Abstract
Staphylococci are associated with both humans and animals. While most are non-pathogenic colonizers, Staphylococcus aureus is an opportunistic pathogen capable of causing severe infections. S. aureus virulence is controlled by the agr quorum sensing system responding to secreted auto-inducing peptides (AIPs) sensed by AgrC, a two component histidine kinase. agr loci are found also in other staphylococcal species and for Staphylococcus epidermidis, the encoded AIP represses expression of agr regulated virulence genes in S. aureus. In this study we aimed to better understand the interaction between staphylococci and S. aureus, and show that this interaction may eventually lead to the identification of new anti-virulence candidates to target S. aureus infections. Here we show that culture supernatants of 37 out of 52 staphylococcal isolates representing 17 different species inhibit S. aureus agr. The dog pathogen, Staphylococcus schleiferi, expressed the most potent inhibitory activity and was active against all four agr classes found in S. aureus. By employing a S. aureus strain encoding a constitutively active AIP receptor we show that the activity is mediated via agr. Subsequent cloning and heterologous expression of the S. schleiferi AIP in S. aureus demonstrated that this molecule was likely responsible for the inhibitory activity, and further proof was provided when pure synthetic S. schleiferi AIP was able to completely abolish agr induction of an S. aureus reporter strain. To assess impact on S. aureus virulence, we co-inoculated S. aureus and S. schleiferi in vivo in the Galleria mellonella wax moth larva, and found that expression of key S. aureus virulence factors was abrogated. Our data show that the S. aureus agr locus is highly responsive to other staphylococcal species suggesting that agr is an inter-species communication system. Based on these results we speculate that interactions between S. aureus and other colonizing staphylococci will significantly influence the ability of S. aureus to cause infection, and we propose that other staphylococci are potential sources of compounds that can be applied as anti-virulence therapy for combating S. aureus infections.
Keywords: Staphylococcus aureus, Staphylococcus schleiferi, quorum sensing, agr, quorum sensing inhibition, auto-inducing peptide, cross-talk, anti-virulence therapy
Introduction
At least 40 different Staphylococcus species have been described to date (Harris et al., 2002). While a large number of these are found to colonize humans, many also colonize animals (Nagase et al., 2002). Staphylococcus aureus is by far the best characterized staphylococcal species. This opportunistic pathogen resides on skin and mucosal membranes in humans and animals, and it can cause a variety of infections ranging from mild skin and soft tissue infections to severe conditions such as septicemia (Lowy, 1998). Expression of the majority of S. aureus virulence factors is controlled by the accessory gene regulator (agr) quorum sensing system. agr is composed of two divergent transcripts: one (P2) encoding the AgrAC two component signal transduction system that responds to autoinducing peptides (AIPs) encoded and secreted by the products of agrBD; and the other (P3) encoding a regulatory RNA, RNAIII, the effector molecule of agr. AIPs bind to the agrC-encoded histidine kinase (AgrC), and via phosphorylation of the AgrA response-regulator, stimulate the expression of RNAIII (Wang et al., 2014). At high cell densities, AIP accumulation results in up-regulation of exoprotein expression including the hla-encoded virulence factor α-hemolysin, and down-regulation of surface-associated proteins such as spa-encoded protein A (Queck et al., 2008; Shoham, 2011).
Staphylococcus aureus strains can be divided into at least four agr classes (groups I–IV), with distinct AIPs inducing virulence gene expression within each group, but repressing expression in strains of other groups by acting as competitive inhibitors to AgrC binding (Ji, 1997; Otto et al., 2001; Geisinger et al., 2012). The agr specificity is determined by AgrB, AgrC, and AgrD, which vary among the four groups. agr has also been identified in other staphylococcal species although with much higher sequence divergence. Despite this diversity, functional agr loci have been demonstrated in Staphylococcus lugdunensis and Staphylococcus epidermidis (Vandenesch et al., 1993; Wamel et al., 1998). S. epidermidis is another clinically important opportunistic pathogen whose virulence is largely controlled via the agr system (Olson et al., 2014), and the S. epidermidis AIP is a potent inhibitor of the S. aureus agr system (Otto et al., 2001). Although, the biological importance of this cross-inhibition of quorum sensing remains unknown, it has been suggested that it contributes to niche competition between S. epidermidis and S. aureus, resulting in the predominance of S. epidermidis on the skin and in indwelling device-associated chronic infections (Otto et al., 2001; Thoendel et al., 2011). The frequent isolation of other staphylococci together with S. aureus also point to a possible interaction between staphylococcal species with a role in niche competition and colonization (Lina et al., 2003; Kozioł-Montewka et al., 2006).
The aim of this study was to examine the extent to which agr cross-inhibitory activity occurs between S. aureus and other staphylococcal species, and to elucidate the mechanism behind this cross-talk. Understanding this interaction between staphylococci may help in the identification of new anti-virulence strategies targeting S. aureus infections.
Materials and Methods
Bacterial Strains and Growth Conditions
Staphylococcus aureus strains used in this study include: Strain 8325-4 (Novick, 1967) was used as a source of AIP-I containing supernatant. For the β-galactosidase plate assays PC203 (S. aureus 8325-4, spa::lacZ), PC322 (S. aureus 8325-4, hla::lacZ), SH101F7 (S. aureus 8325-4, rnaIII::lacZ) (Chan and Foster, 1998) and strain MOZ53 S. aureus agr type III (Horsburgh et al., 2002) were used. For the β-lactamase liquid assay strain RN10829 WT and RN10829 Const (Geisinger et al., 2009) a β-lactamase reporter strain substituting the native agr locus with a chromosomal integration of P2-agrA and P3-blaZ and a plasmid from which a constitutive active variant of AgrC (agrC-I-R238H) is expressed, was used to assess AgrC-dependent effects of staphylococcal supernatants. Fluorescent reporters AH1677, AH430, AH1747, and AH1872 (Hall et al., 2013) were used to evaluate agr induction of the four different S. aureus agr groups. We cloned the Staphylococcus schleiferi agrBD genes into the BglII/EcoRI sites of expression vector pRAB12-lacZ (Helle et al., 2011) using primers 5′-GATACAAGATCTGTTAAGGAGGAGGGCTATTTG-3′ and 5′-GATACAGAATTCCGCTCTCTAAACATTATTTTATTATTC-3′ and chromosomal DNA from S. schleiferi strain 2898 as template generating plasmid pRAB12-agrBDSs. This construct or the vector itself was expressed in the S. aureus agr deletion strain 8325-4Δagr [constructed by transduction (φ11) from strain RN6911 (Novick et al., 1993) into strain 8325-4] by growing the strains overnight under induction (0.2 μg/ml anhydrotetracycline) generating AIPSs containing or AIP negative supernatants respectively. All other staphylococcal strains used are listed in Table 1. Unless otherwise stated, bacteria were grown in Tryptone Soya Broth (TSB), Oxoid (1:10 volume/flask ratio), at 37°C with shaking at 200 rpm.
Table 1.
Staphylococcal strains, their origin, and RNAIII-regulation activity.
| Id. | Name | Host isolation source | rnaIII downregulation | Id. | Name | Host isolation source | rnaIII downregulation |
|---|---|---|---|---|---|---|---|
| 27472 | Staphylococcus intermedius | Dog wound | ++ | 450 | Staphylococcus equorum | Horse nose | X |
| 30755 | Staphylococcus pseudintermedius | Dog skin | ++ | 1312 | Staphylococcus haemolyticus | Dog ear | + |
| C-31106 | Staphylococcus pseudintermedius | Dog wound | ++ | 2181 | Staphylococcus haemolyticus | Bird trachea | X |
| C- 31304 | Staphylococcus pseudintermedius | Dog nose | ++ | 28993 | Staphylococcus haemolyticus | Bird trachea | + |
| 30510 | Staphylococcus pseudintermedius | Dog urine | + | 9525 | Staphylococcus hominis | Not reported | + |
| 30665 | Staphylococcus pseudintermedius | Dog skin | ++ | 1084 | Staphylococcus hominis | Cat urine | + |
| 30703 | Staphylococcus pseudintermedius | Dog wound | ++ | 218 | Staphylococcus hyicus | Pig joint | + + ++ |
| 30106 | Staphylococcus warneri | Dog wound | + | 2808 | Staphylococcus hyicus | Not reported | + + ++ |
| 29927 | Staphylococcus warneri | Dog urine | + | 4948 | Staphylococcus hyicus | Not reported | + + ++ |
| 30331 | Staphylococcus warneri | Dog skin | + | 6948 | Staphylococcus lentus | Mink skin | X |
| 28351 | Staphylococcus xylosus | Cow ear | X | 6949 | Staphylococcus lentus | Mink Skin | X |
| L136 | Staphylococcus xylosus | Horse abscess | X | C-30966-5a | Staphylococcus lentus | Mink skin | X |
| 9529 | Staphylococcus capitis | Not recorded | ++ | 62 | Staphylococcus vitulinus | Horse trachea | + |
| 2877 | Staphylococcus capitis | Dog ear | X | 96 | Staphylococcus vitulinus | Horse wound | + |
| 6994 | Staphylococcus capitis | Dog tonsil | X | 128 | Staphylococcus vitulinus | Horse wound | + |
| 52 | Staphylococcus capitis | Horse uterus | ++ | 30689-20 | Staphylococcus schleiferi | Mink skin | + + ++ |
| 53 | Staphylococcus chromogenes | Horse uterus | +++ | 30743 | Staphylococcus schleiferi | Dog ear | + + ++ |
| 2890 | Staphylococcus chromogenes | Cow wound | ++ | 30219 | Staphylococcus schleiferi | Dog skin | + + ++ |
| 313 | Staphylococcus chromogenes | Cow milk | ++ | 2319 | Staphylococcus schleiferi | Cat | ++ |
| 1069 | Staphylococcus epidermidis | Dog urine | X | 2862 | Staphylococcus schleiferi | Dog ear | ++ |
| 352 | Staphylococcus epidermidis | Ox milk | X | 2898 | Staphylococcus schleiferi | Mink skin | + + ++ |
| 389 | Staphylococcus epidermidis | Not recorded | + | 9468 | Staphylococcus sciuri | Not reported | X |
| 30575 | Staphylococcus epidermidis | Dog kindney | X | 9470 | Staphylococcus sciuri | Not reported | X |
| 29886 | Staphylococcus delphini | Horse wound | + + + | 9505 | Staphylococcus sciuri | Not reported | + |
| C-30966-8a | Staphylococcus delphini | Mink skin | + + + | 1457 | Staphylococcus simulans | Dog | + + + |
| C-31232-2 | Staphylococcus delphini | Mink wound | X | 312 | Staphylococcus simulans | Ox milk | + + + |
Down-regulation was rated according to the size of the inhibition halo around the well where: X: no effect; + slight effect; ++: moderate effect; +++: severe effect and ++++: very severe effect.
β-Galactosidase Plate Assay
The reporter assay was conducted as described by Nielsen et al. (2010). Test supernatants, and control supernatants of strains 8325-4 (AIP-I) and M0Z53 (AIP-III) were used. Incubation until blue color appeared in plates varied from 9 to 48 h.
In vitro Competition As Assessed Using the β-Galactosidase Liquid Assay
The assay was based on the liquid β-galactosidase assay (Miller, 1972) with some modifications. Briefly, Overnight (ON) cultures of our SH101F7 S. aureus RNAIII reporter strain and our S. schleiferi strains (grown at 37°C while shaking at 200 rpm) were diluted 100x in 15 mL TSB and allowed to reach an OD600 of 0.5 and after adjusting each strain to an OD600 of 0.1 in TSB the competition was started at a ratio of 1:1 and followed over time. From each culture, 1 mL was taken at each time interval and the samples were sonicated for 10 s to disrupt aggregates formed. The OD600 was measured and serial dilutions for each sample were made and plated on TSA with X-gal (150 μg/mL). The remaining samples were centrifuged for 3 min at 8000 rpm and 4°C. After centrifugation, the supernatants were removed and the pellets resuspended in 1 mL of TRIS 50 mM, pH 8 and 3 μL of lysostaphin. The mix was incubated at 37°C for 30 min to allow for cell lysis. Z-buffer (400 μL) was then added to each sample which were further incubated for 5 min at 28°C. Lastly, 100 μL of ONPG (ortho-Nitrophenyl-β-galactoside) (4 mg/mL) was added to the mix and the time necessary for the solution to turn yellow was controlled. The OD at 420 and 550 nm for each sample was measured. The activity of the samples was calculated in Miller units using the following formula as described by Miller (1972):
Where: T = time of reaction in min;
V = ml cells added to the assay tubes.
β-Lactamase Assay and Inhibitory Concentration (IC50)
The method used is described by Nielsen et al. (2014). Briefly the RN10829 (P2-agrA:P3-blaZ)/pagrC-I (WT) and RN10829(P2-agrA:P3-blaZ)/pagrC-I-R23H (AgrC const.) reporter strains were grown to an OD600 of 0.4–0.5 where a 1/10 volume of AIP-I containing supernatant (obtained from strain 8325-4) and 1/10 S. schleiferi supernatants were added to the reporter strain culture. In assays using heterologously expressed AIPSs 1/20 volume of AIP-I containing supernatant was challenged with 1/5 volume supernatant from expression cultures. Samples obtained at 30 min time intervals after addition of test solutions were analyzed for β-lactamase activity by nitrocefin conversion. The IC50 of the selected S. schleiferi supernatants was also tested using the β-lactamase assay, where a 1/10 volume (0.5 mL) of supernatant was added to the total volume of 5 mL of the reporter strain culture (RN10829-WT) representing the undiluted supernatant (100%). Then, 80, 60, 40, 20, 10, 5, 2.5, and 2% of the initial volume of the selected supernatant was added to obtain the IC50 curve.
Assessment of agr Inhibition Across agr Groups
Fluorescent S. aureus reporter strains of agr types I-IV (P3-yfp) were used to evaluate the inhibitory potential of staphylococcal supernatants on respective agr types. Individual reporter strains were grown to early exponential phase (non-fluorescent, OD approximately 0.01), then added 20% S. schleiferi 2898 stationary phase supernatant or TSB as a control and grown to late stationary phase to allow for full agr induction. P3 promoter activity of the bacterial population was monitored as accumulated YFP by flow cytometry on a BD AccuriTM C6 flow cytometer using the FL1 channel.
Reverse Transcriptase-Quantitiative PCR
Overnight culture of the strain 8325-4 was diluted 100x in 15 mL of TSB in a 300 mL flask. Three replicate cultures were prepared and once the cultures reached an OD600 of 0.35 they were split into two different conditions; one with 10% 8325-4 supernatant alone was added and the other with an additional 10% of S. schleiferi 2898 supernatant. Cultures were allowed to grow until sample withdrawal at 30 and 60 min. Samples were centrifuged and the pellet was immediately frozen at -80°C. The disruption of the cell membranes was performed with FastPrep and the the QIAGEN RNeasy kit was used to purify the RNA as per manufacturer’s instructions. Genomic DNA was then removed from the samples using DNase-I, RNase-free from Fermentas. The samples were first incubated for 60 min at 37°C, followed by 10 min incubation with 50 mM of EDTA at 65°C. To generate cDNA from the RNA samples, the High Capacity cDNA RT kit from Applied Biosystems was used. 10 μL of RNA and 10 μL of RT-master mix were added to each reaction. The master mix contained RT Buffer, RT random primers, dNTP mix, Nuclease-free water, and reverse transcriptase. The negative controls consisted of samples with no reverse transcriptase added. The samples were run for 10 min at 25°C, 120 min at 37°C, 5 min at 85°C in a standard PCR machine. Finally, 1.5 μL of cDNA was assayed by real-time PCR on a LightCycler® 96 Instrument using FastStart Essential DNA Green Master FastStart Essential DNA Green Master (Roche) and primers for the reference genes (ileS, pyk) and the target gene (rnaIII) as presented in Table 2.
Table 2.
Real time PCR primers used in this study for the assessment of reference (ileS, pyk) and target gene (rnaIII) expression.
| Gene | Sequence forward | Sequence reverse |
|---|---|---|
| rnaIII | GCACTGAGTCCAAGGAAACTAAC | AAGCCATCCCAACTTAATAACC |
| ileS | ACATACAGCACCAGGTCACG | CGCCTTCTTCAGTAAATACACC |
| pyk | AGGTTGAACTCCCCAAACAA | GCAGCCCAAGATTACAAAAA |
In vivo Competition Assay with Galleria mellonella
The Galleria mellonella infection was carried out as described by Pollitt et al. (2014) with minor modifications. Briefly, fifth-instar G. mellonella larvae were inoculated (in a proleg) with a total of 2 × 107 CFU/mL of S. aureus (SH101F7 rnaIII::lacZ), S. schleiferi (2898 erythromycin resistant) or a co-culture of the two strains (to a combined final CFU/mL of 2 × 107) and split into groups for CFU counting (35 per group) and survival benefit observation (20 per group). Two control groups were included; one injected with phosphate buffered saline (PBS) and the other not handled at all. The larvae were then incubated at 37°C. At 24, 48, and 72 h post-inoculation, the hemolymph of the larvae was collected for CFU determination. Colonies were counted after O/N incubation at 37°C on TSA containing erythromycin (5 μg/mL) and X-gal (150 μg/mL). Survival of the larvae was monitored at the same time points as hemolymph collection. This experiment was repeated three times using different batches of larva purchased from a local pet store (HPReptiles, Copenhagen) with similar results.
AIP Sequencing
Several Staphylococcus spp. were sequenced to look for the agrD and the amino acid sequence of the AIP. DNA from the selected staphylococci isolates was extracted using the DNeasy Blood and Tissue kit as described by the manufacturer (Qiagen, Valencia, CA, USA). For sequencing preparation fragment libraries were constructed using the Nextera kit (Illumina, San Diego, CA, USA) followed by 251-bp paired-end sequencing on a MiSeq sequencer (Illumina) according to the manufacturer’s instructions. The sequencing reads were assembled using CLC Genomics Work- bench 8.0 (Qiagen, Aarhus, Denmark) with default parameters to include only contigs of 500 nucleotides and with over 30-fold average coverage. The sequence of S. schleiferi 2898 has been deposited in GenBank (accession number PRJEB15874). All other sequence contigs are available upon request. Extracted AIP sequences were aligned using Muscle as implemented in MEGAv 6.06, where the phylogeny was constructed using the maximum parsimony approach with 100 bootstraps and represented with midpoint rooting.
Chemical Synthesis of AIP
The AIP was synthesized by adopting a protocol based on linear peptide hydrazides, which was previously reported by Liu and coworkers (Zheng et al., 2013) The linear peptide hydrazide was synthesized by standard 9-fluorenylmethyloxycarbonyl (Fmoc) solid-phase peptide synthesis (SPPS) using HCTU/i-Pr2EtN for amino acid activation on an automated peptide synthesizer (SyroWave XP, Biotage). A hydrazide-2-chlorotrityl resin was applied to afford the desired C-terminal peptide hydrazide [YPFCIAYF-NHNH2] after nine coupling/deprotection cycles, followed by concomitant deprotection of side chain functionalities and cleavage from the solid support with CF3COOH-i-Pr3SiH-water (95:2.5:2.5, 5 mL, 3 h, room temperature). The crude peptide was obtained by trituration with cold ether and used without any further purification. ESI-MS m/z calcd for C53H68N10O10S 1037.5, found 1037.2 [M+H+].
The crude linear peptide (10 mg, 0.01 mmol) in DMF (20 μL) was added to a solution of NaNO2 (6 mg, 0.09 mmol) in sodium phosphate buffer (0.1 mM, pH 3.0) containing guanidinium chloride (6 M) at -20°C. The reaction mixture was kept at -20°C for 20 min and was then quenched by addition of DMF (0.2 mL) and cyclization buffer [sodium phosphate buffer (0.6 mL, 0.1 mM, pH 6.5) containing thiophenol (44 mg, 0.4 mmol)]. This reaction mixture was agitated on a rocking table for 16 h at room temperature. The desired AIP was then purified by preparative reversed-phase HPLC on a C18 Phenomenex Luna column (250 mm × 20 mm, 5 μm, 100 Å) using an Agilent 1260 LC system equipped with a diode array UV detector, applying a gradient of eluent I (water-MeCN-TFA, 95:5:0.1) and eluent II (0.1% TFA in acetonitrile) with a flow rate of 20 mL/min. Lyophilization of the fractions containing product, provided a white fluffy solid (∼0.5 mg, 5%) at >98% homogeneity as determined by UPLC–MS analysis at 254 nm. The compound was reconstituted in DMSO and accurate concentration was determined by UV spectroscopy (5 mM) before use. ESI-MS m/z calcd for C53H64N8O10S 1005.5, found 1005.2 [M+H+]. MALDI-TOF MS found 1005.6 [M+H+].
Statistics
Where applicable for the β-lactamase assays (Figures 2B and 4A) statistical analysis was performed using the multiple t-test allowing unequal variance. For the qPCR data (Figure 2A) an unpaired t-test was performed on normalized and log transformed data. For survival curve analysis, the Kaplan–Meier method was applied and statistical analysis was carried out using the Log Rank (Mantel–Cox) Chi square test. Any value above P = 0.05 was considered as not significant. All statistical tests were performed with GraphPad Prism v. 7.0.
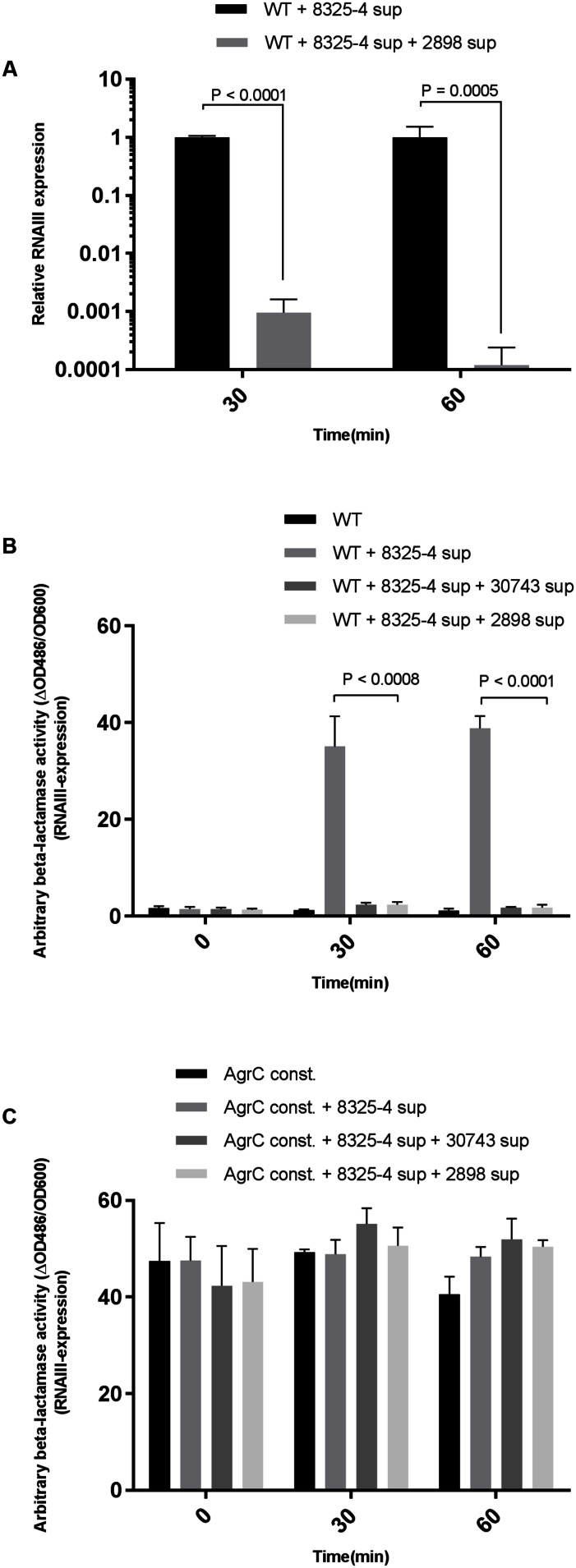
FIGURE 2.
Quantification of interference by S. schleiferi on RNAIII expression in S. aureus. (A) RT-qPCR quantitative verification of RNAIII downregulation in the S. aureus 8325-4 strain. S. aureus in 15 mL TSB was grown to an OD600 = 0.35 and supplemented with a 1:10 volume of S. schleiferi 2898 supernatant. Samples were collected 30 and 60 min post-exposure to S. schleiferi supernatant, RNA was isolated, cDNA prepared (and RNAIII levels were quantified. Interference of S. schleiferi supernatant on S. aureus agr was monitored using the strains (B) RN10829(P2-agrA:P3-blaZ)/pagrC-I (WT) and (C) RN10829(P2-agrA:P3-blaZ)/pagrC-I-R23H (AgrC const.) Reporter strains were grown to an OD600 of 0.4–0.5 where a 1/10 volume of AIP-I containing supernatant from strain 8325-4 and 1/10 S. schleiferi supernatant were added to the reporter strain culture. Samples obtained at 30 min time intervals after addition of test solutions were analyzed for β-lactamase activity by nitrocefin conversion (Nielsen et al., 2014). Each bar represents the average of 3 biological replicates and the error bars represent the standard deviation.)
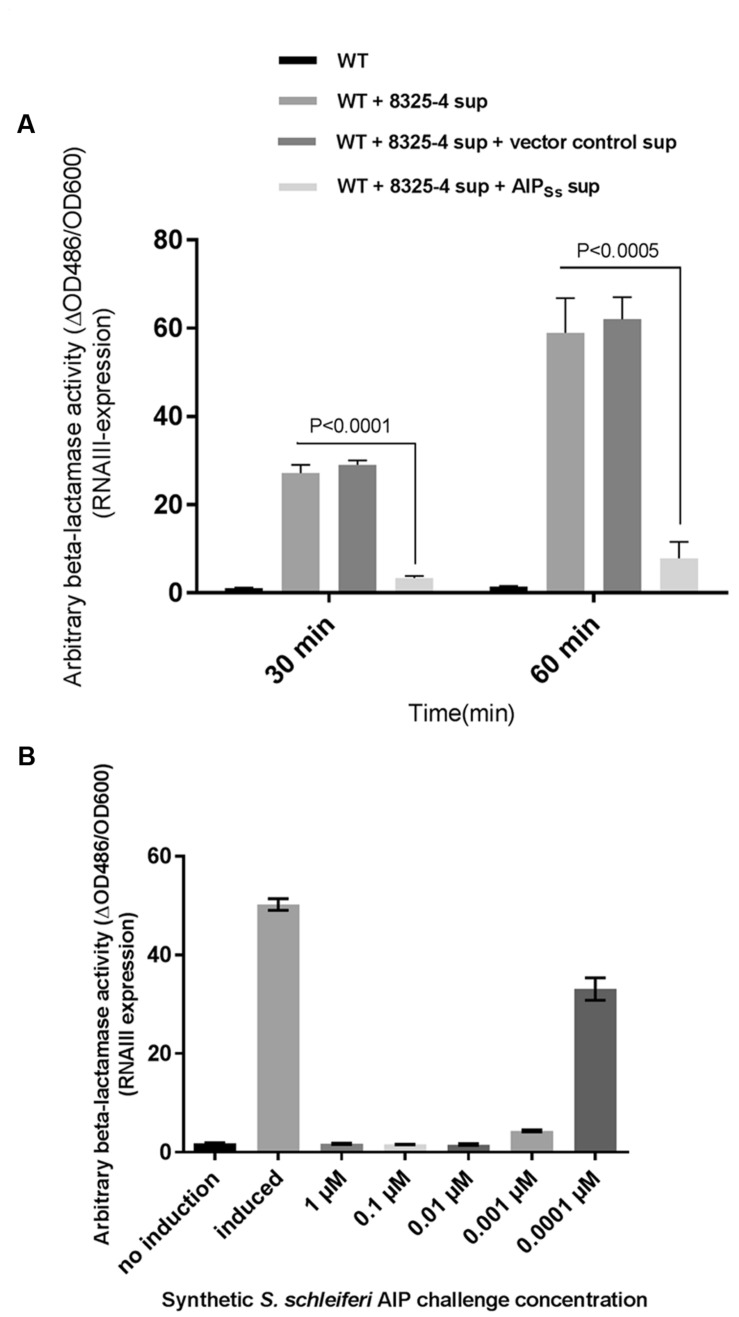
FIGURE 4.
Staphylococcus schleiferi AIP interferes with S. aureus agr. (A) RNAIII expression was recorded as β-lactamase expressed from the P3-blaZ reporter fusion in S. aureus RN10829(P2-agrA:P3-blaZ)/pagrC-I (WT) with addition of 5% AIP-I supernatant and 20% supernatant from either strain 8325-4Δagr/pRAB12-agrBDSs (AIPSs) or the control strain 8325-4Δagr/pRAB12-lacZ (vector control) that had been grown and induced (0.2 μg/ml anhydrotetracycline) overnight. Each bar represents the average of three biological replicates and the error bars represent the standard deviation. (B) P3-blaZ expression recorded from S. aureus RN10829(P2-agrA:P3-blaZ)/pagrC-I (WT) when the inducing AIP-I containing supernatant (10%) is challenged for 45 min with different concentrations of synthetic S. schleiferi AIP at indicated concentrations. No induction and AIP-I containing supernatant alone was included as controls. Each bar represents the average of three biological replicates and the error bars represent the standard deviation.
Results
S. aureus Virulence Gene Expression is Modulated by Staphylococcal Culture Supernatants
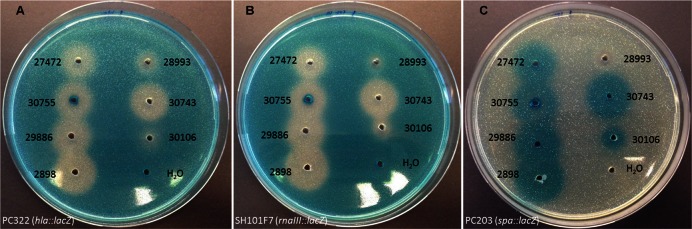
A total of 52 staphylococcal isolates representing 17 species obtained from a variety of different animal hosts (Table 1) were examined for their ability to interfere with the agr quorum sensing system of S. aureus using a previously established reporter assay (Nielsen et al., 2010). Staphylococcal strains to be tested were grown overnight to stationary phase in tryptone soy broth (TSB), and cell-free supernatants were added to wells formed in tryptone soy agar (TSA) plates containing reporter strains carrying lacZ fusions to either one of the agr controlled virulence genes hla, spa, or rnaIII as well as the β-galactosidase substrate, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal). For the majority (37 out of 52) of the staphylococcal supernatants tested we observed a reduced expression of hla and rnaIII but increased spa expression (Figure 1; Table 1). The 37 supernatents exhibiting this expression pattern represents 14 of the 17 staphylococcal species, indicating that these secrete substances that interfere with the agr system of S. aureus.
FIGURE 1.
Modulation of Staphylococcus aureus virulence gene expression by staphylococcal culture supernatants. TSA agar plates (with erythromycin and X-gal) containing (A) the hla-lacZ (PC322; Eryr), (B) the rnaIII-lacZ (SH101F7; Eryr), or (C) the spa-lacZ (PC203; Eryr) reporter strain of S. aureus were exposed to 20 μL (in pre-drilled wells) of supernatants from centrifugation (8000 rpm for 60 s) of overnight cultures of strains 27472 (Staphylococcus intermedius), 28993 (Staphylococcus haemolyticus), 30755 (Staphylococcus pseudintermedius), 30743 (Staphylococcus schleiferi), 29886 (Staphylococcus delphini), 30106 (Staphylococcus warneri) and 2898 (Staphylococcus schleiferi). H2O was used as a control. Zones appeared between 9 and 36 h of incubation at 37°C. This figure is representative of one set of screening plates.
The Dog Pathogen Staphylococcus schleiferi is a Potent Inhibitor of the S. aureus agr Quorum Sensing System
The magnitude of agr-interference caused by staphylococcal supernatants was monitored in S. aureus 8325-4 by RT-qPCR using previously described primers (Nielsen et al., 2012). Using the supernatant of the notably active S. schleiferi strain 2898 we observed that the relative RNAIII expression in S. aureus 8325-4 decreased by 300 and 3000-fold at 30 and 60 min respectively, when compared to unexposed cells (Figure 2A). Furthermore, the 50% inhibitory concentration (IC50) was reached with S. schleiferi supernatant constituting only 6% of the total S. aureus culture volume.
To address if the observed effect of staphylococcal supernatants on virulence gene expression in S. aureus is mediated via direct interference with the S. aureus agr regulatory system, we examined if the effect could be mitigated in a S. aureus strain encoding a constitutively active AgrC sensor histidine kinase (Geisinger et al., 2009). Using the reporter strains RN10829 WT (expressing the WT AgrC) and RN10829 Const. (isogenic mutant expressing the constitutively active AgrC) containing a blaZ gene fused to the P3 promoter as previously described by Nielsen et al. (2014), we examined the culture supernatants obtained from S. schleiferi strains 2898 and 30743. As predicted, in the presence of the WT AgrC in the reporter strain, supernatants of both of the S. schleiferi strains significantly down-regulated RNAIII expression (Figure 2B) but when the S. schleiferi supernatants were added to the strain expressing the constitutively active AgrC (RN10829 Const.) we observed no difference in RNAIII expression (Figure 2C). These results suggest that the effect is AgrC mediated.
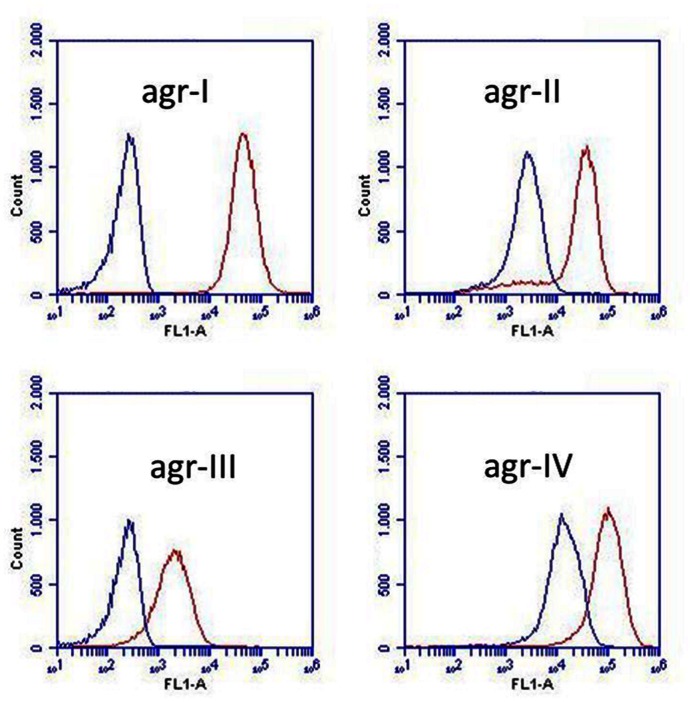
Under the presumption that S. schleiferi produces AIPs, which cross-inhibit the agr system of S. aureus, we assessed the specificity with respect to the different S. aureus agr types. Hence, we examined expression from a previously described P3-yfp reporter construct present in S. aureus strains of known agr types (Hall et al., 2013) that had been grown in the presence or absence of supernatant from S. schleiferi strain 2898. Evidently, the supernatant confered a considerable degree of repression across all agr groups of S. aureus (Figure 3) giving rise to a 10–100 fold reduction in observed fluorescence intensity of all reporters. We note that the agr system appears almost completely repressed in the types I and III reporters even at the late time point of the assay (24 h), while the repression of the types II and IV reporters display a delay in the response. This difference may stem from different cross-inhibitory affinities of the suspected AIP molecule of S. schleiferi against the different AIP-AgrC receptor pairs in S. aureus, or simply reflect a difference in the agr activation kinetics and/or auto-fluorescence of the reporters employed.
FIGURE 3.
Staphylococcus schleiferi supernatant affects the activity of all four S. aureus agr types. Activity of four S. aureus agr group reporters (agr-I, agr-II, agr-III, and agr-IV) assessed by P3 expression (P3-yfp) measured as YFP accumulation by flow cytometry. Non-fluorescent, exponential phase cells were grown with (blue) or without (red) 20% S. schleiferi 2898 supernatant until stationary phase. Depicted fluorescence histograms originate from cells analyzed at the 24 h time point.
S. schleiferi Inhibition of S. aureus agr is AIP-Mediated
Having demonstrated that S. schleiferi supernatant is a potent inhibitor of RNAIII expression in S. aureus we investigated the hypothesis that the inhibition was due to the presence of non-native AIP in the supernatant of S. schleiferi. S. aureus AIP was previously biosynthesized in S. aureus by cloning the agrBD genes in an otherwise agr negative background (Thoendel and Horswill, 2009). Here, after genome sequencing of the S. schleiferi strain 2898, we cloned the 2898 agrBD genes into the expression vector pRAB12-lacZ (Helle et al., 2011) generating plasmid pRAB12-agrBDSs and expressed it in the S. aureus agr deletion strain 8325-4Δagr. In contrast to the inactive vector control strain the supernatant of the S. schleiferi-AIP (AIPSs) producing strain clearly inhibited RNAIII expression as measured by β-lactamase activity from the P3-blaZ reporter strain both 30 and 60 min after addition of the supernatant (Figure 4A). These results confirm that the AIP produced by S. schleiferi is inhibiting agr of S. aureus. To further support that it is in fact the S. schleiferi AIP that is responsible for inhibition of S. aureus RNAIII via AgrC agonist activity, we synthesized the proposed S. schleiferi AIP and tested the synthetic material in the same P3-blaZ reporter strain. Our results show indisputably that the S. schleiferi AIP is a potent inhibitor of S. aureus RNAIII expression and that it acts antagonistically on the reporter strain at low nanomolar concentrations (Figure 4B).
S. aureus agr Repression by S. schleiferi is Observed during Niche Competition Both In vitro and In vivo
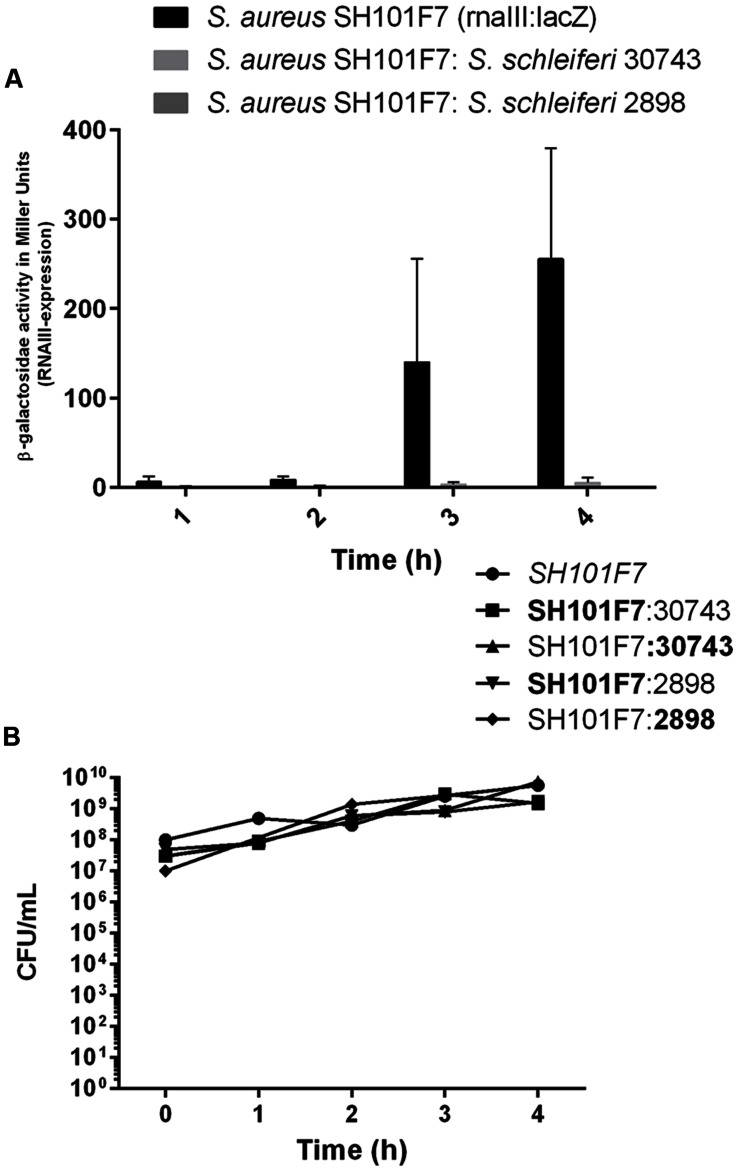
To address if the agr inhibitory activity of S. schleiferi also is expressed when both strains share the same niche environment we co-cultured S. aureus SH101F7 rnaIII::lacZ reporter strain together with the S. schleiferi 2898 or 30743 strains at a 1:1 ratio in TSB. After 4 h where bacterial growth had reached stationary phase and the agr quorum sensing system in S. aureus is normally fully activated, we observed almost complete inhibition of S. aureus RNAIII expression in cells co-cultured with either of the S. schleiferi strains (Figure 5A). Within the time frame of the experiment, the growth proportion of S. aureus to S. schleiferi cells remained essentially unchanged suggesting that neither strain exert a growth inhibitory effect toward each other (Figure 5B). Thus, during co-culture the presence of S. schleiferi efficiently represses the S. aureus agr quorum sensing system.
FIGURE 5.
Staphylococcus schleiferi inhibits S. aureus agr during co-culture without influencing growth. (A) S. aureus rnaIII:lacZ reporter strain SH101F7 grown alone or in co-culture (1:1) with S. schleiferi strain 30743 or 2898 in TSB. β-galactosidase activity under each condition was determined to monitor agr induction. Each bar represents the average of three replicates and the error bars represent the standard error of the mean. (B) Growth of the bacterial cultures measured as colony forming units (CFUs)/mL monitored in parallel for each time point. Distinction between the two species in the co-culture was made by plating on TSA substituted with X-gal, resulting in the SH101F7 reporter strain growing as blue colonies, while the S. schleiferi growing as white.
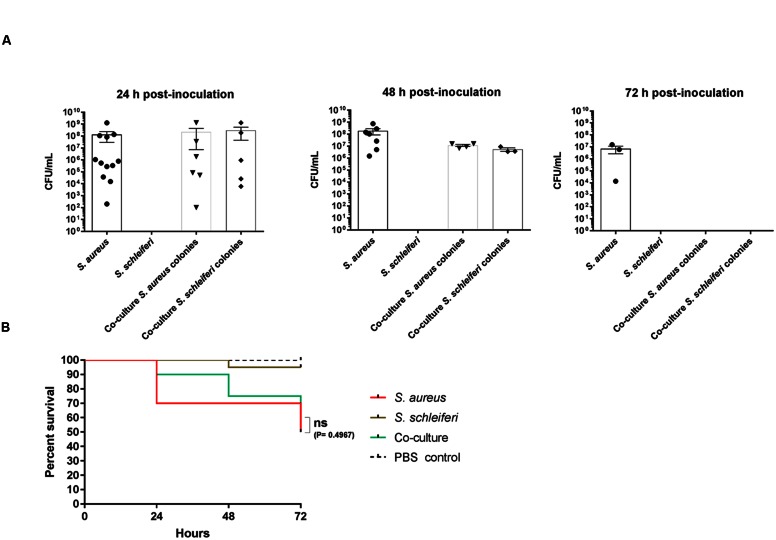
Given the obvious interaction between the two bacterial species during co-culture in laboratory medium we investigated if a similar phenomenon is observed in G. mellonella wax moth larvae, a model previously reported for studies of S. aureus virulence (Desbois and Coote, 2011). Applying the same growth conditions as for the in vitro co-culture experiment we inoculated 2 × 107 colony forming units (CFUs) into the prolegs of fifth-instar larvae and followed the CFU counts and larval survival over a period of 3 days. When single species were inoculated, S. aureus was recovered at 1 × 108 cells 24 h post-inoculation and only declined slightly at 72 h. We were unable to detect any S. schleiferi in the larval hemolymph at any of the time points (Figure 6A), where survival of this group was comparable to the PBS control larvae group (Figure 6B). In contrast, when S. schleiferi was inoculated together with S. aureus both species were present in equal numbers at 24 h, followed by a decline at 48 h and complete eradication of both species after 72 h (Figure 6A). The presence of S. schleiferi 24 and 48 h after inoculation with S. aureus suggests that factors produced by S. aureus allow maintenance of both species in the larva, while inversely the clearance of both species at 72 h may be suggestive that there is a S. schleiferi-mediated repression of S. aureus virulence factors via agr down-regulation, and that this repression allows the larvae to eradicate the combined staphylococcal population. Despite the clearance of both species at 72 h, the co-culture did not offer a significant larval survival benefit over the single S. aureus inoculant group (Figure 6B). Nevertheless, our data show that in the presence of S. aureus, S. schleiferi is maintained for an extended period of time in the larvae and that ultimately their presence leads to eradication of both species.
FIGURE 6.
Dual presence of S. schleiferi and S. aureus influence colonization capabilities in a Galleria mellonella infection model. Fifth-instar G. mellonella larvae were inoculated with a total of 2 × 107 CFU/mL of S. aureus (SH101F7), S. schleiferi (2898) or a co-culture of the two strains (to a combined final CFU/mL of 2 × 107) and split into groups for CFU counting (35 per group) and survival benefit observation (20 per group). (A) At 24, 48, and 72 h post-inoculation, the hemolymph of the larvae was collected for CFU determination. Colonies were counted after O/N incubation at 37°C on TSA containing erythromycin and X-gal, as both strains are erythromycin resistant. (B) Survival of the larvae was monitored at the same time points as hemolymph collection. This experiment was repeated three times with similar results.
AIP Homology in Staphylococci
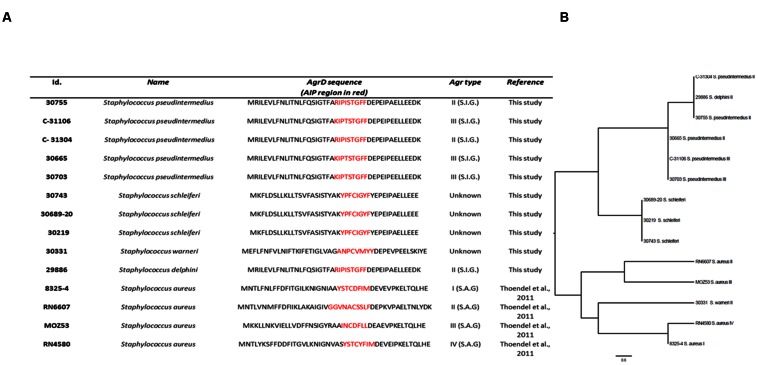
The AIP amino acid composition, positioning and properties have been reported to play an important role in the type of AgrC interaction (Mayville et al., 1999; Lyon et al., 2000; Wright et al., 2004; Tal-Gan et al., 2013a). Accordingly, we analyzed sequence homology of AIPs from a selection of staphylococci for which supernatant activity ranged from no effect to severe inhibition of S. aureus agr (Figure 7A). Alignment of AIP sequences between different staphylococcal species revealed less than 30–40% amino acid conservation. Within the same species, the AIPs were highly conserved; a phenomenon that has also been observed for already reported staphylococcal AIP sequences (Dufour et al., 2002; Thoendel and Horswill, 2009). For our sequenced S. schleiferi AIPs there was 100% conservation and we noted that the most highly conserved amino acids between species were those with specific properties needed for correct interaction or folding of AIP. For example, the C-terminal amino acid of most of the ascertained AIP sequences is tyrosine or phenylalanine which provides the hydrophobicity essential for binding of the AIP to a putative hydrophobic pocket in the AgrC receptor, but is not required for the antagonistic or the activation activity (Wright et al., 2004). Also they retained the central cysteine necessary for AIP cyclization (Thoendel and Horswill, 2009). Interestingly, despite their strong inhibitory activity toward S. aureus agr, neither S. schleiferi nor Staphylococcus delphini AIP contain an alanine residue, which has been reported to play a role in non-native AIP-AgrC binding antagonism (Lyon et al., 2000; Tal-Gan et al., 2013a). Inferred relationships of the various AIPs revealed that they clustered according to species and for the most part also according to AIP group (Figure 7B). The differences in amino acid conservation and the difficulties in locating specific amino acid sequences responsible for non-cognate AIP-AgrC interactions, highlights that spatial arrangement and secondary structure of the AIPs play an important role in the interactions between the non-cognate AIPs and the S. aureus Agr-C (Mayville et al., 1999; Lyon et al., 2000; Wright et al., 2004). We speculate, that AgrC is promiscuous in terms of susceptibility to AIP inhibitory structures and this promiscuity allows inhibition by multiple staphylococcal species.
FIGURE 7.
agrD homology of selected Staphylococci. Isolates were analyzed for agrD homology by Illumina sequencing; (A) Table showing AIP-alignment. The AIP region is highlighted in red. S.I.G.: Staphylococcus intermedius Group; S.A.G.: S. aureus Group; Strains 8325-4, RN6607, MOZ53, and RN4580 were included as S. aureus AIP sequence references. (B) Phylogenetic analysis based on a Muscle alignment of the AIP sequences with midpoint rooting. Scale bar indicate substitutions per site.
Discussion
This study looks at the ecological aspects of regulatory cross-talk and niche sharing between staphylococci as mediated via the agr quorum sensing system, and delves into the possible effects this cross-talk may have on aspects of virulence and colonization for the human pathogen Staphylococcus aureus.
Cell to cell communication via quorum sensing is very common amongst bacteria and promotes both inter and intra-species interactions (LaSarre and Federle, 2013). The agr QS system of S. aureus is especially sensitive to pheromones produced by S. aureus strains of other agr groups (Thoendel and Horswill, 2009; Thoendel et al., 2011) or produced by other staphyloccocal species, namely S. epidermidis (Otto et al., 2001), and has even displayed sensitivity to compounds produced by other microorganisms of unrelated niche environments such as the marine Photobacterium halotolerans-derived QS inhibitor solonamide B (Nielsen et al., 2010, 2014). Despite this well-documented agr cross-inhibition between the different S. aureus agr groups and between S. aureus and other staphylococcal species, we aimed to explore further the communication between S. aureus and staphylococci with little niche overlap, and what repercussions this communication might have in terms of virulence adaptation. While our findings corroborate those of others with regards to staphylococcal cross-talk via agr (Otto et al., 2001; Lina et al., 2003; Thoendel et al., 2011), we also show extensive AIP-mediated cross-talk between previously untested staphylococci and S. aureus, with 82% of the previously untested staphylococci exerting ability to interfere with agr of S. aureus. Importantly, we show that staphylococci from varying environmental niches not only have the capacity to interfere with S. aureus agr, but even influence virulence and colonization capabilities of S. aureus when forced to share a niche, as was shown in the co-infection of G. mellonella with S. schleiferi and S. aureus. Cross-communication involving agr interference has previously been observed as a result of co-habitual competition within the same ecological niche, and we speculate that this also is a reason for the observed interference in our study. A few known examples are the interactions between S. aureus and S. epidermidis on human skin and between S. aureus and Pseudomonas aeruginosa in the lungs of patients suffering from cystic fibrosis (Otto et al., 2001; Qazi et al., 2006). In the first example, S. epidermidis AIP pheromone is capable of inhibiting agr activity of S. aureus groups I, II and III, but not that of group IV, which interestingly is the only S. aureus AIP capable of inhibiting S. epidermidis agr activity. It has been suggested that this cross-talk is one of the reasons why S. epidermidis predominates over S. aureus on human skin (Otto et al., 2001). In the second example, the cross-talk between P. aeruginosa and S. aureus is a case of cross-communication between two unrelated microorganisms. It has been shown that N-acylhomoserine lactone 3-oxo-C12-HSL produced by Pseudomonas is capable of inhibiting S. aureus growth, and that this compound at sub-inhibitory concentrations interferes with S. aureus agr and sarA expression in a manner involving cytoplasmic membrane saturation (Qazi et al., 2006). In cystic fibrosis patients, the interaction between them in addition to host factors tends to favor the predominance of S. aureus colonization in young patients and P. aeruginosa in adult patients, though co-isolation of both organisms is still found in 50% of adult CF patients (Qazi et al., 2006). The co-existence of these two organisms in CF patients suggests an evolutionary role of cross-talk on influencing virulence (Qazi et al., 2006; Pastar et al., 2013). Our data on the effect of S. schleiferi on S. aureus virulence in the G. mellonella host suggests that this influence on virulence via QS cross-talk may be a common mechanism, and offers a good model for further exploration of this evolutionary role and a possible tool to study virulence adaptation. One limitation of our study is the lack of isogenic S. schleiferi agr mutants, which would allow us to firmly establish agr interference as the main cause of the characteristic co-colonization pattern observed in the in vivo model. At present, we cannot exclude the involvement of other factors during niche-sharing. Nevertheless, it still remains baffling that so much cross-talk or interference via QS is observed between S. aureus and distant staphylococci. Although an attempt to understand this was made by sequencing several AIP-encoding genes, the exact mechanisms of this AgrC promiscuity that allows susceptibility to non-cognate AIP are yet to be determined. Further studies that apply a systematic analysis of the S. schleiferi AIP structure and characterization of key structure-activity-relationship functions necessary for S. aureus AgrC activation/modulation or inhibition are needed. Following in the footsteps of the pioneering work described by Blackwell colleagues on S. aureus and S. epidermidis AgrC modulation (Tal-Gan et al., 2013a,b, 2016; Yang et al., 2016), would certainly shed light on the key features responsible for AgrC promiscuity.
Apart from the insight into ecological aspects of regulatory cross-talk and niche sharing, our findings could also be of potential therapeutic interest. The observation that even at nanomolar concentrations the S. schleiferi AIP is a potent inhibitor of S. aureus agr and is active across all agr specificity groups I to IV is exciting, offers a potential new avenue for exploring staphylococci as sources for quorum sensing inhibitors that can be used to target S. aureus agr-related infections. As we are now faced with the ever increasing burden of antimicrobial resistance, alternative therapeutic avenues are being explored to treat infections with antibiotic resistant pathogens, such as methicillin-resistant S. aureus (MRSA) (Boucher et al., 2009). One such alternative approach is anti-virulence therapy, that targets virulence rather than viability of the pathogen (Rasko and Sperandio, 2010). For MRSA, QS inhibition via the application of compounds that target the agr system has been shown to be a promising anti-virulence approach (Nielsen et al., 2014; Sully et al., 2014; Baldry et al., 2016). The potent S. aureus agr-inhibitory activity of S. schleiferi AIP fits perfectly within the scope of anti-virulence therapy targeting S. aureus, and warrants further in vivo investigations to address the true therapeutic potential of staphylococcal derived anti-virulence candidates targeting S. aureus. Furthermore, the significance of our findings are very much in line with the very rescent discovery that human nasal commensal staphylococci are also potential sources of novel antibiotics to combat MRSA (Zipperer et al., 2016).
Conclusion
We show that there is extensive cross-talk between S. aureus and other staphylococcal species mediated via the agr quorum sensing system. We have (i) identified several staphylococcal species with the ability of cross-interfering with S. aureus agr, (ii) identified novel AIP sequences, some of which exhibiting broad antagonistic activity on S. aureus agr, and (iii) provided a method for expressing foreign AIPs in S. aureus. As staphylococcal species are often found in the same habitat, our results suggest that the agr quorum sensing system is important for inter-species communication, that it may play a crucial role in determining the local population structure, and that this communication may influence S. aureus virulence. Lastly, we suggest that AIPs from other staphylococci have the potential to be exploited as anti-virulence compounds targeting S. aureus infections.
Author Contributions
MB, MSB, PD, CO, and HI planned the experiments. JC, MB, and MSB performed the microbiological work. BG and PG performed chemical synthesis. MS did the bioinformatic analysis. PA supervised the bioinformatic analysis and PD and HI the microbiological work. CO supervised the chemical research. JC, MB, MSB, PA, PD, CO, and HI contributed to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that theresearch was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Jakob Krause Haaber for guidance regarding the G. mellonella wax moth larva model. We are grateful to Dr. Alexander Horswill for providing the fluorescent agr type reporters.
Footnotes
Funding. This work was supported by the European Union’s Seventh Framework Program for research, technological development and demonstration under grant agreement N° 289285, and by grants from the Danish Council for Independent Research – Technology and Production (1337-00129 and 1335-00772).
References
- Baldry M., Kitir B., Frokiaer H., Christensen S. B., Taverne N., Meijerink M., et al. (2016). The agr inhibitors solonamide B and analogues alter immune responses to Staphylococccus aureus but do not exhibit adverse effects on immune cell functions. PLoS ONE 11:e0145618 10.1371/journal.pone.0145618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! an update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Chan P. F., Foster S. J. (1998). The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144 2469–2479. 10.1099/00221287-144-11-3229 [DOI] [PubMed] [Google Scholar]
- Desbois A. P., Coote P. J. (2011). Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 66 1785–1790. 10.1093/jac/dkr198 [DOI] [PubMed] [Google Scholar]
- Dufour P., Jarraud S., Vandenesch F., Greenland T., Novick R. P., Bes M., et al. (2002). High genetic variability of the agr locus in Staphylococcus Species. J. Bacteriol. 184 1180–1186. 10.1128/jb.184.4.1180-1186.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E., Chen J., Novick R. P. (2012). Allele-dependent differences in quorum-sensing dynamics result in variant expression of virulence genes in Staphylococcus aureus. J. Bacteriol. 194 2854–2864. 10.1128/JB.06685-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E., Muir T. W., Novick R. P. (2009). Agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc. Natl. Acad. Sci. U.S.A. 106 1216–1221. 10.1073/pnas.0807760106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall P. R., Elmore B. O., Spang C. H., Alexander S. M., Manifold-Wheeler B. C., Castleman M. J., et al. (2013). Nox2 modification of LDL is essential for optimal apolipoprotein B-mediated control of agr Type III Staphylococcus aureus quorum-sensing. PLoS Pathog. 9:e1003166 10.1371/journal.ppat.1003166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L. G., Foster S. J., Richards R. G. (2002). An introduction to Staphylococcus aureus, and techniques for identifying and quantifying S. aureus adhesins in relation to adhesion to biomaterials: review. Eur. Cell Mater. 4 39–60. [DOI] [PubMed] [Google Scholar]
- Helle L., Kull M., Mayer S., Marincola G., Zelder M. E., Goerke C., et al. (2011). Vectors for improved Tet repressor-dependent gradual gene induction or silencing in Staphylococcus aureus. Microbiology 157 3314–3323. 10.1099/mic.0.052548-0 [DOI] [PubMed] [Google Scholar]
- Horsburgh M. J., Aish J. L., White I. J., Shaw L., Lithgow J. K., Foster S. J. (2002). B modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184 5457–5467. 10.1128/JB.184.19.5457-5467.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G. (1997). Bacterial interference caused by autoinducing peptide variants. Science 276 2027–2030. 10.1126/science.276.5321.2027 [DOI] [PubMed] [Google Scholar]
- Kozioł-Montewka M., Szczepanik A., Baranowicz I., Jóźwiak L., Książek A., Kaczor D. (2006). The investigation of Staphylococcus aureus and coagulase-negative staphylococci nasal carriage among patients undergoing haemodialysis. Microbiol. Res. 161 281–287. 10.1016/j.micres.2005.10.002 [DOI] [PubMed] [Google Scholar]
- LaSarre B., Federle M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77 73–111. 10.1128/MMBR.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G., Boutite F., Tristan A., Bes M., Etienne J., Vandenesch F. (2003). Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69 18–23. 10.1128/AEM.69.1.18-23.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339 520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Lyon G. J., Mayville P., Muir T. W., Novick R. P. (2000). Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc. Natl. Acad. Sci. U.S.A. 97 13330–13335. 10.1073/pnas.97.24.12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayville P., Ji G., Beavis R., Yang H., Goger M., Novick R. P., et al. (1999). Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. U.S.A. 96 1218–1223. 10.1073/pnas.96.4.1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. (1972). Assay of β-galactosidase. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press. [Google Scholar]
- Nagase N., Sasaki A., Yamashita K., Shimizu A., Wakita Y., Kitai S., et al. (2002). Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 64 245–250. 10.1292/jvms.64.245 [DOI] [PubMed] [Google Scholar]
- Nielsen A., Månsson M., Bojer M. S., Gram L., Larsen T. O., Novick R. P., et al. (2014). Solonamide B inhibits quorum sensing and reduces Staphylococcus aureus mediated killing of human neutrophils. PLoS ONE 9:e84992 10.1371/journal.pone.0099151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A., Nielsen K. F., Frees D., Larsen T. O., Ingmer H. (2010). Method for screening compounds that influence virulence gene expression in Staphylococcus aureus. Antimicrob. Agents Chemother. 54 509–512. 10.1128/AAC.00940-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L., Roggenbuck M., Haaber J., Ifrah D., Ingmer H. (2012). Diverse modulation of spa transcription by cell wall active antibiotics in Staphylococcus aureus. BMC Res. Notes 5:457 10.1186/1756-0500-5-457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. (1967). In vivo transmission of drug resistance factors between strains of Staphylococcus aureus. J. Exp. Med. 125 45–59. 10.1084/jem.125.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. E., Todd D. A., Schaeffer C. R., Paharik A. E., Van Dyke M. J., Buttner H., et al. (2014). Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J. Bacteriol. 196 3482–3493. 10.1128/JB.01882-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M., Echner H., Voelter W., Gotz F. (2001). Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69 1957–1960. 10.1128/IAI.69.3.1957-1960.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar I., Nusbaum A. G., Gil J., Patel S. B., Chen J., Valdes J., et al. (2013). Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 8:e56846 10.1371/journal.pone.0056846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt E. J. G., West S. A., Crusz S. A., Burton-Chellew M. N., Diggle S. P. (2014). Cooperation, quorum sensing, and evolution of virulence in Staphylococcus aureus. Infect. Immun. 82 1045–1051. 10.1128/IAI.01216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi S., Middleton B., Muharram S. H., Cockayne A., Hill P., O’Shea P., et al. (2006). N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 74 910–919. 10.1128/IAI.00500-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T.-H. L., Khan B. A., Sturdevant D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell. 32 150–158. 10.1016/j.molcel.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Sperandio V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9 117–128. 10.1038/nrd3013 [DOI] [PubMed] [Google Scholar]
- Shoham M. (2011). Antivirulence agents against MRSA. Future Med. Chem. 3 775–777. 10.4155/fmc.11.43 [DOI] [PubMed] [Google Scholar]
- Sully E. K., Malachowa N., Elmore B. O., Alexander S. M., Femling J. K., Gray B. M., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174 10.1371/journal.ppat.1004174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y., Ivancic M., Cornilescu G., Cornilescu C. C., Blackwell H. E. (2013a). Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an AgrC quorum sensing receptor in Staphylococcus aureus. J. Am. Chem. Soc. 135 18436–18444. 10.1021/ja3112115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y., Stacy D. M., Foegen M. K., Koenig D. W., Blackwell H. E. (2013b). Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J. Am. Chem. Soc. 135 7869–7882. 10.1021/ja3112115 [DOI] [PubMed] [Google Scholar]
- Tal-Gan Y., Ivancic M., Cornilescu G., Yang T., Blackwell H. E. (2016). Highly stable, amide-bridged autoinducing peptide analogues that strongly inhibit the AgrC quorum sensing receptor in Staphylococcus aureus. Angew. Chem. Int. 55 8913–8917. 10.1002/anie.201602974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M., Horswill A. R. (2009). Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J. Biol. Chem. 284 21828–21838. 10.1074/jbc.M109.031757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M., Kavanaugh J. S., Flack C. E., Horswill A. R. (2011). Peptide signaling in the staphylococci. Chem. Rev. 111 117–151. 10.1021/cr100370n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenesch F. O., Projan S. J., Kreiswirth B., Etienne J., Novick R. P. (1993). Agr-related sequences in Staphylococcus lugdunensis. FEMS Microbiol. Lett. 111 115–122. 10.1111/j.1574-6968.1993.tb06370.x [DOI] [PubMed] [Google Scholar]
- Wamel W. J. B., Rossum G., Verhoef J., Vandenbroucke-Grauls C. M. J. E., Fluit A. C. (1998). Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol. Lett. 163 1–9. 10.1016/S0378-1097(98)00132-3 [DOI] [PubMed] [Google Scholar]
- Wang B., Zhao A., Novick R. P., Muir T. W. (2014). Activation and inhibition of the receptor histidine kinase AgrC occurs through opposite helical transduction motions. Mol. Cell. 53 929–940. 10.1016/j.molcel.2014.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. S., Lyon G. J., George E. A., Muir T. W., Novick R. P. (2004). Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 101 16168–16173. 10.1073/pnas.0401825101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Tal-Gan Y., Paharik A. E., Horswill A. R., Blackwell H. E. (2016). Structure-function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for AgrC-type receptor modulation. ACS Chem. Biol. 11 1982–1991. 10.1021/acschembio.6b00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.-S., Tang S., Qi Y.-K., Wang Z.-P., Liu L. (2013). Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat. Protoc. 8 2483–2495. 10.1038/nprot.2013.152 [DOI] [PubMed] [Google Scholar]
- Zipperer A., Konnerth M. C., Laux C., Berscheid A., Janek D., Weidenmaier C., et al. (2016). Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535 511–516. 10.1038/nature18634 [DOI] [PubMed] [Google Scholar]