Significance
Transcription addiction is a hallmark of cancer and a potential therapeutic target. RNA polymerase II (pol II) is responsible for synthesizing precursor mRNA in all eukaryotic cells and can be blocked by obstacles, such as DNA lesions and nucleosomes on the DNA template. In this study, we demonstrate that sequence-specific minor groove binding pyrrole-imidazole polyamides can sterically block an elongating polymerase at the targeted binding site. We find this blockage is persistent and cannot be rescued by transcription factor IIS. We further show pyrrole-imidazole polyamides are detected in the minor groove via two conserved residues in the Switch 1 region of pol II. Collectively, these results provide mechanistic insights on how a noncovalent minor groove binder can obstruct pol II elongation.
Keywords: Py-Im polyamide, transcription inhibition, minor groove, DNA
Abstract
RNA polymerase II (pol II) encounters numerous barriers during transcription elongation, including DNA strand breaks, DNA lesions, and nucleosomes. Pyrrole-imidazole (Py-Im) polyamides bind to the minor groove of DNA with programmable sequence specificity and high affinity. Previous studies suggest that Py-Im polyamides can prevent transcription factor binding, as well as interfere with pol II transcription elongation. However, the mechanism of pol II inhibition by Py-Im polyamides is unclear. Here we investigate the mechanism of how these minor-groove binders affect pol II transcription elongation. In the presence of site-specifically bound Py-Im polyamides, we find that the pol II elongation complex becomes arrested immediately upstream of the targeted DNA sequence, and is not rescued by transcription factor IIS, which is in contrast to pol II blockage by a nucleosome barrier. Further analysis reveals that two conserved pol II residues in the Switch 1 region contribute to pol II stalling. Our study suggests this motif in pol II can sense the structural changes of the DNA minor groove and can be considered a “minor groove sensor.” Prolonged interference of transcription elongation by sequence-specific minor groove binders may present opportunities to target transcription addiction for cancer therapy.
In eukaryotes, precursor mRNA synthesis is catalyzed by the RNA polymerase II holoenzyme (pol II), which frequently pauses during transcription elongation (1–3). In addition to regulatory factors that control pol II processivity, various obstacles encountered by pol II can also lead to stalling, and even backtracking, on the DNA template (4). Factors that affect pol II transcription elongation dynamics include intrinsic DNA sequences or structures (5, 6), endogenous epigenetic DNA modifications (7), embedded ribonucleotides (8), DNA lesions (9, 10), small-molecule DNA-binders (11, 12), DNA-binding proteins including nucleosomes (13), and even pol II itself (14, 15). Transient transcriptional pausing or short-lived transcriptional blockage can be rescued by the recruitment of transcription factor IIS (TFIIS) (16, 17), a transcription factor that facilitates the cleavage of backtracked transcript. In contrast, prolonged transcriptional arrest by some bulky DNA lesions triggers either ubiquitination and degradation of pol II, or transcription-coupled nucleotide excision repair, a special DNA repair pathway that preferentially repairs DNA lesions in the transcribed strand (17, 18). Structural, genetic, and biochemical studies have greatly advanced our understanding of how pol II copes with different kinds of covalent DNA lesions caused by oxidation (19, 20), alkylation (21–24), and photocyclo-addition (10). Less well understood are the interactions of the pol II machinery when confronted with a steric blockade by small molecules bound noncovalently in the minor groove of DNA (25–27).
Pyrrole-imidazole (Py-Im) polyamides are a class of small molecules that can be programmed to selectively target specific DNA sequences in the minor groove with high binding affinity (28, 29). This sequence-specificity arises from unique pairs of aromatic amino acids, which distinguish the edges of the four Watson–Crick base pairs (30, 31). Eight-ring hairpin oligomers linked by a central aliphatic γ-aminobutyric acid unit have binding affinity comparable to TFs (32). Early studies suggested that cell-permeable Py-Im polyamides modulate gene expression by targeting TF binding (25, 33, 34). Importantly, inhibition of pol II occupancy at gene start sites leading to Rpb1 degradation has been observed (35). The question arises as to how polyamide occupancy in the DNA minor groove prevents pol II elongation. Do Py-Im polyamides pose a direct impediment to pol II elongation and for how long? In this study, we investigate the mechanism of how pol II transcription elongation is affected by Py-Im polyamide binding at a discrete site downstream from the pol II active site. From molecular modeling, a conserved pol II motif appears to function as a minor groove sensor and could be responsible for pol II elongation inhibition. This finding was verified by site-specific mutations of this region in pol II. These results suggest direct interaction between residues of the conserved motif and the bound Py-Im polyamides. In short, we have identified a sensor function of a conserved motif in pol II.
Results
Py-Im Polyamides Are Roadblocks for Pol II Transcription Elongation.
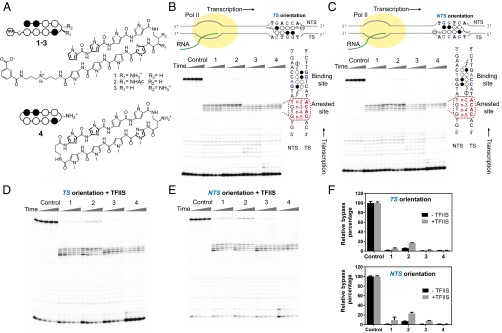
To investigate the impact of a site-specific DNA-binding small molecule on pol II transcription elongation, we assembled a purified pol II elongation complex with a DNA scaffold (78 bp in length) containing a full transcription bubble upstream and a specific 6-bp binding site downstream (59 bp from the scaffold end). The 6-bp site is bound specifically by Py-Im polyamides 1–4 that code for 5′-WGGWCW-3′ according to the pairing rules (Fig. 1). To address the potential differences of hairpin Py-Im polyamide binding orientation relative to transcriptional directionality (upper black arrow in Fig. 1B), we also designed a second scaffold with the downstream polyamide binding site in an opposite orientation (upper black arrow in Fig. 1C). We termed the orientation in which the pol II encounters the γ-turn moiety of the hairpin Py-Im polyamide first (Fig. 1B) the “template strand (TS) binding orientation,” and the reverse orientation where pol II would encounter the “C-terminal linker” first the “nontemplate strand (NTS) binding orientation” (Fig. 1C).
Fig. 1.
Py-Im polyamides are strong roadblocks for RNA pol II transcription elongation. (A) Four Py-Im polyamides share the same 6-bp DNA target sequence (5′-WGGWCW-3′). The black solid circle refers to N-methylimidazole (Im), and the hollow circle refers to N-methylpyrrole (Py). (B) RNA pol II transcription blockage by Py-Im polyamides with a TS binding orientation and (C) with a NTS binding orientation. (D and E) PAGE gel analysis of TFIIS effect on Py-Im polyamide induced pol II arrest. The Py-Im polyamide binds DNA in a TS orientation (D) or NTS orientation (E). In these gel images, control lanes are RNA pol II transcription in the absence of Py-Im polyamides. Time points for transcription assays were 10 min, 20 min, 30 min, 1 h, and 2 h. The concentration of Py-Im polyamides was 0.5 µM. (F) Quantitative analysis of TFIIS effect on rescuing Py-Im polyamide-induced pol II arrest.
Substitutions on the γ-turn or the C terminus of Py-Im polyamides influence DNA affinity as well as cellular uptake (36). To evaluate the effects of these substitutions on pol II transcription elongation, we compared Py-Im polyamides 1–4 (Fig. 1A). Hairpins 1, 2, and 3 have identical C terminus and vary on the γ-turn, where 1 has a chiral (R)-α-amino group, 2 has an acetylated (R)-α-amino group, and 3 has a (R)-β-amino substitution. Compound 4 is a cycle flanked by the same β-amino γ-turn as 3 and an unsubstituted γ-turn.
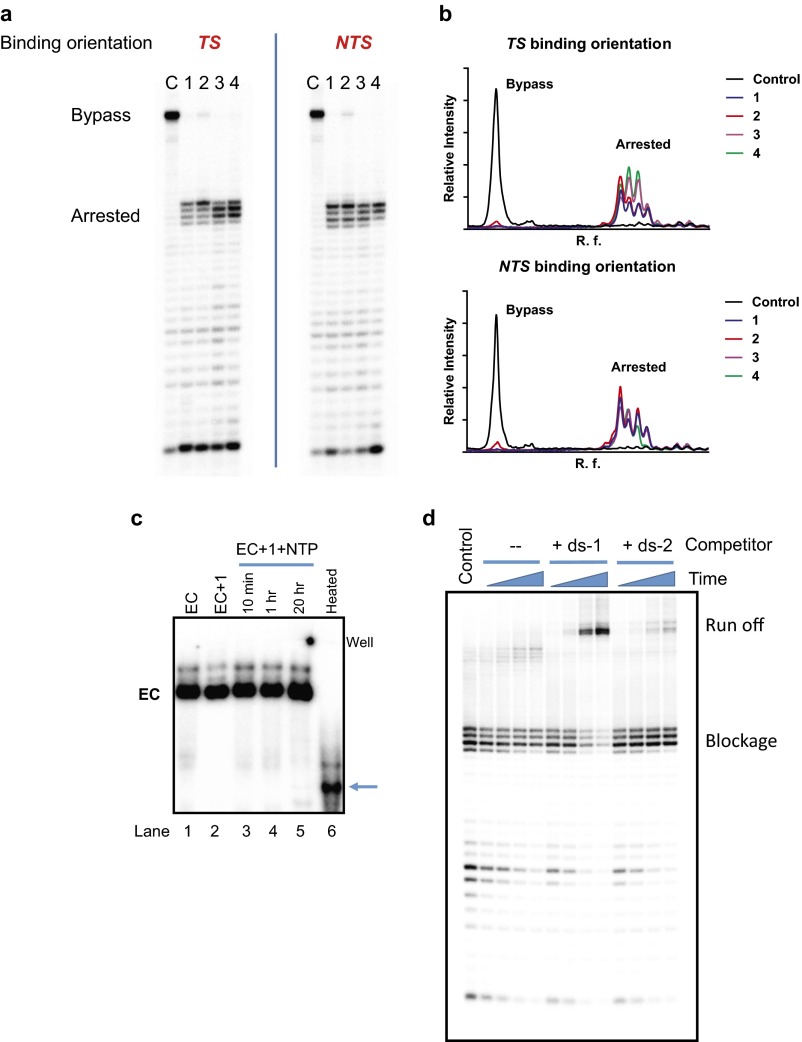
All four Py-Im polyamides (1–4) block RNA pol II transcription elongation at specific positions upstream of their binding sites in both the TS and NTS orientations (Fig. 1 B and C), whereas RNA pol II can transcribe through the DNA template to obtain full-length RNA transcripts in the absence of Py-Im polyamide treatment (see control lanes in Fig. 1 B and C). Importantly, pol II cannot bypass the polyamide blockage even after prolonged incubation (2–20 h) (Fig. 1 B and C and Fig. S1), implying that binding of Py-Im polyamides can induce prolonged arrest of pol II at the transcription elongation phase in a sequence-specific manner. To exclude the possibilities that pol II could fall off from DNA, or RNA could be released during the long time incubation in the presence of polyamides, we performed a native gel analysis to test stability of the polyamide-arrested pol II complex and we observed that the complex is very stable and no obvious RNA transcripts are released (Fig. S1C). To determine the stability of the polyamide-arrested pol II complex (without RNA transcript release) and whether transcription can recover upon the removal of polyamide, we designed competition experiments to remove the polyamide from the elongation complex by addition of competitor duplex DNA with the same binding site, and found that the elongation can be slowly recovered (18 h) and reach the full transcripts in the presence of high concentration of competitor duplex DNA (Fig. S1D). We found that the arrested region generally has two to four pausing bands that are located 2–5 bp upstream of the 5′-WGGWCW-3′ binding site (see the dashed box on the right of the gel image in Fig. 1 B and C, named the n-2, n-3, n-4, and n-5 bands, from top to bottom). These results reveal that pol II is able to sense the barrier and becomes arrested several base pairs upstream of the bound Py-Im polyamide.
Fig. S1.
RNA pol II was persistently arrested by Py-Im polyamides. (A) RNA pol II transcription elongation is blocked by Py-Im polyamides in both binding orientations (TS and NTS). C, control lanes in the absence of Py-Im polyamides; 1, 2, 3, and 4 represent the transcription inhibition results by four compounds listed in Fig. 1A. The incubation time for this transcription assay was 20 h. RNA pol II failed to bypass the blockage with prolonged incubation. (B) Traces of lanes in the presence of different compounds. (C) Pol II elongation complex (EC) was persistently arrested by the polyamides and no obvious free RNA transcript is released. The pol II EC stability experiment was performed in 4% (wt/vol) native PAGE with 32P labeled in the RNA transcript. Lane 1: pol II EC assembled as described in the method in the absence of Py-Im polyamide 1; lane 2: pol II EC in the presence of Py-Im polyamide 1; lanes 3–5: Pol II transcription elongation in the presence of 1 after the designated time points (10 min, 1 h, and 20 h, respectively); lane 6: the 20-h time point sample heated at 95 °C for 10 min before loading. Free RNA transcript is released upon heat treatment (indicated by the blue arrow). (D) The arrested pol II EC was slowly recovered upon treatment of the DNA duplex competitor. The control lane is the arrested EC in the presence of compound 1 after 1 h transcription elongation. The duplex competitor with 5′-WGGWCW-3′ binding site (ds-1) was subsequently added into the arrested EC mixture. As comparison, the DNA duplex without the designed binding site (ds-2) was incubated with the arrested EC as well. The sample without any competitor (–) was also analyzed. The EC was 50 nM, the compound 1 was 1 µM, and the competitor DNA duplex was 10 µM. Time points were 1, 4, 18, and 42 h.
Quantitative analysis of the pol II pausing bands reveals specific patterns of polyamide-induced pol II arrest. The pattern of pausing bands depends on the chemical nature of the polyamides, the concentration of the compounds, the binding orientations, and the incubation time (Fig. 1 and Figs. S1–S5). In the TS binding orientation, where the γ-turn faces toward oncoming pol II, acetylation of the α-amine group in hairpin 2 causes more dominant pausing at the n-2 site than hairpin 1 (Fig. 1 and Fig. S1). Interestingly, moving the amine group into the β-position in hairpin 3 causes the major pausing sites to shift to n-3 and n-4 sites (Fig. 1 and Fig. S1). Finally, cycle 4 shares a similar pattern with hairpin 3 as might be expected, with a minor increase of the n-2 site with prolonged incubation (Fig. S1). In the NTS binding orientations where the C terminus faces toward the oncoming pol II, hairpins 1, 2, and 3 show similar stall patterns, which is consistent with their identical C-terminus moieties. In contrast, cycle 4 only has two major pausing sites (n-2 and n-3) and one minor pausing site (n-4), consistent with a shorter γ-turn facing pol II rather than a longer C-terminus linker. Despite the subtle pattern differences, both binding orientations had a common pausing region (n-2 to n-5) relative to the Py-Im polyamide binding site.
Fig. S5.
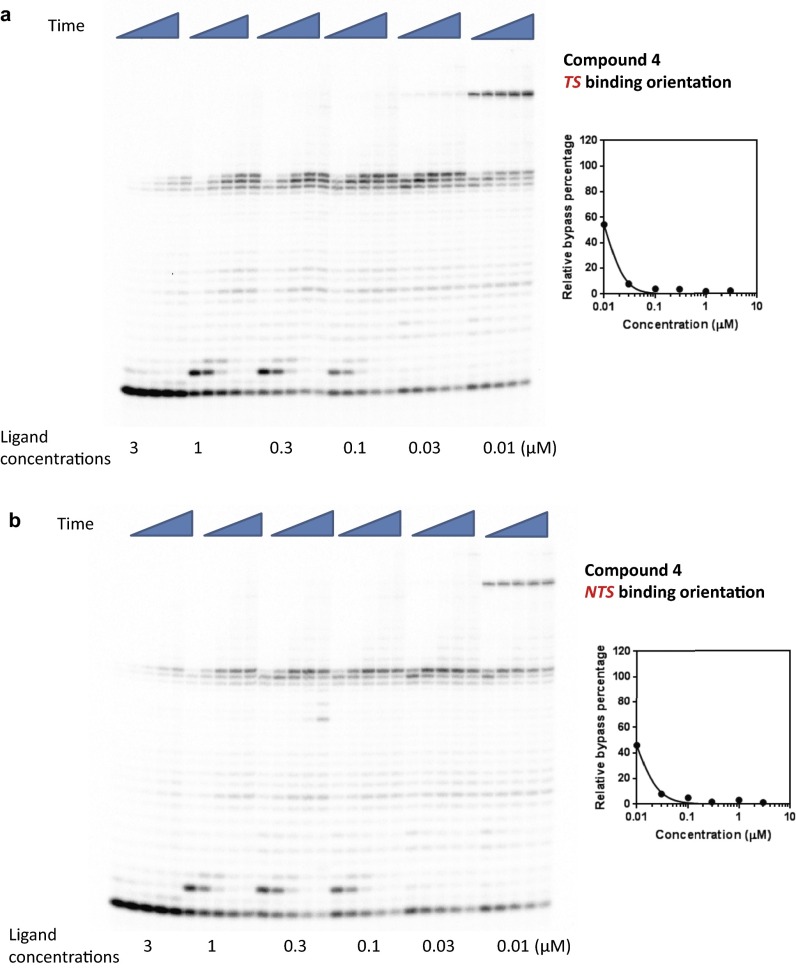
Concentration-dependent transcriptional inhibition by Py-Im polyamide 4 in TS (A) and NTS (B) binding orientations. Time points were 1 min, 5 min, 20 min, 1 h, and 3 h.
Fig. S3.
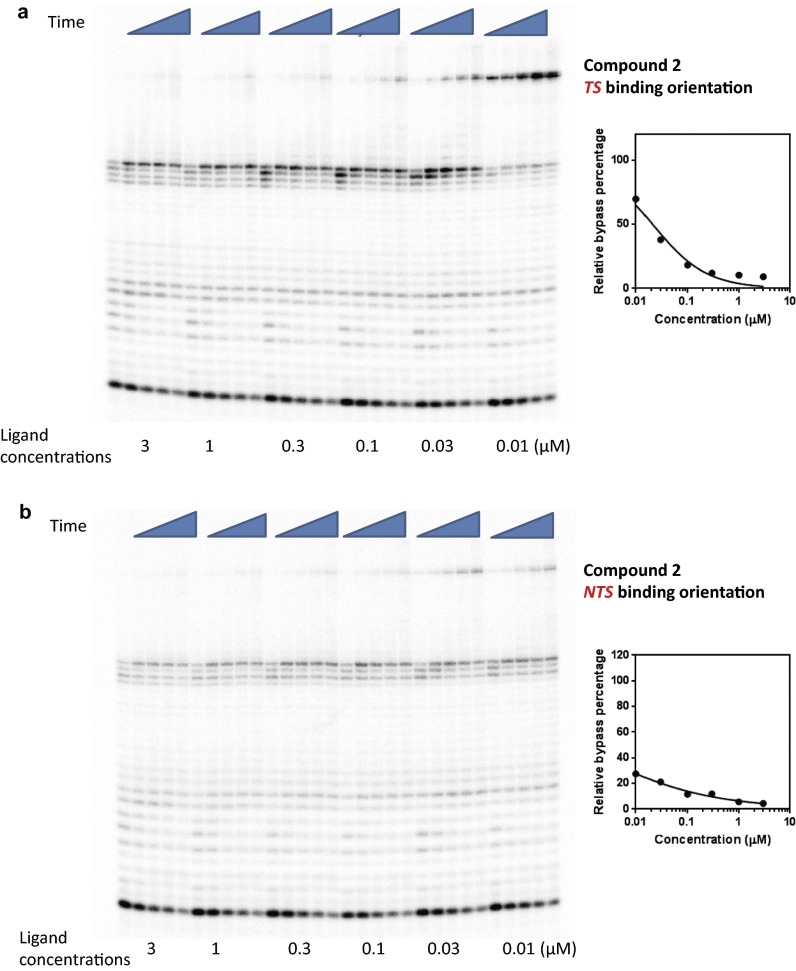
Concentration-dependent transcriptional inhibition by Py-Im polyamide 2 in TS (A) and NTS (B) binding orientations. Time points were 1 min, 5 min, 20 min, 1 h, and 3 h.
Fig. S4.
Concentration-dependent transcriptional inhibition by Py-Im polyamide 3 in TS (A) and NTS (B) binding orientations. Time points were 1 min, 5 min, 20 min, 1 h, and 3 h.
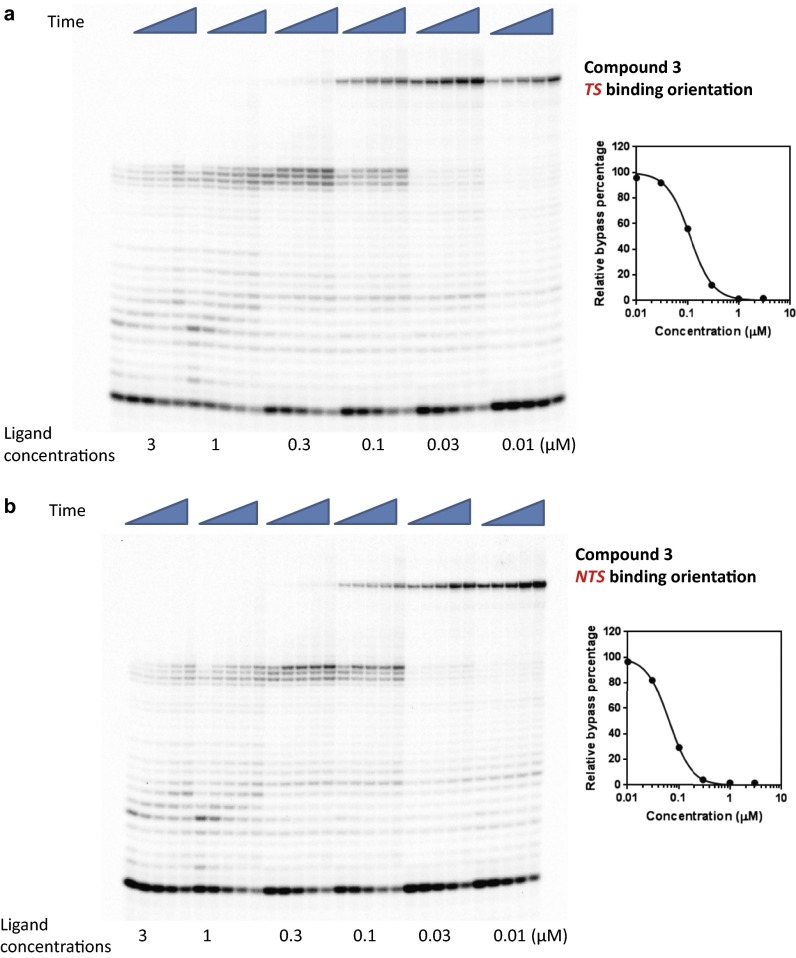
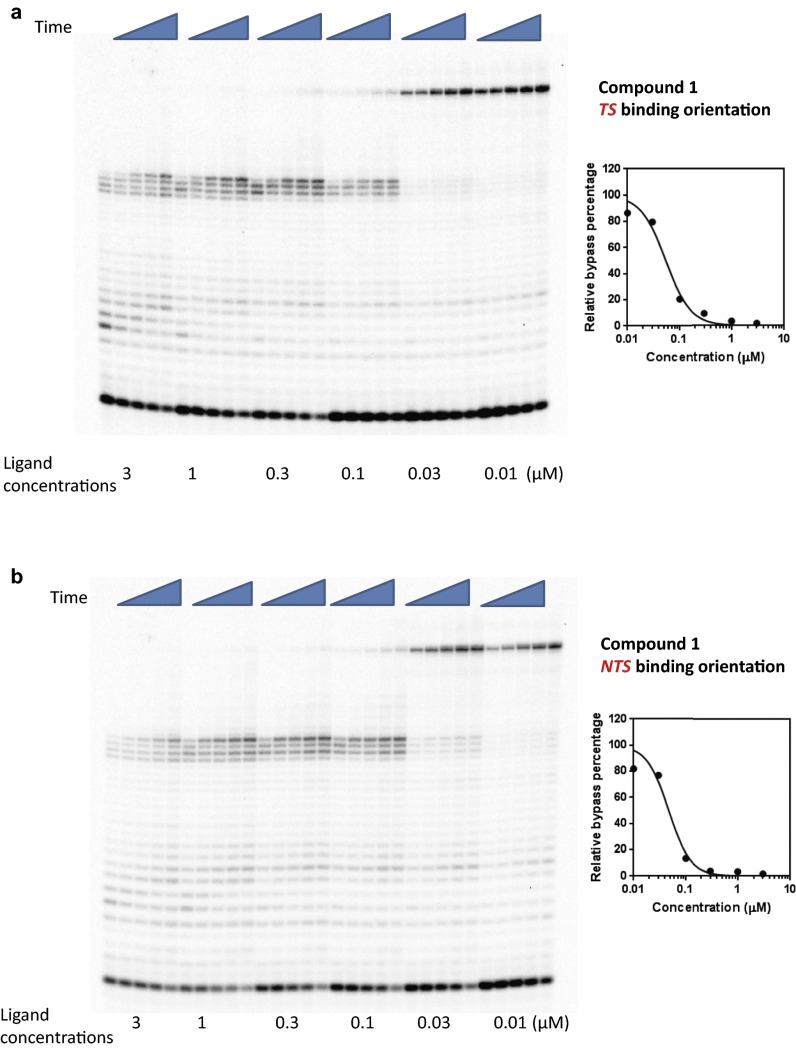
To further characterize the strength of Py-Im polyamides as inhibitors of pol II elongation, we performed concentration-dependent pol II inhibition analysis for the four polyamides 1–4 in both binding orientations (Figs. S2–S5). The full-length pol II transcripts are diminished and the arrested bands accumulate as polyamide concentration increases (Figs. S2–S5). To quantitatively evaluate the concentration-dependent inhibition data, we systematically determined the inhibition parameter Ki (the concentration of ligand that can inhibit 50% of pol II elongation) (see SI Materials and Methods for details) for each Py-Im polyamide and binding orientation (Table 1 and Figs. S2–S5). For each polyamide, the NTS binding orientation generally leads to a stronger inhibition effect than the TS binding orientation, suggesting an asymmetric inhibition of pol II transcription elongation. The Ki concentrations did not correlate with the binding affinity of these polyamides observed from duplex melting experiments (Table S1). A comparison of these four ligands reveals that 2 and 4 have the strongest inhibition. Comparison between hairpins 1, 2, and 3 clearly shows the influence of substitutions on the γ-turn, which faces the approaching enzyme in the TS binding mode. Acetylation of the γ-turn α-amine group in 2, which decreases DNA binding affinity, increases the inhibition ability by ∼threefold, whereas moving the α-amine group to the β-position (3) decreases the inhibition ability by ∼threefold. Interestingly, cyclization of 4 greatly increases the inhibition ability by ∼16-fold over 3. Although modifications in the γ-turn and the C terminus can modulate the inhibition abilities of these molecules, all four molecules cause strong inhibition of pol II elongation with Ki values at nanomolar concentrations.
Fig. S2.
Concentration-dependent transcriptional inhibition by Py-Im polyamide 1 in TS (A) and NTS (B) binding orientations. Time points were 1 min, 5 min, 20 min, 1 h, and 3 h.
Table 1.
Concentration of Py-Im polyamides for 50% inhibition of RNA pol II bypass (Ki, nM)
| Orientation | 1 | 2 | 3 | 4 |
| TS | 50 ± 10 | ∼17 ± 2 | 160 ± 30 | ∼10 ± 2 |
| NTS | 45 ± 12 | <5 | 63 ± 17 | ∼8 ± 2 |
Table S1.
DNA thermal stabilization analysis of compounds 1–4 on TS match and mismatch sequence
| Polyamide | 5′-GACT TGGTCA TACA -3′ | 5′-GACT TGTTCA TACA -3′ | ||
| Tm/°C | ΔTm/°C | Tm/°C | ΔTm/°C | |
| — | 56.4 ± 0.7 | — | 52.9 ± 0.5 | — |
| 1 | 71.3 ± 0.8 | 14.9 | 53 ± 0.8 | 0.1 |
| 2 | 66.6 ± 0.7 | 10.2 | 55.9 ± 0.2 | 3.0 |
| 3 | 71.1 ± 0.8 | 14.7 | 54.2 ± 1.0 | 1.3 |
| 4 | 71.8 ± 0.9 | 15.4 | 53.1 ± 0.3 | 0.2 |
The compound binding site is indicated in bold font.
TFIIS Is Insufficient to Fully Rescue Py-Im Polyamide-Arrested Pol II.
Transcription elongation factor TFIIS can rescue arrested pol II and enhance pol II bypass of a variety of translocation barriers, such as nucleosomes, some DNA lesions, small-molecule DNA binders, and intrinsic pausing DNA sequences (12, 13, 16, 37). To investigate whether TFIIS can stimulate pol II bypass of polyamide-induced pol II arrest, we performed transcription elongation experiments in the absence or presence of TFIIS. As shown in Fig. 1 D and E, the majority of pol II remains arrested by Py-Im polyamides even in the presence of TFIIS, regardless of the binding orientation (TS or NTS), with only a small portion of transcriptional bypass observed for hairpins 1 and 2. These results indicate that TFIIS fails to effectively rescue polyamide-induced pol II arrest. The presence of TFIIS altered the patterns of the pausing bands, as the major arrested bands are shifted toward the upstream positions (Fig. 1 D and E, n-4 or n-5 positions). This finding suggests that pol II may adopt pretranslocation or backtracked states during Py-Im polyamide-induced arrest. TFIIS facilitates cleavage of short RNA oligos from the 3′-end of RNA transcripts, leading to the shortened transcripts and shifted pausing patterns.
Quantitative comparison of bypass percentage in the absence (Fig. 1 B and C) and presence of TFIIS (Fig. 1 D and E) reveals distinct responses to TFIIS among the four polyamides (Fig. 1F). For example, the lowest-affinity hairpin 2 is more sensitive to TFIIS treatment with ∼12% transcript bypass of the blockage and ∼88% arrest in the TS binding orientation. In contrast, the higher-affinity cycle 4 is resistant to TFIIS treatment. We observed no transcriptional bypass in the presence of TFIIS even after 2-h incubation. Similar TFIIS sensitivity profiles were also observed for NTS binding orientation, with cycle 4 most resistant to TFIIS treatment (Fig. 1F). These results demonstrate that TFIIS is insufficient to rescue Py-Im polyamide-induced pol II transcriptional arrest, indicating that Py-Im polyamide-induced transcriptional blockage is strong and stable. This finding is in contrast with other translocation barriers, such as nucleosomes or other small-molecule binders (Distamycin and DAPI) (12, 13).
Sequence-Specific RNA Pol II Transcriptional Blockage by Py-Im Polyamides.
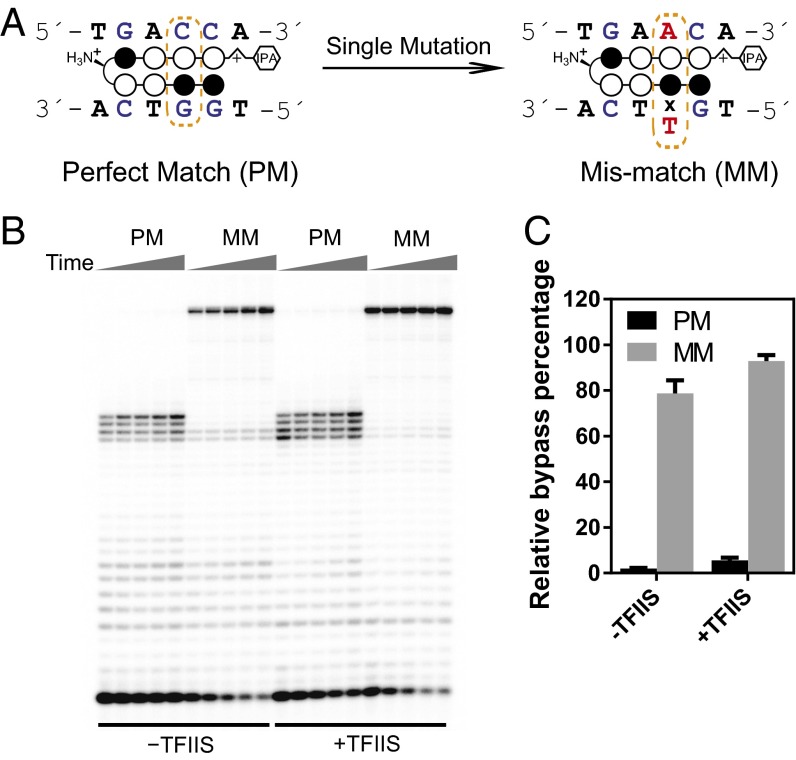
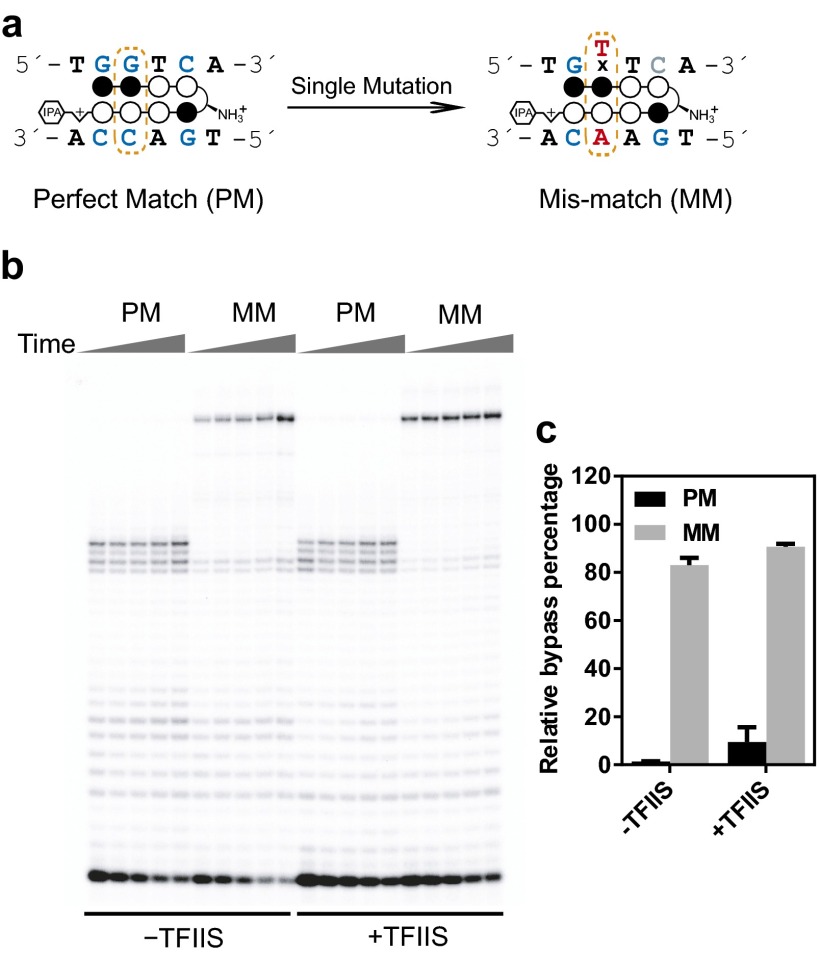
Our results support the DNA target specificity of Py-Im polyamides (32), as evidenced by these polyamides only causing pol II stalling specifically 2–5 bp upstream of the match binding site but not in other sites in the template DNA. To further directly verify that pol II transcription stalling by polyamides is indeed a result of its discrete binding to the 5′-WGGWCW-3′ sequence, we compared the transcriptional inhibitory effect of hairpin 1 upon its binding to DNA with a perfectly matched (PM) sequence (5′-TGGTCA-3′) or with a single mismatched (MM) target sequence (5′-TGTTCA-3′) (single mutation underlined) (Fig. 2A and Fig. S6). As shown in Fig. 2, a single mutation in the binding sequence (MM) abolished Py-Im polyamide-induced pol II transcriptional arrest in the TS binding mode (both in the presence and absence of TFIIS). A similar effect is observed for the scaffold containing a single mutation in the NTS binding orientation (Fig. S6). Furthermore, DNA duplex melting experiments confirm that a single mutation significantly diminishes ligand binding affinity (Table S1), consistent with transcriptional inhibition results. These results demonstrate that polyamide-induced pol II transcriptional arrest is dependent upon the matched DNA sequence in both directions.
Fig. 2.
Py-Im polyamides cause sequence-specific inhibition of pol II transcription elongation. The Py-Im polyamide binding site is in a TS orientation. (A) Scheme of single mutation of the designed target sequence. The PM sequence is 5′-WGGWCW-3′, and the single-base MM sequence is 5′-WGWWCW-3′ (mutation underlined). (B) Comparison of inhibition of 1 on pol II transcription elongation from the PM or MM sequence. The effect of TFIIS was also investigated as shown in PAGE gel. Time points were 10 min, 20 min, 30 min, 1 h, and 2 h. Py-Im polyamide concentration was 0.5 µM. (C) Quantitative analysis of selective transcriptional inhibition by 1.
Fig. S6.
Py-Im polyamide cause sequence-specific inhibition of pol II transcription elongation. The polyamide binding site is in a NTS orientation. (A) Scheme of single mutation of the designed target sequence. The PM sequence is 5′-WGGWCW-3′, whereas the single base MM sequence is 5′-WGWWCW-3′ (mutation underlined). (B) Comparison of inhibition of polyamide 1 on pol II transcription elongation from the PM sequence or MM sequence. The effects of TFIIS were also investigated as shown in PAGE gel. Time points were 10 min, 20 min, 30 min, 1 h, and 2 h. Py-Im polyamide concentration was 0.5 µM. (C) Quantitative analysis of selective transcriptional inhibition by polyamide 1.
Structural Insight into Py-Im Polyamide-Induced Pol II Arrest Revealed by Molecular Modeling.
To gain further mechanistic understanding of how pol II stalls upstream of the polyamide binding sites (2–5 bp), we modeled a series of pol II-arrested complexes in the presence of Py-Im polyamide binding to mimic pol II progression into the pausing region in a stepwise manner. The models are built based on the superposition of crystal structures of the downstream DNA duplex of pol II elongation complex (PDB ID code 3M3Y) (23) with the structure of the polyamide bound DNA duplex (PDB ID code 3I5L) (31). Here we modeled each of the four major pausing states observed in our biochemical studies. Details for model set-up can be found in SI Materials and Methods.
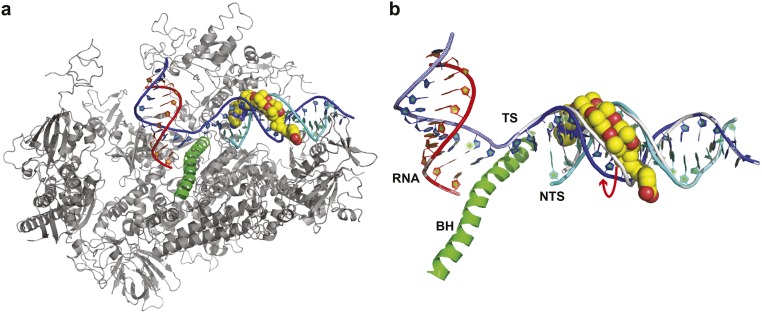
These models provide structural insights into the molecular mechanism of polyamide-induced pol II transcriptional arrest. First, as shown in Fig. S7A, the polyamide binding site remains within the intact duplex DNA region several base pairs downstream of the transcription bubble edge when pol II pauses at the first couple of pausing sites (states i and ii, or n-5 and n-4). This result suggests that pol II senses polyamide binding and pauses several base pairs before the actual polyamide binding site reaches the transcription bubble downstream edge. This result may rule out the simple model that pol II blockage results exclusively from prevention of the unwinding of DNA duplex at the transcription bubble downstream edge by polyamides. From high-resolution X-ray studies, it is known that the polyamide widens the minor groove of DNA, increasing the distance between the two DNA phosphodiester backbones (Fig. S7B) (31). These structural changes may cause misalignment of pol II residues involved in interactions with the DNA minor groove or phosphodiester backbone.
Fig. S7.
Molecular modeling of RNA pol II transcriptional pausing and arrest by Py-Im polyamide. (A) Overall view of an arrested pol II complex in the presence of Py-Im polyamide. This model view is derived from 10 subunit RNA pol II elongation complex with Rpb2 omitted. The protein residues are colored gray; RNA, template DNA, and nontemplate DNA are shown in red, blue, and cyan, respectively. The small DNA binding molecule is highlighted in yellow CPK mode. (B) Py-Im polyamide binding causes a widened DNA duplex minor groove. The Py-Im polyamide bound downstream duplex (blue and cyan) was aligned with regular downstream DNA duplex with pol II elongation complex (gray). The arrow indicates the shifted strand caused by Py-Im polyamide binding.
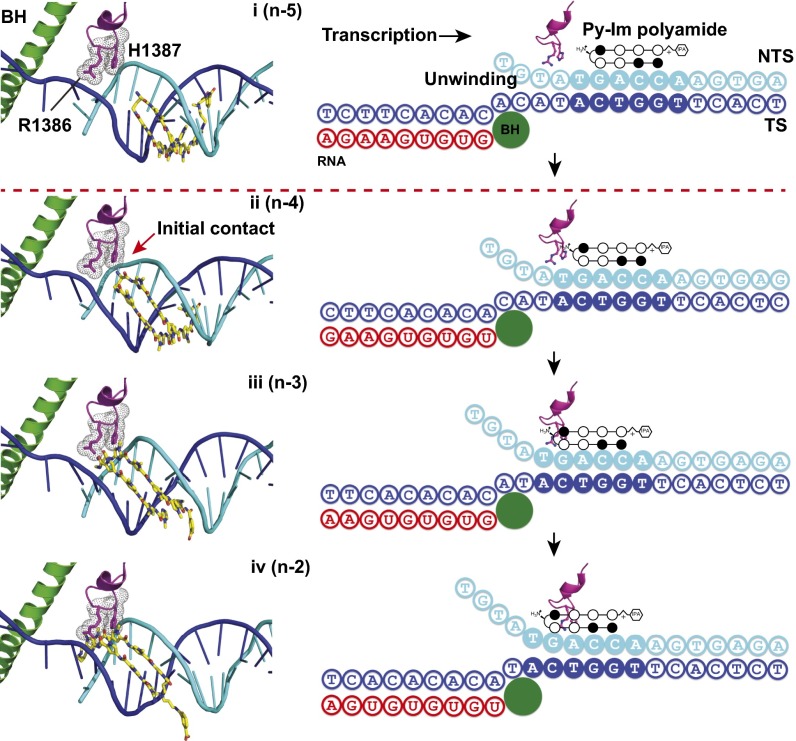
Structural analysis further reveals that the Switch 1 region in the Rpb1 subunit (38, 39) is likely to be involved in the interactions with polyamide-bound DNA. The Switch 1 region is a highly conserved motif of pol II that is located at the base of the clamp and serves as a hinge for clamp closure in the presence of a DNA–RNA hybrid in the active center (39). We identified two critical residues in the Switch 1 region of Rpb1, Arg1386 and His1387, which insert into the DNA minor groove in the wild-type pol II elongation complex and are likely either directly interacting with polyamide backbones (state ii, or n-4, for example) or sterically clashing with the Py-Im polyamide (states iii and iv) during RNA pol II translocation toward the Py-Im polyamide binding site (Fig. 3). Therefore, these two residues may play an interrogative role in sensing or scanning the DNA minor groove conformation and are likely to contribute to polyamide-induced pol II stalling. Indeed, these models are consistent with the data that revealed two to four pausing bands with increasing difficulty to reach beyond the n-2 position (severe steric clash in state iv). Similar steric clash is expected between these two residues and the polyamide backbone for all four compounds regardless of their binding orientations (TS or NTS).
Fig. 3.
Molecular modeling of RNA pol II transcriptional pausing and arrest by Py-Im polyamide. The functional interplay between two critical residues (R1386 and H1387) from the conserved Switch 1 region of Rpb1 and the polyamide-bound minor groove during RNA pol II progression. During translocation, the pol II may move from step i to step iv with distinct steric interactions. In step i, the Py-Im polyamide and these two residues are in positions without obvious steric interaction; in step ii, the polyamide and the residues establish direct contact; in steps iii and iv, strong steric clashes were observed between pol II Switch 1 residues and Py-Im polyamide, and duplex unwinding was also prevented, both of which lead to strong inhibition for RNA pol II elongation complex.
Conserved Pol II Switch 1 Motif Functions as an Inspector for the DNA Minor Groove.
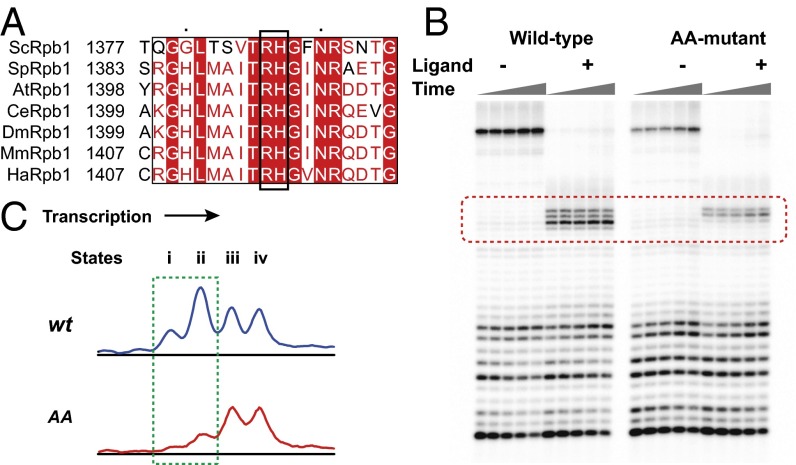
Our structural modeling revealed that two critical residues in the Switch 1 region of Rpb1 (Arg1386 and His1387) may serve as a sensor for the DNA minor groove and could stall pol II elongation via steric clash with minor groove binders. These two residues are conserved across eukaryotes from yeast to human (Fig. 4A). To verify the role of these two residues, we generated pol II with site-specific substitutions of Rpb1, R1386A and H1387A (AA-mutant) (see SI Materials and Methods for details) and performed transcription elongation assays in the presence and absence of hairpin 1 for both wild-type pol II and the AA-mutant. We theorized that the AA-mutant would have ablated steric clash with minor groove binders in early pausing states i and ii. Indeed, as shown in the dashed box in Fig. 4B, the pausing pattern of the AA-mutant is significantly different from that of the wild-type. The two stalled bands corresponding to states i and ii (Fig. 4B) were absent in the AA-mutant, revealing that these two residues are responsible for the early inspection of the downstream DNA minor groove environment before DNA unwinding. The downstream pol II pausing (states iii and iv or n-2 and n-3 position) was still observed upon polyamide binding, which was most likely caused by a direct blockage to DNA unwinding. Quantitative analysis (Fig. 4C) clearly indicated distinct blockage behaviors of the AA-mutant, in which the pausing at states i and ii (band n-4 and n-5) was abolished. This pattern difference between the wild-type and AA-mutant confirmed the inspective role of Arg1386 and His1387 of the Rpb1 switch 1 motif in sensing the DNA minor groove before duplex unwinding. In addition, it is worth mentioning that compared with the wild-type pol II, the AA-mutant has a relatively strong sequence pausing effect during elongation (see the bottom part of gel in Fig. 4B), which may indicate possible roles of these two residues during normal transcriptional progression.
Fig. 4.
Effects of Py-Im polyamide on the elongation of wild-type and AA-mutant are distinct. (A) Sequence alignment of Switch 1 region of Rpb1 of RNA pol II in different species. (B) Gel data of transcriptional elongation of the wild-type and AA-mutant. Time points were 10 min, 20 min, 30 min, 1 h, and 2 h. The different pausing patterns in the presence of 0.5 µM Py-Im polyamide 1 are highlighted in the red dashed box. (C) Quantitative analysis of pausing patterns between the wild-type and AA-mutant. Different intensity peaks refer to different pausing states as described in Fig. 3. The major difference was highlighted in the green dashed box.
SI Materials and Methods
RNA Pol II Purification.
Wild-type RNA pol II used for transcription was purified from Saccharomyces cerevisiae, as previously described unless otherwise stated (51, 52). The AA-mutant was introduced into a protease-deficient yeast strain GRY3175 by plasmid shuffle (56). Hexahistidine-tagged Rpb3 was used to pull down pol II from the whole cell lysate essentially as previously described (56, 57). In parallel, the wild-type RNA pol II used as comparison in the mutation study was also generated and purified via the same method as that for the AA-mutant. The DNA template and nontemplate oligonucleotides were purchased from IDT. RNA primers were purchased from TriLink Biotechnologies and radiolabeled using [γ-32P] ATP and T4 Polynucleotide Kinase (New England Biolabs).
DNA Template Design.
DNA sequences of template and nontemplate strand used for TS binding orientation were 5′-GACTCTTCTGACTTGGTCATACACACACTTCTCTCTCTCGTTGTTCCTCTCGATTGTTAAGTGATGTGTCGTTGTAAT-3′ and 5′-ATTACAACGACACATCACTTATGTCAGATCTACGCTC ACGAGAGAGAGAAGTGTGTGTATGACCAAGTCAGAAGAGTC-3′, and the binding site for compounds 1–4 is underlined. Sequences of template and nontemplate strand used for NTS binding orientation are 5′-GACTCTTCTGACTTGACCATACACACACTTCTCTCTCTCGTTGTTCCTCTCGATTGTTAAGTGATGTGTCGTTGTAAT-3′ and 5′-ATTACAACGACACATCACTTATGTCAGATCTACGCTC ACGAGAGAGAGAAGTGTGTGTATGGTCAAGTCAGAAGAGTC-3′, and the binding site for compounds 1–4 is underlined. The single mutation was introduced in both TS binding orientation and NTS binding orientation. The single mutated template strand for the TS binding orientation is 5′-GACTCTTCTGACTTGTTCATACACACACTTCTCTCTCTCGTTGTTCCTCTCGATTGTTAAGTGATGTGTCGTTGTAAT-3′ and the corresponding nontemplate strand is 5′-ATTACAACGACACATCACTTATGTCAGATCTACGCTCACGAGAGAGAGAAGTGTGTGTA TGAACAAGTCAGAAGAGTC-3′, and the mutated binding site is underlined. The single mutated template and nontemplate strand for the template strand for NTS binding orientation are 5′-GACTCTTCTGACTTGAACATACACACACTTCTCTCTCTCGTTGTTCCTCTCGATTGTTAAGTGATGTGTCGTTGTAAT-3′ and 5′-ATTACAACGACACATCACTTATGTCAGATCTACGCTCACGAGAGAGAGAAGTGTGTGT ATGTTCAAGTCAGAAGAGTC-3′, and the mutated binding site is underlined. The RNA primer used in all these scaffolds was 5′-AUCGAGAGGA-3′.
RNA Pol II Elongation Complex Assembly with a Full Transcription Bubble.
The pol II elongation complexes in the full transcription bubble were assembled based on previous reported methods with slight modifications (7, 58). Briefly, an aliquot of 5′-32P-labeled RNA was annealed with a 1.2-fold amount of template DNA and 1.5-fold amount of nontemplate DNA to form the RNA/DNA bubble scaffold in elongation buffer [20 mM Tris⋅HCl (pH 7.5), 40 mM KCl, 5 mM MgCl2]. An aliquot of annealed scaffold of RNA/DNA was then incubated with a fourfold excess amount of pol II at room temperature for 10 min to ensure the formation of a pol II elongation complex.
In Vitro RNA Pol II Transcription Assay.
The assay was carried out as previously described (7, 58–60). Briefly, the preincubated polymerase/scaffold complex was mixed with an equal volume of solution containing 40 mM KCl, 20 mM Tris⋅HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, and 2 mM nucleotide triphosphate (NTP). Final reaction concentrations after mixing were 25 nM scaffold, 100 nM pol II, 5 mM MgCl2, and 1 mM NTP in elongation buffer unless stated differently somewhere else. Reactions were quenched at various times by addition of one volume of 0.5 M EDTA (pH 8.0). For the TFIIS treatment, the elongation complex is mixed with equal volume of solution containing 40 mM KCl, 20 mM Tris⋅HCl (pH 7.5), 10 mM DTT, 10 mM MgCl2, 2 mM NTP, and 2 µM TFIIS. Final reaction concentrations after mixing were 25 nM scaffold, 100 nM pol II, 5 mM MgCl2, 1 mM NTP, and 1 µM TFIIS in the elongation buffer. The transcript was analyzed by 12% (wt/vol) denatured PAGE.
Transcription Inhibition Assay by Py-Im Polyamide.
For the Py-Im polyamide transcription inhibition assay, the RNA/DNA scaffold was preformed and incubated with various Py-Im polyamides for 3 h before the addition of pol II. The in vitro transcription was performed as described above. For the concentration titration assays, the final concentration of elongation complex is 2 nM.
Ki values for these compounds were determined by fitting the curves of the percentage of pol II transcriptional full-length bypass in the presence of the Py-Im polyamide with concentrations of the Py-Im polyamide in a log scale. The percentage of bypass was calculated by comparing the full-length transcript with the actively elongated transcript. Relative transcriptional bypass percentage was normalized against the maximal percentage of full-length transcript in the absence of ligand. Inhibition curves were determined using GraphPad Prism 6 by variable-slope, nonlinear regression fit to a dose–response model with a bottom constraint of 0 and a top constraint of 100.
Py-Im Polyamide Synthesis.
Py-Im polyamides 1, 2, and 3 were synthesized by microwave-assisted, solid-phase synthesis according to previously described protocols (54). Boc protected N-methylpyrrole and N-methylimidazole monomers were attached on oxime resin (Peptides International) using published PyBOP coupling conditions. All hairpin polyamides were cleaved from resin with 3,3′-diamino-N-methyldipropylamine, precipitated with Et2O, and coupled to isophthalic acid before purification by reverse phase HPLC. Polyamide 4 was synthesized on 2-chlorotrityl chloride resin (Bachem) using FmocPyOH and FmocImOH monomers (Wako), as previously reported (55). Purity and identity of compounds were verified by analytical HPLC and MALDI-TOF mass spectrometry.
MALDI-TOF characterization for polyamides 1–4 are summarized as follows:
-
i)
MALDI-TOF [M + H]+ calculated for C64H76N23O12+ = 1358.6, observed = 1358.9
-
ii)
MALDI-TOF [M + H] + calculated for C66H77N23O13+ = 1399.6, observed = 1400.7
-
iii)
MALDI-TOF [M + H] + calculated for C64H76N23O12+ = 1358.6, observed = 1358.9
-
iv)
MALDI-TOF [M + H] + calculated for C53H61N22O10+ = 1165.5, observed = 1165.5.
Thermal Denaturation Analysis.
DNA thermal stabilization analysis of polyamides 1–4 was performed on a thermos-controlled cell holder equipped Varian Cary 100 spectrophotometer. The match oligo (5′-GACT TGGTCA TACA-3′) and mismatch oligo (5′-GACT TGTTCA TACA -3′) were purchased from Valuegene. DNA duplexes and polyamides were added to a final concentration of 2 µM and 3 µM, respectively, to a degassed solution of 10 mM sodium cacodylate, 10 mM KCl, 10 mM MgCl2, and 5 mM CaCl2 at pH 7.0, as previously described (53). Denaturation profiles were recorded at λ = 260 nm from 25 °C to 95 °C with a heating rate of 0.5 °C/min. Thermal stabilization (ΔTm) by polyamides was calculated relative to naked control oligonucleotides. Results are reported as an average of three experiments.
Molecular Modeling.
The 3D model of Py-Im polyamide 1 was oriented from the crystal structure of cyclic Py-Im polyamide inhibitor with DNA (PDB ID code 3I5L) (31), and modified by Chemdraw3D software. The DNA sequence was mutated according to the Py-Im polyamide 1-binding sequence with COOT (61). The docking of ligand with the DNA-binding site was optimized to get the Py-Im polyamide 1-DNA complex using the software ICM-browser-pro (23, 62). To get the register of the first pol II pausing state (state i), part of Py-Im polyamide 1-DNA complex (from the 1st base to the 10th base of Chain A) was first aligned with the downstream NTS DNA region of the pol II complex (from i+5 site to i+14 site) (PDB ID code 3M3Y). Then, the portion of downstream DNA duplex of pol II elongation complex (from i+5 to i+14) was substituted by the superimposed 10 bp Py-Im polyamide 1-binding DNA duplex structure and extended with the ideal B-form DNA duplex downstream (beyond i+15 site) added with COOT. Finally, the upstream DNA and RNA sequences were modified accordingly to generate the model of the arrested state i. To mimic the stepwise pol II progression toward Py-Im polyamide binding site, we aligned and substituted a portion of downstream DNA duplex of pol II elongation complex with the superimposed 10 bp Py-Im polyamide 1-binding DNA duplex structure to generate the other three arrested states (from i+4 site to i+13 site for state ii, from i+3 site to i+12 site for state iii, from i+2 site to i+11 site for state iv, respectively) in a similar manner. All structure figures were rendered in PyMOL (www.pymol.org).
Discussion
Minor-Groove Sensor Motif Signals Transcription Pause and Blockage.
Although extensive mechanistic studies have been focused on how pol II recognizes and processes covalent DNA lesions (such as UV damage), or bypasses nucleosome barriers, less is known about the molecular mechanism of how pol II processes noncovalent DNA-binding small molecules. In this study, we investigated the effect of site-specific binding by Py-Im polyamides on pol II elongation in two binding orientations using biochemical assays, modeling, and mutation studies.
Our data from these studies revealed that an elongating pol II can sense polyamide binding, and pause as a stable complex immediately upstream of the binding site for over 20 h. This event is sequence-specific, where a single base pair mutation in the polyamide target sequence leads to the abolition of pol II arrest (Fig. 2). We further show the persistent pol II arrest induced by Py-Im polyamides cannot be rescued by TFIIS. This finding is remarkable given the fact that pol II itself has a strong capability to unwind the downstream DNA duplex to form the transcription bubble. This result is also in sharp contrast with the fact that TFIIS is able to rescue nucleosome-induced pol II arrest and a variety of pol II pausing/arrest events (13, 16, 40), considering that the binding affinity of histone–DNA interaction is very strong, within the similar nanomolar range as polyamides (41, 42). This observation may be explained by the kinetics of Py-Im polyamide DNA binding; the rate of dissociation is slow, comparable to DNA binding proteins, but the rate of association is much faster (43). Hence, the resistance to the effect of TFIIS may be attributed to the long-lifetime bound state of Py-Im polyamides during transcriptional elongation. These results support the idea that polyamides can block pol II elongation in a sequence-specific manner.
Furthermore, we find that substitutions of the functional moieties of Py-Im polyamides cause varied inhibitory potencies toward pol II elongation with Ki values in the 1- to 100-nM range (Table 1). Intriguingly, the magnitude of inhibitory effects does not correlate directly with the polyamides’ DNA binding affinities. This difference may reflect the difference between the aqueous solution where DNA binding affinity is measured and the amino acid-rich environment of the pol II downstream main channel where transcription inhibition is measured (Fig. S7).
Additionally, our results revealed that pol II senses polyamide binding in the minor groove via its conserved Switch 1 region and becomes stalled 2–5 bp upstream. The early polyamide-induced blockage of pol II elongation (n-4, n-5) is because of a steric clash with minor groove residues Arg1386 and His1387 during transcriptional progression toward the binding site (before formation of the transcriptional bubble), whereas the latter arrest (n-2, n-3) is attributed by direct prevention of DNA unwinding (Figs. 3 and 4 and Fig. S7).
Biological Implications and Consequences.
Unlike DNA damaging agents, such as cisplatin, that covalently modify DNA in a global manner, Py-Im polyamides bind to the DNA minor groove noncovalently in a sequence-specific manner. Previous studies have indicated that polyamides can inhibit pol II elongation and trigger the degradation of Rpb1 in vivo (35). Our studies provide a mechanistic explanation of these observations. We found that polyamide-induced pol II arrest is stable for hours and cannot be rescued by TFIIS. Such prolonged stalled pol II is likely a target for ubiquitination and degradation in a similar manner to pol II stalled by cisplatin-induced DNA damage, and subsequent apoptosis (44, 45). The molecular processing by pol II transcription machinery may provide important insights into the understanding of how cells respond to this kind of noncovalent DNA minor groove binder in vivo. Given the potent sequence-specific inhibition by polyamide interference, and the demonstrated antitumor activity in xenografts with low host toxicity, it would be very attractive to consider whether there is a role in cancer therapy for these molecules. Transcription inhibition has been explored as a therapeutic strategy in cancer treatment (46–49). Inhibition of a ubiquitous and essential pathway in cancer could overcome, in part, the drug resistance conferred through the genetic heterogeneity of the disease. Indeed, a recent study has shown that many cancers have a concomitant hemizygous loss of POLR2A (RPB1) and p53, rendering them more vulnerable to transcription inhibition (49). Development of small molecules that selectively obstruct the pol II transcription machinery in a sequence-specific manner would present a therapeutic strategy to target transcription addiction in cancer cells.
Finally, this study reveals a sensor function of the Switch 1 region to detect downstream DNA minor groove variations during pol II elongation. A highly conserved motif in pol II (Arg1386 and His1387 of Rpb1) has an inspective role within the DNA minor groove and can sense DNA minor groove binders. During transcription elongation, this motif likely interrogates the downstream duplex DNA before the formation of the transcription bubble. This motif and our previously identified major groove “epi-loop” (50) both exhibit DNA-sensing roles. Working together, these motifs can detect abnormalities in the downstream DNA template from both the major and minor grooves, which may play important roles in terms of controlling transcriptional fidelity and DNA damage recognition. Future studies may aim to reveal the function roles of this motif in the normal pol II transcription process, which may possibly unveil new insights into RNA pol II transcription regulation.
Materials and Methods
RNA Pol II Purification and in Vitro Transcription Assay.
Wild-type RNA pol II was purified from Saccharomyces cerevisiae as previously described, unless otherwise stated (51, 52). For the transcription inhibition assay, the RNA/DNA scaffold is preformed and incubated with various Py-Im polyamides for 3 h before the addition of pol II.
DNA Template Design.
DNA sequences of template strand used for TS and NTS binding orientation were 5′-GACTCTTCTGACTTGGTCATACACACACTTCTCTCTCTCGTTGTTCCTCTCGATTGTTAAGTGATGTGTCGTTGTAAT-3′ and 5′-GACTCTTCTGACTTGACCATACACACACTTCTCTCTCTCGTTGTTCCTCTCGATTGTTAAGTGATGTGTCGTTGTAAT-3′ (1–4 binding site is underlined).
Py-Im Polyamide Synthesis.
Compounds 1–4 were synthesized on solid support as previously described (53–55). All compounds were purified by RP-HPLC and correct masses were verified by MALDI-TOF.
Additional experimental details including RNA pol II assembly, DNA template design, Py-Im polyamide synthesis, and molecular modeling can be found in SI Materials and Methods.
Acknowledgments
This work was supported in part by NIH Grants GM102362 (to D.W.) and GM27681 (to P.B.D.); D.G. and J.N.S. were supported in part by the Intramural Research Program of the NIH, National Cancer Institute, and the Center for Cancer Research; and A.A.H. was supported in part by a postdoctoral Fellowship F32CA173977 from the NIH.
Footnotes
Conflict of interest statement: P.B.D. and F.Y. own shares in Gene Sciences, Inc., a biotechnology company that is developing therapeutics for prostate cancer.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612745113/-/DCSupplemental.
References
- 1.Cheung AC, Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149(7):1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Svetlov V, Nudler E. Basic mechanism of transcription by RNA polymerase II. Biochim Biophys Acta. 2013;1829(1):20–28. doi: 10.1016/j.bbagrm.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, Bushnell DA, Kornberg RD. RNA polymerase II transcription: structure and mechanism. Biochim Biophys Acta. 2013;1829(1):2–8. doi: 10.1016/j.bbagrm.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript elongation by RNA polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 5.Dedrick RL, Kane CM, Chamberlin MJ. Purified RNA polymerase II recognizes specific termination sites during transcription in vitro. J Biol Chem. 1987;262(19):9098–9108. [PubMed] [Google Scholar]
- 6.Kerppola TK, Kane CM. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19(8):831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, et al. Impact of template backbone heterogeneity on RNA polymerase II transcription. Nucleic Acids Res. 2015;43(4):2232–2241. doi: 10.1093/nar/gkv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tornaletti S, Patrick SM, Turchi JJ, Hanawalt PC. Behavior of T7 RNA polymerase and mammalian RNA polymerase II at site-specific cisplatin adducts in the template DNA. J Biol Chem. 2003;278(37):35791–35797. doi: 10.1074/jbc.M305394200. [DOI] [PubMed] [Google Scholar]
- 10.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315(5813):859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 11.Hardenbol P, Van Dyke MW. In vitro inhibition of c-myc transcription by mithramycin. Biochem Biophys Res Commun. 1992;185(2):553–558. doi: 10.1016/0006-291x(92)91660-i. [DOI] [PubMed] [Google Scholar]
- 12.Mote J, Jr, Ghanouni P, Reines D. A DNA minor groove-binding ligand both potentiates and arrests transcription by RNA polymerase II. Elongation factor SII enables readthrough at arrest sites. J Mol Biol. 1994;236(3):725–737. doi: 10.1006/jmbi.1994.1185. [DOI] [PubMed] [Google Scholar]
- 13.Kireeva ML, et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18(1):97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 14.Saeki H, Svejstrup JQ. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol Cell. 2009;35(2):191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA polymerase II collision interrupts convergent transcription. Mol Cell. 2012;48(3):365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlet-Berguerand N, et al. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25(23):5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LH. DNA damage response and transcription. DNA Repair (Amst) 2011;10(7):743–750. doi: 10.1016/j.dnarep.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 19.Walmacq C, et al. Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions. Proc Natl Acad Sci USA. 2015;112(5):E410–E419. doi: 10.1073/pnas.1415186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damsma GE, Cramer P. Molecular basis of transcriptional mutagenesis at 8-oxoguanine. J Biol Chem. 2009;284(46):31658–31663. doi: 10.1074/jbc.M109.022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cline SD, Riggins JN, Tornaletti S, Marnett LJ, Hanawalt PC. Malondialdehyde adducts in DNA arrest transcription by T7 RNA polymerase and mammalian RNA polymerase II. Proc Natl Acad Sci USA. 2004;101(19):7275–7280. doi: 10.1073/pnas.0402252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung Y, Lippard SJ. RNA polymerase II blockage by cisplatin-damaged DNA. Stability and polyubiquitylation of stalled polymerase. J Biol Chem. 2006;281(3):1361–1370. doi: 10.1074/jbc.M509688200. [DOI] [PubMed] [Google Scholar]
- 23.Wang D, Zhu G, Huang X, Lippard SJ. X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct. Proc Natl Acad Sci USA. 2010;107(21):9584–9589. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellinger MW, Park GY, Chong J, Lippard SJ, Wang D. Effect of a monofunctional phenanthriplatin-DNA adduct on RNA polymerase II transcriptional fidelity and translesion synthesis. J Am Chem Soc. 2013;135(35):13054–13061. doi: 10.1021/ja405475y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Regulation of gene expression by small molecules. Nature. 1997;387(6629):202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 26.Dickinson LA, et al. Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc Natl Acad Sci USA. 1998;95(22):12890–12895. doi: 10.1073/pnas.95.22.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majumder P, et al. Minor groove binder distamycin remodels chromatin but inhibits transcription. PLoS One. 2013;8(2):e57693. doi: 10.1371/journal.pone.0057693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trauger JW, Baird EE, Dervan PB. Recognition of DNA by designed ligands at subnanomolar concentrations. Nature. 1996;382(6591):559–561. doi: 10.1038/382559a0. [DOI] [PubMed] [Google Scholar]
- 29.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391(6666):468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 30.Kielkopf CL, et al. A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science. 1998;282(5386):111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- 31.Chenoweth DM, Dervan PB. Allosteric modulation of DNA by small molecules. Proc Natl Acad Sci USA. 2009;106(32):13175–13179. doi: 10.1073/pnas.0906532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu CF, et al. Completion of a programmable DNA-binding small molecule library. Tetrahedron. 2007;63(27):6146–6151. doi: 10.1016/j.tet.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc Natl Acad Sci USA. 2007;104(25):10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muzikar KA, Nickols NG, Dervan PB. Repression of DNA-binding dependent glucocorticoid receptor-mediated gene expression. Proc Natl Acad Sci USA. 2009;106(39):16598–16603. doi: 10.1073/pnas.0909192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang F, et al. Antitumor activity of a pyrrole-imidazole polyamide. Proc Natl Acad Sci USA. 2013;110(5):1863–1868. doi: 10.1073/pnas.1222035110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edelson BS, et al. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res. 2004;32(9):2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belotserkovskii BP, Mirkin SM, Hanawalt PC. DNA sequences that interfere with transcription: Implications for genome function and stability. Chem Rev. 2013;113(11):8620–8637. doi: 10.1021/cr400078y. [DOI] [PubMed] [Google Scholar]
- 38.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292(5523):1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 39.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292(5523):1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 40.Kuraoka I, et al. RNA polymerase II bypasses 8-oxoguanine in the presence of transcription elongation factor TFIIS. DNA Repair (Amst) 2007;6(6):841–851. doi: 10.1016/j.dnarep.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Cotton RW, Hamkalo BA. Nucleosome dissociation at physiological ionic strengths. Nucleic Acids Res. 1981;9(2):445–457. doi: 10.1093/nar/9.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ausio J, Seger D, Eisenberg H. Nucleosome core particle stability and conformational change. Effect of temperature, particle and NaCl concentrations, and crosslinking of histone H3 sulfhydryl groups. J Mol Biol. 1984;176(1):77–104. doi: 10.1016/0022-2836(84)90383-8. [DOI] [PubMed] [Google Scholar]
- 43.Baliga R, et al. Kinetic consequences of covalent linkage of DNA binding polyamides. Biochemistry. 2001;40(1):3–8. doi: 10.1021/bi0022339. [DOI] [PubMed] [Google Scholar]
- 44.Somesh BP, et al. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121(6):913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Wilson MD, Harreman M, Svejstrup JQ. Ubiquitylation and degradation of elongating RNA polymerase II: The last resort. Biochim Biophys Acta. 2013;1829(1):151–157. doi: 10.1016/j.bbagrm.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Stellrecht CM, Chen LS. Transcription inhibition as a therapeutic target for cancer. Cancers (Basel) 2011;3(4):4170–4190. doi: 10.3390/cancers3044170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511(7511):616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell. 2015;163(1):174–186. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520(7549):697–701. doi: 10.1038/nature14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, et al. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature. 2015;523(7562):621–625. doi: 10.1038/nature14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127(5):941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324(5931):1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dose C, Farkas ME, Chenoweth DM, Dervan PB. Next generation hairpin polyamides with (R)-3,4-diaminobutyric acid turn unit. J Am Chem Soc. 2008;130(21):6859–6866. doi: 10.1021/ja800888d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puckett JW, Green JT, Dervan PB. Microwave assisted synthesis of Py-Im polyamides. Org Lett. 2012;14(11):2774–2777. doi: 10.1021/ol3010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li BC, Montgomery DC, Puckett JW, Dervan PB. Synthesis of cyclic Py-Im polyamide libraries. J Org Chem. 2013;78(1):124–133. doi: 10.1021/jo302053v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irvin JD, et al. A genetic assay for transcription errors reveals multilayer control of RNA polymerase II fidelity. PLoS Genet. 2014;10(9):e1004532. doi: 10.1371/journal.pgen.1004532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kireeva ML, Lubkowska L, Komissarova N, Kashlev M. Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 2003;370:138–155. doi: 10.1016/S0076-6879(03)70012-3. [DOI] [PubMed] [Google Scholar]
- 58.Kellinger MW, Ulrich S, Chong J, Kool ET, Wang D. Dissecting chemical interactions governing RNA polymerase II transcriptional fidelity. J Am Chem Soc. 2012;134(19):8231–8240. doi: 10.1021/ja302077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu L, Plouffe SW, Chong J, Wengel J, Wang D. A chemical perspective on transcriptional fidelity: Dominant contributions of sugar integrity revealed by unlocked nucleic acids. Angew Chem Int Ed Engl. 2013;52(47):12341–12345. doi: 10.1002/anie.201307661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu L, et al. Strand-specific (asymmetric) contribution of phosphodiester linkages on RNA polymerase II transcriptional efficiency and fidelity. Proc Natl Acad Sci USA. 2014;111(32):E3269–E3276. doi: 10.1073/pnas.1406234111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Abagyan R, Totrov M, Kuznetsov D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15:488–506. [Google Scholar]