Significance
Human major depressive disorder is a chronic remitting syndrome that affects millions of individuals worldwide; however, the molecular mechanisms mediating this syndrome remain elusive. Here, using a unique combination of epigenome-wide and behavioral analyses, we demonstrate a role for histone variant dynamics in the nucleus accumbens (NAc)—a critical brain center of reward and mood—contributing to stress susceptibility in mice. These studies, which also demonstrate that molecular blockade of aberrant dynamics in the NAc promotes resilience to chronic stress, promise to aid in the identification of novel molecular targets (i.e., downstream genes displaying altered expression as the result of stress-induced histone dynamics) that may be exploited in the development of more effective pharmacotherapeutics.
Keywords: H3.3, nucleus accumbens, depression, chronic social defeat stress, histone dynamics
Abstract
Human major depressive disorder (MDD), along with related mood disorders, is among the world’s greatest public health concerns; however, its pathophysiology remains poorly understood. Persistent changes in gene expression are known to promote physiological aberrations implicated in MDD. More recently, histone mechanisms affecting cell type- and regional-specific chromatin structures have also been shown to contribute to transcriptional programs related to depressive behaviors, as well as responses to antidepressants. Although much emphasis has been placed in recent years on roles for histone posttranslational modifications and chromatin-remodeling events in the etiology of MDD, it has become increasingly clear that replication-independent histone variants (e.g., H3.3), which differ in primary amino acid sequence from their canonical counterparts, similarly play critical roles in the regulation of activity-dependent neuronal transcription, synaptic connectivity, and behavioral plasticity. Here, we demonstrate a role for increased H3.3 dynamics in the nucleus accumbens (NAc)—a key limbic brain reward region—in the regulation of aberrant social stress-mediated gene expression and the precipitation of depressive-like behaviors in mice. We find that molecular blockade of these dynamics promotes resilience to chronic social stress and results in a partial renormalization of stress-associated transcriptional patterns in the NAc. In sum, our findings establish H3.3 dynamics as a critical, and previously undocumented, regulator of mood and suggest that future therapies aimed at modulating striatal histone dynamics may potentiate beneficial behavioral adaptations to negative emotional stimuli.
Major depressive disorder (MDD) affects ∼17% of the population, making it the most prominent and debilitating psychiatric disease worldwide (1). Current antidepressants (ADs) can take weeks to months to produce an effective therapeutic response, with about one-third of patients remaining nonresponsive to existing treatment strategies. Although the prevalence of MDD and a lack of sufficient treatment options highlight the importance of identifying new drug targets, progress has been hindered by a general lack of understanding of the precise molecular mechanisms underlying this disorder.
The nucleus accumbens (NAc), a critical component of the brain’s limbic reward circuitry, has become increasingly implicated in depression, as well as in mechanisms of antidepressant action (2, 3). In recent years, numerous studies in both human MDD and in animal models of depression have identified changes in gene expression, along with related alterations in chromatin structure and function, that contribute to aberrant forms of transcriptional and behavioral plasticity associated with depressive-like phenotypes (4–9). Although multiple studies have successfully linked alterations in histone posttranslational modifications (PTMs) or chromatin-remodeling events to chronic stress susceptibility or resilience (e.g., using a model of chronic social defeat stress—CSDS—an etiologically valid rodent model of human depression), the precise histone regulatory phenomena involved in stress-mediated behavioral responses remain poorly understood.
Recently, we demonstrated a mechanistic role for histone variant turnover/dynamics in the central nervous system, a process that is dissociated from histone PTMs in adult brain, in the regulation of activity-dependent gene transcription, synaptic connectivity, and behavioral plasticity (10). Moreover, the histone variant H3.3, which can be actively incorporated into neural chromatin, was found to accumulate to near saturating levels in neurons by midadolescence, but remains constitutively dynamic (i.e., it is continuously turned over and replaced by newly transcribed/translated H3.3) throughout life. Stalling histone dynamics using viral-mediated reductions in H3.3 transcription was found to attenuate activity-dependent gene expression in neurons and reduce behavioral flexibility. Although the elucidation of histone turnover as a critical mediator of neuronal function has begun to shed light on the precise chromatin-templated processes contributing to normal neurodevelopment and cognition, this phenomenon has yet to be explored in the context of adult psychiatric illness.
Here, using a combination of genome-wide and behavioral approaches, we demonstrate a role for heightened H3.3 dynamics in NAc in the mediation of susceptibility to depression-related phenotypes. Specifically, we show that the stimulus-dependent H3.3 gene, H3f3b, is significantly increased in expression—a proxy for enhanced dynamics—in NAc of both humans with MDD and in animal models of depression (e.g., CSDS) and stress susceptibility (e.g., maternal separation). These increased dynamics are reversed by chronic AD treatments in both humans and rodents and can be attenuated by early life environmental enrichment (EE) before chronic stress exposure in rodent models. In agreement with the hypothesis that increased dynamics contribute to susceptibility to depressive-like behaviors, we further demonstrate that viral-mediated stalling of H3.3 turnover is sufficient to produce resilience to CSDS. Finally, using chromatin immunoprecipitation coupled to massively parallel next-generation sequencing (ChIP-seq), we identify a series of genomic loci displaying altered H3.3 dynamics in response to stress, which contribute to deficits in transcriptional and behavioral plasticity observed in depression models. In sum, our data demonstrate a role for aberrant H3.3 dynamics in MDD and stress susceptibility, and we identify potential targets for future pharmacotherapeutic interventions.
Results
H3.3 Dynamics Are Increased in NAc of Depressed Humans and in Animal Models of Depression.
First, to investigate if histone dynamics in NAc may be involved in depression, we began by quantifying the expression of genes encoding H3.3 (H3F3A and H3F3B) in human postmortem NAc of subjects diagnosed with MDD—on or off ADs at their time of death—vs. appropriately matched nondepressed controls (see Tables S1 and S2 for demographic information). H3F3A exists as the more abundant of the two transcripts; however, H3F3B is activity-dependent, and increases in its transcript levels have been tightly linked to potentiated rates of histone turnover in brain (10). Following quantitative PCR (qPCR), we observed a significant increase in H3F3B, but not in H3F3A, mRNA levels in nonmedicated depressed subjects compared with controls, an effect that is reversed in individuals with MDD on ADs (Fig. 1A). H3F3A levels were also found to decrease in response to ADs in MDD subjects, perhaps indicating a role for global reductions in histone turnover in NAc in the alleviation of depressive symptoms.
Table S1.
Human demographic data in controls
| Controls | Sex | Age (y) | Race | RIN | PMI (h) |
| 1 | M | 63 | B | 6.1 | 15 |
| 2 | M | 34 | C | 9.7 | 21.47 |
| 3 | M | 48 | C | 8.6 | 17.2 |
| 4 | F | 50 | C | 9 | 10.45 |
| 5 | M | 51 | C | 9.8 | 17.1 |
| 6 | M | 57 | C | 5.5 | 7 |
| 7 | M | 77 | C | 8.9 | 13.4 |
| 8 | M | 47 | C | 8.6 | 22.3 |
| 9 | M | 72 | C | 6.2 | 23 |
| 10 | M | ? | C | 8.5 | 23 |
| 11 | M | 55 | C | 9.1 | 11 |
PMI, postmortem interval; RIN, RNA integrity number.
Table S2.
Human demographic data in MDD
| MDD | Sex | Age (y) | Race | RIN | PMI (h) |
| −ADs | |||||
| 1 | F | 59 | C | 7.8 | 19.8 |
| 2 | M | 65 | C | 5.6 | 20.3 |
| 3 | M | 40 | C | 7.3 | 21 |
| 4 | M | 61 | C | 9.1 | 21.45 |
| +ADs | |||||
| 5 | M | 61 | C | 7.5 | 19 |
| 6 | F | 41 | B | ? | 13 |
| 7 | M | 25 | C | 8.6 | 21 |
| 8 | M | 42 | C | 9.1 | 17 |
PMI, postmortem interval; RIN, RNA integrity number.
Fig. 1.
H3.3 dynamics in NAc are increased in human MDD and in response to chronic stress in rodents. (A) H3F3B, but not H3F3A, expression is increased in NAc of nonmedicated subjects with MDD vs. controls, an effect that is reversed with ADs (*P < 0.05, ***P < 0.0001 by one-way ANOVA followed by Dunnett’s post hoc test; n = 4–11 per group). (B) Mice were segregated into susceptible vs. resilient populations following CSDS using SI testing for subsequent molecular analyses (***P < 0.0001 by one-way ANOVA followed by Dunnett’s post hoc test; n = 8 per group). (C) H3f3b, but not H3f3a, is significantly increased in expression in NAc of susceptible, but not resilient, mice following CSDS. This effect is reversed by chronic imipramine treatments in behaviorally responsive susceptible animals (*P < 0.05 by one-way ANOVA followed by Dunnett’s post hoc test; n = 3–13 per group; in some cases, tissues from multiple animals were pooled/biological replicate). (D) ELS (maternal separation) in mice results in faster rates of H3.3 chromatin accumulation in NAc with age in comparison with SR animals. LC-MS/MS analysis [*P < 0.05, **P < 0.01 by unpaired Student’s t test; n = 3 (pooled/biological replicate) per group]. (E) EE during adolescence promotes reduced expression of H3f3b, but not H3f3a, in NAc in comparison with normally housed mice (*P < 0.05 by unpaired Student’s t test; n = 7 per group) and (F) renders animals resilient to subsequent bouts of CSDS (**P < 0.01 by unpaired Student’s t test; n = 5 per group). Data are represented as means ± SEM.
Next, to understand the role of H3.3 dynamics in the development of depressive-like behaviors, we used a well-established rodent model of depression, CSDS (11, 12), to behaviorally segregate animals into stress-susceptible vs. resilient populations for subsequent molecular analyses (Fig. 1B). Similar to our studies of human postmortem tissue, H3.3 gene expression in NAc was analyzed following CSDS, where again H3f3b, but not H3f3a, levels were significantly elevated in susceptible mice in comparison with resilient and nonstressed controls (Fig. 1C). Furthermore, chronic treatments with the classic tricyclic AD imipramine in behaviorally susceptible animals—treatments that fully attenuate behavioral deficits associated with CSDS in a subset of animals (so-called “responders”) (13)—resulted in a complete reversal of increased H3f3b expression observed in susceptible mice, with no effects seen on H3f3a expression (Fig. 1C).
Although chronic stress in adult animals is well established to elicit depressive-like behaviors, early life stress (ELS) paradigms (e.g., maternal separation during postnatal life), which mimic aspects of early life adversity in humans, are known to disrupt normal patterns of brain development and can promote greater susceptibility to later life stressors (14, 15). Therefore, we next explored whether aberrant histone dynamics may similarly be affected by stressful events experienced early in life. To do so, mice were subjected to early life maternal separation vs. standard rearing (SR), followed by longitudinal proteomic analyses of H3.3 (vs. canonical H3.1 and H3.2) dynamics at various developmental stages post-ELS. Consistent with previous work from our laboratory, we found that H3.3 levels constitute only a small portion of the total H3 pool in embryonic day 16.5 striatal chromatin, and we confirmed that H3.3 accumulates in NAc with age, similar to that observed in other brain regions. In line with our CSDS data in Fig. 1C, ELS was similarly found to promote a more rapid accumulation—a measure of turnover kinetics—of H3.3 levels in neuronal chromatin (with concomitant loss of H3.2) in comparison with SR controls (Fig. 1D). These data indicate that ELS has a stimulatory effect on histone turnover, which may contribute to later life stress susceptibilities.
Next, to examine if positive life experiences may result in opposing patterns of H3.3 dynamics in NAc, we used an EE paradigm that has previously been shown to promote enhanced synaptic plasticity and cognition and can prevent or reverse some aspects of depressive-like behaviors, such as anhedonia and anxiety (16–18). We observed that EE results in a significant down-regulation of H3f3b, but not of H3f3a, expression in mouse NAc (Fig. 1E), a phenomenon associated with the potentiation of resilience to subsequent stress-induced behavioral deficits (Fig. 1F).
Reducing H3.3 Dynamics in NAc Promotes Resilience to CSDS.
To investigate causal roles for H3.3 dynamics in mediating depressive-like behaviors, we next used a recently described H3f3a/b viral knockdown strategy in NAc to stall histone turnover in neuronal chromatin by way of depletion of soluble nuclear “reserve” pools of H3.3 (10). Briefly, adeno-associated virus (AAV) vectors expressing synthetic microRNAs (miRs) targeting either both H3.3 gene copies [AAV-miR (H3f3a/b)] or a sequence not present in the vertebrate genome as a control [AAV-miR (–)] were injected bilaterally into NAc of wild-type mice. Following a 28-d incubation to allow for maximal viral expression and knockdown of both genes (Fig. 2 A and B, respectively), animals were subjected to CSDS. As predicted, animals infected with AAV-miR (–) exhibited significantly lower social interaction (SI) with a novel mouse following CSDS vs. that observed in nonstressed controls. Animals injected with AAV-miR (H3f3a/b), however, spent significantly more time with the novel mouse in comparison with their stressed controls (Fig. 2C). To further assess anxiety-like behavior, an additional phenotype elicited by CSDS in rodents, mice were examined in an open field test post-CSDS to assess their time spent in the center of the arena. As expected, AAV-miR (–) stressed mice spent significantly less time in the center of the novel arena in comparison with nonstressed controls, indicative of an anxiogenic response. Knockdown of H3f3a/b, however, was found to partially rescue this anxiety-like behavior in stressed animals (Fig. 2D). These data indicate that increased histone dynamics in NAc may function as a global mediator of both social aversion and anxiety. No differences in general locomotor behavior were observed between groups (Fig. 2E).
Fig. 2.
Blockade of H3.3 dynamics in NAc promotes resilience to CSDS. (A) Viral targeting of AAV-miR (H3f3a/b)-IRES-GFP in NAc. (Magnification: 20×.) (B) Confirmation of H3f3a and H3f3b mRNA expression in NAc following viral knockdown (**P < 0.01, ***P < 0.001 by unpaired Student’s t test; n = 3 per group). (C) Reducing H3.3 dynamics in NAc promotes increased SI following CSDS (**P < 0.01 by two-way ANOVA followed by Bonferroni post hoc test; n = 7–10 per group; (#P < 0.05 by planned Student’s t test postdetermination of main-effect significance). (D) Reducing H3.3 dynamics in NAc is anxiolytic post-CSDS (*P < 0.05 by two-way ANOVA followed by Bonferroni post hoc test; n = 7–9 per group). (E) Stalling H3.3 dynamics in NAc does not affect locomotor behavior. Data are represented as means ± SEM.
Chronic Stress-Induced H3.3 Dynamics in NAc Contribute to Aberrant Transcriptional Plasticity.
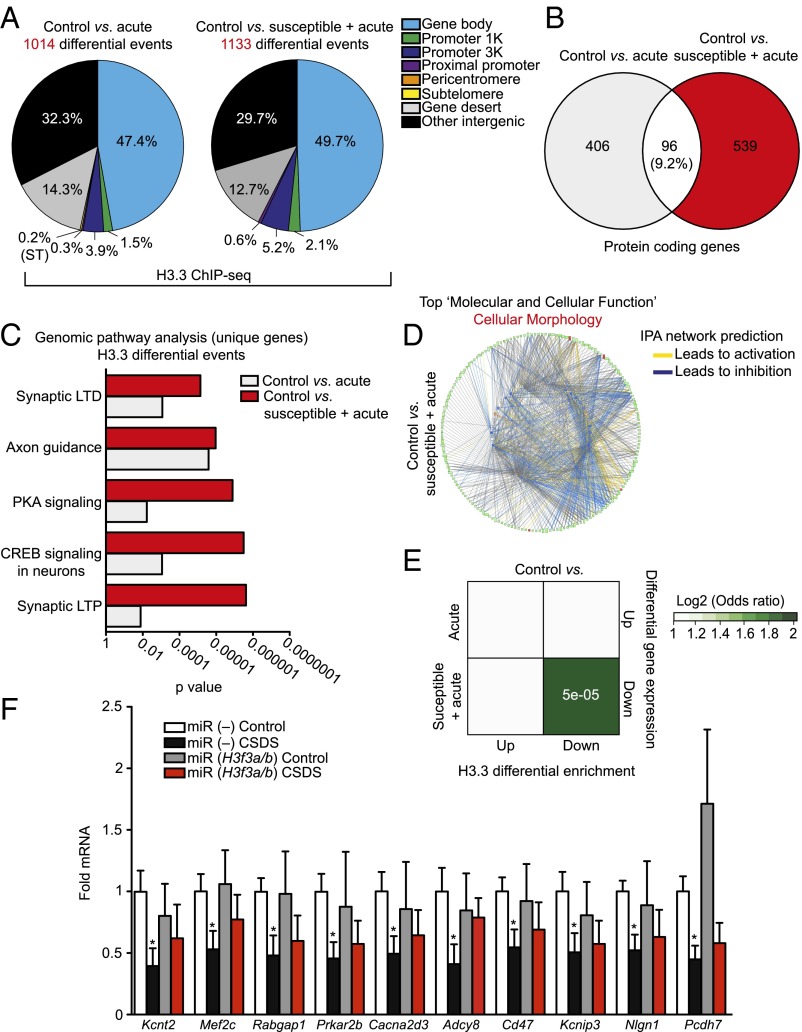
In an attempt to better understand the contribution of H3.3 dynamics to transcriptional dysregulation in response to stress, we next sought to profile H3.3 enrichment genome-wide using ChIP-seq. Because H3.3 dynamics can be most easily interpreted in the context of a stimulus (i.e., as an activity-dependent response), H3.3 enrichment profiles were analyzed in NAc from three groups of animals––controls (never exposed to a social stress), acutely stressed animals (exposed to a single stressful experience––one that does not elicit depressive-like behaviors in mice), and susceptible + acute animals (animals deemed susceptible post-CSDS, followed by 28 d of recovery and re-exposure to a single stressful event)––the overarching goal of which was to identify how alterations in H3.3 dynamics segregate between stress naive animals vs. susceptible mice experiencing a subsequent stressful stimulus. Following ChIP-seq and differential analyses of H3.3 dynamics (note that previous data from our laboratory indicate that alterations in H3.3 enrichment act as a proxy for nucleosomal turnover), we assessed both the number and the genomic distribution of differential events between acute or susceptible + acute animals in comparison with their controls. Interestingly, we observe that both a single stressful event and re-exposure to social stress in susceptible mice results in approximately equal numbers of differential H3.3 enrichment events (1,014 vs. 1,133, respectively) with perhaps a modest increase in the number of dynamics observed in susceptible + acute mice (Datasets S1 and S2). Furthermore, the genomic distribution of these events was found to be similar between the two stress groups, with the majority of differential sites occurring in gene bodies (Fig. 3A), a observation that is consistent with our current understanding of H3.3 turnover in neurons (10).
Fig. 3.
Chronic stress-mediated H3.3 dynamics promote transcriptional dysregulation of synaptic-related genes in NAc. (A) Genomic distribution (and numbers) of H3.3 differential enrichment events in NAc in response to either an acute stress (control vs. acute) or following re-exposure to stress in susceptible mice (control vs. susceptible + acute). (B) Venn diagrams showing minimal overlap between protein-coding genes displaying differential H3.3 enrichment in control vs. acute and control vs. susceptible + acute stress groups. (C) IPA-based pathway analysis of unique protein-coding genes displaying differential H3.3 enrichment in control vs. acute and control vs. susceptible + acute stress groups. (D) IPA-based network prediction diagram showing assumed relationships between unique genes displaying differential H3.3 enrichment in control vs. susceptible + acute NAc. Focus on most heavily enriched molecular and cellular function category of cellular morphology indicates a role for these genes in the inhibition of morphological/dendritic plasticity. (E) Odds ratio analyses of differential enrichment events for H3.3 (both control vs. acute and control vs. susceptible + acute stress groups) vs. stress-mediated gene expression in NAc indicates a significant relationship between H3.3 dynamics in susceptible mice only and reduced gene expression, effects that are predicted to disinhibit cellular morphology-associated gene pathways. (F) qPCR validation of candidate genes (predicted a priori from sequencing data) displaying H3.3 dynamics and reduced gene expression in NAc following CSDS. Knockdown of H3f3a and H3f3b results in a partial renormalization of transcriptional deficits post-CSDS (*P < 0.05 by unpaired Student’s t test vs. miR (–) control; n = 5 per group). Data are represented as means ± SEM. See Materials and Methods, H3.3 ChIP-seq Analysis and Correlations with Gene Expression for statistical comparisons, FC cutoffs, and numbers.
Next, to examine if H3.3 dynamics between these two stress groups occur at similar vs. unique genomic loci, we focused our attention on protein-coding genes. Despite similar numbers of differential events observed, we identified very little overlap (<10%) between genes displaying H3.3 dynamics in response to a single stressor vs. those observed after re-exposure to a subsequent stress (Fig. 3B). Such limited overlap prompted us to then examine specific properties of these genes that may function to promote stress susceptibility. To do so, we performed Ingenuity Pathway Analysis (IPA) on unique genes displaying H3.3 dynamics in acute vs. susceptible + acute groups and found that susceptible animals display considerably higher levels of statistical enrichment within genic pathways associated with synaptic plasticity, including those associated with synaptic long-term potentiation, synaptic long-term depression, CREB signaling in neurons, etc. (Fig. 3C). Furthermore, IPA-based “molecular and cellular function” enrichment analyses indicate strong statistical enrichment for susceptible + acute, but not acute, associated genes (primarily displaying decreased enrichment for H3.3) as putative regulators of “cellular morphology” (P value: 3.73E-03–4.10E-16; no. of molecules = 186 of 539), a category that includes functional annotations such as “density of dendritic spines” (1.74E-05) and “morphology of dendritic spines” (P value: 1.28E-03) (Fig. 3D). These data are intriguing in light of published studies demonstrating strong associations between alterations in NAc dendritic spine density and type and behavioral susceptibility to CSDS (19, 20). Within the context of cellular morphology-enriched genes in susceptible + acute animals, IPA was further used to predict network relationships between affected genes and molecular outcomes constrained by functional annotations within the functional enrichment category. IPA analyses indicate strong associations between differentially enriched genes and functional inhibition of cellular morphology. Given such predictions, we sought to examine the precise relationship between H3.3 differential enrichment and alterations in gene expression that may influence the activation or inhibition of these functional biological processes. To do so, odds ratio analyses were performed comparing H3.3 ChIP-seq analyses with corresponding published mRNA expression data (21). We found that, although acute stress leads to roughly as many changes in H3.3 dynamics as re-exposure in susceptible animals, only those dynamics observed in susceptible mice exhibit strong correlations with differential gene expression patterns observed in these animals (Fig. 3E). Furthermore, our data indicate that such dynamics in susceptible + acute NAc associate with reduced, but not activated, gene expression following stress. These findings, in concert with those presented in Fig. 3D, indicate that repression of differentially enriched genes in susceptible mice may promote disinhibition of cellular morphology-enriched pathways leading to enhanced synaptic plasticity and behavioral susceptibility to stress.
Finally, to validate assumptions regarding potential roles for H3.3 dynamics in stress-mediated gene expression, we performed qPCR on candidate genes enriched in IPA functional categories falling under the category of cellular morphology ± CSDS and transduction with either AAV-H3f3a/b or AAV-miR (–) in NAc. As predicted from our genome-wide analyses, we validated numerous genes associated with inhibition of cellular morphology to be down-regulated in response to CSDS, effects that we could partially rescue following H3f3a/b knockdown (Fig. 3F). In total, these data indicate that reducing histone dynamics in NAc is sufficient to inhibit aberrant transcriptional plasticity that likely contributes to stress susceptibility in both rodents and humans.
Discussion
Here, we demonstrate that histone dynamics in NAc are bidirectionally mediated by negative vs. positive environmental stimuli to mediate stress susceptibility or resilience, respectively. In addition, we found that molecular blockade of aberrant H3.3 dynamics in NAc can efficiently promote resilience to subsequent bouts of CSDS, with AD treatments poststress similarly reversing aberrant increases in H3f3b expression in behaviorally responsive animals. Furthermore, we have identified, at least in part, the molecular consequences of stress-mediated histone dynamics in NAc, in which stress-mediated chromatin alterations resulting from H3.3 turnover act to potentiate aberrant forms of transcriptional, and possibly synaptic, plasticity. Collectively, our results provide a molecular mechanism contributing to the pathophysiology of depressive-like behaviors and indicate the need for future studies aimed at delineating both the upstream mechanisms and the downstream consequences of this chromatin regulatory phenomenon in brain.
In previous studies of H3.3 turnover in brain (10), we identified robust correlations between H3.3 dynamics and activity-dependent gene expression in both cortex and hippocampus, in which increased turnover (irrespective of enrichment directionality) was often correlated with increased gene expression, specifically within the context of late-response synaptic genes. In NAc, on the other hand, we find, in response to chronic social stress, that increased turnover more strongly associates with reduced gene expression leading to disinhibition of functional networks (e.g., cellular morphology) regulating depression-associated morphological and synaptic plasticity. Such opposite effects are intriguing in that they suggest that altering chromatin accessibility—via H3.3 dynamics—is not, in and of itself, sufficient to dictate the directionality of cognate gene expression. Rather, it seems that a concerted effort between nucleosomal turnover and recruitment of specific transcription factors (TFs), both activating and repressive, is needed to guide transcriptional outcomes in response to stress. Future studies aimed at uncovering the precise relationship between histone dynamics, chromatin accessibility, and TF binding in brain are needed to more fully understand the complex interplay between these molecular processes and MDD.
Furthermore, although our data indicate an integral role for H3.3 dynamics in NAc in the regulation of mood-related behaviors, it remains unclear whether these dynamics occur globally in NAc or are more specifically affected within the various cell types expressed in this region (e.g., D1- vs. D2-type medium spiny neurons, glia, etc.). Although well beyond the scope of the current study, it will be necessary in the future to investigate the contributions of these numerous cell types to transcriptional, synaptic, and behavioral abnormalities observed following CSDS. Indeed, previous work has already provided a wealth of data demonstrating that distinct cell types and circuits in striatum are segregated, both molecularly and electrophysiologically, in their responses to chronic stress, thereby contributing differentially to distinct phenotypes observed across numerous psychiatric disease models (22, 23).
Another intriguing finding of the current study is the observation that not only can histone dynamics be reversed by AD treatments in both humans with MDD and in susceptible mice exposed to CSDS—a molecular phenomenon strongly associated with the reversal of depressive-like behaviors—but also by positive early life experiences before stress, such as those observed after a period of juvenile enrichment resulting in protective-like molecular and behavioral effects. Conversely, extended periods of early life trauma, such as those modeled by maternal separation in rodents, lead to increased stress susceptibility later in life, likely through stimulation of this common molecular pathway. These findings are significant in that they suggest that molecular responses to stress (e.g., increased H3.3 turnover) can be influenced, and perhaps even “trained,” by positive early life experiences to be less responsive to later life stress. Although the depression literature tends to focus most heavily on identification of molecular targets of susceptibility for the development of reversal therapies (i.e., ADs), it is becoming increasingly clear that a greater focus on the molecular mechanisms of resilience may prove beneficial in the prevention of depression-like symptomatic emergence.
In conclusion, this study demonstrates a critical and previously undocumented role for H3.3 dynamics in the NAc in stress susceptibility and depression. These events likely function to: (i) alter cell-type–specific chromatin accessibility profiles, (ii) differentially alter the transcriptome within distinct neural subpopulations of the NAc, and (iii) promote circuit-wide changes that ultimately converge to promote behavioral sensitivity to chronic stress. Understanding how these rather granular mechanisms, in concert with previously identified chromatin processes associated with mood, contribute to MDD will be essential to the future development of more targeted and effective ADs.
Materials and Methods
Human Tissue Collection.
Postmortem human NAc tissue was obtained from the Dallas Brain Collection (DBC), in compliance with The University of Texas Southwestern’s Institutional Review Board, as previously described (4). All tissues for the DBC were collected from the Dallas Medical Examiner’s Office and The University of Texas Southwestern’s Tissue Transplant Program provided that consent from the next-of-kin was granted. Tissue was analyzed from both males and females matched for age, RNA integrity number, postmortem interval and pH; case demographics are given in Tables S1 and S2. Samples were blindly dispensed for analysis.
Animals.
Male 6- to 8-wk-old C57BL/6J mice and 6-mo-old CD1 retired breeders were maintained on a 12-h light–dark cycle (lights on at 7:00 AM) at 22–25 °C with ad libitum access to food and water. C57BL/6J mice were housed five per cage except following defeat experiments, at which point mice were singly housed. All experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai. All behavioral testing occurred during the animals’ light cycle. Experimenters were blinded to experimental group, and the order of testing was counterbalanced during behavioral experiments.
CSDS and Social Interaction Testing.
All experiments used an established 10-d (5 min per session) CSDS protocol to induce depressive-like behaviors (or resilience) in mice followed by social interaction testing (11, 12).
For gene expression studies involving chronic imipramine treatments in behaviorally susceptible animals, NAc RNA was obtained from behavioral “responders,” as determined in a recently published study (13). For ChIP-seq experiments, control and susceptible mice (determined in SI test 48 h postdefeat) were killed 28 d following the final defeat episode. Mice were either taken directly from their home cage (baseline) or following a stress re-exposure (5 min of acute aggression, followed by 55 min housed adjacent to aggressor, i.e., stress-primed).
Open Field.
Exploration of an open field arena (44 × 44 cm) was assessed during a 10-min test in red light. A video-tracking system (Ethovision 3.0, Noldus) measured locomotor activity, as well as the time spent in the center (34 × 34 cm) and periphery of the test arena as an index of anxiety.
Early Life Stress.
Two C57/BL6 females were mated with one male in our animal facilities. The male was removed after 1 wk, and the females were separated into individual cages 1–3 d before giving birth. Litters were weighed and counted, and cages were cleaned on the day of birth [postnatal day (P) 0] but were otherwise undisturbed.
For the ELS paradigm, from P10 to P17, an experimentally determined sensitive period for early life stress-enhanced vulnerability to depressive-like behaviors (15, 24), nesting material (crinkled Enviro-Dri paper) was reduced to one-fourth the amount, and whole litters were removed to clean cages for 4 h/day (i.e., maternal separation). Nesting material was restored at P17, and pups were weaned at P21. Punches from three animals were pooled for each sample for LC-MS/MS, and siblings were pooled whenever possible.
Environmental Enrichment.
Juvenile C57BL/6 male mice were weaned into an enriched environment or standard housing at P21. Enrichment consisted of mice being housed in large hamster cages with enrich-o-cob bedding (Andersons Laboratory) and “enrichment” devices such as mouse tunnels, wheels, domes, huts, crawl balls (Bio Serv), and so forth. Mice remained in these housing conditions for 4 wk until P49 when tissue was collected for further processing. For animals undergoing subsequent bouts of CSDS, normally housed vs. EE animals were immediately transferred from their respective housing to defeat cages, where CSDS was carried out as described above. Social interaction testing occurred 24 h following the final defeat.
Viral Constructs and Stereotaxic Surgery.
AAV expressing chained miR constructs targeting H3f3a and H3f3b was generated as previously described (10) and injected into NAc at a rate of 0.1 μL/min for 5 min (bregma coordinates: anterior/posterior, 1.6 mm; medial/lateral, 1.5 mm; dorsal/ventral, –4.4 mm; 10° angle). Stereotaxic surgeries occurred ∼4 wk before CSDS to allow for maximal viral expression and gene knockdown. The rationale for targeting both H3f3a and H3f3b genes, instead of H3f3b alone, is based upon previous studies in which we found that, although H3f3b is by far the most dynamic of the two H3.3 genes in response to environmental stimuli, knockdown of either H3f3a or H3f3b in isolation in brain is not sufficient to block stimulus-dependent H3.3 protein turnover. In other words, knockdown of one gene copy appears to be compensated for by the presence of the other gene copy. Although different in sequence and promoter structure, both genes encode identical H3.3 protein products. Knockdown of both genes simultaneously, however, results in the depletion of soluble nuclear pools of H3.3 leading to a global stalling of H3.3 turnover in neuronal chromatin (10).
RNA Isolation and qPCR.
Bilateral 14-gauge punches were taken from mouse NAc (16-gauge punches were taken from virally infected NAc under a fluorescent microscope). RNA extractions and reverse transcription reactions were performed exactly as described in Maze et al. (10). Gapdh or 18S were used as normalization controls. See Table S3 for a list of qPCR primers used.
Table S3.
Human and mouse qPCR primers
| Primer | Sequence |
| mKcnt2F | TTGCTCAGGTGTGGAGTGAC |
| mKcnt2R | GCGCGAGAGAGTAACAATCC |
| mMef2cF | ATCTGCCCTCAGTCAGTTGG |
| mMef2cR | CGTGGTGTGTTGTGGGTATC |
| mRabgap1lF | CAAAGAGATACCGGGCAGAG |
| mRabgap1lR | AGTTCATGGGCAAGGTCATC |
| mPrkar2bF | ACCTCTCCTGGTGCTCTGTG |
| mPrkar2bR | GTCTGCCAAATCTCCCTGAG |
| mCacna2d3F | GTGAGGTGGAAGGAGCTGTC |
| mCacna2d3R | TAGCTGTCATGTCGGAGTGC |
| mAdcy8F | ATGGACAGCACGGGAGTAAG |
| mAdcy8R | AAGATGAATGGGTTGGGTTG |
| mCd47F | GCTCACAGTCATCGTGGTTG |
| mCd47R | GACCAAAGCAAGGACGTAGC |
| mKcnip3F | GGCTCAGACAGCAGTGACAG |
| mKcnip3R | CGAAGGCATTGAAGAGGAAG |
| mNlgn1F | CGGTGGATCAGAGGGACTAC |
| mNlgn1R | TCTGGAGCATGGGTTAGGTC |
| mPcdh7F | GCCTACTGTCCAGCTTCACC |
| mPcdh7R | TACGAAATGGCTGTTTGCTG |
| mGapdhF | AGGTCGGTGTGAACGGATTTG |
| mGapdhR | TGTAGACCATGTAGTTGAGGTCA |
| m18sF | TGCACAATAGCTGCCAAATC |
| m18sR | GGCCTCTTCTGACCTCTGTG |
| mH3f3aF | CAAAAGCCGCTCGCAAGA |
| mH3f3aR | GGCCTGTAACGATGAGGTTTCT |
| mH3f3bF | GCCATCCACGCCAAGAGA |
| mH3f3bR | GGCGAGCCAACTGGATGT |
| hH3F3AF | TGATTCGCAAACTTCCCTTC |
| hH3F3AR | TTGGCATGGATAGCACACAG |
| hH3F3BF | AGAAGCCTCATCGCTACAGG |
| hH3F3BR | GGCACACAGGTTGGTATCTTC |
Mass Spectrometry.
LC-MS/MS analyses were performed exactly as described in Maze et al. (10).
Immunohistochemistry.
Brains were prepared for GFP immunohistochemistry exactly as described in Maze et al. (10).
H3.3 ChIP-seq Preparation.
H3.3 cross-linking ChIP assays on NAc tissues (bilateral NAc from five animals was pooled/biological replicate) were performed exactly as previously described (10). All experiments were analyzed in biological triplicates.
H3.3 ChIP-seq Analysis and Correlations with Gene Expression.
Detection of regions of differential H3.3 ChIP binding was performed using diffReps v1.55.4 (25) with the parameter setting “–pval 0.001–frag 250–window 250” as an initial run. Detected differential events were then further filtered with additional stringent cutoffs of P value < 0.0001 and an absolute fold change >3. This procedure was performed for comparisons of (i) control vs. acute and (ii) control vs. susceptible + acute samples. The diffReps analysis performed also provides annotation of the differential events into categories of genomic features (proximal promoter, gene body, intergenic etc.), from which their genome-wide distributions were tabulated. Genes containing differential event(s) in their promoters and gene bodies were deemed as being associated with the differential event(s) and compiled into lists for each comparison (including directionality) of enrichment. Gene lists of differential H3.3 enrichment/events were then analyzed against gene lists of differential gene expression (21) using the R GeneOverlap package (shenlab-sinai.github.io/shenlab-sinai/), which calculates P value significance and the odds ratio of the overlap between two gene lists.
Statistical Analyses.
One- or two-way ANOVAs were performed to determine significance for qPCR and behavioral experiments involving multiple treatment conditions––Dunnett’s (one-way) and Bonferroni (two-way) post hoc tests were used if significant main effects were observed via ANOVA. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.0001. In instances where ANOVAs resulted in significant main effects without post hoc significance (or where a priori assumptions could be made based upon previous data, for example, Fig. 3F), planned two-tailed Student’s t tests were used to compare control conditions to respective treatment groups. #/*P ≤ 0.05. Two-tailed Student’s t tests were used for all other statistical comparisons. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. All data are presented as means ± SEM.
Supplementary Material
Acknowledgments
This work was supported by a Brain & Behavior Research Foundation (NARSAD) Young Investigator Award (to I.M.); an MQ Research Fellowship MQ15FIP100011 (to I.M.); an Alfred P. Sloan Foundation Fellowship in Neuroscience (to I.M.); a Rosen Family Research Scholar Award (to I.M.); and an NIH/National Institute of Mental Health Grant P50 MH096890 (to E.J.N., C.D.A., and I.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE85900). Differential ChIP-seq data are provided as Datasets S1 and S2.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608270113/-/DCSupplemental.
References
- 1.Kessler RC, et al. National Comorbidity Survey Replication The epidemiology of major depressive disorder: Results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289(23):3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 2.Nestler EJ, Carlezon WA., Jr The mesolimbic dopamine reward circuit in depression. Biol Psychiatry. 2006;59(12):1151–1159. doi: 10.1016/j.biopsych.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Charney DS, Manji HK. Life stress, genes, and depression: Multiple pathways lead to increased risk and new opportunities for intervention. Sci STKE. 2004;2004(225):re5. doi: 10.1126/stke.2252004re5. [DOI] [PubMed] [Google Scholar]
- 4.Covington HE, III, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71(4):656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covington HE, III, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29(37):11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13(9):1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renthal W, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 8.Sun H, et al. ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat Med. 2015;21(10):1146–1153. doi: 10.1038/nm.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter RG, Gagnidze K, McEwen BS, Pfaff DW. Stress and the dynamic genome: Steroids, epigenetics, and the transposome. Proc Natl Acad Sci USA. 2015;112(22):6828–6833. doi: 10.1073/pnas.1411260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maze I, et al. Critical role of histone turnover in neuronal transcription and plasticity. Neuron. 2015;87(1):77–94. doi: 10.1016/j.neuron.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berton O, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311(5762):864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 12.Krishnan V, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131(2):391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 13.Bagot RC, et al. Ketamine and imipramine reverse transcriptional signatures of susceptibility and induce resilience-specific gene expression profiles. Biol Psychiatry. June 18, 2016 doi: 10.1016/j.biopsych.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermes G, Li N, Duman C, Duman R. Post-weaning chronic social isolation produces profound behavioral dysregulation with decreases in prefrontal cortex synaptic-associated protein expression in female rats. Physiol Behav. 2011;104(2):354–359. doi: 10.1016/j.physbeh.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Der-Avakian A, Markou A. Neonatal maternal separation exacerbates the reward-enhancing effect of acute amphetamine administration and the anhedonic effect of repeated social defeat in adult rats. Neuroscience. 2010;170(4):1189–1198. doi: 10.1016/j.neuroscience.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447(7141):178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 17.Vega-Rivera NM, et al. The neurogenic effects of an enriched environment and its protection against the behavioral consequences of chronic mild stress persistent after enrichment cessation in six-month-old female Balb/C mice. Behav Brain Res. 2016;301:72–83. doi: 10.1016/j.bbr.2015.12.028. [DOI] [PubMed] [Google Scholar]
- 18.Koe AS, Ashokan A, Mitra R. Short environmental enrichment in adulthood reverses anxiety and basolateral amygdala hypertrophy induced by maternal separation. Transl Psychiatry. 2016;6:e729. doi: 10.1038/tp.2015.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christoffel DJ, et al. IκB kinase regulates social defeat stress-induced synaptic and behavioral plasticity. J Neurosci. 2011;31(1):314–321. doi: 10.1523/JNEUROSCI.4763-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golden SA, et al. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat Med. 2013;19(3):337–344. doi: 10.1038/nm.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagot RC, et al. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron. 2016;90(5):969–983. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maze I, et al. G9a influences neuronal subtype specification in striatum. Nat Neurosci. 2014;17(4):533–539. doi: 10.1038/nn.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobo MK, et al. ΔFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J Neurosci. 2013;33(47):18381–18395. doi: 10.1523/JNEUROSCI.1875-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raineki C, Cortés MR, Belnoue L, Sullivan RM. Effects of early-life abuse differ across development: Infant social behavior deficits are followed by adolescent depressive-like behaviors mediated by the amygdala. J Neurosci. 2012;32(22):7758–7765. doi: 10.1523/JNEUROSCI.5843-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen L, et al. diffReps: Detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8(6):e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.