Abstract
Background
De novo mutations are a frequent cause of disorders related to brain development. We report the results of screening patients diagnosed with both epilepsy and intellectual disability (ID) using exome sequencing to identify known and new causative de novo mutations relevant to these conditions.
Methods
Exome sequencing was performed on 39 patient–parent trios to identify de novo mutations. Clinical significance of de novo mutations in genes was determined using the American College of Medical Genetics and Genomics standard guidelines for interpretation of coding variants. Variants in genes of unknown clinical significance were further analysed in the context of previous trio sequencing efforts in neurodevelopmental disorders.
Results
In 39 patient–parent trios we identified 29 de novo mutations in coding sequence. Analysis of de novo and inherited variants yielded a molecular diagnosis in 11 families (28.2%). In combination with previously published exome sequencing results in neurodevelopmental disorders, our analysis implicates HECW2 as a novel candidate gene in ID and epilepsy.
Conclusions
Our results support the use of exome sequencing as a diagnostic approach for ID and epilepsy, and confirm previous results regarding the importance of de novo mutations in this patient group. The results also highlight the utility of network analysis and comparison to previous large-scale studies as strategies to prioritise candidate genes for further studies. This study adds knowledge to the increasingly growing list of causative and candidate genes in ID and epilepsy and highlights HECW2 as a new candidate gene for neurodevelopmental disorders.
Keywords: Intellectual disability, Epilepsy, Exome sequencing, HECW2, ERC2
Introduction
Intellectual disability (ID) has a prevalence of 1%–3% and is defined by an IQ <70 with an onset before the age of 18.1 It has been estimated that 20%–30% of patients with ID also have epilepsy, pointing to a drastic over-representation of epilepsy in patients with ID compared with the general population (prevalence of 0.5%–1%).2 The prevalence of epilepsy is even higher with increased severity of ID, and epilepsy co-occurring with ID is also more commonly treatment resistant and displays a higher mortality rate than epilepsy in the general population.3 4 In both ID and epilepsy it is well established that a large fraction of cases have a genetic cause, and there are numerous genetic syndromes where ID and epilepsy are part of the phenotype. These facts together indicate that there is a strong genetic correlation between ID and epilepsy and gives incentive to further identify and investigate genes with causative mutations in patients with both conditions.
Investigations into the genetic aetiology of ID and epilepsy have primarily been performed using chromosomal microarray analysis (CMA) as a first genetic test, resulting in clinically significant findings in 15%–20% of patients.5 With the rapid development of high-throughput sequencing technologies, exome sequencing of trios to identify de novo mutations (DNMs) has been introduced in genetic diagnostics, typically resulting in clinically significant findings in 20%–30% of patients already screened by CMA.6–9 The limitations in clinical yield include a lack of molecular understanding, resulting in many variants labelled as being of uncertain clinical significance. In addition, genetic interaction effects and environmental causes may play roles in a significant fraction of patients. To address the problem of lacking molecular understanding, major exome sequencing projects, such as the Deciphering Developmental Disorders (DDD), have screened large cohorts in order to identify novel disease-causing genes.10 By screening 1133 cases the DDD project identified 12 novel disease genes, increasing the proportion of cases with a molecular diagnosis by 10%.11 Exome sequencing in epileptic encephalopathies has also implicated DNMs as a major cause and has led to identification of several new candidate genes.7 This underlines the importance of adding to the growing list of causative genes. For frequently co-occurring conditions such as ID and epilepsy, an expanded list of causative genes may also greatly help to understand the pathophysiology and genetic aetiology as well as the connection between these conditions.
In this study we used exome sequencing in 39 patient–parent trios, where the patients have ID in combination with epilepsy. We report the identification of 29 DNMs and one pathogenic inherited single nucleotide variant (SNV) in coding sequence of 23 trios, of which 16 were found in genes previously known to cause epilepsy and/or ID. For 11 families we identified variants determined to be pathogenic, giving a clinical yield of 28.2% in this cohort. Our results also lend further support to previously identified candidate genes in ID and epilepsy, and highlight HECW2 as a novel candidate gene in neurodevelopmental disorders based on network analysis and a combined analysis with previous exome sequencing efforts.
Methods
Study design and patients
The participating patients and parents were recruited between 2012 and 2015 in collaboration with the Genetic Diagnostics Unit at Uppsala University Hospital. Ethical approval for exome sequencing was received from the Uppsala Ethical Review Board and informed consent was received from the parents of all patients. The selection criteria for patients included ID and epilepsy, while parents had to be healthy with no family history of neurodevelopmental disorders. All patients had previously been screened with CMA (250K Nsp Array, Genome-Wide SNP Array V.6.0 or CytoScan HD (Affymetrix, Santa Clara, California, USA)) and no pathogenic CNVs had been detected. Genomic DNA was extracted from peripheral blood leucocytes according to standard procedures.
Sequencing
Exome enrichment was performed using SureSelect (Agilent) versions 2–5 and samples were sequenced on either SOLiD, Illumina or IonProton platforms. The sequencing was performed to achieve at least 30× coverage of the captured regions. Mapping of SOLiD reads was performed using Bioscope (Life Technologies) until the release of Life Scope (Life Technologies), which was then used. Illumina reads were mapped using Burrows-Wheeler Aligner (BWA)12 and IonProton reads were mapped using the Torrent suit software (Life Technologies). All reads were mapped to the Hg19 version of the human reference genome. Programs used for mapping were run using default settings.
Data analysis
After alignment of SOLiD and Illumina reads, variants were called using the Genome Analysis Toolkit (GATK) HaplotypeCaller and the standard GATK workflow (Broad Institute). For IonProton, variants were called using the Torrent suit software (LifeTechnologies) and standard settings. To identify DNMs all called SNVs were filtered against our in-house database containing previously identified variants from 170 exomes and the Database of SNP (dbSNP) V.42 (non-flagged).13 To identify inherited disease-causing variants all SNVs with a frequency >0.001 in the Exome Aggregation Consortium (ExAC) database were removed from our results. After this all variants homozygous in the patient and heterozygous in each parent were identified among the filtered variants. To retrieve genes with compound heterozygous variants, each gene containing two or more variants with one inherited from each parent, and where no parent carried both variants, was identified. To calculate the probability of mutations in the HECW2 gene the R library denovolyzeR was used.14
Validation and comparison to previous studies
DNMs were validated by Sanger sequencing using standard protocols. Each validated variant was interpreted using the American College of Medical Genetics and Genomics (ACMG) guidelines.15 For genes where a DNM was validated, the number of DNMs identified in cases (ID, epilepsy and autism) and controls in a selected set of previous exome sequencing studies were counted.6 16–23
Network generation
Network generation was performed using GeneMania adding all genes where DNMs were found in the present study together with a compiled list of genes reported to be associated to both ID and epilepsy.24 To compile the list of genes previously associated to both ID and epilepsy, first all genes that were categorised as confirmed ID genes by the DDD project were collected. After this the Human Phenotype Ontology (HPO) terms associated with the findings in each of these genes were filtered so that only genes with at least one HPO term associated with epileptic seizures was included (terms included were HP:0002184, HP:0010818, HP:0011171, HP:0002384, HP:0002373, HP:0007294, HP:0006902, HP:0006869, HP:0007075, HP:0007284, HP:0007202, HP:0002123, HP:0002306, HP:0002182, HP:0002348, HP:0001275, HP:0002466, HP:0002125, HP:0002417, HP:0010520, HP:0006997, HP:0002391, HP:0002437, HP:0002434, HP:0001303, HP:0002479, HP:0002432, HP:0002279, HP:0002430, HP:0002431, HP:0002794, HP:0001250). The network was then compiled using only protein–protein and pathway interactions, and only genes with at least one connection to any other gene was included in the final network.
Results
Trio sequencing
By exome sequencing of 39 trio families we identified a total of 29 DNMs within protein-coding regions. All variants were validated using Sanger sequencing. The number of DNMs ranged from 0 to 3 per trio and DNMs were identified in 22 of the 39 families. Among the coding DNMs identified four were stopgain mutations, 20 were non-synonymous and five were synonymous mutations. Out of the 29 genes with DNMs, 13 have previously been associated both with ID and epilepsy, one associated with ID only and one only with epilepsy (table 1). Three of the DNMs were identified in genes previously known to cause recessive or x-linked forms of ID (AAAS, MED12 and CERS1). In none of these cases could a second mutation or CNV be identified despite careful review of alignments and array probe intensities across the genes. Using the ACMG guidelines for classification of variants, we identified pathogenic and likely pathogenic DNMs in known causative genes in 10 families.15 This results in a diagnostic yield of 25.6% based on DNMs. The full list of patient phenotypes and the mutations identified in each patient are described in online supplementary table S1. Each parent–offspring trio was also investigated for homozygous and compound heterozygous SNVs of clinical relevance in order to identify recessive candidate genes. This analysis led to the identification of one additional gene (ADSL) determined to be causative. Including both DNMs and inherited variants, we thus identify pathogenic variants in 11 of 39 families (28.2%).
Table 1.
A list of disease-associated genes with DNMs identified in this study
| Gene | Position | Family | Mutation type | ID | Epilepsy | Associated disorder (OMIM designation and number) | Inheritance |
|---|---|---|---|---|---|---|---|
| CDKL5 | chrX:18598085:C/T | Fam2 | Stopgain | Yes | Yes | Epileptic encephalopathy, early infantile, 2, MIM:300672 | XD |
| KCNQ2 | chr20:62044879:C/A | Fam3 | Non-synonymous | Yes | Yes | Epileptic encephalopathy, early infantile, 7, MIM:613720, Seizures, benign neonatal, 1, MIM: 121200 | AD |
| SYNGAP1 | chr6:33400477:C/T | Fam4 | Stopgain | Yes | Yes | Mental retardation, autosomal dominant 5, MIM:612621 | AD |
| SETD5 | chr3:9490126:G/T | Fam5 | Stopgain | Yes | Yes | Mental retardation, autosomal dominant 23, MIM:615761 | AD |
| SMC1A | chrX:53423489:G/A | Fam6 | Stopgain | Yes | Yes | Cornelia de Lange syndrome 2, MIM:300590 | XD |
| ZMYND11 | chr10:298399:C/T | Fam7 | Non-synonymous | Yes | No | Mental retardation, autosomal dominant 30, MIM:616083 | AD |
| EFTUD2 | chr17:42931953:T/G | Fam7 | Non-synonymous | Yes | Yes | Mandibulofacial dysostosis, Guion-Almeida type, MIM:610536 | AD |
| AAAS | chr12:53702981:C/T | Fam8 | Non-synonymous | Yes | Yes | Achalasia-addisonianism-alacrimia syndrome, MIM:231550 | AR |
| GABRG2 | chr5:161576159:G/A | Fam8 | Non-synonymous | No | Yes | Epilepsy, generalised, with febrile seizures plus, type 3, MIM:611277, Epilepsy, childhood absence, susceptibility to, 2, MIM:607681 | AD |
| GRIN1 | chr9:140053150:A/C | Fam9 | Non-synonymous | Yes | Yes | Mental retardation, autosomal dominant 8, MIM:614254 | AD |
| SCN2A | chr2:166166923:C/T | Fam10 | Non-synonymous | Yes | Yes | Epileptic encephalopathy, early infantile, 11, MIM:613721, Seizures, benign familial infantile, 3, MIM:607745 | AD |
| ST5 | chr11:8752629:G/C | Fam11 | Non-synonymous | Yes | Yes | Mental retardation, MIM:140750 | AD |
| KCNA1 | chr12:5021751:C/T | Fam12 | Non-synonymous | Yes | Yes | Episodic ataxia/myokymia syndrome, MIM:160120 | AD |
| CERS1 | chr19:18990105:A/T | Fam14 | Non-synonymous | Yes | Yes | Epilepsy, progressive myoclonic, 8, MIM:616230 | AR |
| MED12 | chrX:70349234:G/A | Fam16 | Non-synonymous | Yes | Yes | Lujan-Fryns syndrome, MIM:309520, Ohdo syndrome, X-linked, MIM:300895, Opitz-Kaveggia syndrome, MIM:305450 | XR |
For each gene it is noted if it has been associated with ID, epilepsy or both, as well as OMIM IDs for each specific disease it has been associated with.
AD, autosomal dominant; AR, autosomal recessive; ID, intellectual disability; XD, X-linked dominant; XR, X-linked recessive.
A list of each family in the study showing phenotypes, mutations identified and pathogenicity of each mutation. Key to abbreviations: DD=developmental delay, ID=intellectual disability, GUS=gene of uncertain significance, VOUS=variant of uncertain significance, M=male, F=female.
jmedgenet-2016-103814supp_table1.pdf (47.4KB, pdf)
To measure the deleteriousness of the identified DNMs we calculated combined annotation-dependent depletion (CADD) scores for all mutations.25 CADD scores are to be interpreted as a relative measurement of pathogenicity of genetic variants, and higher CADD scores indicate higher pathogenicity. The CADD scores showed a distribution where the synonymous mutations all showed a low score (<10), while stopgains mutations and nonsynonymous variants (with one exception) had scores >10 (see online supplementary table S2). Approximately half of the identified DNMs had a CADD score higher than 20. It has previously been shown that disease-causing variants in the OMIM database are enriched for CADD scores higher than 20.26
A list of all DNMs with calculated CADD, GERP, SIFT, Polyphen2 scores, MutationTaster predictions and allele frequencies from the Exome Sequencing Project (ESP) and 1000 genome project. For MutationTaster predictions, A=Disease causing automatic, D=Disease causing, N=Polymorphism, 0=No prediction. The last column shows the Pubmed ID associated to sequencing projects where the exact same mutation has previously been found.
jmedgenet-2016-103814supp_table2.pdf (92.5KB, pdf)
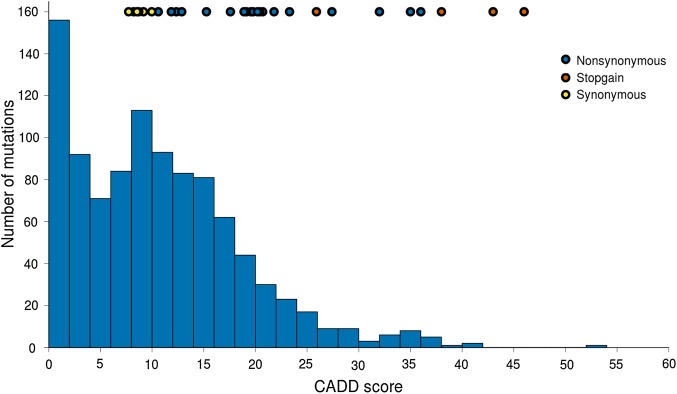
To investigate the magnitude of the impact of the DNMs, the CADD scores of the DNMs were compared with the CADD scores of 1000 randomly chosen SNVs. The analysis was performed by randomly picking the same fraction of synonymous, non-synonymous and stopgain SNVs from the ExAC database as the set of mutations identified in this study. The comparison showed that 50% of the DNMs found in this study had a higher CADD score than 89% of the randomly chosen SNVs from the ExAC database, indicating a clear shift towards more deleterious variants identified in our patient cohort (figure 1). To further categorise the mutations the level of conservation was assessed using genomic evolutionary rate profiling (GERP) scores for the non-synonymous and stopgain mutations. These scores measure the level of constraint of each base. The results showed that 62% of the mutations could be considered to be in positions that are subjected to evolutionary constraint (GERP >3) (see online supplementary table S2). To complement the CADD scores described above, other commonly used prioritisation scores (SIFT, PolyPhen2 and MutationTaster) are also listed (see online supplementary table S2).
Figure 1.
A histogram showing the distribution of combined annotation-dependent depletion (CADD) scores from the Exome Aggregation Consortium (ExAC) project, with the CADD score of de novo mutations found in this study shown as coloured circles at the top of the figure.
Due to the genetic heterogeneity of neurodevelopmental disorders, DNMs in causative genes are expected to be individually rare, and gathering data from several studies may therefore be one way to find further support for the involvement of candidate genes. To evaluate the potential pathogenicity of the DNMs identified in our families in the context of previous exome sequencing studies in neurodevelopmental disorders we collated data from 13 studies in ID, epilepsy, autism spectrum disorder and control trios (see Methods). The patient categories were chosen as considerable overlap has been shown in the genes implicated in these disorders, and based on the fact that there is commonly overlap in phenotype between these patient groups. In total, this amounted to 5338 patient trios and 2181 control trios. Of the 29 genes with DNMs in this study, 15 genes were found to have DNMs reported in patients in previous studies. In total, these 15 genes contained 63 previously reported DNMs, of which six were synonymous mutations and therefore unlikely to cause disease. Of the 15 genes, 10 have previously been linked to ID and/or epilepsy. The genes with the highest number of DNMs in the patient group are established causative genes such as SCN2A and SYNGAP1, identified in 20 and 12 cases, respectively (table 2). Of genes previously not implicated in neurodevelopmental disorders we find that DNMs in the gene HECW2, identified in one of our trios, has also been identified in patients in five previous studies, while no DNMs have been found in controls. Among the previously reported variants in HECW2 all had a CADD score >15 (range 15–27). To more formally evaluate the finding of DNMs in HECW2 in our trios and previous studies, we used the statistical framework developed by Samocha et al.14 Using the 5338 trio families collated above together with our 39 trios yields an expected number of DNMs (non-synonymous and stopgains) in HECW2 of 0.7, while we observed a total of six non-synonymous mutations (p-value = 6.11×10e-5). These results suggest that DNMs in HECW2 are associated with neurodevelopmental phenotypes.
Table 2.
Showing the number of DNMs identified in cases and controls in previous exome sequencing studies, as well as the OMIM designation number, for genes with DNMs found in this study
| Gene | Cases | Controls | ID | Epilepsy | Associated disorder (OMIM designation and number) |
|---|---|---|---|---|---|
| SCN2A | 20 | 0 | Yes | Yes | Epileptic encephalopathy, MIM:613721, seizures, MIM: 607745 |
| SYNGAP1 | 12 (1) | 1 (1) | Yes | Yes | Mental retardation, MIM:612621 |
| SETD5 | 6 | 1 | Yes | No | Mental retardation, MIM:615761 |
| HECW2 | 5 | 0 | No | No | None |
| CDKL5 | 3 | 0 | Yes | Yes | Epileptic encephalopathy, MIM:300672 |
| KCNQ2 | 3 (1) | 0 | Yes | Yes | Epileptic encephalopathy, MIM:613720, Myokymia, MIM:121200 |
| ZMYND11 | 2 | 0 | Yes | No | Mental retardation, MIM:616083 |
| KIAA1244 | 1 | 1 | No | No | None |
| TBC1D4 | 1 (1) | 0 | No | No | None |
| KCNA1 | 1 | 0 | Yes | Yes | Episodic ataxia/myokymia syndrome, MIM:160120 |
| BAZ1A | 1 | 1 (1) | No | No | None |
| ERC2 | 1 (1) | 1 | No | No | None |
| GABRG2 | 1 | 0 | No | Yes | Epilepsy, MIM:611277, MIM:607681 |
| GRIN1 | 1 | 1 | Yes | Yes | Mental retardation, MIM:614254 |
| SMC1A | 1 | 0 | Yes | Yes | Cornelia de Lange syndrome, MIM:300590 |
The number of synonymous DNMs for each gene and category is noted in parenthesis. One DNM in each of these genes was found in this study, including four stopgains (SYNGAP1, SETD5, CDKL5, SMC1A), ten non-synonymous (SCN2A, HECW2, KCNQ2, ZMYND11, TBL1D4, KCNA1, BAZ1A, ERC2, GABRG2, GRIN1) and one synonymous (KIAA1244). Variants identified in the present study are not included in this table (listed in table 1)
ID, intellectual disability; DNM, de novo mutation.
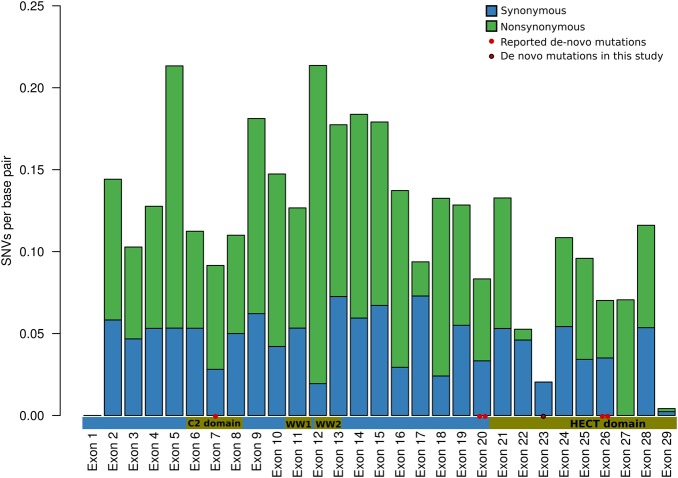
Of the five mutations in HECW2 detected in previous studies, one DNM was identified in an epilepsy cohort, two in patients from autism cohorts and two in patients with ID and seizures. Although detailed phenotype descriptions are not available for most of these patients, we note that one of the autism cases also had a low IQ (<65), while the second patient had febrile seizures reported. The HECW2 gene is a HECT-type ubiquitin ligase and is known to regulate the stability of p73.27 The DNM in our study was identified in the Homologous to the E6-AP Carboxyl Terminus (HECT) domain of the protein. Out of the five previously reported mutations, four were located in exons associated with the HECT domain, which displays a lower number of non-synonymous mutations in the general population (figure 2).
Figure 2.
A bar plot showing the number of SNVs per base pair in each exon of the HECW2 gene, blue bars show silent mutations and green bars show non-synonymous mutations. Values shown for each exon are normalised for exon length. The horizontal bar shows the different domains in the HECW2 protein and their correlation to each HECW2 exon. The red dots show the coordinates of de novo mutations (DNMs) identified in the present and previous exome sequencing projects in intellectual disability, autism or epilepsy (see materials and methods). The DNM identified in the present study is located in exon 23. Exons 1 and 29 are mainly untranslated with only short coding sequence, explaining the low number of coding variants in these exons.
Network categorisation
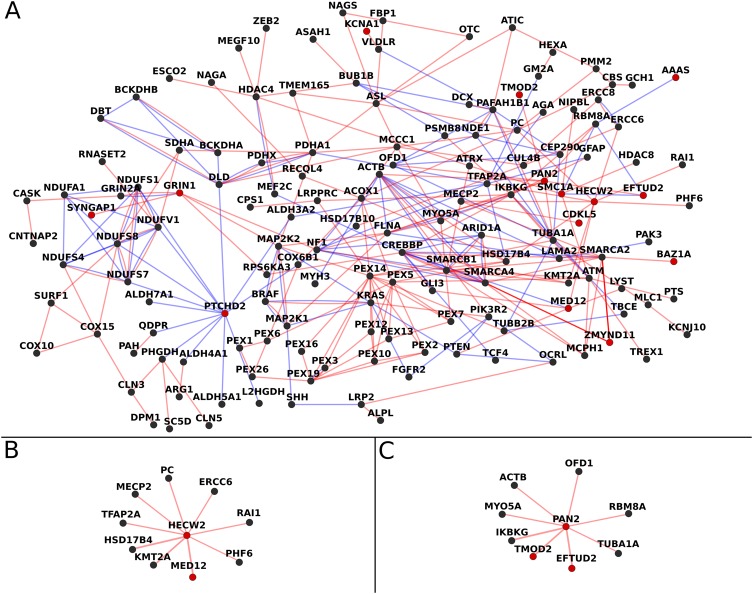
To further categorise the genes with DNMs in this study in the context of previously established causative genes, we extracted all genes with causative DNMs in patients with ID and epilepsy from the DDD project data. The genes extracted from DDD were then used together with the genes identified in this study to construct a network based on protein–protein interactions and known pathways (figure 3). The resulting network shows that 48% of the genes with DNMs in this study interact with at least one other gene in the network, while the remaining DNMs showed no connections. Of the genes with interactions, PTCHD2, TMOD2, BAZ1A, PAN2 and HECW2 have not previously been shown to be associated with ID and/or epilepsy. The variants detected in PTCHD2 and TMOD2 were silent mutations and therefore not considered candidate causative mutations in this study. The BAZ1A gene codes for a chromatin remodelling factor, providing another potential candidate gene to the list of known causative chromatin remodelling genes in ID and epilepsy.28 The PAN2 protein interacts with several DNA-binding proteins, and is a subunit of the PAN with the function to shorten the poly(A)-tails of RNA. Mice homozygous for pan2 mutations exhibit embryonic lethality, while seizures have been reported in mice carrying a heterozygous deletion of pan2.29 In our network analysis PAN2 was connected to eight other genes, making it one of the most highly interconnected genes among the genes found in this study. The network analysis also points to a central role for HECW2, showing interaction with nine other known causative genes, furthering strengthening the candidacy of HECW2 as a new causative gene in neurodevelopmental disorders.
Figure 3.
(A) Interaction network based on genes with de novo mutations (DNMs) found in this study together with genes previously implicated in intellectual disability (ID) and epilepsy. Red lines show protein–protein interactions and blue lines show pathway interactions. Red dots mark the genes with DNMs identified in this study. Genes not connected to any other gene were removed from the figure. Four of the genes we identified with DNMs were previously reported as causative in the Deciphering Developmental Disorders (DDD) project, while the remaining genes identified by us and present in the figure could be linked by known interactions. (B) Cutout of HECW2 and connected genes from the network in A. (C) Cutout of poly(A)-nuclease 2 (PAN2) and connected genes from the network in A.
De novo mutations in genes previously associated with ID or epilepsy
Among the 15 genes with DNMs identified in this study and reported to be involved in ID and/or epilepsy, the mutations detected in SETD5, CDKL5, SYNGAP1 and SMC1A were stopgains, and the remaining genes carried non-synonymous mutations. Five of the nonsense and non-synonymous mutations identified had previously been reported as causative in dbSNP (ZMYND11, SCN2A, SETD5, GABRG2, CDKL5).6 30–33 In addition, the mutation in KCNQ2 was found in the same position as a previously reported causative mutation, but with a different base change leading to another amino acid substitution.34 The identification of DNMs already present in public databases are in line with recently published data from a deep sequencing of 10 trios where 3.5% of the identified DNMs were already present in dbSNP.35 The symptoms reported for the patients carrying mutations in SYNGAP1, EFTUD2, KCNQ2, GRIN1, SMC1A and ADSL in this study all mirrored the phenotypes previously reported for patients with mutations in these genes. The patient carrying a mutation in EFTUD2 also had a second DNM in ZMYND11. The two mutations were determined to be likely pathogenic (EFTUD2) and pathogenic (ZMYND11).
The ST5 non-synonymous mutation occurred in a region outside any known motifs or domains present in the ST5 protein. The mutation was found in a patient presenting with ID, seizures, delayed speech, slight dysmorphic features, frequent infections and a benign teratoma. The function of the ST5 protein is relatively unknown; however, studies show that the protein can function as a tumour suppressor in cultured cells.36 To our knowledge, only a single patient has previously been reported to carry a translocation interrupting the ST5 gene.37 This patient presented with a similar phenotype, including ID, epilepsy, recurrent infections and a partially overlapping facial gestalt, although more severely affected. Altogether this adds convincing evidence for ST5 being causative in our patient, strengthening the evidence of ST5 as causative in ID and epilepsy. An interesting feature, however, is the benign teratoma in infancy present in our patient, as ST5 has been described as a tumour suppressor.
The de novo stopgain mutation in SETD5 was discovered in a patient with ID and myoclonic seizures. Recent calculations show that loss-of-function mutations in SETD5 might explain up to 0.7% of ID cases identified.38 Seizures have been reported in a subset of patients. It is therefore interesting to note that the patient with an SETD5 mutation also carries a DNM in ERC2. The ERC2 gene encodes a protein with a central role in the presynaptic active zone. In mice, conditional knockout of ERC2 has been shown to lead to a large increase in inhibitory synaptic strength by increasing the size of releasable vesicles at inhibitory neurons.39 It is therefore possible that the mutation in ERC2, potentially in concert with the mutation in SETD5, further exacerbates the myoclonus phenotype in this patient.
Discussion
Our exome study identified clinically significant DNMs in 10 of 39 patients with ID and epilepsy. In one case we found a recessive cause for the patient phenotype. The diagnostic yield (28.2%) is similar to previous exome sequencing studies in ID, reporting a diagnostic yield ranging from 16% to 29% with the majority explained by DNMs.8 11 40 Of the genes with DNMs that we identified, approximately half have previously been associated with both ID and epilepsy, indicating that the patients selected for this study represent a genetically well-defined group. In one trio (2.5% of patients) we identified a pathogenic recessive mutation. The number of recessive mutations identified in previous exome sequencing projects differ significantly and range from 0 in one study investigating 245 families to 20% in a recent study investigating 45 patients.7 10 40 In several of the known causative genes the specific mutations we identify have not been previously reported, adding to the catalogues of clinically relevant mutations in these genes. The identification of mutations in previously reported genes also adds further evidence to their causative nature and contributes to the description of the clinical spectrum of mutation carriers.
Our analysis shows that many of the genes with DNMs are interconnected through protein–protein interactions or exist in the same pathway together with genes previously linked to ID and epilepsy. As networks are constructed using proof and knowledge from previous studies, it is interesting to notice that several of the genes independently linked to ID and epilepsy are also interconnected in the networks generated. This indicates that the knowledge of gene interactions accumulated to date are sufficient to identify pertinent connections between the genes identified in studies where the patients are selected by a well-defined and delimited set of symptoms. From the network analysis it is interesting to note that genes with DNMs, not previously linked to ID or epilepsy, are connected to several other genes in the network. Even though interaction cannot be considered a proof of clinical significance this makes them interesting candidates for further study and shows the strength of network analysis as a tool for prioritisation of candidate genes.
Two genes, PAN2 and HECW2, stand out in the network analysis by showing connections to multiple other known causative ID and epilepsy. Of these, HECW2 is the most interesting as five DNMs in HECW2 have been identified previous exome sequencing projects in neurodevelopmental disorders, including ID and epilepsy.10 18 19 23 We show that this represents a significantly higher number of DNMs than would be expected in the number of trios included in the survey. Using residual variation intolerance scores we further notice that the HECW2 gene is among the 0.98% genes most intolerant to functional variation in the human genome,41 which is also evident from the plot of synonymous and non-synonymous variants reported in the ExAC database. Looking into distribution of variation across the exons of the gene we see that most DNMs reported cluster in the exons that are depleted in coding variation, with five of six reported DNMs located within (n=3) or immediately adjacent to (n=2) the HECT domain of the HECW2 protein. Taken together, these lines of evidence indicate that DNMs in HECW2 are associated with neurodevelopmental phenotypes. Interestingly, a search of social media led to the discovery of additional patients with de novo HECW2 mutations, displaying overlapping phenotypes. Knockout mice for this gene do not show a similar phenotype, with partial preweaning lethality, lean body mass and lowered mean platelet volume as major symptoms.42 In light of the mouse knockout phenotype, it is important to note that all DNMs reported are non-synonymous, potentially pointing to a gain of function or dominant negative role. Using BrainSpan data,43 we find that HECW2 is expressed at moderate levels in the brain throughout development, with the highest expression in frontal cortex. Interestingly, the gene that shows the highest correlation in expression in frontal cortex during brain development is CDKL5, a well-established causative gene in ID and epilepsy. The expression pattern therefore lends further support to HECW2 as a highly interesting novel candidate gene. Our results show that the systematic use of interaction data can be used as an effective tool for candidate prioritisation. However, results must also be interpreted with caution, as the effectiveness of this strategy will be dependent on the criteria used in the patient selection and to what extent the gene in question has been studied previously.
The identification of mutations in known causative genes provides the opportunity to refine and expand on previous reports on associated clinical symptoms. In our data, we identify several patients that provide potential new insight into the genotype–phenotype correlation. For example, the patient carrying a GABRG2 mutation found in this study had several phenotypes not present in previously confirmed carriers. At the same time, these results must be interpreted with caution, as it is possible that the additional symptoms are the result of a second mutation. For example, in the patient with the EFTUD2 mutation a second pathogenic mutation was found in the ZMYND11 gene, making it probable that both genes contribute to phenotype. The finding of a second causative mutation is in accordance to a recent study where it was calculated that about 1.4% of patients had a second mutation contributing to the phenotype.44 The fact that second mutations may have an impact on the resulting phenotype is further highlighted by the stopgain SETD5 mutation. In this patient a second mutation was found in ERC2, a gene known to be involved in the presynaptic active zone where it has an effect on inhibitory synaptic strength. Previous studies show that even modest changes in synaptic plasticity at inhibitory neurons may trigger epileptic activity.45 This raises the possibility that ERC2 contributes to the epileptic phenotype in our patient, but additional patients with ERC2 mutations or further functional studies are needed to confirm its involvement in epilepsy aetiology.
An explanation for additional symptoms identified in a patient with mutation in a known causative gene may be that the disease phenotype is poorly defined due to a limited number of patients. In such cases the identification of additional patients is crucial, underlining the importance of this and similar studies. It is also important to point out that epileptic episodes have in many cases been observed to cause brain damage, and when investigating patients with both ID and epilepsy it is possible that the ID reported may be a consequence of an early epileptic episode.
One drawback of our study is that patients have been run sequentially over a longer time period, with a concurrent development in technology and analysis tools. Trios have therefore been sequenced with different capture kits and different sequencing approaches. We do not find statistically significant differences between these different technologies due to the limited size of our study, but there is a trend towards identification of more DNMs and more likely pathogenic mutations in more recent analyses. Still, we find an average of 0.79 DNMs per trio, which is similar or better than several previous large-scale trio exome sequencing studies,46 47 but lower than studies that have performed much deeper sequencing.6 40 It is therefore likely that several causative DNMs have been missed, especially in the trios sequenced first. Future whole-genome resequencing of these patients will hopefully provide a molecular diagnosis for additional families.
In summary, we identify variants likely to be pathogenic in 11 genes previously linked to ID and/or epilepsy, resulting in a molecular diagnostic yield of 28%. We also identified several mutations that point to candidate causative genes such as the PAN2, HECW2 and ERC2 genes. HECW2 is the strongest novel candidate as DNMs affecting a specific domain of the protein have been identified in several studies in closely related disorders. Additional patients, better clinical phenotype information or functional studies will be required to conclusively determine the potential role of HECW2 in brain development. All in all this study underlines the potential and possibilities of using exome sequencing as a tool for identification of disease genes in a stringently selected group of patients, and the utility of using previous knowledge of protein interaction and biological pathways to prioritise candidate genes.
Acknowledgments
We are very grateful to the participating families for their cooperation. Sequencing was performed using the SciLifeLab National Genomics Infrastructure at Uppsala Genome Center and the Uppsala SNP & Seq Facility. Computational analyses were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX).
Footnotes
Contributors: JH, A-CT and LF designed and planned the study. JH and JJZ analysed the data. JJZ and AZ performed experimental lab work and validated variants. PG-H, CSZ, EM, HES, GB and A-CT diagnosed the patients, performed clinical assessments and collected patient samples. JH, A-CT and LF wrote the manuscript. All authors read, commented on and approved the manuscript.
Funding: This work was supported by grants from the Föreningen Margarethahemmet, the Sävstaholm Society and the ‘Regionala forskningsrådet’ (to A-CT), and the European Research Council ERC Starting Grant Agreement no. 282330 and the Swedish Medical Research Council (to LF).
Competing interests: None declared.
Ethics approval: Uppsala Ethical Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet 2010;11:161–87. 10.1146/annurev-genom-082509-141640 [DOI] [PubMed] [Google Scholar]
- 2.Bell GS, Sander JW. The epidemiology of epilepsy: the size of the problem. Seizure 2002;11(Suppl A):306–14; quiz 315–16. [PubMed] [Google Scholar]
- 3.Lhatoo SD, Sander JWAS. The epidemiology of epilepsy and learning disability. Epilepsia 2001;42(Suppl 1):6–9; discussion 19–20 10.1046/j.1528-1157.2001.00502.x [DOI] [PubMed] [Google Scholar]
- 4.McGrother CW, Bhaumik S, Thorp CF, Hauck A, Branford D, Watson JM. Epilepsy in adults with intellectual disabilities: prevalence, associations and service implications. Seizure 2006;15:376–86. 10.1016/j.seizure.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 5.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010;86:749–64. 10.1016/j.ajhg.2010.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, Cremer K, Hempel M, Horn D, Hoyer J, Joset P, Röpke A, Moog U, Riess A, Thiel CT, Tzschach A, Wiesener A, Wohlleber E, Zweier C, Ekici AB, Zink AM, Rump A, Meisinger C, Grallert H, Sticht H, Schenck A, Engels H, Rappold G, Schröck E, Wieacker P, Riess O, Meitinger T, Reis A, Strom TM. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012;380:1674–82. 10.1016/S0140-6736(12)61480-9 [DOI] [PubMed] [Google Scholar]
- 7.Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, Epstein MP, Glauser T, Goldstein DB, Han Y, Heinzen EL, Hitomi Y, Howell KB, Johnson MR, Kuzniecky R, Lowenstein DH, Lu Y-F, Madou MRZ, Marson AG, Mefford HC, Esmaeeli Nieh S, O'Brien TJ, Ottman R, Petrovski S, Poduri A, Ruzzo EK, Scheffer IE, Sherr EH, Yuskaitis CJ, Abou-Khalil B, Alldredge BK, Bautista JF, Boro A, Cascino GD, Consalvo D, Crumrine P, Devinsky O, Fiol M, Fountain NB, French J, Friedman D, Geller EB, Glynn S, Haut SR, Hayward J, Helmers SL, Joshi S, Kanner A, Kirsch HE, Knowlton RC, Kossoff EH, Kuperman R, McGuire SM, Motika PV, Novotny EJ, Paolicchi JM, Parent JM, Park K, Shellhaas RA, Shih JJ, Singh R, Sirven J, Smith MC, Sullivan J, Lin Thio L, Venkat A, Vining EPG, Von Allmen GK, Weisenberg JL, Widdess-Walsh P, Winawer MR. De novo mutations in epileptic encephalopathies. Nature 2013;501:217–21. 10.1038/nature12439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ligt J, Willemsen MH, van Bon BWM, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, del Rosario M, Hoischen A, Scheffer H, de Vries BBA, Brunner HG, Veltman JA, Vissers LELM. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 2012;367:1921–9. 10.1056/NEJMoa1206524 [DOI] [PubMed] [Google Scholar]
- 9.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, Yuen RKC, Uddin M, Roberts W, Weksberg R, Woodbury-Smith M, Zwaigenbaum L, Anagnostou E, Wang Z, Wei J, Howe JL, Gazzellone MJ, Lau L, Sung WWL, Whitten K, Vardy C, Crosbie V, Tsang B, D'Abate L, Tong WWL, Luscombe S, Doyle T, Carter MT, Szatmari P, Stuckless S, Merico D, Stavropoulos DJ, Scherer SW, Fernandez BA. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA 2015;314:895–903. 10.1001/jama.2015.10078 [DOI] [PubMed] [Google Scholar]
- 10.Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, Bevan AP, Bragin E, Chatzimichali EA, Gribble S, Jones P, Krishnappa N, Mason LE, Miller R, Morley KI, Parthiban V, Prigmore E, Rajan D, Sifrim A, Swaminathan GJ, Tivey AR, Middleton A, Parker M, Carter NP, Barrett JC, Hurles ME, FitzPatrick DR, Firth HV. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 2015;385:1305–14. 10.1016/S0140-6736(14)61705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 2015;519:223–8. 10.1038/nature14135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–11. 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, Wall DP, MacArthur DG, Gabriel SB, DePristo M, Purcell SM, Palotie A, Boerwinkle E, Buxbaum JD, Cook EH, Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Neale BM, Daly MJ. A framework for the interpretation of de novo mutation in human disease. Nat Genet 2014;46:944–50. 10.1038/ng.3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med 2008;10:294–300. 10.1097/GIM.0b013e31816b5cae [DOI] [PubMed] [Google Scholar]
- 16.Jiang YH, Yuen RKC, Jin X, Wang M, Chen N, Wu X, Ju J, Mei J, Shi Y, He M, Wang G, Liang J, Wang Z, Cao D, Carter MT, Chrysler C, Drmic IE, Howe JL, Lau L, Marshall CR, Merico D, Nalpathamkalam T, Thiruvahindrapuram B, Thompson A, Uddin M, Walker S, Luo J, Anagnostou E, Zwaigenbaum L, Ring RH, Wang JJ, Lajonchere C, Wang JJ, Shih A, Szatmari P, Yang H, Dawson G, Li Y, Scherer SW. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet 2013;93:249–63. 10.1016/j.ajhg.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Fu S-C, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Crooks L, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Sean Hill R, Ionita-Laza I, Jimenez Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei J, Lehtimäki T, Lin C-F, Ma'ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnström K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Rüther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Wang L-S, Weiss LA, Jeremy Willsey A, Yu TW, Yuen RKC, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barrett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014;515:209–15. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee Y, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014;515:216–21. 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, Leal SM, Bernier R, Eichler EE. Excess of rare, inherited truncating mutations in autism. Nat Genet 2015;47:582–8. 10.1038/ng.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roach JC, Glusman G, Smit AFA, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 2010;328:636–9. 10.1126/science.1186802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BWM, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, Tearle R, Bo T, Pfundt R, Yntema HG, de Vries BBA, Kleefstra T, Brunner HG, Vissers LELM, Veltman JA. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014;511:344–7. 10.1038/nature13394 [DOI] [PubMed] [Google Scholar]
- 22.The Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 2014;46:818–25. 10.1038/ng.3021 [DOI] [PubMed] [Google Scholar]
- 23.Appenzeller S, Balling R, Barisic N, Baulac S, Caglayan H, Craiu D, De Jonghe P, Depienne C, Dimova P, Djémié T, Gormley P, Guerrini R, Helbig I, Hjalgrim H, Hoffman-Zacharska D, Jähn J, Klein KM, Koeleman B, Komarek V, Krause R, Kuhlenbäumer G, Leguern E, Lehesjoki A-E, Lemke JR, Lerche H, Linnankivi T, Marini C, May P, Møller RS, Muhle H, Pal D, Palotie A, Pendziwiat M, Robbiano A, Roelens F, Rosenow F, Selmer K, Serratosa JM, Sisodiya S, Stephani U, Sterbova K, Striano P, Suls A, Talvik T, von Spiczak S, Weber Y, Weckhuysen S, Zara F, Abou-Khalil B, Alldredge BK, Andermann E, Andermann F, Amron D, Bautista JF, Berkovic SF, Bluvstein J, Boro A, Cascino G, Consalvo D, Crumrine P, Devinsky O, Dlugos D, Epstein MP, Fiol M, Fountain NB, French J, Friedman D, Geller EB, Glauser T, Glynn S, Haas K, Haut SR, Hayward J, Helmers SL, Joshi S, Kanner A, Kirsch HE, Knowlton RC, Kossoff EH, Kuperman R, Kuzniecky R, Lowenstein DH, McGuire SM, Motika PV, Novotny EJ, Ottman R, Paolicchi JM, Parent J, Park K, Poduri A, Sadleir L, Scheffer IE, Shellhaas RA, Sherr E, Shih JJ, Singh R, Sirven J, Smith MC, Sullivan J, Thio LL, Venkat A, Vining EPG, Von Allmen GK, Weisenberg JL, Widdess-Walsh P, Winawer MR, Allen AS, Cossette P, Delanty N, Eichler EE, Goldstein DB, Han Y, Heinzen EL, Johnson MR, Marson AG, Mefford HC, Nieh SE, O'Brien TJ, Petrou S, Petrovski S, Ruzzo EK. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet 2014;95:360–70. 10.1016/j.ajhg.2014.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214–20. 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–5. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shyr C, Tarailo-Graovac M, Gottlieb M, Lee JJY, van Karnebeek C, Wasserman WW. FLAGS, frequently mutated genes in public exomes. BMC Med Genomics 2014;7:64 10.1186/s12920-014-0064-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu L, Hu S, Wei R, Qiu X, Lu K, Fu Y, Li H, Xing G, Li D, Peng R, He F, Zhang L. The HECT type ubiquitin ligase NEDL2 is degraded by anaphase-promoting complex/cyclosome (APC/C)-Cdh1, and its tight regulation maintains the metaphase to anaphase transition. J Biol Chem 2013;288:35637–50. 10.1074/jbc.M113.472076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J 2002;21:2231–41. 10.1093/emboj/21.9.2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winawer MR, Klassen TL, Teed S, Shipman M, Leung EH, Palmer AA. A locus on mouse Ch10 influences susceptibility to limbic seizure severity: fine mapping and in silico candidate gene analysis. Genes Brain Behav 2014;13:341–9. 10.1111/gbb.12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Tippin Davis B, Baxter RM, Zeng W, Mroske C, Parra MC, Gandomi SK, Lu I, Li X, Lu H-MH, Lu HMH, Salvador D, Ruble D, Lao M, Fischbach S, Wen J, Lee S, Elliott A, Dunlop CLM, Tang S. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med 2015;17:578–86. 10.1038/gim.2014.154 [DOI] [PubMed] [Google Scholar]
- 31.Liao Y, Anttonen AK, Liukkonen E, Gaily E, Maljevic S, Schubert S, Bellan-Koch A, Petrou S, Ahonen VE, Lerche H, Lehesjoki AE. SCN2A mutation associated with neonatal epilepsy, late-onset episodic ataxia, myoclonus, and pain. Neurology 2010;75:1454–8. 10.1212/WNL.0b013e3181f8812e [DOI] [PubMed] [Google Scholar]
- 32.Carvill GL, Heavin SB, Yendle SC, McMahon JM, O'Roak BJ, Cook J, Khan A, Dorschner MO, Weaver M, Calvert S, Malone S, Wallace G, Stanley T, Bye AME, Bleasel A, Howell KB, Kivity S, Mackay MT, Rodriguez-Casero V, Webster R, Korczyn A, Afawi Z, Zelnick N, Lerman-Sagie T, Lev D, Møller RS, Gill D, Andrade DM, Freeman JL, Sadleir LG, Shendure J, Berkovic SF, Scheffer IE, Mefford HC. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet 2013;45:825–30. 10.1038/ng.2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rademacher N, Hambrock M, Fischer U, Moser B, Ceulemans B, Lieb W, Boor R, Stefanova I, Gillessen-Kaesbach G, Runge C, Korenke GC, Spranger S, Laccone F, Tzschach A, Kalscheuer VM. Identification of a novel CDKL5 exon and pathogenic mutations in patients with severe mental retardation, early-onset seizures and Rett-like features. Neurogenetics 2011;12:165–7. 10.1007/s10048-011-0277-6 [DOI] [PubMed] [Google Scholar]
- 34.Kato M, Yamagata T, Kubota M, Arai H, Yamashita S, Nakagawa T, Fujii T, Sugai K, Imai K, Uster T, Chitayat D, Weiss S, Kashii H, Kusano R, Matsumoto A, Nakamura K, Oyazato Y, Maeno M, Nishiyama K, Kodera H, Nakashima M, Tsurusaki Y, Miyake N, Saito K, Hayasaka K, Matsumoto N, Saitsu H. Clinical spectrum of early onset epileptic encephalopathies caused by KCNQ2 mutation. Epilepsia 2013;54:1282–7. 10.1111/epi.12200 [DOI] [PubMed] [Google Scholar]
- 35.Besenbacher S, Liu S, Izarzugaza JMG, Grove J, Belling K, Bork-Jensen J, Huang S, Als TD, Li S, Yadav R, Rubio-García A, Lescai F, Demontis D, Rao J, Ye W, Mailund T, Friborg RM, Pedersen CNS, Xu R, Sun J, Liu H, Wang O, Cheng X, Flores D, Rydza E, Rapacki K, Damm Sørensen J, Chmura P, Westergaard D, Dworzynski P, Sørensen TIA, Lund O, Hansen T, Xu X, Li N, Bolund L, Pedersen O, Eiberg H, Krogh A, Børglum AD, Brunak S, Kristiansen K, Schierup MH, Wang J, Gupta R, Villesen P, Rasmussen S. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat Commun 2015;6:5969 10.1038/ncomms6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ioannou MS, Bell ES, Girard M, Chaineau M, Hamlin JNR, Daubaras M, Monast A, Park M, Hodgson L, McPherson PS. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J Cell Biol 2015;208:629–48. 10.1083/jcb.201407068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Göhring I, Tagariello A, Endele S, Stolt CC, Ghassibé M, Fisher M, Thiel CT, Trautmann U, Vikkula M, Winterpacht A, FitzPatrick DR, Rauch A. Disruption of ST5 is associated with mental retardation and multiple congenital anomalies. J Med Genet 2010;47:91–8. 10.1136/jmg.2009.069799 [DOI] [PubMed] [Google Scholar]
- 38.Grozeva D, Carss K, Spasic-Boskovic O, Parker MJ, Archer H, Firth HV, Park S-M, Canham N, Holder SE, Wilson M, Hackett A, Field M, Floyd JAB, Hurles M, Raymond FL. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet 2014;94:618–24. 10.1016/j.ajhg.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaeser PS, Deng L, Chávez AE, Liu X, Castillo PE, Südhof TC. ELKS2alpha/CAST deletion selectively increases neurotransmitter release at inhibitory synapses. Neuron 2009;64:227–39. 10.1016/j.neuron.2009.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helbig KL, Farwell Hagman KD, Shinde DN, Mroske C, Powis Z, Li S, Tang S, Helbig I. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med 2016. Published Online First: 21 January 2016. 10.1038/gim.2015.186 10.1038/gim.2015.186 [DOI] [PubMed] [Google Scholar]
- 41.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013;9:e1003709 10.1371/journal.pgen.1003709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown SDM, Moore MW. The international mouse phenotyping consortium: past and future perspectives on mouse phenotyping. Mamm Genome 2012;23:632–40. 10.1007/s00335-012-9427-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Funded by ARRA Awards 1RC2MH089921-01, 1RC2MH090047-01 and 1RC2MH089929-01. © 2011. BrainSpan: Atlas of the Developing Human Brain. http//developinghumanbrain.org.
- 44.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–9. 10.1001/jama.2014.14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chagnac-Amitai Y, Connors BW. Horizontal spread of synchronized activity in neocortex and its control by GABA-mediated inhibition. J Neurophysiol 1989;61:747–58. [DOI] [PubMed] [Google Scholar]
- 46.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha RC, Choi M, Overton JD, Bjornson RD, Carriero NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Günel M, Roeder K, Geschwind DH, Devlin B, State MW. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012;485:237–41. 10.1038/nature10945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet 2011;43:864–8. 10.1038/ng.902 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A list of each family in the study showing phenotypes, mutations identified and pathogenicity of each mutation. Key to abbreviations: DD=developmental delay, ID=intellectual disability, GUS=gene of uncertain significance, VOUS=variant of uncertain significance, M=male, F=female.
jmedgenet-2016-103814supp_table1.pdf (47.4KB, pdf)
A list of all DNMs with calculated CADD, GERP, SIFT, Polyphen2 scores, MutationTaster predictions and allele frequencies from the Exome Sequencing Project (ESP) and 1000 genome project. For MutationTaster predictions, A=Disease causing automatic, D=Disease causing, N=Polymorphism, 0=No prediction. The last column shows the Pubmed ID associated to sequencing projects where the exact same mutation has previously been found.
jmedgenet-2016-103814supp_table2.pdf (92.5KB, pdf)