Abstract
Staphylococcus aureus is a major pathogen in humans and causes serious problems due to antibiotic resistance. We investigated the antimicrobial effect of glycyrrhetinic acid (GRA) and its derivatives against 50 clinical S. aureus strains, including 18 methicillin-resistant strains. The minimum inhibitory concentrations (MICs) of GRA, dipotassium glycyrrhizate, disodium succinoyl glycyrrhetinate (GR-SU), stearyl glycyrrhetinate and glycyrrhetinyl stearate were evaluated against various S. aureus strains. Additionally, we investigated the bactericidal effects of GRA and GR-SU against two specific S. aureus strains. DNA microarray analysis was also performed to clarify the mechanism underlying the antibacterial activity of GR-SU. We detected the antimicrobial activities of five agents against S. aureus strains. GRA and GR-SU showed strong antibacterial activities compared to the other three agents tested. At a higher concentration (above 2x MIC), GRA and GR-SU showed bactericidal activity, whereas at a concentration of 1x MIC, they showed a bacteriostatic effect. Additionally, GRA and GR-SU exhibited a synergistic effect with gentamicin. The expression of a large number of genes (including transporters) and metabolic factors (carbohydrates and amino acids) was altered by the addition of GR-SU, suggesting that the inhibition of these metabolic processes may influence the degree of the requirement for carbohydrates or amino acids. In fact, the requirement for carbohydrates or amino acids was increased in the presence of either GRA or GR-SU. GRA and GR-SU exhibited strong antibacterial activity against several S. aureus strains, including MRSA. This activity may be partly due to the inhibition of several pathways involved in carbohydrate and amino acid metabolism.
Introduction
Staphylococcus aureus is a commensal bacterium in humans that can also be pathogenic primarily as an opportunistic infectious agent. S. aureus causes various suppurative diseases, food poisoning, pneumonia and sepsis [1, 2, 3]. Chemotherapeutic treatment is generally applied to S. aureus infectious diseases. However, the emergence of methicillin-resistant Staphylococcus aureus (MRSA) represents a serious problem for the treatment of S. aureus infections. Since most clinical MRSA strains exhibit a high level of multidrug resistance, the treatment of MRSA infections is now a serious medical concern worldwide [4, 5].
Glycopeptides such as vancomycin and teicoplanin are often used to treat MRSA infections [6]. However, vancomycin-intermediate resistant S. aureus (VISA) emerged in the late 1990s [7, 8]. Furthermore, vancomycin-resistant S. aureus (VRSA) was first reported in Michigan in 2007 [9]. Daptomycin, which is an antibacterial agent against MRSA, has been used recently, but daptomycin-resistant strains have also been reported [10]. Thus, it is very likely that chemotherapy against S. aureus infections will become more difficult in the future.
Some plant extracts have long been known to exert antibacterial effects. Among these, polyphenols, which are classified into flavonoids (e.g., catechin, flavonol, and tannin) and nonflavonoids (e.g., phenolic acid, neolignan), have been especially well studied [11, 12, 13, 14]. For example, it has been shown that tannins from tea leaves or persimmon have antibacterial effects against Streptococcus mutans, S. aureus and Escherichia coli. [15, 16]. Polyphenols are thought to be less likely to induce the emergence of resistant strains than general antibiotics because the mode of action of polyphenols has been demonstrated to cause cytoplasmic membrane destabilization and/or permeabilization [11]. The side effects of these agents are also thought to be less severe than those of traditional antibiotics.
Licorice is a Leguminosae perennial found in the Mediterranean region, south Russia, central Asia, northern China and America. This plant has been traditionally used as a medicinal agent due to its antibacterial, anti-viral, anti-inflammatory and calming effects [17–20]. Glycyrrhetinic acid is a major component in licorice extract. Previous studies have shown that glycyrrhetinic acid has many properties, including anti-inflammatory, anti-allergic, anti-peptic ulcer, and anti-viral activities [21–25]. Its anti-inflammatory effect is particularly well known and has been applied to clinical therapy. It has been reported that the structure of glycyrrhetinic acid is similar to that of cortisone, a steroid hormone that exerts a strong anti-inflammatory effect [23]. In Japan, a preparation of monoammonium glycyrrhizinate (Stronger Neo-Minophagen C) is used to treat chronic hepatitis [26, 27]. Glycyrrhetinic acid has also been reported to have antibacterial effects against some bacterial species, but the mechanism underlying this activity remains unknown [28, 29]. Additionally, several derivatives of glycyrrhetinic acid exhibit anti-inflammatory activities, but their antibacterial activities have not been elucidated [30, 31, 32].
The aims of this study are to evaluate the antibacterial effects of glycyrrhetinic acid and its derivatives against clinical S. aureus strains and to investigate the mechanism underlying their antibacterial effect against S. aureus.
Materials and Methods
Bacterial strains and culture
S. aureus was cultured in tryptic soy broth (TSB) at 37°C with shaking at 50 rpm. Fifty clinical S. aureus strains, including 31 methicillin-sensitive S. aureus (MSSA) strains and 19 MRSA strains, were used in this study (S1 Table). These strains were from laboratory collection of S. aureus clinical isolates at Department of Bacteriology, Hiroshima University Graduate School of Biomedical and Health Sciences. A chemically defined medium (CDM) supplemented with glucose (50 mM) as the sole carbon source was prepared [33]. When necessary, glucose was replaced with other sugars (lactose, trehalose, and sucrose).
Glycyrrhetinic acid derivatives
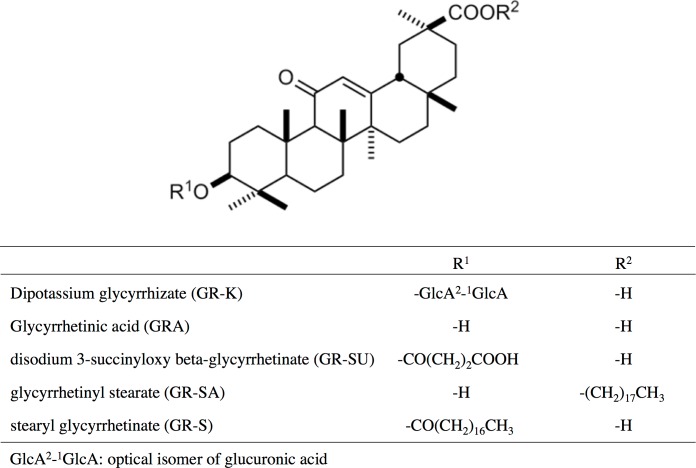
Glycyrrhetinic acid (GRA) and its derivatives used in this study are shown in Fig 1. These agents were obtained from Maruzen Pharmaceuticals Co., Ltd., Hiroshima, Japan. Dipotassium glycyrrhizate (GR-K) and disodium succinoyl glycyrrhetinate (GR-SU) were solubilized in distilled water. Glycyrrhetinic acid (GRA) was solubilized in 100% dimethyl sulfoxide (DMSO). Stearyl glycyrrhetinate (GR-S) and glycyrrhetinyl stearate (GR-SA) were solubilized in 100% ethanol. Stock solutions were prepared at a concentration of 20 mg/ml and were diluted in medium to the appropriate concentrations indicated in each experiment.
Fig 1. Structures of GRA and its derivatives.
Determination of the minimum inhibitory concentration (MIC)
The MICs were determined by using the micro-dilution method as previously described [33]. Briefly, each GRA derivative was adjusted to 4,096 mg/L in TSB, and 2-fold serial dilutions were prepared in a 96-well microplate (Thermo Fisher Scientific, Roskilde, Denmark). Overnight bacterial cultures were adjusted to an OD660 of 1.0 (1x109 cells/ml) and diluted to 1:100 with TSB (1x107 cells/ml). Ten microliters of the bacterial culture (1x105 cells/well) was applied to each well (100 μl total volume). The MICs of glycyrrhetinic acid and its derivatives were determined after the plate was incubated for 24 h at 37°C.
S. aureus growth curve
Overnight cultures of S. aureus MW2 were adjusted to an OD660 of 1.0. Then, 100 μl of bacterial culture was inoculated into 5 ml of TSB and incubated at 37°C with shaking. When the OD660 reached 0.3, various concentrations of either GRA or GR-SU were added to the medium, and the growth was monitored. To investigate cell viability, appropriate dilutions of TSB after the addition of either GRA or GR-SU were inoculated onto a tryptic soy agar (TSA) plate. After overnight incubation at 37°C, the colony forming units (CFUs) were counted. Three independent experiments were performed, and the mean ± SD was calculated. The data were analyzed for statistically significant differences compared to untreated control at each timepoint by a two-way ANOVA followed by Dunnett’s post hoc test.
Effects of GRA and GR-SU on cell viability
Overnight cultures of S. aureus MW2 cells were washed with 10 mM sodium phosphate buffer (PB; pH 6.8) and suspended in PB. The bacterial suspension was diluted to 107 cells/ml with PB, and 10 μl of the diluted suspension was inoculated into 500 μl of PB containing various concentrations of GR-SU (1x MIC: 128 mg/L) or GRA (1x MIC: 512 mg/L) and incubated for 1 or 2 hours at 37°C. Dilutions of the reaction mixture (100 μl) were plated onto TSA plates and incubated at 37°C overnight. The antibacterial effect was calculated as the ratio of the number of surviving cells (survival rate %) to the total number of bacteria incubated in PB. To verify the cell number of each strain inoculated in PB, the dilutions of the bacterial suspension prior to inoculation were plated onto TSA and incubated at 37°C overnight. Three independent experiments were performed, and the mean ± SD was calculated. The data were analyzed for significant differences compared to untreated control using two-way ANOVA followed by Dunnett’s post hoc test.
Effects of GRA and GR-SU in combination with antibiotics
The effects of combination treatments of gentamicin (Wako Pure Chemical Industries Ltd, Osaka, Japan), chloramphenicol (Sigma-Aldrich Corporation, Tokyo, Japan), tetracycline (Wako Pure Chemical Industries Ltd), oxacillin (Sigma-Aldrich Corporation) or ofloxacin (Daiichi Sankyo Company, Tokyo, Japan) with either GRA or GR-SU were investigated. Overnight cultures of S. aureus MW2 were adjusted to 107 cells/ml, and 10 μL of the bacterial suspension was applied to each well in a 96-well plate. Antibiotics were dissolved in TSB alone or TSB containing either GRA or GR-SU (1/2, 1/4 or 1/8 MICs for each agent). Stock solutions (1 g/L) of gentamicin, oxacillin or ofloxacin were prepared in TSB or TSB containing either GRA or GR-SU. Stock solutions (10 g/L) of chloramphenicol and tetracycline were prepared in ethanol. The final concentrations of gentamicin, chloramphenicol, tetracycline, oxacillin and ofloxacin were 32 mg/L, 32 mg/L, 64 mg/L, 128 mg/L, and 4 mg/L, respectively, and these solutions were then serially diluted 2-fold with the same media. After a 24 h incubation at 37°C, the MICs were determined as described above. The fractional inhibitory concentration (FIC) was calculated with a method previously described [34]. FIC indexes of ≤0.5, >0.5 – ≤4.0 and >4.0 were defined as synergy, no interaction and antagonism, respectively.
Microarray analysis
Overnight cultures of S. aureus (108 cells) were inoculated into 30 ml of fresh TSB and cultured at 37°C with shaking. When the OD660 reached 0.3, GR-SU (1x MIC: 128 mg/L) was added to the medium. After 2 h of incubation, the bacterial cells were collected by centrifugation at 5,000 x g for 5 min at 4°C and stored at -80°C until further use. Additionally, bacterial cells without GR-SU treatment were prepared as the control. Total RNA was extracted from the bacterial cells using the FastRNA Pro Blue Kit (MP Biomedicals, Cleveland, OH, USA) according to the manufacturer’s protocol. Then, cDNA was synthesized from 10 μg of total RNA using the FairPlay III Microarray Labeling Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. The Agilent eArray platform (Agilent Technologies) was used to design the microarray; 13,939 probes (60-mers) were designed for the 2,628 protein-coding genes of S. aureus MW2 (up to five probes per gene). For microarray analyses, the test (GR-SU treatment) and control cDNAs were labeled with Cy3 mono-Reactive Dye and Cy5 mono-Reactive Dye (GE Healthcare, UK), respectively. The fluorescently labeled cDNAs were purified using the FairPlay III Microarray Labeling Kit (Agilent Technologies) The Cy3-labeled and Cy5-labeled cDNAs were mixed and hybridized onto an array using the Hi-RPM Gene Expression Hybridization Kit (Agilent Technologies). The arrays were scanned with an Agilent scanner, and data extraction, filtering and normalization were conducted using the Agilent Feature Extraction software according to the manufacturer’s instructions. The experiments were performed using two biological replicates (two technical replicates for each condition), and the expression data were deposited into the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession GSE80500. We compared difference between control and test sample for each gene by student t-test using Cyber-T (http://cybert.microarray.ics.uci.edu/). Factors that exhibited a greater than 2-fold change (increase/decrease) and P<0.05 were defined as being altered by GR-SU.
Gene expression analysis by quantitative PCR
Overnight cultures of S. aureus MW2 (108 cells) were inoculated into 5 ml of TSB and aerobically incubated at 37°C with shaking. When the OD660 reached 0.3, either 1/4 or 1x MIC of GR-SU or GRA was added to the culture. The bacterial cells were collected after 1, 2, 4 and 8 h of incubation. RNA extraction and cDNA synthesis were performed as described above. Quantitative PCR was performed with the LightCycler Nano system (Roche, Tokyo, Japan) using cDNA as the template DNA. The primers used are shown in Table 1. The results were normalized to the housekeeping gene gyrB. Three independent experiments were performed, and the mean ± SE was calculated.
Table 1. Primers used in this study.
| Gene name | Upper primer | Lower primer |
|---|---|---|
| For quantitative PCR | ||
| gyLB | 5'- AGGTCTTGGAGAAATGAATG 3' | 5' -CAAATGTTTGGTCCGCTT 3' |
| seh | 5' -TCAAGGTGATAGTGGCAAT 3' | 5' -CCAATCACCCTTTCCTGT 3' |
| clfB | 5' -TGCAAGTGCAGATTCCGA 3' | 5' -TCATTTGTTGAAGCTGGCTC 3' |
| sea | 5' -GATCAATTTATGGCTAGACG 3' | 5' -CGAAGGTTCTGTAGAAGTATGA 3' |
| sbi | 5' -CCAAGATAACAAAGCACCAC 3' | 5' -TGCTGATTTCACACGCTC 3' |
| sak | 5' -ACAGGCCCGTATTTGATG 3' | 5' -GCCCATTCGACATAGTATTC 3' |
| clfA | 5' -TACAAGTGCGCCTAGAATGA 3' | 5' -TTTGACATAGCCTGCTTGGT 3' |
| hlgC | 5' -ATTTCGTTCCAGACAGTGAG 3' | 5' -CCGTCTAAATAACTGTTGCC 3' |
| hla | 5' -GGGGACCATATGATAGAGATT 3' | 5' -TGTAGCGAAGTCTGGTGAAA 3' |
| coa | 5' -ACAGGGCACAATTACAGGT 3' | 5' -GGTGTCGTCGGTTGAGTAT 3' |
| lukS-PV | 5' -ATGAGGTGGCCTTTCCAATAC 3' | 5' -CCTGTTGATGGACCACTATTA 3' |
| nuc | 5' -AATCATACGGGTCCTTTCA 3' | 5' -CCGTTTCTGGCGTATCAA 3' |
| fnbA | 5' -TGAGGGTGGTTATGTTGATG 3' | 5' -CAGTGTATCCTCCAACGTGA 3' |
| icaA | 5' -AGTTGTCGACGTTGGCTAC 3' | 5' -CCAAAGACCTCCCAATGT 3' |
Effect of GRA and GR-SU on carbohydrate and amino acids requirements
Chemically defined medium (CDM) and CDM containing 1/2 MIC or 1/4 MIC of GRA-K, GR-SU or GRA were used in this experiment. Overnight cultures of S. aureus MW2 were adjusted to 1x107 cells/ml. Glucose, trehalose, sucrose or fructose (each 50 mM) was serially diluted 2-fold with either TSB or TSB containing GRA, K-GRA or GR-SU (1/2 MIC or 1/4 MIC at the final concentration for each agent). Then, 10 μl of the bacterial culture (1x105 cells) was applied to each well. After incubation for 24 h at 37°C, the minimum concentration of each sugar that showed visible bacterial growth was determined.
To investigate the effect on amino acid requirement, we used CDM containing 18 amino acids necessary for S. aureus growth. The amino acids used in this experiment and their concentrations have been previously described [33]. The CDM was prepared using 2-fold serial dilutions with CDM depleted of all amino acids. Then, 10 μl of the bacterial culture (1x105 cells) was applied to each well. After incubation for 24 h at 37°C, the minimum dilution rate that showed visible bacterial growth was determined.
Results
Antimicrobial activities of GRA and its derivatives against S. aureus strains
The MICs of GRA and its 4 derivatives are shown in Table 2. The MICs were evaluated using 50 S. aureus strains (19 MRSA strains and 31 MSSA strains). The MICs of GR-K and GR-S were higher than GA, GR-SU and GR-SA. The MICs of GR-K and GR-S against all strains were more than 256 mg/L. While GR-SA exhibited an intermediate level of MICs on the tested strains (ranged from 64 to 4,096 mg/L except 2 strains [more than 4,096 mg/L]), the MICs of GRA and GR-SU ranged from 16 to 512 mg/L and from 16 to 256 mg/L, respectively. The distribution of MICs in the MRSA and MSSA strains is shown in Table 2. We found no differences in susceptibility between the MRSA and MSSA strains.
Table 2. MIC of glycyrrhetinic acid and its derivatives against S.aureus strains.
| MIC (mg/L) | |||||
|---|---|---|---|---|---|
| strain | GR-Ka | GR-SUb | GRAc | GR-Sd | GR-SAe |
| MRSA | |||||
| SA5001 | 2048 | 128 | 64 | >4096 | 1024 |
| SA5002 | 256 | 16 | 64 | >4096 | 512 |
| SA5003 | 1024 | 32 | 32 | >4096 | 1024 |
| SA5004 | >4096 | 64 | 64 | >4096 | 1024 |
| SA5007 | 2048 | 128 | 256 | >4096 | 512 |
| SA5008 | 4096 | 128 | 256 | >4096 | 1024 |
| SA5012 | 4096 | 256 | 512 | >4096 | 2048 |
| SA5013 | 4096 | 128 | 64 | >4096 | 512 |
| SA5021 | 2048 | 256 | 128 | 1024 | 512 |
| SA5046 | 1024 | 64 | 512 | 512 | 512 |
| SA5052 | 4096 | 128 | 512 | 512 | 512 |
| SA5053 | 1024 | 16 | 32 | 2048 | 2048 |
| SA5056 | >4096 | 128 | 512 | >4096 | 1024 |
| SA5057 | >4096 | 128 | 128 | 4096 | 512 |
| SA5059 | 256 | 32 | 32 | 1024 | 1024 |
| SA5060 | 4096 | 128 | 128 | 2048 | 2048 |
| SA5065 | 4096 | 64 | 32 | 256 | 128 |
| SA5070 | 4096 | 32 | 512 | 2048 | 512 |
| MW2 | 2048 | 128 | 512 | 2048 | 1024 |
| MSSA | |||||
| SA5006 | >4096 | 32 | 64 | >4096 | 512 |
| SA5014 | 1024 | 32 | 128 | 1024 | 256 |
| SA5015 | 2048 | 128 | 128 | >4096 | 256 |
| SA5016 | 1024 | 64 | 64 | >4096 | 1024 |
| SA5018 | 512 | 16 | 16 | 256 | 128 |
| SA5019 | 1024 | 64 | 64 | 256 | 64 |
| SA5020 | 2048 | 256 | 256 | 4096 | 4096 |
| SA5022 | 2048 | 64 | 512 | 2048 | 512 |
| SA5024 | 4096 | 256 | 512 | 4096 | 4096 |
| SA5025 | 256 | 64 | 256 | 256 | 64 |
| SA5026 | 1024 | 256 | 512 | 4096 | 2048 |
| SA5027 | 512 | 256 | 512 | 4096 | 2048 |
| SA5029 | 4096 | 256 | 512 | 2048 | 2048 |
| SA5031 | 4096 | 64 | 128 | >4096 | >4096 |
| SA5032 | >4096 | 128 | 512 | >4096 | 1024 |
| SA5033 | 2048 | 256 | 512 | 2048 | 4096 |
| SA5034 | 2048 | 128 | 512 | 256 | 512 |
| SA5038 | 4096 | 256 | 512 | 4096 | 512 |
| SA5039 | 2048 | 128 | 512 | 512 | 1024 |
| SA5040 | 2048 | 64 | 256 | 512 | 512 |
| SA5042 | 2048 | 32 | 256 | 1024 | 256 |
| SA5043 | 1024 | 32 | 512 | 512 | 512 |
| SA5044 | 4096 | 256 | 512 | 2048 | 2048 |
| SA5062 | 4096 | 256 | 256 | 2048 | 2048 |
| SA5063 | 4096 | 256 | 512 | 512 | 256 |
| SA5064 | 4096 | 256 | 512 | 1024 | 1024 |
| SA5066 | 2048 | 64 | 128 | 256 | 128 |
| SA5067 | 1024 | 64 | 512 | 2048 | 512 |
| SA5069 | 2048 | 128 | 256 | >4096 | 1024 |
| SA5071 | 2048 | 64 | 128 | >4096 | >4096 |
| SA5073 | 2048 | 256 | 256 | 2048 | 1024 |
aDipotassium glycyrrhizate
bdisodium succinoyl glycyrrhetinate
cGlycyrrhetinic acid
dStearyl glycyrrhetinate
eglycyrrhetinyl stearate
Effects of GRA and GR-SU on S. aureus growth
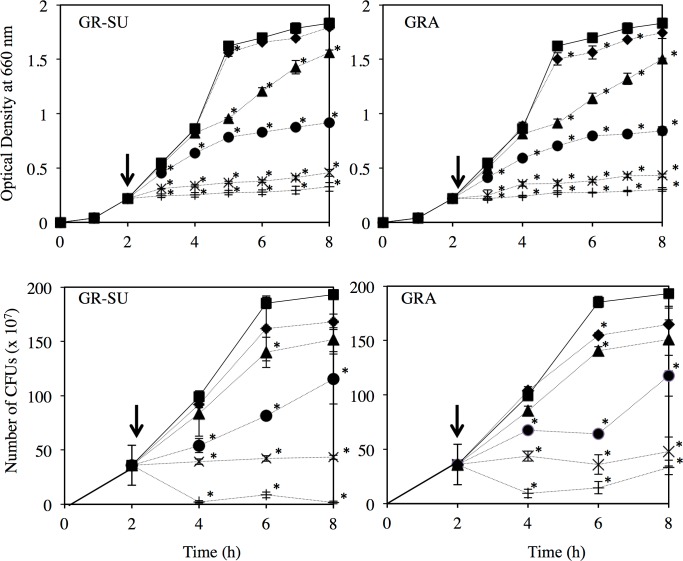
As shown in Fig 2, S. aureus MW2 growth was inhibited by the addition of either GRA or GR-SU in a dose-dependent manner. The addition of more than 1x MIC of GRA or GR-SU fully inhibited the growth of S. aureus MW2 as expected, but growth was already inhibited by both agents at sub-MIC concentrations (1/16 and 1/4 MIC). We also investigated the effects of GRA and GR-SU on cell viability in a time course experiment (Fig 2). The result was similar to that obtained with the OD. The viable cell number was suppressed by the addition of either GRA or GR-SU in a dose-dependent manner. The bacterial cell numbers were decreased after 4 h by the addition of 2x MIC of GRA or GR-SU. Additionally, we analyzed a methicillin-sensitive strain (MS23513) and obtained similar findings (S1 Fig).
Fig 2. Effect of GRA and GRA derivatives on S. aureus MW2 growth.
Overnight cultures of S. aureus MW2 were adjusted to an OD of 1.0. Then, 100 μl of bacterial culture was inoculated into 5 ml of TSB. The bacterial culture was incubated at 37°C with shaking. When the OD at 660 nm reached 0.3, various concentrations (■: control, ♦: 1/64 MIC, ▲: 1/16 MIC, ●: 1/4 MIC, ×: 1x MIC, +: 2x MIC) of GR-SU or GRA were added to the medium. The growth and colony counts were monitored. Three independent experiments were performed, and the mean ± SD was calculated. The data were analyzed for statistically significant differences compared to untreated control by a two-way ANOVA followed by Dunnett’s post hoc tests. *P<0.05.
Effects of GRA and GR-SU on cell viability
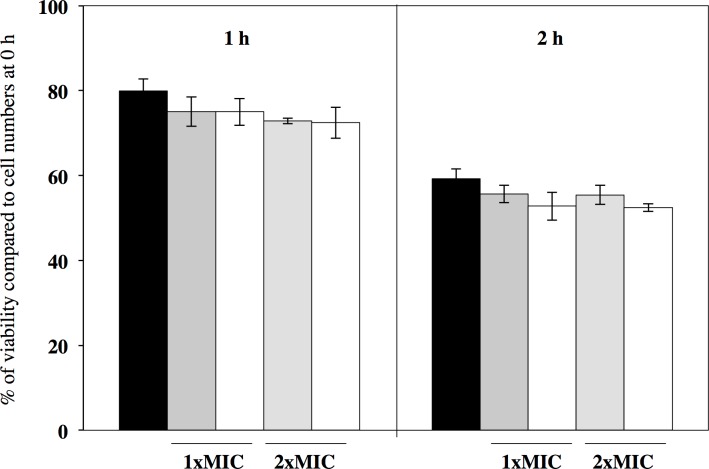
Because GRA and GR-SU exhibited antimicrobial activity against S. aureus strains during active growth, we investigated whether this activity was observed under non-growing conditions. We treated S. aureus MW2 cells with either GRA or GR-SU in 10 mM PB. As shown in Fig 3, the viable cell number of the MW2 strain treated with 1x MIC and 2x MIC of GRA or GR-SU showed no significant difference compared to the control (no treatment).
Fig 3. Effect of GRA and GRA derivatives on S. aureus cell viability under non-growing conditions.
Cell viability was evaluated under non-growing conditions following the addition of GRA or GR-SU. The methods are described in the Materials and Methods section. S. aureus MW2 was treated with 1x MIC and 2x MIC of GR-SU (grey bar) or GRA (white bar) in 10 mM sodium phosphate buffer. An untreated bacterial culture (black bar) was used as a control. After 1 and 2 h of incubation, the reaction mixtures (100 μl) were plated on trypticase soy agar and incubated at 37°C overnight. The colony forming units (CFUs) were determined as the total number of colonies identified on each plate. The antibacterial effect was calculated as the ratio of the number of surviving cells (survival rate %) to the total number of bacteria incubated. Three independent experiments were performed, and the mean ± SD was calculated. The data were analyzed for statistically significant differences compared to the untreated control by a two-way ANOVA followed by Dunnett’s post hoc test. Significant differences were not observed.
Synergistic effects of GRA and GR-SU against 8 S. aureus strains
Next, we evaluated the synergistic effects of GRA and GR-SU in combination with other antibiotics. As shown in Table 3, all strains tested showed synergy with the combination of gentamicin with either GRA or GR-SU, while one strain showed synergy with the combination of tetracycline or chloramphenicol with GR-SU, and 2 strains showed synergy with the combination of oxacillin with GRA. Oxacillin, ofloxacin and erythromycin had no effect (no interaction) when combined with GR-SU, and chloramphenicol, ofloxacin and erythromycin had no effect when combined with GRA. Furthermore, the MICs of antibiotics in the absence or presence of GRA or GR-SU are shown in S2 Table. A synergistic effect of GRA and GR-SU with gentamycin against both gentamicin-resistant (4 to 128 mg/L) and susceptible strains (3 strains with MIC of 1 mg/L) was found (S2 Table). In the presence of 1/2 MIC of GRA, 4 strains exhibited a 2- to 8-fold decrease in the MIC of oxacillin, while 4 other strains exhibited no change. In the presence of the 1/4 MIC of GRA, 2 strains also exhibited an 8-fold decrease in the MIC of oxacillin. In the presence of 1/2 and 1/4 MICs of GRA or GR-SU, all strains exhibited a 4- to 8-fold decrease in the MIC of gentamicin. In the presence of the 1/8 MICs of GRA or GR-SU, increased MICs were not observed in any of the antibiotics tested in this study (S2 Table).
Table 3. FICIs in combination of various antibiotics with GRA or GR-SU.
| Number of strains | ||||||
|---|---|---|---|---|---|---|
| FICIa (GR-SU) | FICI (GRA) | |||||
| Antibiotics | ≤0.5 | >0.5~≤4.0 | >4.0 | ≤0.5 | >0.5~≤4.0 | >4.0 |
| Oxacillin | 0 | 8 | 0 | 2 | 6 | 0 |
| tetracycline | 1 | 7 | 0 | 0 | 8 | 0 |
| gentamicin | 8 | 0 | 0 | 8 | 0 | 0 |
| chloramphenicol | 1 | 7 | 0 | 0 | 8 | 0 |
| ofloxacin | 0 | 8 | 0 | 0 | 8 | 0 |
| erythromycin | 0 | 8 | 0 | 0 | 8 | 0 |
FICI: ≤0.5; synergy, >0.5~≤4.0; no interaction, >4.0; antagonism
Microarray analysis
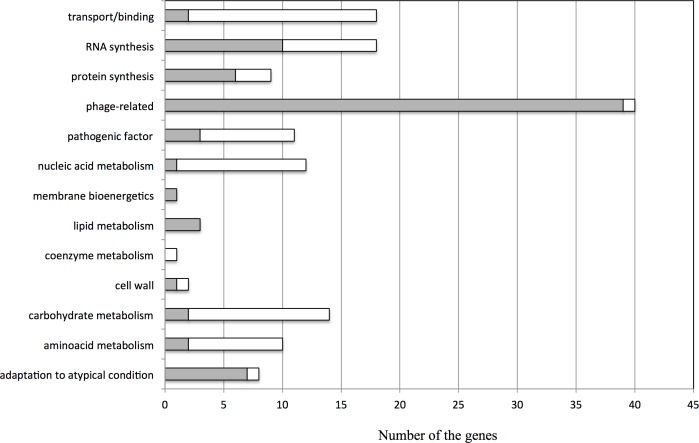
We performed a microarray analysis to identify genes with altered expression patterns in response to GR-SU. As summarized in Fig 4, more than 200 genes exhibited altered expression levels in response to GR-SU. Among these, 39 phage-related genes were increased by the addition of GR-SU. In the S. aureus MW2 genome, two phage-related operons (MW1380-MW1451 and MW1882-MW1952) were found. Specifically, in the operon of MW1380-MW1451 (72 genes), the expression of 31 genes was increased. Many genes responsible for metabolic and biosynthetic pathways also exhibited altered expression patterns (S3–S6 Tables). The expression levels of 6 genes involved in protein synthesis, 10 genes in RNA synthesis and 2 genes in amino acid metabolism were increased, whereas the levels of 8 genes for RNA synthesis, 12 genes for carbohydrate metabolism and 8 genes for amino acid metabolism were decreased. In addition, the expression levels of 2 transporter and substrate-binding protein genes were increased, and 16 genes were decreased after GR-SU treatment. Regarding pathogenic factors, the expression of 3 factors was increased while that of 8 factors was decreased.
Fig 4. Microarray analysis of genes with altered expression levels in response to GR-SU.
The microarray analysis was performed as described in the Materials and Methods section. The numbers of genes with 2-fold upregulated (grey bar) or downregulated (white bar) expression are indicated.
Effect of GRA and GR-SU on virulence factor expression
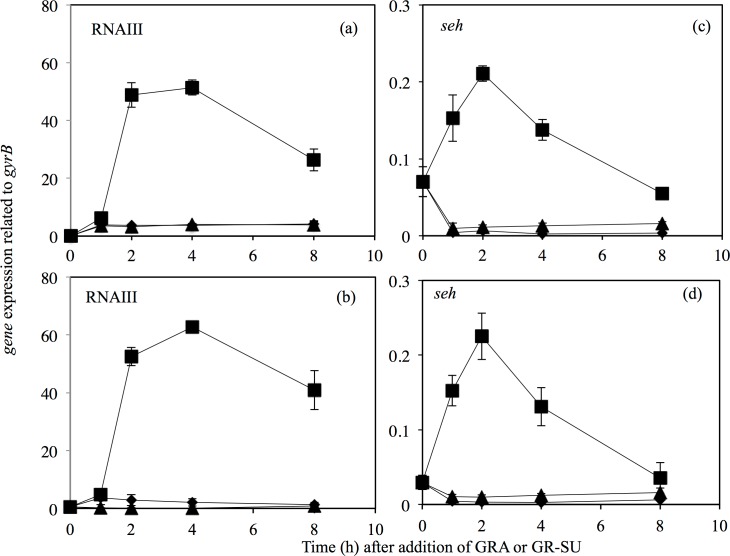
As we found in the microarray analysis, the expression levels of several virulence-related genes were altered in response to GR-SU. We analyzed the gene expression of RNAIII, which is a key factor involved in the expression of many virulence genes, in more detail by quantitative PCR. As shown in Fig 5A and 5B, RNAIII expression was significantly increased during the exponential phase in the absence of GRA or GR-SU, whereas GRA and GR-SU both fully suppressed RNAIII expression at 1/4 MIC. The expression of seh, which is a RNAIII-dependent virulence factor, exhibited a pattern similar to that of RNAIII in response to GRA and GR-SU (Fig 5C and 5D). These results suggest that the repression of RNAIII by GR-SU may explain the alterations in gene expression of several virulence-related genes, which were observed in the microarray analysis (Fig 4).
Fig 5.
Effects of GRA and GRA derivatives on the expression of RNAIII (a, b) and seh (c, d). Overnight cultures of S. aureus MW2 (108 cells) were inoculated into 5 ml of TSB and aerobically incubated at 37°C with shaking. When the OD at 660 nm reached 0.3, 1/4 MIC (a, c) or 1x MIC (b, d) of GR-SU or GRA was added to the culture (■: control, ♦: GR-SU, ▲: GRA). Bacterial cells were collected after 1, 2, 4 and 8 h of incubation. Then, RNA extraction and cDNA synthesis were performed. Quantitative PCR was performed using cDNA as the template DNA. Three independent experiments were performed and the mean ± SE was calculated.
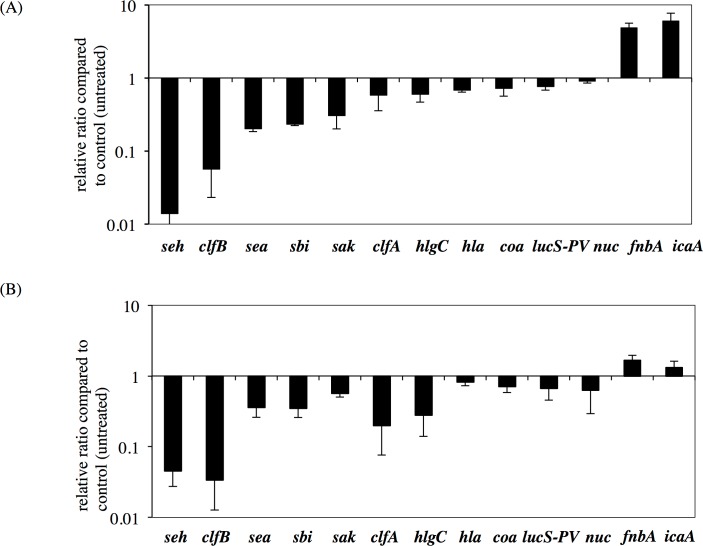
Therefore, we investigated the expression levels of 12 virulence factors and found that several toxins such as sea, clfAB and hlgC were downregulated by GRA, whereas the cell surface factors fnbA and ica were upregulated (Fig 6). The effect of GR-SU on virulence factor expression was similar to that of GRA.
Fig 6. Effects of GRA and GRA derivatives on virulence factor expression.
Overnight cultures of S. aureus MW2 (108 cells) were inoculated into 5 ml of TSB and aerobically incubated at 37°C with shaking. When the OD at 660 nm reached 0.3, 1x MIC of GR-SU (a) or GRA (b) was added to the culture. The bacterial cells were collected after 2 h. Then, RNA extraction and cDNA synthesis were performed. A quantitative PCR was performed using cDNA as the template DNA. Three independent experiments were performed and the mean ± SE was calculated.
Effects of GRA and GR-SU on sugar and amino acid requirements
Because amino acid and carbohydrate metabolism tended to decrease following a 2-hour exposure to GRA and GR-SU based on the microarray analysis, we hypothesized that GRA and GR-SU may inhibit the growth of S. aureus by influencing amino acid and carbohydrate metabolism. Therefore, we investigated sugar and amino acid requirements in the presence of GRA, GR-SU or GRA-K (1/2 MIC or 1/4 MIC at the final concentration for each agent). As shown in Table 4, the minimum concentrations of each sugar for the growth of S. aureus MW2 in CDM containing glucose, sucrose, lactose or trehalose was increased by 2- or 4-fold in the presence of 1/2 MIC of GRA or GR-SU, whereas GRA-K did not increase the sugar requirement. Additionally, we investigated the amino acid requirements. GRA and GR-SU treatment caused a 2-fold higher requirement compared to untreated bacteria. When 1/4 MIC of GRA or GR-SU was used, the minimal concentration of each sugar or amino acids was not altered.
Table 4. Effect of GRA and GR-SU on sugar and amino acids requirement.
| Minimum concentration for growth (mM: except for amino acids) | |||||
|---|---|---|---|---|---|
| GRA derivatives | glucose | sucrose | lactose | trehalose | Amino acidsa |
| none | 0.78 | 0.39 | 0.39 | 0.39 | 1/32b |
| GR-K | 0.78 | 0.39 | 0.39 | 0.39 | 1/32 |
| GR-SU | 3.13 | 1.56 | 1.56 | 1.56 | 1/16 |
| GRA | 3.13 | 0.78 | 0.78 | 0.78 | 1/16 |
a: dilution ratio from CDM (1x amino acids)
Discussion
In this study, we demonstrated the antibacterial activities of GRA and GR-SU against S. aureus strains. These two compounds exhibited strong antibacterial activities, but the antibacterial activities of the other GRA derivatives (GR-S, GR-K and GR-SA) were much weaker than those of GRA and GR-SU. GR-SU is synthesized from GRA by adding a succinic acid moiety to increase water solubility. GR-K (dipotassium glycyrrhizate) is a potassium salt of glycyrrhizin (GRA with two glucuronic acids) and also has high water solubility. Both GR-SU and GR-K have high water solubility, but the antibacterial effect of GR-K is much lower than that of GR-SU. Thus, difference in water solubility is not related to the difference in antibacterial activity among these compounds. Instead, some other structural differences between the two agents (Fig 1) may affect their antibacterial activities against S. aureus strains. GRA and GR-SU have been clinically used as anti-inflammatory agents [21, 32]. Taken together with our findings, GRA and GR-SU are expected to exert two biological activities of clinical importance: antibacterial and anti-inflammatory activities.
Although several investigations regarding the antibacterial effects of GRA have been reported [28, 29], this is the first report of the use of GR-SU as an antibacterial agent. Glycyrrhetinic acid derivatives are known anti-inflammatory drugs [30, 31, 32]. Stronger Neo-Minophagen C, which is the most well-known glycyrrhetinic acid derivative drug, contains 2 mg/ml of monoammonium glycyrrhizinate hydrate at the dose used to clinically treat chronic hepatitis; this dose is approximately 4- to 20-fold higher than the concentration used in this study [26, 27]. Therefore, GRA and GR-SU may have a potential use against S. aureus infectious diseases caused by antibacterial and anti-inflammatory effects.
Long, D.R. et al. previously reported the bacteriostatic activity of GRA and the bactericidal effect of GRA at a high concentration (above 62.5 mg/L) against S. aureus [28]. In this study, we also observed bactericidal activity of GRA and GR-SU at a 2x MIC dose (GR-SU: 256 mg/L, GRA: 1,024 mg/L). As shown in the microarray analysis (Fig 4), the expression levels of many phage-related genes were increased by the addition of 1x MIC of GR-SU, and we speculated that the bactericidal effect of a high dose of GR-SU (2xMIC) was due to phage induction-mediated cell lysis.
Among the genes that exhibited altered expression following the addition of GR-SU, many genes involved in sugar (S3 Table), amino acid (S4 Table) and nucleic acid (S5 Table) metabolism were significantly decreased. Moreover, the expression levels of many genes that encoded transporters were also decreased (S6 Table). These results suggest that GRA and GR-SU-treatments inhibit the uptake of nutrients, which then causes alterations in the expression of genes related to nutrient metabolism. Supporting this hypothesis, S. aureus MW2 required a higher concentration of sugars and amino acids in the presence of GRA and GR-SU than in the absence of these compounds (Table 4). Based on these findings, we propose that GRA and GR-SU have no specific mechanism for antibacterial activity but induce antimicrobial effects by inhibiting nutrient acquisition and affecting bacterial metabolism. Additionally, the expression of several transporters responsible for the efflux and/or influx of compounds was increased in the microarray analysis. The transporters with altered expression levels in the presence of either GRA or GR-SU included some transporters that might be involved in the high susceptibility to gentamicin in the presence of either GRA or GR-SU. GRA and GR-SU had weak synergistic activities with other protein synthesis inhibitors, speculating that transporters specific for gentamicin influx may be influenced by GRA and GR-SU.
In this study, we found that the expression of RNAIII, which is responsible for the regulation of many S. aureus virulence factors, was fully repressed by GRA and GR-SU [35, 36]. Because RNAIII is related to the agr quorum sensing system, RNAIII expression is growth dependent [35]. We found that GRA and GR-SU suppressed growth in a dose-dependent manner, suggesting that RNAIII expression was inhibited. Then, we investigated the expression of several virulence factors in the presence or absence of GRA or GR-SU. As shown in Fig 6, the expression levels of toxins such as enterotoxins (seh and sea), staphylokinase (sak) and hlgC were decreased, whereas the expression levels of cell surface factors such as fnbA and ica were increased. From these results, we hypothesize that GRA and GR-SU can modulate virulence in addition to their antimicrobial effects on S. aureus.
In conclusion, we have found indications that the antibacterial activity of GRA and GR-SU are mostly occurring via effects on nutrient metabolism. Additionally, these agents suppressed the expression of several virulence genes. Taken together with previous reports of inflammatory effects, GRA and GR-SU may have potential for clinical use against S. aureus infectious diseases.
Supporting Information
Overnight cultures of S. aureus MS23513 were adjusted to an OD at 660 nm of 1.0. Then, 100 μl of bacterial culture was inoculated in 5 ml TSB. Bacterial cultures were incubated at 37°C with shaking. When the OD at 660 nm reached 0.3, various concentrations (■: control; ♦: 1/64 MIC; ▲:1/16 MIC; ●: 1/4 MIC; ×: 1x MIC; 十: 2x MIC) of GR-SU or GRA were added to the medium. Growth and colony counts were monitored during growth. Three independent experiments were performed, and the mean ± SD was calculated. The data were analyzed for statistically significant differences compared to untreated control for each condition by a two-way ANOVA followed by Dunnett’s post hoc tests. *P<0.05.
(PPTX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Dr. Kaori Yasuda, Cell Innovator Inc. for assistance with microarray data acquisition. This study was supported in part by Grants-in-Aid for Scientific Research (C) (Grant No: 25462868) from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan.
Data Availability
Microarray data are deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE80500.
Funding Statement
This study was supported in part by Grants-in-Aid for Scientific Research (C) (Grant No: 25462868) from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Foster TJ. The Staphylococcus aureus “superbug”. J Clin Invest 2004; 114: 1693–6. 10.1172/JCI23825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. New Engl J Med 1998; 339: 520–32. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 3.Manders SM. Toxin-mediated streptococcal and staphylococcal disease. J Am Acad Dermatol 1998; 39: 383–8. [DOI] [PubMed] [Google Scholar]
- 4.Deurenberg RH, Vink C, Kalenic S Friedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect 2007; 13: 222–35. 10.1111/j.1469-0691.2006.01573.x [DOI] [PubMed] [Google Scholar]
- 5.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet 2006; 368: 874–5. 10.1016/S0140-6736(06)68853-3 [DOI] [PubMed] [Google Scholar]
- 6.Shimada K, Kobayashi H, Sunagawa K, Inamatsu T, Yamaguchi K. Clinical summary of intravenous use of vancomycin hydrochloride for severe infection caused by methicillin-resistant Staphylococcus aureus. Jpn J Chemother 1995; 43: 1048–61. [Google Scholar]
- 7.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococci aureus clinical strains with reduced vancomycin susceptibility. J Antimicrob Chemother 1997; 40: 135–6. [DOI] [PubMed] [Google Scholar]
- 8.Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced Vancomycin Susceptibility in Staphylococcus aureus, Including Vancomycin-Intermediate and Heterogeneous Vancomycin-Intermediate Strains: Resistance Mechanisms, Laboratory Detection, and Clinical Implications. Clin Microbiol Rev 2010; 23: 99–139. 10.1128/CMR.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finks J, Wells E, dyke TL, Husain N, Plizga L, Heddurshetti R et al. Vancomycin-resistant Staphylococcus aureus, Michigan, USA, 2007. Emerg Infect Dis 2009; 15: 943–5. 10.3201/eid1506.081312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marty FM, Yeh WW, Wennersten CB, Venkataraman L, Albano E, Alyea EP et al. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J Clin Microbiol 2006; 44: 595–7. 10.1128/JCM.44.2.595-597.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor PW, Hamilton-Miller JM, Stapleton PD. Antimicrobial properties of green tea catechins. Food Sci Technol Bull 2005; 2: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green AE, Rowland Rs, Cooper RA, Maddocks SE. The effect of the flavonol morin on adhesion and aggregation of Streptococcus pyogenes. FEMS Microbiol Lett 2012; 333: 54–8. 10.1111/j.1574-6968.2012.02598.x [DOI] [PubMed] [Google Scholar]
- 13.Chung KT, Wong TY, Wei CI, Huang YW, Lin Y. Tannins and human health: a review. Crit Rev Food Sci Nutr 2008; 38: 421–64. [DOI] [PubMed] [Google Scholar]
- 14.Corthout J, Pieters L, Claeys M, Geerts S, Vanden Berghe D, Vlietinck A. Antibacterial and molluscicidal phenolic acids from Spondias mombin. Planta Med 1994; 60: 460–3. 10.1055/s-2006-959532 [DOI] [PubMed] [Google Scholar]
- 15.Taleb H, Maddocks SE, Morris RK, Kanekanian AD. The Antibacterial activity of date syrup polyphenols against S. aureus and E. coli. Front Microbiol 2016; 7: 198 10.3389/fmicb.2016.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatano T, Kusuda M, Inada K, Ogawa TO, Shiota S, Tsuchiya T et al. Effects of tannins and related polyphenols on methicillin-resistant Staphylococcus aureus. Phytochemistry 2005; 66: 2047–55. 10.1016/j.phytochem.2005.01.013 [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Yang R, Yuan B, Liu Y, Liu C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm Sin B 2015; 5: 310–5. 10.1016/j.apsb.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JK, Oh SM, Kwon HS, Oh YS, Lim SS, Shin HK. Anti-inflamatory effect of roasted licorice extracts on lipopolysaccharide-induced inflammatory responses in murine macrophages. Biochem Biophys Res Commun 2006; 345: 1215–23. 10.1016/j.bbrc.2006.05.035 [DOI] [PubMed] [Google Scholar]
- 19.Thyagarajan SP, Jayaram S, Gopalakrishnan V, Hari R, Jeyakumar P, Sripathi MS. Herbal medicines for liver diseases in India. J Gastroenterol Hepatol 2002; 17: 370–6. [DOI] [PubMed] [Google Scholar]
- 20.Fukai T, Marumo A, Kaitou K, Kanda T, Terada S, Nomura T. Antimicrobial activity of licorice flavonoids against methicillin-resistant Staphylococcus aureus. Fitoterapia 2002; 73: 536–9 [DOI] [PubMed] [Google Scholar]
- 21.Ming LJ, Yin AC. Therapeutic effects of glycyrrhizic acid. Nat Prod Commun 2013; 8: 415–8. [PubMed] [Google Scholar]
- 22.Hoever G, Baltina L, Michaelis M, Kondratenko R, Baltina L, Tolstikov GA et al. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J Med Chem 2005; 48: 1256–9. 10.1021/jm0493008 [DOI] [PubMed] [Google Scholar]
- 23.Kao TC, Shyu MH, Yen GC. Glycyrrhizic acid and 18beta-glycyrrhetinic acid inhibit inflammation via P13k/Akt/GSK3beta signaling and glucocorticoid receptor activation. J Agric Food Chem 2010; 58: 8623–9. 10.1021/jf101841r [DOI] [PubMed] [Google Scholar]
- 24.Park HY, Park SH, Yoon HK, Han MJ, Kim DH. Anti-allergic activity of 18beta-glycyrrhetinic acid-3-O-beta-D-glucuronide. Arch Pharm Res 2004; 27: 57–60. [DOI] [PubMed] [Google Scholar]
- 25.Yano S, Harada M, Watanabe K, Nakamaru K, Hatakeyama Y, Shibata S et al. Antiulcer activities of glycyrrhetinic acid derivatives in experimental gastric lesion models. Chem Pharm Bull 1989; 37: 2500–4. [DOI] [PubMed] [Google Scholar]
- 26.Lino S, Tango T, Matsushima T Toda G, Miyake K, Hino K et al. Therapeutic effects of stronger neo-minophagen C at different doses on chronic hepatitis and liver cirrhosis. Hepatol Res 2001; 19: 31–40. [DOI] [PubMed] [Google Scholar]
- 27.Kumada H. Long-term treatment of chronic hepatitis C with glycyrrhizin [stronger neo-minophagen C(SNMC)] for preventing liver cirrhosis and hepatocellular carcinoma. Oncology 2002; 62: 94–100. [DOI] [PubMed] [Google Scholar]
- 28.Long DR, Mead J, Hendricks JM, Hardy ME, Voyich JM. 18β-Glycyrrhetinic acid inhibits methicillin-resistant Staphylococcus aureus survival and attenuates virulence gene expression. Antimicrob Agents Chemother 2013; 57: 241–7. 10.1128/AAC.01023-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li HE,Qiu JZ, Yang ZQ, Dong J, Wang JF, Luo MJ et al. Glycyrrhetinic acid protects mice from Staphylococcus aureus pneumonia. Fitoterapia 2012; 83: 241–248. 10.1016/j.fitote.2011.10.018 [DOI] [PubMed] [Google Scholar]
- 30.Inoue H, Mori T, Shibata S, Koshihara Y. Modulation by glycyrrhetinic acid derivatives of TPA-induced mouse ear oedema. Br J Pharmacol 1989; 96: 204–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue H, Inoue K, Takeuchi T, Nagata N, Shibata S. Inhibition of rat acute inflammatory paw oedema by dihemiphthalate of glycyrrhetinic acid derivatives: comparison with glycyrrhetinic acid. J Pharm Pharmacol 1993; 34: 1067–71. [DOI] [PubMed] [Google Scholar]
- 32.Finney RS, Somers GF. The anti-inflammatory activity of glycyrrhetinic acid and derivatives. J Pharm Pharmacol 1958; 10: 613–620. [DOI] [PubMed] [Google Scholar]
- 33.Komatsuzawa H, Fujiwara T, Nishi H, Yamada S, Ohara M, McCallum N et al. The gate controlling cell wall synthesis in Staphylococcus aureus. Mol Microbiol 2004; 53: 1221–31. 10.1111/j.1365-2958.2004.04200.x [DOI] [PubMed] [Google Scholar]
- 34.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 2003; 52:1 10.1093/jac/dkg301 [DOI] [PubMed] [Google Scholar]
- 35.Yarwood JM, Schlievert PM. Quorum sensing in Staphylococcus infections. J Clin Invest 2003; 112: 1620–5. 10.1172/JCI20442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003; 48: 1429–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overnight cultures of S. aureus MS23513 were adjusted to an OD at 660 nm of 1.0. Then, 100 μl of bacterial culture was inoculated in 5 ml TSB. Bacterial cultures were incubated at 37°C with shaking. When the OD at 660 nm reached 0.3, various concentrations (■: control; ♦: 1/64 MIC; ▲:1/16 MIC; ●: 1/4 MIC; ×: 1x MIC; 十: 2x MIC) of GR-SU or GRA were added to the medium. Growth and colony counts were monitored during growth. Three independent experiments were performed, and the mean ± SD was calculated. The data were analyzed for statistically significant differences compared to untreated control for each condition by a two-way ANOVA followed by Dunnett’s post hoc tests. *P<0.05.
(PPTX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
Microarray data are deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE80500.