Significance
Efficient clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9)-mediated mutagenesis is necessary for robust genetic screening in primary cells and requires sufficiently high levels of Cas9 and reliable single guide RNAs (sgRNAs). We provide a sgRNA design tool that selects high-fidelity sgRNAs and a Cas9 transgenic mouse line that expresses Cas9 at high levels in all cells analyzed. Using this system, we achieved an average knockout efficiency of 80% in primary B cells and established a robust CRISPR/Cas9-mediated genetic screening system that can be adapted to other primary cell types.
Keywords: CRISPR/Cas9, sgRNA design, knockout efficiency, primary cells, B cells
Abstract
Applying clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9)-mediated mutagenesis to primary mouse immune cells, we used high-fidelity single guide RNAs (sgRNAs) designed with an sgRNA design tool (CrispRGold) to target genes in primary B cells, T cells, and macrophages isolated from a Cas9 transgenic mouse line. Using this system, we achieved an average knockout efficiency of 80% in B cells. On this basis, we established a robust small-scale CRISPR-mediated screen in these cells and identified genes essential for B-cell activation and plasma cell differentiation. This screening system does not require deep sequencing and may serve as a precedent for the application of CRISPR/Cas9 to primary mouse cells.
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) technology is a powerful tool for gene editing (1–3). Application of this technology to primary cells is still in its initial phase, but promises to accelerate research. In many instances, such as the primary hematopoietic cells studied here, high knockout efficiencies are essential for studying gene function, as the limited life span of the cells often does not allow for selection and expansion of knockout clones before in vitro experiments. Moreover, screens that are based on readouts that do not affect survival require high knockout efficiencies to identify true positive hits. The two most decisive factors for high CRISPR/Cas9 knockout efficiencies are sufficient nuclear Cas9 levels and the selection of potent sgRNAs (4, 5). To overcome the requirement of Cas9 delivery to primary cells, transgenic mice expressing Cas9 and GFP from the Rosa26 locus have been generated (ref. 6; a related approach, targeting the hematopoietic system, has been lentiviral Cas9 transduction into hematopoietic stem cells, ref. 7). Whereas primary dendritic cells of these mice have been successfully used for functional screening in vitro (6, 8), their lymphocytes express only low levels of GFP, and their use in CRISPR-mediated screens has not been reported so far. In addition, these mice were not generated on a pure C57BL/6 background. We thus generated a C57BL/6 Cas9-transgenic line with an improved Cas9 expression cassette (9). As we were interested in small-scale screening, a key consideration was to use only few, but reliable single guide RNAs (sgRNAs) without the necessity of sgRNA-testing experiments before screening. Previous studies have shown large variations in sgRNA efficiencies (8, 10–12). Multiple sgRNA design programs have been developed, but none of them combines all of the criteria that we developed in laboratory to reach optimal sgRNA activity and specificity, including on-target efficiency, genome-wide off-target predictions taking into account the impact of mismatch type and position, targeting of all isoforms of genes, and conservation of functional secondary structure of the sgRNA (13). We thus developed a sgRNA design tool that selects sgRNAs having high specificity, high on-target efficiency, and intact secondary structure important for the recognition by Cas9. Combining this program with our Cas9 transgenic C57BL/6 mice, we established an easy and robust screening method to study gene function in primary hematopoietic (and likely other) cells with high signal-to-noise ratio.
Results
Design of Specific and Efficient sgRNAs.
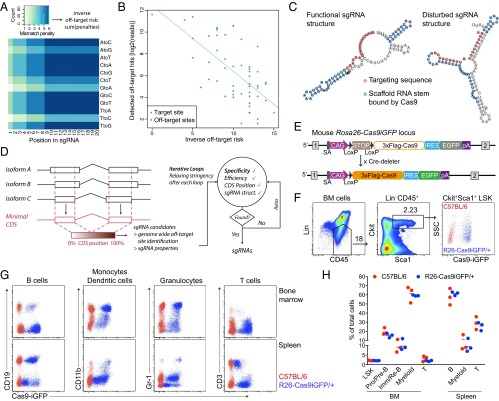
Coding genes typically contain hundreds of protospacer adjacent motif (PAM) sequences, allowing for a wide choice among possible sgRNAs. To design the most specific and efficient sgRNAs to target a gene or set of genes, we implemented our sgRNA design rules in a tool that we termed “CrispRGold.” CrispRGold first projects the coding sequences (CDSs) of the different isoforms of a gene to a single “minimal CDS,” to identify sgRNAs that would target most or all isoforms of that gene. For each sgRNA candidate, the position in the minimal CDS is stored and the sgRNA is aligned to the genome using the Burrows–Wheeler aligner (BWA) (14) to identify potential off-target sites. Based on the quantitative measurement of off-target site activity (15), we defined a type- and position-dependent mismatch-penalty matrix, such that the sum of mismatch penalties (or mutation distance) for the various off-target sites would inversely correlate with the frequency of detected off-target site activities (Fig. 1 A and B). This mismatch penalty matrix especially weighs the increased impact of mismatches close to the 3′-end of the sgRNA and the decreased relevance of mutations causing wobble base pairing. CrispRGold uses this matrix to calculate the risk of each individual predicted off-target site genome-wide for a given sgRNA, sorts these sites according to their risk, and attributes a global off-target risk (or specificity score) to each sgRNA based on the three off-target sites with the highest risk. In addition, CrispRGold computes sgRNA-intrinsic properties, such as the GC content and the folding energy between the targeting sequence and the scaffold RNA of the sgRNA (16). A high binding energy between the targeting sequence and the scaffold may alter the secondary structure of the sgRNA and potentially disturb recognition of the sgRNA by Cas9 or binding of the targeting sequence to the DNA (Fig. 1C). Finally, CrispRGold goes through iterative loops of decreasing stringency to find the required number of sgRNAs per gene or sequence. In each loop, sgRNAs are prioritized according to their specificity (Fig. 1D).
Fig. 1.
Design of efficient sgRNAs and characterization of Cas9-transgenic animals. (A) Heatmap showing mismatch penalties depending on the position within the targeting sequence of the sgRNA and the type of mismatch. Scores were chosen to fit the off-target frequencies detected by Tsai et al. (15). (B) Dotplot of the predicted inverse off-target risk of each off-target site of the VEGFA_3 sgRNA versus the reported GUIDEseq read numbers. Read numbers were extracted from Tsai et al. (15). (C) Predicted secondary structure of two sgRNAs targeting mouse Btk. sgRNAs with low binding energy between the targeting sequence and the scaffold RNA do not disturb the secondary structure of the scaffold RNA (Left), in contrast to sgRNAs with high binding energies between the targeting sequence and the scaffold RNA (Right). (D) Scheme of the CrispRGold sgRNA design tool. (E) Scheme of the R26-LSL-Cas9iGFP allele before (Top) and after crossing to Cre-deleter mice (Bottom). Cre-mediated removal of the LSL cassette leads to ubiquitous expression of Cas9 and GFP in the entire mouse. (F) FACS analysis of GFP expression in HSCs, defined as the Lin−,CD45+,Sca1+cKit+ LSK population. LSK cells from wild-type C57BL/6 mice (red) and R26-Cas9iGFP/+ mice (blue) are shown on the same FACS plot (Right). (G) FACS analysis of GFP expression in B cells (CD19+), monocytes/dendritic cells (CD11b+), granulocytes (Gr-1+), and T cells (CD3+). Cells were isolated from the BM (Top) or spleen (Bottom). Cells from wild-type C57BL/6 mice (red) and R26-Cas9iGFP/+ mice (blue) are shown in the same FACS plots. (H) Percentages of the indicated cell populations within all cells in the BM (Left) or the spleen (Right) of wild-type animals (red) or R26-Cas9iGFP/+ animals (blue). Each dot represents one mouse.
Characterization of the Cas9 Transgenic Mouse Line.
In the mice created by Platt et al. (6) (hereafter called R26-Cas9p2aGFP), the Cas9 protein is fused to eGFP via a self-cleaving P2A peptide. In our Rosa26-LSL-Cas9iGFP mice, generated by CRISPR-mediated editing of C57BL/6 zygotes, Cas9 is also expressed from the Rosa26 locus in a Cre-dependent manner, but with the Cas9 coding sequence linked to eGFP via an internal ribosomal entry site (IRES) (9). To apply the CRISPR/Cas9 technology to primary immune cells, we generated Cas9-transgenic mice with ubiquitous expression of Cas9 by crossing Rosa26-LSL-Cas9iGFP with Cre-deleter mice (Fig. 1E). We then evaluated the expression of Cas9 in the resulting R26-Cas9iGFP/+ animals by monitoring GFP reporter levels by FACS. All hematopoietic subpopulations analyzed exhibited high GFP levels, including hematopoietic stem cells (HSCs) in the bone marrow (BM), and various hematopoietic lineages, such as T, B, and myeloid cells, independent of their anatomical location (Fig. 1 F and G and Fig. S1 A–C). The percentages of HSCs, B cells, T cells, and myeloid cells were similar between R26-Cas9iGFP/+ and control mice, indicating that the expression of Cas9iGFP is not toxic to any of these cell types (Fig. 1H). GFP levels in R26-Cas9iGFP/+ mice were higher than in R26-Cas9p2aGFP/+ mice (6), likely due to the presence of the IRES instead of the P2A peptide between Cas9 and GFP (Fig. S2 A and B). To confirm that Cas9 is indeed present in the GFP+ cells, we performed Western blotting on splenic B cells that were activated in vitro and on BM-derived macrophages (BMDMs). In both cases, we detected high levels of Cas9 protein (Fig. S3A). Moreover, Cas9 was localized in the nucleus in the BMDM (Fig. S3B). Thus, primary immune cells of R26-Cas9iGFP/+ animals express high levels of GFP and nuclear Cas9 to nontoxic levels, allowing for genome editing and easy discrimination of Cas9+ and Cas9− cells in subsequent experiments.
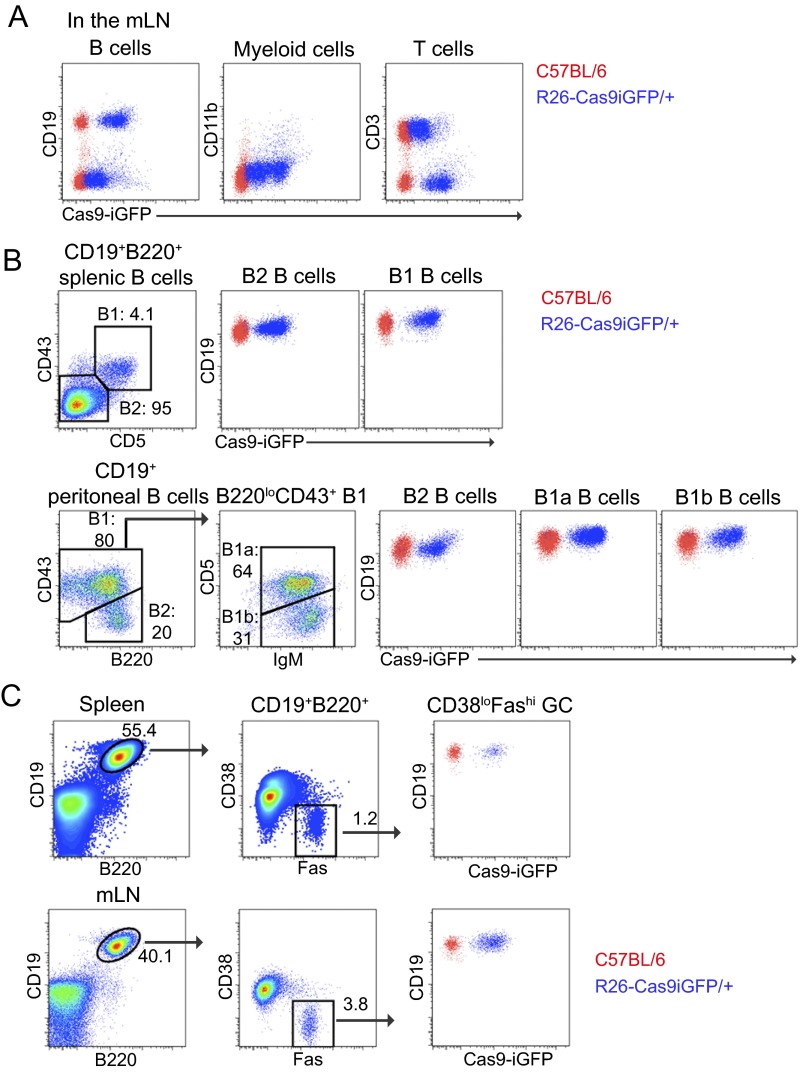
Fig. S1.
Characterization of heterozygous R26-Cas9iGFP transgenic mice. (A) FACS plots of T cells (CD3+), myeloid cells (CD11b+), and B cells (CD19+) isolated from the mLNs of Cas9 transgenic or wild-type animals. (B) FACS plots of B cells (CD19+) isolated from the peritoneal cavity and spleen of Cas9 transgenic or wild-type animals. Gates were set on B1 and B2 cells in the spleen and B2, B1a, and B1b in the peritoneal cavity based on CD19, B220, CD43, CD5, and IgM markers as indicated. (C) FACS plots of B220+CD19+FashiCD38low GC B cells isolated from the spleen and mLNs. Data are representative of four independent experiments. Cells from wild-type C57BL/6 animals (red) and R26-Cas9iGFP/+ animals (blue) are shown in the same FACS plots.
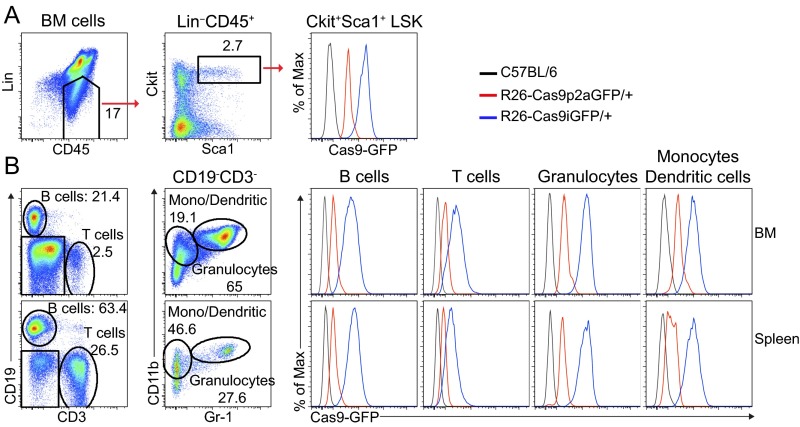
Fig. S2.
Comparison of GFP expression levels in HSCs of R26-Cas9iGFP and R26-Cas9p2aGFP mice. (A) FACS plots of HSCs (defined as Lin−Sca1+c-Kit+ or LSK), isolated from the BM of R26-Cas9iGFP/+ (blue), R26-Cas9p2aGFP/+ (red), and C57BL/6 (black) mice. (B) FACS plots of B cells (CD19+), T cells (CD3+), granulocytes (Gr1+CD11b+), and monocytes/dendritic cells (CD11b+Gr1−) isolated from the BM and spleen of R26-Cas9iGFP (blue), R26-Cas9p2aGFP (red), and wild-type (black) mice. The data are representative of two independent experiments.
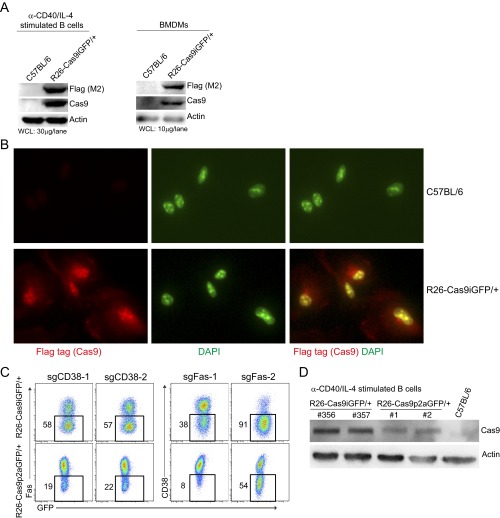
Fig. S3.
Cas9 levels and localization in primary cells of R26-Cas9iGFP/+ mice and Cas9-dose-dependent knockout efficiency. (A) Western blots of Cas9 proteins (detected by anti-Cas9 and anti-Flag antibodies) in anti-CD40/IL-4–activated B cells and BMDMs isolated from R26-Cas9iGFP/+ and wild-type animals. β-Actin was used as a loading control. (B) Immunostaining of Cas9 in BMDMs derived from R26-Cas9iGFP/+ and wild-type animals. Cas9 was stained using anti-Flag (M2) antibodies (red). DAPI was used for counterstaining of nuclei (green). (C) Knockout efficiencies in B cells from R26-Cas9iGFP/+ (Top) and R26-Cas9p2aGFP/+ (Bottom) mice that were activated and transduced with retroviruses expressing sgCD38-1, sgCD38-2, sgFas-1, and sgFas-2 side by side. The gates in the FACS plots show the percentage of cells that lost the surface marker 4 d after transduction. (D) Western blots of Cas9 proteins in anti-CD40/IL-4–activated B cells isolated from R26-Cas9iGFP/+, R26-Cas9p2aGFP/+, and wild-type animals. The data are representative of two independent experiments.
Efficient CRISPR/Cas9-Mediated Mutagenesis in Primary Immune Cells.
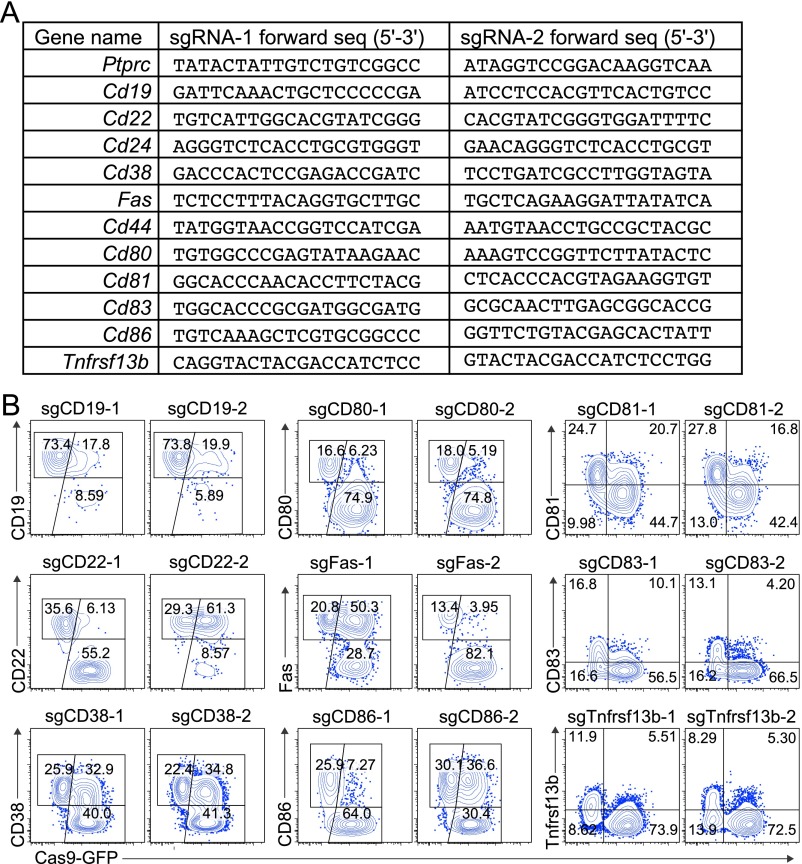
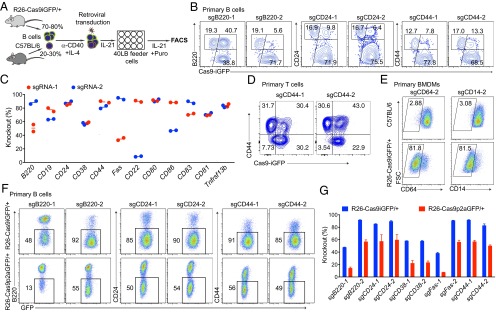
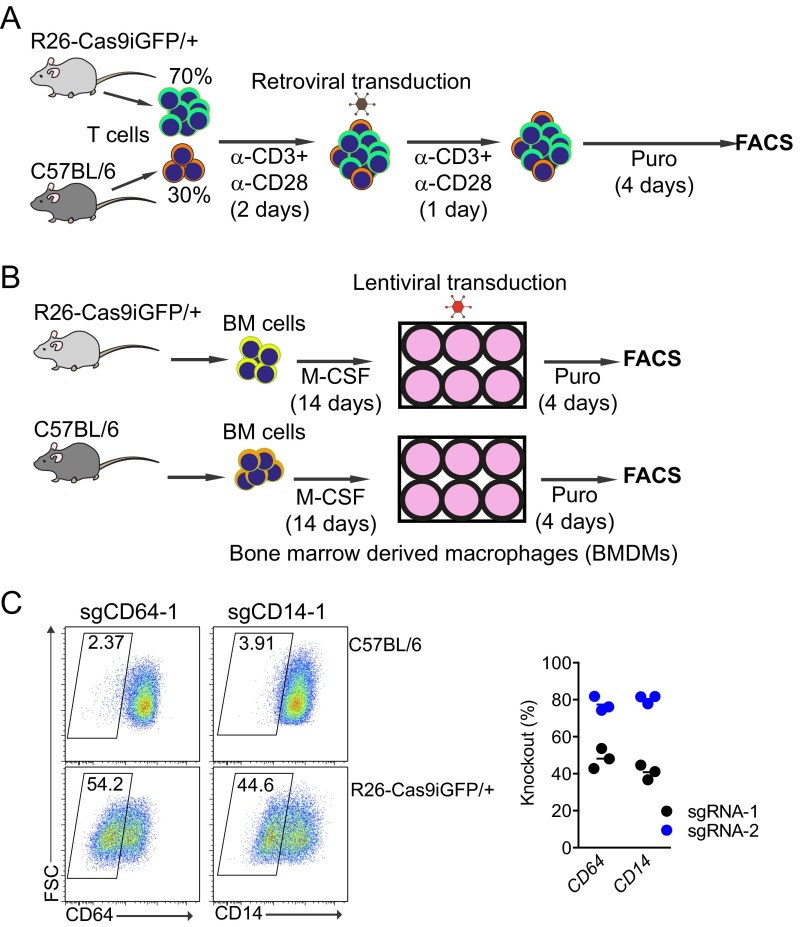
To assess the knockout efficiency of the sgRNAs designed by CrispRGold in primary immune cells, we first selected 12 genes encoding B-cell surface markers and designed two sgRNAs per gene (Fig. S4A). These sgRNAs were individually cloned into retroviruses bearing a blue fluorescent protein (BFP) reporter and a puromycin resistance gene. We then isolated B cells from wild-type and R26-Cas9iGFP/+ mice and mixed them in a 1:4 ratio, such that we could use the Cas9− (GFP−) cells as internal controls. These mixed B cells were activated with anti-CD40 antibodies and IL-4 for 2 days and then transduced with retroviral particles. The transduced cells were further cultured on 40LB feeder cells (17) (NIH3/T3 cells stably expressing CD40 ligand and BAFF) in the presence of IL-21 and puromycin (Fig. 2A). Four days after transduction, all sgRNAs had led to cell surface marker knockouts, mostly with very high efficiency and an average knockout frequency above 70% (Fig. 2 B and C and Fig. S4B). Only one sgRNA led to significantly lower knockout efficiencies due to a T-rich sequence adjacent to the PAM, now implemented in CrispRGold. To confirm that these high knockout efficiencies are not unique to B cells, we performed a similar experiment using primary T cells that we activated with anti-CD3 and anti-CD28 (Fig. S5A). Four days after transduction with retroviral particles encoding sgRNAs against CD44, about 50% of the cells showed a complete knockout of CD44 (Fig. 2D). Finally, to determine whether we reached similarly high knockout frequencies in quiescent primary myeloid cells, we differentiated bone marrow cells into resting BMDMs using macrophage colony-stimulating factor (M-CSF) and transduced them with lentiviral vectors expressing sgRNAs targeting CD64 and CD14 (Fig. S5B). Four days after transduction, 40–80% of the cells showed a knockout of the targeted genes (Fig. 2E and Fig. S5C). Thus, primary immune cells of R26-Cas9iGFP/+ mice are suitable for CRISPR/Cas9-mediated genome editing, and CrispRGold designs reliably highly efficient sgRNAs. In addition to the intrinsic properties of sgRNAs, the on-target dosage of sgRNA/Cas9 complexes is essential for efficient mutagenesis. The lower GFP levels in lymphocytes of R26-Cas9p2aGFP/+ mice (6) suggested that they have lower levels of Cas9 than the R26-Cas9iGFP/+ mice here described. To compare the impact of Cas9-dosage on knockout efficiency, we thus repeated the experiment depicted in Fig. 2A, using the same sgRNAs and B cells of both Cas9-transgenic lines side by side. Indeed, all sgRNAs tested led to significantly lower knockout efficiencies in B cells from R26-Cas9p2aGFP/+ mice compared with R26-Cas9iGFP/+ mice (Fig. 2 F and G and Fig. S3C), consistent with lower Cas9 levels in these cells (Fig. S3D). Thus, the combination of optimal sgRNAs and high levels of Cas9 in primary cells of R26-Cas9iGFP/+ mice led to the high knockout efficiencies observed in primary cells isolated from these animals.
Fig. S4.
Knockout efficiencies in Cas9-expressing B cells. (A) Selection of sgRNAs designed with CrispRGold targeting 12 B-cell surface makers. Targeting sequences are shown 5′–3′. (B) FACS plots showing the knockout efficiencies in Cas9-expressing B cells 4 d after transduction with sgRNAs against the indicated surface markers. Note that some surface markers are only induced upon activation (CD83 and Tnfrsf13b). The sgRNAs used are indicated in the title of each FACS plot. The data are representative of three independent experiments.
Fig. 2.
Efficient CRISPR-mediated gene inactivation in primary immune cells. (A) Scheme of CRISPR-mediated gene knockout in Cas9-expressing B cells. Splenic B cells isolated from wild-type and Cas9 transgenic mice were mixed at a 1:4 ratio. These cells were activated with anti-CD40 and IL-4 for 2 d and transduced with retroviral particles expressing specific sgRNAs and a puromycin resistance gene. One day after transduction, the cells were transferred into 40LB feeder cell plates and cultured in the presence of IL-21 and puromycin. (B) FACS plots of CD19+BFP+ cells targeted with sgRNAs against B220 (Left), CD24 (Middle), and CD44 (Right) 4 d after transduction. Knockout efficiencies were calculated based on the GFP+ cells that lost the surface marker (subgates of the GFP+ population). (C) Summary of the knockout efficiencies for the 12 indicated surface marker genes with two sgRNAs per gene. Knockout efficiencies were determined by FACS 4 d after transduction of Cas9-expressing primary B cells with the respective sgRNAs. (D) FACS plots showing the CD44 knockout efficiency in Cas9-expressing primary T cells. Mixed Cas9 (GFP+) and wild-type (GFP−) splenic T cells were activated and transduced with retroviral particles expressing sgCD44-1 and sgCD44-2. The gates indicate the knockout efficiency in GFP+ T cells. (E) FACS plots showing the knockout efficiency of CD64 and CD14 in nondividing primary BMDMs. BMDMs from C57BL/6 (Top) and R26-Cas9iGFP/+ (Bottom) mice were transduced with lentiviruses expressing sgCD64-2 and sgCD14-2. (F) FACS plots of knockout efficiencies in primary B cells from R26-Cas9iGFP/+ (Upper) and R26-Cas9p2aGFP/+ (Lower) mice, using the same setup as in A and sgRNAs against B220, CD24, and CD44. (G) Summary of knockout efficiencies using primary B cells from R26-Cas9iGFP/+ (blue) and R26-Cas9p2aGFP/+ (red) mice.
Fig. S5.
CRISPR-mediated gene knockout in primary T cells and BMDMs. (A) Cas9-expressing T cells were mixed with wild-type T cells. The cells were activated with anti-CD3 and anti-CD28 antibodies for 2 d. The activated T cells were transduced with retroviral particles expressing sgRNAs. The transduced T cells were selected with puromycin for 4 d before assessment of knockout efficiencies. (B) Scheme of CRISPR-mediated gene knockout in primary BMDMs. BM cells from R26-Cas9iGFP/+ mice and wild-type mice were differentiated for 2 wk in the presence of M-CSF. BMDMs were transduced with lentiviruses expressing sgRNAs targeting mouse CD64 and CD14 genes. The transduced cells were selected with puromycin for 4 d and the gene knockout efficiency was analyzed by flow cytometry. (C) FACS plots of the transduced BMDMs derived from control animals (Top) and R26-Cas9iGFP/+ animals (Bottom) stained with antibodies against the targeted surface markers. The summary of the experiments is shown at Right. The data are representative of three individual Cas9 transgenic mice (each representing a data point in the summary).
Robust Inactivation of Transcription Factors Known To Be Important for B-Cell Differentiation.
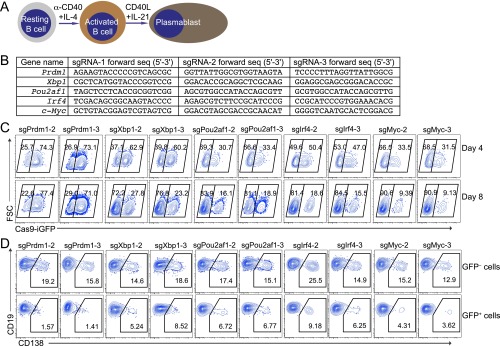
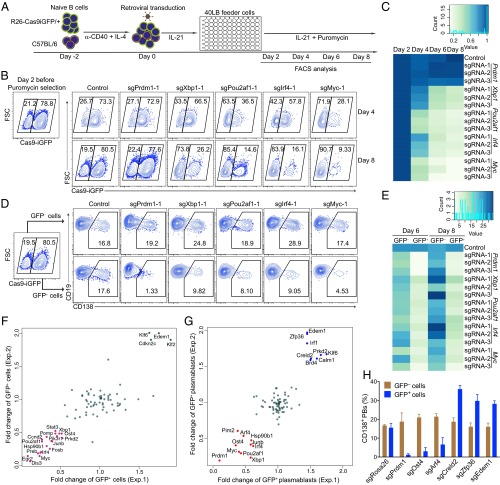
Extending the analysis to transcription factors (TFs), we selected five TFs, Prdm1, Irf4, Myc, Xbp1, and Pou2af1, that are known to be important for B-cell survival, proliferation, and differentiation (18–24) (Fig. S6A). We designed three sgRNAs per TF (Fig. S6B) and performed retroviral transduction experiments as before, but this time analyzing B-cell survival and plasma cell differentiation (Fig. 3A). Expression of sgRNAs targeting Xbp1, Irf4, Pou2af1, and Myc led to a strong survival disadvantage of Cas9-expressing cells, indicated by the decreased percentage of GFP+ cells (Fig. 3 B and C and Fig. S6C). As for plasma cell differentiation, monitored by CD138 expression, targeting of all five TFs led to a strong block of differentiation (Fig. 3 D and E and Fig. S6D). These findings are consistent with previous reports, showing that Xbp1, Irf4, Pou2af1, and Myc are important for both B-cell survival, proliferation, and terminal differentiation (18–22), whereas Prdm1 is only important for the latter (23, 24). Moreover, in all cases, the three individual sgRNAs showed a strong and consistent effect on the biological readout, further demonstrating that sgRNAs designed by CrispRGold work with high efficiency and consistency.
Fig. S6.
Robust detection of transcription factors important for B-cell activation and differentiation. (A) Scheme of B-cell activation and differentiation. Splenic B cells were activated with anti-CD40+IL4 and subsequently differentiated into plasmablasts using IL-21 and CD40-ligand expressing feeder cells. (B) sgRNAs used to target Prdm1, Xbp1, Pou2af1, Irf4, and Myc. (C) Analysis of the percentage of GFP+ Cas9-expressing B cells and GFP− wild-type B cells. These cells were mixed, activated, and transduced (individually) with three sgRNAs per gene targeting Prdm1, Xbp1, Pou2af1, Irf4, and Myc. The FACS plots show the GFP+ Cas9-expressing B cells 4 and 8 d after transduction. (D) Analysis of the induction of CD138 in GFP+ Cas9-expressing and GFP− wild-type B cells 6 d after transduction. The data are representative of three independent experiments.
Fig. 3.
Identification of genes involved in B-cell activation and differentiation using robust CRISPR-mediated screening. (A) Scheme of CRISPR-mediated screening in primary B cells. GFP+ splenic B cells from R26-Cas9iGFP/+ mice were mixed with GFP− wild-type B cells and treated as indicated in the 96-well system. The cells were analyzed by flow cytometry at the indicated time points. (B) FACS analysis of mixed GFP+/GFP− B cells 4 or 6 d after transduction with empty vector (control), sgPrdm1-1, sgXbp1-1, sgPou2af1-1, sgIrf4-1, or sgMyc-1. The gates show the percentage of GFP+ and GFP− B cells after gating on BFP+ cells. (C) Heatmap of the percentages of GFP+ B cells at day 4, 6, or 8 normalized to day 2. One control sgRNA and three different sgRNAs per TF were used to knockout Prdm1, Xbp1, Pou2af1, Irf4, or Myc. (D) FACS analysis of plasma cell differentiation, indicated by the expression of CD138, in the same experiment as in B. Gates are set on CD138+ plasmablasts in GFP− (Top) and GFP+ (Bottom) cells. (E) Heatmap of the percentages of CD138+ plasmablasts within the GFP+ and GFP− at the indicated time points. (F) Fold changes of the percentages of GFP+ B cells at day 8 after transduction with sgRNAs against the candidate genes in two independent experiments. The repressor genes are indicated in green and the enhancer genes are shown in magenta. Fold changes were normalized to the initial percentage of GFP+ cells. (G) Fold changes of the percentages of CD138+GFP+ plasmablasts at day 8 in the same experiment as in F. The repressor genes are indicated in blue and the enhancer genes are shown in red. (H) Validation of the newly identified genes important in plasma cell differentiation. Histogram showing the percentages of GFP+CD138+ and GFP−CD138+ plasmablasts 6 d after transduction with sgRNAs targeting Rosa26, Prdm1, Ost4, Arf4, Creld2, Zfp36, and Edem1.
Identification of Genes Important for B-Cell Activation and Plasma Cell Differentiation.
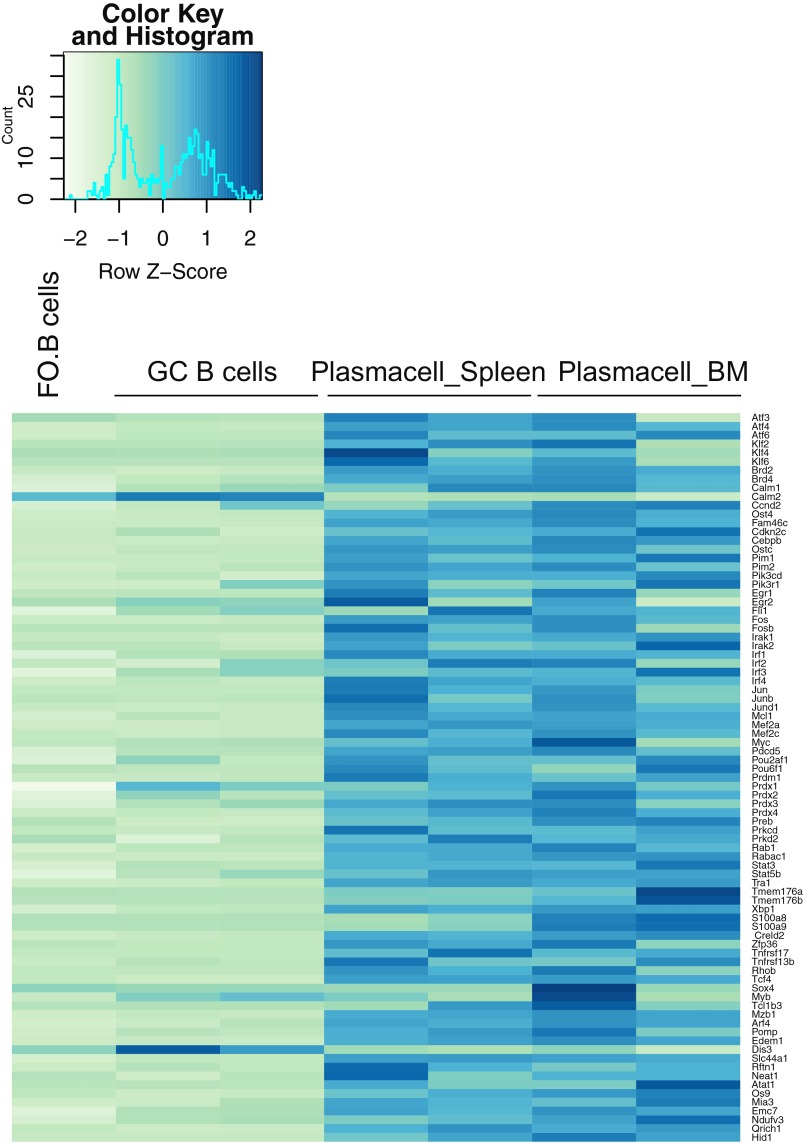
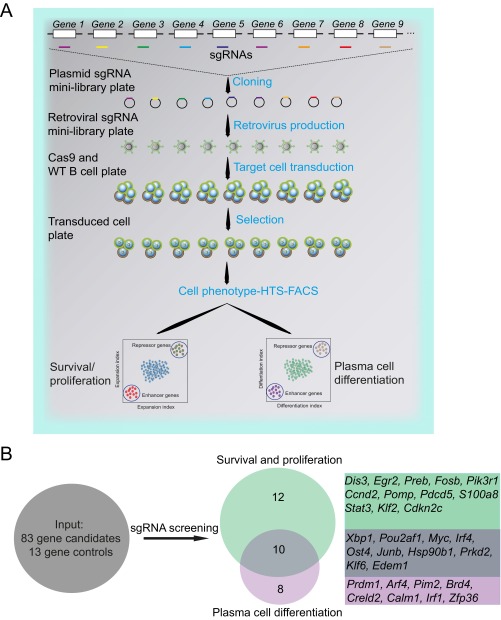
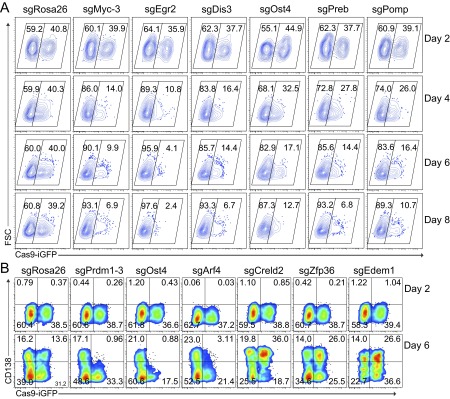
Benefitting from the robustness of our system and the consistently high knockout frequencies, we performed a small-scale screen to identify genes important for B-cell proliferation and terminal differentiation. Based on a microarray analysis of B cells and plasma cells isolated from immunized mice, we selected 83 candidate genes up-regulated during plasma cell differentiation in vivo (Fig. S7). We designed one sgRNA against each of these genes and 13 control genes. We then performed the same experiment as before in a 96-well system, using the percentage of GFP+ and CD138+ cells as readout for survival/proliferation and plasma cell differentiation, respectively (Fig. 3A and Fig. S8A). Based on two experiments, which led to highly consistent results, we identified 22 genes to have a strong impact on survival/proliferation and 8 genes to selectively affect plasma cell differentiation (Fig. 3 F and G and Fig. S8B). The 22 genes important for B-cell survival/proliferation contained genes whose function in this context was previously unknown, such as Egr2, Dis3, Ost4, Preb, and Pomp, which we validated in further experiments (Fig. S9A). Dis3 is potentially involved in Ig class switch recombination via targeting AID (25), whereas Pomp might be involved in plasma cell differentiation (26). Furthermore, we identified Arf4, Creld2, and Zfp36 among the genes enhancing or blocking plasma cell differentiation (Fig. 3H and Fig. S9B). Of note, mice deficient for Zfp36 have been shown earlier to develop autoimmune disease, a finding that could connect to our observation of enhanced plasma cell differentiation in its absence (27). These results show that the screening system as described here leads to clear and consistent functional results, permitting small-scale screens in primary mouse cells without the need of high numbers of sgRNAs per gene or deep sequencing.
Fig. S7.
Gene set used for the small-scale screen. Total RNA was isolated from follicular B, GC, and plasma cells that were isolated from the spleen and BM of immunized animals. Microarrays were performed and data were normalized before analysis. The heatmap shows the expression levels of the selected genes with differential expression in the plasma cell populations.
Fig. S8.
Small-scale CRISPR-mediated screening to detect novel genes important for B-cell activation and plasma cell differentiation. (A) Scheme of the high-throughput 96-well-based screening approach. One sgRNA was designed for each gene of interest. Each sgRNA was separately cloned into a retroviral vector. The cloning, retrovirus production, and transduction of B cells were performed in 96-well plates. The transduced cells were selected with puromycin. Survival/proliferation was analyzed based on monitoring the percentage of GFP+ Cas9-expressing B cells at different time points. The fold changes were calculated by comparing to the percentage of GFP+ cells at day 2, allowing for the identification of repressor or enhancer genes. The plasmablast assay was based on the normalization of GFP+-derived CD138+ plasmablasts to GFP−-derived CD138+ plasmablasts. (B) Diagram showing the screen set and identification of important genes for survival and proliferation (Middle, Top) and plasma cell differentiation (Middle, Bottom). A total of 22 genes were crucial for activated B-cell survival and proliferation, and 8 genes were essential for plasma cell generation.Ten genes were essential for both survival/proliferation and differentiation.
Fig. S9.
Validation of candidates important for B-cell survival/proliferation and plasma cell differentiation. (A) FACS data showing the percentage of GFP+ Cas9-expressing and GFP− wild-type B cells 2, 4, 6, and 8 d after transduction with the indicated sgRNAs. As before, Cas9-expressing B cells were mixed with wild-type B cells, activated, and transduced with retroviral particles expressing sgRNA targeting Rosa26 (as control), Myc, Egr2, Dis3, Ost4, Preb, and Pomp. (B) Analysis of the induction of CD138 in GFP+ Cas9-expressing and GFP− wild-type B cells. Cas9-expressing B cells were mixed with wild-type B cells, activated, and transduced with retroviral particles expressing sgRNAs targeting Rosa26 (as control), Prdm1, Ost4, Arf4, Creld2, Zfp36, and Edem1.
Discussion
We provide a flexible sgRNA design tool that reliably identifies highly specific and efficient sgRNAs for a given set of genes or DNA sequences. CrispRGold can also be used for other organisms and will be available on a web platform (crisprgold.mdc-berlin.de). Primary cells of R26-Cas9iGFP/+ mice express high levels of GFP and Cas9 to nontoxic levels, allowing for efficient CRISPR-mediated gene editing and the use of GFP− internal controls. We show that these tools can be combined in an easy and robust screening assay of CRISPR mutagenesis in primary hematopoietic cells and identify genes important for B-cell activation and terminal differentiation. The same approach can be easily applied to other types of primary cells.
Methods
Mice.
Animal care and mouse work were conducted according to the guidelines of the Institutional Animal Care and Use Committee of the Max Delbrück Center for Molecular Medicine, the Landesamt für Gesundheit und Soziales, and the Bundesministerium für Wissenschaft und Forschung. Cas9 knockin mice were reported previously (9). Briefly, we knocked a Cas9-IRES-GFP cassette preceded by a LoxP-flanked Stop cassette into the mouse Rosa26 locus. The LSL-Cas9 mice were bred with Cre-deleter mice to generate Cas9 transgenic mice (available from The Jackson Laboratory, stock no. 028555). The C57BL/6 mice were obtained by local breeding. The heterozygous R26-Cas9p2aGFP/+ mice were purchased from The Jackson Laboratory (stock no. 026179). BALB/c mice were from Charles River Laboratories. For the microarray of in vivo generated plasma cells, BALB/c mice were immunized intraperitoneally with 100 µg 2-phenyl oxazolone coupled to chicken serum albumin (CSA), precipitated with aluminum hydroxide. After 6–8 wk, mice were boosted i.v. with 100 µg soluble antigen. Plasma cells were sorted from spleen and the BM at day 6 and day 60 after secondary immunization. Animal experiments were approved by the Institutional Animal Care and Use Committee.
CrispRGold.
Minimal CDS (minCDSs) are generated based on the refseq tables (mm9). sgRNA candidates are defined by the presence of NGG and cutting site inside the minCDS. sgRNA candidates are mapped to the mouse genome (mm9) using BWA (14) (version 0.7.12) to identify potential off-target sites, ignoring the first base of the targeting sequence and allowing for four mismatches. The folding energy between the targeting sequence and the scaffold RNA is calculated using RNAduplex (Vienna Package) (16). CrispRGold is written in Perl, as all scripts are that were implemented in CrispRGold if not otherwise stated. Briefly, sgRNA candidates are processed and scored as described. The first loop considers sgRNAs within the first 45% of the minCDS, with Tm ≤ 60 °C, the lowest off-target risk score >11, a scaffold-folding energy ≤20 kcal/mol, targeting the max N isoforms, without low-efficiency features and distance to the CDS-start ≥50 nt. The second loop considers sgRNAs as the first loop, but within the first 60% and with the lowest off-target risk score >6. The third loop considers sgRNAs as the second loop, but with Tm ≤ 65 °C and distance to CDS-start ≥10 nt. The fourth loop considers sgRNAs as the third loop, but with distance to the CDS-start ≥1 nt and neglecting Tm, scaffold-folding energy, and low-efficiency features. The last loop considers sgRNAs as the fourth loop, but extending the search space to 90% of the minCDSs.
Ninety-Six-Well Cloning Approach.
The MSCV_hU6_CcdB_PGK_Puro_T2A_BFP vector was generated by cloning the PCR-amplified hU6-BbsI-CcdB-BbsI-gRNA fragment into the SalI and XhoI sites of the murine stem cell virus (MSCV) vector. The PGK-puromycin-T2A-BFP fragment was amplified by overlapping PCR and cloned into the MluI site of the MSCV-hU6-BbsI-CcdB-BbsI-gRNA vector. For generating the minilibrary, forward and reverse oligos were separately ordered in 96-deep-well plates. Each forward and reverse oligo was mixed and phosphorylated individually. Then annealed oligo duplexes were cloned into the BbsI sites of the MSCV_U6_CcdB_PGK_Puro_T2A_BFP vector. The plasmids were transformed into DH5α bacteria using a heat-shock 96-well system. After a 30-min preculture at 37 °C, the transformed bacteria were transferred into 96-deep-well plates containing 1.5 mL LB liquid medium and sealed with PCR seals (Thermo Scientific). These plates were cultured for 12 h then split into two new 96-deep-well plates and further cultured for 10–12 h. Bacteria were collected by centrifugation at 4,000 rpm (Rotor A-4-81, Centrifuge 5810R, Eppendorf, in all following steps) for 1 min and plasmids were isolated using the NucleoSpin 96 plasmid core kit (Macherey-Nagel).
Cell Culture.
Retroviral Plat-E packaging cells were maintained in DMEM (Gibco) supplied with 10% (vol/vol) FCS (Gibco), 2 mM l-glutamine (Gibco), and 2 mM sodium pyruvate (Gibco). 40LB feeder cells, producing BAFF and CD40L, were previously generated by Nojima et al. (17) and maintained in completed DMEM. To prepare the feeder layer, 40LB feeder cells were irradiated with 12 Gy and plated at 5 × 104 cells per centimeter. Naïve B cells were isolated from the spleen of R26-Cas9iGFP/+, R26-Cas9p2aGFP/+, or C57BL/6 mice by depletion of CD43+ cells using CD43 microbeads (Miltenyi Biotec). Resting B cells were plated at 106 cells per milliliter in DMEM (Gibco) supplied with 10% FCS (Gibco), 2 mM l-glutamine, 2 mM sodium pyruvate, 2 mM Hepes (Gibco), 1× NAA (Gibco), β-mercaptoethanol (Sigma), and 10 μg/mL gentamicin (Lonza). Cas9 (GFP+)-expressing B cells were mixed wild-type (GFP−) B cells and stimulated with anti-CD40 antibody (Biolegend) and IL-4 (Perprotech) for 48 h before retroviral transduction.
High-Throughput Retroviral Production.
Plat-E cells were plated at 2.2 × 104 cells per 100 μL medium in each well of 96-well plates at day 0. One day later, the cells were transfected with 100 ng DNA using FugeneHD transfection reagents (Promega). Twelve hours after transfection, the supernatant was discarded and the cells were cultured with 150 μL new complete medium. The transfected cells were cultured in a 32 °C incubator for 24 h. The retroviral supernatant was collected 48 h and 72 h after transfection into 96-deep-well plates. The 96-deep-well plates with supernatant were centrifuged at 1,300 rpm for 5 min and the supernatant was transferred into new 96-deep-well plates and stored at −80 °C.
High-Throughput Retroviral Transduction.
At 48 h after stimulation, GFP+/GFP− mixed B cells were harvested and resuspended at a density of 2 × 105 cells per milliliter in completed B cell medium supplemented with 16 μg/mL Polybrene (Sigma). A total of 100 μL cell suspension was transferred into each well of the 96-well plates and 100 μL of retroviral supernatant was added, then activated B cells were spin transduced at 2,500 rpm for 90 min. The 96-well plates were placed in a 37 °C incubator and on the next day the transduced GFP+/GFP− B cells were transferred into 96-well 40LB feeder cell plates in the presence of 20 ng/mL of IL-21 (Perprotech) and selected with 1.25 μg/mL puromycin (Sigma). The transduced GFP+/GFP− B cells were harvested and analyzed at different time points by using BD Fortessa with a 96-well high-throughput sample unit.
T-Cell Activation and Transduction.
Total T cells were isolated from R26-Cas9iGFP/+ and C57BL/6 mice using Pan T Cell Isolation Kit II; mouse (Miltenyi Biotec) and subsequently CD4 and CD8 T cells were sorted by FACS; and the Cas9 (GFP+)-expressing T cells were mixed with wild-type (GFP−) T cells and activated with plate-bound anti-CD3 (145-2C11, Biolegend) and anti-CD28 (37.51, Biolegend) in the presence of 25 ng/mL recombinant IL-2 (Peprotech) for 2 d. The activated T cells were spin transduced with retroviral particles expressing sgRNAs. The transduced T cells were selected with puromycin for 4 d. After puromycin selection, the transduced T cells were analyzed by flow cytometry or sorted.
BMDM Generation and Lentiviral Transduction.
BMDMs were differentiated from total mouse BM cells for 14 d in macrophage growth medium (DMEM) with 10% FCS, 20% l-cell supernatant, 1% sodium pyruvate, 1% l-glutamine). sgRNAs targeting CD64 and CD14 surface makers were cloned into BbsI sites of pLKV-U6gRNA(BbsI)-PGKPuroP2ABFP vector (Addgene, 50946). Lentiviral particles were produced at the Centre International de Recherche en Infectiologie (CIRI) U1111/UMR5308 Institut National de la Santé et de la Recherche Médicale-Centre National de la Recherche Scientifique-Université Claude-Bernard Lyon 1–École Normale Supérieure de Lyon (Lyon, France). BMDMs were seeded at a density of 2 × 105 cells per well of 24-well plates and transduced with lentiviral supernatant for 4 h. The medium was then replaced, and 2 d later, transduced cells were selected with 2 μg/mL puromycin for 4 d. The transduced BMDMs were analyzed by FACS.
FACS Analysis and Sorting.
For FACS analysis, single cell suspensions were prepared from the BM, spleen, peritoneal cavity, and mesenteric lymph nodes (mLNs) from R26-Cas9iGFP/+, R26-Cas9p2aGFP/+, or C57BL/6 mice and cells were blocked with FcγR antibody (Biolegend) for 10 min. The surface antigens were stained with fluorescent-conjugated antibodies for 15 min. The cells were washed with FACS buffer (PBS/1% BSA) and analyzed by BD Fortessa. The data were analyzed using FlowJo software.
For cell sorting, the BFP+ (sgRNA) GFP+ (Cas9) cells were sorted into 15-mL Falcon tubes and centrifuged before DNA isolation for further analaysis. For single cell sequencing analysis, single BFP+GFP+ cells were sorted into 96-well plates containing 5 μL of DNA quick extraction buffer (Epicenter). The plates were briefly centrifuged, DNA was denatured, and PCR was performed.
Immunofluorescent Staining and Western Blotting.
The 1 × 105 BMDMs were transferred into lysine-coated coverslips and further cultured for 24 h. The cells were washed with cold PBS and fixed with 4% paraformaldehyde for 20 min at room temperature (RT). The cells were washed and permeabilized with PBS/0.25% Triton X-100 for 10 min at RT. The cells were washed and stained overnight with anti-Flag tag M2 antibody at 4 °C. Anti-Flag tag antibody was developed with rat anti-mouse IgG1 PE (Biolegend) for 1 h at RT. The nuclei were counterstained with DAPI (Sigma). The images were analyzed by Keyence fluorescence microscopy. To detect Cas9 protein, activated B-cell and BMDM lysates were run on SDS/PAGE gel and transferred into PVDF membrane (GE Healthcare). The Cas9 protein was developed with anti-Flag tag and anti-Cas9 antibodies; β-actin was used for loading controls.
Antibodies
The following anti-mouse antibodies were purchased from Biolegend: anti-B220 (RA3-6B2), anti-CD19 (6D5), anti-CD138 (281-2), anti-CD22 (OX-97), anti-CD24 (M1/69), anti-CD44 (IM7), anti-CD80 (16-10A1), anti-CD81 (Eat-2), anti-CD83 (Michel-19), anti-CD86 (GL-1), anti-CD45 (30-F11), anti-CD11b (M1/70), anti-Gr-1 (RB6-8C5), anti-CD3ε (145-2C11), anti-Ter-119 (Ter-119), anti-Ckit (2B8), anti-Sca1 (D7), and anti-CD64 (clone X54-5/7.1). Anti-CD95 (15A7), anti-CD14 (clone Sa2-8), and anti-Tnfrsf13b (ebio8F10-3) antibodies were ordered from eBioscience. Anti-Cas9 (7A9-3A3) antibody was purchased from Cell Signaling Technology, and anti-Flag (M2) antibody and anti–β-actin were purchased from Sigma. HRP-conjugated secondary anti-mouse Ig antibody was ordered from Rockland Immunochemicals.
Acknowledgments
We thank H. P. Rahn for excellent FACS-related support. This work was supported by the European Research Council (ERC Advanced Grant 268921, to K.R.), the German Ministry of Education and Research within the Validation of the Innovation Potentials of Academic Research (VIP) program (03V0261, to R.K.), and a Berlin Institute of Health Einstein fellowship (to M.H.S.). M.H.S. is Institut National de la Santé et de la Recherche Médicale–Helmholtz group leader and received grants from the Agence Nationale de la Recherche (ANR-11-BSV3-026-01) and Institut National du Cancer (13-10/405/AB-LC-HS).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613884113/-/DCSupplemental.
References
- 1.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32(12):1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osakabe Y, et al. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep. 2016;6:26685. doi: 10.1038/srep26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aubrey BJ, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Reports. 2015;10(8):1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Parnas O, et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162(3):675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu VT, et al. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 2016;16(1):4. doi: 10.1186/s12896-016-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MdelC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 11.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham DB, Root DE. Resources for the design of CRISPR gene editing experiments. Genome Biol. 2015;16:260. doi: 10.1186/s13059-015-0823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenz R, et al. ViennaRNA package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nojima T, et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2:465. doi: 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- 18.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383(6600):538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 19.Lin KI, Lin Y, Calame K. Repression of c-myc is necessary but not sufficient for terminal differentiation of B lymphocytes in vitro. Mol Cell Biol. 2000;20(23):8684–8695. doi: 10.1128/mcb.20.23.8684-8695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 21.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7(7):773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 22.Corcoran LM, et al. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J Exp Med. 2005;201(9):1385–1396. doi: 10.1084/jem.20042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapiro-Shelef M, et al. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276(5312):596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 25.Basu U, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144(3):353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, et al. MicroRNA-101 suppresses tumor cell proliferation by acting as an endogenous proteasome inhibitor via targeting the proteasome assembly factor POMP. Mol Cell. 2015;59(2):243–257. doi: 10.1016/j.molcel.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 27.Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]