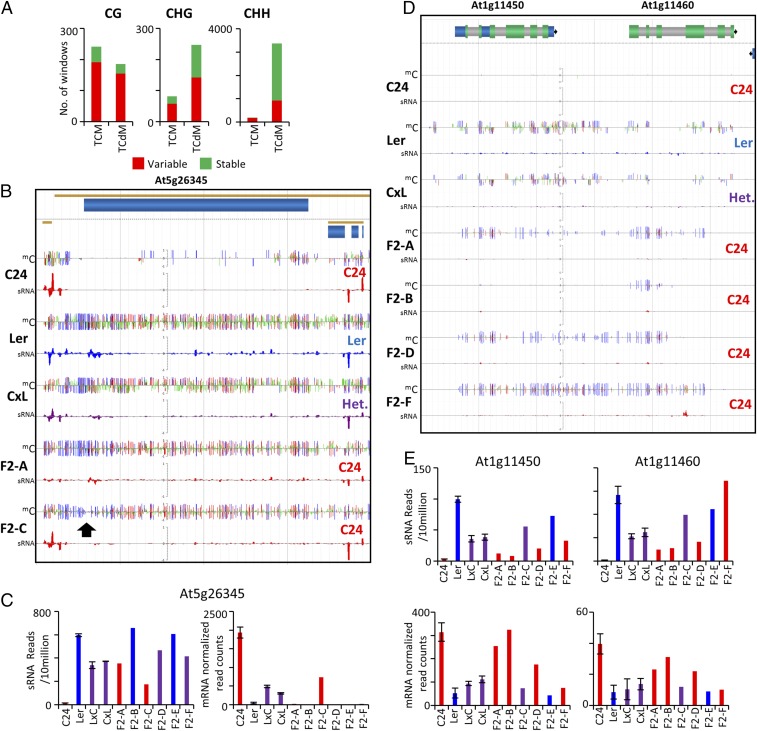
DNA methylation displays variable levels of stability across generations dependent on the molecular mechanisms of initiation and maintenance (15, 16). We determined how variable the altered mC states were between individual F2 plants and if their stability differed between loci. We analyzed all inherited windows in which at least two F2 individuals were homozygous for the allele from the parent of interest. Individual F2 plants varied in their ability to retain altered mCG and mCHG states (Fig. 2A). The majority of mCHH-inherited windows showed a decreased level of mCHH, that once lost in the F1 hybrid, was not regained (Fig. 2A). Stable inheritance occurred at the previously described region At3g43340/50 and at At5g26345 (9; Fig. 2B). At5g26345 is heavily methylated in Ler with only low levels of mC in C24. Both F1 hybrids show high levels of mC through a TCM event. All F2 individuals, including those homozygous for the unmethylated parental allele C24, retained a high level of mC initiated in the F1 (Fig. 2B and SI Appendix, Fig. S4A). This demonstrates that at At5g26345 the newly methylated C24 alleles in the F2 maintain the altered methylated state even when the inducing Ler allele has been segregated away. The increase in mC over the C24 chromosomal segments also correlated with increased levels of sRNAs and decreased levels of gene expression (Fig. 2C and SI Appendix, Fig. S4A). In the F2-C plant there is a lower level of mCG and mCHG directly over the transcription start site, which may account for the increased expression observed in this plant (Fig. 2B, black arrow).
Fig. 2.
Mode of inheritance between F2 individuals. (A) F2-inherited windows with two or more F2 individuals homozygous for the parent of interest were classified as either stable (all individuals had the same altered mC states) or variable (mC states were different between individuals of the same genotype). (B) Stable inheritance at At5g26345. Genome browser shot of mC and sRNA levels in parents, C24 × Ler F1 hybrid (C×L), and two individuals homozygous for the low methylated parent C24. Black arrow highlights a slight loss of mC at the transcription start site in the F2-C individual, which may result in a low level of expression. (C) sRNA and mRNA levels of parents, F1 hybrids, and the six individual F2 plants at At5g26345. (D) Variable inheritance at At1g11450 and At1g11460. Genome browser shot of mC and sRNA levels in parents, C24 × Ler F1 hybrid (C×L), and four individuals homozygous for the low methylated parent C24. (E) sRNA and mRNA levels of parents, F1 hybrids, and the six individual F2 plants at At1g11450 and At1g11460. Genotype of each track is defined on the Right with the color of sRNA tracks also representing genotypes C24 homozygous (red), Ler homozygous (blue), and heterozygous (purple). For mC tracks, blue represents mCG, red represents mCHG, and green represents mCHH. In C and E, genotypes are colored red (C24 homozygous), blue (Ler homozygous), and purple (heterozygous).
A TCM event overlapping the two genes At1g11450 and At1g11460 (At1g11450/60) displays variable inheritance of TCM-mediated mC patterns (Fig. 2D and SI Appendix, Fig. S4B). At these genes, the C24 parent is unmethylated, whereas the Ler parent is methylated in all mC contexts. In the F2 population, four of the six individuals (F2-A, F2-B, F2-D, and F2-F) were homozygous for the C24 allele (Fig. 2D and SI Appendix, Fig. S4B). The mC pattern varied between these individuals, with mC levels corresponding to the level of sRNAs over the region in the F2 individuals (Fig. 2 D and E and SI Appendix, Fig. S4B). As in At5g26345, mRNA levels were altered in a manner consistent with high levels of mC and sRNAs suppressing gene expression (Fig. 2E). These results demonstrate that gene expression levels in F2 individual plants do not necessarily follow the pattern seen in the parental allele but instead change in relation to sRNA and mC levels.
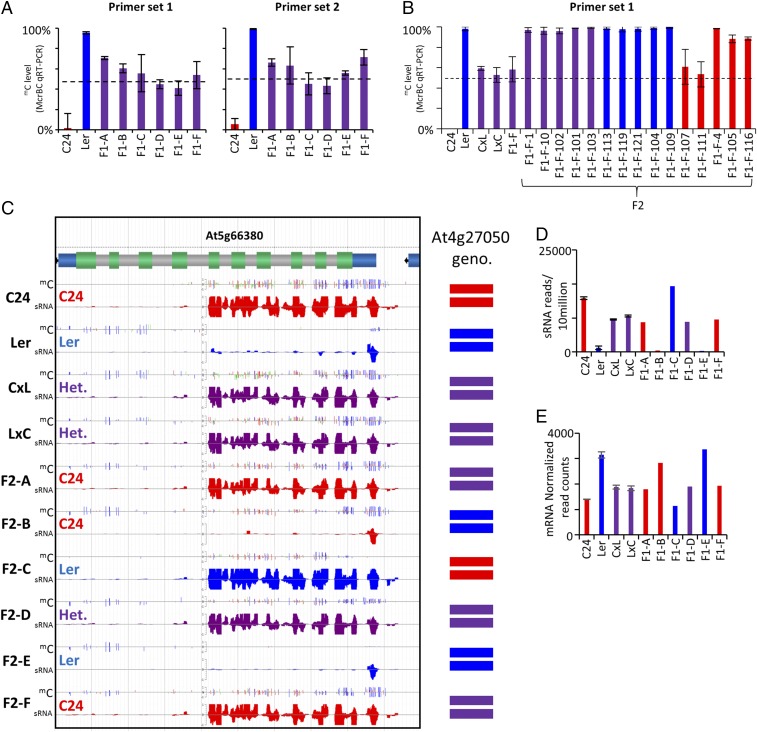
The variable inheritance observed at At1g11450 and At1g11460 in the F2 individuals could reflect varying levels of mC in different parental F1 plants. mC levels at At1g11450 varied between F1 individuals, with some above the midparent value (MPV), consistent with TCM, whereas others were at or below the MPV (Fig. 3A). However, the level of mC in the F1 did not explain the level of mC observed in the F2 progeny (e.g., parental F1-B versus progeny F2-B; Figs. 2D and 3A). To better examine TCM/TCdM stability at this region, we examined 15 F2 siblings derived from a single F1 plant (F1-F). Individuals homozygous for Ler retained the high level of mC observed in the original Ler parent (Fig. 3B). Heterozygous F2 individuals all had mC levels at the high parent Ler, higher than observed in the original heterozygous F1 parent (Fig. 3B). This finding suggests that the F1-acquired mC level is reinforced, potentially through the feedback mechanism of the RNA-dependent DNA methylation pathway (RdDM; reviewed in ref. 17), when present in a heterozygous state for two generations. Individuals homozygous for the unmethylated C24 parent varied in their ability to retain the mC patterns over the C24 chromosomal segment.
Fig. 3.
Mechanisms of variable inheritance into the F2 generation. (A) McrBC qRT-PCR of At1g11450 in the six parental F1 plants each individual F2 plant was derived from. Positions of primer sets 1 and 2 can be seen in SI Appendix, Fig. S4 B (black arrows). (B) McrBC qRT-PCR parents, F1 hybrids, and an additional 15 F2 plants for At1g11450. Dotted line represents MPV. (C) Browser shot of mC and sRNA over At5g66380 in parents, F1 hybrids (C×L and L×C) and six F2 individuals (ATFOLT1). Beside the browser is the genotype at At4g27050, a region that may carry a rearranged copy of ATFOLT1 (ATFOLT2), which initiates mC at At5g66380 (18, 20). (D) sRNA levels across At5g66380. (E) mRNA levels of At5g66380. In graphs, sRNA tracks, and the browser, red represents C24 homozygous, blue represents Ler homozygous, and purple represents heterozygous. For mC tracks, blue represents mCG, red represents mCHG, and green represents mCHH.
At5g66380 (ATFOLT1) also shows variable inheritance of altered mC states with examples of no inheritance (F2-E), TCM (F2-C), and TCdM (F2-B; Fig. 3C). mC and the associated sRNA at this region are due to extra copies of ATFOLT1 present on chromosome 4 at region At4g27050 (18–20). The presence of extra copies in C24 and not Ler (20) results in the different patterns of mC observed at ATFOLT1. Inheritance of the altered mC state in F2 individuals correlates with the presence of the chromosome 4 C24 segment located at the At4g27050 region (Fig. 3C). The absence of this C24 chromosome 4 region, as observed in C24 homozygous F2-B, results in a loss of sRNAs, TCdM over parts of ATFOLT1, and a gene expression level above that observed in the C24 parent (Fig. 3 C–E).
We defined thousands of inherited windows localized to either upstream, downstream, or gene body for each mC context. A comparison of expression levels of these genes suggested that in some cases inherited mCG and mCHG TCM correlate with changes in gene expression (SI Appendix, Fig. S5). We identified 34 potential candidate genes whose expression patterns in the F2 may be a result of changes to mCG and mCHG patterns in their genic regions (SI Appendix, Table S2 and SI Materials and Methods) of which close to one-third were TE genes (SI Appendix, Table S2). Due to the strict parameters used to define these candidate genes, it is likely that 34 is an underestimation of genes in the F2, whose altered expression is linked to epigenetic changes.
sRNAs Are Required for TCM.
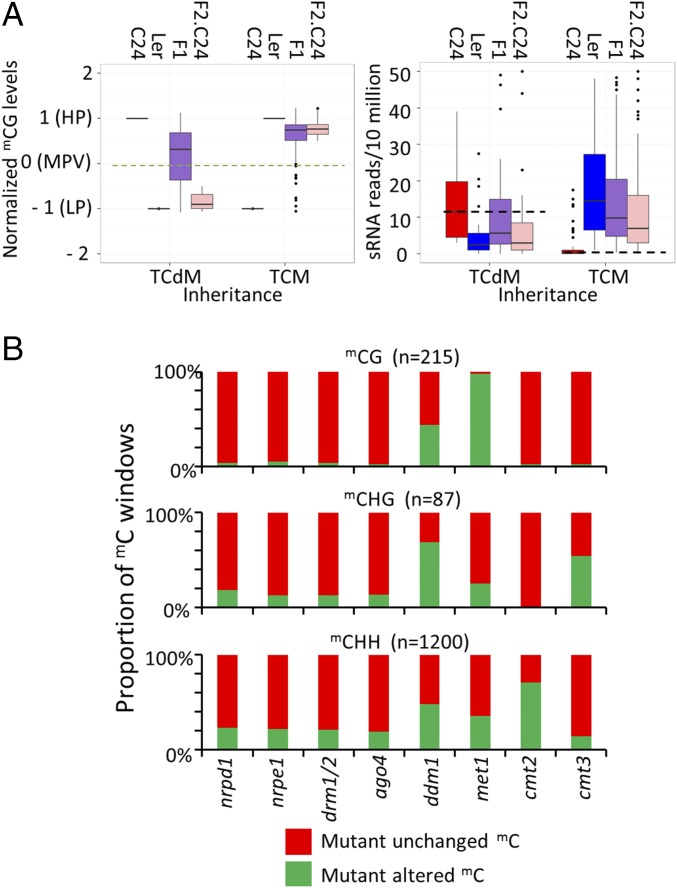
Based on the results at individual loci, sRNAs may play an integral role in the interaction of alleles to produce an altered mC state via TCM. The RdDM pathway provides sequence-specific targeting of de novo methylation to both alleles. To examine the role of sRNAs in TCM events, we determined how many of the total DMRs were associated with sRNAs (sRNA associated ≥5 reads per 10 million). mCG DMRs were largely independent of sRNAs (82.6%), whereas both mCHG and mCHH DMRS were associated with sRNAs (88.5% and 80.0%, respectively; SI Appendix, Fig. S6A). mCG-inherited windows showed a greater association with sRNAs than total mCG DMRs, consistent with sRNAs playing a role in the inheritance of altered mCG patterns (SI Appendix, Fig. S6B). A total of 50% of mCG-inherited windows were sRNA independent, suggesting that at some loci, sRNA-independent mechanisms may result in the altered mC state. At most sRNA-associated inherited windows, sRNA levels correlated with the level of mC (Fig. 4A and SI Appendix Fig. S6C). For example, at TCM-inherited windows, the homozygous chromosomal segment in the F2 is now associated with sRNAs not present in the parental accession (Fig. 4A and SI Appendix, Fig. S6C). This association could result from either the newly methylated segment producing sRNAs or from segregation of another allele in the genome that produces sRNAs.
Fig. 4.
Role of sRNAs in altered mC states. (A) sRNA levels correlate with altered mC states in the F2 populations. Dotted green line represents median mC level of the two parents. The black line represents the median sRNA level in the parental accessions. (B) Enzymes involved in maintaining mC at the F2-inherited windows. n = the number of inherited windows that had coverage and were methylated in Col (21).
To determine the importance of sRNAs in maintaining the altered mC state in the F2 generation, we examined mC patterns at these regions in Columbia (Col) ecotype and in Col plants defective for key regulators of DNA mC (21). mCG-inherited windows in Col were maintained by DECREASE IN DNA METHYLATION 1 (DDM1) and DNA METHYLTRANSFERASE 1 (MET1) (Fig. 4B). Maintenance of mCHG required DDM1 and CHROMOMETHYL TRANSFERASE (CMT)3 with a small number of windows also dependent on components of the RdDM pathway (Fig. 4B). Maintenance of mCHH-inherited windows occurred through DDM1, CMT2, and components of the RdDM pathway (Fig. 4B). These data suggest that sRNA are not essential to maintain the altered mC states observed in the F2 generation.
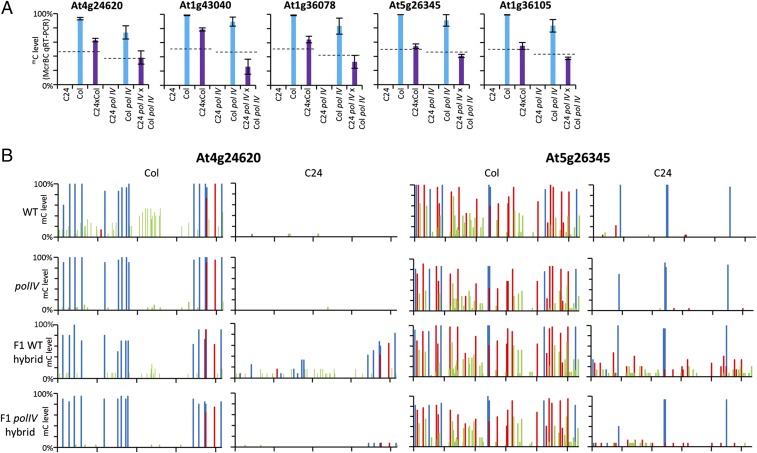
We determined whether sRNAs are required to initiate the altered mC state in the F1 generation. In plants, the majority of 24-nt siRNAs are derived from a noncoding RNA transcript produced by RNA polymerase IV (POL IV) which, through the RdDM pathway, initiates de novo mC. Hybrids devoid of 24-nt siRNAs were produced by crossing a Col polIV mutant (Col polIV) with a C24 polIV mutant (SI Appendix, Fig. S7). Five regions known to undergo TCM in C24 × Col WT hybrids were analyzed in C24 polIV × Col polIV hybrids (polIV hybrid) to determine whether removing 24-nt siRNAs prevented TCM. In all five regions (At4g24620, At1g43040, At1g36078, At5g26345, and At1g36105) Col is methylated, whereas C24 is unmethylated (Fig. 5A). The Col polIV parental line retained most of the mC sites present in WT plants, indicating that 24-nt siRNAs are not required to maintain the majority of mC at these regions. This was important for our analysis, as we required one polIV parent to remain methylated so it could act as the donor in a TCM event. At these regions, mC is maintained by a combination of DDM1, MET1, and CMT3 (21) (SI Appendix, Fig. S8). WT hybrids display mC levels above the MPV, consistent with these regions undergoing a TCM event. Unlike the WT hybrids, the polIV hybrids showed no evidence of TCM at any of the five regions with mC levels at or below the MPV (Fig. 5A). These results were confirmed at At4g24620 and At5g26345 by bisulphite PCR. In both cases the C24 allele gains TCM-mediated mC in the WT hybrid, but not in the polIV hybrid (Fig. 5B). At these regions, POLIV-dependent sRNAs are the inducing molecules initiating de novo methylation of the unmethylated parental allele in the F1 hybrid. The fact that the polIV parents can maintain the mC state independent of sRNAs suggests the sRNAs are not required to maintain the new mC state.
Fig. 5.
sRNAs are required for many TCM events. (A) McrBC qRT-PCR in immature inflorescences of C24 × Col WT and polIV mutants. Dotted line represents MPV. (B) Bisulphite PCR of hybrid and parental lines for a TCM event upstream of At4g24620 and over the gene At5g26345. (Left) Col alleles. (Right) C24 alleles. Blue columns represent mCG, red represent mCHG, and green represent mCHH.
Discussion
Hybrid systems can undergo epigenetic changes that alter gene expression patterns (1, 3–5, 22–24). The epigenetic reprogramming observed in F1 hybrids may happen when the two genomes first come together in the one nucleus. However, in the individual loci we analyzed and on a genome-wide level, TCM events normally occur later in development (9). Only one of the four loci we examined, At5g26345, had TCM-generated levels of mC early in the hybrid life cycle (3 DAS). Similar patterns occur in the maize paramutation locus BOOSTER1 and in Arabidopsis at the PHOSPHORIBOSYLANTHRANILATE ISOMERASE (PAI) locus (25, 26). The mC level at the booster1 locus increases during early development and peaks when the plant has 10 leaves (26). The altered mC state initiated in Ws × Col hybrids at the PAI locus increases over several generations (25). In Arabidopsis VIGS-FWA transgene experiments, initially weak mC silencing of the FWA gene in the infected plants became stronger in subsequent generations (27). These cases are consistent with the requirement for an inducing molecule that increases in concentration over time and triggers the mC change.
Thousands of regions undergo TCM in the F1 hybrid (5). Some of these regions have changes in gene expression potentially affecting the plant phenotype in the F1 generation (1, 5, 9). We have shown that hundreds of these TCM/TCdM events are inherited to the F2 generation. Maize recombinant inbred lines (RILs) derived from a B73 × Mo17 cross displayed a large number of regions that retained an altered mC state compared with the parental conditions and could result in altered gene expression patterns reminiscent of paramutation (28, 29). Soybean RIL populations also have hundreds of regions with mC patterns not matching the mC states of the parental genomes, suggestive of TCM events (30). In epihybrids between Col WT and met1 mutants, thousands of DMRs show the retention of altered mC states. Further changes to these altered mC states can occur in subsequent generations, resulting in plants with novel epigenetic patterns not present in either original parental line (16).
There are differences in the inheritance of altered mC states; in some cases the transmission of altered mC states was high, with At3g43340/50 and At5g26345 maintaining the altered mC states in progeny derived from different F1 hybrids (9, 10). In these cases, the altered mC state correlated with changes in sRNA and mRNA levels. These changes can last for several generations independently of the presence of the original methylated allele (10). Previously we established that mCHH methylation was decreased in the F1 hybrid (9). The loss of mCHH was retained in F2 individuals, suggesting that these regions are unable to restore the mC state in the next generation. In the case of mCG and mCHG, the inheritance of altered mC states differed between F2 individuals derived from different F1 plants. This finding could be the result of different TCM events in the individual F1 plants, segregation of other genomic sequences required to establish and maintain the altered mC state, and/or stochastic variability in the maintenance of the altered mC state across individuals. At1g11450 and At1g11460 had different mC levels in F2 plants derived from different F1 parents, but also between sibling F2 plants derived from the same parent. sRNA levels over these two genes mirrored the differences in mC, with highly methylated individuals having the highest level of sRNAs. Variability in mC states has also been observed in a TE adjacent to FLC in Col/Ler F1 hybrids (31). Loci that vary in mC states in different F1 individuals may not have any impact on the uniform increase in plant biomass associated with F1 hybrid populations. It is likely that given enough samples, even stably inherited windows may have a low level of variability between individuals.
In Sha/Col hybrids, the epigenetic state and correlated change in gene expression of ATFOLT1 (At5g66380) is dependent on its cosegregation with DNA sequences on chromosome 4 (20). In RIL populations derived from the Sha/Col crosses, the silencing and mC of ATFOLT1 was maintained for six generations after the inducing locus was segregated away (20). The C24/Ler system shows the same mC and silencing of ATFOLT1 and is maintained in subsequent generations. However, unlike Sha/Col RILs, the silencing in C24/Ler F2 plants is only maintained if the associated C24 chromosome 4 locus is present. Some of the variable inheritance may be the result of variability between F1 individuals and cosegregation of other genetic elements involved in mediating the mC change. To predict plant phenotype, both the genotype and the overlapping epigenotype need to be considered. This was evident at At1g11450 and At1g11460, which had four F2 individuals with the same genotype but different epigenotypes resulting in differential gene expression.
Two different RdDM pathways involved in mC have been described (27, 32, 33). One pathway is dependent on POLII transcription and RNA DEPENDENT RNA POLYMERASE 6 (RDR6) to generate 21- to 22-nt sRNAs used in initiating mC (RDR6-RdDM). The second pathway involves POLIV transcription, with RDR2 and 24-nt siRNAs directing mC (POLIV-RdDM). Recently a model has been suggested where de novo mC is initiated by RDR6-RdDM, then maintained through POLIV-RdDM (reviewed in ref. 17). Our analysis, in Arabidopsis, of TCM events suggests that POLIV-RdDM alone is sufficient to establish a new mC pattern on the previously unmethylated or low methylated parental allele, which can then be maintained through other pathways.
The ability for a region to undergo TCM may be dependent on a number of factors such as chromatin state, DNA structure, or gene expression levels. These factors may result in a threshold level of sRNAs required to initiate a mC change at a locus. In the case of TCM, the interaction of sRNAs transcribed from one allele in the hybrid is required to initiate mC on the previously low or unmethylated parental allele. In TCdM, the dilution of sRNA molecules over both alleles in the F1 hybrid may fall below a threshold required to establish and/or maintain mC over both alleles, particularly for mCHH (reviewed in ref. 13). The timing of TCM events may reflect an increase in concentration of particular 24-nt siRNAs to a level required for the TCM event. The immature inflorescence has an increased level and diversity of siRNAs, which may result in the higher frequency of TCM events observed in immature inflorescence (34).
In our genome-wide studies, the presence of sRNA-independent inherited windows suggests that mechanisms and pathways other than POLIV-RdDM can be involved in generating the altered mC state (7). A total of 50% of mCG-inherited windows were not associated with sRNAs. Many mCG-inherited windows are located in gene bodies, and as gene body mCG is correlated with gene expression levels (35), it is possible that changes in gene expression in the F2 result in the altered gene body mCG patterns. Another possibility is that the altered mC state may be induced by sRNAs expressed at a specific time of development not analyzed in this paper. Once present, mC maintenance enzymes would maintain the altered mC state without the requirement for continued sRNA production.
The mC changes we observed could alter plant phenotypes and traits important for crop production. Epigenetic RILs derived from crosses between Col ddm1 or met1 mutants with Col WT plants show a variety of phenotypic changes, including flowering time, stem height, and biomass (36–39). Crossing WT Col plants with Col plants deficient in MutS HOMOLOG 1 (MSH1), a plastid and mitochondrial targeting protein, also have genome-wide mC changes (40). Selection of F4 lines derived from this cross show enhanced growth, suggesting that altered epigenetic states can influence the potential for growth to produce vigor-like characteristics (40, 41). Through our genome-wide analysis, we identified 34 genes with altered epigenetic states and changes to gene expression. As with the above examples, there is no clear evidence that the epigenetic changes and their impact on gene expression have a functional significance for the vigor phenotype of F1 hybrid plants.
We have established that the epigenetic reprogramming undertaken in the F1 hybrid requires, in many cases, changes in sRNA activity to establish the new mC pattern. Once these patterns are established, they can be retained in subsequent generations where they continue to correlate with gene activity changes. It is possible that the heterogeneity in the F2 phenotypes seen in hybrid systems is a result of gene expression patterns associated with genetic segregation but also with the altered epigenetic state of loci in F2 plants.
A recently published paper (42) studied DNA methylomes of reciprocal C24/Col hybrids and their parents. TCM and TCdM events were identified in the same genomic features as we reported in C24/Ler hybrids. Five loci we identified had TCM in both the C24/Ler and C24/Col hybrids but there could be many differences in the TCM patterns in the two hybrids as a consequence of sampling at different developmental times and the unique methylomes of Col and Ler. Both analyses conclude that 24nt siRNAs are important for the initiation of TCM and TCdM events. Zhang et al. showed that hybrid vigor is not affected by the abolition of the POLIV/POLV-directed TCM and TCdM events (42). We showed that the altered mC states can affect gene expression but had no evidence that hybrid vigor was affected.
Materials and Methods
Plants were grown in long-day conditions with material collected and frozen in liquid nitrogen. Total RNA was extracted using the Direct-zol RNA Miniprep kit (Zymo Research). DNA was extracted using the Plant DNeasy Mini kit (Qiagen). mRNA and sRNA libraries were prepared following manufacturer instructions and run on an Illumina Hi-Seq platform. Methyl-sequencing (methyl-seq) libraries were prepared as previously described (12). Methylcytosine restricting system BC (McrBC) qRT-PCR and bisulphite PCR were performed as previously described (5, 9). Reads were aligned using Biokanga v 2.95.0 for mRNA and sRNA with methyl-seq reads mapped using Bismark v v0.13.0. See SI Appendix, SI Materials and Methods for more details.
Supplementary Material
Acknowledgments
We thank Dr. Jen Taylor for help with initial discussions in the methylome analysis and Dr. Ming Bo Wang and Mr. Neil Smith for the Northern blot, demonstrating no 24-nt siRNA activity in the C24 polIV mutant.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data including raw sequencing files and processed files reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE85551).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613623113/-/DCSupplemental.
References
- 1.Shen H, et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell. 2012;24(3):875–892. doi: 10.1105/tpc.111.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groszmann M, et al. Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc Natl Acad Sci USA. 2015;112(46):E6397–E6406. doi: 10.1073/pnas.1519926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groszmann M, et al. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA. 2011;108(6):2617–2622. doi: 10.1073/pnas.1019217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha M, et al. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA. 2009;106(42):17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaves IK, et al. Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA. 2012;109(9):3570–3575. doi: 10.1073/pnas.1201043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Varala K, Moose SP, Hudson ME. The inheritance pattern of 24 nt siRNA clusters in Arabidopsis hybrids is influenced by proximity to transposable elements. PLoS One. 2012;7(10):e47043. doi: 10.1371/journal.pone.0047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greaves I, Groszmann M, Dennis ES, Peacock WJ. Trans-chromosomal methylation. Epigenetics. 2012;7(8):800–805. doi: 10.4161/epi.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greaves IK, et al. Epigenetic changes in hybrids. Plant Physiol. 2015;168(4):1197–1205. doi: 10.1104/pp.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES. Inheritance of Trans Chromosomal Methylation patterns from Arabidopsis F1 hybrids. Proc Natl Acad Sci USA. 2014;111(5):2017–2022. doi: 10.1073/pnas.1323656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, et al. Hybrid mimics and hybrid vigor in Arabidopsis. Proc Natl Acad Sci USA. 2015;112(35):E4959–E4967. doi: 10.1073/pnas.1514190112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groszmann M, et al. Epigenetics in plants-vernalisation and hybrid vigour. Biochim Biophys Acta. 2011;1809(8):427–437. doi: 10.1016/j.bbagrm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Vaughn MW, et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5(7):e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334(6054):369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigal M, et al. Epigenome confrontation triggers immediate reprogramming of DNA methylation and transposon silencing in Arabidopsis thaliana F1 epihybrids. Proc Natl Acad Sci USA. 2016;113(14):E2083–E2092. doi: 10.1073/pnas.1600672113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15(6):394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 18.Törjék O, et al. Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor Appl Genet. 2006;113(8):1551–1561. doi: 10.1007/s00122-006-0402-3. [DOI] [PubMed] [Google Scholar]
- 19.Simon M, et al. Quantitative trait loci mapping in five new large recombinant inbred line populations of Arabidopsis thaliana genotyped with consensus single-nucleotide polymorphism markers. Genetics. 2008;178(4):2253–2264. doi: 10.1534/genetics.107.083899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durand S, Bouché N, Perez Strand E, Loudet O, Camilleri C. Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr Biol. 2012;22(4):326–331. doi: 10.1016/j.cub.2011.12.054. [DOI] [PubMed] [Google Scholar]
- 21.Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1–2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He G, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22(1):17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barber WT, et al. Repeat associated small RNAs vary among parents and following hybridization in maize. Proc Natl Acad Sci USA. 2012;109(26):10444–10449. doi: 10.1073/pnas.1202073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chodavarapu RK, et al. Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci USA. 2012;109(30):12040–12045. doi: 10.1073/pnas.1209297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luff B, Pawlowski L, Bender J. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol Cell. 1999;3(4):505–511. doi: 10.1016/s1097-2765(00)80478-5. [DOI] [PubMed] [Google Scholar]
- 26.Haring M, et al. The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J. 2010;63(3):366–378. doi: 10.1111/j.1365-313X.2010.04245.x. [DOI] [PubMed] [Google Scholar]
- 27.Bond DM, Baulcombe DC. Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2015;112(3):917–922. doi: 10.1073/pnas.1413053112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, et al. Mendelian and non-Mendelian regulation of gene expression in maize. PLoS Genet. 2013;9(1):e1003202. doi: 10.1371/journal.pgen.1003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regulski M, et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 2013;23(10):1651–1662. doi: 10.1101/gr.153510.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz RJ, et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013;23(10):1663–1674. doi: 10.1101/gr.152538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai J, et al. Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet. 2008;4(4):e1000056. doi: 10.1371/journal.pgen.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng B, et al. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23(24):2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuthikattu S, et al. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 2013;162(1):116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C, et al. Elucidation of the small RNA component of the transcriptome. Science. 2005;309(5740):1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 35.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39(1):61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 36.Teixeira FK, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323(5921):1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 37.Reinders J, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23(8):939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roux F, et al. Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics. 2011;188(4):1015–1017. doi: 10.1534/genetics.111.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dapp M, et al. Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids. Nat Plants. 2015;1:15092. doi: 10.1038/nplants.2015.92. [DOI] [PubMed] [Google Scholar]
- 40.Virdi KS, et al. Arabidopsis MSH1 mutation alters the epigenome and produces heritable changes in plant growth. Nat Commun. 2015;6:6386. doi: 10.1038/ncomms7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, et al. MutS HOMOLOG1-derived epigenetic breeding potential in tomato. Plant Physiol. 2015;168(1):222–232. doi: 10.1104/pp.15.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, et al. Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci USA. 2016;113(29):E4248–E4256. doi: 10.1073/pnas.1607851113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.