SUMMARY
The primary cilium is an antenna-like, microtubule-based organelle on the surface of most vertebrate cells for receiving extracellular information. Although primary cilia form in the quiescent phase, ciliary disassembly occurs when quiescent cells re-enter the proliferative phase. It was shown that a mitotic kinase, Polo-like kinase 1 (PLK1), is required for cell-proliferation-coupled primary cilia disassembly. Here, we report that kinesin superfamily protein 2A (KIF2A), phosphorylated at T554 by PLK1, exhibits microtubule-depolymerizing activity at the mother centriole to disassemble the primary cilium in a growth-signal-dependent manner. KIF2A-deficient hTERT-RPE1 cells showed the impairment of primary cilia disassembly following growth stimulation. It was also found that the PLK1-KIF2A pathway is constitutively active in cells from patients with premature chromatid separation (PCS) syndrome and is responsible for defective ciliogenesis in this syndrome. These findings provide insights into the roles of the PLK1-KIF2A pathway in physiological cilia disassembly and cilia-associated disorders.
INTRODUCTION
The primary cilium is a microtubule (MT)-based, nonmotile projection located on the surface of most vertebrate cells that senses extracellular information to transduce signals required for cell proliferation, embryogenesis, and tissue homeostasis (Ishikawa and Marshall, 2011). Dysfunctions of primary cilia cause ciliopathies characterized by a range of clinical symptoms including polycystic kidney, obesity, neuronal, and other developmental abnormalities (Nigg and Raff, 2009). The dynamics of primary cilia assembly and disassembly is tightly controlled throughout the cell cycle (Tucker et al., 1979). In the G0 quiescent phase, the centrosome migrates to the apical cell surface, where the mother centriole is transformed to a basal body, with subsequent assembly of the primary cilium from the basal body (Kobayashi and Dynlacht, 2011; Nigg and Stearns, 2011). Upon re-entry into the cell cycle, the primary cilium disassembles quickly to release the centrioles to form a bipolar spindle assembly, suppressing primary cilium formation during the proliferative phase (Kobayashi and Dynlacht, 2011; Nigg and Stearns, 2011). In contrast to primary cilia assembly, little is known about the molecular basis underlying the induction of primary cilia disassembly and blocking of inappropriate ciliogenesis in the proliferative phase. Recent studies, however, have revealed that a mitotic kinase Polo-like kinase1 (PLK1) plays a pivotal role in growth-signal-dependent primary cilia disassembly (Lee et al., 2012; Seeger-Nukpezah et al., 2012; Wang et al., 2013a). PLK1 phosphorylates histone deacetylase 6 (HDAC6) to promote tubulin deacetylation and to destabilize the axonemal MTs of primary cilia (Wang et al., 2013a). PLK1 also interacts with a Wnt signal component Dishevelled 2 (DVL2) to initiate primary cilia disassembly through the activation of HDAC6 following noncanonical Wnt5a ligand stimulation (Lee et al., 2012). PLK1 regulates many substrates to orchestrate mitotic spindle formation and chromosomal stability (Bruinsma et al., 2012). However, as yet, HDAC6 is the only major PLK1 substrate identified in PLK1-mediated primary cilia disassembly.
The human kinesin-13 gene family consists of KIF2A, KIF2B, KIF2C/MCAK, and KIF24 (Walczak et al., 2013). Unlike conventional kinesin motor proteins involved in intracellular transport, kinesin-13 proteins do not “walk” along MTs but have the unique activity of ATP-dependent MT depolymerization (Walczak et al., 2013). The MT-depolymerizing activity of kinesin-13 proteins operates in a range of physiological contexts such as spindle assembly, chromosome segregation, and axonal growth (Ganem and Compton, 2004; Homma et al., 2003; Jang et al., 2009). Recently, it was reported that mammalian KIF24 blocks ciliogenesis by recruiting CP110 at the mother centrioles and remodeling centriolar MTs through its MT-depolymerizing activity. In addition, small interfering RNA (siRNA)-mediated knockdown of KIF24 causes inappropriate ciliogenesis even in cycling cells (Kobayashi et al., 2011). However, whether other kinesin-13 proteins are involved in primary cilia formation remains unclear. Previous studies have demonstrated that PLK1-mediated phosphorylation of KIF2A, KIF2B, and KIF2C controls their MT-depolymerizing activity for faithful chromosome segregation and spindle assembly, and the PLK1-phosphorylation sites on KIF2B and KIF2C, but not KIF2A, were identified (Hood et al., 2012; Jang et al., 2009; Zhang et al., 2011). PLK1-related biological links between ciliary disassembly and tuning of kinesin-13-mediated MT depolymerization led us to explore how PLK1 and kinesin-13s cooperate to regulate primary cilia disassembly in the proliferative phase. In this study, we report that PLK1 phosphorylates KIF2A at T554 in the subdistal appendages of the mother centriole to activate its MT-depolymerizing activity and disassemble primary cilia following growth stimulation.
Premature chromatid separation (PCS) syndrome (Mendelian Inheritance in Man [MIM] ID: 176430), also known as mosaic variegated aneuploidy (MVA) syndrome (MIM ID: 257300), is a rare autosomal recessive disorder caused by germline mutations in the budding uninhibited by benzimidazoles 1 homolog beta (BUB1B) gene, encoding BUBR1, a central player in the mitotic spindle assembly checkpoint (Hanks et al., 2004; Matsuura et al., 2006). The clinical symptoms of PCS (MVA) syndrome patients include a high risk of cancer and symptoms that overlap with the ciliopathy disease spectrum, including polycystic kidney, Dandy-Walker complex, and infantile obesity (Hanks et al., 2004; Matsuura et al., 2006; Miyamoto et al., 2011). We previously demonstrated that skin fibroblasts from these patients show reduced ciliogenesis, thus PCS (MVA) syndrome is defined as a ciliopathy (Miyamoto et al., 2011). Here, we show that the PLK1-KIF2A pathway is dysregulated in cells from PCS (MVA) syndrome patients and that this is a pathological mechanism of the observed ciliopathy disease in this syndrome.
RESULTS
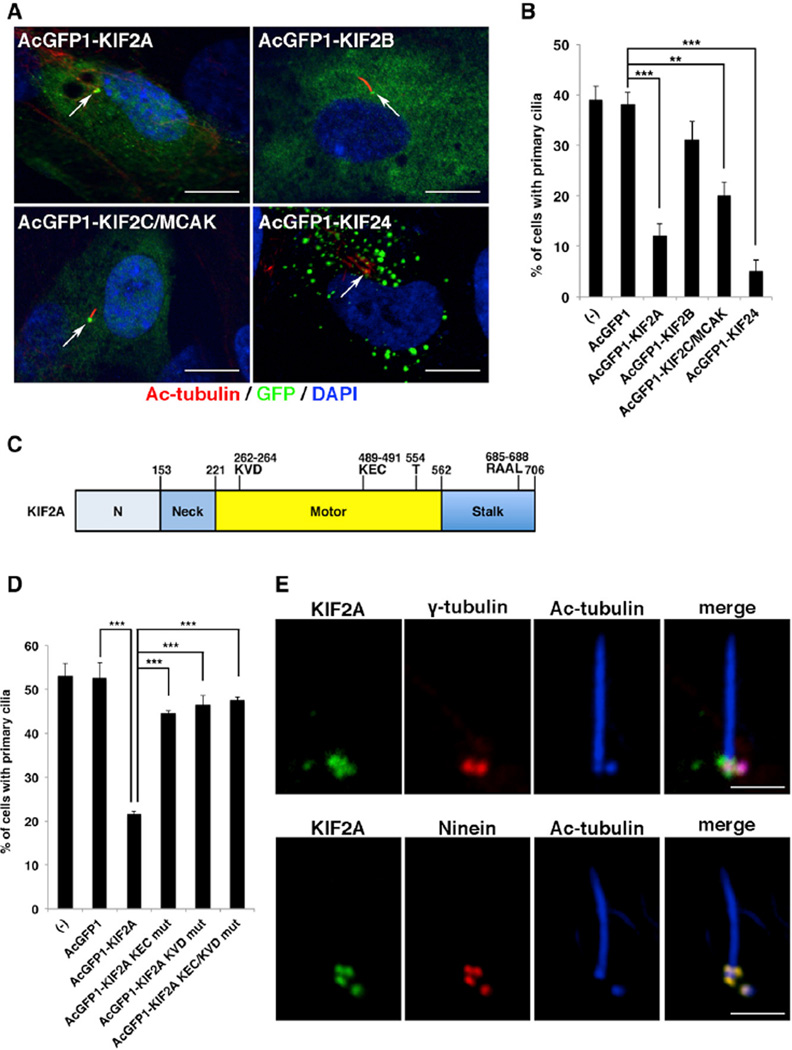
To investigate which kinesin-13 family members have primary cilia disassembly function, we analyzed the effects of exogenous AcGFP1-tagged kinesin-13s on primary cilia formation in serum-starved hTERT-RPE1cells (Figures 1A and 1B). In agreement with a previous report (Kobayashi et al., 2011), exogenous KIF24 formed aggregates in the cytoplasm and suppressed primary cilia to less than 5% (Figures 1A and 1B). We found that exogenous KIF2A, KIF2B, and KIF2C all localized to the centrosomes, and KIF2A and KIF2C, but not KIF2B, reduced ciliated cells to around 10%–20% compared with the control AcGFP1 (Figures 1A and 1B). Given that KIF2A suppressed primary cilia formation more efficiently than KIF2C, we focused on the KIF2A molecule as an effector of PLK1-dependent primary cilia disassembly in this study. KIF2A contains a central catalytic motor domain, an N-terminal neck domain, and a C-terminal stalk domain. The catalytic motor domain contains a MT-depolymerization motif (KVD) and a MT-binding motif (KEC), which are required for MT depolymerization (Figure 1C). The neck domain contains a kinesin-13 family-specific stretch of ~60 highly charged amino acids. The stalk domain is essential for KIF2A dimerization (Figure 1C). We transfected serum-starved hTERT-RPE1 cells with AcGFP1-tagged KIF2A loss-of-function mutants for KEC and KVD (Walczak et al., 2013) and evaluated the effect on ciliogenesis. Mutants for KEC, KVD, and KEC/KVD all localized to the centrosome but failed to reduce ciliated cells (Figure 1D). These results suggested that the MT-depolymerizing activity of KIF2A at the basal body is required for primary cilia suppression.
Figure 1. KIF2A Promotes Primary Cilia Disassembly through Microtubule-Depolymerizing Activity.
(A) hTERT-RPE1 cells transfected with plasmids expressing AcGFP1-tagged KIF2A, KIF2B, KIF2C/MCAK, or KIF24 and incubated for 24 hr without serum were immunostained with anti-GFP (green) and anti-acetylated tubulin (red) antibodies. DNA was stained with DAPI (blue). Arrow indicates the centrosome/basal body. The scale bar represents 10 µm.
(B) The percentage of hTERT-RPE1 cells with primary cilium from (A) was significantly reduced upon exogenous KIF2A, KIF2C, or KIF24 expression (means ± SD; **p < 0.01; ***p < 0.001; t test; n = 3; >200 cells per experiment).
(C) Schematic of KIF2A structure. The neck domain is a kinesin-13-family-specific stretch of ~60 amino acids. The central catalytic motor domain contains a MT-depolymerization motif (KVD), a MT-binding motif (KEC), and the PLK1-phosphorylation residue (T554). An APC/C recognition motif, D-box (RAAL), is located in the stalk domain (see also Figure S1). The stalk domain is necessary for KIF2A dimerization.
(D) Mutations in either or both the microtubule-binding motif (KEC) and microtubule-depolymerizing motif (KVD) of KIF2A decreased the negative activity of ciliogenesis in quiescent hTERT-RPE1 cells (means ± SD; ***p < 0.001; t test; n = 3; >200 cells per experiment).
(E) Immunofluorescence staining of hTERT-RPE1 cells in quiescent G0 phase with anti-KIF2A (green), anti-acetylated-tubulin (blue), and anti-ninein (red) or anti-γ-tubulin (red) antibodies. KIF2A colocalized with ninein, but not with γ-tubulin. The scale bar represents 2.5 µm.
To precisely determine the localization of KIF2A in the quiescent G0 phase, we stained endogenous KIF2A in hTERT-RPE1 cells with anti-KIF2A antibody. Three KIF2A-positive spots at the mother centriole and one KIF2A-positive spot at the daughter centriole were observed. These colocalized with ninein signals, which correspond to the subdistal appendages of the mother centriole and the proximal ends of both centrioles, but did not exactly colocalize with the centrosomal signals of γ-tubulin (Figure 1E). KIF24 also localized to both centrioles during the G0 phase (Figure S3I). The signals of KIF24 at the mother centriole were partially overlapped with the distal spots of ninein and detected at the more-distal portion residing at subdistal appendages (Figure S3I).
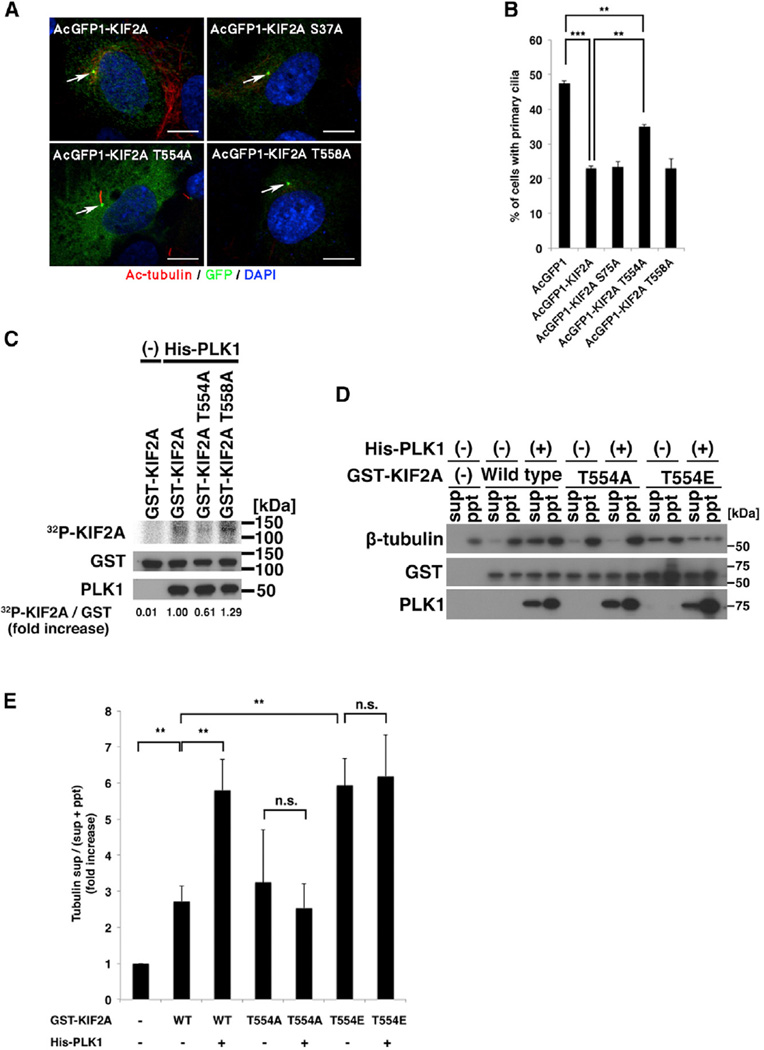
Next, we attempted to identify the PLK1-phosphorylation site on KIF2A associated with primary cilia disassembly. KIF2A contains three conserved motifs for PLK1 phosphorylation (D/E-X-S/T-X1–3-X-D/E; Nakajima et al., 2003). To test whether any of these PLK1 motifs are required for primary cilia suppression, we constructed AcGFP1-tagged alanine-substituted mutants (S37A, T554A, or T558A), which were expected to reduce the MT-depolymerizing activity of KIF2A. We introduced them into serum-starved hTERT-RPE1 cells and counted ciliated cells. Although the S37A and T558A mutants showed similar activity to wild-type KIF2A, the T554A KIF2A mutant did not reduce the numbers of primary cilia observed to the same extent (Figures 2A and 2B). These results suggested that the T554 residue is involved in the KIF2A-mediated primary cilia suppression. The percentage of ciliated cells expressing KIF2A-T554A was also significantly different from the control AcGFP1 (Figure 2B), suggesting a residual (basal) activity of this mutant for primary cilia suppression independently of Plk1 phosphorylation. To confirm that this residue is indeed the PLK1-phosphorylation target, we performed an in vitro kinase assay with His-tagged PLK1 and glutathione S-transferase (GST)-tagged KIF2A, the T558A mutant or the predicted phosphoresistant T554A mutant. Wild-type KIF2A and the T558A mutant were phosphorylated by PLK1. On the other hand, the T554A mutant showed intermediate level of PLK1 phosphorylation between control and the wild-type KIF2A (Figure 2C). These results suggested that T554 on KIF2A is at least one of the PLK1-phosphorylation sites. We then examined whether the MT-depolymerizing activity of KIF2A is regulated by PLK1 phosphorylation of T554 on KIF2A using an in vitro MT-depolymerization assay. Recombinant KIF2A, KIF2A T554A, or KIF2A phosphomimetic mutant (T554E) mutant were purified and mixed with PLK1 and ATP and then with Taxol-polymerized MTs. The reaction mixtures were fractionated using ultracentrifugation, with the remaining MTs found in pellets and depolymerized tubulins in supernatants. Recombinant KIF2A and PLK1 did not exhibit pelleting in the absence of MTs (data not shown), whereas they were coprecipitated with MTs (Figure 2D). KIF2A alone induced the depolymerization of MT, suggesting KIF2A has a basal and PLK1-independent activity for MT depolymerization. Incubation of KIF2A with PLK1 enhanced the MT-depolymerizing activity of KIF2A, whereas KIF2A T554A did not effectively depolymerize Taxol-polymerized MTs even in the presence of PLK1 (Figures 2D and 2E). KIF2A T554E alone showed the MT-depolymerizing activity similar to that of PLK1-treated wild-type KIF2A (Figures 2D and 2E). We concluded that PLK1 phosphorylates KIF2A at T554 to promote MT depolymerization.
Figure 2. PLK1 Phosphorylates KIF2A at Thr 554 to Enhance Microtubule Depolymerization and Primary Cilia Disassembly.
(A) hTERT-RPE1 cells were transfected with AcGFP1-tagged KIF2A or AcGFP1-tagged KIF2A with alanine substitutions at PLK1 phosphorylation consensus sites, serum starved for 24 hr, and then immunostained with anti-GFP (green) and anti-acetylated-tubulin (red) antibodies. DNA was stained with DAPI (blue). Arrow indicates the centrosome/basal body. The scale bar represents 10 µm.
(B) Percentage of cells with primary cilia in GFP-positive cells from (A). The T554A mutation of KIF2A significantly diminished the negative effect on ciliogenesis by KIF2A (means ± SD; **p < 0.01; ***p < 0.001; t test; n = 3; >200 cells per experiment).
(C) In vitro kinase assay to validate the PLK1-mediated phosphorylation site on KIF2A. GST-tagged KIF2A, KIF2A T554A, or KIF2A T558A were incubated with His-tagged PLK1 and γ-32P-ATP and analyzed by autoradiography. Western blotting indicates the loading level of PLK1 and GST-tagged proteins. The level of 32P-uptake into GST-tagged KIF2A recombinants was normalized to GST.
(D) GST-tagged KIF2A, KIF2A T554A, and KIF2A T554E were phosphorylated with His-PLK1 and incubated with Taxol-stabilized microtubules for 10 min. The reaction mix was centrifuged at 100,000 g for 30 min to separate depolymerized tubulin (sup) and microtubule (ppt) fractions, followed by detection of tubulin, KIF2A, and PLK1 with western blotting.
(E) The fold increase of the ratios of tubulin in the supernatant to total input of tubulin [tubulin sup/(sup + ppt)] was calculated. Data from three independent experiments were shown. Error bars represent SD. PLK1 enhanced the MT-depolymerizing activity of wild-type KIF2A, but not KIF2A T554A (means ± SD; **p < 0.01; t test). KIF2A T554E effectively depolymerized MTs even in the absence of PLK1. n.s., not significant; WT, wild-type.
To examine how PLK1-mediated phosphorylation of KIF2A is regulated in the context of primary cilia disassembly, we raised anti-phospho-KIF2A (T554) polyclonal antibody and analyzed the centrosomal localization of phosphorylated KIF2A. We first validated that the antibody specifically recognized KIF2A phosphorylated at T554, but not the KIF2A T554A mutant (Figure 3A), and that the level of phosphorylated KIF2A in the whole-cell lysate of BI2536-treated hTERT-RPE1 cells was decreased (Figure S4A). We next examined the precise localization of KIF2A by immunostaining at 0 hr and 4 hr following serum stimulation of 24 hr serum-starved hTERT-RPE1 cells. Total KIF2A signals were detected at both the mother and daughter centriole at all time points (Figure 3B). In contrast, phospho-KIF2A (T554) signals became increasingly enriched in the subdistal appendages of the mother centriole following serum stimulation and were decreased by treatment with BI3536, a PLK1 inhibitor (Figure 3C). Phospho-PLK1 (T210) signals showed similar temporal and spatial localization to those of phospho-KIF2A (T554; Figure 3D). These data demonstrate that PLK1 at the subdistal appendages phosphorylates KIF2A at T554 in a growth-signal-dependent manner. BI2536 treatment impaired suppression of primary cilia by wild-type KIF2A (Figures 3E and 3F). To evaluate the KIF2A T554 phosphorylation effect on ciliogenesis, we introduced KIF2A phosphomimetic mutant (T554E) into serum-starved hTERT-RPE1 cells. Even in the presence of BI2536, exogenous KIF2A T554E reduced ciliated hTERT-RPE1 cells compared with KIF2A and its phosphoresistant mutant (T554A; Figures 3E and 3F), suggesting that the T554E substitution enhances the basal activity of KIF2A for suppressing primary cilia formation. Mutations in the MT-binding motif (KEC) and MT-depolymerizing motif (KVD) abolished the phosphomimetic mutation effect on ciliogenesis (Figures 3E and 3F). Taken together, KIF2A is phosphorylated at T554 by PLK1 in the subdistal appendages of the mother centriole, which enhances MT depolymerization to disassemble primary cilia in a growth-signal-dependent manner.
Figure 3. PLK1-Mediated KIF2A Phosphorylation Occurs at the Mother Centriole following Growth Stimulation.
(A) PLK1-mediated phosphorylation of KIF2A at T554 in vitro was probed by western blotting with the anti-phospho-KIF2A (T554) rabbit polyclonal antibody. The loading levels of His-PLK1 and GST-tagged KIF2A proteins were also detected by western blotting.
(B–D) After 24 hr serum starvation, hTERT-RPE1 cells were stimulated with 10% serum for 4 hr in the presence or absence of 100 nM BI2536 and then immunostained with anti-ninein (red), anti-acetylated tubulin (blue), and anti-KIF2A (B); anti-phospho-T554-KIF2A (C); or anti-phospho-T210 PLK1 (D) (green) antibodies. In the presence of 100 nM BI2536, the phospho-T554-KIF2A and phospho-T210-PLK1 signals at the mother centriole were rarely detected at 4 hr post-serum stimulation. The scale bar represents 2 µm.
(E) hTERT-RPE1 cells were transfected with AcGFP1-tagged KIF2A, PLK1 phosphorylation mimic KIF2A mutant (T554E), or KIF2A T554E mutant with KEC/KVD motif double mutations and cultured without serum for 24 hr in the presence or absence of 100 nM BI2536 before immunostaining with anti-GFP (green) and anti-acetylated- tubulin (red) antibodies. DNA was stained with DAPI (blue). Arrow indicates the centrosome/basal body. The scale bar represents 10 µm.
(F) Percentage of cells with primary cilia in GFP-positive cells from (E). The phospho-mimic T554E KIF2A mutant significantly inhibited ciliogenesis even in the presence of 100 nM BI2536. KEC/KVD motif mutations of KIF2A abrogated the T554E enhancement of primary cilia disassembly (means ± SD; ***p < 0.001; t test; n = 3; >200 cells per experiment).
We investigated the levels of phospho-KIF2A (T554) and total KIF2A in the quiescent or proliferating hTERT-RPE1 cells by western blot analysis. Both the levels of phospho-KIF2A (T554) and total KIF2A normalized by GAPDH were increased in a growth-stimulation-dependent manner (Figure S1A), but the increased rates were different. These results suggested that the half-life of KIF2A is distinctly regulated during the cell cycle. Therefore, we speculated that KIF2A is differently ubiquitinated in quiescent compared with proliferating hTERT-RPE1 cells. As expected, KIF2A was polyubiquitinated in the quiescent G0 phase, whereas the ubiquitination level in the cycling phase was decreased (Figure S1B). Because kinesin-13s contain consensus sequences for the destruction box (D-box: RXXL), which is recognized by anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase, we explored a physical interaction between kinesin-13s and an APC/C component, CDC27, using immunoprecipitation analysis with anti-FLAG antibody in HEK293T cells transfected with 3× FLAG-tagged kinesin-13s. All members of the kinesin-13 family were able to bind to CDC27 (Figure S1C), suggesting that KIF2A is polyubiquitinated by APC/C. We constructed three D-box mutants for KIF2A by substituting RXXL in the D-box with AXXA. Mutation of RAAL in the D-box 3 at 685–688 amino acids (aas) inhibited its ubiquitination (Figure S1E), indicating that APC/C recognizes the D-box 3 of KIF2A. The D-box mutation did not alter the centrosomal localization but enhanced the ciliary disassembly activity of KIF2A, whereas this effect was abrogated by mutations within the MT-binding and depolymerizing motifs (Figures S1F and S1G). We thus concluded that KIF2A is degraded through the APC/C-mediated ubiquitin/proteasome system in the quiescent G0 phase.
To explore the physiological function of human KIF2A more precisely, we examined the effect of disruption of KIF2A on primary cilia disassembly following growth stimulation and suppression of ciliogenesis during the proliferative phase. We constructed expression vectors encoding transcription-activator-like effector nucleases (TALENs) to introduce DNA double-strand breaks into human KIF2A exon 10, corresponding to 291–321 aas in the functional P loop (Figures S2A and S3A). Targeting vectors were designed to disrupt the KIF2A gene by replacing exon 10 with a gene cassette of a herpes simplex virus thymidine kinase gene and neomycin- or puromycin-resistant genes separated by a 2A peptide sequence, allowing expression of the distinct proteins from a single open reading frame. After transfection and selections by puromycin and neomycin in hTERT-RPE1 cells, one heterozygous (KIF2A +/−) and four homozygous (KIF2A −/−) mutant cell clones were obtained. Southern blot analysis confirmed the disruption of the KIF2A gene without random integration of the targeting vectors (Figures S2B and S3B). RT-PCR, western blotting, and immunostaining analyses using anti-KIFA2A and anti-phospho-KIF2A (T554) antibodies all showed no KIF2A products in the two KIF2A −/− cells (Figures S2C–S2E, S3C, S4A, S4C, and S4D). Therefore, the two KIF2A −/− clones were used for subsequent studies.
The frequency and length of primary cilia were examined in 24 hr serum-starved KIF2A −/− cells. They showed no significant change in ciliogenesis compared with those of KIF2A +/+ cells (Figures S2F, S2G, S3D, and S3E). After serum stimulation, primary cilia disassembly was delayed in KIF2A −/− cells (Figures S2F, S2G, S3D, and S3E), indicating that KIF2A is required for primary cilia disassembly following growth stimulation. KIF2A −/− cells also showed impaired blocking of inappropriate primary cilia formation during the cycling phase (Figures S3F and S3G). However, KIF2A-deficient cells were not able to completely attenuate primary cilia disassembly and blocking of inappropriate ciliogenesis (Figures S2F, S2G, S3F, and S3G). An immunoprecipitation experiment indicated that KIF2A potentially interacts with CP110, which is a binding molecule of KIF24 for blocking aberrant ciliogenesis (Figure S1D). The centriolar localizations of KIF24 and CP110 were not altered in KIF2A −/− cells (Figures S3H and S3I), suggesting that KIF24 in KIF2A −/− cells redundantly plays a role of blocking ciliogenesis during the cycling phase. Most of KIF2A −/− hTERT-RPE1 cells showed normal bipolar spindle formation, whereas a small fraction of them had multipolar spindle (Figures S4E and S4F). Depletion of KIF2A in hTERT-RPE1 cells did not significantly affect cell-cycle progression, as determined by flow cytometry (Figure S4G). These data suggest that ciliary phenotypes in KIF2A −/− cells are not secondary to the abnormalities of mitosis and cell-cycle progression.
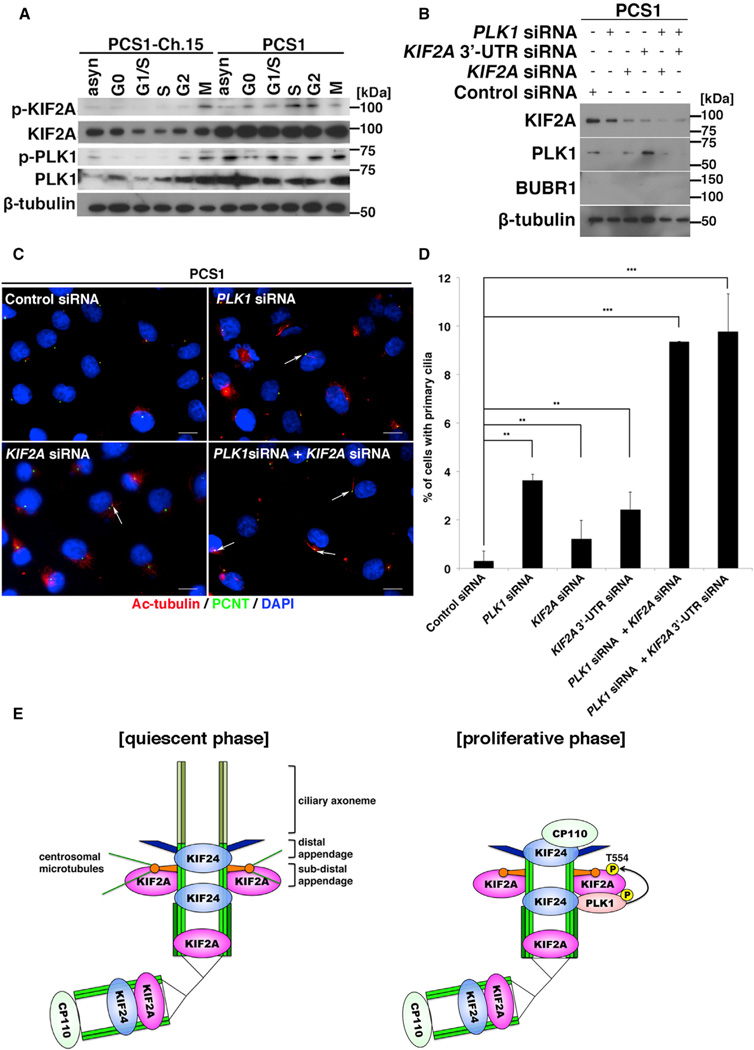
In PCS (MVA) patient cells, PLK1 was aberrantly activated throughout the cell cycle (Figure 4A; Izumi et al., 2009). We therefore examined whether reduced ciliogenesis in the patient cells is owing to the constitutive activation of PLK1. PLK1 inhibition with siRNA or BI2536 partially restored primary cilia formation in the patient cells (data not shown), suggesting that aberrant PLK1 activity contributes to defective ciliogenesis in PCS (MVA) syndrome. To address whether the deregulation of KIF2A in the patient cells contributes to the pathological mechanism, the expression and phosphorylation levels of KIF2A were examined by western blotting analysis. In patient (PCS1) cells transferred with a whole chromosome 15 containing the BUB1B locus (PCS1-Ch.15 cells), the expression level of KIF2A gradually increased from G1 to M phase, with T554 phosphorylation strongly detected in M phase, correlating with PLK1 dynamics in the cell cycle (Figure 4A). In contrast, in PCS1 cells, KIF2A was highly expressed and phosphorylated at T554 throughout the cell cycle (Figure 4A), suggesting that KIF2A is involved in the defective ciliogenesis observed in patient cells. Consistent with this, siRNA-mediated knockdown of KIF2A in PCS1 cells resulted in partial recovery of primary cilia formation (Figures 4B–4D). A double knockdown of PLK1 and KIF2A increased cilia restoration compared with PLK1 or KIF2A siRNA-mediated knockdown alone (Figures 4B–4D), suggesting that a constitutively active PLK1-KIF2A molecular pathway is underlying the ciliopathy spectrum in PCS (MVA) syndrome. Based on these results, we propose a model where (1) in the quiescent G0 phase, the enzymatic activity of PLK1 and KIF2A is negatively regulated to allow primary cilia assembly and (2) growth signals are transmitted to the PLK1-KIF2A cascade to enhance the MT-depolymerizing activity of KIF2A at the mother centriole for cell-proliferation-coupled primary cilia disassembly (Figure 4E).
Figure 4. Inhibition of the PLK1-KIF2A Pathway Rescues Ciliogenesis in PCS Syndrome.
(A) Phosphorylation levels of PLK1 (T210) and KIF2A (T554) during the cell cycle in immortalized fibroblast cells from a patient (PCS1) with PCS (MVA) syndrome and PCS1 cells transferred with a whole chromosome 15 containing the BUB1B locus (PCS1-Ch.15 cells) were analyzed by western blotting.
(B) siRNA-mediated knockdown of PLK1 and/or KIF2A in PCS1 cells was evaluated by western blotting. β-tubulin served as a loading control.
(C) PCS1 cells transfected with control siRNA, PLK1 siRNA, KIF2A siRNA, or a combination of PLK1 siRNA and KIF2A siRNA were cultured without serum for 24 hr and then immunostained with anti-acetylated tubulin (red) and anti-pericentrin (green) antibodies. DNA was stained with DAPI (blue). Arrows indicate primary cilia. The scale bar represents 10 µm.
(D) Quantification of the percentage of ciliated cells from (C). Knockdown of PLK1 and/or KIF2A in PCS1 cells significantly restored ciliogenesis (means ± SD; **p < 0.01; ***p < 0.001; t test; n = 3; >200 cells per experiment).
(E) Model for the physiological activation of the PLK1-KIF2A pathway in the context of primary cilia disassembly activated by cell-cycle re-entry. During the proliferative phase, KIF2A phosphorylated at T554 byPLK1 enhances the MT disassembly at the subdistal appendage for primary cilia disassembly, whereas KIF24 at the more-distal end than the subdistal appendage recruits CP110 to prevent inappropriate ciliogenesis (Tsang and Dynlacht, 2013). In the pathological context of PCS (MVA) syndrome, germline mutations of BUB1B gene encoding BUBR1 causes the constitutive activation of the PLK1-KIF2A pathway to impair ciliogenesis. See also Figures S2–S4.
DISCUSSION
Primary cilia disassembly following growth stimulation is a critical step in cell proliferation because the basal body/centrosome beneath the cell membrane must be released to act as a mitotic bipolar spindle pole. However, the mechanism by which primary cilia disassembly is coupled with cell proliferation is less well understood. Here, we found that KIF2A promotes primary cilia disassembly via its MT-depolymerizing activity in a growth-signal-dependent manner. It has been established that metazoan KIF2A localizes to mitotic spindle MTs and poles (Figure S3C) and controls spindle assembly and poleward MT flux for proper chromosome segregation through its MT-depolymerizing activity (Ganem and Compton, 2004; Jang et al., 2009). Studies in KIF2A-knockout mice showed that KIF2A in postmitotic neurons regulates the number and length of collateral branches from an axon (Homma et al., 2003; Maor-Nof et al., 2013), indicating the existence of nonmitotic functions of KIF2A. Interestingly, ancient kinesin-13 members in Giardia, Leishmania, and Chlamidomonas regulate flagellar length (Blaineau et al., 2007; Dawson et al., 2007; Piao et al., 2009; Wang et al., 2013b). Importantly, it was reported that human KIF24 suppresses inappropriate ciliogenesis in proliferating cells by stabilizing CP110 at the distal end of mother centrioles and by remodeling centriolar MTs via its MT-depolymerizing activity (Kobayashi et al., 2011). Therefore, whether the other kinesin-13s play a role in the control of primary cilia disassembly during the proliferative phase remains an open question. In this study, overexpression of each kinesin-13 protein in serum-starved hTERT-RPE1 cells indicated that all members did not equally reduce ciliated cells in the quiescent G0 phase and that KIF2A was able to mediate primary cilia disassembly through its MT-depolymerizing activity with a similar potency to KIF24 (Figures 1A and 1B). Loss of KIF2A by TALEN-mediated knockout in hTERT-RPE1 cells caused impairment of primary cilia disassembly following growth stimulation and inappropriate ciliogenesis in the proliferative phase (Figures S2 and S3); thereby, the MT-depolymerizing activity of KIF2A is essential for primary cilia disassembly coupled with cell proliferation and blocking inappropriate ciliogenesis during the cycling phase. However, depletion of KIF2A did not completely impair primary cilia disassembly and suppression (Figures S2F, S2G, S3F, and S3G). The mother centriole localization of KIF24, which has the strongest potency to suppress ciliogenesis among kinesin-13 family members (Figures 1A and 1B), is not changed in KIF2A-deficient cells (Figure S3I). KIF2A potentially binds to CP110 in vitro as well as KIF24 does (Figure S1D). These data imply that KIF24 redundantly acts in the contexts of KIF2A-mediated cilia disassembly and suppression. Further analyses of all kinesin-13 family members will be required to elucidate how primary cilia disassembly is orchestrated.
How does KIF2A disassemble primary cilia via the MT depolymerizaton? Here, we demonstrate that KIF2A localizes to the subdistal appendages of the mother centrioles during the interphase. However, the subdistal appendages topologically seem unlikely to be the relevant site of microtubule depolymerization for primary cilia disassembly, because they are not the free ends of axonemal microtubules. Recently, it was reported that the subdistal appendages are required for the stabilization of centrosomal (cytoplasmic) MTs during the quiescent phase (Tateishi et al., 2013). One possible mechanism for primary cilia disassembly is that KIF2A at the subdistal appendages might depolymerize centrosomal (cytoplasmic) MTs to arrest transport of materials such as tubulin into cilium (Bhogaraju et al., 2013). Alternatively, it was proposed that cytosolic tubulin concentration modulates cilia length and number (Sharma et al., 2011). Depletion of KIF2A did not increase the total amount of tubulin (Figure S2D) but promoted to polymerize cytoplasmic microtubules (Figures S4B–S4D), suggesting that KIF2A controls the cytoplasmic tubulin concentration to regulate primary cilia disassembly. Although further studies are required to clarify how the MT-depolymerizing activity of KIF2A is transmitted to primary cilia disassembly, this study demonstrates that the MT-depolymerizing activity of KIF2A at the mother centriole is a driving force of primary cilia disassembly during the proliferative phase.
Our results indicate that KIF2A is quantitatively controlled by APC/C ubiquitin ligase during the cell cycle. KIF2A levels were relatively low in the quiescent G0 phase but increased during the proliferative phase (Figure S1A). APC/C activity is tightly regulated during the cell cycle through the binding of CDC20 or another coactivator CDH1 (Peters, 2006). CDC20 binds to APC/C in a CDK1/cyclin B-dependent manner to initiate the metaphase-anaphase transition through ubiquitin/proteasome-mediated degradation of securin and cyclin B. By the end of mitosis, APC/CCDC20 is no longer active. From late M to G1 and G0 phases, CDH1 preferentially interacts with APC/C to activate the APC/CCDH1 ubiquitin ligase activity. Previous studies indicated that APC/CCDH1 activity is indispensable for apical docking of the basal body via tuning of the level of its substrate DVL in the context of ciliogenesis during the G0 phase (Ganner et al., 2009; Miyamoto et al., 2011). To the contrary, a recent study demonstrated that CDC20 during the G0 phase localizes to centrosomes to regulate the proper ciliary length and that it is released from centrosomes during the proliferative phase (Wang et al., 2014). We found that KIF2A is ubiquitinated by APC/C during the quiescent G0 phase and its ubiquitination is inhibited to increase the total amount of KIF2A in the proliferative phase. Interestingly, KIF24 expression is also low in the G0/G1 phase (Kobayashi et al., 2011) and this study demonstrates that it binds to an APC/C subunit (Figure S1C), implying that APC/C in the quiescent G0 phase ubiquitinates both KIF2A and KIF24 to robustly suppress premature initiation of cilia disassembly. Thus, PLK1 and APC/C-mediated dual regulation connect the MT-depolymerizing activity of KIF2A to a physiological primary cilia disassembly during the proliferative phase.
Previously, we showed that BUBR1 in the G0 phase mediates ubiquitin-mediated proteasome degradation of CDC20 to regulate the optimal level of DVL proteins for apical docking of the basal body in ciliogenesis and, consequently, high levels of DVLs in the PCS (MVA) syndrome patient cells block primary cilia formation (Miyamoto et al., 2011). We also showed that, in the interphase, BUBR1 localizes to centrosomes to inhibit PLK1 activity (Izumi et al., 2009) and that the PCS (MVA) syndrome patient cells had an aberrant activation of PLK1 throughout the cell cycle (Figure 4A). In this study, we found an additional pathological mechanism of impaired ciliogenesis in PCS (MVA) syndrome. Our data demonstrated that the aberrant PLK1-KIF2A pathway contributes to the ciliopathy disease spectrum in PCS (MVA) syndrome.
In conclusion, we demonstrate that the PLK1-KIF2A pathway is essential for primary cilia disassembly induced by growth signals and that constitutive activation of this pathway is responsible for the defective ciliogenesis of the PCS (MVA) syndrome.
EXPERIMENTAL PROCEDURES
Cell Culture
hTERT-RPE1, HEK293T, an immortalized fibroblast cell line from a patient with the PCS (MVA) syndrome (PCS1; Matsuura et al., 2006; Miyamoto et al., 2011), and chromosome-15-transferred PCS1 cells (PCS1-Ch.15) (Matsuura et al., 2006; Miyamoto et al., 2011) were maintained in DMEM supplemented with 10% fetal bovine serum at 37°C with 5% CO2. Transfection of plasmids or siRNAs into cells was performed using Lipofectamine LTX reagent (Life Technologies) or Lipofectamine RNAi MAX reagent (Life Technologies), respectively, according to the manufacturer’s protocol. At 24 hr after transfection, the medium was replaced with serum-free DMEM and the cells were incubated for 24 hr to become Ki-67-negative and achieve quiescent G0 phase to observe ciliogenesis. To avoid the possibility of ciliogenesis induced by contact inhibition, cells were maintained at <100% confluence in all experiments. Cell-cycle profile was analyzed with Muse Cell Analyzer (Millipore).
Detailed experimental procedures are included in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Drs. A. Inoko and Y. Saito for critical reading and helpful discussions. We also thank Dr. H. Hosoya for technical support and Ms. Y. Tonouchi for assistance. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to S.M. and T.M.); a Grant-in-Aid for Scientific Research from the Ministry of Health, Labour and Welfare (to S.M.); research grants from Naito Foundation (to S.M.), Takeda Science Foundation (to T.M.), and Tsuchiya Medical Foundation (to T.M.); a research grant from Platform for Dynamic Approach to Living Systems from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to H.O.); and an NIH grant R01HD069647 (to B.D.D.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.01.003
REFERENCES
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, Lorentzen E. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pagès M, Bastien P. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr. Biol. 2007;17:778–782. doi: 10.1016/j.cub.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Bruinsma W, Raaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem. Sci. 2012;37:534–542. doi: 10.1016/j.tibs.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Dawson SC, Sagolla MS, Mancuso JJ, Woessner DJ, House SA, Fritz-Laylin L, Cande WZ. Kinesin-13 regulates flagellar, interphase, and mitotic microtubule dynamics in Giardia intestinalis. Eukaryot. Cell. 2007;6:2354–2364. doi: 10.1128/EC.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganner A, Lienkamp S, Schäfer T, Romaker D, Wegierski T, Park TJ, Spreitzer S, Simons M, Gloy J, Kim E, et al. Regulation of ciliary polarity by the APC/C. Proc. Natl. Acad. Sci. USA. 2009;106:17799–17804. doi: 10.1073/pnas.0909465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Méhes K, Nash R, Robin N, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- Homma N, Takei Y, Tanaka Y, Nakata T, Terada S, Kikkawa M, Noda Y, Hirokawa N. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell. 2003;114:229–239. doi: 10.1016/s0092-8674(03)00522-1. [DOI] [PubMed] [Google Scholar]
- Hood EA, Kettenbach AN, Gerber SA, Compton DA. Plk1 regulates the kinesin-13 protein Kif2b to promote faithful chromosome segregation. Mol. Biol. Cell. 2012;23:2264–2274. doi: 10.1091/mbc.E11-12-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Izumi H, Matsumoto Y, Ikeuchi T, Saya H, Kajii T, Matsuura S. BubR1 localizes to centrosomes and suppresses centrosome amplification via regulating Plk1 activity in interphase cells. Oncogene. 2009;28:2806–2820. doi: 10.1038/onc.2009.141. [DOI] [PubMed] [Google Scholar]
- Jang CY, Coppinger JA, Seki A, Yates JR, 3rd, Fang G. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J. Cell Sci. 2009;122:1334–1341. doi: 10.1242/jcs.044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J. Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 2011;145:914–925. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- Lee KH, Johmura Y, Yu LR, Park JE, Gao Y, Bang JK, Zhou M, Veenstra TD, Yeon Kim B, Lee KS. Identification of a novel Wnt5a-CK1ε-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J. 2012;31:3104–3117. doi: 10.1038/emboj.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor-Nof M, Homma N, Raanan C, Nof A, Hirokawa N, Yaron A. Axonal pruning is actively regulated by the microtubule-destabilizing protein kinesin superfamily protein 2A. Cell Rep. 2013;3:971–977. doi: 10.1016/j.celrep.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Matsuura S, Matsumoto Y, Morishima K, Izumi H, Matsumoto H, Ito E, Tsutsui K, Kobayashi J, Tauchi H, Kajiwara Y, et al. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am. J. Med. Genet. A. 2006;140:358–367. doi: 10.1002/ajmg.a.31069. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Porazinski S, Wang H, Borovina A, Ciruna B, Shimizu A, Kajii T, Kikuchi A, Furutani-Seiki M, Matsuura S. Insufficiency of BUBR1, a mitotic spindle checkpoint regulator, causes impaired ciliogenesis in vertebrates. Hum. Mol. Genet. 2011;20:2058–2070. doi: 10.1093/hmg/ddr090. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Piao T, Luo M, Wang L, Guo Y, Li D, Li P, Snell WJ, Pan J. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc. Natl. Acad. Sci. USA. 2009;106:4713–4718. doi: 10.1073/pnas.0808671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger-Nukpezah T, Liebau MC, Höpker K, Lamkemeyer T, Benzing T, Golemis EA, Schermer B. The centrosomal kinase Plk1 localizes to the transition zone of primary cilia and induces phosphorylation of nephrocystin-1. PLoS ONE. 2012;7:e38838. doi: 10.1371/journal.pone.0038838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Kosan ZA, Stallworth JE, Berbari NF, Yoder BK. Soluble levels of cytosolic tubulin regulate ciliary length control. Mol. Biol. Cell. 2011;22:806–816. doi: 10.1091/mbc.E10-03-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J. Cell Biol. 2013;203:417–425. doi: 10.1083/jcb.201303071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Dynlacht BD. CP110 and its network of partners coordinately regulate cilia assembly. Cilia. 2013;2:9. doi: 10.1186/2046-2530-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Gayek S, Ohi R. Microtubule-depolymerizing kinesins. Annu. Rev. Cell Dev. Biol. 2013;29:417–441. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]
- Wang G, Chen Q, Zhang X, Zhang B, Zhuo X, Liu J, Jiang Q, Zhang C. PCM1 recruits Plk1 to the pericentriolar matrix to promote primary cilia disassembly before mitotic entry. J. Cell Sci. 2013a;126:1355–1365. doi: 10.1242/jcs.114918. [DOI] [PubMed] [Google Scholar]
- Wang L, Piao T, Cao M, Qin T, Huang L, Deng H, Mao T, Pan J. Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. J. Cell Sci. 2013b;126:1531–1540. doi: 10.1242/jcs.124255. [DOI] [PubMed] [Google Scholar]
- Wang W, Wu T, Kirschner MW. The master cell cycle regulator APC-Cdc20 regulates ciliary length and disassembly of the primary cilium. eLife. 2014;3:e03083. doi: 10.7554/eLife.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Shao H, Huang Y, Yan F, Chu Y, Hou H, Zhu M, Fu C, Aikhionbare F, Fang G, et al. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 2011;286:3033–3046. doi: 10.1074/jbc.M110.165340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.