Significance
Inositol polyphosphates and pyrophosphates are ubiquitous eukaryotic messengers and are involved in numerous cellular processes, including insulin signaling and cell migration. Nevertheless, annotation of the specific signaling events underlying these processes has been stymied by the lack of suitable tools. By applying chemically synthesized affinity reagents, we comprehensively annotated inositol polyphosphate binding proteins in the model organism Saccharomyces cerevisiae, revealing a role for these molecules in nucleotide metabolism, ribosome biogenesis, and phosphorylation-based signaling. Furthermore, the reagents enabled the magnesium-dependent isolation of targets of protein pyrophosphorylation, a posttranslational modification mediated by the inositol pyrophosphates. The protein targets are diverse in function and highlight the complex regulation of cellular signaling and metabolic networks by inositol pyrophosphates.
Keywords: inositol pyrophosphates, affinity reagents, protein pyrophosphorylation, signal transduction, metabolism
Abstract
Inositol-based signaling molecules are central eukaryotic messengers and include the highly phosphorylated, diffusible inositol polyphosphates (InsPs) and inositol pyrophosphates (PP-InsPs). Despite the essential cellular regulatory functions of InsPs and PP-InsPs (including telomere maintenance, phosphate sensing, cell migration, and insulin secretion), the majority of their protein targets remain unknown. Here, the development of InsP and PP-InsP affinity reagents is described to comprehensively annotate the interactome of these messenger molecules. By using the reagents as bait, >150 putative protein targets were discovered from a eukaryotic cell lysate (Saccharomyces cerevisiae). Gene Ontology analysis of the binding partners revealed a significant overrepresentation of proteins involved in nucleotide metabolism, glucose metabolism, ribosome biogenesis, and phosphorylation-based signal transduction pathways. Notably, we isolated and characterized additional substrates of protein pyrophosphorylation, a unique posttranslational modification mediated by the PP-InsPs. Our findings not only demonstrate that the PP-InsPs provide a central line of communication between signaling and metabolic networks, but also highlight the unusual ability of these molecules to access two distinct modes of action.
Signal transduction pathways and metabolic circuits are essential for cell homeostasis and survival. These two types of networks have historically been viewed as separate entities, but it is becoming increasingly clear that they must be coordinately regulated. Indeed, growth factor-stimulated signaling pathways can promote the metabolic activity of the cell (1). Conversely, the activity of signaling proteins can be controlled by specific metabolites (2), either by allosteric mechanisms or via nutrient-sensitive covalent modifications, such as acetylation (3) and glycosylation (4).
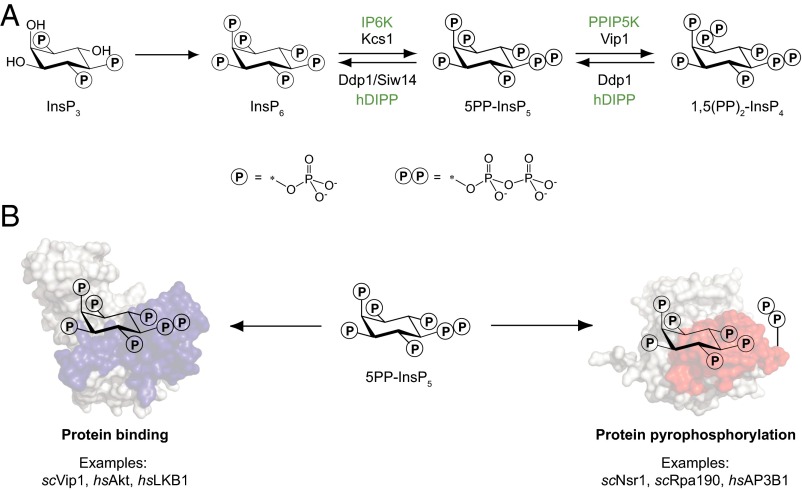
The highly phosphorylated inositol polyphosphates (InsPs), and in particular the inositol pyrophosphates (PP-InsPs), are primed to provide additional junctures between signaling and metabolic networks (5–7). A cascade of phosphorylation reactions converts the secondary messenger inositol trisphosphate (InsP3) to the fully phosphorylated inositol hexakisphosphate (InsP6) (8). Subsequent action of inositol hexakisphosphate kinases (IP6Ks) and diphosphoinositol pentakisphosphate kinases (PPIP5Ks) furnishes the PP-InsP messengers, a unique class of signaling molecules containing one or two high-energy phosphoanhydride bonds (Fig. 1A) (6, 7, 9). A number of studies have indicated a central role for PP-InsPs in metabolic reprogramming and phosphorylation-based signaling at the cellular and organismal level. For example, the biochemical properties of the IP6Ks confer an “energy sensing” function onto these enzymes (10). Because the IP6Ks have a Km for ATP between 1.0 and 1.4 mM—concentrations that are similar to the intracellular ATP levels—the biosynthesis of 5PP-InsP5 is a direct reflection on cellular ATP concentration (11–13). In the eukaryotic model organism Saccharomyces cerevisiae, PP-InsPs were shown to regulate cellular energy expenditure by altering the metabolic flux through glycolysis vs. oxidative phosphorylation, a mechanism that appears to be conserved in mammalian systems (14). In addition, mice lacking the enzyme IP6K1 manifest insulin sensitivity and are resistant to high-fat-diet–induced obesity (15, 16). The observed phenotypes were explained, in part, by an inhibitory activity of 5PP-InsP5, the enzymatic product of IP6K1, toward the protein kinase Akt (16).
Fig. 1.
Biosynthesis and signaling mechanisms of PP-InsPs. (A) Abbreviated biosynthetic pathway for PP-InsPs, involving the action of IP6K and PPIP5K in mammals (green). The corresponding enzymes in S. cerevisiae are shown in black. The pyrophosphate moiety can be cleaved by the mammalian phosphohydrolase hDIPP and the yeast proteins Ddp1 and Siw14. (B) PP-InsPs transmit information via two proposed mechanisms, protein binding and protein pyrophosphorylation, a covalent posttranslational modification. Specific examples for either mechanism are listed. hs, Homo sapiens; sc, S. cerevisiae.
Although this last example provides an intriguing mechanistic hypothesis, in most cases, the PP-InsP–dependent molecular mechanisms and their direct targets have not been identified. Analogous to the phosphatidylinositol lipid messengers, the InsPs and PP-InsPs can exert their pleiotropic functions by binding to protein targets. This mechanism is well characterized for InsP3 (17), and also for inositol tetrakisphosphate (InsP4), inositol pentakisphosphate (InsP5), and InsP6, a number of examples exist in which the small molecules serve as important signals or structural cofactors (18–20). Two recent examples of PP-InsP–interacting proteins include the regulation of casein kinase 2 (CK2) (21) and synaptotagmin (Syt1) (22) by 5PP-InsP5, but canonical binding domains have not been elucidated.
Adding an additional layer of complexity, the PP-InsPs have the ability to transfer their β-phosphoryl group onto phosphoproteins, a process termed protein pyrophosphorylation (Fig. 1B) (23, 24). Pyrophosphorylation is thought to be enzyme-independent, but requires the presence of divalent cations, preferably magnesium ions. This novel posttranslational modification has proven difficult to detect and can presently only be visualized by using radiolabeled PP-InsPs in an in vitro pyrophosphorylation reaction. As a result, a mere handful of pyrophosphorylation targets have been characterized (14, 23, 25, 26).
To identify the cellular targets of highly phosphorylated inositols, we describe the development of specific InsP/PP-InsP affinity reagents, by coupling InsP6 and a nonhydrolyzable analog of 5PP-InsP5 to a solid phase resin. Application of these reagents to S. cerevisiae cell lysates uncovered >150 proteins as putative InsP6 and 5PP-InsP5 interaction partners. Remarkably, the presence or absence of magnesium ions had a profound effect on the protein-binding properties of InsP6 and 5PP-InsP5: When cellular concentrations of magnesium were present, the affinity reagents isolated known and novel substrates of protein pyrophosphorylation. Gene Ontology (GO) analysis of InsP6 and 5PP-InsP5 targets revealed a significant enrichment of proteins involved in ribosome biogenesis and nucleotide metabolism. Our results provide a picture in which the InsPs/PP-InsPs function to establish communication between phosphorylation-based signaling pathways and the cell’s metabolic machinery.
Results
Synthesis of InsP6 and 5PP-InsP5 Affinity Reagents.
Affinity-based probes have been widely used by the inositol community, in particular, to identify interactions between phosphatidyl inositols and their protein targets (27–31). Furthermore, immobilized InsP6 has enabled the identification of two enzymes involved in InsP biosynthesis (12, 32, 33) and allowed for affinity capture of the Ku protein, a required factor for nonhomologous end joining (34). Most recently, tethered InsP6 was used to characterize putative cytosolic targets from a colonic carcinoma cell line (35). However, a comprehensive annotation of cytosolic and nuclear InsP6– and 5PP-InsP5–interacting proteins has been lacking. We therefore pursued the synthesis of resin-bound InsP6 and 5PCP-InsP5, a diphosphoinositol polyphosphate analog containing a nonhydrolyzable bisphosphonate group in the 5-position. Our group previously reported the synthesis of 5PCP-InsP5, and we showed that the bisphosphonate analog closely resembled the natural molecule, both structurally and biochemically, while exhibiting increased stability toward hydrolysis in a cell lysate (36, 37).
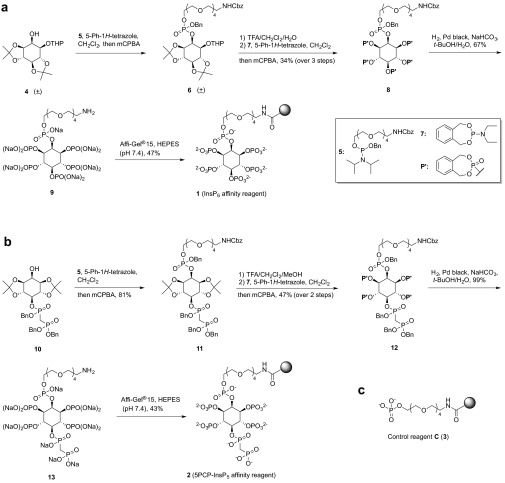
The synthesis of InsP6 affinity reagent 1 started from a racemic mixture of alcohol 4, which can be obtained from myo-inositol in three steps (Scheme 1A and SI Appendix, Scheme S1A) (38, 39). Next, phosphoramidite 5 (SI Appendix, Scheme S1B) was coupled to the inositol ring, followed by oxidation with meta-chloroperoxybenzoic acid. The acid labile protecting groups of 6 were hydrolyzed, and the resulting pentol was phosphitylated with o-xylylene N,N-diethylphosphoramidite (7) and oxidized. Subsequent hydrogenolysis in the presence of sodium bicarbonate removed the o-xylyl, benzyl, and carboxybenzyl protecting groups and afforded sodium salt 9: a molecule of InsP6 with a polyethylene glycol (PEG) linker attached at the 2-position. The primary amine on the PEG linker was coupled to N-hydroxysuccinimide (NHS)-activated Affi-Gel 15 beads in buffered Hepes solution to yield InsP6 affinity reagent 1 (Scheme 1A).
Scheme 1.
Synthesis of affinity reagents. (A) Synthetic procedure to obtain InsP6 affinity reagent 1. (B) Synthetic procedure to obtain 5PCP-InsP5 affinity reagent 2. (C) Control reagent C. See also SI Appendix, Scheme S1.
To obtain the corresponding 5PCP-InsP5 affinity reagent, we used compound 10, which we reported as an intermediate in the synthesis of 5PCP-InsP5 (Scheme 1B) (36). Attachment of the PEG linker proceeded smoothly to afford 11. Removal of the ketals was followed by phosphitylation/oxidation, and the o-xylyl, carboxybenzyl, and benzyl groups were cleaved via hydrogenolysis. The resulting primary amine was coupled to NHS-activated Affi-Gel 15 to provide the 5PCP-InsP5 affinity reagent 2 (Scheme 1B). Lastly, to account for the effects of nonspecific binding to either the resin or the linker, we synthesized control reagent C, which contains the same Affi-Gel 15 resin and PEG linker as reagents 1 and 2 (Scheme 1C). In addition, control reagent C bears a negatively charged phosphate head group, so that nonspecific electrostatic interactions can be detected in subsequent experiments (SI Appendix, Scheme S1C).
Validation of Affinity Reagents and Application to S. cerevisiae Cell Lysates.
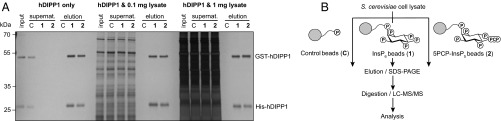
We next evaluated the binding specificity of reagents 1, 2, and C, using the protein human diphosphoinositol polyphosphate phosphohydrolase 1 (hDIPP1). hDIPP1 is a phosphatase that removes the β-phosphoryl group from various PP-InsP substrates (40), and hDIPP1 is known to interact strongly with 5PP-InsP5, 5PCP-InsP5, and InsP6 (37, 41). When recombinant GST- and His-hDIPP1 were exposed to reagents 1, 2, and C, the proteins were retained by reagents 1 and 2, but not by control reagent C (Fig. 2A). After three washes with buffer, the beads were treated with excess InsP6 (or 5PCP-IP5; SI Appendix, Fig. S1) for effective elution of the bound proteins. In a subsequent experiment, 5 μg of GST- and His-hDIPP1 were premixed with a cell lysate from S. cerevisiae at two different concentrations (0.1 and 1 mg/mL). The mixtures were applied to reagents 1, 2, and C, followed by buffer washes and elution. Reagents 1 and 2 remained efficient at binding and retaining the GST- and His-hDIPP1 proteins in the presence of an excess of cell lysate, confirming a strong and specific interaction between the hDIPP1 proteins and the affinity reagents.
Fig. 2.
Validation of affinity reagents and application to cell lysates. (A) hDIPP1 fusion proteins were retained by reagents 1 and 2 in the presence of cell lysate. A total of 5 μg of GST-hDIPP1 and 5 μg of His-hDIPP1 were applied to reagents 1, 2, or C, either in pure form or premixed with a cell lysate from S. cerevisiae at two different concentrations (0.1 and 1 mg/mL). After incubation at 4 °C for 2 h and removal of supernatants, the bound fraction was eluted with InsP6 (10 mM). Samples were separated by SDS/PAGE and visualized by using silver staining. The results were replicated in two independent experiments. (B) General workflow for enrichment procedure from cell lysates. The head groups of the affinity reagents are drawn, and phosphate groups (OPO32−) are depicted as P; the linker and resin are shown as a cartoon.
After validation, we wanted to identify native InsP6– and 5PP-InsP5–interacting partners from a eukaryotic proteome. S. cerevisiae was our model organism of choice, because much of the initial characterization of inositol polyphosphate and pyrophosphate biology relied on the use of this system. Equal amounts of yeast cell lysate were applied to reagents 1, 2, and C, followed by three washes with buffer (Fig. 2B). Because the InsPs have a high propensity for coordinating metal cations, and in particular form strong complexes with magnesium ions (42–44), magnesium concentrations were kept constant at 1 mM (corresponding to the approximate concentration of freely available cellular Mg2+) during affinity enrichment, wash steps, and elution. The eluted fractions were resolved by SDS/PAGE, and the gel lanes were excised. After digestion with trypsin, the samples were analyzed by high-resolution mass spectrometry, and peptide and protein assignments were made through searching the liquid chromatography–tandem MS data against the Saccharomyces Genome Database (45). In total, we found 89 proteins that were enriched fourfold or higher, as assessed by their relative spectral counts, by reagent 1 or 2 compared with control reagent C (Dataset S1). Of those 89 proteins, 53 were not detected in the control sample at all. We observed no correlation between the estimated cellular concentrations of these proteins and their relative abundance following isolation in our experiments (SI Appendix, Fig. S2), further indicative of selective enrichment. In addition, a replicate run showed a high degree of overlap (70 of 89) between two fully independent experiments (Dataset S1).
Interestingly, a distinct preference for InsP6 or 5PCP-InsP5 binding was only apparent in a few cases, and the majority of isolated proteins appeared to display comparable affinities toward 1 and 2. It is possible that the protein-binding properties of these two molecules are highly alike because of their similar charge and structure and that the major difference in signaling properties between InsP6 and 5PP-InsP5 stems from the fact that 5PP-IP5 can participate in phosphoryl-transfer chemistry. Alternatively, because InsP6 has long been viewed as an important structural cofactor (46), the majority of InsP6 binding sites may already be occupied in a cell lysate, and structurally closely related 1 and 2 would predominantly isolate 5PP-InsP5 binding proteins, accounting for their similar proteomic profiles in our assay. Because our objective was to provide a first global picture of the protein-interacting partners of the highly phosphorylated inositols, the results of enrichments with reagents 1 and 2 were evaluated together in subsequent experiments.
InsP6– and 5PCP-InsP5–Interacting Proteins Are Enriched for Nuclear Processes.
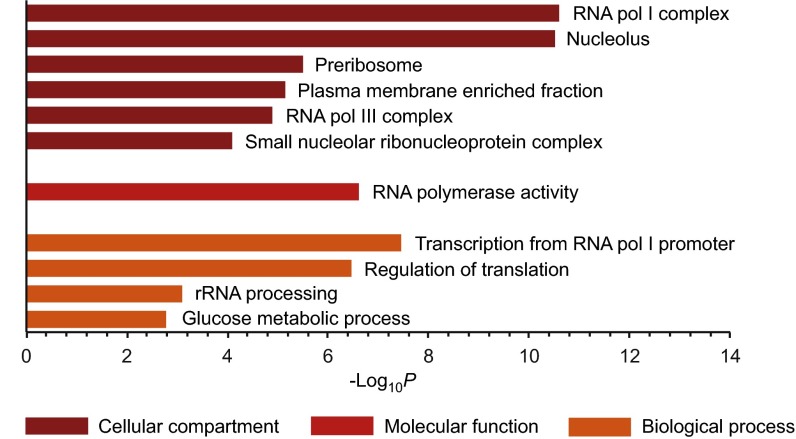
GO analysis of the InsP6 and 5PCP-InsP5 protein targets using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (47, 48) showed a significant overrepresentation (P < 10−3) of nuclear and nucleolar processes. Proteins that localize to the nucleolus were highly enriched (P = 3.1 × 10−11), including components of RNA polymerase I complex, RNA polymerase III complex, and small nucleolar ribonucleoproteins (P = 2.5 × 10−11, 1.3 × 10−5, and 8.1 × 10−5, respectively; Fig. 3 and Dataset S2). Among the overrepresented biological processes were the regulation of translation and rRNA processing (P = 3.4 × 10−7 and 8.0 × 10−4), consistent with the data on cellular localization. This analysis strongly implies a role for the highly phosphorylated inositols in ribosome biogenesis. Indeed, a recent report described a direct function of the PP-InsPs in ribosome biogenesis, by controlling RNA polymerase I activity (25).
Fig. 3.
The affinity reagents isolate a specific set of proteins in the presence of magnesium ions. GO analysis of proteins enriched with magnesium ions present, using GO terms related to cellular compartment, molecular function, and biological process, is shown (see also Dataset S2).
The overrepresentation of GO terms related to RNA biology may be artificially inflated by the possible predilection of the negatively charged inositol head groups to interact electrostatically with arginine-rich motifs (ARMs) in RNA-binding proteins (49, 50). Therefore, we determined the percent composition of arginine of all 89 proteins and compared it to the whole yeast proteome (Dataset S3). We observed no significant difference between the two groups (SI Appendix, Table S1), indicating that the affinity reagents did not preferentially interact with arginine-rich proteins. In addition, DAVID provides information on common protein folds or domains. We explored whether structural commonalities between the isolated InsP6/5PP-InsP5 binding proteins existed, but no such domains, nor ARM domains, were identified within the current dataset. Lastly, 70 of the 89 proteins are characterized phosphoproteins (P = 3.8 × 10−13; SI Appendix, Fig. S3), suggesting a connection between the InsPs and phosphorylation-based cellular signaling pathways. A search for motifs within these 89 proteins that are likely to be phosphorylated by specific kinases revealed a broad range of consensus sites; among the most frequent ones were sites for ribosome biogenesis and tRNA synthetase-associated kinase (Rtk1), casein kinase alpha subunit (Cka1), and Pkb-activating Kinase homolog (Pkh2; Dataset S4).
InsP6 and 5PCP-InsP5 Protein Interactions Reveal Targets of Pyrophosphorylation.
Among the enriched proteins were the known InsP6/5PP-InsP5–interacting proteins Ddp1, the yeast diphosphoinositol polyphosphate hydrolase, and casein kinase 2 (51, 52), although with low abundance. Much more pronounced was the isolation of proteins that are known to undergo pyrophosphorylation, a posttranslational modification that is mediated by the PP-InsPs (23). In fact, five of the seven characterized pyrophosphorylation targets from S. cerevisiae were among the proteins most highly enriched by our reagents (Dataset S1). Because reagents 1 and 2 were effective at isolating established pyrophosphorylation targets, we wondered whether the protein-interaction partners comprised additional substrates of protein pyrophosphorylation.
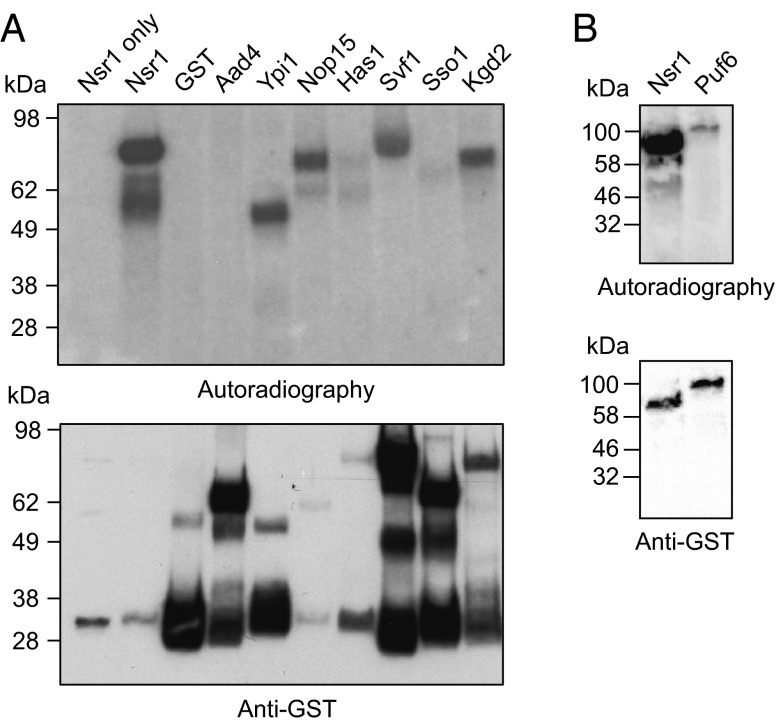
Six proteins (Ypi1, yeast phosphatase inhibitor 1; Nop15, nucleolar protein 15; Has1, helicase associated with Set1; Svf1, survival factor 1; Sso1, plasma membrane t-SNARE; and Kgd2, α-ketoglutarate dehydrogenase 2) were selected from the list of InsP/PP-InsP-protein targets, to explore their proclivity to undergo pyrophosphorylation in vitro. These particular proteins were chosen because they cover a wide range of cellular functions and processes, and their interactions with reagents 1 and 2 were exclusively observed in the presence of magnesium cations (for magnesium-free conditions, see Affinity Reagents Devoid of Magnesium Ions Isolate a Distinct Set of Protein Targets). The six recombinant GST-fusion proteins were expressed and purified from S. cerevisiae to ensure the necessary prephosphorylation by endogenous protein kinases. In addition, we expressed GST-Nsr1, a readily pyrophosphorylated protein (23), as a positive control, and GST-Aad4 and GST alone as negative controls (Aad4 was arbitrarily chosen, it was not identified in any of the proteomic datasets). To see whether in vitro pyrophosphosphorylation could occur on these substrates, the proteins were treated with β[32P]5PP-InsP5 for 40 min at 37 °C. The reaction mixtures were boiled in sample buffer, resolved by SDS/PAGE, and transferred to a PVDF membrane, and the 32P signals were detected by autoradiography. As expected, a strong signal was observed for GST-Nsr1, consistent with covalent modification by β[32P]5PP-InsP5, whereas the control proteins GST and GST-Aad4 were not modified with the radiolabel (Fig. 4A, lanes 2–4). In contrast to the negative control proteins, four of the six tested proteins displayed a robust signal from radiolabeling (Fig. 4A, lanes 5–10), indicative of protein pyrophosphorylation. Alignment of the bands from the autoradiogram with the signals from accompanying anti-GST Western blots confirmed that the fusion proteins GST-Ypi1, GST-Svf1, and GST-Kgd2 corresponded to the pyrophosphorylated species (Fig. 4A and SI Appendix, Fig. S4). However, for GST-Nop15, the major band in the autoradiogram exhibited a higher molecular mass than the confirmed migration of GST-Nop15. MS-based proteomic analysis of the region of the gel displaying strong radioactivity revealed that the Nop15-interacting protein pumilio-homology domain family (Puf6) is the most abundant species, suggesting that Puf6, rather than Nop15, is a substrate of pyrophosphorylation by β[32P]5PP-InsP5. We validated this hypothesis by expressing and purifying GST-Puf6 from S. cerevisiae and subjecting it to treatment with β[32P]5PP-InsP5. As predicted, a band in the autoradiogram was visible for GST-Puf6, migrating at the correct molecular mass (Fig. 4B).
Fig. 4.
The affinity reagents isolate substrates of protein pyrophosphorylation. (A) In vitro pyrophosphorylation assays reveal targets of pyrophosphorylation. GST-fusion proteins purified from S. cerevisiae were treated with 1 μM β[32P]5PP-InsP5 (15 μCi) in 25 mM Tris (pH 7.4, 50 mM NaCl, 6 mM MgCl2, and 1 mM DTT) at 37 °C for 40 min, while still on beads. Reactions were quenched, heated, resolved by SDS/PAGE, transferred to PVDF membranes, and visualized by autoradiography. Protein loading was analyzed by Western blot of the PVDF membrane with an anti-GST antibody (see also SI Appendix, Fig. S4). The predicted molecular masses are as follows: GST-Nsr1, 71 kDa; GST, 26 kDa; GST-Aad4, 63 kDa; GST-Ypi1, 44 kDa; GST-Nop15, 51 kDa; GST-Has1, 83 kDa; GST-Svf1, 80 kDa; GST-Sso1, 59 kDa; and GST-Kgd2, 76 kDa. All pyrophosphorylation reactions were confirmed in at least one additional independent experiment. (B) GST-Puf6 is a pyrophosphorylation substrate, confirmed by the experimental procedure outlined in A.
Previous work by our group showed that the nonhydrolyzable analog 5PCP-InsP5 can inhibit 5PP-InsP5 mediated pyrophosphorylation of Nsr1 (37). In addition, in the case of GST-Puf6 and GST-Svf1, the pyrophosphorylation reaction was effectively inhibited by the addition of excess 5PCP-InsP5 (SI Appendix, Fig. S5). Because InsP6-affinity reagent 1 isolated these proteins as well (Dataset S1), we tested the ability of InsP6 to inhibit pyrophosphorylation, which was indeed the case. Overall, we discovered several targets of protein pyrophosphorylation, which include a phosphatase regulatory subunit (Ypi1), a protein relevant to cell survival and diauxic shift (Svf1), a ribosomal protein (Puf6), and an important mitochondrial enzyme (Kgd2). Contrary to all other characterized pyrophosphorylation targets, Kgd2 does not contain a casein kinase consensus motif, indicating that pyrophosphorylation may not be limited to substrates primed by the constitutively active kinase CK2.
Affinity Reagents Devoid of Magnesium Ions Isolate a Distinct Set of Protein Targets.
Although affinity enrichment with reagents 1 and 2 isolated the known InsP6-interacting proteins Ddp1 and Cka1 from a yeast cell lysate, these first experiments failed to produce other characterized binding partners, including the kinases involved in InsP biosynthesis. Inspection of several crystal structures of InsP6–protein complexes exposed a theme in which large, highly positively charged regions bound to the negatively charged inositol phosphate, completely devoid of any metal cations (SI Appendix, Fig. S6) (19, 53, 54). Consequently, we repeated the affinity enrichments using “metal-free” conditions, where lysis, washing, and elution buffers contained 1 mM EDTA. The metal chelator will remove divalent and trivalent cations, which interact tightly with the highly phosphorylated inositols (44, 45).
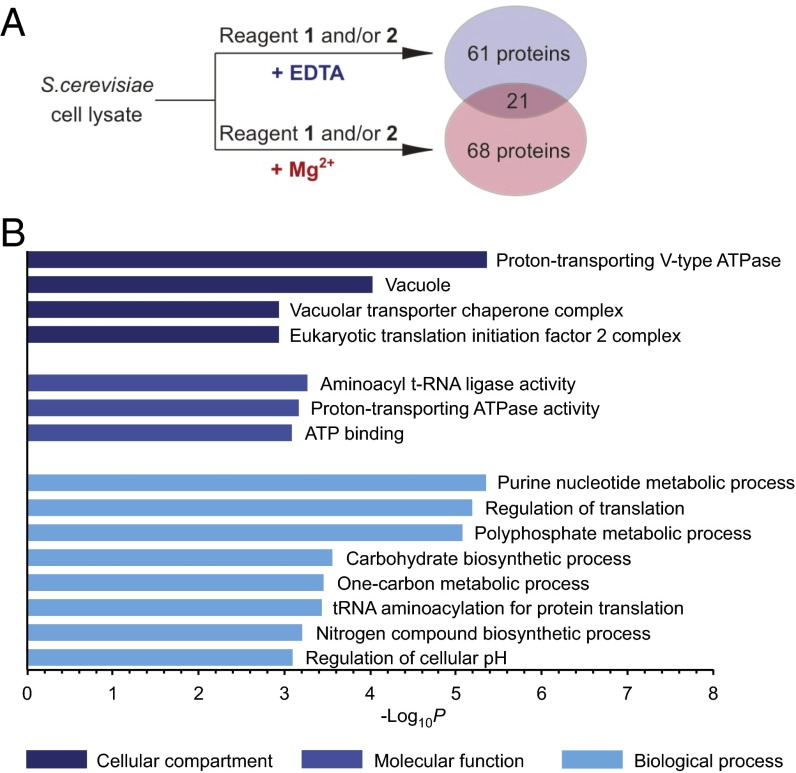
Addition of EDTA significantly altered the protein interactions of reagents 1 and 2, and the majority of the 82 proteins that were isolated with EDTA present had not been enriched in the presence of magnesium ions (Fig. 5A and Dataset S1). Among the isolated proteins were the enzymes involved in PP-InsP synthesis, Kcs1 (protein kinase C1 suppressor), and Vip1 kinase. Furthermore, Gpm1 (phosphoglycerate mutase 1), a protein known to bind to InsP6 (54), was retained by reagents 1 and 2, as well as Pho81, a protein proposed to interact with 1PP-InsP5 (55), a less abundant PP-InsP5 isomer, to mediate phosphate sensing in yeast. Identification of several known InsP– and PP-InsP–interacting partners confirmed the efficiency of the affinity reagents under conditions controlling metal cation availability. However, it should be noted that some of these InsP–protein interactions may be more transient in a cellular setting, where InsPs are likely to form strong complexes with magnesium (43, 44).
Fig. 5.
Enrichment in the absence of divalent ions uncovers a distinct set of protein targets of InsP6 and 5PP-InsP5. (A) Reagents 1 and 2 isolate different proteins, depending on the presence or absence of magnesium ions during affinity enrichment. (B) GO analysis of proteins enriched under conditions restricting metal ion availability, using GO terms related to cellular compartment, molecular function, and biological process (see also SI Appendix, Table S2).
GO analysis of this set of InsP6 and 5PCP-InsP5 targets showed that the highly phosphorylated inositols preferentially interacted with proteins involved in nucleotide metabolism, polyphosphate metabolism, and carbohydrate metabolism (P = 4.5 × 10−6, 8.4 × 10−6, and 2.8 × 10−4 respectively; Fig. 5B and Dataset S2). Furthermore, the highly phosphorylated inositols appear to regulate processes in tRNA acylation and maintenance of cellular pH (P = 3.7 × 10−4 and 8.0 × 10−4). Finally, 67 of the 82 proteins are known phosphoproteins (P = 4.0 × 10−14; SI Appendix, Fig. S3 and Dataset S4), again highlighting a close connection between the InsPs and the cellular signaling machinery.
InsP6 and 5PCP-InsP5 Bind to the Catalytic Core of Vtc4 Inorganic Polyphosphate Polymerase.
Evaluation of the structural commonalities among the set of InsP6/5PP-InsP5 binding proteins that were isolated in the presence of EDTA revealed an overrepresentation of SPX and vacuolar transporter chaperone (VTC) domains (P = 3.7 × 10−4, and P = 8.4 × 10−4, respectively; Dataset S2). This observation is substantiated by a recent report that InsP6/5PP-InsP5 can target basic binding surfaces of SPX domain-containing proteins, thereby triggering the association of SPX proteins with specific transcription factors to regulate phosphate homeostasis in plants (56). Whether the highly phosphorylated inositols could also target VTC domains was a question that had not been addressed; therefore, Vtc4 (vacuolar membrane polyphosphate polymerase) was chosen for additional characterization. Vtc4 catalyzes the synthesis of inorganic polyphosphate (polyP), and, in complex with its VTC complex components Vtc1, Vtc2, and Vtc3, promotes the subsequent transfer of polyP to the vacuole. In Vtc4, the VTC domain interacts with the growing polyphosphate chain and could potentially provide a binding site for the highly phosphorylated inositols (SI Appendix, Fig. S3) (57). Vtc4, together with Vtc2 and Vtc3, were highly enriched in the eluted fractions from affinity reagents 1 and 2 (Dataset S1).
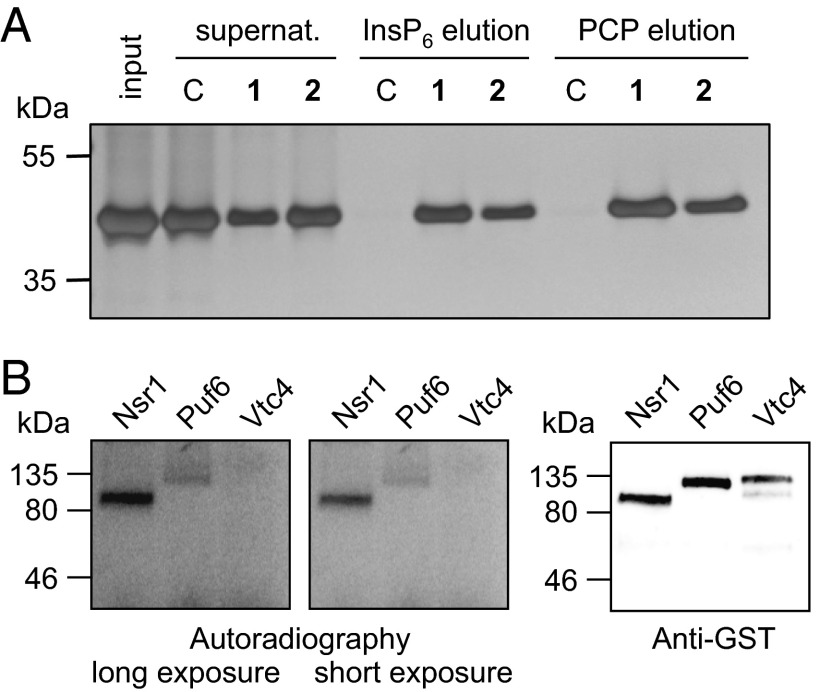
We expressed and purified a recombinant truncated version of Vtc4, His-Vtc4*, that contained the VTC domain, but not the SPX domain (60). Despite lacking the SPX domain, the purified protein bound strongly to reagents 1 and 2, but not to the control resin (Fig. 6A). The interaction of the VTC domain of Vtc4 with 1 and 2 could be disrupted by subsequent elution with either InsP6 or 5PCP-InsP5, and qualitatively, reagents 1 and 2 were equally effective at retaining the truncated His-Vtc4*. Interestingly, Mayer and coworkers observed similar affinities of InsP6 and 5PP-InsP5 toward the SPX domains in biochemical experiments; however, 5PP-InsP5 was a much stronger activator of polyP synthesis when isolated yeast vacuoles—containing intact VTC complexes—were used (56). To investigate whether pyrophosphorylation is involved in the regulation of Vtc4, we expressed and purified full-length Vtc4 from S. cerevisiae. Although the high efficacy of 5PP-InsP5 to stimulate the catalytic activity of Vtc4 may point to a role for protein pyrophosphorylation, we deemed that mechanism unlikely because the Vtc4–PP-InsP interaction did not depend on the presence of magnesium ions, which are a required cofactor for pyrophosphorylation. Indeed, when full-length Vtc4 was treated with β[32P]5PP-InsP5 in the presence of magnesium ions, we observed no labeling of the protein substrate (Fig. 6B).
Fig. 6.
Inositol polyphosphates interact with the VTC domain of Vtc4. (A) The interaction between a truncated version of Vtc4, His-Vtc4*, and reagents 1 and 2 was confirmed. Reagents 1, 2, and C were exposed to 20 μg of His-Vtc4*, incubated for 2 h at 4 °C, and eluted with either 10 mM InsP6 or 10 mM 5PCP-InsP5 solution (PCP). His-Vtc4* input (lane 1), supernatants (lanes 2–4), and eluents (elution with InsP6: lanes 5–7; elution with 5PCP-InsP5, lanes 8–10) were separated by SDS/PAGE and visualized by using silver staining. (B) Vtc4 is not pyrophosphorylated. Recombinant GST-Vtc4 was expressed and purified from S. cerevisiae and treated with 1 μM β[32P]5PP-InsP5 (15 μCi) in 25 mM Tris (pH 7.4, 50 mM NaCl, 6 mM MgCl2, and 1 mM DTT) at 37 °C for 40 min, while still on beads. GST-Nsr1 and GST-Puf6 were included as positive control experiments.
In the future, a more stringent characterization of the sites of interaction, the effect of the different PP-InsP isomers, and the cooperation of the VTC and SPX domain binding sites in the context of the whole Vtc complex will be of great interest, because it can provide a mechanistic rationale for cellular phosphate homeostasis in yeast.
Discussion
Enormous efforts to elucidate the function of lipid-anchored phosphoinositides have uncovered the code among these secondary messengers over the past decades. In contrast, annotating the signaling functions of the soluble inositol phosphates has been slow, stymied by the lack of methods to decipher their mechanisms of action. The synthesis of affinity reagents 1 and 2 is a first step toward meeting this need. Using the affinity reagents, we were able to isolate numerous proteins from an S. cerevisiae cell lysate that were known to interact with InsP6 and 5PP-InsP5, including several proteins involved in PP-InsP metabolism. Furthermore, a large additional set of putative InsP6/5PP-InsP5–interacting proteins was identified, providing an initial survey of InsP6– and 5PP-InsP5–dependent signaling events. The majority of InsP6– and 5PP-InsP5–interacting proteins were phosphoproteins, suggesting an important regulatory role for the InsPs within cellular signaling pathways. Compared with a recently published study on mammalian InsP6-interacting proteins (35), we observed little overlap. Burgess and coworkers focused on cytosolic extracts from mammalian cells and consequently enriched a large number of phosphatidyl inositol binding proteins as potential InsP6 targets. In our experiments, whole-cell lysates from yeast were used, and a theme of nuclear centrality emerged among the putative InsP6– and 5PP-InsP5–interacting proteins. For example, the InsPs/PP-InsPs affinity reagents isolated a disproportionally large number of proteins important for ribosome biogenesis and maturation. Ribosome biogenesis is controlled by the activity of the protein kinase complex TORC1 and constitutes a heavy toll on cellular energy consumption (58). It therefore appears logical that several cellular surveillance mechanisms cooperate, including one that reports both on cellular energy status and phosphate availability, via the InsPs/PP-InsPs. Cellular energy status also needs to be communicated to central metabolic pathways, and the InsPs/PP-InsPs provide critical connections with proteins involved in nucleotide metabolism, polyphosphate metabolism, and carbohydrate metabolism, as revealed by our affinity reagents.
Although a few examples of specific protein interactions with either InsP6 or 5PCP-InsP5 exist within our data, the majority of proteins appeared to have comparable affinities for InsP6 and 5PCP-InsP5. Considering that the cellular concentration of 5PP-InsP5 amounts to only 2–5% of the concentration of InsP6 (5, 6), the lack of specific 5PP-InsP5–interacting proteins was surprising, because it suggests that InsP6 may compete with 5PP-InsP5 binding sites within cells. How can PP-InsPs then act as specific signaling molecules to regulate downstream signaling events without interference from the structurally similar InsP6? We posit that various factors could be responsible for the observed phenomenon. First, the specificity of our reagents may be partially compromised because of the linker attachment at the 2-position. Certain proteins may require a phosphate monoester at the 2-position for efficient binding. As a result, our data may be biased toward proteins that are more tolerant of substitution at the 2-position; hence, the observed overlap between reagents. To address this issue in the future, we are investigating alternative immobilization strategies that will present the InsPs in multiple orientations.
The observed overlap could also be attributed to the fact that InsP6 serves as a structural cofactor, permanently occupying the binding sites on protein targets, and thereby making these sites unavailable for reagents 1 and 2. Consequently, reagents 1 and 2 would principally isolate 5PP-InsP5 binding proteins, which could explain the similar proteomic profiles in our assay. Alternatively, Shears et al. have recently put forward the interesting idea that signaling by PP-InsPs is evolutionarily ancient, and, as a result, the molecules InsP6 and 5PP-InsP5 may have partially overlapping functions (7). Very similar affinities of InsP6 and 5PP-InsP5 were, for example, observed toward PtdInsP3-binding domains (59). It was proposed that more stereospecific interactions of PP-InsP messengers may have evolved more recently with the advent of PPIP5 kinases (7), which add a β-phosphoryl group to InsP6 and 5PP-InsP5 at the 1-position, yielding the metabolites 1PP-InsP5 and 1,5(PP)2-InsP4, respectively. It will be interesting to test this hypothesis in the future, by designing the corresponding affinity reagents. Finally, the major difference in signaling properties between InsP6 and 5PP-InsP5 could stem from the fact that 5PP-InsP5 can participate in phosphoryl-transfer chemistry, whereas InsP6 cannot.
With magnesium ions present in the washing and elution buffers, we isolated the majority of known pyrophosphorylation substrates, an observation that is important for a number of reasons. First, the capture of pyrophosphorylation substrates directly implies a specific interaction between 5PP-InsP5 and its protein targets and suggests that specificity in pyrophosphorylation reactions can, in part, be achieved by selective recognition between the PP-InsP species and the protein substrate. Secondly, the magnesium dependence for isolating pyrophosphorylation targets implies that the interactions between the highly phosphorylated inositols and the protein substrates are metal-mediated. Although a physical interaction of the negatively charged InsP with a negatively charged polyacidic sequence (which is typically encountered around pyrophosphorylation sites) (23) on the protein surface may have seemed counterintuitive, a PP-InsP–magnesium complex could be recognized by a flexible stretch of acidic amino acids on the pyrophosphorylation substrates. Lastly, given the significant enrichment of known pyrophosphorylation targets, our list of binding proteins may contain novel substrates of protein pyrophosphorylation. Indeed, when we tested six of these putative pyrophosphorylation targets, four of them yielded a robust signal for pyrophosphorylation. Certainly, new analytical methodologies for direct identification and characterization of this posttranslational modification are much needed, but our current data provide an unexpected, yet promising, starting point for exploring protein pyrophosphorylation.
To date, no crystallographic data are available for any of the previously characterized pyrophosphorylation sites, but presumably these polyacidic regions recognize and bind to PP-InsP–magnesium complexes. These magnesium-dependent binding sites must differ starkly from the structurally characterized InsP/PP-InsP binding pockets that do not require magnesium. The latter group of InsP/PP-InsP–interacting partners are likely better captured by the group of proteins we isolated in the presence of EDTA, in which the high negative charge of the InsPs/PP-InsPs is neutralized by lysine and arginine side chains within the positively charged binding pockets. A picture thus emerges in which the InsPs/PP-InsPs can interact with two different types of binding sites on proteins, depending on their complexation with magnesium. Whether the basic sites are typically targeted by the PP-InsPs for allosteric regulation and the acidic stretches predominantly constitute pyrophosphorylation sites remains to be seen. Surely, however, the PP-InsPs can access two distinct modes of action, to mediate such a wide array of signaling and metabolic functions.
Conclusions
Overall, the InsPs and PP-InsPs interact with signaling and metabolic networks via two different mechanisms, protein binding and protein pyrophosphorylation. The large majority of proteins that were isolated with affinity reagents 1 and 2 were previously not characterized to exhibit high affinity toward the InsPs/PP-InsPs, and thus constitute a useful resource for the community. Interestingly, most newly identified proteins are known phosphoproteins, immediately implying an important link between the highly phosphorylated inositols, and phosphorylation-based signaling networks. Furthermore, ribosome biogenesis, a process that consumes a large amount of energy, appears to be tightly regulated by the PP-InsPs. Here the PP-InsPs may serve as “metabolic messengers” (60), to report on cellular energy status and phosphate availability. Energy status also needs to be evaluated by the metabolic circuits of the cell, and the InsPs/PP-InsPs interact with multiple proteins involved in nucleotide metabolism, polyphosphate metabolism, and carbohydrate metabolism. Lastly, protein pyrophosphorylation, underappreciated in the past, has the potential to be a widespread signaling mechanism. In addition to known nucleolar pyrophosphorylation substrates, we characterized additional targets, illustrating the broad range of processes that are potentially regulated by this unusual modification. In nature’s ever expanding repertoire of posttranslational modifications, the direct modification of phosphoproteins via a reactive metabolite would provide an elegant regulatory mechanism to communicate cellular energy status to phosphorylation-based signaling pathways.
Materials and Methods
Validation of Reagents 1, 2, and C Using hDIPP1.
A 100-μL suspension of the beads 1, 2, and C in storage solution [2% (wt/vol) sodium azide in water] was washed three times with 1.0 mL of buffer M (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Triton X-100, and 1 mM MgCl2), and a solution of 5 μg of GST-hDIPP1 and 5 μg of His-hDIPP1 in buffer M was added at 4 °C. After incubation for 2 h, the supernatants were removed, and the beads were washed with buffer M (three times with 1.0 mL). For elution, the beads were treated with a solution of either 10 mM InsP6 or 10 mM 5PCP-InsP5 in buffer A (0.2 mL; 50 mM Tris, pH 7.5, 150 mM NaCl, and 0.05% Triton X-100) at 4 °C for 4 h. The eluents were mixed with SDS sample buffer, and all samples were boiled for 5 min and resolved by SDS/PAGE, followed by silver staining. For experiments involving cell lysate, S. cerevisiae cell lysate was prepared from a log phase culture. Briefly, a pellet from a 50-mL culture was treated with 600 μL of buffer A with protease and phosphatase inhibitors; glass beads were added, and cells were lysed by FastPrep (MP Biomedicals). The protein concentration was determined by Bradford assay and a solution (900 μL containing 1 mg or 0.1 mg of protein) was prepared. A total of 5 μg of GST-hDIPP1, 5 μg of His-hDIPP1, and 10 μL of a 100 mM MgCl2 solution were added to the lysate, and the mixture was exposed to a 100-μL suspension of the beads 1, 2, and C in buffer A. After binding and washing, hDIPP1 was eluted with a solution of InsP6 (10 mM), resolved by SDS/PAGE, and visualized by silver staining.
Affinity Enrichment from S. cerevisiae Proteome Using Reagents 1, 2, and C.
A 200-μL suspension of reagents 1, 2, and C in storage solution [2% (wt/vol) sodium azide in water] was washed with buffer M (three times with 1.0 mL) or buffer E (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Triton X-100, and 1 mM EDTA, three times with 1.0 mL) at 4 °C before treatment with 1 mL of cell lysate from S. cerevisiae (1 mg/mL in buffer A), which was prepared from a log-phase culture (see above). An EDTA-free protease inhibitor mixture was used to prepare the cell lysates, to ensure accurate control over the amounts of EDTA and magnesium ions in the samples. The mixtures were shaken for 2 h at 4 °C before removal of the supernatants. The beads then were washed three times with the buffer M or buffer E and eluted with InsP6 elution buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Triton X-100, 1 mM EDTA, and 20 mM InsP6). The eluted fractions were mixed with 4× SDS sample buffer, boiled, and resolved by SDS/PAGE.
Identification of Pyrophosphorylation Substrates.
S. cerevisiae strains containing the constructs for GST-Nsr1, GST, GST-Aad4, GST-Nop15, GST-Has1, GST-Svf1, GST-Sso1, GST-Puf6, GST-Vtc4, GST-Ypi1, and GST-Kgd2 were grown in synthetic dextrose lacking uracil with 2% raffinose and induced by the addition of galactose (to a final concentration of 4%) for 4 h. The cells were lysed as described above, and the GST-tagged proteins were purified by using glutathione beads (GE Healthcare) following the supplier instructions, except for using buffer A for bead washing, and containing protease inhibitors and phosphatase inhibitors. While still on beads, the purified recombinant proteins were treated with ∼1 μM β[32P]5PP-InsP5 (15 μCi) in 25 mM Tris (pH 7.4, 50 mM NaCl, 6 mM MgCl2, and 1 mM DTT) at 37 °C for 40 min. The reactions were quenched by addition of 20 mM EDTA and heated in sample buffer for 10 min at 70 °C. The samples were resolved by SDS/PAGE and transferred to PVDF membranes. Membranes were dried for 1 h and then exposed to MS Film for 4 d at −80 °C. Alternatively, pyrophosphorylation bands were visualized by using a storage phosphor screen exposed for 4 d at −20 °C and imaged with a Typhoon scanner. The protein concentration was analyzed by anti–GST-HRP antibody detection from immunoblotting.
Chemical Synthesis, Cloning and Expression of hDIPP1, Peptide Sample Preparation, Mass-Spectrometric Data Analysis, SI Schemes and Figures, and NMR Spectra.
Detailed procedures for additional experiments are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Muir, Doyle and Sorensen groups for use of their chemicals and instruments; S. Kyin and H. Shwe of the Princeton Proteomics and Mass Spectrometry Core for assistance with sample preparation and data acquisition; Marc Wippich (Leibniz-Institut für Molekulare Pharmakologie) for help with GO analysis; the Mayer group at Université de Lausanne for sharing the Vtc4 construct; and the Andrews group at the University of Toronto for the GST-Ypi1 and GST-Kgd2 constructs. This work was supported by Princeton University and NIH Grant R00 GM087306.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606853113/-/DCSupplemental.
References
- 1.Wellen KE, Thompson CB. A two-way street: Reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13(4):270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 2.Hardie DG, Ross FA, Hawley SA. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdin E, Ott M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16(4):258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 4.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson MSC, Livermore TM, Saiardi A. Inositol pyrophosphates: Between signalling and metabolism. Biochem J. 2013;452(3):369–379. doi: 10.1042/BJ20130118. [DOI] [PubMed] [Google Scholar]
- 6.Wundenberg T, Mayr GW. Synthesis and biological actions of diphosphoinositol phosphates (inositol pyrophosphates), regulators of cell homeostasis. Biol Chem. 2012;393(9):979–998. doi: 10.1515/hsz-2012-0133. [DOI] [PubMed] [Google Scholar]
- 7.Shears SB. Inositol pyrophosphates: Why so many phosphates? Adv Biol Regul. 2015;57:203–216. doi: 10.1016/j.jbior.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monserrate JP, York JD. Inositol phosphate synthesis and the nuclear processes they affect. Curr Opin Cell Biol. 2010;22(3):365–373. doi: 10.1016/j.ceb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty A, Kim S, Snyder SH. Inositol pyrophosphates as mammalian cell signals. Sci Signal. 2011;4(188):re1. doi: 10.1126/scisignal.2001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wundenberg T, Grabinski N, Lin H, Mayr GW. Discovery of InsP6-kinases as InsP6-dephosphorylating enzymes provides a new mechanism of cytosolic InsP6 degradation driven by the cellular ATP/ADP ratio. Biochem J. 2014;462(1):173–184. doi: 10.1042/BJ20130992. [DOI] [PubMed] [Google Scholar]
- 11.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9(22):1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 12.Voglmaier SM, et al. Purified inositol hexakisphosphate kinase is an ATP synthase: Diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc Natl Acad Sci USA. 1996;93(9):4305–4310. doi: 10.1073/pnas.93.9.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J Biol Chem. 2000;275(32):24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- 14.Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334(6057):802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- 15.Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc Natl Acad Sci USA. 2008;105(7):2349–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakraborty A, et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143(6):897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Streb H, Irvine RF, Berridge MJ, Schulz I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature. 1983;306(5938):67–69. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- 18.Watson PJ, Fairall L, Santos GM, Schwabe JWR. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481(7381):335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montpetit B, et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472(7342):238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulsen CE, Armache J-P, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520(7548):511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao F, et al. Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2. Mol Cell. 2014;54(1):119–132. doi: 10.1016/j.molcel.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee TS, et al. Inositol pyrophosphates inhibit synaptotagmin-dependent exocytosis. Proc Natl Acad Sci USA. 2016;113(29):8314–8319. doi: 10.1073/pnas.1521600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhandari R, et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci USA. 2007;104(39):15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306(5704):2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- 25.Thota SG, Unnikannan CP, Thampatty SR, Manorama R, Bhandari R. Inositol pyrophosphates regulate RNA polymerase I-mediated rRNA transcription in Saccharomyces cerevisiae. Biochem J. 2015;466(1):105–114. doi: 10.1042/BJ20140798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc Natl Acad Sci USA. 2009;106(50):21161–21166. doi: 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catimel B, et al. PI(3,4,5)P3 Interactome. J Proteome Res. 2009;8(7):3712–3726. doi: 10.1021/pr900320a. [DOI] [PubMed] [Google Scholar]
- 28.Conway SJ, et al. Synthesis and biological evaluation of phosphatidylinositol phosphate affinity probes. Org Biomol Chem. 2010;8(1):66–76. doi: 10.1039/b913399b. [DOI] [PubMed] [Google Scholar]
- 29.Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nat Chem Biol. 2010;6(7):507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Best MD. Global approaches for the elucidation of phosphoinositide-binding proteins. Chem Phys Lipids. 2014;182:19–28. doi: 10.1016/j.chemphyslip.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 31.Jungmichel S, et al. Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry. Cell Reports. 2014;6(3):578–591. doi: 10.1016/j.celrep.2013.12.038. [DOI] [PubMed] [Google Scholar]
- 32.Marecek JF, Prestwich GD. Synthesis of tethered phytic acid. Tetrahedron Lett. 1991;32(16):1863–1866. [Google Scholar]
- 33.Abdullah M, et al. Purification and characterization of inositol-1,3,4-trisphosphate 5/6-kinase from rat liver using an inositol hexakisphosphate affinity column. J Biol Chem. 1992;267(31):22340–22345. [PubMed] [Google Scholar]
- 34.Jiao C, Summerlin M, Bruzik KS, Hanakahi L. Synthesis of biotinylated inositol hexakisphosphate to study DNA double-strand break repair and affinity capture of IP6-binding proteins. Biochemistry. 2015;54(41):6312–6322. doi: 10.1021/acs.biochem.5b00642. [DOI] [PubMed] [Google Scholar]
- 35.Yin MX, et al. Synthesis of an inositol hexakisphosphate (IP6) affinity probe to study the interactome from a colon cancer cell line. Integr Biol (Camb) 2016;8(3):309–318. doi: 10.1039/c5ib00264h. [DOI] [PubMed] [Google Scholar]
- 36.Wu M, Dul BE, Trevisan AJ, Fiedler D. Synthesis and characterization of non-hydrolysable diphosphoinositol polyphosphate second messengers. Chem Sci (Camb) 2013;4(1):405–410. doi: 10.1039/C2SC21553E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu M, et al. Elucidating diphosphoinositol polyphosphate function with nonhydrolyzable analogues. Angew Chem Int Ed Engl. 2014;53(28):7192–7197. doi: 10.1002/anie.201402905. [DOI] [PubMed] [Google Scholar]
- 38.Chung SK, Chang YT, Kwon YU. Syntheses of all regioisomers of myo-inositol bisphosphate. J Carbohydr Chem. 1998;17(3):369–384. [Google Scholar]
- 39.Godage HY, Riley AM, Woodman TJ, Potter BVL. Regioselective hydrolysis of myo-inositol 1,3,5-orthobenzoate via a 1,2-bridged 2′-phenyl-1′,3′-dioxolan-2′-ylium ion provides a rapid route to the anticancer agent Ins(1,3,4,5,6)P5. Chem Commun (Camb) 2006;(28):2989–2991. doi: 10.1039/b605392k. [DOI] [PubMed] [Google Scholar]
- 40.Safrany ST, et al. A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998;17(22):6599–6607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kilari RS, Weaver JD, Shears SB, Safrany ST. Understanding inositol pyrophosphate metabolism and function: Kinetic characterization of the DIPPs. FEBS Lett. 2013;587(21):3464–3470. doi: 10.1016/j.febslet.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jahnen-Dechent W, Ketteler M. Magnesium basics. Clin Kidney J. 2012;5(Suppl 1):i3–i14. doi: 10.1093/ndtplus/sfr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veiga N, et al. Coordination, microprotonation equilibria and conformational changes of myo-inositol hexakisphosphate with pertinence to its biological function. Dalton Trans. 2014;43(43):16238–16251. doi: 10.1039/c4dt01350f. [DOI] [PubMed] [Google Scholar]
- 44.Hager A, et al. Cellular cations control conformational switching of inositol pyrophosphate analogues. Chemistry. 2016;22(35):12406–12414. doi: 10.1002/chem.201601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherry JM, et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012;40(Database issue) D1:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macbeth MR, et al. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309(5740):1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bayer TS, Booth LN, Knudsen SM, Ellington AD. Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA. 2005;11(12):1848–1857. doi: 10.1261/rna.2167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Varani G. Protein families and RNA recognition. FEBS J. 2005;272(9):2088–2097. doi: 10.1111/j.1742-4658.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- 51.Safrany ST, et al. The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase. Overlapping substrate specificities in a MutT-type protein. J Biol Chem. 1999;274(31):21735–21740. doi: 10.1074/jbc.274.31.21735. [DOI] [PubMed] [Google Scholar]
- 52.Lee W-K, et al. Structural and functional insights into the regulation mechanism of CK2 by IP6 and the intrinsically disordered protein Nopp140. Proc Natl Acad Sci USA. 2013;110(48):19360–19365. doi: 10.1073/pnas.1304670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008;322(5899):265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rigden DJ, Walter RA, Phillips SEV, Fothergill-Gilmore LA. Polyanionic inhibitors of phosphoglycerate mutase: Combined structural and biochemical analysis. J Mol Biol. 1999;289(4):691–699. doi: 10.1006/jmbi.1999.2848. [DOI] [PubMed] [Google Scholar]
- 55.Lee Y-S, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316(5821):109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wild R, et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science. 2016;352(6288):986–990. doi: 10.1126/science.aad9858. [DOI] [PubMed] [Google Scholar]
- 57.Hothorn M, et al. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science. 2009;324(5926):513–516. doi: 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- 58.Fingar DC, Blenis J. Target of rapamycin (TOR): An integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23(18):3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 59.Gokhale NA, Zaremba A, Janoshazi AK, Weaver JD, Shears SB. PPIP5K1 modulates ligand competition between diphosphoinositol polyphosphates and PtdIns(3,4,5)P3 for polyphosphoinositide-binding domains. Biochem J. 2013;453(3):413–426. doi: 10.1042/BJ20121528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bennett M, Onnebo SM, Azevedo C, Saiardi A. Inositol pyrophosphates: Metabolism and signaling. Cell Mol Life Sci. 2006;63(5):552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.