Abstract
Triethylene glycol dimethacrylate (TEGDMA) is a diluent monomer used pervasively in dental composite resins. Through hydrolytic degradation of the composites in the oral cavity it yields a hydrophilic biodegradation product, triethylene glycol (TEG), which has been shown to promote the growth of Streptococcus mutans, a dominant cariogenic bacterium. Previously it was shown that TEG up-regulated gtfB, an important gene contributing to polysaccharide synthesis function in biofilms. However, molecular mechanisms related to TEG’s effect on bacterial function remained poorly understood. In the present study, S. mutans UA159 was incubated with clinically relevant concentrations of TEG at pH 5.5 and 7.0. Quantitative real-time PCR, proteomics analysis, and glucosyltransferase enzyme (GTF) activity measurements were employed to identify the bacterial phenotypic response to TEG. A S. mutans vicK isogenic mutant (SMΔvicK1) and its associated complemented strain (SMΔvicK1C), an important regulatory gene for biofilm-associated genes, were used to determine if this signaling pathway was involved in modulation of the S. mutans virulence-associated genes. Extracted proteins from S. mutans biofilms grown in the presence and absence of TEG were subjected to mass spectrometry for protein identification, characterization and quantification. TEG up-regulated gtfB/C, gbpB, comC, comD and comE more significantly in biofilms at cariogenic pH (5.5) and defined concentrations. Differential response of the vicK knock-out (SMΔvicK1) and complemented strains (SMΔvicK1C) implicated this signalling pathway in TEG-modulated cellular responses. TEG resulted in increased GTF enzyme activity, responsible for synthesizing insoluble glucans involved in the formation of cariogenic biofilms. As well, TEG increased protein abundance related to biofilm formation, carbohydrate transport, acid tolerance, and stress-response. Proteomics data was consistent with gene expression findings for the selected genes. These findings demonstrate a mechanistic pathway by which TEG derived from commercial resin materials in the oral cavity promote S. mutans pathogenicity, which is typically associated with secondary caries.
Introduction
Over the past few decades, resin composites have been widely used as dental restorative materials. This is due to their superior aesthetics, excellent adhesive strength to dentin and enamel, minimal intervention approaches to restore the posterior teeth and concern related to possible adverse effects of mercury released from dental amalgam [1]. Since their development in the 1960s, the basic properties of resin composites such as mechanical, physical and bonding properties have been remarkably improved. However, a number of clinical studies have reported higher failure rates, increased frequency of replacement and shorter longevity for composite restorations compared to amalgams [1–6]. One of the main reasons for resin composite restoration failure is secondary or recurrent caries [1–3, 5, 7–10]. Furthermore, the result of two most recent systematic reviews suggested that resin composite restorations in posterior teeth have less longevity and a higher number of secondary caries when compared to amalgam restorations [11, 12]. Based on one of these systematic reviews, the incidence of secondary caries around amalgams varied between 0% and 4.9%, but composite restorations tend to exhibit markedly more secondary caries with incidences varying between 0% and 12.7% [12]. These findings necessitate more fundamental research to unravel all underlying causes promoting secondary caries, as premature replacement of resin composite restorations due to secondary caries imposes a tremendous burden on health care expenditure [13]. The total cost of dental restoration is approximately $46 billion/year in the U.S.A and 70% of this cost is related to the replacement of failed restorations [14, 15]. Failed composite restorations are responsible for more than half of all dental restorations [16].
The polymeric matrix of commonly used composite resin restorations typically contain a viscous dominant hydrophobic monomer, bis-phenyl glycidyl dimethacrylate (BisGMA), as well as dilutive hydrophilic monomer such as triethylene glycol dimethacrylate (TEGDMA) [17]. While TEGDMA has many advantages such as rapid conversion and setting in the oral cavity and ease of manipulation [17], it is highly susceptible to hydrolytic biodegradation, catalyzed by human and bacterial esterases [18, 19]. The reason for this susceptibility is the presence of an unprotected ester linkage within its structure. The degradation of TEGDMA results in the production of a biodegradation by-product called tri-ethylene-glycol (TEG) [20–24].
The degradation process contributes to the deterioration of the interface and ingress of cariogenic microorganisms such as Streptococcus mutans that can potentially result in the development of secondary caries [25]. S. mutans is one of the leading species associated with human dental caries and is considered to be the most cariogenic of all the oral streptococci [26]. A previous study by the authors reported that TEG can up-regulate the expression of a virulence gene, gtfB, which encodes a glucosyltransfrerase (GTF) enzyme in S. mutans NG8. TEG up-regulated gtfB in both planktonic and biofilm cells and at both cariogenic (5.5) and neutral (7.0) pH [27]. However, the downstream impact of this upregulation on molecular pathways that control metabolism, acid production, potential for biofilm formation, and other cell functions remained unknown. Despite the limited time frame in which TEGDMA is being released from new resin composite restorations [23], most studies to date have focused on the interactions of this monomer with biological systems in the oral cavity, investigating the cytotoxic, genotoxic and estrogenic effects on both mammalian and bacterial cells. Yet, there are only very limited studies that examined the biological effects of the degradation by-product from this monomer, TEG, which is released over a long term, throughout the life of the restoration [23].
In the present study, we used a well-characterized reference strain of S. mutans, UA159, with its fully annotated genome sequence available to enable a more thorough investigation of individual gene expression analysis and the synthesis of its proteins [28]. S. mutans biofilms were exposed to clinically-relevant concentrations of TEG (0.001–1.0 mM) [29] at cariogenic and neutral pH.
The objectives of the present study were: i) to elucidate the effects of TEG on the expression of known S. mutans virulence genes, i.e. gtfB, gtfC, gbpB, comC, comD, comE and atpH using qRT-PCR; ii) to identify potential molecular pathway(s) involved in those effects; iii) to quantify global protein synthesis and identify other molecular pathways that were affected by interaction of TEG with the bacteria using proteomics, and finally; iv) to measure the effect of TEG on S. mutans GTF enzyme activity, associated with biofilm formation, to confirm that the effect of TEG on gtfB and gtfC expression could result in increased production of water-insoluble exopolysaccharide glucan, a known virulence factor in S. mutans [30]. The seven selected genes in this study have unique virulence properties in S. mutans and have been linked to its cariogenicity by previous preclinical and clinical studies [31–38].
S. mutans isogenic mutants deleted in a key regulatory gene, vicK, (SMΔvicK1) and its associated complemented strain (SMΔvicK1C) were used to identify the molecular pathway(s) involved in the TEG-mediated cellular effect. VicRK is one of the 14 two-component signal transduction systems (TCSTSs) in S. mutans [28] that regulates the comCDE system, gtfB, gtfC, gbpB expression and acid tolerance in S. mutans [39–43]. TCSTSs in S. mutans respond to environmental stimuli by transmitting signals from a membrane-associated histidine kinase (HK) to an intracellular response regulator (RR) protein via transphosphorylation, which in turn, regulates the transcription of its target genes [44]. Therefore, the current study investigated whether the effects of TEG on S. mutans gene expression are regulated by the vicRK system.
To the best of the authors’ knowledge, the current investigation provides the first proteomics analysis of cariogenic S. mutans UA159 after exposure to the dental composite degradation product TEG. The implications of the work are significant given that TEGDMA is pervasively used in oral restorations around the world.
Materials & Methods
Bacterial Strain and Growth Conditions
The S. mutans UA159 wild-type strain was obtained from Dr. Arnold Bleiweis (University of Florida) and stored in 20% glycerol at -80°C. To construct the vicK-deficient mutant (SMΔvicK1), a PCR-ligation mutagenesis strategy was used as previously described [45, 46]. The vicK complemented strain (SMΔvicK1C) was made using pIB166 to replace the vicK gene with an erm resistant gene cassette by a double crossover recombination event. Then a constitutive vector pIB166 harboring the full length vicK gene between BamHI and XhoI cutting sites was transformed into a vicK-deletion mutant to generate the complemented strain. The primers used for the vicK deletion and complementation constructs are listed in Table 1 (Operon, AL, USA). The S. mutans wild-type strain was sub-cultured on a Todd-Hewitt agar plate supplemented by 0.3% yeast extract (THYE) (BBL; Becton Dickenson, Cockeysville, MD, USA), whereas the vicK mutant was maintained on a THYE agar containing 10 μg/mL of erythromycin [45]. All S. mutans overnight cultures were routinely grown in THYE broth at 37°C in a 5% CO2−95% air mixtures.
Table 1. Primers used for construction of vicK knock-out and complemented strains.
| Primer use and name | Nucleotide Sequence |
|---|---|
| Primers for vicK deletion: | |
| VicK-P1 | GTTACCAGAACTAGACGGTCTTGAAG |
| VicK-P2 | GG^CGCGCCTTAGTCATATGATTTCATGTAATAACCAAC |
| VicK-P3 | GGCCGG^CCATGACAGAAACAGGTTTTAGATAC |
| VicK-P4 | AAACTCCAAGGGTAGATACCTG |
| Erm cst-F | GG^CGCGCCCCGGGCCCAAAATTTGTTTGAT |
| Erm cst-B | GGCCGG^CCAGTCGGCAGCGACTCATAGAAT |
| Primers for VicK complementation: | |
| VicK-F | 5' ggattcATGACTAATGTGTTTGAATCAAG 3' |
| VicK-R | 5' ctcgagTCATGATTCGTCTTCATCTTC 3' |
Preparation of Planktonic Cells
Overnight cultures of S. mutans UA159 were diluted (1/10) in TYEG medium containing tryptone (10 g/L), yeast extract (5 g/L) and glucose (5 mM) buffered either at pH 5.5 with 100 mM MES (2-(N-Morpholino)-ethanesulfonic acid, Sigma-Aldrich, St. Louis, MO, USA) or at pH 7.0 with 100 mM MOPS (3-(N-Morpholino) propanesulfonic acid, Bioshop, Burlington, ON, Canada) to control pH, supplemented with 0.1% glucose and 500 μL of the appropriate amount of sterile filtered TEG (99.9% pure, Sigma-Aldrich) yielding final concentrations of 0, 0.001, 0.01, 0.1 mM TEG in the solutions [29]. The culture tubes were incubated for 18 hours (37°C, 5% CO2). The pH of sample cultures was verified (H135 minilabTM pro, HACH, Germany). Bacterial cell density was monitored using a UV spectrophotometer (Ultraspec 3000, Biotech) at 600 nm in order to ensure that the cultures were harvested once they reached mid-logarithmic phase (optical density or OD = 0.4). This was followed by centrifugation at 2300 g for 10 minutes (4°C). The supernatants were discarded and the pellets were snap frozen in liquid nitrogen and stored at -80°C until required for RNA isolation.
Preparation of Biofilm Cells
Overnight cultures of S. mutans UA159 were diluted (1:60) in TYEG medium and added to six-well polystyrene microtiter plates (Fisher Scientific). Each well containing 3 mL of ¼ strength TYEG medium buffered to pH 5.5 or 7.0 and 50 μL of overnight culture were supplemented with 0.1% glucose and 500 μL of TEG stock solutions as described for planktonic conditions above. Cells were then incubated for 18 hours (37°C, 5% CO2) after which the pH of sample cultures was measured. Then, the liquid contents were removed and 3 mL of phosphate-buffered saline (PBS: 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4 and 0.24 g KH2PO4 dissolved in distilled water to make a 1 L solution, pH 7.4) was slowly added to each well and gently stirred to remove loosely attached cells, leaving only adhered biofilm cells. The remaining PBS was then removed and replaced with 1 mL of fresh PBS. Biofilm cells from each well were scraped and the resulting cell suspensions were transferred to 50 mL tubes and centrifuged at 2300 g for 10 minutes (4°C). The supernatants of each sample were then removed/discarded and the pellets were snap frozen in liquid nitrogen and stored at -80°C until required for RNA isolation.
Gene Expression Analysis Using qRT-PCR
Total RNA was isolated by disruption of S. mutans UA159 cells (Thermo Savant, Fast-Prep FP 101), followed by DNase treatment of the RNA samples and cDNA synthesis as described before [27]. QRT-PCR was used to quantify the relative gene expression of selected genes: gtfB, gtfC, gbpB, comC, comD, comE and atpH as previously described [27]. The primers used are presented in Table 2 (Operon, AL, USA). Quantitative gene expression data were then normalized to the 16S rRNA, a well-established housekeeping gene [47]. The level of 16S rRNA message was not affected by TEG within the concentration range studied (data not shown). QRT-PCR gene expression analysis was also employed to investigate the involvement of the vicRK system in regulation of the S. mutans virulence genes of interest in the presence TEG. Using vicK knock-out (SMΔvicK1) and complemented (SMΔvicK1C) strains, gene expression analysis was repeated for three representative genes, gtfB (biofilm formation), comD (quorum sensing) and atpH (acid tolerance), encompassing different functional roles within the virulence character. For statistical analysis, one-way analysis of variance (ANOVA) and Tukey post hoc analyses were performed to determine changes in gene expression between different concentrations of TEG and the no-TEG control within each growth mode (p<0.05). Two-way ANOVA and Tukey post-hoc analyses were conducted to validate differences in gene expression between growth modes (biofilm vs. planktonic) at the same concentration (p<0.05). A 2-fold or greater change in gene expression (up-regulation) and ≤ 0.5-fold (down-regulation) with a P-value cut-off of < 0.05 were considered physiologically significant [48]. All qRT-PCR reactions were run in triplicate for each experimental condition and the experiments were reproduced four separate times using four different overnight cultures of S. mutans UA159.
Table 2. Nucleotide sequence of primers used for qRT-PCR.
| Gene | Description | Primer Sequence (5'-3') | |
|---|---|---|---|
| Forward | Reverse | ||
| gtfB | GTF I, glucan production | ACACTTTCGGGTGGCTTG | GCTTAGATGTCACTTCGGTTG |
| gtfC | GTF II, glucan production | CCAAAATGGTATTATGGCTGTCG | GAGTCTCTATCAAAGTAACGCAGT |
| gbpB | Glucan binding protein | AGCAACAGAAGCACAACCATCAG | CCACCATTACCCCAGTAGTTTCC |
| comC | Competence-stimulating peptide | GACTTTAAAGAAATTAAGACTG | AAGCTTGTGTAAAACTTCTGT |
| comD | Two-component regulatory system | CTCTGATTGACCATTCTTCTGG | CATTCTGAGTTTATGCCCCTC |
| comE | Two-component regulatory system | CCTGAAAAGGGCAATCACCAG | GGGGCATAAACTCAGAATGTGTCG |
| atpH | Acid tolerance | ACCATACATTTCAGGCTG | TTTTAGCACTTGGGATTG |
| 16SrRNA | Normalizing internal standard | CTTACCAGGTCTTGACATCCCG | ACCCAACATCTCACGACACGAG |
Sample Preparation for Mass Spectrometry
Biofilms from both experimental (0.01, 0.1 and 1.0 mM) and control (no TEG) groups were washed twice in cold PBS and re-suspended in 1 mL of PBS buffer. The cells were disrupted using a homogenizer (Thermo Savant, FastPrep FP 101) for 45 minute and then centrifuged at 15700 x g for 1 minute. Supernatant were carefully removed, separated in aliquots of 50 μL and stored at -80°C. The total protein concentration in each sample was assessed by the micro bicinchoninic acid (Micro BCA) assay [49]. Equal amounts of protein (20 μg) from both experimental and control groups were dried by a rotary evaporator, denatured and reduced for 2 hours by the addition of 200 μL of 4 M urea, 10 mM dithiothreitol (DTT), and 50 mM NH4HCO3, pH 7.8. After four-fold dilution with 50 mM NH4HCO3, pH 7.8, tryptic digestion was carried out for 18 h at 37°C, following the addition of 2% (w/w) sequencing-grade trypsin (Promega, Madison, WI, USA).
Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS) and Relative Proteome Quantitation
Peptide separation and mass spectrometric analyses were carried out as described previously [49]. The MS/MS spectra were analysed against the streptococcal protein database (Swiss Prot and TrEMBL, Swiss Institute of Bioinformatics, Geneva, Switzerland, http://ca.expasy.org/sprot/) using SEQUEST algorithm in Proteome Discoverer 1.3 software (Thermo Scientific, San Jose, CA, USA) [49].
For quantitative proteome analysis, three MS raw files from each group (control and experimental groups) were analyzed using SIEVE software (Version 2.0 Thermo Scientific, San Jose, CA, USA). For the alignment step, a single MS raw file belonging to the control group (no TEG) was selected as the reference file and all of the other files were adjusted to generate the best correlation to this reference file. After alignment, the feature detection and integration (or framing) process was performed using the MS level data. For statistical analyses of protein abundance, peak integrations were summarized into protein-level annotation in SIEVE using a weighted average of intensities for the LC-ESIMS/MS for each protein. In addition, a statistical model based on an ANOVA framework with Tukey’s post hoc test was carried out. All study groups were run 3 separate times to increase coverage of the samples and identify more proteins, with 4 independent samples (using 4 independent S. mutans cultures) in each group. Relative abundance of an individual protein from different TEG concentration groups was considered significantly different from the control group (no TEG) when the values observed were < 0.5 for decreased abundance and >1.5 for increased abundance with a P-value cut-off of < 0.05 [49, 50].
GTF Enzyme Activity Assay
GTF enzyme activity, related to the cleavage of sucrose into fructose and glucose and then added to the growing exopolysaccharides, was measured by determining the rate that [14C]-sucrose was converted to glucan polymers, [51]. Briefly, S. mutans UA159 biofilms were grown in a 6-well polystyrene microtiter plate containing ¼ TYEG medium buffered to pH 5.5. Defined amounts of TEG were added to the medium to yield the target final concentrations (0.00, 0.01, 0.1 and 1.0 mM). Overnight cultures were added to the mixture and incubated for 18 hours. Bacterial cells were collected, transferred to 50 mL tubes and pelleted by centrifugation at 2300 X g at 4°C for 10 minutes. The supernatant was then removed/discarded and the pellets were washed twice in cold PBS and re-suspended in 1 mL of PBS buffer. The cells were disrupted using a homogenizer (Thermo Savant, FastPrep FP 101) for 45 min and then centrifuged at 15,700 X g for 1 minute. Different aliquots of the supernatant were carefully removed and stored separately at -80°C. Total protein concentration in each sample was assessed by the micro bicinchoninic acid (Micro BCA) assay. Fifteen μg of protein from both the experimental and control groups was added to 0.2 M potassium phosphate buffer (pH 6.8) for a total volume of 20 μL, this solution was mixed with 20 μL of 14C-radiolabelled reaction buffer containing 0.2 M KPO4 (pH6.8), 20 mM sucrose, and 10 μL/mL 14C-sucrose (24.4 GBq/mmol; Amersham). The mixture was incubated at 37°C for 60 minutes after which the reaction mixtures were adsorbed onto 25mm filters (0.22 μm GVWP; Millipore). The samples were then air dried for 20 minutes and washed three times with 2 mL distilled water to remove the water-soluble glucan and serve only as water-insoluble glucan samples. The water-insoluble glucan, mainly synthesized by GtfB and GtfC enzymes. The samples were then placed in 5 mL scintillation fluid (ScintiSafe Econo 2 Cocktail; Fisher) and synthesized [14C]-glucan was measured using a liquid scintillation counter (Beckman LS6500). One way ANOVA and Tukey post hoc analyses were used to determine significance in GTF activity.
Results
Effects of TEG on S. mutans UA159 Gene Expression at pH 5.5 and 7.0
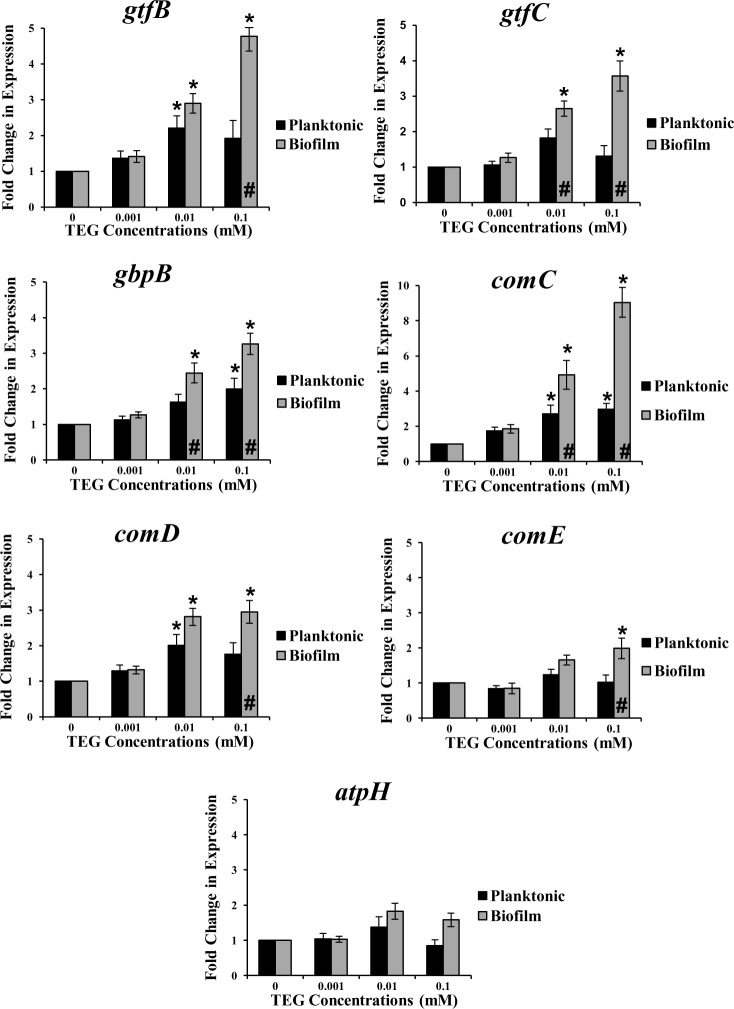
Exposure of S. mutans UA159 to different concentrations of TEG at pH 5.5 significantly increased the expression of the four virulence genes, gtfB, gbpB, comC and comD at the concentration of 0.01 and/or 0.1 mM of TEG in both planktonic and biofilm cultures when compared to the no TEG control (P<0.05) (Fig 1). The fold change expression for the biofilm cultures was higher than the planktonic cells and was different for each gene, ranging from 2- to 9-fold (P<0.001). GtfC and comE were only up-regulated in biofilms (p<0.05) at 0.01 and 0.1 mM for gtfC and 0.1 mM for comE, respectively. AtpH, was not affected by TEG at any concentration in either growth mode (Fig 1). In contrast, at pH 7.0, expression of the selected genes in biofilms was not affected by TEG at any concentration, but, gtfB, gbpB, comC and comE were significantly (P<0.05) up-regulated in planktonic cultures at 0.01 mM TEG, and comD was up-regulated at both 0.01 and 0.1 mM (p<0.05) (Fig 2). The fold change levels were substantially lower at pH 7.0 when compared to the pH 5.5. Similarly to pH 5.5, atpH was not affected by TEG in either growth modes (Fig 2). Based on these findings, all genes were up-regulated more significantly in biofilms at cariogenic pH (5.5) in the presence of TEG. Hence, the remaining experiments, including knock-out and complemented experiments, proteomics and GTF enzyme activity, were conducted only with biofilms under acidic pH (5.5). Since 0.001 mM TEG had no effect on S. mutans virulence gene expression, this concentration was dropped from the remaining studies and a new higher concentration, 1.0 mM, was added to several of the subsequent studies [29].
Fig 1. Relative expression of the S. mutans virulence genes: gtfB, gtfC, gbpB, comC, comD, comE and atpH for planktonic and biofilm growth conditions with different concentrations of TEG (0.001, 0.01 and 0.1 mM) at pH 5.5 relative to the no TEG control.
* represents significant difference between individual TEG concentrations compared to the control (no TEG) in either growth mode (P<0.05). # Represents significant difference between biofilm and planktonic cultures at the same TEG concentration (P<0.001). Data are plotted with standard error of the mean (±SE), n = 4.
Fig 2. Relative expression of the S. mutans virulence genes: gtfB, gtfC, gbpB, comC, comD, comE and atpH for planktonic and biofilm growth conditions with different concentrations of TEG (0.001, 0.01 and 0.1 mM) at pH 7.0 relative to the no TEG control.
* represents significant difference between individual TEG concentrations compared to the control (no TEG) in either growth mode (P<0.05). # represents significant difference between biofilm and planktonic cultures at the same TEG concentration (P<0.001). Data are plotted with standard error of the mean (±SE), n = 4.
Effects of TEG on S. mutans UA159 vicK Knock-Out and Complemented Strains’ Gene Expression
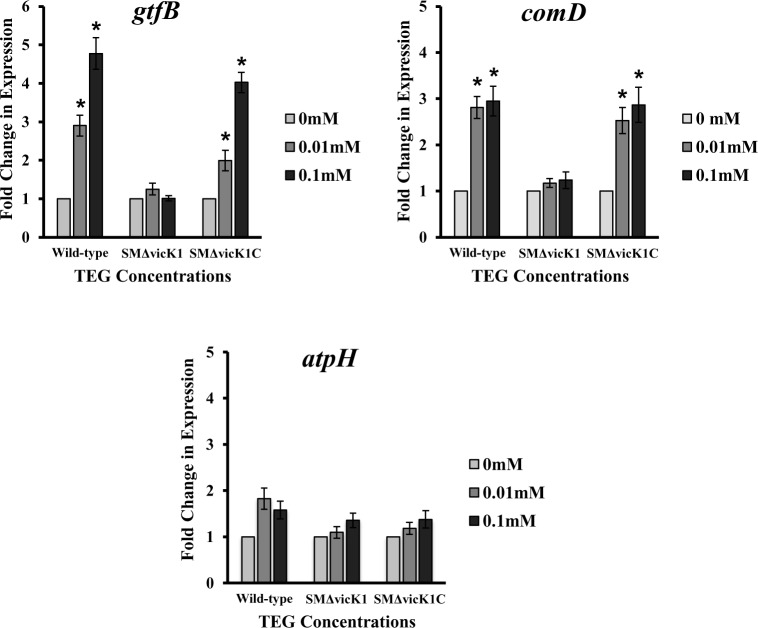
Exposure of S. mutans UA159 vicK knock-out strain (SMΔvicK1) to different concentrations of TEG resulted in no significant change (P>0.05) in the expression level for any of the selected virulence genes relative to the no TEG control. Data are presented for three representative genes, gtfB (implicated in biofilm formation), comD (associated with quorum sensing) and atpH (important to acid tolerance) in Fig 3. Exposure of S. mutans UA159 vicK complemented strain (SMΔvicK1C) to different concentrations of TEG resulted in a similar expression pattern of the aforementioned three representative genes to that of the wild-type strain (Fig 3). Both gtfB and comD were up-regulated by TEG at the same concentrations as the parent strain of 0.01 & 0.1 mM, while atpH was not affected by TEG at any concentration similar to what was observed for the wild-type strain (Fig 3).
Fig 3. Relative expression of gtfB, comD and atpH in wild-type, knock-out (SMΔvicK1) and complemented (SMΔvicK1C) vicK strains of S. mutans UA159 in the presence of different concentrations of TEG (0.01 and 0.1 mM) at pH 5.5.
One-way analysis of variance (ANOVA) and Tukey post hoc analyses were performed to determine the differences in gene expression between individual TEG concentration and the no-TEG control (P<0.05). Expression of the related genes in complemented strain was similar to that of wild-type. Data are plotted with standard error of the mean (±SE), n = 4.
S. mutans Global Proteome Analysis in Response to TEG
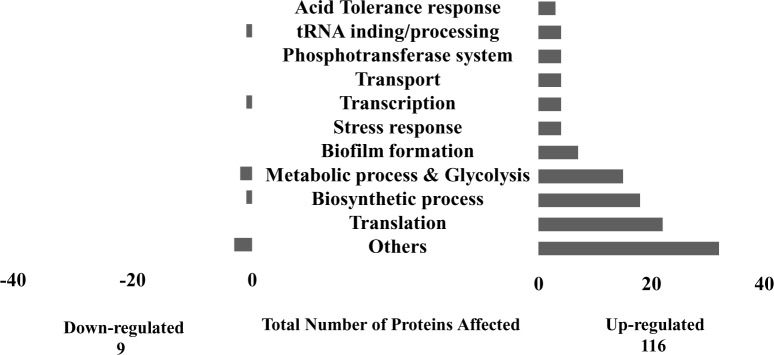
The base-peak chromatogram for reversed-phase chromatography monitored by the mass spectrometer for all concentrations of TEG showed a consistent elution of protein/peptides in the range of 20 to 50 min (S1 Fig). A total number of 314 proteins have been identified (S1 Table), of which 125 proteins were differentially expressed in S. mutans biofilm grown in the presence of TEG, among which 116 proteins had enhanced expression (≥1.5 fold) and 9 proteins had diminished expression (≤ 0.5) fold compared to the control. The first step in the quantitative proteomic analysis by SIEVE was to promote an alignment of all mass spectrometry chromatograms (S1 Fig). Then the correlation coefficient score values were acquired for each mass spectrometric chromatogram and mean score values were calculated for each group. In the current study, one of the chromatograms from the no-TEG group was selected as the benchmark. All other chromatograms were compared with this chromatogram. The correlation coefficient mean values were 0.901 for 0.01 mM TEG group, 0.895 for 0.1 mM TEG group and, 0.841 for 1.0 mM TEG group. Interestingly, it was observed that by increasing the TEG concentration in each sample group, the alignment value (correlation coefficient value) became more distant from the default chromatogram group (S1 Fig). This observation suggested a change in quantity and quality of protein/peptides in a dose-dependent manner relative to the TEG concentrations, which means both number and amount of the proteins as wells as their time of elution were changing by increasing TEG concentrations. The distribution of proteins identified by gene ontology (GO) predicted and categorized a specific biological function for each protein and showed that the majority of proteins were associated with important functions such as energy metabolism, amino acid biosynthesis, transcription, translation, transport and binding (Fig 4). The organization was performed via proteomics website http://www.uniprot.org [52].
Fig 4. Distribution of S.mutans UA159 proteins into specific gene ontology (GO) of biological processes aftter exposure to TEG.
A total number of 125 proteins were differentially expressed in S. mutans biofilm grown in the presence of different TEG concentrations (0.01, 0.1 and 1.0 mM), among which 116 proteins were more abundant and 9 proteins were less abundant compared to the no-TEG control.
Exposure of S. mutans UA159 to different TEG concentrations resulted in a significantly higher (P<0.05) abundance of proteins involved in biofilm formation, specifically GtfB, GtfC, GbpB, ComD, ComE and SpaP (Table 3). As well, there was a TEG effect on proteins involved in acid-tolerance, which are different subunits of the F1F0-ATPase proton translocating pump (ATPA, subunit α, ATPD, subunit β and AtpH, subunit C) (Table 4). The findings also identified TEG effects on proteins involved in stress response (DnaK, RecA, GroL and ClpX) (Table 5), and proteins encoding the Phosphoenolpyruvate: phosphotransferase sugar transport system (PEP:PTS), including: ptsI encodes EI; mtlF encodes EII; SMU1879 and SMU1960c encodes EIID man and EIIB man respectively (Table 6). Further, another sixty-six individual proteins differing in abundance by two-fold or more including sixteen proteins involved in metabolic processes and glycolysis, five proteins involved in transcription, five proteins involved in RNA binding and processing, twenty-two proteins involved in translation, nineteen proteins involved in amino acid biosynthesis, four proteins involved in transport, and finally thirty-four proteins with their functions either uncharacterized or that did not belong to any identified categories (Tables A-G in S1 File).
Table 3. Biofilm-related proteins from S. mutans UA159 grown in the presence of TEG at pH 5.5.
| Gene name | Protein name | Protein function | Ratio 1mM/0mM | P | Ratio 0.1mM/0mM | P | Ratio 0.01mM/0mM | P |
|---|---|---|---|---|---|---|---|---|
| gtfB | GtfB | Glucan biosynthetic process | 2.34* | 0.01 | 1.57* | 0.01 | 2.54* | 0.01 |
| gtfC | GtfC | Glucan biosynthetic process | 1.55* | 0.01 | 1.41 | 0.12 | 0.79 | 0.07 |
| gbpB | GbpB | single-species biofilm formation on inanimate substrate | 1.54* | 0.01 | 1.09 | 0.09 | 0.65 | 0.85 |
| comC | ComC | Multiorganism process, biological adhesion | 1.12 | 0.02 | 1.10 | 0.03 | 0.99 | 0.02 |
| comD | ComD | Single-organism process, cellular process, metabolic process | 0.90 | 0.06 | 1.90* | 0.04 | 0.99 | 0.04 |
| comE | ComE | signaling,biological regulation, single-organism process | 1.76* | 0.01 | 1.87 | 0.06 | 2.40* | 0.05 |
| spaP | Cell-surface antigen I/II | (Surface protein antigen implicated in dental caries) | 1.92* | 0.05 | 0.61 | 0.02 | 0.69 | 0.01 |
*Represents P<0.05
Table 4. Acid tolerance-response proteins from S. mutans UA159 grown in the presence of TEG at pH 5.5.
| Gene name | Protein name | Protein function | Ratio 1mM/0mM | P | Ratio 0.1mM/0mM | P | Ratio 0.01mM/0mM | P |
|---|---|---|---|---|---|---|---|---|
| atpA | ATP synthase F1, alpha subunit | ATP hydrolysis coupled proton transport,plasma membrane ATP synthesis coupled proton transport | 0.91 | 0.05 | 1.94* | 0.01 | 0.62 | 0.01 |
| atpD | ATP synthase F1, beta subunit | ATP hydrolysis coupled proton transport,plasma membrane ATP synthesis coupled proton transport | 0.96 | 0.01 | 1.58* | 0.05 | 0.53 | 0.02 |
| atpH | F1F0 membrane-bound proton-translocating ATPase c subunit | hydrogen ion transmembrane transporter activity | 2.13* | 0.02 | 1.57 | 0.13 | 2.56 | 0.21 |
*Represents P<0.05
Table 5. Stress-response proteins from S. mutans UA159 grown in the presence of TEG at pH 5.5.
| Gene name | Protein name | Protein function | Ratio 1mM/0mM | P | Ratio 0.1mM/0mM | P | Ratio 0.01mM/0mM | P |
|---|---|---|---|---|---|---|---|---|
| DnaK | Chaperone protein DnaK | protein folding,response to stress | 1.30 | 0.92 | 1.65* | 0.005 | 2.20* | 0.05 |
| recA | Protein RecA | DNA recombination, DNA repair, SOS response | 1.98* | 0.02 | 1.71* | 0.028 | 1.67* | 0.01 |
| groL | 60 kDa chaperonin | protein refolding | 1.67* | 0.01 | 1.82* | 0.000 | 2.36* | 0.04 |
| clpX | ATP-dependent Clp protease, ATP-binding subunit | protein folding | 2.15* | 0.01 | 1.61 | 0.088 | 2.07* | 0.01 |
*Represents P<0.05
Table 6. Carbohydrate transport proteins from S. mutans UA159 grown in the presence of TEG at pH5.5.
| Gene name | Protein name | Protein function | Ratio 1mM/0mM | P | Ratio 0.1mM/0mM | P | Ratio 0.01mM/0mM | P |
|---|---|---|---|---|---|---|---|---|
| ptsI | Phosphoenolpyruvate-protein phosphotransferase | PEP:PTS Core phosphotransfer protein | 2.09* | 0.01 | 2.10* | 0.05 | 1.43 | 0.01 |
| mtlF | Phosphotransferase system enzyme II | PEP:PTS permease | 1.79* | 0.02 | 1.84* | 0.02 | 2.21* | 0.01 |
| SMU_1879 | Mannose-specific phosphotransferase system component IID | PEP:PTS permease/regulatory protein | 1.99* | 0.01 | 1.99* | 0.01 | 0.86 | 0.04 |
| SMU_1960c | PTS system protein, mannose-specific IIB component | PEP:PTS regulatory protein | 1.84* | 0.01 | 1.85* | 0.01 | 2.36* | 0.01 |
*Represents P<0.05
Effects of TEG on GTF Enzyme Activity
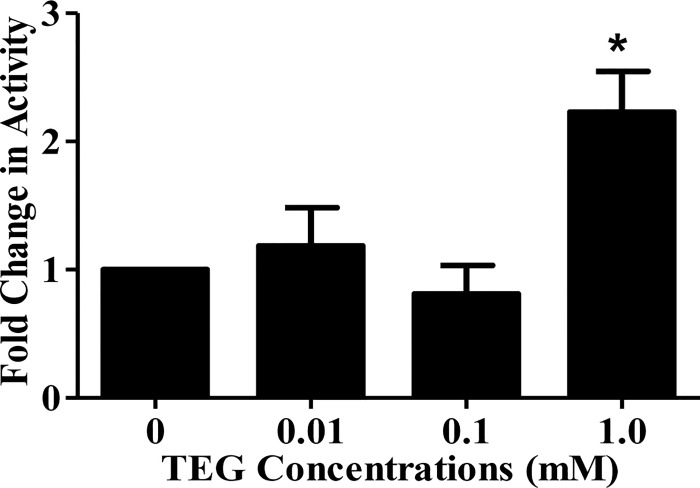
The total amount of insoluble glucan synthesized by S. mutans UA159 cell-associated GTF grown in the presence or absence of different concentrations of TEG was significantly higher P<0.05) for cells grown in the presence of 1.0 mM TEG vs. no TEG (Fig 5). There was no effect related to the 0.01 and 0.1 mM concentrations of TEG on GTF enzyme activity (Fig 5).
Fig 5. Effects of different concentrations of TEG (0.01, 0.1 and 1.0 mM) on S. mutans UA159 glucosyltransferase activity.
The total amount of insoluble glucan synthesized by biofilm cells grown in the presence of 1 mM TEG was significantly increased compared to the no-TEG control (P<0.05). Data are plotted with standard error of the mean (±SE), n = 4.
Discussion
The findings of the present study suggest that the degradation product from the most common diluent resin composite monomer, TEGDMA, in concentrations relevant to in vivo conditions [29], may contribute to S. mutans cariogenic potential by up-regulating several virulence genes/related proteins and activating specific adaptation mechanisms such as biofilm formation, acid tolerance, optimization/modulation of the PEP:PTS system, and stress response pathways. At the cariogenic pH of 5.5, up-regulation of gtfB, gtfC, gbpB, comC, comD and comE was more significant for biofilms when compared to the planktonic cells. These are relevant findings since both biofilm and pH 5.5 are conditions for development of dental caries in vivo. The data also provided further insight into the molecular pathways involved in S. mutans response to the hydrophilic TEGDMA derived degradation product, TEG. This was emphasized by evaluating the role of a two-component signal transduction system sensor protein encoded by vicK, which has been shown to have a regulatory control on the comCDE system and also influences gtfB, gtfC, gbpB expression and acid tolerance in S. mutans [40–43].
TEG is a small molecule with a low molecular weight of 150.17 g/mol and is the ether glycol portion of the TEGDMA monomer. It is characterised by two hydroxyl groups along with two ether linkages, which contribute to its high water solubility, hygroscopicity, solvent properties and reactivity with many organic compounds [53]. Due to its lipophilic nature, TEGDMA, the precursor of TEG, can easily penetrate the cytosol and lipid compartment of mammalian cells [54, 55]. There have been a few studies regarding the metabolism of polyethylene glycol monomers in bacteria, indicating the oxidation of these monomers to an aldehyde and carboxylic acid by aldehyde or alcohol dehydrogenase, followed by cleavage of ether bond resulting in a polyglycol molecules that are reduced by one glycol unit [54, 56, 57].
TEG has been used for a variety of applications in industry including natural gas dehydration and as a humectant, solvent, and chemical intermediate in the synthesis of resins, plasticizers, lubricants and polyurethanes [53]. In the 1940s TEG vapor or mist was first introduced for disinfection purposes, especially in hospital wards [58]. A few studies have shown TEG to be bactericidal for Beta hemolytic Streptococcus, Streptococcus pneumoniae, Streptococcus pyogenes, Influenza A virus and Penicillium notatum fungi [59–64]. It was also found that concentrated polyethylene glycol 400 (PEG 400) solutions have antibacterial activity against several pathogenic bacteria including Klebsiella pneumoniae, Pseudomonas aeruginosa, Escherichia coli, and Staphylococcus aureus [65].
Although these studies indicated that TEG had some bactericidal activity towards several bacterial species, most of the works have only demonstrated the antimicrobial activity of TEG in its vapor form against airborne, solution suspended, and surface bound microbes rather than cariogenic bacterial biofilms such as S. mutans. In addition, higher dosages of bactericide are needed to eradicate bacteria existing in biofilms compared to their planktonic counterparts due to the protective nature of biofilms [66–69]. The in vivo-relevant concentrations of TEG that have been used in the current study (0.001–1 mM) are far from the concentrations that were used in cases where TEG demonstrated antibacterial activity against non-cariogenic bacteria (e.g. Robertson et al. 250 mM or Chirife et al. 4000 mM) [62, 65]. Further, Khalichi et al. [70] reported that TEG at the concentration range of 0.5–10.0 mM significantly stimulated the growth of two S. mutans strains (NG8 and JH1005) only at pH 5.5. These findings suggest that the effect of TEG on bacteria is dosage- and pH- dependant and varies among different bacterial species.
It has been shown that anaerobic bacteria are capable of degrading glycol monomers using oligo- and polyethylene glycols and ether alcohols as their carbon source especially in minimum media [56, 71]. In addition, the small size and hydrophilic nature of TEG can facilitate its mobility within the biofilms’ exopolysaccharide matrix, activating TCSTs and the resultant gene expression response. The knock-out and complemented strain experiments in the present study, strongly suggest that exposure of S. mutans biofilms to TEG results in up-regulation of virulence-associated genes via vicRK signalling pathway.
The more pronounced up-regulation of the virulence-associated genes at acidic pH vs. neutral pH by TEG could be related to the fact that low pH (5.5) triggers changes in the bacterial cell membrane fatty acid composition that can affect membrane permeability [72] and would results in even easier penetration of TEG from the environment. Further, TEG exhibits the properties of a weak acid (pKa of 15.12), therefore, its undissociated form at acidic pH might facilitate its penetration into the bacterial membrane compared to neutral pH [73, 74], where it can further react and degrade, affecting various signaling pathways such as stress response, carbohydrates transport and acid tolerance. The greater effect of TEG on biofilms vs. planktonic growth mode can be explained by the close proximity of the cells in the former growth mode, shortening and enhancing the response time to BBP, as well as making the effective concentration of the BBP higher due to a diffusion-barrier effect of the biofilm [12].
Among the S. mutans virulence attributes, the ability of this bacterium to produce extracellular polysaccharide from dietary carbohydrates has been demonstrated to significantly enhance its cariogenicity [30]. GtfB and gtfC encode the enzymes glucosyltransferase-B (GTF-I) and-C (GTF-SI) respectively, which are responsible for the synthesis of water-insoluble glucans that function in the adhesion and accumulation of the bacteria on the tooth surfaces. Mutant strains of S. mutans defective in gtf genes, especially gtfB and gtfC, are far less cariogenic than the parent strains in vivo [38]. Oral bacterial aggregation is also mediated by interactions between surface-associated glucan-binding proteins (GBPs) that adhere to glucans, thereby promoting plaque formation [39, 75]. GbpB is also associated with S. mutans virulence because immunization with this protein mediates protection against dental caries in animal models [76]. In addition, GbpB appears to play an essential function in S. mutans viability and has also been described as a stress-response protein [77]. Hence, the upregulation of these genes by TEG suggests that there may be an increase in the virulence of the bacteria.
The proteomics findings demonstrated the enhanced expression of biofilm-related proteins in the presence of TEG in S. mutans biofilms at cariogenic pH (5.5). Up-regulation of gtfB, gtfC and gbpB and their corresponding proteins by TEG could have a significant impact on S. mutans cariogenicity by making thicker and stickier biofilm and potentially contributes in resin composite restoration failure [78–80].
The gene expression analysis demonstrated the greatest change in fold expression for comC with a nine-fold increase in mRNA levels in the presence of 0.1 mM TEG when compared to the no-TEG control in biofilm grown cells at pH 5.5. In addition, comD and comE were up-regulated by TEG at 0.01 and/or 0.1 mM in biofilms at pH 5.5 relative to the control. In S. mutans comDE TCSTS together with comC, which encodes competence stimulating peptide (CSP), regulate cellular processes and many virulence factors including genetic transformation, biofilm formation, acidogenicity, aciduricity, cell viability and bacteriocin production in response to environmental cues [81]. Importantly, the proteomics findings regarding the biofilm-associated proteins, GtfB, GtfC, GbpB, ComC, ComD, ComE are in agreement with the gene expression results for the respective genes. The fold change in protein synthesis and gene expression was not at the same magnitude in all cases, however the trends were conserved. The discordance in specific dose responses could be due to differences in the sensitivity of the methods, post-translational modifications of proteins and the status of mRNA stability and degradation [80, 82, 83]. Of particular note, the presence of 1.0 mM TEG also increased the protein abundance of SpaP, which is the major S. mutans surface receptor (known as antigen I/II and wall-associated antigen), that contributes to sucrose-independent adhesion [28, 84–86]. SpaP was one of the first gene products linked to adherence of S. mutans to saliva coated surfaces [86].
A very significant finding in this work was the GTF enzyme activity results, which indicated that the total rate of insoluble glucan synthesis by S. mutans UA159 biofilms grown in the presence of 1.0 mM TEG was significantly higher (P<0.05) compared to the no-TEG control biofilms. These findings suggest that TEG can affect S. mutans virulence factors at three different molecular levels (mRNA, protein and enzyme activity), which can eventually result in production of thicker exopolysaccharide glucan as a major contributor in S. mutans virulence [78–80].
The findings of this study provide a mechanistic explanation to the role of biomaterials-bacterial interaction in the pathogenesis of secondary caries, by uncovering underlying signalling pathways, specifically the vicRK regulatory system and its dependence on TEG with respect to the S. mutans virulence genes expression. This TCSTS plays important roles in the regulation of genetic competence, biofilm formation, and acid tolerance by regulating comCDE system and biofilm associated genes, gtfB, gtfC, gbpB, in S. mutans [39–43].
Oral biofilms are regularly subjected to acid stress as a result of the accumulation of acidic end-products from bacterial carbohydrate metabolism. S. mutans, the major cariogenic bacteria in plaque, is adapted to function well at low pH values and its cariogenicity is closely related to its acid tolerance [87]. The membrane-bound F1F0-ATPase pump is the primary mechanism of proton extrusion in S. mutans. To maintain pH homeostasis, the F1F0-ATPase export H+ as the growth conditions become more acidic [88]. The proteomic data indicated that three subunits of F1F0-ATPase pump [AtpA (subunit α), AtpD (subunit β) and AtpH (subunit C)] had a significantly increased abundance of related proteins upon exposure to different concentrations of TEG. While AtpA and AtpD were induced by 0.1 mM TEG, AtpH was only induced at 1.0 mM TEG, which is in agreement with the gene expression analysis indicating no significant effect at lower concentrations of TEG (0.01 and 0.1 mM) for AtpH. On the other hand, both AtpA and AtpD were downregulated at 0.01 mM. The observed variances for the different protein products could reflect the heterogeneity of biofilm cells, which might have resulted in different response with different concentrations of TEG in some cases [50]. Such findings suggest that S. mutans may have the ability to enhance acid adaptation and survival after exposure to dental composite degradation product.
To cope with the environmental stresses, S. mutans has developed several strategies that allow it to grow under harsh conditions. For example S. mutans induces the expression of a group of proteins that are either chaperones or proteases [89]. Unfolded or misfolded proteins accumulate inside the cell due to stress, and molecular chaperones are required to proper folding and assembly of those aberrant proteins [89]. Proteases are involved in the degradation of the proteins, not only under stress conditions, but also under normal growth conditions [89]. Stress responsive proteins, DnaK, RecA, GroL and ClpX were among the proteins that were more abundant (by two-fold or greater) in at least one of the three TEG concentrations when compared to the no TEG control. The production of stress proteins including DnaK, RecA, GroL and ClpX is central to the tolerance of environmental insults by microorganisms [90]. DnaK is known to be induced in response to multiple stresses such as acid shock and carbon starvation [91]. Lowering the DnaK level results in impaired capacity of S. mutans to form biofilm in the presence of glucose [92]. In other work it has been shown that the recombinase A (RecA) protein is involved in the homologous recombination of GtfB and GtfC in S. mutans [93]. RecA is also contributor to late steps of competence development as well as biofilm formation and acid stress survival [87, 94]. Both DnaK and GroL can regulate signal transduction pathways by controlling the stability and activities of transcriptional regulators and protein kinases [95, 96].
CLP proteins are responsible for degrading proteins that cannot be folded by molecular chaperones [97, 98]. ClpX has been shown to modulate the expression of important virulence attributes of S. mutans including biofilm formation, viability and acid survival [99]. Expression of genes involved in biofilm formation, competence and mutacin production was down-regulated in ClpX deficient mutant strain [99]. The data in the current study suggests that ClpX presents a high level of abundance (more than two fold) when 0.01 and 1.0 mM of TEG were added to the bacterial cultures. Collectively, these findings suggest that TEG can enhance S. mutans ability to adapt to environmental stresses by modulating stress-response pathways in order to survive and persist in cariogenic biofilms.
The proteomics results also demonstrated that TEG has the ability to induce the expression of the PEP:PTS, which is the main carbohydrate transport system in oral streptococci [90]. Carbohydrate transport and metabolism can be used as fuel for the downstream energy generating pathways such as glycolysis [100]. Given the important role of the PEP:PTS system in S. mutans sugar transport, metabolism and global regulatory effect on gene expression, specifically controlling biofilm formation and acid tolerance [101, 102], up-regulation of different components of this system by TEG is a very significant finding.
In general, the findings indicate global changes in protein synthesis by S. mutans biofilms in response to the dental composite degradation product TEG, which may contribute to S. mutans successful fitness and establishment in diverse cariogenic biofilm. These findings may provide an explanation for the higher failure rate due to increased presence of secondary caries, and increased frequency of replacement of resin composite restorations compared to amalgam [103]. More importantly, the findings signal a call to the clinical community to insist that commercial manufacturers for resin composites exercise more due diligence on the biological characterization of their products and related degradation by-products, and to refrain from referring to current materials as biologically inert dental materials.
This in vitro study used only one of the multiple bacterial species found in dental plaque and growth conditions for biofilms were simplified, and as such, the data cannot reproduce all of the complex in vivo conditions of plaque. However, the findings of the current study provide much new insight into the potential mechanisms behind recurrent caries and contribution of resin composite degradation products to the failure of resin composite restorations. In addition, this simplified in vitro system can provide the opportunity to examine direct effect(s) of specific molecules such as TEG and other resin composite components and/or degradation products on bacterial physiological responses, independent of other complex interactions in vivo. Future studies could investigate the influence of the resin composite degradation products on the pathogenic potential of cariogenic oral bacteria in a multi-species biofilm under both in vitro and in vivo conditions.
Supporting Information
Blue (1.0 mM), red (0.1 mM), violet (0.01mM) and green (0.00 mM).
(TIF)
Tables A-G: Other differentially expressed proteins following exposure to TEG.
(PPTX)
(XLSX)
Acknowledgments
The authors thank Dr. Shida Wang, Kirsten Krastel, Yizhi Xiao and Eduardo B. Moffa, for their technical assistance. This investigation was funded by National Institute of Dental & Craniofacial Research R01DE021385; Canadian Institute of Health Research MOP115113, CIHR grant # 97577 and Canada Foundation for Innovation grant # 25116. This is an independent study free of conflict of interest.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Institute of Dental & Craniofacial Research R01DE021385 (YF); Canadian Institute of Health Research MOP115113 (YF); Canadian Institute of Health Research grant #97577 (WLS); and Canada Foundation for Innovation grant #25116 (WLS).
References
- 1.Kopperud SE, Tveit AB, Gaarden T, Sandvik L, Espelid I. Longevity of posterior dental restorations and reasons for failure. Eur J Oral Sci. 2012;120(6):539–48. 10.1111/eos.12004 [DOI] [PubMed] [Google Scholar]
- 2.Bernardo M, Luis H, Martin MD, Leroux BG, Rue T, Leitao J, et al. Survival and reasons for failure of amalgam versus composite posterior restorations placed in a randomized clinical trial. J Am Dent Assoc. 2007;138(6):775–83. [DOI] [PubMed] [Google Scholar]
- 3.Rho YJ, Namgung C, Jin BH, Lim BS, Cho BH. Longevity of direct restorations in stress-bearing posterior cavities: a retrospective study. Oper Dent. 2013;38(6):572–82. 10.2341/12-432-C [DOI] [PubMed] [Google Scholar]
- 4.Roumanas ED. The frequency of replacement of dental restorations may vary based on a number of variables, including type of material, size of the restoration, and caries risk of the patient. J Evid Based Dent Pract. 2010;10(1):23–4. 10.1016/j.jebdp.2009.11.009 [DOI] [PubMed] [Google Scholar]
- 5.Soncini JA, Maserejian NN, Trachtenberg F, Tavares M, Hayes C. The longevity of amalgam versus compomer/composite restorations in posterior primary and permanent teeth: findings From the New England Children's Amalgam Trial. J Am Dent Assoc. 2007;138(6):763–72. [DOI] [PubMed] [Google Scholar]
- 6.Sunnegardh-Gronberg K, van Dijken JW, Funegard U, Lindberg A, Nilsson M. Selection of dental materials and longevity of replaced restorations in Public Dental Health clinics in northern Sweden. Journal of dentistry. 2009;37(9):673–8. 10.1016/j.jdent.2009.04.010 [DOI] [PubMed] [Google Scholar]
- 7.Chrysanthakopoulos NA. Reasons for Placement and Replacement of Resin-based Composite Restorations in Greece. J Dent Res Dent Clin Dent Prospects. 2011;5(3):87–93. 10.5681/joddd.2011.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordan VV, Riley JL 3rd, Geraldeli S, Rindal DB, Qvist V, Fellows JL, et al. Repair or replacement of defective restorations by dentists in The Dental Practice-Based Research Network. J Am Dent Assoc. 2012;143(6):593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opdam NJ, Bronkhorst EM, Roeters JM, Loomans BA. A retrospective clinical study on longevity of posterior composite and amalgam restorations. Dent Mater. 2007;23(1):2–8. 10.1016/j.dental.2005.11.036 [DOI] [PubMed] [Google Scholar]
- 10.Palotie U, Vehkalahti MM. Reasons for replacement of restorations: dentists' perceptions. Acta Odontol Scand. 2012;70(6):485–90. 10.3109/00016357.2011.640274 [DOI] [PubMed] [Google Scholar]
- 11.Moraschini V, Fai CK, Alto RM, Dos Santos GO. Amalgam and resin composite longevity of posterior restorations: A systematic review and meta-analysis. Journal of dentistry. 2015;43(9):1043–50. 10.1016/j.jdent.2015.06.005 [DOI] [PubMed] [Google Scholar]
- 12.Nedeljkovic I, Teughels W, De Munck J, Van Meerbeek B, Van Landuyt KL. Is secondary caries with composites a material-based problem? Dent Mater. 2015;31(11):247–77. [DOI] [PubMed] [Google Scholar]
- 13.A.D.A. Survey of Dental Services Rendered. Chicago: the American Dental Association, ISBN 1-60122-027-8. 2007. [Google Scholar]
- 14.Beazoglou T, Eklund S, Heffley D, Meiers J, Brown LJ, Bailit H. Economic impact of regulating the use of amalgam restorations. Public Health Rep. 2007;122(5):657–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frost PM. An audit on the placement and replacement of restorations in a general dental practice. Primary dental care: journal of the Faculty of General Dental Practitioners. 2002;9(1):31–6. [DOI] [PubMed] [Google Scholar]
- 16.Simecek JW, Diefenderfer KE, Cohen ME. An evaluation of replacement rates for posterior resin-based composite and amalgam restorations in U.S. Navy and marine corps recruits. J Am Dent Assoc. 2009;140(2):200–9 [DOI] [PubMed] [Google Scholar]
- 17.Ferracane JL. Resin composite—state of the art. Dent Mater. 2011;27(1):29–38. 10.1016/j.dental.2010.10.020 [DOI] [PubMed] [Google Scholar]
- 18.Bourbia M, Ma D, Cvitkovitch DG, Santerre JP, Finer Y. Cariogenic bacteria degrade dental resin composites and adhesives. J Dent Res. 2013;92(11):989–94. 10.1177/0022034513504436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shokati B, Tam LE, Santerre JP, Finer Y. Effect of salivary esterase on the integrity and fracture toughness of the dentin-resin interface. J Biomed Mater Res B Appl Biomater. 2010;94(1):230–7. 10.1002/jbm.b.31645 [DOI] [PubMed] [Google Scholar]
- 20.Finer Y, Santerre JP. Biodegradation of a dental composite by esterases: dependence on enzyme concentration and specificity. Journal of biomaterials science Polymer edition. 2003;14(8):837–49. [DOI] [PubMed] [Google Scholar]
- 21.Finer Y, Santerre JP. Salivary esterase activity and its association with the biodegradation of dental composites. J Dent Res. 2004;83(1):22–6. [DOI] [PubMed] [Google Scholar]
- 22.Finer Y, Santerre JP. The influence of resin chemistry on a dental composite's biodegradation. J Biomed Mater Res A. 2004;69(2):233–46. 10.1002/jbm.a.30000 [DOI] [PubMed] [Google Scholar]
- 23.Finer Y, Santerre JP. Influence of silanated filler content on the biodegradation of bisGMA/TEGDMA dental composite resins. J Biomed Mater Res A. 2007;81(1):75–84. 10.1002/jbm.a.31004 [DOI] [PubMed] [Google Scholar]
- 24.Santerre JP, Shajii L, Leung BW. Relation of dental composite formulations to their degradation and the release of hydrolyzed polymeric-resin-derived products. Critical reviews in oral biology and medicine: an official publication of the American Association of Oral Biologists. 2001;12(2):136–51. [DOI] [PubMed] [Google Scholar]
- 25.Kermanshahi S, Santerre JP, Cvitkovitch DG, Finer Y. Biodegradation of resin-dentin interfaces increases bacterial microleakage. J Dent Res. 2010;89(9):996–1001. 10.1177/0022034510372885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50(4):353–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalichi P, Singh J, Cvitkovitch DG, Santerre JP. The influence of triethylene glycol derived from dental composite resins on the regulation of Streptococcus mutans gene expression. Biomaterials. 2009;30(4):452–9. 10.1016/j.biomaterials.2008.09.053 [DOI] [PubMed] [Google Scholar]
- 28.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(22):14434–9. 10.1073/pnas.172501299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaffer F, Finer Y, Santerre JP. Interactions between resin monomers and commercial composite resins with human saliva derived esterases. Biomaterials. 2002;23(7):1707–19. [DOI] [PubMed] [Google Scholar]
- 30.Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries research. 2011;45(1):69–86. 10.1159/000324598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li YH, Lau PC, Tang N, Svensater G, Ellen RP, Cvitkovitch DG. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol. 2002;184(22):6333–42. 10.1128/JB.184.22.6333-6342.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, et al. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002;184(10):2699–708. 10.1128/JB.184.10.2699-2708.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li YH, Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors. 2012;12(3):2519–38. 10.3390/s120302519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munro C, Michalek SM, Macrina FL. Cariogenicity of Streptococcus mutans V403 glucosyltransferase and fructosyltransferase mutants constructed by allelic exchange. Infection and immunity. 1991;59(7):2316–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroeder VA, Michalek SM, Macrina FL. Biochemical characterization and evaluation of virulence of a fructosyltransferase-deficient mutant of Streptococcus mutans V403. Infection and immunity. 1989;57(11):3560–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith DJ, Taubman MA. Experimental immunization of rats with a Streptococcus mutans 59-kilodalton glucan-binding protein protects against dental caries. Infection and immunity. 1996;64(8):3069–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vacca Smith AM, Scott-Anne KM, Whelehan MT, Berkowitz RJ, Feng C, Bowen WH. Salivary glucosyltransferase B as a possible marker for caries activity. Caries research. 2007;41(6):445–50. 10.1159/000107930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infection and immunity. 1993;61(9):3811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duque C, Stipp RN, Wang B, Smith DJ, Hofling JF, Kuramitsu HK, et al. Downregulation of GbpB, a component of the VicRK regulon, affects biofilm formation and cell surface characteristics of Streptococcus mutans. Infection and immunity. 2011;79(2):786–96. 10.1128/IAI.00725-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senadheera D, Krastel K, Mair R, Persadmehr A, Abranches J, Burne RA, et al. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J Bacteriol. 2009;191(20):6415–24. 10.1128/JB.00793-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senadheera DB, Cordova M, Ayala EA, Chavez de Paz LE, Singh K, Downey JS, et al. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol. 2012;194(6):1307–16. 10.1128/JB.06071-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, et al. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 2005;187(12):4064–76. 10.1128/JB.187.12.4064-4076.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senadheera MD, Lee AW, Hung DC, Spatafora GA, Goodman SD, Cvitkovitch DG. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J Bacteriol. 2007;189(4):1451–8. 10.1128/JB.01161-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annual review of biochemistry. 2000;69:183–215. 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- 45.Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. Journal of microbiological methods. 2002;49(2):193–205. [DOI] [PubMed] [Google Scholar]
- 46.Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;183(3):897–908. Epub 2001/02/24. 10.1128/JB.183.3.897-908.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stipp RN, Goncalves RB, Hofling JF, Smith DJ, Mattos-Graner RO. Transcriptional analysis of gtfB, gtfC, and gbpB and their putative response regulators in several isolates of Streptococcus mutans. Oral microbiology and immunology. 2008;23(6):466–73. 10.1111/j.1399-302X.2008.00451.x [DOI] [PubMed] [Google Scholar]
- 48.Shemesh M, Tam A, Steinberg D. Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology. 2007;153(Pt 5):1307–17. 10.1099/mic.0.2006/002030-0 [DOI] [PubMed] [Google Scholar]
- 49.Siqueira WL, Bakkal M, Xiao Y, Sutton JN, Mendes FM. Quantitative proteomic analysis of the effect of fluoride on the acquired enamel pellicle. PLoS One. 2012;7(8):e42204 10.1371/journal.pone.0042204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svensater G, Welin J, Wilkins JC, Beighton D, Hamilton IR. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS microbiology letters. 2001;205(1):139–46. [DOI] [PubMed] [Google Scholar]
- 51.Terao Y, Isoda R, Murakami J, Hamada S, Kawabata S. Molecular and biological characterization of gtf regulation-associated genes in Streptococcus mutans. Oral microbiology and immunology. 2009;24(3):211–7. 10.1111/j.1399-302X.2008.00497.x [DOI] [PubMed] [Google Scholar]
- 52.Len AC, Cordwell SJ, Harty DW, Jacques NA. Cellular and extracellular proteome analysis of Streptococcus mutans grown in a chemostat. Proteomics. 2003;3(5):627–46. 10.1002/pmic.200300391 [DOI] [PubMed] [Google Scholar]
- 53.Ballantyne B, Snellings WM. Triethylene glycol HO(CH2CH2O)3H. J Appl Toxicol. 2007;27(3):291–9. 10.1002/jat.1220 [DOI] [PubMed] [Google Scholar]
- 54.Engelmann J, Leyhausen G, Leibfritz D, Geurtsen W. Metabolic effects of dental resin components in vitro detected by NMR spectroscopy. J Dent Res. 2001;80(3):869–75. [DOI] [PubMed] [Google Scholar]
- 55.Geurtsen W, Leyhausen G. Chemical-Biological Interactions of the resin monomer triethyleneglycol-dimethacrylate (TEGDMA). J Dent Res. 2001;80(12):2046–50. [DOI] [PubMed] [Google Scholar]
- 56.Kawai F. Microbial degradation of polyethers. Appl Microbiol Biotechnol. 2002;58(1):30–8. [DOI] [PubMed] [Google Scholar]
- 57.Kawai F, Kimura T, Fukaya M, Tani Y, Ogata K, Ueno T, et al. Bacterial oxidation of polyethylene glycol. Applied and environmental microbiology. 1978;35(4):679–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolman A, Fair GM, Hardenbergh WA, Lawrence RE, Maxcy KF, Tiedeman WD, et al. Recent Studies on Disinfection of Air in Military Establishments. Am J Public Health Nations Health. 1947;37(2):189–98. [PMC free article] [PubMed] [Google Scholar]
- 59.Bigg E, Jennings BH, Olson FC. Epidemiologic Observations on the Use of Glycol Vapors for Air Sterilization. Am J Public Health Nations Health. 1945;35(8):788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamburger M Jr., Hurst V, et al. The effect of triethylene glycol vapor on airborne beta hemolytic streptococci in hospital wards; the action of glycol vapors at low relative humidities. J Infect Dis. 1945;77:177–80. [DOI] [PubMed] [Google Scholar]
- 61.Lester W Jr., Robertson OH, et al. The rate of bactericidal action of triethylene glycol vapor on microorganisms dispersed into the air in small droplets. Am J Hyg. 1949;50(2):175–88. [DOI] [PubMed] [Google Scholar]
- 62.Robertson OH, Appel EM, et al. A study of the bactericidal activity in vitro of certain glycols and closely related compounds. J Infect Dis. 1948;83(2):124–37. [DOI] [PubMed] [Google Scholar]
- 63.Robertson OH, Lester W Jr., Smith M. The lethal effect of triethylene glycol vapor on dried air-borne bacteria. Am J Hyg. 1951;53(1):69–79. [DOI] [PubMed] [Google Scholar]
- 64.Rosebury T, Meiklejohn G, Kingsland LC, Boldt MH. Disinfection of Clouds of Meningopneumonitis and Psittacosis Viruses with Triethylene Glycol Vapor. J Exp Med. 1947;85(1):65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chirife J, Herszage L, Joseph A, Bozzini JP, Leardini N, Kohn ES. In vitro antibacterial activity of concentrated polyethylene glycol 400 solutions. Antimicrob Agents Chemother. 1983;24(3):409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000;44(3):640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falsetta ML, Klein MI, Lemos JA, Silva BB, Agidi S, Scott-Anne KK, et al. Novel antibiofilm chemotherapy targets exopolysaccharide synthesis and stress tolerance in Streptococcus mutans to modulate virulence expression in vivo. Antimicrob Agents Chemother. 2012;56(12):6201–11. 10.1128/AAC.01381-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koo H, Hayacibara MF, Schobel BD, Cury JA, Rosalen PL, Park YK, et al. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. J Antimicrob Chemother. 2003;52(5):782–9. 10.1093/jac/dkg449 [DOI] [PubMed] [Google Scholar]
- 69.Koo H, Seils J, Abranches J, Burne RA, Bowen WH, Quivey RG Jr. Influence of apigenin on gtf gene expression in Streptococcus mutans UA159. Antimicrob Agents Chemother. 2006;50(2):542–6. 10.1128/AAC.50.2.542-546.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khalichi P, Cvitkovitch DG, Santerre JP. Effect of composite resin biodegradation products on oral streptococcal growth. Biomaterials. 2004;25(24):5467–72. 10.1016/j.biomaterials.2003.12.056 [DOI] [PubMed] [Google Scholar]
- 71.Fincher EL, Payne WJ. Bacterial utilization of ether glycols. Appl Microbiol. 1962;10:542–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fozo EM, Quivey RG Jr. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Applied and environmental microbiology. 2004;70(2):929–36. 10.1128/AEM.70.2.929-936.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casal M, Queiros O, Talaia G, Ribas D, Paiva S. Carboxylic Acids Plasma Membrane Transporters in Saccharomyces cerevisiae. Advances in experimental medicine and biology. 2016;892:229–51. 10.1007/978-3-319-25304-6_9 [DOI] [PubMed] [Google Scholar]
- 74.Ji C, Stockbridge RB, Miller C. Bacterial fluoride resistance, Fluc channels, and the weak acid accumulation effect. J Gen Physiol. 2014;144(3):257–61. 10.1085/jgp.201411243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujita K, Takashima Y, Inagaki S, Nagayama K, Nomura R, Ardin AC, et al. Correlation of biological properties with glucan-binding protein B expression profile in Streptococcus mutans clinical isolates. Archives of oral biology. 2011;56(3):258–63. 10.1016/j.archoralbio.2010.09.018 [DOI] [PubMed] [Google Scholar]
- 76.Smith DJ. Caries vaccines for the twenty-first century. J Dent Educ. 2003;67(10):1130–9. [PubMed] [Google Scholar]
- 77.Mattos-Graner RO, Porter KA, Smith DJ, Hosogi Y, Duncan MJ. Functional analysis of glucan binding protein B from Streptococcus mutans. J Bacteriol. 2006;188(11):3813–25. 10.1128/JB.01845-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klein MI, Xiao J, Lu B, Delahunty CM, Yates JR 3rd, Koo H. Streptococcus mutans protein synthesis during mixed-species biofilm development by high-throughput quantitative proteomics. PLoS One. 2012;7(9):45795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 2010;192(12):3024–32. 10.1128/JB.01649-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR 3rd, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8(4):1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cvitkovitch DG, Li YH, Ellen RP. Quorum sensing and biofilm formation in Streptococcal infections. The Journal of clinical investigation. 2003;112(11):1626–32. 10.1172/JCI20430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14(11):2308–18. 10.1101/gr.2523904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–73. 10.1016/j.febslet.2009.10.036 [DOI] [PubMed] [Google Scholar]
- 84.Sciotti MA, Chatenay-Rivauday C, Yamodo I, Ogier J. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct functional domains. Advances in experimental medicine and biology. 1997;418:699–701. [DOI] [PubMed] [Google Scholar]
- 85.Sciotti MA, Yamodo I, Klein JP, Ogier JA. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct binding domains. FEMS microbiology letters. 1997;153(2):439–45. [DOI] [PubMed] [Google Scholar]
- 86.Tamura H, Yamada A, Kato H. Molecular characterization of the dextran-binding lectin B gene dblB of Streptococcus criceti in Streptococcus mutans strain GS-5 with mutations in both gbpC and spaP genes. Genes Genet Syst. 2014;89(2):41–50. [DOI] [PubMed] [Google Scholar]
- 87.Quivey RG Jr., Faustoferri RC, Clancy KA, Marquis RE. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS microbiology letters. 1995;126(3):257–61. [DOI] [PubMed] [Google Scholar]
- 88.Bender GR, Sutton SV, Marquis RE. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infection and immunity. 1986;53(2):331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chattoraj P, Banerjee A, Biswas S, Biswas I. ClpP of Streptococcus mutans differentially regulates expression of genomic islands, mutacin production, and antibiotic tolerance. J Bacteriol. 2010;192(5):1312–23. 10.1128/JB.01350-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr Issues Mol Biol. 2005;7(1):95–107. [PubMed] [Google Scholar]
- 91.Jayaraman GC, Burne RA. DnaK expression in response to heat shock of Streptococcus mutans. FEMS microbiology letters. 1995;131(3):255–61. [DOI] [PubMed] [Google Scholar]
- 92.Lemos JA, Luzardo Y, Burne RA. Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J Bacteriol. 2007;189(5):1582–8. 10.1128/JB.01655-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Inagaki S, Fujita K, Takashima Y, Nagayama K, Ardin AC, Matsumi Y, et al. Regulation of recombination between gtfB/gtfC genes in Streptococcus mutans by recombinase A. Scientific World Journal. 2013:405075 10.1155/2013/405075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Inagaki S, Matsumoto-Nakano M, Fujita K, Nagayama K, Funao J, Ooshima T. Effects of recombinase A deficiency on biofilm formation by Streptococcus mutans. Oral microbiology and immunology. 2009;24(2):104–8. 10.1111/j.1399-302X.2008.00480.x [DOI] [PubMed] [Google Scholar]
- 95.Erbse A, Mayer MP, Bukau B. Mechanism of substrate recognition by Hsp70 chaperones. Biochem Soc Trans. 2004;32(Pt 4):617–21. 10.1042/BST0320617 [DOI] [PubMed] [Google Scholar]
- 96.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295(5561):1852–8. 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- 97.Len AC, Harty DW, Jacques NA. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology. 2004;150(Pt 5):1353–66. 10.1099/mic.0.26888-0 [DOI] [PubMed] [Google Scholar]
- 98.Len AC, Harty DW, Jacques NA. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology. 2004;150(Pt 5):1339–51. 10.1099/mic.0.27008-0 [DOI] [PubMed] [Google Scholar]
- 99.Kajfasz JK, Martinez AR, Rivera-Ramos I, Abranches J, Koo H, Quivey RG Jr., et al. Role of Clp proteins in expression of virulence properties of Streptococcus mutans. J Bacteriol. 2009;191(7):2060–8. 10.1128/JB.01609-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van den Bogert B, Boekhorst J, Herrmann R, Smid EJ, Zoetendal EG, Kleerebezem M. Comparative genomics analysis of Streptococcus isolates from the human small intestine reveals their adaptation to a highly dynamic ecosystem. PLoS One. 2013;8(12):83418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abranches J, Chen YY, Burne RA. Characterization of Streptococcus mutans strains deficient in EIIAB Man of the sugar phosphotransferase system. Applied and environmental microbiology. 2003;69(8):4760–9. 10.1128/AEM.69.8.4760-4769.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Loo CY, Mitrakul K, Voss IB, Hughes CV, Ganeshkumar N. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J Bacteriol. 2003;185(21):6241–54. 10.1128/JB.185.21.6241-6254.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spencer P, Ye Q, Misra A, Goncalves SE, Laurence JS. Proteins, pathogens, and failure at the composite-tooth interface. J Dent Res. 2014;93(12):1243–9. 10.1177/0022034514550039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blue (1.0 mM), red (0.1 mM), violet (0.01mM) and green (0.00 mM).
(TIF)
Tables A-G: Other differentially expressed proteins following exposure to TEG.
(PPTX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.