Significance
Despite being the simplest organisms, bacteria have complex subcellular anatomies. How does such organization occur in openly diffusive cytoplasm? We find that the cell pole organizing protein PopZ facilitates network formation by binding directly to at least eight other proteins. The binding region in PopZ is intrinsically disordered, suggesting PopZ has a flexible structure that adopts a different interface for each partner protein. In this way, PopZ resembles eukaryotic hub proteins, such as p53 and BRCA1, which coordinate complex signaling networks. Rapid cycles of binding, unbinding, and rebinding within PopZ networks indicate that bacterial cell poles, similar to their eukaryotic counterparts, are highly dynamic structures.
Keywords: PopZ, intrinsically disordered protein, bacteria, Caulobacter, hub protein
Abstract
Despite their relative simplicity, bacteria have complex anatomy at the subcellular level. At the cell poles of Caulobacter crescentus, a 177-amino acid (aa) protein called PopZ self-assembles into 3D polymeric superstructures. Remarkably, we find that this assemblage interacts directly with at least eight different proteins, which are involved in cell cycle regulation and chromosome segregation. The binding determinants within PopZ include 24 aa at the N terminus, a 32-aa region near the C-terminal homo-oligomeric assembly domain, and portions of an intervening linker region. Together, the N-terminal 133 aa of PopZ are sufficient for interacting with all binding partners, even in the absence of homo-oligomeric assembly. Structural analysis of this region revealed that it is intrinsically disordered, similar to p53 and other hub proteins that organize complex signaling networks in eukaryotic cells. Through live-cell photobleaching, we find rapid binding kinetics between PopZ and its partners, suggesting many pole-localized proteins become concentrated at cell poles through rapid cycles of binding and unbinding within the PopZ scaffold. We conclude that some bacteria, similar to their eukaryotic counterparts, use intrinsically disordered hub proteins for network assembly and subcellular organization.
The bacterial cytoplasm is highly organized, despite the absence of cytoskeletal motors for directed transport or, in most species, internal membranes for compartmentalization. Within this openly diffusing environment, complex structures are formed as individual protein components are added onto existing structures. Well-characterized examples of this include the assembly of the flagellum and the division plane, both of which comprise dozens of distinct protein subunits. In rod-shaped bacteria, the cell poles are also sites for the assembly of complex structures. Electron microscopy has shown that the cytoplasm at the cell poles in Caulobacter is visibly distinct from other areas of the cytoplasm (1), and a genome-wide screen identified more than 80 different proteins that are localized to these locations (2). Although some of these proteins are known to require the presence of an upstream recruitment factor, a fully ordered system of polar assembly has not been established.
An emerging theme for the organization of bacterial cell poles is the recruitment of multiple binding partner proteins through hub-like organizing proteins. A hub-like protein in Gram-positive bacteria called DivIVA self-assembles into a 3D scaffold at sites of negative membrane curvature (3), thereby forming a localized platform that interacts directly with multiple binding partners. Bacillus subtilis DivIVA interacts directly with at least five different binding proteins, including a tethering factor that is important for partitioning the chromosomal centromere into the developing forespore (4). In Gammaproteobacteria, a hub-like transmembrane protein called HubP is targeted to cell poles via a periplasmic domain, and the cytoplasmic region of HubP recruits flagellar assembly components, chemotaxis arrays, and the chromosome centromere partitioning protein ParA (5, 6).
In the Alphaproteobacterium Caulobacter crescentus, polar organizing protein Z (PopZ) is required for the polar localization of at least 11 different proteins. These include cell cycle regulatory proteins that control gene expression and the timing of chromosome replication, although it is not known whether any of these proteins bind directly to PopZ (1). The only two proteins that have been shown to interact directly with PopZ are ParA and ParB, which are responsible for chromosome segregation and anchoring to the cell pole (7). Thus, a common function among DivIVA, HupB, and PopZ is that they all recruit the centromere to the cell pole through direct binding with ParA and/or ParB.
DivIVA, HubP, and PopZ are not homologous in sequence, but they do have overlapping structural characteristics. The C-terminal region of PopZ is analogous to the C-terminal region of DivIVA, in that both form homo-oligomers that undergo higher-order assembly into interconnected scaffold lattices (8, 9). The middle region of PopZ is analogous to a cytoplasmic portion of HubP, in that both are acidic and proline-rich. This region of HubP is required for interacting with ParA, but the protein-binding activity of the analogous region in PopZ has not been tested. As highly charged proline-rich sequences tend to be intrinsically disordered (10), a structural similarity between HubP and PopZ may, in fact, be a lack of structure.
In eukaryotes, proteins with intrinsically disordered domains often act as interaction hubs in multiprotein networks (11). A well-characterized example is p53, which has intrinsically disordered domains in its C and N termini that interact specifically with at least 60 different proteins (12). The sites for protein interaction within intrinsically disordered hub regions are called molecular recognition features (MoRFs), which include a few evolutionarily conserved residues within a larger region of lower conservation and structural disorder (13). Because of their conformational flexibility, MoRFs acquire different structures as they bind to each target protein in their network (12). Such “many-to-one” binding is thought to simplify multicomponent networks by enabling organization around a single hub protein. Eukaryotic genomes, which produce many complex networks, contain a relatively high amount of protein disorder (14), and this has led to the hypothesis that the evolution of higher organisms has favored intrinsically disordered protein interactions as a means of handling complexity (15).
In this work, we find that a 133-aa region within the N terminus of PopZ interacts directly with at least eight other proteins (six from this work and two previously known). Through structural and functional characterizations, we present evidence that this region in PopZ includes a MoRF that facilitates multiprotein complex assembly at the cell poles. In combining an N-terminal network assembly domain with a C-terminal domain that assembles into macromolecular superstructures, PopZ provides a simple mechanism for cell pole organization within the openly diffusing environment of the bacterial cytoplasm.
Results
PopZ Interacts Directly with Multiple Proteins.
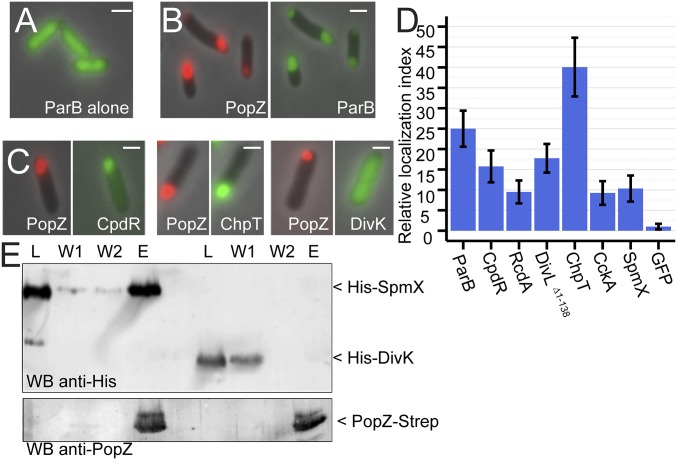
We used Escherichia coli cells to screen a pool of candidate proteins for the ability to interact with Caulobacter PopZ (Fig. 1 A–C). This was possible because PopZ self-assembles into macromolecular complexes that accumulate at E. coli cell poles (16), and these can be visualized as brightly fluorescent foci by expressing PopZ as a fusion with mCherry (mChy). When a PopZ binding protein such as ParA-GFP or GFP-ParB is coexpressed with mChy-PopZ, both proteins colocalize in polar foci (7, 16, 17).
Fig. 1.
Interactions between PopZ and binding partners. (A) Localization of ParB-GFP (green) when expressed independent of PopZ in E. coli. (B) Localization of ParB-GFP (green, Right) when coexpressed with mChy-PopZ (red, Left) in E. coli. (C) mChy-PopZ exhibits selective recruitment of Caulobacter proteins in this assay. (D) The polar localization of each GFP-tagged candidate protein was normalized with respect to free GFP and plotted as a “relative localization index.” (E) Direct interaction between PopZ and SpmX in vitro. Purified 6His-SUMO-SpmX or 6His-SUMO-DivK (lane L) was passed through a column with PopZ bound to the matrix. After washing (lanes W1 and W2), PopZ and other bound proteins were eluted (lane E). (Scale bars, 1 μm.)
We screened 26 Caulobacter proteins for colocalization with mChy-PopZ in E. coli (SI Appendix, Fig. S1). To enrich for potential PopZ binding partners, we selected candidates that have polar localization in Caulobacter or a role in cell cycle regulation. Seven of the candidate proteins formed bright foci independent of PopZ, but in these cases, the GFP-labeled foci either did not colocalize with mChy-PopZ or were only partially overlapping (SI Appendix, Fig. S1). As these candidates appeared to form aggregates that did not interact with PopZ, they were not pursued further. Six other candidates exhibited the expected localization patterns for a PopZ binding protein: disperse when expressed without mChy-PopZ, and localized in polar foci with mChy-PopZ when the proteins were expressed simultaneously (Fig. 1C and SI Appendix, Figs. S1–S4).
To confirm that the colocalizing proteins are in close physical proximity, we performed a bacterial two-hybrid assay. Those candidate proteins that colocalized with PopZ also exhibited elevated galactosidase expression, suggesting a physical interaction (SI Appendix, Fig. S5). To demonstrate that PopZ can interact directly with a candidate protein, we performed an in vitro binding assay (Fig. 1E). We purified epitope-labeled forms of a positive candidate, SpmX, and a negative candidate, DivK, and passed them through a column that contained PopZ bound to a solid matrix. DivK was detected in the column wash fractions, indicating it did not bind to PopZ. SpmX was retained during the washes and eluted together with PopZ, indicating direct binding. In summary, our E. coli colocalization screen yielded six additional candidate binding partners for PopZ.
Localization Determinants Within PopZ-Interacting Proteins.
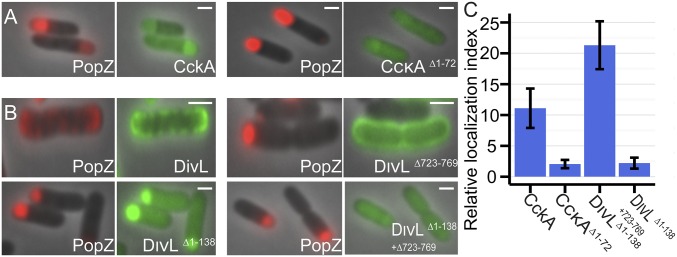
To test the physiological relevance of our E. coli coexpression assay, we asked whether the molecular determinants for polar localization in Caulobacter are also important for PopZ colocalization and recruitment in E. coli. In Caulobacter, the histidine kinase CckA is normally found at cell poles, but truncation of a predicted transmembrane region in the first 72 aa results in disperse localization (18). In our E. coli coexpression system, the same truncation reduced the level of CckAΔ1–72-GFP recruitment to near-background levels (Fig. 2 A and C).
Fig. 2.
Interaction determinants in PopZ binding partners. An E. coli coexpression assay was used to characterize interactions among PopZ and mutant variants of CckA (A) and DivL (B). mChy-PopZ (red, Left) and the CckA-GFP or DivL-GFP variant (green, Right) are shown. (C) Polar recruitment of CckA-GFP and DivL-GFP variants was quantified as in Fig. 1E. (Scale bars, 1 μm.)
Expression of the histidine kinase DivL disrupted the typical PopZ localization patterns we observed in E. coli, although the results still indicate a direct interaction between these proteins (Fig. 2 B and C). When full-length DivL-GFP was coexpressed with mChy-PopZ, both proteins became concentrated in membrane patches at the cell periphery. In Caulobacter, the polar localization determinant in DivL lies in the C-terminal 46 aa of the protein (19), and in our E. coli coexpression system, the same 46-aa region was required for recruiting PopZ into polar patches. To observe interactions without interference from membrane anchoring, we truncated the N-terminal transmembrane region of DivL. We found that DivLΔ1–138-GFP colocalized strongly with mChy-PopZ in polar foci. In combination with truncation of the C-terminal polar localization determinant, interaction with PopZ was lost, and this fragment of DivL was diffuse in the cytoplasm.
All Binding Partners Require Common Elements Within PopZ.
A well-conserved region, the N-terminal 23 aa of PopZ, is required for interaction with the chromosome segregation proteins ParA and ParB (9, 7). We found that the same region is required for interacting with each of the PopZ binding partners we identified in this study (Fig. 3A and SI Appendix, Fig. S3). Previous studies suggest that the middle region of PopZ (named PED for proline, glutamate, aspartate rich), which is not well conserved at the sequence level but is negatively charged and proline-rich in most Alphaproteobacteria, is a flexible linker that serves as a bridge between two conserved domains (9). Consistent with this, we found that the middle region of PopZ could be truncated to less than one-third of its original size without affecting binding partner recruitment in the E. coli coexpression system. Complete removal of PED, however, inhibited recruitment of binding partners to PopZ foci, indicating that a part of this region is required for allowing PopZ to interact with other proteins.
Fig. 3.
Interaction determinants within PopZ. (A) Interactions between PopZ variants and ParB in E. coli. The mChy-PopZ variant (red, Left) and ParB-GFP (green, Right) are shown. We also observed interaction among PopZ and CpdR, RcdA, DivLΔtm, ChpT, CckA, and SpmX (SI Appendix, Fig. S2). (B) PopZ variants tested in these experiments. Potential α-helices, as predicted by JPred3, are represented as orange bars. (C) PopZ variants in A were tested for their ability to recruit CckA-CFP to cell poles in Caulobacter. The eYFP-PopZ variant (yellow, Upper) and CckA-CFP (cyan, Bottom) are shown. (D) Quantification of CckA-CFP localization in C. Green bars represent the percentage of cells with bipolar foci, red bars unipolar foci, and blue bars no foci. (Scale bars, 1 μm.)
To ask whether there are sequence requirements for PED function, we tested the binding activities of PopZ variants that had altered PED sequences (SI Appendix, Fig. S6). We found that the amino acids within PED can be scrambled without affecting binding activity, as previously observed (9), but that changing the negatively charged residues to proline, replacing Glu/Pro with Gly/Ser, or further truncating PED to 12 aa eliminated protein binding. Overall, the results suggest that the identities of individual amino acids at specific positions in PED are not important, but that it requires a minimal length and must also have certain biochemical qualities, perhaps negative charge, to support binding activity.
The C-terminal 75 aa of PopZ are necessary and sufficient for oligomerization and higher-order assembly into macromolecular structures (9). This region includes a predicted helical sequence, termed H2, which is dispensable for subcellular localization, suggesting this portion of PopZ has a different function (20). We tested a ΔH2 variant, mChy-PopZΔ102–133, in our E. coli coexpression system (Fig. 3A), and found that this variant is unable to interact with binding partners. Overall, we conclude that PopZ uses sequences within the conserved N-terminal region, the PED region, and H2 to interact with other proteins (Fig. 3 A and B).
To test the binding activities of PopZ variants in a native context, we produced modified Caulobacter strains in which the wild-type popZ sequence was replaced by a mutant variant and CckA was labeled by expressing it as a CFP-fusion protein (Fig. 3C). We found that the nonbinding mutants in Fig. 3A were defective in recruiting CckA-CFP to Caulobacter cell poles, and they failed to complement the filamentous cell morphology phenotype of the ∆popZ mutant, suggesting loss of function. Conversely, the active variants of PopZ in our E. coli coexpression assay supported normal Caulobacter cell morphology, and they were capable of recruiting CckA-CFP to cell poles.
Higher-Order Assembly and Polar Localization Are Not Required for Interactions with Binding Partners.
Without modification, the E. coli coexpression system cannot be used to test the binding activity of PopZ variants that are defective in polar localization (Fig. 4A). To overcome this limitation, we fused one of the two candidate binding partners to DivIVA, which has been used to target substrate proteins to cell poles based on its tendency to assemble at areas of high membrane curvature (20). DivIVA could be fused to full-length mChy-PopZ without affecting the recruitment of binding partners to cell poles (Fig. 4B). Removing H3 and H4 (Δ134–177) from the DivIVA-mChy-PopZ fusion protein had no effect on the recruitment of binding partners (Fig. 4C), indicating that the homo-oligomerization and higher-order assembly determinants that enable polar localization of wild-type PopZ are not required for interactions with other proteins. However, when we extended the C-terminal truncation by 10 aa (Δ124–177), interactions with binding partners were eliminated (Fig. 4D). Taken together with the results from Fig. 3A, we conclude that H2 and the sequence between H2 and H3 are required for PopZ’s interactions with other proteins.
Fig. 4.
Uncoupling PopZ self-assembly from interactions with other proteins. (A–F) Localization of mChy-PopZ variants (red, Left) and ParB-GFP (green, Right) coexpressed in E. coli. (Scale bars, 1 μm.) (G) Diagrams of the fusion proteins expressed in A–F.
We could also reverse the order of polar recruitment by placing the DivIVA polar localization tag on one of PopZ’s binding partners. We found that mChy-PopZΔ134–177, a cytoplasmic monomer (9), could be recruited to polar foci composed of DivIVA-ParB-GFP (Fig. 4E), and that ParB was required for this interaction (Fig. 4F). To ask whether PopZΔ134–177 can interact with its binding partners outside any polar assemblage, we used a bacterial 2-hybrid assay in which neither component had a polar localization tag and found that the interaction also occurs in this context (SI Appendix, Fig. S5). Overall, we conclude that amino acids 1–133 of PopZ are sufficient for interacting with other proteins, independent of higher-order assembly.
PopZ Is Similar to Intrinsically Disordered Hub Proteins in Eukaryotes.
The protein interaction domains of many eukaryotic hub proteins are intrinsically disordered (12). We used four separate disorder prediction algorithms to analyze PopZ, and all predicted disorder over the protein-binding region in amino acids 1–133 (Fig. 5A). To determine the extent of structural disorder in this region of PopZ experimentally, we purified the PopZΔ134–177 fragment and analyzed it using solution NMR spectroscopy and circular dichroism. The 2D 1H-15N heteronuclear single quantum correlation (HSQC) spectrum (Fig. 5C, red signal, and SI Appendix, Fig. S7) shows well-defined peaks with roughly uniform intensities and line shapes for the majority of resonances. Notably, all the backbone proton peaks were clustered in the 7.80–8.85-ppm region. This narrow dispersion of proton chemical shifts is a strong indicator of an intrinsically disordered protein, compared with well-ordered proteins, which exhibit more broadly distributed peaks in the proton dimension (21). Circular dichroism analysis suggests that PopZ∆134–177 is mostly composed of random coil (Fig. 5B), providing further evidence of structural disorder.
Fig. 5.
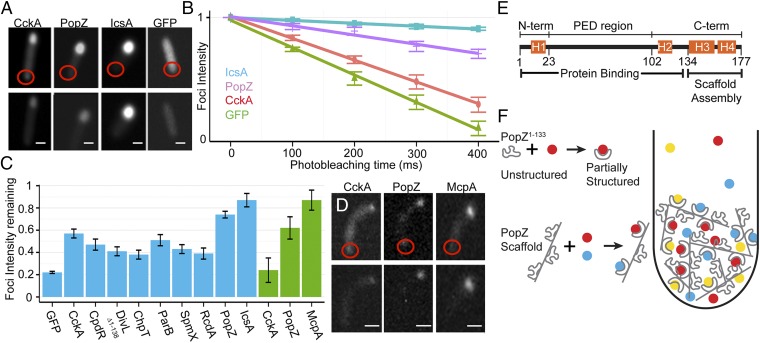
Investigation of PopZ structure. (A) The probability of intrinsic disorder over the primary sequence of PopZ (red line), represented as the average scores from Metadisorder MD2, DnDisorder, MFDp2, and SPINE-D. MoRF probability (blue line), represented as average score from ANCHOR, MoRFpred, and MoRFCHiBi_WEB. Results of individual programs are shown in SI Appendix, Fig. S8. (B) Analysis of PopZ∆134–177 by circular dichroism. Strong negative signal at 195–200 nm indicates random coil. (C) Two-dimensional 1H-15N HSQC NMR spectra of PopZΔ134–177, before mixing (red peaks) and after mixing (black peaks) with the indicated excess unlabeled protein. The two outlier peaks at <7.7 ppm (1H) and 112 ppm (15N) are signals from glutamine side chains and are not an indication of local structure.
In many eukaryotic intrinsically disordered hub proteins, the binding interface includes a short sequence, generally less than 25 aa in length, called a MoRF (13). MoRFs are metastable sequences that often acquire a regular structure on interacting with a binding partner, but they are unable to form strong intrachain interactions on their own (22). They are embedded within larger regions of structural disorder, and they exhibit a high degree of evolutionary conservation in comparison with surrounding sequences (23). This common set of characteristics among experimentally verified MoRFs can be recognized by computational algorithms. Three different prediction algorithms identified a strong MoRF signature near the N-terminal region of PopZ (Fig. 5A) in a sequence that contains the most highly conserved amino acids in the protein (9). As secondary structure prediction programs suggest this sequence also has alpha helical character (Fig. 3B), this is an indication of metastability that is consistent with MoRF-like functionality during interactions with other proteins.
To ask whether the structure of PopZ is affected by interaction with another protein, we repeated our NMR analysis on isotopically labeled PopZΔ134–177, after mixing with RcdA, ChpT, or a noninteracting control protein (Fig. 5C). Whereas the addition of the control protein had little effect on the spectra, the addition of RcdA or ChpT induced significant changes in some of the peaks. Notably, mixing with RcdA or ChpT affected the same set of resonances, suggesting these proteins interact with the same amino acids in PopZΔ134–177. Future analyses will determine whether the affected amino acids are in the N-terminal MoRF-like region, and whether this sequence adopts helical structure on binding.
PopZ Interaction Partners Have Rapid Binding Kinetics in Vivo.
We used a fluorescence loss in photobleaching (FLIP) assay to measure the dynamic behavior of PopZ and PopZ binding proteins in live E. coli cells. To do this, we used a diffraction-limited laser beam to illuminate an area of the cytoplasm opposite the polar focus (Fig. 6A, Upper). Photobleaching occurred as the fluorescent proteins left the polar focus and diffused through the laser-illuminated area (Fig. 6A, Lower). Thus, the observed level of photobleaching is proportional to the rate at which the protein dissociates from the polar focus.
Fig. 6.
Protein dynamics in vivo. (A) Examples of FLIP in E. coli cells, shown before and after photobleaching. The targeted area is shown as a red circle. CckA-GFP was coexpressed with mChy-PopZ. (B) Loss of polar fluorescence intensity over successive rounds of photobleaching, plotted on a log scale. (C) Levels of polar fluorescence remaining after 200 ms photobleaching. Data from E. coli and Caulobacter cells are shown as blue and green bars, respectively. In E. coli, PopZ binding partners were coexpressed with mChy-PopZ. (D) Examples of the FLIP assay in Caulobacter cells. (E) Functional regions in the primary sequence of PopZ. (F) A model of PopZ’s role in polar organization. Colored circles represent proteins that bind to the intrinsically disordered region. The diagram shows a macromolecular complex at a cell pole. PopZ binding partners become concentrated as they undergo repeated cycles of binding, unbinding, diffusion, and rebinding within the PopZ network. (Scale bars, 1 μm.)
To obtain an upper boundary for the dissociation rate, we measured the decrease in polar fluorescence intensity of freely soluble GFP over multiple rounds of photobleaching (Fig. 6B). Because soluble GFP has no affinity for the cell pole, its residence time at this location is limited by the rate of diffusion. To obtain a lower boundary for the rate of polar dissociation, we performed the FLIP assay on IcsA507-620-mChy, a protein that forms aggregates in E. coli (24). We expected that individual proteins within an aggregate would undergo very little exchange with the cytoplasm, and this was supported by our analysis.
Next, we performed FLIP analysis on the PopZ binding protein CckA-GFP and on PopZ itself. We found that mChy-PopZ and PopZ-GFP polar foci were stable compared with free GFP, which is expected for a polymeric assembly. CckA-GFP polar foci were bleached relatively quickly compared with PopZ, with approximately half of the signal lost after 200 ms of photobleaching. Given that the dissociative half-life of a molecular interaction is equal to ln2/koff (25), this provides a minimum value of ∼3.5 s−1 as the koff for the dissociation of CckA-GFP from polar foci of PopZ. This is necessarily an underestimation of the true value of koff because the bleaching rate is further limited by the rate of cytoplasmic diffusion and the area of nonilluminated cytoplasm that released protein must travel across before passing through the laser beam. Still, the lower bound of 3.5 s−1 is above the mean koff for general protein interactions, which is consistent with molecular interactions at intrinsically disordered interfaces (26).
We compared the bleaching of CckA-GFP with other pole-localized PopZ binding proteins, and found that they all exhibited the same highly dynamic behavior (Fig. 6C). We also asked whether CckA and PopZ exhibit dynamic activity in Caulobacter cells (Fig. 6 C and D). Again, CckA-GFP was more rapidly bleached than PopZ-GFP, indicating it is more dynamically associated with the cell pole. The dynamic behavior of CckA-GFP was increased in Caulobacter relative to E. coli cells, possibly because of competition with other proteins for binding to PopZ. Both of these proteins were more dynamic than McpA-GFP, a transmembrane chemoreceptor that forms stable clusters (27).
Discussion
Here, we show that ∼75% of Caulobacter PopZ is intrinsically disordered, and that this region of the protein is necessary and sufficient for interacting with several binding partners. According to our analyses of mutations within this region, we propose that sequences at the N- and C-terminal ends of the disordered region (H1 and H2) are responsible for binding specificity, and that the intervening sequence (PED) is an unstructured linker. Whether H1 and H2 work together to form a binding interface or whether one of these provides indirect support for the other is unknown. The remaining portion of PopZ, amino acids 134–177, is likely to be structured, and is both necessary and sufficient for assembly into a macromolecular scaffold (1, 9, 20). This combination of features allows a single protein to partition the bacterial cytoplasm into distinct spaces with different protein compositions.
In cells, interactions with PopZ normally occur within a 3D scaffold superstructure. It is possible that a protein that enters the network and binds to PopZ will dissociate and then have a significant chance of rebinding to another molecule of PopZ before it escapes from the network. Such rebinding events increase the dwell time of SH2-domain proteins at concentrated patches of activated receptors in plasma membranes (28). The chances of rebinding increase with faster association and dissociation rates, even compared with other interactions that are of equal affinity but are kinetically slower (29). Thus, the rapid binding kinetics we have observed are compatible with a rebinding model. We propose that rapid cycles of binding, detachment, short distance diffusion, and rebinding act as a “speed trap” to slow the rate of movement of PopZ binding proteins after they move through the PopZ network. Just as the concentration of racing cars on a track will increase in low-speed corners, the concentration of PopZ’s binding partners increases as they enter the network.
In Caulobacter cells, PopZ’s direct binding partners are not usually distributed proportionally between the foci of PopZ at the two cell poles. Here, localization may also be influenced by other proteins. An example is TipN, which is localized to the new pole, independent of PopZ, and interacts with the PopZ-binding protein ParA (7). In the absence of TipN, the distribution of ParA changes from being highly concentrated at the new pole to being more evenly distributed between PopZ foci at both poles (30). Other mechanisms for producing asymmetry in polar localization are signaling through phosphorylation and c-di-GMP binding. Five of the six PopZ binding proteins identified in this study are affected, either directly or indirectly, by one or both of these signaling mechanisms (31, 32). An example is the switch from monopolar to bipolar distribution in CpdR mutants that cannot be phosphorylated (33). The E. coli platform we use in this study may be useful in determining which, if any, of these signals affects interaction with PopZ.
We describe a bacterial hub protein that uses an intrinsically disordered domain for organizing complex multiprotein networks at cell poles. Other polar scaffolding proteins that have similar organizational mechanisms may be discovered on the basis of having similarity to PopZ, DivIVA, HupB, and related proteins. In many ways, PopZ’s role in cell pole organization is comparable to FtsZ’s central role as a scaffold in divisome assembly. In particular, FtsZ has an intrinsically disordered region that works in concert with its short, MORF-like C-terminal domain to interact directly with other proteins (34, 35). Ongoing research on the roles of unstructured protein regions in bacteria may reveal a more generalized mechanism for the organization of complex bacterial networks.
Among eukaryotes, there are many well-characterized examples of intrinsically disordered hub proteins, including some, such as p53 and BRCA1, that lie at the center of complex networks (36, 37). Some have hypothesized that intrinsically disordered hub proteins are advantageous in complex systems because they simplify multicomponent networks by allowing many binding partners to be organized around a single hub (15, 14). Dozens of different proteins are localized specifically to Caulobacter cell poles (2). In this light, the role of PopZ in organizing such a complex network in bacteria is consistent with the functions of other intrinsically disordered hub proteins.
Methods
Caulobacter and E. coli strains and culture methods, the construction of plasmids for protein expression, and full details on experimental methods are provided in the SI Appendix.
Imaging was performed on live log phase cells immobilized on an agarose pad. PopZ-dependent polar recruitment was quantified by tracing the outlines of the whole cell, the GFP focus, and diffuse GFP in the cytoplasm. After subtracting background signal, the amount of diffuse fluorescence in the whole cell (defined as quantity B) was calculated by finding the average pixel intensity in the diffuse area of the cytoplasm and multiplying that value by the total area of the cell. The amount of fluorescence enrichment at the cell pole (quantity A) was calculated by measuring the total fluorescence of the polar focus and subtracting the amount of diffuse fluorescence signal that would otherwise be present in that area of cytoplasm. Given these quantities, the percentage of GFP fluorescence attributed to PopZ colocalization is given by the formula (A)/(A + B). A minimum of 200 cells were used for quantification in every experiment. The figures in the main text compare strains that were grown under identical conditions on the same day. Averaging the results from three independent experiments gave very similar results (SI Appendix, Fig. S4). We automated this analysis with computer software and obtained very similar results in cells with visible polar foci (SI Appendix, Fig. S3).
In FLIP experiments, images were collected prebleach and 5-s postbleach. Photobleaching caused by the collection of fluorescence images was measured by calculating the average loss of signal in five nonlaser-targeted foci during image capture, and this value was applied to cells during image processing. A minimum of 50 cells were analyzed in every experiment. Comparisons are between cells that were grown under identical conditions on the same day. For quantifying diffuse GFP, we used a user-defined circle based on the size of mChy-PopZ foci.
For NMR analysis, purified PopZ∆134–177-6His was buffer exchanged into buffer HMK (20 mM Hepes at pH 7.5, 2 mM MgCl2, 100 mM KCl). D2O, NaN3, and DSS were added to a 10% (vol/vol), 4 mM, and 1 mM concentration, respectively, for a final PopZ∆134–177-6His concentration of 150 μM. Mixed samples were prepared by adding purified RcdA, ChpT, or 6His-Sumo-DivK at final concentrations of 1,150, 610, and 600 μM, respectively. Two-dimensional 1H-15N HSQC spectra were collected at 25 °C on a Bruker 600-MHz spectrometer equipped with a 5-mm SmartProbe.
Supplementary Material
Acknowledgments
We thank Ginka Kubelka, Miroslav Tomschik, Adam Perez, and Jay Gatlin for technical expertise; Johan Paulson for providing msfgp; Mark Gomelsky for guidance; and Jennifer Aaberg for technical assistance. This work was supported in part by National Science Foundation Grants MCB-1518171 (to G.R.B.) and CHE-1413696 (to K.V.), and NIH Grant P20GM103432 (to K.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602380113/-/DCSupplemental.
References
- 1.Bowman GR, et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol. 2010;76(1):173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Werner JN, et al. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA. 2009;106(19):7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106(32):13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Baarle S, et al. Protein-protein interaction domains of Bacillus subtilis DivIVA. J Bacteriol. 2013;195(5):1012–1021. doi: 10.1128/JB.02171-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaichi Y, et al. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26(20):2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossmann F, et al. The role of FlhF and HubP as polar landmark proteins in Shewanella putrefaciens CN-32. Mol Microbiol. 2015;98(4):727–742. doi: 10.1111/mmi.13152. [DOI] [PubMed] [Google Scholar]
- 7.Ptacin JL, et al. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci USA. 2014;111(19):E2046–E2055. doi: 10.1073/pnas.1405188111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliva MA, et al. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29(12):1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowman GR, et al. Oligomerization and higher-order assembly contribute to sub-cellular localization of a bacterial scaffold. Mol Microbiol. 2013;90(4):776–795. doi: 10.1111/mmi.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero P, et al. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 12.Oldfield CJ, et al. Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhowmick P, Guharoy M, Tompa P. Bioinformatics approaches for predicting disordered protein motifs. Adv Exp Med Biol. 2015;870:291–318. doi: 10.1007/978-3-319-20164-1_9. [DOI] [PubMed] [Google Scholar]
- 14.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: Disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 15.Schlessinger A, et al. Protein disorder—A breakthrough invention of evolution? Curr Opin Struct Biol. 2011;21(3):412–418. doi: 10.1016/j.sbi.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell. 2008;134(6):956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman GR, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134(6):945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs C, Domian IJ, Maddock JR, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97(1):111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 19.Iniesta AA, Hillson NJ, Shapiro L. Cell pole-specific activation of a critical bacterial cell cycle kinase. Proc Natl Acad Sci USA. 2010;107(15):7012–7017. doi: 10.1073/pnas.1001767107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laloux G, Jacobs-Wagner C. Spatiotemporal control of PopZ localization through cell cycle-coupled multimerization. J Cell Biol. 2013;201(6):827–841. doi: 10.1083/jcb.201303036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyson HJ, Wright PE. Unfolded proteins and protein folding studied by NMR. Chem Rev. 2004;104(8):3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 22.Mészáros B, Simon I, Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLOS Comput Biol. 2009;5(5):e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rokney A, et al. E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J Mol Biol. 2009;392(3):589–601. doi: 10.1016/j.jmb.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Tummino PJ, Copeland RA. Residence time of receptor-ligand complexes and its effect on biological function. Biochemistry. 2008;47(20):5481–5492. doi: 10.1021/bi8002023. [DOI] [PubMed] [Google Scholar]
- 26.Shammas SL, Rogers JM, Hill SA, Clarke J. Slow, reversible, coupled folding and binding of the spectrin tetramerization domain. Biophys J. 2012;103(10):2203–2214. doi: 10.1016/j.bpj.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulmeister S, et al. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci USA. 2008;105(17):6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh D, et al. Fast rebinding increases dwell time of Src homology 2 (SH2)-containing proteins near the plasma membrane. Proc Natl Acad Sci USA. 2012;109(35):14024–14029. doi: 10.1073/pnas.1203397109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagerholm BC, Thompson NL. Theory for ligand rebinding at cell membrane surfaces. Biophys J. 1998;74(3):1215–1228. doi: 10.1016/S0006-3495(98)77836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29(18):3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lori C, et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523(7559):236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 32.Smith SC, et al. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc Natl Acad Sci USA. 2014;111(39):14229–14234. doi: 10.1073/pnas.1407862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iniesta AA, Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc Natl Acad Sci USA. 2008;105(43):16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du S, Park KT, Lutkenhaus J. Oligomerization of FtsZ converts the FtsZ tail motif (conserved carboxy-terminal peptide) into a multivalent ligand with high avidity for partners ZipA and SlmA. Mol Microbiol. 2015;95(2):173–188. doi: 10.1111/mmi.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher MA, Zeng W. Structures of the nucleoid occlusion protein SlmA bound to DNA and the C-terminal domain of the cytoskeletal protein FtsZ. Proc Natl Acad Sci USA. 2016;113(18):4988–4993. doi: 10.1073/pnas.1602327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uversky AV, Xue B, Peng Z, Kurgan L, Uversky VN. On the intrinsic disorder status of the major players in programmed cell death pathways. F1000 Res. 2013;2:190. doi: 10.12688/f1000research.2-190.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadav LR, Rai S, Hosur MV, Varma AK. Functional assessment of intrinsic disorder central domains of BRCA1. J Biomol Struct Dyn. 2015;33(11):2469–2478. doi: 10.1080/07391102.2014.1000973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.