Significance
Local translation in dendrites of neurons has been shown to be important for neuronal function and synaptic biology. We imaged changes in the localization of β-actin mRNA and protein in dendritic spines. Our results showed that activating specific synapses can drive changes in the localization of endogenous mRNA and the translation of reporter RNA in dendrites of hippocampal neurons. Enhancing our understanding of the spatial and temporal kinetics of mRNA localization in dendrites informs local protein synthesis in neurons. These results provide direct evidence of protein synthesis away from the soma and allow us to determine how the kinetics of mRNA localization and translation could influence synaptic physiology and plasticity.
Keywords: single molecule, glutamate uncaging, β-actin, RNA localization, HaloTag
Abstract
Localization of mRNA is required for protein synthesis to occur within discrete intracellular compartments. Neurons represent an ideal system for studying the precision of mRNA trafficking because of their polarized structure and the need for synapse-specific targeting. To investigate this targeting, we derived a quantitative and analytical approach. Dendritic spines were stimulated by glutamate uncaging at a diffraction-limited spot, and the localization of single β-actin mRNAs was measured in space and time. Localization required NMDA receptor activity, a dynamic actin cytoskeleton, and the transacting RNA-binding protein, Zipcode-binding protein 1 (ZBP1). The ability of the mRNA to direct newly synthesized proteins to the site of localization was evaluated using a Halo-actin reporter so that RNA and protein were detected simultaneously. Newly synthesized Halo-actin was enriched at the site of stimulation, required NMDA receptor activity, and localized preferentially at the periphery of spines. This work demonstrates that synaptic activity can induce mRNA localization and local translation of β-actin where the new actin participates in stabilizing the expanding synapse in dendritic spines.
Subcellular localization of mRNA allows control of protein synthesis with respect to space and time (1). By sorting mRNAs to their respective compartments, neurons can regulate translation in response to extracellular signal at the place of protein function (2). Many mRNAs have been shown to be present in dendrites and axons (3), and efforts to characterize mRNA transport revealed that depolarization can lead to detectable increases in alpha calcium calmodulin kinase II (αCaMKII), BDNF, or β-actin mRNAs in dendrites (4–6). Likewise, studies in local translation have shown that dendrites can synthesize the necessary complement of proteins for synaptic plasticity (7). Furthermore, electron microscopy observations of polyribosomes within dendrites and synaptic spines have confirmed that translation occurs readily in neuronal subdomains far from the soma (8, 9). The development and use of fluorescent protein-based translation reporters were pivotal in visualizing local translational output within dendrites (10–12). However, missing from these findings was the high-resolution detection of spatial and kinetic events that result in dynamic repositioning of individual mRNAs and translated proteins within dendrites in response to locally defined input.
Actin is the major cytoskeletal component of dendritic spines where filamentous actin (F-actin) dynamics confer motility and structural plasticity (13). Interestingly, the mRNA that encodes for the most abundant actin isoform in neurons, β-actin, is also present in dendrites in relatively large numbers (3). Similar to β-actin, the mRNAs for PSD-95 and αCaMKII (among many) also are found at high levels, with the latter considered one of the most abundant mRNAs in dendrites. Why such abundant synaptic proteins need to be locally synthesized has raised questions about the relevance of dendritic protein synthesis in neuronal function. Although it has been speculated that the central role these gene products play in synaptic function underlies the critical need for rapid local synthesis, the sheer number of preexisting proteins argues against the production of more proteins for the purpose of increasing quantity. A better understanding of the roles of newly translated synaptic proteins in spines is needed to establish a clear molecular link between local protein synthesis and changes at the synapse.
To investigate whether β-actin mRNA localization and local translation in dendrites is a stimulus-dependent process, we applied high-resolution techniques such as single-molecule imaging and glutamate uncaging to probe the locations of single particles (RNA or protein) at the level of a single spine. First, we assessed how individual endogenous β-actin mRNAs move within dendrites by imaging cultured neurons from the transgenic knock-in mouse in which 24 tandem MS2 aptamers (MBS) were incorporated into the 3′ UTR of the β-actin gene locus (14, 15). To establish the causal relationship between stimulation and mRNA localization, we applied an optical method that could selectively deliver neurotransmitters to dendritic spines. Photolytic uncaging uses focused light to unmask a caged compound and to convert it to its active form with fine spatial and temporal precision and facilitates the efficient delivery of biomolecules to specific subcellular domains (16). Next, to characterize the local synthesis of β-actin in dendrites, we devised a translation reporter system that allows the visualization of both the transcript and the translated product at single-particle resolution using nonrepetitive MS2 aptamers (17) and the self-labeling HaloTag (18), respectively. When combined with tetraalkylrhodamine derivatives conjugated to the Halo-ligand, the HaloTag-labeled fusion proteins can be detected at high resolution in living cells (19). By sequential labeling of Halo-actin fusion proteins with spectrally distinct Halo-ligands, it is possible to distinguish between new and preexisting populations of proteins across time.
In this report, we have characterized the changes in RNA-mediated mechanisms in response to specific input using techniques that provide positional information about single molecules. We show that stimulated spines can capture β-actin mRNAs trafficking along the dendrite. The mRNAs that are recruited to the base of the spine subsequently undergo translation. The newly synthesized actin is translocated to the periphery of the spine head to stabilize the structure of the enlarged spine. Moreover, the time course of mRNA localization and translation is consistent with the process of local protein synthesis participating in the late phase of spine remodeling. Our approach combines targeted postsynaptic stimulation of dendritic spines and single-particle detection of endogenous mRNA or protein to reveal the spatial and temporal resolution of RNA localization and protein synthesis.
Results
Endogenous β-Actin mRNAs Move Dynamically Along the Dendrite.
The prevailing view of dendritic mRNA localization was that transcripts are sorted to specific compartments (i.e., dendrite or axonal growth cone) where mRNAs function as outposts awaiting regulatory cues for translation while remaining anchored (15). However, in distal dendrites (>100 μm from soma) the abundance of any one species of mRNA can be quite low, with even the highly abundant β-actin mRNA reaching levels below 0.1 mRNA/μm (20). Because β-actin mRNA is not present in every dendritic spine, it was proposed that an mRNA would have to scan through the dendrite (21) for recruitment by an activated spine for the targeted delivery of new proteins. Our goal was to characterize the trafficking behavior of β-actin mRNAs in dendrites of live neurons to discern whether mRNAs remained stationary as outposts or had the capacity to survey the dendrite for active synapses.
Dissociated hippocampal neuron cultures were prepared from β-actin–MS2 transgenic mice (14) expressing the MS2 capsid protein (MCP) fused to GFP (22) and were imaged between 14–21 d in vitro (DIV) at 32–35 °C. We performed fast, continuous imaging at 20 frames/s (fps) to track mRNA using widefield microscopy (Fig. S1). From the time-series images, we generated (x–t) kymographs to detect the positions of individual β-actin mRNAs along the dendrite (Fig. S1 A and B and Movie S1). Single mRNA particles moved in both anterograde and retrograde directions (∼1–1.3 μm/s), but most were stationary over the 25-s duration of acquisition, consistent with previous reports (15, 23). Next, we acquired 100 time points at increasing durations (5, 50, 500, and 5,000 s) to determine the optimal time resolution for detecting mRNA transport within dendrites (Movies S2–S5). The comparison of kymographs at increasing durations within individual dendrites suggested that more mRNA movement was progressively detectable as imaging duration increased (Fig. 1 A–D). Notably at 500 s (0.2 fps), several β-actin mRNAs were observed trafficking to and persisting at sites previously not occupied by mRNA (Fig. 1C, red asterisks). At 5,000 s (0.02 fps), almost all mRNAs changed positions throughout acquisition (Fig. 1D), suggesting that mRNAs continually reposition themselves along the dendrite by cycling between mobile and stationary phases. Next, we analyzed random dendritic segments imaged at the durations noted above to quantify the length of time an mRNA was stationary over consecutive time points (Fig. S1 C–F). A stationary mRNA was defined as a particle that remained within a 1-μm radius of its initial position for 90 or more consecutive time points to include stationary particles as well as particles that exhibited limited displacement. Although 71.5 ± 2.6% of mRNAs were stationary after 5 s (20 fps), only 6.3 ± 2.2% were stationary after 5,000 s (0.02 fps). Comparison of stationary events from dendrites imaged for 5 versus 5,000 s showed a significant difference in the population of immobile particles (Fig. 1E). The analysis suggested that steady-state β-actin mRNA movement can be summarized as repetitions of a stationary period (∼7 min) (Fig. 1G) followed by a short mobile period (in seconds) (Fig. 1F) leading to a redistribution of mRNA within dendrites over a timescale of minutes.
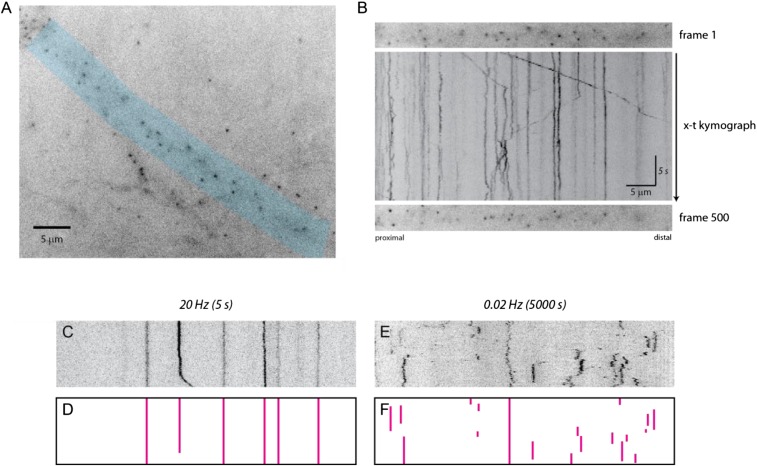
Fig. S1.
Kymograph representation of β-actin mRNA trafficking over time. (A) Streaming (20-Hz) image of dendrites expressing tdMCP-GFP. The dendrite shaded in blue was straightened to generate a kymograph. (Scale bar, 5 μm.) (B) Time-series images were converted to an (x–t) kymograph. The Top and Bottom panels show (x–y) images of the first and last time points of the dendrite, respectively. The Middle panel shows the kymograph where mRNA motion within dendrites can be traced and visualized. RNAs travel in both anterograde and retrograde directions, and corralled and stationary mRNAs are also present. (Horizontal scale bar, 5 μm; vertical scale bar, 5 s.) (C) A representative kymograph of the image acquired at 20 Hz. (D) The traces of stationary particles from C. (E) A representative kymograph of the image acquired at 0.02 Hz. (F) The traces of stationary particles from E.
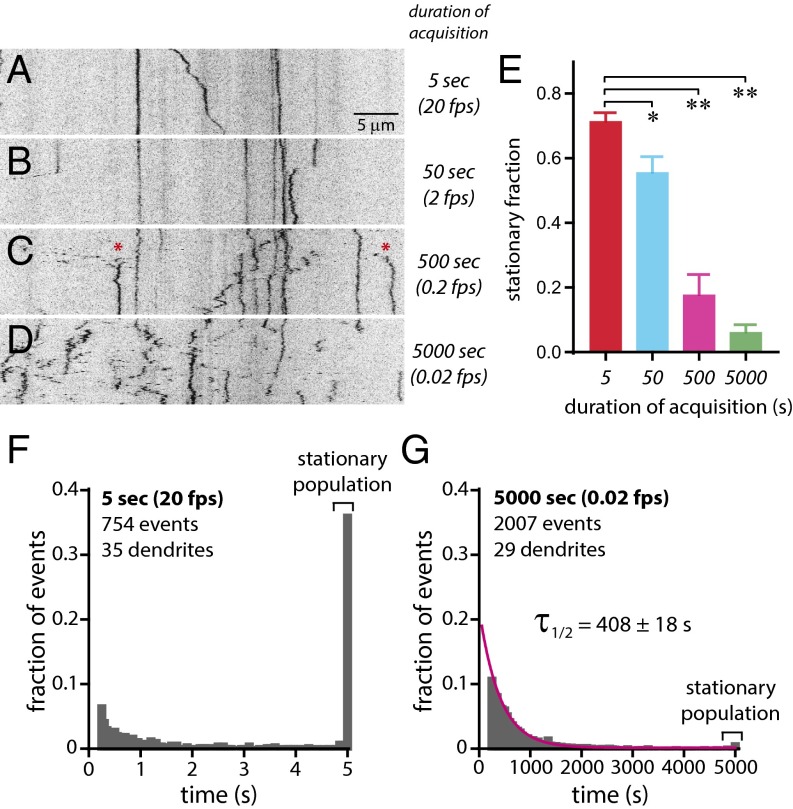
Fig. 1.
Dendritic β-actin mRNA dynamics over time. (A–D) Kymographs (x–t) of dendritic β-actin mRNA imaged for 100 time points over 5 s (20 fps) from a single plane (A), over 50 s (2 fps) from a single plane (B), z-stacks over 500 s (0.2 fps) (C), and z-stacks over 5,000 s (0.02 fps) (D). (Scale bar, 5 μm.) Asterisks indicate mRNA particles in transit. (E) Analysis of stationary mRNA populations from different imaging durations (5 s, n = 35 dendrites, red bar; 50 s, n = 16 dendrites, blue bar; 500 s, n = 15 dendrites, magenta bar; 5,000 s, n = 29 dendrites, green bar). *P < 0.01; **P < 0.0001; unpaired Student’s t test. All error bars indicate SEM. (F) Histogram of stationary events from dendritic β-actin mRNAs imaged for 5 s normalized to total events. Brackets denote the fraction of stationary events. A single mRNA can have multiple events (35 dendrites, n = 754 events). (G) Histogram of stationary events from dendritic β-actin mRNAs imaged for 500 s normalized to total events. Brackets denote the fraction of stationary events. The magenta curve is fit to a single exponential Ae−t/τ (duration τ1/2 = 408 ± 18 s; 29 dendrites; n = 2,007 events).
Glutamate Uncaging Induces β-Actin mRNA Capture.
To determine whether the movement of endogenous β-actin mRNA in dendrites was in response to postsynaptic activity, we devised a photolytic uncaging method to deliver the neurotransmitter glutamate near spines (Fig. S2 and Movies S6–S8). Uncaging was performed on neurons at DIV14–21 (32–35 °C) in the absence of extracellular magnesium and in the presence of 1.5 μM TTX (24–26). We selected distal dendritic regions more than 100 μm from the soma where β-actin mRNA was in low abundance (≤0.1 mRNA/μm) (Table S1). To measure recruitment to stimulated spines, we selected dendrites where β-actin mRNA was absent within a 10-μm region and applied low-frequency stimulation (3 × 10 pulses, 5 mW, 0.5 Hz) to six spots (three on each side) encompassing a 6-μm dendritic segment (Fig. 2A) to increase the probability of β-actin mRNA capture by stimulated spines. Images were acquired every 10 s. In the dendrite shown, two β-actin mRNAs can be seen localizing to the stimulated region previously devoid of mRNA (Fig. 2B and Movie S9). A kymograph of the dendrite was generated to show the temporal dynamics of the β-actin mRNAs within 15 min following stimulation (Fig. 2B, Middle). It also was possible to observe an increase in the fluorescence of the cell-volume marker (red) as an indication of changes in spine volume at the site of stimulation (Movie S9).
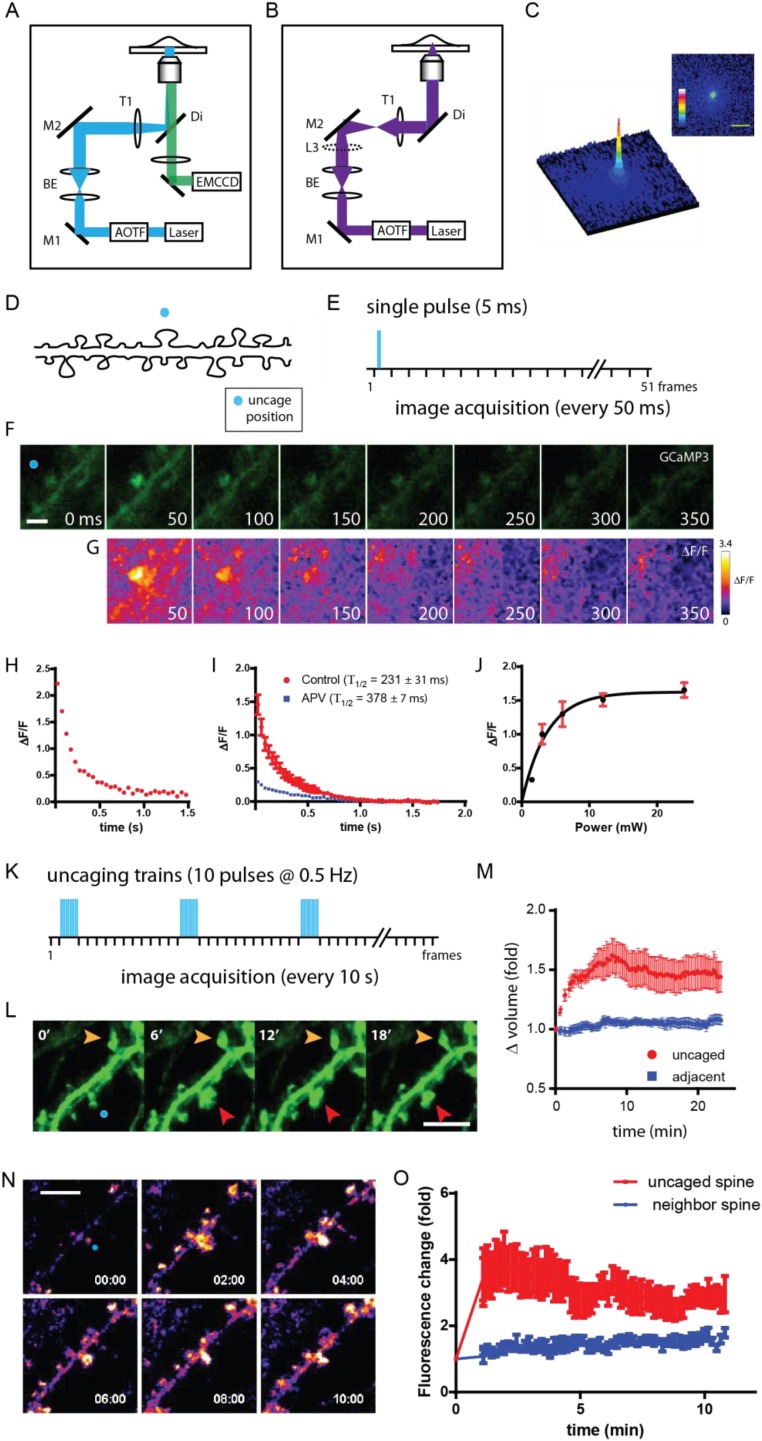
Fig. S2.
Glutamate uncaging-dependent calcium entry into spines and structural remodeling. (A) The optical path for fluorescence imaging. (B) The optical path for the focused 405-nm laser. The collimating lens (L3) is introduced into the optical path, thereby focusing the light into a diffraction-limited spot for photolytic uncaging. (C, Right) The beam profile generated by L3. (Left) The surface plot of the laser profile. (Scale bar, 5 μm.) (D) Schematic depiction of the uncaging position (blue circle) along a dendrite. (E) Schematic of the uncaging event (blue bar) and image acquisitions. (F) Time series images of GCaMP3 following glutamate uncaging (blue circle). Time (in milliseconds) is noted at the bottom right of each image. (Scale bar, 2 μm.) (G) Change in GCaMP3 fluorescence as determined by ΔF/F. (H) Plot of average ΔF/F within the spine head of the dendrite shown in D. (I) Plot of average ΔF/F from uncaged spines (n = 13; red circles) and APV-treated spines (n = 20; blue squares). Fluorescence decay constants (Τ1/2) are shown for both conditions. (J) Plot of average change in GCaMP3 fluorescence at various 405-nm laser power levels. (K) Schematic of uncaging trains (3 × 10; blue bars) and image acquisitions. (L) Time-series images of GFP following glutamate uncaging (blue circle). Time (in minutes) is noted at the upper left of each image. Red arrowheads indicate the uncaged spine; orange arrowheads indicate the adjacent spine. (Scale bar, 5 μm.) (M) Plot of the change in GFP fluorescence in glutamate uncaged and adjacent spines (n = 10). All error bars indicate SEM. (N) Time-lapse images of GFP-actin at 2-min intervals. The blue circle indicates the uncaging spot. (Scale bar, 5 μm.) (O) Plot of the average change in fluorescence intensity following uncaging (n = 6) in the targeted spines (uncaged spine) and the adjacent spines (neighbor spine). All error bars indicate SEM.
Table S1.
β-Actin mRNA counts and localization efficiency at the end of the glutamate uncaging assay
| Treatment | Uncaged | Mock | Low power | +mRNA | APV | CNQX | CHX | jasp | cytoD | ZBPKO |
| n* | 43 | 28 | 24 | 20 | 24 | 27 | 23 | 22 | 23 | 20 |
| mRNA (dendrites)† | 28 (22) | 4 (4) | 5 (5) | 2 (2) | 3 (3) | 9 (8) | 15 (12) | 2 (2) | 2 (2) | 3 (3) |
| Efficiency (%)‡ | 67 (52) | 14 (14) | 21 (21) | 10 (10) | 13 (13) | 33 (30) | 68 (55) | 9 (9) | 9 (9) | 15 (15) |
Total number of dendrites tested for each condition.
Total number of β-actin mRNAs detected within the stimulated region for each condition at the last time point of the assay. Numbers in parentheses indicate the total number of dendrites that successfully localized mRNA.
Total number of localized mRNAs or total dendrites which localized mRNA divided by total number of dendrites tested (n) represented as percent.
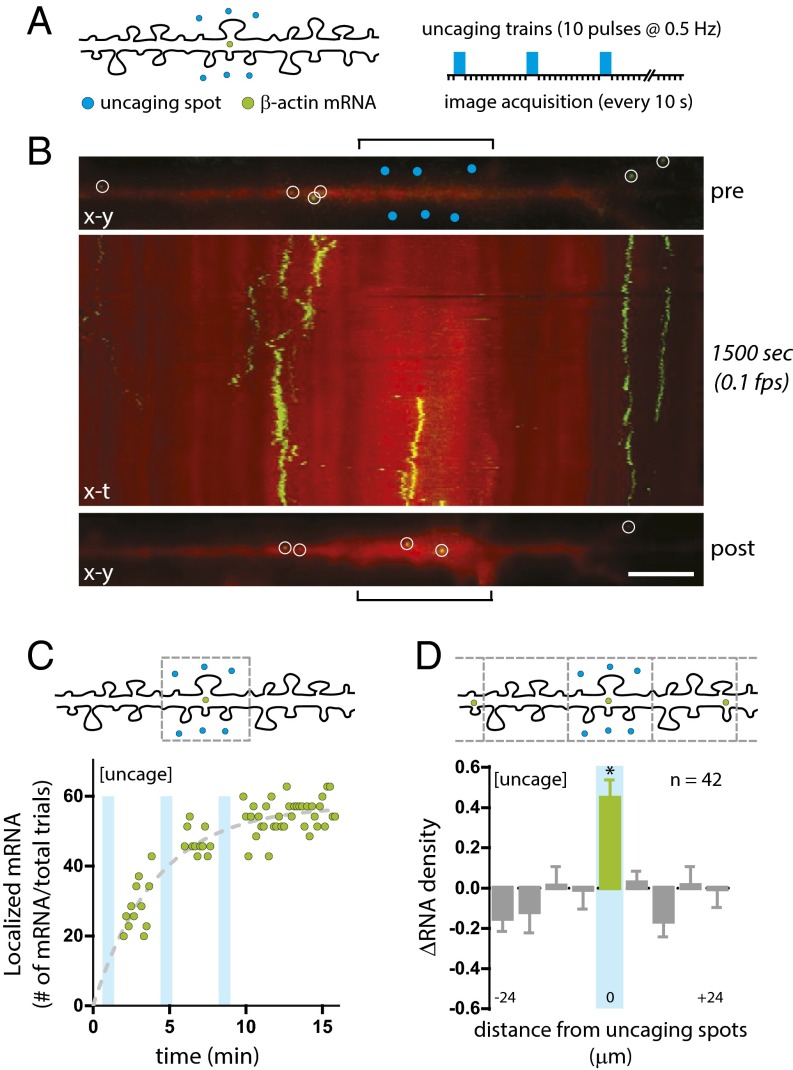
Fig. 2.
Glutamate uncaging leads to β-actin mRNA localization. (A, Left) Schematic depiction of the uncaging positions (blue circles) and mRNA (green circles) along a dendrite. (Right) Uncaging events (3 × 10 blue bars) and image acquisitions. (B, Top) An (x–y) image of β-actin mRNA (green, MCP-GFP) and dendrite (red, mCherry) before uncaging (pre). Open white circles represent detected mRNA; blue circles indicate uncaging spots; brackets indicate the stimulated region. (Middle) An (x–t) kymograph of β-actin mRNA localization. Imaging duration and interval are shown on the right. (Bottom) An (x–y) image of the last time point (post). (Scale bar, 5 μm.) (C) Quantification of β-actin mRNA localization. (Upper) Schematic of the stimulation region for mRNA counting. (Lower) Plot of β-actin mRNA localization efficiency at the stimulated region over time (green circles). Cyan bars represent trains of uncaging stimulation. The dashed trend line (gray) fits to f(t) = A(1 − e−t/τ), where τ was globally determined. (D) Quantification of the change in β-actin mRNA density (ΔRNA) along dendrites. (Upper) Schematic of dendritic segments as bins for mRNA counting. (Lower) Plot of the change in β-actin mRNA density (ΔRNA) in 6-μm dendritic segments (n = 42). *P < 0.05 relative to all other segments; ANOVA and Dunnett’s post hoc analysis. All error bars indicate SEM.
Dynamics of mRNA Localization to Stimulated Spines.
Evaluation of the efficiency of β-actin mRNA localization showed that in 52% of the glutamate uncaging trials we detected one or more β-actin mRNAs localized within 15 min (Table S1). A plot of cumulative mRNA arrival times at the stimulated region showed a gradual increase in mRNAs after each uncaging train (Fig. 2C). In addition, we analyzed the changes in the local positions of β-actin mRNAs along the dendrite. We binned 6-μm segments centered at the uncaging spots and determined changes in mRNA density of the dendrite by averaging mRNA counts from the last 10 time points and normalized these counts to mRNA counts before stimulation. The uncaging assay led to an increase in mRNA density coincident with the uncaging segment over time (ΔRNA = 0.45 ± 0.09), indicating that mRNA localization was specific to the targeted region (Fig. 2D). The increased mRNA density at the stimulated segment was highly significant as compared with the flanking segments, demonstrating that the enrichment was dependent on the location of the stimulus. Dendritic segments flanking the stimulation region exhibited a net aggregate decrease in density, suggesting that mRNA from nearby segments most likely contributed to the mRNA enrichment at the stimulated region. This observation was supported by the fact that we observed almost no change in the average number of mRNAs per dendrite before and after stimulation (Table S2).
Table S2.
β-Actin mRNA counts in dendrites
| Treatment | Uncaged | Mock | Low power | +mRNA | APV | CNQX | CHX | jasp | cytoD |
| n* | 43 | 28 | 24 | 20 | 24 | 27 | 23 | 22 | 23 |
| Total pre† | 163 | 90 | 97 | 118 | 55 | 61 | 106 | 87 | 80 |
| Total post‡ | 164 | 91 | 108 | 129 | 71 | 64 | 105 | 109 | 84 |
| Δ§ | 1 | 1 | 11 | 11 | 16 | 3 | −1 | 22 | 4 |
| Avg pre ± SD¶ | 3.7 ± 2.4 | 4.1 ± 2.7 | 4.0 ± 2.1 | 5.9 ± 3.3 | 2.3 ± 1.9 | 2.3 ± 2.6 | 3.8 ± 3.1 | 4.0 ± 2.0 | 3.5 ± 2.3 |
| Avg post ± SD# | 3.7 ± 2.5 | 4.1 ± 2.8 | 4.5 ± 2.3 | 6.5 ± 3.3 | 3.0 ± 1.9 | 2.4 ± 2.6 | 3.8 ± 3.4 | 5.0 ± 2.4 | 3.7 ± 2.9 |
| Avg Δ ± SD|| | 0.0 ± 2.2 | 0.0 ± 2.5 | 0.5 ± 1.6 | 0.6 ± 1.8 | 0.7 ± 1.5 | 0.1 ± 1.3 | 0.0 ± 1.4 | 1.0 ± 1.6 | 0.2 ± 1.6 |
Average changes in β-actin mRNA for each condition.
Total number of dendrites tested for each condition.
Total number of mRNA detected from all dendrites for each condition at the start of the assay (before uncaging).
Total number of mRNAs detected from all dendrites for each condition at the end of the assay (last imaging time point).
The difference in total mRNA counts between Total pre and Total post.
Average counts and SD of detected mRNA per dendrite before uncaging.
Average counts and SD of detected mRNA per dendrite after uncaging (last time point).
Average difference and SD of mRNA counts per dendrite.
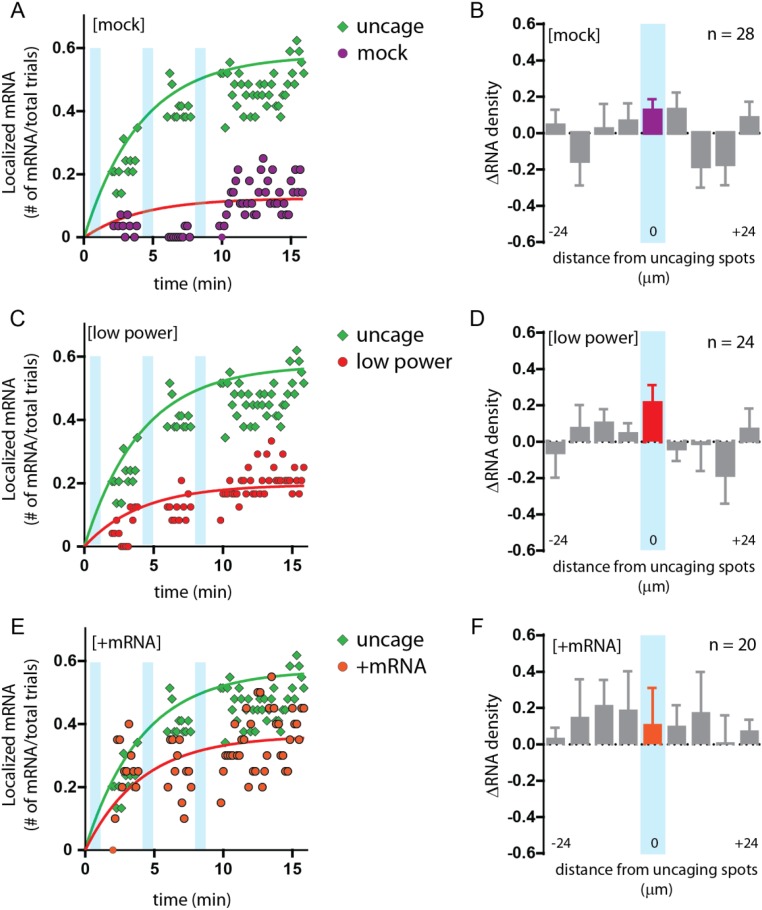
To determine whether the localization of β-actin mRNA was dependent on glutamate, we also performed the assay in the absence of caged glutamate (mock). In 14% of the mock uncaged trials we observed mRNA in the stimulated segment with some fluctuations in mRNA counts in the center bin throughout the latter half of the time course (ΔRNA = 0.13 ± 0.06), suggesting that, as is consistent with dendritic trafficking, β-actin mRNAs move randomly through the dendrite but do not dock or persist at the mock uncaged segments (Fig. S3 A and B). Furthermore, the result demonstrated that the focused uncaging light did not play a role in mRNA localization or impair the trafficking of mRNAs through the stimulated region. When the assay was carried out using only 10% of the original laser power (low power), localization efficiency was reduced (21%) (Fig. S3 C and D). The reduction in laser power led to fewer uncaged molecules, proportionately decreasing receptor activation and mRNA localization and thereby underscoring the tunable response of RNA recruitment by subthreshold stimulation.
Fig. S3.
Characterization of β-actin mRNA localization under various control conditions. (A, C, and E) Plots of β-actin mRNA localization at the uncaged region over time for each condition (green circles). Cyan bars represent trains of uncaging stimulation. The red trend lines are fit to f(t) = A(1 − e−t/τ), where τ was globally determined. Conditions: no MNI-glutamate (mock, purple circles) (A), 10% uncaging power (low power, red circles) (B), and mRNA present (+mRNA, orange circles) (C). (B, D, and F) Changes in mRNA density (ΔRNA) in dendritic segments following uncaging under the conditions shown in A, C, and E. Each segment is 6 μm, centered on the uncaging segment (cyan). All error bars indicate SEM.
Next, we performed glutamate uncaging in dendritic regions that already had mRNAs docked at the base of spines (+mRNA) to determine whether β-actin mRNAs would remain docked following stimulation and whether stimulation could recruit more mRNAs to spines. Results did not show a significant change, suggesting that the stimulation of spines already harboring a β-actin mRNA does not recruit additional mRNAs (Fig. S3 E and F). More importantly, we observed only a 10% change in mRNAs following stimulation but saw a large fluctuation in mRNA counts in the last 10 time points. The results suggested that the additional mRNAs scanning through the dendrite were less likely to dock and remain within a stimulated region in which an mRNA was already present.
NMDA Receptor Activity Is Required for β-Actin mRNA Localization.
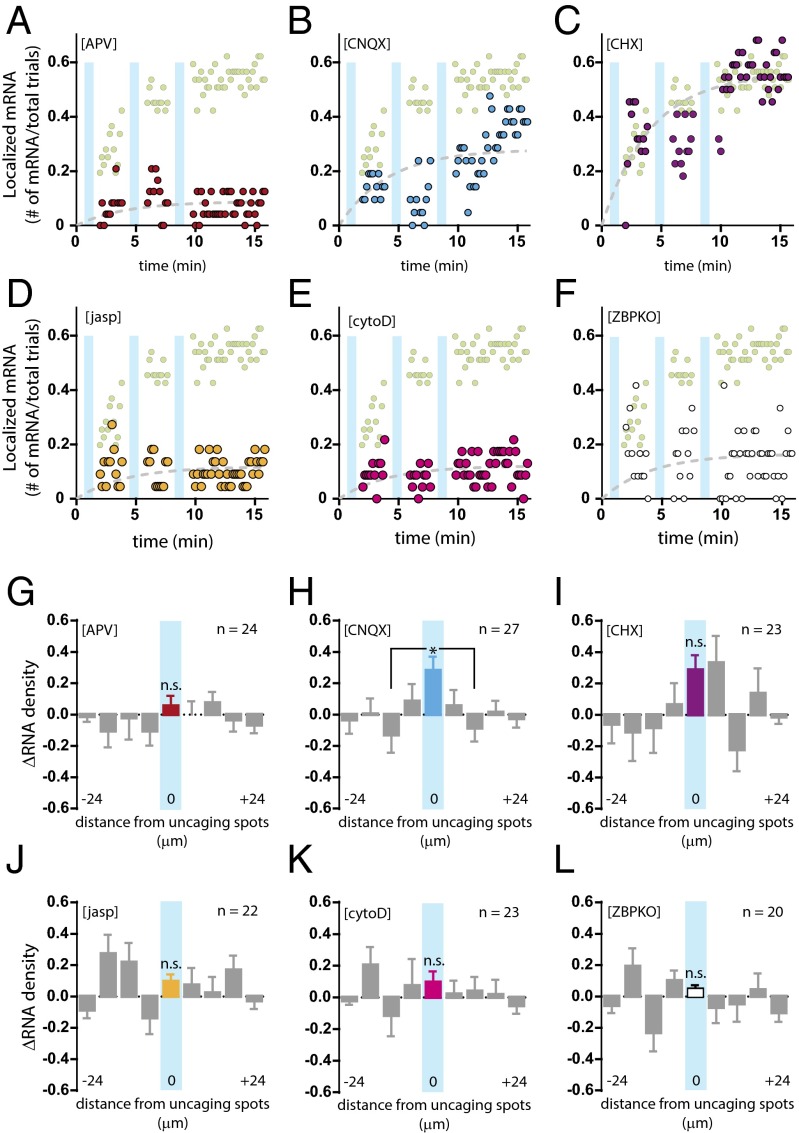
NMDA receptors play a central role in synaptic activity (27). To assess the role of NMDA receptor activity on β-actin mRNA localization, we applied the specific antagonist APV [(2R)-amino-5-phosponovaleric acid] before stimulation and observed localization in only 13% of the trials, i.e., and efficiency similar to that in the absence of glutamate (Fig. 3 A and G). The results suggested that NMDA receptor activity is critical for activity-dependent mRNA localization in dendrites and also appeared to reduce overall β-actin mRNA trafficking in dendrites relative to other conditions. To determine whether the effect on localization might be caused by sample bias, we counted the average number of mRNAs per dendrite before and after uncaging and also across different conditions and detected similar amounts of mRNA in the dendrites assayed (Table S2). On the other hand, inhibition of AMPA receptors by CNQX (6-cyano-7-nitroquinoxaline-2,3-dione disodium) did not lead to a significant reduction in localization relative to the glutamate trials (33%) (Fig. 3 B and H). Because AMPA receptor activation is required to relieve the magnesium block of NMDA receptors, one possible interpretation is that AMPA receptor blockade led to insufficient depolarization of spines and thus to a modest reduction of localized mRNAs.
Fig. 3.
β-Actin mRNA localization requires NMDA receptors, dynamic actin, and ZBP1. (A–F) Plots of β-actin mRNA localization efficiency at the uncaged region over time for each condition are shown overlaid with uncaging trials (light green circles): (A) 20 μM APV (red circles); (B) 40 μM CNQX (blue circles); (C) 60 μM CHX (brown circles); (D) 2 μM jasp (yellow circles); (E) 100 nM cytoD (magenta circles); and (F) ZBP1 knockout (open black circles). Cyan bars represent trains of uncaging stimulation. The dashed trend lines (gray) are fit to f(t) = A(1 − e−t/τ), where τ was globally determined. (G–L) Changes in mRNA density (ΔRNA) in dendritic segments after uncaging for each condition are shown: (G) 20 μM APV (red bar; n = 24); (H) 40 μM CNQX (blue bar; n = 27); (I) 60 μM CHX (brown bar; n = 23); (J) 2 μM jasp (yellow bar; n = 22); (K) 100 nM cytoD (magenta bar; n = 23); and (L) ZBP1 knockout (open black bar; n = 20). Each segment is 6 μm, and the distances were centered from the uncaged segment. The central bin, which received glutamate, is color-coded and overlaid with a cyan bar. All flanking segments are shown in gray. *P < 0.05 relative to center segment; n.s., not significant; ANOVA and Dunnett’s post hoc analysis. All error bars indicate SEM.
Transport of β-Actin mRNAs Is Independent of Translation.
There is evidence that translation is repressed for the duration of transport to facilitate spatial specificity and the regulation of local translation (20, 28, 29). To determine whether β-actin mRNA translation in dendrites plays a role in trafficking or localization, we performed the uncaging assay in the presence of a translation inhibitor, cycloheximide (CHX). Blocking translation had no effect on localization: 55% of trials exhibited localization of β-actin mRNAs within the stimulated segment (Fig. 3 C and I), suggesting that dendritic control of mRNA localization and local translation are decoupled.
A Dynamic Actin Network and Zipcode-Binding Protein 1 Are Necessary for β-Actin mRNA Localization.
Dynamic actin filaments are enriched in synaptic spines (30), and actin network remodeling underlies much of our understanding of spine structural plasticity (13). To probe the function of the actin cytoskeleton in β-actin mRNA localization, we stimulated dendrites in the presence of the F-actin–stabilizing or –destabilizing agents jasplakinolide (jasp) and cytochalasin D (cytoD), respectively. Under both conditions we observed very limited localization efficiency. When we stabilized F-actin, 10% localization was detected upon stimulation (Fig. 3 D and J), and when F-actin was destabilized, we obtained similar 10% localization efficiency (Fig. 3 E and K). The data indicated that β-actin mRNA localization was sensitive to perturbations in spine actin dynamics. In other words, stabilization of F-actin in all spines led to ectopic or mislocalization of β-actin mRNA to spines with drug-stabilized F-actin and limited the availability of scanning mRNAs to the stimulated segment (Fig. 3J). Conversely, destabilization of F-actin led to the loss of anchoring sites for mRNAs recruited to the spine (Fig. 3K). Notably, cytoD treatment did not inhibit the overall movement of β-actin mRNAs along the dendrites, as demonstrated by changes in the density of β-actin mRNAs in the flanking segments. These results show that the dynamic F-actin network is required for capturing and anchoring mRNAs at dendritic spines.
Zipcode-binding protein 1 (ZBP1) is an RNA-binding protein that interacts directly with the zipcode localization element within the 3′ UTR of β-actin mRNA (31) and is required for localization in fibroblasts (32). In neurons, ZBP1 has been shown to play an important role in the localization of β-actin mRNA and other mRNAs in axonal growth cones (33) and in dendrites (5, 34, 35). To determine the effect of ZBP1 on the localization of β-actin mRNA, we performed glutamate uncaging on hippocampal neurons generated from mice crossed between the ZBP1 knock-out (35, 36) and β-actin MBS mice (ZBPKO). The results suggested that although β-actin mRNAs initially trafficked to the stimulated dendritic segment at levels comparable to those in the glutamate uncaging trials, the localized β-actin mRNAs failed to persist (Fig. 3F). Unlike other conditions that exhibited an overall reduction in mRNA detection at the stimulation site (i.e., administration of APV or actin drugs), the loss of ZBP1 appeared to impair persistence but did not affect localization or transport to the region. The average ΔRNA density from the last 10 time points did not show a significant change at the center bin (Fig. 3L), suggesting that RNA counts continued to decrease toward the end of the time course. Results from the ZBP1-knockout neurons underscore the physiological role of an RNA-binding protein in mediating the postsynaptic recruitment of mRNAs following stimulation.
Localized β-Actin mRNAs Persist at Stimulated Spines.
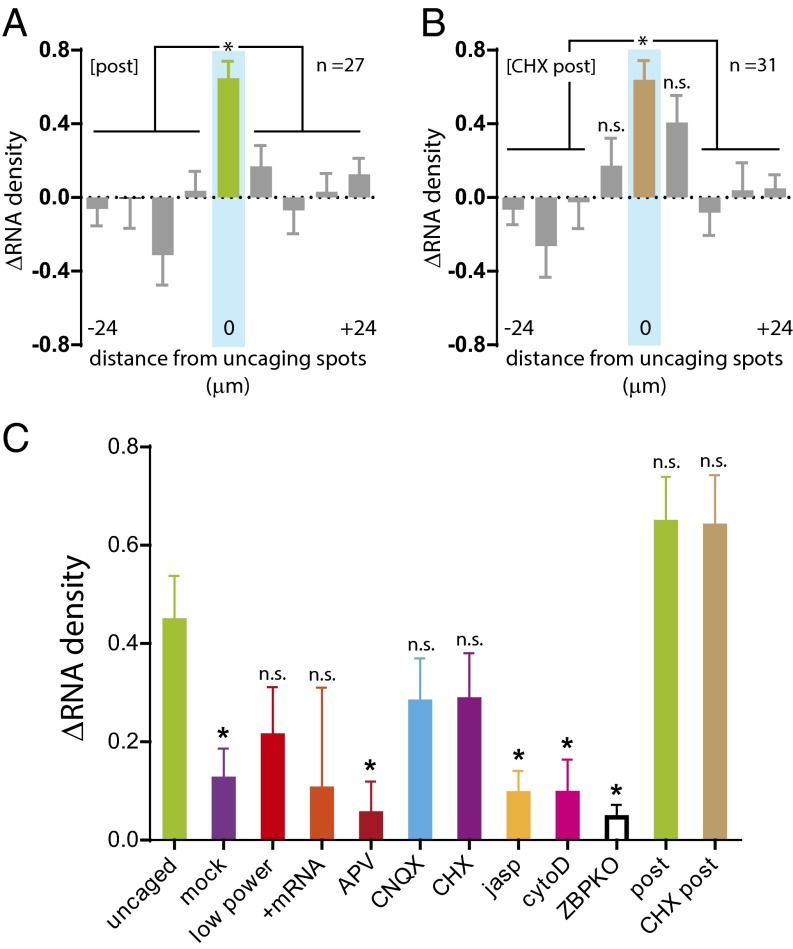
In neurons, synaptic changes can last for several hours to days and could require an mRNA to persist at a specific synaptic site. To determine whether β-actin mRNAs remained anchored at stimulated spines, we imaged dendrites 2 h after stimulation and found that the mRNA count within the stimulated region had increased (ΔRNA = 0.65 ± 0.09) (Fig. 4A). These results showed that the recruitment of β-actin mRNAs at stimulated spines was not a transient event but continued for at least 2 h. In steady-state dendrites, mRNAs traffic through the dendrite, docking transiently at dendritic spines for a short time (7 min). In contrast, upon local application of glutamate, we observed mRNAs at the stimulated sites for 2 h, and mRNAs became more persistently docked at stimulated spines. Although our time resolution (10 s per frame or after 2 h) cannot unequivocally determine whether they are the same β-actin mRNA molecules, it is clear that more mRNAs were detectable in the stimulated region over time. We observed similar levels of increased localization in CHX-treated dendrites after 2 h (ΔRNA = 0.61 ± 0.10), further emphasizing that mRNA localization and capture functions independently of translation (Fig. 4B). One interesting feature of the CHX treatment was the localization of β-actin mRNAs to the flanking segments immediately adjacent to the stimulation site. Perhaps local synthesis of an additional protein factor(s) is necessary to mediate positional specificity of β-actin mRNA anchoring at stimulated spines.
Fig. 4.
Long-lasting localization of β-actin mRNA to stimulated dendrites. (A and B) Plots of the change in β-actin mRNA density (ΔRNA) from glutamate-uncaged (A) and CHX-treated (B) dendrites imaged 2 h after uncaging. Each segment is 6 μm, and the distances were centered from the uncaged segment (cyan). The center bin is green, and flanking segments are gray. For post (n = 27), ΔRNA at the uncaged segment was significant relative to all other segments. For CHX-post (n = 31), ΔRNA at the uncaged segment was significant except for the two adjacent flanking segments. *P < 0.05 relative to the center segment; n.s., not significant; ANOVA and Dunnett’s post hoc analysis. All error bars indicate SEM. (C) Comparison of the change in β-actin mRNA density (ΔRNA) in the stimulated segment of all conditions tested. *P < 0.05 relative to the uncaged group; ANOVA and Dunnett’s post hoc analysis. All error bars indicate SEM.
Comparison of RNA density at the stimulated segment in uncaged trials and with APV administration, F-actin inhibition, or ZBP1 knockout showed that the ΔRNA was statistically significant (summarized in Fig. 4C). The analysis demonstrates that local glutamate uncaging can be an effective means to induce input-specific β-actin mRNA localization. Moreover, the sensitivity of β-actin mRNA localization to NMDA receptor activity, actin cytoskeleton, and ZBP1 mirrors the requirements for synaptic plasticity, reinforcing the hypothesis that localization of β-actin mRNA is linked to synaptic function.
A Translation Reporter System for High-Resolution Detection of RNA and Protein.
To achieve spatiotemporal control of translation, dendritic translation must follow mRNA transport to stimulated spines (37). The use of translation reporters in neurons to observe discrete enrichment of proteins takes advantage of the polarized morphology in which dendrites and axons provide subcellular compartments away from the soma (38). Local translational output can be detected within dendrites (10) or in a subpopulation of newly translated proteins (11, 12) after stimulation. In previous studies, the abundance and subcellular localization of the reporter RNA was inferred by FISH of fixed neurons. Our goal was to use our uncaging approach both to direct the position of the reporter RNAs to nearby spines and to detect newly synthesized proteins.
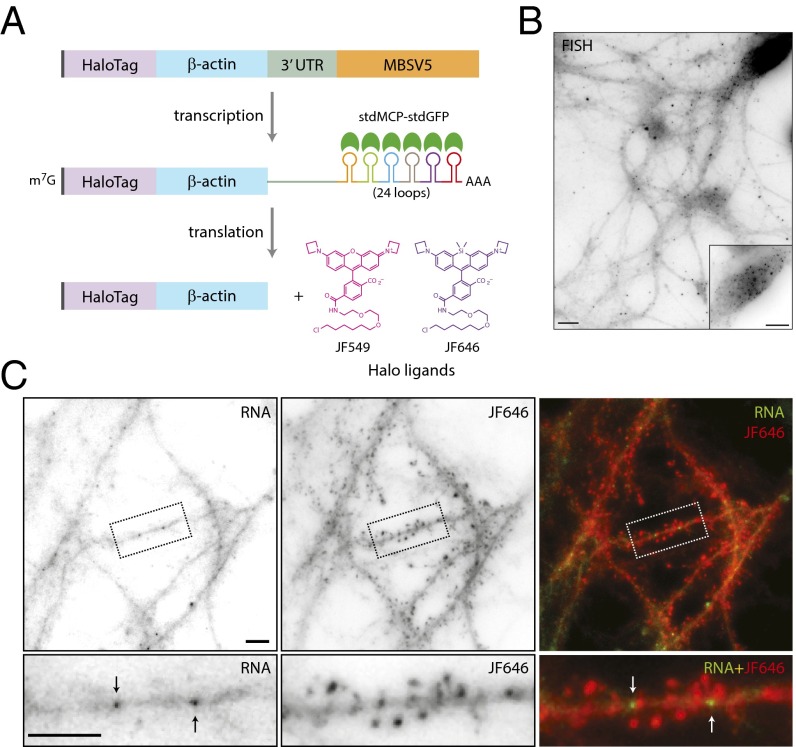
We designed a translation reporter in which the RNA was labeled by MS2 and the translated protein was fused to a HaloTag for simultaneous detection at high resolution in live neurons. The reporter includes FLAG-tag and HaloTag sequences upstream and in-frame of the β-actin coding sequence followed by the 3′ UTR of β-actin and the nonrepetitive MBSV5 aptamers (Fig. 5A). Detection of MBSV5 in living cells requires the coexpression of the tandem-dimer MS2 capsid protein, stdMCP-stdGFP (17). For the detection of translated proteins, cell-permeable Halo-ligand conjugated to a tetraalkylrhodamine derivative was applied to neurons (19). The HaloTag system has been used successfully to detect single proteins in the nucleus (39).
Fig. 5.
Halo-actin translation reporter expression and detection in neurons. (A) Schematic of the Halo-actin translation reporter. The reporter contains the coding sequences of FLAG tag (gray), HaloTag (purple), and β-actin (blue) followed by the 3′ UTR of β-actin (green) and MBSV5 (orange). MBSV5 consists of 24 nonrepetitive MS2 stem–loops (multicolored loops). Upon transcription, the reporter RNA binds stdMCP-stdGFP (green half-circles) on MBSV5 stem–loops. Translated reporter proteins are detected by Halo-ligand conjugated to JF549 or JF646 dyes. (B) Image of reporter RNA in neurons as detected by single-molecule FISH. (Inset) Soma at lower gain. (Scale bars, 10 μm.) (C) Reporter RNAs detected in dendrites by stdMCP-stdGFP (RNA, black arrows) (Left) and the reporter proteins by JF646 Halo-ligand (JF646) (Center) in gray scale. Dotted boxes indicate the dendritic segment containing reporter RNA and the protein, as shown at higher magnification in the lower panels. (Right) The merged images show the overlay of reporter RNAs (green) indicated by white arrows and proteins (red). (Scale bars, 5 μm.)
To validate the expression and localization of the reporter RNA, we performed FISH using probes designed to recognize MBSV5. We detected discrete puncta of reporter RNAs throughout neuronal processes as well as in the soma (Fig. 5B). Next, we assessed detection of RNA and protein in live neurons by coexpression of stdMCP-stdGFP and labeling by Halo-ligand (10 nM), respectively. We observed RNA in dendritic shafts and Halo-actin distributed throughout the dendrites and enriched in heads of dendritic spines, similar to endogenous actin (Fig. 5C). The accumulation of Halo-actin in spine heads suggested that the reporter was functional and able to assemble into spine F-actin cytoskeleton. The results demonstrate that our reporter system can detect the subcellular localization of both RNA and protein in live neurons.
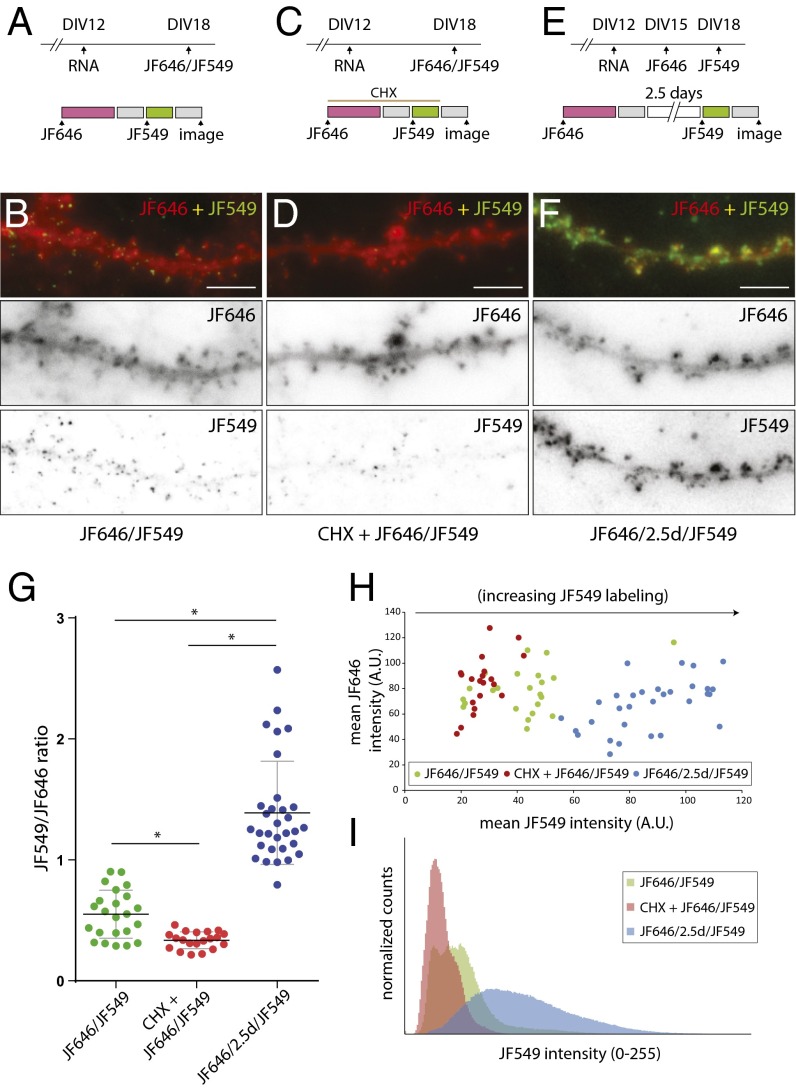
The availability of Halo-ligands conjugated to cell-permeable fluorophores (Janelia Fluor) with distinct spectral properties, JF549 and JF646 (the numbers indicate the maximum excitation wavelength) provided a means to distinguish between preexisting and newly synthesized populations of proteins, similar to a pulse-chase assay. We expressed the reporter RNA at DIV12 followed by Halo-ligand labeling at DIV18 (Fig. 6A). Neurons were sequentially labeled with 200 nM JF646 for 1 h and 10 nM JF549 for 30 min (JF646/JF549). Each labeling step was followed by a 30-min washing to remove excess unbound Halo-ligand before image acquisition. JF646 labeling showed reporter proteins enriched in bulbous spine heads, whereas JF549 labeling exhibited discrete puncta of reporter proteins (JF646/JF549) (Fig. 6B). Notably, almost all the JF549 puncta were colocalized with the globular JF646 signal. When CHX was included along with the Halo-ligands to inhibit translation of the reporter RNA (Fig. 6C), we observed a reduction in the JF549 labeling, suggesting that Halo-ligand binding was specific and required new synthesis of Halo-actin (CHX+JF646/JF549) (Fig. 6D). Next, JF646 and JF549 labeling was separated by 2.5 d to provide time for additional reporter protein synthesis and to determine if Halo-ligand–bound proteins had any negative effects on translation over time (Fig. 6E). After 2.5 d we observed robust labeling by JF549 at dendritic spines (JF646/2.5d/JF549) (Fig. 6F), where the signal no longer appeared punctate but globular within dendritic spines, similar to JF646. More importantly, the results indicated that JF646 labeling did not affect subsequent synthesis of Halo-actin or its localization or assembly into the spine F-actin network.
Fig. 6.
Detection of Halo-actin proteins in dendrites. (A, C, and E) Schematic of experimental (Upper) and JF646/JF549 labeling (Lower) timelines. Neurons were infected with reporter RNA at DIV12. Protein labeling was performed on DIV18 for the JF646/JF549 or CHX+JF646/JF549 (60 μM CHX) conditions. JF646 labeling was performed on DIV15 for JF646/2.5d/JF549 experiments. The 200-nM JF646 Halo-ligand was applied for 1 h (magenta). The wash step was 30 min (gray). JF549 Halo-ligand labeling was performed on DIV18 at 10 nM for 30 min (green). (B, D, and F) Sample images of dendritic segments following protein labeling as in A, C, and E. Upper panels show merged images of JF646 (red) and JF549 (green). Lower panels show images of JF646 and JF549 in gray scale. (Scale bars, 5 μm.) (G) Plot of the intensity ratio of JF549/JF646 for each dendrite. JF646/JF549 is shown in green, CHX+JF646/JF549 is shown in red, and JF646/2.5d/JF549 is shown in blue. *P < 0.0001; unpaired Student’s t test. Horizontal black lines indicate averages. All error bars indicate SEM. (H) Scatter plot of JF549 and JF646 mean intensities for each dendrite of each condition. (I) Histogram showing normalized counts of JF549 pixel intensities from dendrites for each condition.
The ratio between the mean fluorescence intensities of JF646 and JF549 from dendrites was calculated, and the average translational output for each condition was significantly different (Fig. 6G). A scatter plot of JF549 and JF646 mean intensities for each condition showed that the difference in the ratio was caused by increases in JF549 labeling of new proteins (Fig. 6H). Histograms of JF549 intensity indicated that JF646/JF549 and JF646/2.5d/JF549 conditions exhibited a greater number of high-intensity pixels than the CHX+JF646/JF549 condition, reflecting the increased level of reporter synthesis over time (Fig. 6I).
Actin Is Synthesized and Localized to Spine Heads Following Stimulation.
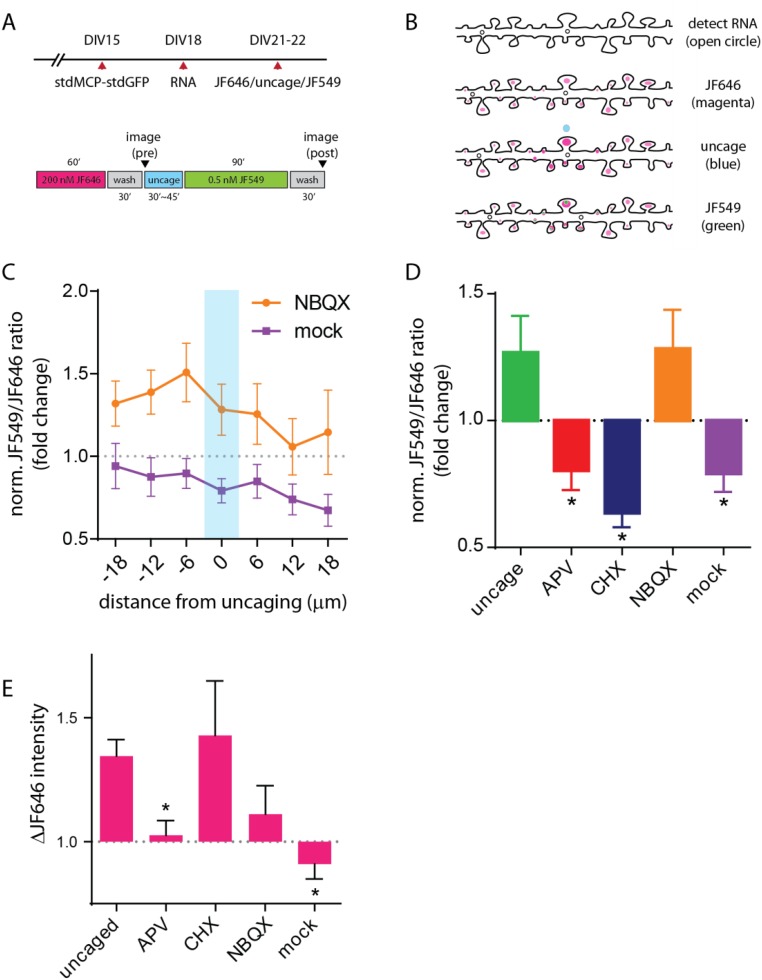
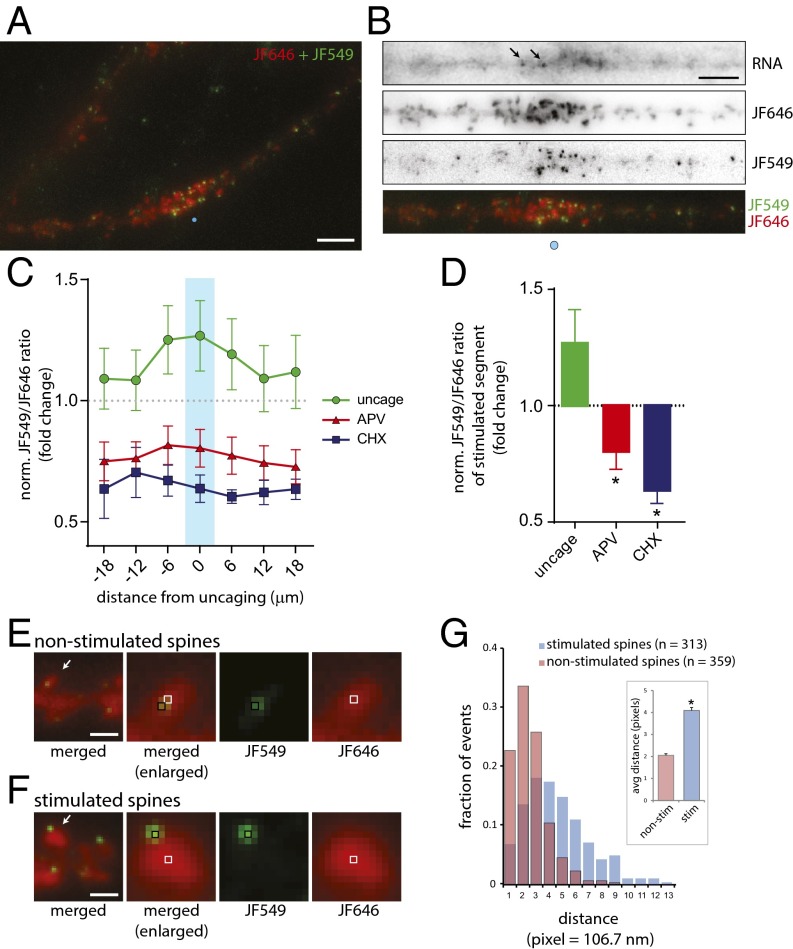
Next, we combined JF646/JF549 labeling of the Halo-actin with glutamate uncaging to determine whether spot stimulation would result in enrichment of newly translated protein at a specific position along the dendrite (Fig. S4A). Schematically, the approach would require the detection of preexisting proteins and RNAs in the dendrite which then would allow the selection of a target site for the delivery of glutamate specifically to a spine (Fig. S4B). The newly synthesized proteins would be labeled and visualized to determine their intensity and position along the dendrite. Following JF646 labeling (200 nM, 1 h) and image acquisition, low-frequency uncaging stimulation (30 pulses, 0.5 Hz) was delivered to a single spot above the spine with the reporter RNA nearby. Neurons then were labeled with 0.5 nM JF549 for 1.5 h to allow time for protein synthesis and were imaged (Fig. 7A and Fig. S5). In the example shown, reporter RNAs are localized near the site of stimulation in the dendritic shaft (Fig. 7B, RNA). Also, preexisting Halo-actin was present in spine heads, and several spines had undergone structural remodeling (Fig. 7B, JF646). New Halo-actin was enriched near the site of stimulation (Fig. 7B, JF549). Overlay of JF646 and JF549 images shows discrete JF549 puncta localized within a cluster of globular JF646 signal. In addition, we readily observed an increase in JF646 intensity in spines at the site of stimulation (Fig. S5), indicating spine structural remodeling as a result of glutamate uncaging (40, 41).
Fig. S4.
Stimulation leads to synthesis and localization of Halo-actin to synaptic spines. (A) Schematic of the experimental (Upper) and JF646-uncage-JF549 labeling assay (Lower) timelines. Neurons were infected with stdMCP-stdGFP and reporter RNA lentivirus at DIV15 and DIV18, respectively. For pulse labeling, 200 nM JF646 Halo-ligand was applied for 1 h (magenta). Each wash step lasted for 30 min (gray). After washing, images of RNA and JF646 were acquired (pre). After glutamate uncaging (blue), 0.5 nM JF549 Halo-ligand was added and incubated for 1.5 h (green). After washing (gray), images of RNA, JF646, and JF549 were acquired (post). (B) Schematic of the experimental outcome. After pulse labeling, neurons positive for both capsid protein (open circle) and JF646 (magenta circle) are identified. Uncaging pulses (30 pulses, 5 mW, 0.5 Hz) are delivered to a spine with a reporter RNA nearby (blue circle). Newly synthesized reporter proteins (green circles) are detected by JF549 Halo-ligand. (C) Plot of normalized JF646/JF549 ratios for the JF646-uncage-JF549 labeling assay of NBQX (orange trace) and mock (purple trace) conditions along the dendrite. The x axis indicates 6-μm segments centered on the uncaging site. Flanking segments proximal or distal to the uncaging site are indicated by (±) distance from center. The center bin is indicated by a cyan bar. All error bars indicate SEM. (D) Plot of average JF646/JF549 ratios at the center segment for the conditions noted. *P < 0.05 relative to the uncaged group; ANOVA and Dunnett post hoc analysis. (E) Plot of average ΔJF646 at the center segment for all conditions tested. Changes were normalized by comparing pre- and post-JF646 images. *P < 0.05 relative to the uncaged group; ANOVA and Dunnett post hoc analysis.
Fig. 7.
Stimulation leads to new synthesis and localization of Halo-actin proteins to synaptic spines. (A) Image of a dendrite after JF646/JF549 labeling (Materials and Methods). JF646 is shown in red, and JF549 is shown in green. The blue circle indicates the uncaging position. (Scale bar, 5 μm.) (B) Image of the stimulated dendrite in A showing reporter RNA, JF646, and JF549 in gray scale and JF646 and JF549 merged (red and green, respectively) as in A. The blue circle indicates the uncaging position. Arrows indicate RNA. (Scale bar, 5 μm.) (C) Plot of normalized JF549/JF646 ratios of uncaged (green trace), APV (red trace), and CHX (blue trace) conditions along the dendrite. The ratios were normalized to the JF549/JF646 ratio of nonstimulated dendrites (dotted line). The x axis indicates 6-μm segments centered on the uncaging site. Flanking segments proximal or distal to the uncaging site are indicated by (±) distance from center. The center bin is indicated by a cyan bar. All error bars indicate SEM. (D) Plot of average JF549/JF646 ratios at the center segment for the conditions noted. *P < 0.05 relative to the uncaged group; ANOVA and Dunnett’s post hoc analysis. (E and F) Images of dendritic spines following Halo-actin labeling by JF646 and JF549. (Left) Merged images of JF646 (red) and JF549 (green) in nonstimulated (E) or stimulated (F) dendrites. (Scale bars, 5 μm.) Arrows in the left panels indicate the spine shown in the adjacent panel. The black and white boxes indicate local maxima of JF549 or JF646, respectively and are shown individually in the center-right and right panels. (G) Histogram of distances between local maxima of JF549 and JF646 within stimulated (n = 313) or nonstimulated (n = 359) spines. The x axis indicates pixels: one pixel = 106.7 nm. (Inset) Average distance of each population. *P < 0.05; unpaired Student’s t test.
Fig. S5.
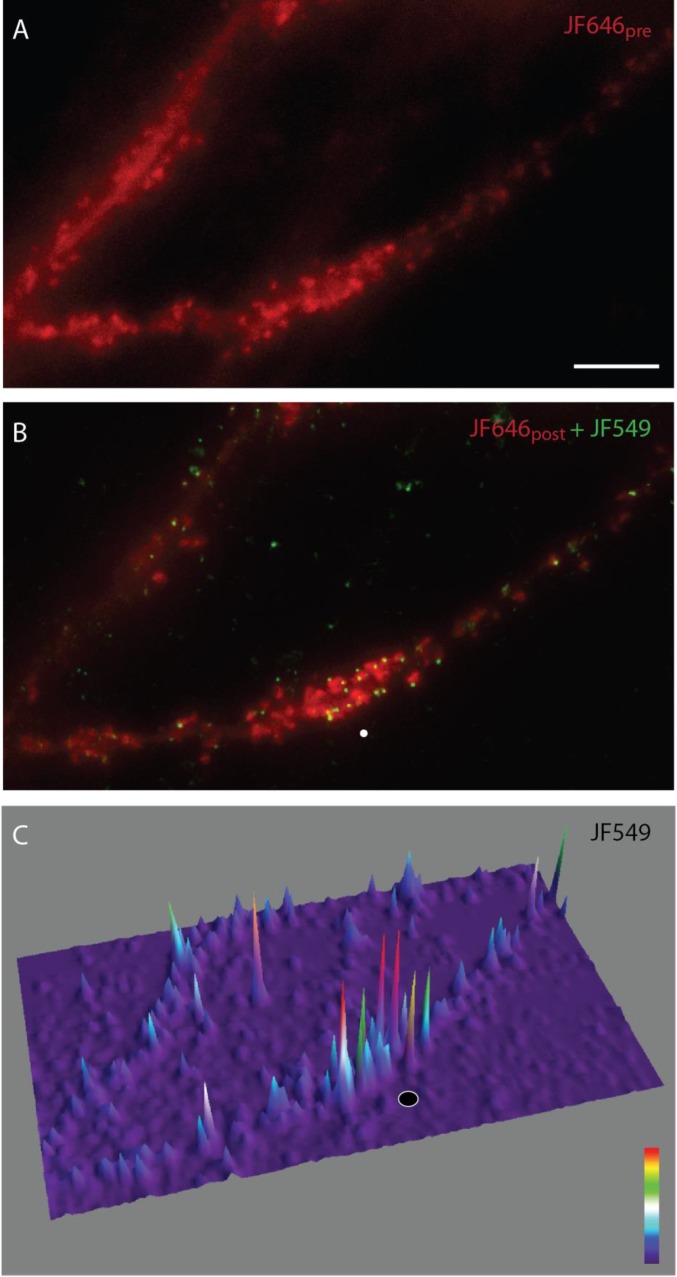
JF646-uncage-JF549 labeling of the Halo-actin reporter. (A) Image of a dendrite labeled with JF646pre (red) before glutamate uncaging. (Scale bar, 5 μm.) (B) Merged image of a dendrite labeled with JF549 (green) and JF646post (red) after glutamate uncaging. The white circle indicates the uncaging spot. (C) 3D image of JF549. The 3D image better represents the relative puncta intensities of JF549 localized to spines. The black circle indicates the uncaging spot. Look-up table is on the lower right.
NMDA and AMPA Receptor Activities Have Opposing Effects on β-Actin Synthesis in Dendrites.
To evaluate the translational output in dendrites, we determined the average fluorescence intensity of JF646 and JF549 for each dendrite in 6-μm segments centered on the site of stimulation. The ratio of JF549 (after stimulation) and JF646 (before stimulation) intensities were quantified for each condition and normalized to the ratio from a control group of 52 dendrites. For the control group, sequential labeling of reporter protein was performed in the imaging medium in the presence of magnesium and without TTX or caged glutamate to determine the basal translation output in dendrites; this output was used as baseline. Glutamate uncaging led to a local increase of 26.8 ± 14.5% in protein synthesis at the center bin with the immediate flanking segments exhibiting similar levels of enrichment (Fig. 7C). In contrast, when neurons were pretreated with APV (20 μM) before uncaging, we observed a reduction in new protein synthesis of 19.7 ± 7.8% relative to the control group with no obvious increase within the center segment. As expected, when we blocked translation with CHX, we observed a greater reduction in translational output along the dendrite with a 36.3 ± 5.7% decrease at the site of uncaging. The addition of the NMDA receptor antagonist or translation inhibitor resulted in an overall reduction in protein synthesis in dendrites as well as the loss of positional information of the stimulus. Blockade of AMPA receptors with 10 μM NBQX (2,3-dioxo-6-nitro-1,2,3,4-tetrahydrobenzo [f] quinoxaline-7-sulfonamide) led to translation at the center bin at levels similar to those in glutamate (Fig. S4D), suggesting that NMDA receptor activity is essential for the induction of translation. However, NBQX treatment resulted in the loss of the spatial specificity of glutamate stimulation, with some variability in proteins throughout dendrites (Fig. S4C), indicating that NMDA and AMPA receptor activities may have opposing effects on translation to modulate protein synthesis at dendritic spines. To validate whether the observed translation was caused by glutamate, we delivered uncaging pulses in the absence of caged glutamate (mock) to determine the specific effect of local stimulation on dendritic translation. Mock stimulation led to a reduction in translation at the stimulated segment of 20.8 ± 7.3%, to levels equal to the APV-treated dendrites; this result further corroborated the importance of NMDA receptor activation for translation induction (Fig. S4 C and D). Comparison of the glutamate uncaged center segments across conditions showed that enrichment of newly synthesized Halo-actin proteins was significant relative to the APV and CHX conditions (Fig. 7D). Taken together, the evidence strongly suggests that the local translation of β-actin is specific to and dependent on NMDA receptor activity.
Newly Synthesized Actin Is Enriched at the Periphery of Stimulated Spines.
New Halo-actin proteins in stimulated spines exhibited two distinct features. First, the proteins accumulated in bright foci within dendritic spines, as demonstrated by the increased presence of proteins in stimulated segments (Fig. 7A). Second, the discrete puncta of new proteins were localized to the periphery of stimulated spines (Fig. 7F). In contrast, JF646/JF549 labeling of nonstimulated control dendrites showed a high degree of overlap between the preexisting and new actin within spines (Fig. 7E). To determine the distance between the preexisting and new populations of Halo-actin for each spine, we calculated the local maxima of JF549- and JF646-labeled Halo-actin proteins. The histogram of distances showed that stimulation led to more instances in which newly synthesized proteins localized to the periphery of the spine than to nonstimulated spines (Fig. 7G). On average, the distance between new and preexisting proteins increased by twofold; this increase was highly significant (Fig. 7G, Inset). The results showed that new actin proteins localized and accumulated preferentially at a specific position within spine heads, perhaps near the postsynaptic density (42). In steady-state spines, F-actin has been observed to treadmill constitutively toward the center of the spine head; however the results suggested that new actin could be translocated to a specific subspine position and that new actin could play a role outside the spine F-actin network. Perhaps spines are capable of detecting and sorting newly synthesized actin. Also, the observed timing of new actin enrichment at the spine is consistent with locally synthesized β-actin having a structural role in stabilizing the expanded the postsynaptic density after initial remodeling (43).
Discussion
In this study, we used high-resolution imaging in live neurons to examine the RNA-mediated processes at dendritic spines. The ability to visualize endogenous β-actin mRNA over time allowed us to correlate the dynamics of individual mRNAs with spatially restricted postsynaptic stimulation. By uncaging glutamate at spines without a β-actin mRNA, we effectively targeted specific spines as sites for activity-dependent capture and translation of RNAs. Although it is difficult to generalize that all dendritic mRNAs behave in a stimulation-dependent manner, β-actin serves as an excellent model to understand how dendritic spines can regulate local protein synthesis of high-abundance mRNAs that encode important synaptic proteins.
All Endogenous β-Actin mRNAs in Dendrites Are on the Move.
The scanning behavior of endogenous β-actin mRNA indicates that mRNAs are not simply targeted to a specific dendritic location to function as outposts for the synthesis of actin protein (15) but instead are capable of repositioning dynamically within distal dendrites over time (Fig. 1) (21). The capture and release of β-actin mRNA by synaptic spines allows spatial specificity of local translation with a relatively fast response time to furnish newly synthesized actin protein to the place of function. Moreover, when stationary, β-actin mRNAs are positioned at the base of spines, and not within the head of a specific spine, to facilitate rapid redistribution. The localization of β-actin mRNA within dendrites reflects an optimal strategy to provide newly translated actin proteins dynamically and efficiently to many dendritic spines in an activity-dependent manner.
Local Postsynaptic Stimulation by Glutamate Uncaging.
The availability of stable caged compounds combined with photolytic uncaging has provided a powerful tool for studying the synaptic physiology of neurons with high spatial and temporal resolution. Examples include mapping of glutamate receptors (25), structural plasticity (24, 44), input specificity (26), calcium-dependent signaling (40, 41, 45), and postsynaptic input integration (46, 47), to name only a few. In addition to visualizing synaptic changes in individual spines (Fig. 2), glutamate uncaging can be applied to dynamic imaging of RNA and proteins. Moving forward, genetically encoded indicators and biosensors that can detect spine calcium or membrane potential can be combined with single-molecule strategies to dissect molecular events that regulate postsynaptic changes with high spatial and temporal resolution (48–51). Such tools will help in the visualization of discrete local changes in gene expression and spine physiology at the single-spine and single-molecule level.
Synaptic Capture of mRNAs Following Uncaging Stimulation.
Following glutamate uncaging, β-actin mRNAs begin accumulating at sites of stimulation after the first uncaging train and reach a maximum by 10 min (Fig. 2). Although the timing of stimulation-dependent localization is similar to the average residency time of mRNAs in steady state (408 ± 18 s), the key difference is the persistence of localized mRNA for at least 2 h at sites of glutamate stimulation (Fig. 4 A and B). Taken together, the localization kinetics is consistent with capture of scanning mRNAs by stimulated spines. Further evidence in support of the synaptic capture model comes from total mRNA counts in the dendrites that show very little change throughout the assay (Table S2). The results suggest that localized β-actin mRNA originated from local pools and was unlikely to have been transported from the soma, and the persistence indicates a long-lasting role of mRNA localization and translation within dendrites.
The sensitivity of β-actin mRNA capture to inhibition of NMDA receptors, perturbation of actin cytoskeleton, or the loss of ZBP1 underscores the significance of each component to achieve stimulation-dependent mRNA localization in dendrites (Figs. 3 and 4C). Postsynaptic input, the actin network, and transacting mRNA localization factor(s) participate in cascade to achieve spatially and temporally coordinated control of gene expression in dendrites. In particular, the link between NMDA receptor activity and the actin cytoskeleton make NMDA receptors a strong candidate for governing downstream signaling for mRNA localization (52). Moreover, the loss of ZBP1 impairs the anchoring of β-actin mRNA at specific synapses following stimulation, as is consistent with previous lower-resolution observations that ZBP1 plays an important role in dendritic RNA targeting (5, 35, 53). We now can quantify the effect of the loss of ZBP1 on mRNA targeting. Our results demonstrate that local postsynaptic stimulation by glutamate can lead to the localization and capture of mRNAs at synaptic spines previously devoid of mRNA.
Designing a Translation Reporter for Visualizing RNA and Protein at High Resolution.
The application of fluorescent protein-based translation reporters in neurons has allowed significant progress in visualizing local protein synthesis within dendrites and axons (38). The modular design of translation reporters has opened a way for investigators to incorporate specific features into a reporter to enhance the spatial and temporal resolution of detection (10). However, the current palette of fluorescent proteins possesses intrinsic properties, such as chromophore maturation, photostability, and brightness, that hinder high-resolution detection in living cells. Also, validation of reporter RNA localization was determined in separate experiments. To overcome such issues, we incorporated features to detect reporter RNAs and proteins in dendrites together in living cells (Fig. 5). The self-labeling HaloTag, derived from bacterial haloalkane dehalogenase, covalently binds to synthetic ligands (Halo-ligands) with high affinity (18). Halo-ligands conjugated to bright cell-permeable dyes allow high-resolution detection of HaloTag fusion proteins (19). Second, each MS2 stem–loop within MBS was redesigned to minimize repetitive sequences among the 24 MBSV5 stem–loops (17). The nonrepetitive nature of MBSV5 reduces recombination during viral transduction and facilitates sequence amplification. The combination of MBSV5 and HaloTag can achieve high-resolution simultaneous detection of reporter RNA and protein in living cells.
Glutamate Uncaging Promotes Actin Synthesis and Incorporation into Spines.
The localization of β-actin mRNAs following glutamate uncaging suggested that the function of localization was to provide newly synthesized actin protein to active synaptic spines. However, the sheer abundance of actin protein in the soma and dendrites raised questions about whether the local translation of β-actin mRNA was necessary or even detectable. In an earlier work, Scheetz et al. (54) identified synthesis of a 42-kDa protein following brief treatment with glutamate and NMDA in metabolically labeled rat synaptosomes. Although the identity of the protein was not pursued, it coincided with the molecular weight of actin.
To detect newly synthesized actin in dendrites, two spectrally distinct dyes, JF549 and JF646, conjugated to the Halo-ligand were applied in a double-labeling assay, similar to pulse-chase, in neurons expressing the Halo-actin translation reporter (Fig. 6). Next, we incorporated local glutamate uncaging (Fig. S2) between each of the labeling steps to determine local changes in translation after stimulation (Fig. 7). The assay revealed that (i) the reporter RNA remained in the stimulated region before and after stimulation; (ii) the spines within the stimulated segment were enlarged; and (iii) newly labeled proteins were enriched in the stimulated region. In addition, the changes observed at the uncaged region required NMDA receptor activity.
The results suggested that newly synthesized actin is enriched in dendritic spines that have experienced glutamate stimulation, with some localized to the tips of the spine head. Because the quantity of newly synthesized actin is unlikely to affect the overall concentration of actin in dendrites over a time scale of minutes, this finding suggested that newly synthesized actin may be qualitatively different from preexisting actin proteins. For example, in many cell types actin has been shown to undergo a variety of posttranslational modifications, such as acetylation, arginylation, phosphorylation, or ubiquitylation, to name a few (55). Perhaps modification of newly synthesized dendritic actin in spines can serve as binding sites or a tag for interactions with other plasticity-related proteins (i.e., FMRP) or inducible dendritic RNAs (i.e., Arc) that may localize temporally downstream of β-actin mRNA. Alternatively, newly synthesized actin protein may mediate gene-expression changes through its interactions with MRTF (myocardin-related transcription factor) and affect transcription in the nucleus (56). The possibility that locally translated actin may have roles beyond the spine cytoskeleton may provide insights into the function of dendritic β-actin mRNA localization and synthesis.
The Missing Link.
There is much evidence linking new protein synthesis with synaptic plasticity and memory (57, 58). However, visualizing the dendritic localization and function of new proteins has proven difficult. One fascinating result from the Halo-actin reporter assay was the distinct localization of new protein to the periphery of dendritic spines. By labeling a subpopulation of proteins, we were able to visualize the translocation of new proteins within spine heads. Because actin is a cytoskeletal protein, the observation that newly synthesized actin could be assembled into a discrete structure in isolation from the bulk F-actin in spine heads suggested that a spine may be capable of distinguishing and sorting new and preexisting actin. Visualizing RNA and protein together allows the detection of changes in dendritic spines by local protein synthesis. Recently, an alternative method for detecting mRNA and protein together was demonstrated using metabolic labeling and proximity ligation assay (59). Seeing when and where mRNA localization and protein synthesis occur within dendrites at high resolution provides strong evidence that translation in dendrites serves neuronal function, synaptic plasticity, and memory.
Materials and Methods
Animal work was performed in accordance with Institutional Animal Care and Use Committee protocols at Albert Einstein College of Medicine. DNA constructs used in this work are available at Addgene. Detailed stepwise protocols for methods and reagent information herein are available upon request.
Constructs and Lentivirus Generation.
Coding sequences for tandem-dimer MCP-mEos2-GFP (tdMCP-mEos2-GFP), stdMCP-stdGFP (17), GCaMP3 (60), mCherry, and EGFP were cloned into the lentivirus expression vector. Coding sequences of FLAG tag and HaloTag (Promega) were cloned in frame with the β-actin ORF and 3′ UTR. The MBSV5 (17) was inserted between the 3′ UTR and the 3′ end of the multicloning site of the lentivirus expression vector. Lentivirus particles were produced as follows: plasmids for ENV (pMD2.VSVG), packaging (pMDLg/pRRE), REV (pRSV-Rev), and the expression vector were mixed and transfected into HEK 293T cells using calcium phosphate (61). Expression of the insert was under the control of the UbC promoter. The virus-containing supernatant was harvested and concentrated using Lenti-X concentrator (Clontech) according to the manufacturer’s instructions. The virus was resuspended in Neurobasal A and stored at −80 °C for subsequent infection of neurons in culture.
Dissociated Mouse Hippocampal Neuron Culture.
Postnatal day 1 (P1) mouse hippocampal tissue was isolated from homozygous MBS knockin (14) or wild-type (Charles River) newborn pups. Hippocampi were placed in 0.25% trypsin for 15 min at 37 °C. Tissue was triturated and plated onto poly-d-lysine (Sigma)–coated glass-bottomed dishes (MatTek) at 75,000 cells per dish (for live cell imaging) and were cultured in normal growth medium [NGM: Neurobasal A medium supplemented with B-27, GlutaMAX, and primocin (InvivoGen)]. Embryonic hippocampal neurons (E18) from ZBP1-knockout mice were prepared as previously described (35). All reagents were acquired from Life Technologies unless noted otherwise.
Sample Preparation and Live Imaging.
Dissociated mouse hippocampal neurons were infected with lentivirus expressing the capsid protein (MCP) on DIV5. Neurons were imaged between 2–3 wk in culture. For live imaging of neurons, cells were washed in prewarmed HBS medium (119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM d-glucose, and 20 mM Hepes at pH 7.4) and were imaged at 35–37 °C. Neurons expressing the tdMCP-FP were imaged under streaming (50 ms; single plane), time-lapse (500 ms; single plane), time-lapse z-series (5 s; seven steps/3 μm), or long time-lapse z-series (50 s; seven steps/3 μm) conditions, and 100 time points were acquired at each condition. For time-lapse z-series images, stacks were maximally projected. For glutamate uncaging experiments, we infected neurons with lentivirus encoding GCaMP3, GFP, mCherry, tdMCP-mEos2-GFP, or tdMCP-TagRFPT and used baculovirus (BacMam) for GFP-actin (Molecular Probes), as noted in the figure legends for Fig. 1 and Figs. S1 and S2. Whenever possible, we double-infected our neurons for two-color imaging with the second lentivirus serving as the volume marker (or as an other marker) as noted in the text. Before the uncaging experiments, cells were washed and placed in Mg2+-free HBS medium (119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 30 mM d-glucose, and 20 mM Hepes at pH 7.4) along with 1.5 μM TTX and 2 mM 4-methoxy-7-nitroindolinyl-l-glutamate (MNI-glutamate). Pharmacological reagents were purchased from Tocris unless noted otherwise. The glutamate receptor antagonists APV (20 μM) or CNQX (40 μM) or the translation inhibitors CHX (60 μM) (Sigma), anisomycin (40 μM), puromycin (100 μM) (Invivogen), or the actin drugs jasp (2 μM) (Enzo) and cytoD (100 nM) (Enzo) were added following medium exchange. Cells were allowed to equilibrate in the medium for 15–20 min before the start of the experiment.
Fluorescence Microscopy and Uncaging Setup.
The uncaging experiments were performed on a fluorescence microscope built around an IX-81 stand (Olympus). The back port of the microscope was removed to allow custom laser illumination. For excitation of fluorescent proteins, a 491-nm laser (Calypso-25; Cobolt), a 561-nm line (LASOS-561-50; Lasertechnik GmbH), and a 640-nm line (CUBE 640-40C; Coherent Inc.) were combined, expanded, and delivered through the back port (Fig. S2A). A size-adjustable iris was used to limit the illumination to an area ∼80 µm in diameter. The lasers were reflected by a four-band excitation dichroics mirror (Di01-R405/488/561/635; Semrock) to a 150× 1.45 NA oil immersion objective (Olympus). The fluorescence was collected by the same objective, passed through the dichroic mirror, a notch filter (NF01-405/488/561/635), and emission filters and was recorded on an electron multiplying CCD camera (iXon3 DU-897E-CS0-#BV; Andor). The emission filters (FF01-525/50 for green and FF01-605/64 for red; Semrock) were mounted on a motorized filter wheel (FW-1000; Applied Scientific Instrumentation) for fast switching between wavelengths. To uncage glutamate, a focused 405-nm laser (LuxX 405-60; Omikron) was used. The 405-nm laser followed the same beam path (Fig. S2A), but a lens was used to collimate the laser before entering the objective (Fig. S2B). An image of the focused laser was acquired (Fig. S2C). Switching between the diffraction-limited and widefield illumination was achieved by flipping a lens (AC254-150-A, L3; Thorlabs) in and out of the light path via a motorized flipping mount (MFF001; Thorlabs). The time required to flip the lens out of the light path was less than 5 ms. All laser power and shuttling were controlled by an acousto-optic tunable filter (AOTF) (AOTFnC-400.650-TN; AA Opto Electronic). The microscope also was equipped with an automated XY stage (MS2000-XY with an extra-fine lead-screw pitch of 0.635 mm and a 10-nm linear encoder resolution; Applied Scientific Instrumentation) and a piezo-Z stage (Applied Scientific Instrumentation) for fast z-stack acquisition. The AOTF, flipping mount, and piezo-Z stage were all controlled by a data acquisition board (DaqBoard/2001; IOtech, Inc.). The cells were kept at 37 °C with a stage top incubator (INUBH-ZILCS-F1; Tokai Hit).
JF646-Uncage-JF549 Labeling Assay.
Dissociated hippocampal neurons were infected with lentivirus expressing the capsid protein (stdMCP-stdGFP) and reporter RNA at DIV15 and DIV18, respectively. The JF646/JF549 labeling assay was performed at DIV21–22. For JF646 labeling, JF646 was added to a final concentration of 200 nM for 1 h and then was washed four times with HBS and replaced with NGM for 30 min. After washout, the medium was exchanged for Mg2+-free HBS medium along with 2 μM TTX and 2 mM MNI-glutamate. Glutamate uncaging was performed and imaged as described above. For chase labeling, the medium was exchanged for HBS with 2 μM TTX and 0.5 nM JF549 followed by incubation for 1.5 h at 35–37 °C. The neurons were washed in HBS for 30 min and imaged. Glutamate receptor antagonists APV (20 μM) or NBQX (10 μM) or the translation inhibitor CHX (60 μM; Sigma) were added to pulse and included in all subsequent steps until the last washout step before imaging. Images of dendrites were sum-projected and straightened as described above for image analysis.
Image Analysis.
Calculation of ΔF/F was previously described (62). The ΔF/F dynamics of the selected spine were calculated by averaging the region of interested at each time point. For the uncaging experiments, 5D image series (x, y, z, color, and time) of selected dendrites were obtained before (pre) and after (post) uncaging to capture the mRNA dynamics. The z-series of each color were acquired sequentially. The z-steps (seven steps with an interval of 0.5 µm) were chosen to minimize the exposure to the laser light and to increase the imaging time. The images were first maximum z-projected. Each time point was registered to the first image using the mCherry channel to remove the long-term drift according to a descriptor-based algorithm (63). The uncaged dendrite then was straightened with the Image J plug-in (64). The uncaged position was mapped to the coordinates of the straightened dendrite by projecting the uncaging spot to the nearest point on the dendrite. The mRNA locations were obtained with an automated spot-detection algorithm, u-track (65). If an mRNA position fell within ±3 µm of the boundary of the uncaged region (within the 6-µm segment), the mRNA was considered to be localized. To analyze the temporal dynamics of mRNA localization, the number of localized mRNAs for each time point was determined for each dendrite. To examine the spatial accumulation of mRNAs in the uncaged region, the mRNA density was derived by counting the number of mRNAs within the specified bins along the dendrite. Different dendrites were aligned together by setting the center of the uncaged region as coordinate zero. The ΔRNA density was obtained by subtracting the post-uncaging mRNA density from the pre-uncaging mRNA density. In both temporal and spatial analyses, each dendrite was processed independently, and the final result was averaged by pooling all dendrites together. For statistical analysis, the data were analyzed by unpaired Student’s t test for comparison of two groups or by ANOVA and Dunnett’s post hoc test for comparison of multiple groups (more than two groups) with control.
For the JF646-uncage-JF549 labeling assay, change in JF646 intensity (ΔJF646) was calculated by comparing JF646 intensities along the length of the dendrite before and after uncaging (Fig. S4E). Average fluorescence intensity was measured from each 6-μm segment (56 pixels at 106.7 nm per pixel) centered on the coordinates of the uncaging spot. The intensity values were normalized to the furthest flanking segment ±24 μm. To assess the translation output of JF646/JF549-labeling experiments, average fluorescence intensities per 6-μm segment were calculated for JF646 labeling before uncaging and for JF549-labeling after uncaging. The ratio of JF646 to JF549 labeling provides the relative abundance of translation output, because neurons will express different amounts of reporter proteins as the result of varying levels of lentivirus expression. The ratios of experimental groups (uncaged, APV, CHX, NBQX, and mock) were normalized to the ratio from dendrites that did not receive uncaging stimulation. Hence the relative changes in JF646-/JF549-labeling ratios reflect the changes in reporter synthesis that are dependent on the effect of each treatment.
To calculate local maxima of JF646 and JF549, images of individual spines were selected and analyzed by the Image J plug-in FociPicker 3D (a 3D and 2D particle counter) to identify the center of fluorescence for each channel. The distances between JF646 and JF549 were calculated and plotted as a histogram, or distances from each spine were averaged for each condition.
SI Materials and Methods
To establish the causal relationship between stimulation and localized gene expression, we applied an optical method that could selectively deliver neurotransmitter to individual spines. Photolytic uncaging uses focused light to unmask a caged compound and convert it to its active form with fine spatial and temporal precision and facilitates the efficient delivery of biomolecules to specific subcellular domains (16). For example, the use of caged neurotransmitter MNI-glutamate has aided in the mapping of glutamate receptors, input specificity, and structural plasticity at the level of individual spines (24–26).
To couple single-molecule imaging with optical stimulation, we focused 405-nm light to a diffraction-limited spot (smaller than the wavelength of light) to uncage MNI-glutamate locally (Fig. S2 A–C). Unless noted otherwise, glutamate uncaging experiments were performed in Hepes buffer containing 2 mM MNI-glutamate, no extracellular magnesium, and TTX. Reduction of magnesium in the medium removed the need for depolarization to facilitate removal of the magnesium block of NMDA receptors. TTX blocks the presynaptic sodium channels necessary for propagation of spontaneous activity. Blocking presynaptic activity with TTX allows the input to be confined to a specific set of glutamatergic synapses by uncaging.
To calibrate uncaging efficiency, we expressed a genetically encoded calcium indicator, GCaMP3, as a readout of glutamate receptor-mediated calcium entry into spines (60). We delivered a single 5-ms uncaging pulse (5 mW, back aperture) to a position adjacent to the selected spine (Fig. S2 D and E) and distal to the dendritic axis and acquired streaming (50-ms) images of GCaMP3 (Fig. S2F). We calculated the fluorescence decay (Fig. S2H), fit to a single exponential (T1/2 = 231 ± 31 ms at 35.5–36.5 °C) and determined the correlation between the intensity of uncaging light and GCaMP3 fluorescence change (ΔF/F), demonstrating that uncaging efficiency can be tuned (Fig. S2J). The addition of the NMDA receptor antagonist APV to cells before uncaging reduced the change in GCaMP3 fluorescence (T1/2 = 378 ± 7 ms) (Fig. S2I). The results demonstrate that NMDA receptors mediate rapid calcium entry into spines in response to glutamate.
We next expressed soluble GFP to assess changes in spine volume following uncaging. We applied three trains of 10 pulses at 0.5 Hz with a 2-min interval between trains (Fig. S2K). We observed a mean volume increase of 50 ± 14% at 5 min poststimulation in the activated spines; the volume change persisted for 20 min (Fig. S2 L and M). In contrast, adjacent spines exhibited a 5.4 ± 3.3% change in volume.
To visualize stimulation-dependent F-actin dynamics in spines, we imaged exogenous GFP-actin–expressing neurons. Previous studies showed that repeated uncaging pulses were necessary to induce structural changes in synaptic spines (40, 41), and we delivered a low-frequency uncaging stimulation (30 pulses at 0.5 Hz) to target spines. Using this protocol (Fig. S2K), we induced dynamic reorganization of actin filaments within spines and elicited structural remodeling (Fig. S2N). We observed an initial fourfold increase in fluorescence which relaxed to a threefold increase that lasted throughout imaging (Fig. S2O). The change in fluorescence was caused by the recruitment of the enlargement pool of globular actin assembling into the F-actin network within the volume of the spine head (43, 44). The overexpressed actin is likely the contributing source of the large increase observed in spines, as well as the adjacent spines undergoing modest increases during the same period. The results demonstrate that our uncaging protocol can induce structural changes in the targeted spine. In sum, these results indicate that glutamate can lead to calcium entry, morphological dynamics, and enlargement of spines, consistent with the changes observed by postsynaptic stimulation of spines. For all subsequent uncaging assays we used this low-frequency uncaging protocol (3 × 10 pulses at 0.5 Hz).
Live Imaging and Analysis.
We used both Fiji (66) and ImageJ for kymograph generation. Time-series images of dendrites were straightened using the Straighten ImageJ plug-in (64), and then the stacks were rotated using Fiji (Plugins>>Transform>>TransformJ>>TransformJ Turn). The rotated images were maximally projected to generate (x–t) kymographs. To determine the immobile population of mRNA from our time-series images, we first detected mRNA using u-track (65). Once mRNA positions were determined, we used a custom-written MATLAB script to calculate whether each successive frame had an mRNA within a 1-μm radius of the originally detected particle. If the position of the mRNA particle persisted for more than 90 consecutive frames (out of 100 total frames), it was determined to be stationary. Under this criterion, the stationary particles were quantified for each time point. Once the number of stationary particles was known, we calculated the fraction of the stationary population from the total number of detected particles at each time point. The analysis also provided events in which the mRNA remained docked at a specific position for less than 90 frames, which we defined as transient events. The minimum criterion for a transient event was the detection of a particle within a 1-μm radius of the originally detected particle for five or more consecutive frames.
Microscopy Software.
The microscope was controlled with the imaging software MetaMorph (Molecular Devices). At the beginning of each experimental session, the focused laser position was recorded and saved in MetaMorph as a global variable. To uncage a specific spine, the stage was moved until the desired spot coincided with the prerecorded laser position. To uncage several spines, the desired uncaging positions were saved in MetaMorph, and the stage was moved to each position sequentially before an uncaging laser pulse was fired. The pulse width and the power of the uncaging laser were controlled by AOTF. After the uncaging pulse was applied, there was a 5-ms delay to allow the flipping lens to move out of the light path before imaging. All the procedures described here were automated with custom-written programs in MetaMorph. To image calcium dynamics following the uncaging of glutamate, a streaming time series from a single plane was acquired with 35-ms exposure time. The same dendrite was imaged immediately before uncaging as reference.
Supplementary Material
Acknowledgments
We thank all past members of the R.H.S. laboratory who contributed to the generation of β-actin MBS-knockin transgenic mice; X. Meng for making many of the constructs used in the study; and Yang Wu and other members of the R.H.S. laboratory for their critical reading of the manuscript. A.T. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation Grant DRG-2220-15. This work was supported by NIH Grant GM84364/NS083085-19 (to R.H.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614267113/-/DCSupplemental.
References
- 1.Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell. 2009;136(4):719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt CE, Schuman EM. The central dogma decentralized: New perspectives on RNA function and local translation in neurons. Neuron. 2013;80(3):648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cajigas IJ, et al. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74(3):453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rook MS, Lu M, Kosik KS. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: Visualization by GFP linkage. J Neurosci. 2000;20(17):6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiruchinapalli DM, et al. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23(8):3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17(24):9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273(5280):1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 8.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21(4):741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 9.Ostroff LE, Fiala JC, Allwardt B, Harris KM. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35(3):535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- 10.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30(2):489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 11.Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314(5796):144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- 12.Wang DO, et al. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324(5934):1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290(5492):754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- 14.Lionnet T, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8(2):165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park HY, et al. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343(6169):422–424. doi: 10.1126/science.1239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callaway EM, Yuste R. Stimulating neurons with light. Curr Opin Neurobiol. 2002;12(5):587–592. doi: 10.1016/s0959-4388(02)00364-1. [DOI] [PubMed] [Google Scholar]
- 17.Wu B, et al. Synonymous modification results in high-fidelity gene expression of repetitive protein and nucleotide sequences. Genes Dev. 2015;29(8):876–886. doi: 10.1101/gad.259358.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Los GV, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3(6):373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 19.Grimm JB, et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;12(3):244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buxbaum AR, Wu B, Singer RH. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343(6169):419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011;30(17):3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu B, Chao JA, Singer RH. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys J. 2012;102(12):2936–2944. doi: 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dynes JL, Steward O. Arc mRNA docks precisely at the base of individual dendritic spines indicating the existence of a specialized microdomain for synapse-specific mRNA translation. J Comp Neurol. 2012;520(14):3105–3119. doi: 10.1002/cne.23073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429(6993):761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuzaki M, et al. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001;4(11):1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey CD, Svoboda K. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 2007;450(7173):1195–1200. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watkins JC, Evans RH. Excitatory amino acid transmitters. Annu Rev Pharmacol Toxicol. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- 28.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81(3):403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 29.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124(3):521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 30.Matus A, Ackermann M, Pehling G, Byers HR, Fujiwara K. High actin concentrations in brain dendritic spines and postsynaptic densities. Proc Natl Acad Sci USA. 1982;79(23):7590–7594. doi: 10.1073/pnas.79.23.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17(4):2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park HY, Trcek T, Wells AL, Chao JA, Singer RH. An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Reports. 2012;1(2):179–184. doi: 10.1016/j.celrep.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26(16):3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang HL, et al. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31(2):261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 35.Patel VL, et al. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012;26(1):43–53. doi: 10.1101/gad.177428.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz ZB, et al. β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 2012;26(17):1885–1890. doi: 10.1101/gad.190413.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutton MA, Schuman EM. Local translational control in dendrites and its role in long-term synaptic plasticity. J Neurobiol. 2005;64(1):116–131. doi: 10.1002/neu.20152. [DOI] [PubMed] [Google Scholar]
- 38.Chao JA, Yoon YJ, Singer RH. Imaging translation in single cells using fluorescent microscopy. Cold Spring Harb Perspect Biol. 2012;4(11):a012310. doi: 10.1101/cshperspect.a012310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156(6):1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harvey CD, Yasuda R, Zhong H, Svoboda K. The spread of Ras activity triggered by activation of a single dendritic spine. Science. 2008;321(5885):136–140. doi: 10.1126/science.1159675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472(7341):100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010;67(1):86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosch M, et al. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 2014;82(2):444–459. doi: 10.1016/j.neuron.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57(5):719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 45.Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458(7236):299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Govindarajan A, Israely I, Huang SY, Tonegawa S. The dendritic branch is the preferred integrative unit for protein synthesis-dependent LTP. Neuron. 2011;69(1):132–146. doi: 10.1016/j.neuron.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Losonczy A, Magee JC. Integrative properties of radial oblique dendrites in hippocampal CA1 pyramidal neurons. Neuron. 2006;50(2):291–307. doi: 10.1016/j.neuron.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 48.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11(8):825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446(7136):633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 51.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. 2014;17(6):884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenmund C, Westbrook GL. Calcium-induced actin depolymerization reduces NMDA channel activity. Neuron. 1993;10(5):805–814. doi: 10.1016/0896-6273(93)90197-y. [DOI] [PubMed] [Google Scholar]
- 53.Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23(32):10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3(3):211–216. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- 55.Terman JR, Kashina A. Post-translational modification and regulation of actin. Curr Opin Cell Biol. 2013;25(1):30–38. doi: 10.1016/j.ceb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knöll B. Actin-mediated gene expression in neurons: The MRTF-SRF connection. Biol Chem. 2010;391(6):591–597. doi: 10.1515/BC.2010.061. [DOI] [PubMed] [Google Scholar]
- 57.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127(1):49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 58.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61(1):10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.tom Dieck S, et al. Direct visualization of newly synthesized target proteins in situ. Nat Methods. 2015;12(5):411–414. doi: 10.1038/nmeth.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6(12):875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Follenzi A, Naldini L. Generation of HIV-1 derived lentiviral vectors. Methods Enzymol. 2002;346:454–465. doi: 10.1016/s0076-6879(02)46071-5. [DOI] [PubMed] [Google Scholar]
- 62.Jia H, Rochefort NL, Chen X, Konnerth A. In vivo two-photon imaging of sensory-evoked dendritic calcium signals in cortical neurons. Nat Protoc. 2011;6(1):28–35. doi: 10.1038/nprot.2010.169. [DOI] [PubMed] [Google Scholar]
- 63.Preibisch S, Saalfeld S, Schindelin J, Tomancak P. Software for bead-based registration of selective plane illumination microscopy data. Nat Methods. 2010;7(6):418–419. doi: 10.1038/nmeth0610-418. [DOI] [PubMed] [Google Scholar]
- 64.Kocsis E, Trus BL, Steer CJ, Bisher ME, Steven AC. Image averaging of flexible fibrous macromolecules: The clathrin triskelion has an elastic proximal segment. J Struct Biol. 1991;107(1):6–14. doi: 10.1016/1047-8477(91)90025-r. [DOI] [PubMed] [Google Scholar]
- 65.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5(8):695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.