Abstract
Colorectal cancer is one of the most common types of inflammation-based cancers and is occurred due to growth and spread of cancer cells in colon and/or rectum. Previously genetic association of cell cycle genes, both proto-oncogenes and the tumor suppressors has been proved. But there were few studies about association of immune related genes such as killer-cell immunoglobulin-like receptors (KIRs). Thus we intend to perform a meta-analysis to find the association of different genes of KIR and susceptibility to be affected by colorectal cancer. The overall population of the four studies investigated in our meta-analysis was 953 individuals (470 individuals with colorectal cancer and 483 individuals in control groups). After the analyses, we concluded that colorectal cancer is affected by KIR2DS5 and also there were no protecting gene. This result shows the inflammatory basis of this cancer. In other words, in contrast to leukemia and blood cancers, colorectal cancers seem to be affected by hyper activity of natural killer-cells (NKs). Whys and therefore of this paradox, is suggested to be investigated further.
Keywords: Colorectal cancer, Killer cell immunoglobulin-like receptor, Meta-analysis
1. Introduction
The most common cancer in males is the prostate [1] cancer whereas the most common cancer and malignancy in female is the breast cancer [2], [3], [4], [5]. Among cancers, colorectal and lung cancers are of the most widespread cancers in both genders [1]. So colorectal cancer has a strategic importance for governments. Although other cancers like ovarian and endometrial cancers are also prevalent in women [6], [7], but the strategic importance of colorectal cancers is higher mortality and affecting both genders.
Colorectal disease like Crohn's disease and ulcerative colitis has very strategic importance from the epidemiological points of view [8] as well as colorectal cancers. Colorectal cancer is one of the most common types of cancers, particularly in developed countries and it is defined as growth and spread of cancer cells in colon or rectum [9]. This disease usually has no specific symptom and often diagnose in latest and dangerous stages of the disease. Most important symptoms are blood existence in stool, weight loss and permanent fatigue [10], [11]. The cause of getting this cancer is related to lifestyle, age and gender (men will getting this cancer more than women) because of obesity, alcohol consumption, high consumption of red meat, absence of fibers in diet, physical inactivity and especially tobacco abuse [12], [13], [14]. Increasing age also has a significant impact on the risk of this cancer so that a lot of people over 70 years of age in Western societies gett adenoma and adenoma increases the possibility of getting this cancer [15], [16]. This disease has several stages. In the first stage, cancer cells are in inner layer of colon or rectum, in the next stage the cancer spreads in muscular layer of colon or rectum, thereafter in a next stage, the cancer enter to a few lymph nodes in the same area and after that in another stage metastasis occurs and the cancer spreads into other tissues and body areas; at this stage the disease is usually not curable [17], [18], [19]. Most colorectal cancers originate from the benign adenomas that forms in colon. Molecular and genetic studies indicate that about 70% of colorectal cancers arise by mutation and inactivation of the gene adenomatous polyposis coli (APC) and other tumors arising as a result of activating mutations in the genes that producing beta-catenin and axin [16], [20], [21], [22]. Also other genetic studies have shown that this cancer usually arises due to the mutations that cause instability of some chromosomes and change the structure of these chromosomes. These chromosomal changes cause not to produce an enough number of copies of the tumor suppressors like APC and P53 [23], [24], [25], [26], [27], [28]. Other than tumor suppressor genes, this and some other cancers can be affected by proto-oncogenes like the ubiquitin-like with PHD and ring-finger domains 1 (UHRF1) [29]. Other researches have indicated that mutation in KRAS and epidermal growth factor (EGF) signaling pathways have an important role in getting this cancer [30], [31].
Immune system responses have important role in controlling and preventing development of colorectal cancers [26]. Colorectal cancer cells have several antigens which recognized by immune system. Among these antigens, Carcinoembryonic antigen (CEA) is the most studied [32], [33]. In addition, several neo-antigens were detected in these tumor cells. These neo-antigens are the cause of high levels of lymphocytes infiltrating to the tumor cells [23]; so the cancer progression can be diagnosed from the level of lymphocyte infiltration. Since generally the cancerous cells fail to express human leukocyte antigen (HLA) class I and therefore bypass cytotoxic T lymphocytes, natural killer-cells (NKs) as important components of the innate immune system, play a key role in this condition [34]. Killer cell immunoglobulin-like receptors (KIRs) (also called as CD158) are polymorphic glycoproteins expressed on cell surface of NKs and T cell subsets [35]. KIR gene family is highly polymorphic and its genomic diversity is achieved through differences in gene content as well as allelic polymorphism [36], [37]. Of course the most polymorphic loci in human genome is HLA [38] which the molecules of its, are in direct contact with KIR molecules. Hereby we intend to investigate the role of the KIR genes in colorectal cancer as a meta-analysis to find the association of different genes of KIR and susceptibility to be affected by colorectal cancer.
2. Material and methods
The present study is a meta-analysis which approximately covers all the original studies on this topic done before. We searched in databases such as google scholar, science direct and Pubmed. Totally six papers were found that four of them had same protocols.
The overall population of this four studies consists of 953 individuals (470 individuals with colorectal cancer and 483 individuals in control groups). The test chi-squared 2 multiplied by 2 with Yate's correction was used to assay each gene separately. Then the results were imported into the software comprehensive meta-analysis version 2. The 5 genes 2DL4, 3DL2, 3DL3, 2DP1 and 3DP1 were excluded from test because of their persistence in all participants of the both groups.
3. Results
3.1. About KIR
Depending on the number of extracellular immunoglobulin domains, KIRs are divided into two distinct groups (2D or 3D). Two types of KIR, i.e. inhibitory and activating, have been distinguished based on length of the intracellular domain. Inhibitory KIRs (iKIRs) are characterized by a long intra-cytoplasmic tail (denoted by an ‘L’ in their name) and presence of at least one immunoreceptor tyrosine-based inhibitory motif (ITIM). Activating KIR (aKIR) are characterized by a short intra-cytoplasmic tail (denoted by an ‘S’ in their name) and the absence of ITIM [37].
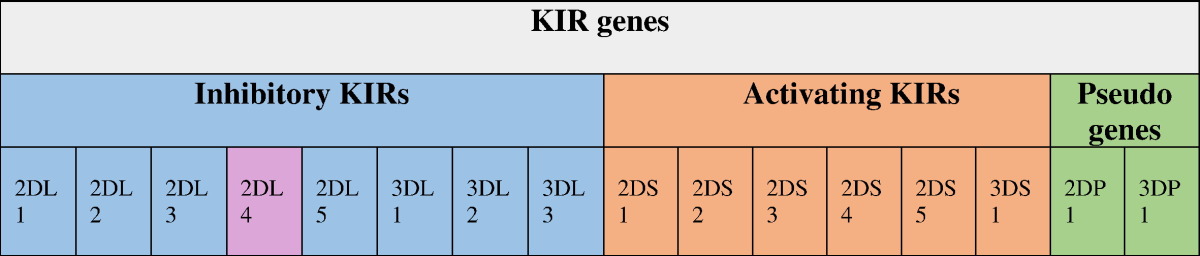
Up to now, fourteen distinct types of KIR have been identified in the human genome [39]. NKs are a subset of lymphocytes comprising around 10–15% of total lymphocytes in peripheral blood [40]. NKs principally contribute to innate immunity and also adaptive immune responses by killing the targeted cells of theirs and production of a variety of cytokines and chemokines [37]. Overall, upon interaction with their ligands which are usually HLA class I, KIR provide inhibitory or activating signals to regulate the activity of NKs, which contributes to pathogenesis of diverse kinds of diseases [41], [42]. Different compounds of KIR-HLA genotypes can induce different thresholds of activation in NK family and such genotypic variations have been found to be associated with a number of human diseases and complications including viral infections, autoimmune disorders and cancers [43] as well as reproduction abnormalities [44], [45]. The KIR gene cluster on chromosome 19q13.4 within the leukocyte receptor complex (LRC) consists of a centromeric and telomeric region [46]. So far, 14 KIR genes and 2 pseudogenes have been described [47] (Table 1). Seven genes of KIR3DL1-3, KIR2DL1-3 and KIR2DL5 encode for the inhibitory KIR (iKIR), six genes of KIR3DS1 and KIR2DS1-5 encode for activating KIRs (aKIR), one gene encodes for KIR2DL4 with both inhibitory and activating functions, but more of inhibitory, and two genes of KIR2DP1 and KIR3DP1 are pseudogenes that do not encode a functional KIR molecule [47].
Table 1.
KIR genes. KIR has 14 discovered genes and 2 discovered pseudo-genes. Seven number of them are inhibitory, 1 of them is both inhibitory and activating and 6 number them are activating. Each gene has different alleles; So KIR is highly polymorphic like HLA.
About NK subsets, we have mainly CD16+ CD56dim and CD16− CD56bright; the dim form has more cytotoxic capacity called as “cytotoxic NK” and the bright form contributes in secretion of inflammatory cytokines called as “immune-regulatory NK”. Both of them express KIR, but the dim form express more. [35], [37], [44], [46], [48], [49], [50], [51], [52].
The KIR gene cluster is flanked by KIR3DL3 at centromeric end and KIR3DL2 at telomeric end; both of which are present on virtually all haplotypes. Two groups of KIR haplotypes have been defined on the basis of gene content and are termed as haplotypes A and B. The A haplotypes are uniform in terms of gene content and the most prevalent form of them is composed of five inhibitory genes (KIR2DL1, 2DL3, 3DL1, 3DL2 and 3DL3), one activating gene (KIR2DS4), and the KIR2DL4 which may have both inhibitory and activating capacity. Interestingly, many A haplotypes possess null variants of both KIR2DS4 and KIR2DL4 that are not expressed on the cell surface. Thus these haplotypes technically possess no functional aKIR gene. The B haplotypes contain variable numbers of activating and inhibitory genes and are the primary contributors to the extraordinary differences in KIR gene profiles observed in distinct ethnic populations across the world. The interaction of inhibitory KIR with HLA class I as their ligands, triggers the signals that turn off the NKs. Therefore, by expressing HLA-A, -B and -C molecules, healthy cells are protected against NK lysis. Down-regulation of HLA class I expression due to tumor transformation or viral infection permits NKs to lyse these unhealthy targeted cells of theirs, a phenomenon first described as the “missing-self” hypothesis. Thus the compound KIR-HLA genotypes that lead to lower inhibition and higher activation appear to be beneficial in resistance to viral infections and cancers. On the other hand, these dominant activating genotypes may constitute a risk for susceptibility to autoimmune and inflammatory diseases [43].
3.2 KIR and colorectal cancer
As a general rule NKs play a key role against the cancerous cells escaped from toxic activity of T cells. So the KIRs expressed on surface of NKs become important. We know that the interaction KIR-HLA has two sides of KIR and HLA. For example overexpression of HLA-E on surface of colorectal cancer cells can result in inhibition of NKs [53] and such patients have better survival from the disease [54]; of course the receptor of HLA-E is CD94/NKG2a [55], [56]. KIR2DL1 and 2DS1 interact with HLA-C2, and KIR2DL2, 2DL3 and 2DS2 interact with HLA-C1 [57]; so every mathematical predict do not occur. Although at the first glance it seems that inhibitory types of each or both sides may be associated with the more susceptibility, but this rule is not true for such inflammation-based cancers. The whys and therefore of this paradox, is not clear; however there are some good data in study of Middleton et al. [58] about loss of HLA expression in colorectal cancer cells giving us insight. Also role of allograft inflammatory factor 1 (Alf1) shows the inflammation base of this disease [59].
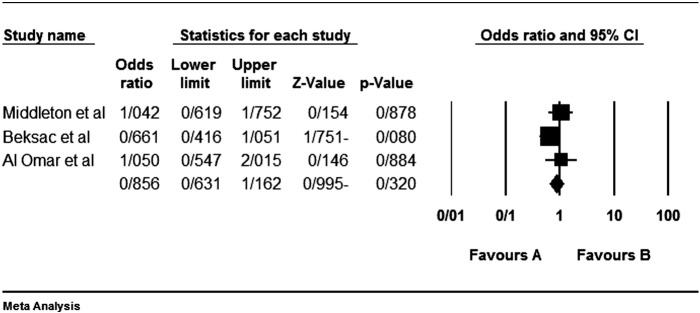
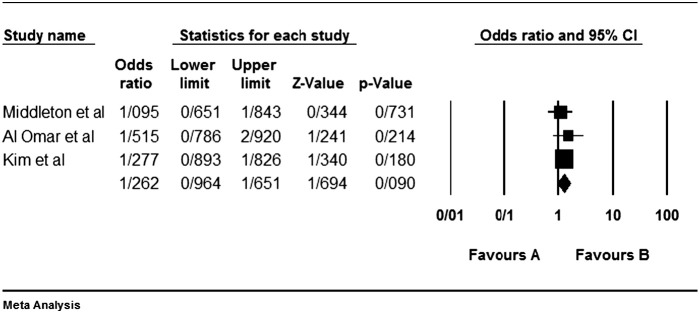
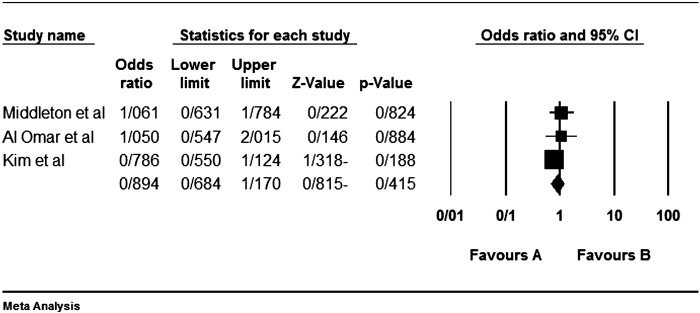
Although the most inhibitory effect is attributed to the interaction KIR2DL1-HLA-C2 [57], but none of our analyzed studies do not show any significant protective effect. Of course we reanalyzed previous articles' data with Yate's correction; so the relation of some genes in some studies were significant in the study of theirs without Yate's correction. For example Beksac et al. [60] had been found a significant protective effect for KIR2DL1 as we did not found so (Fig. 1).
Fig. 1.
Colorectal cancer is not statistically affected or protected by the KIR2DL1. Left side (the favours A) shows protecting effect in all figures.
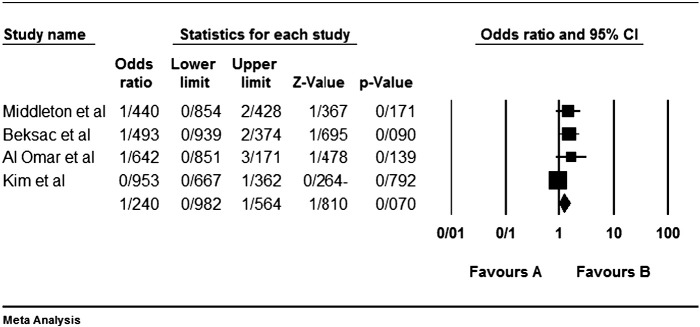
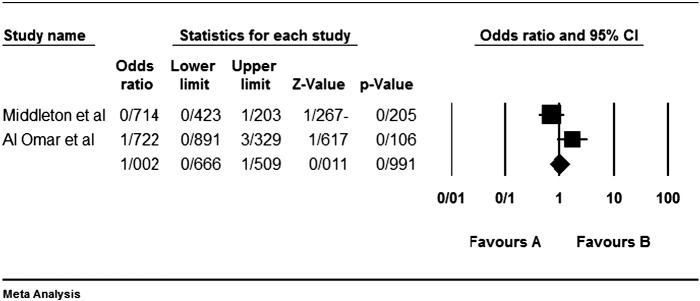
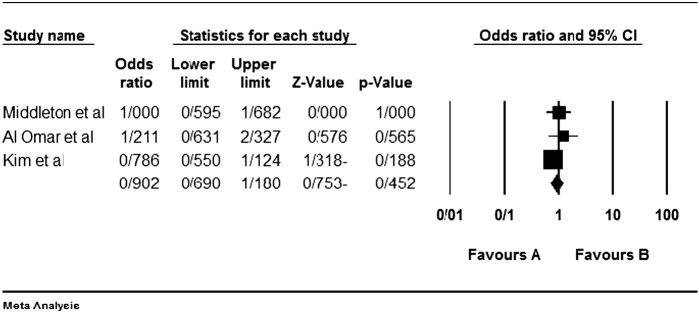
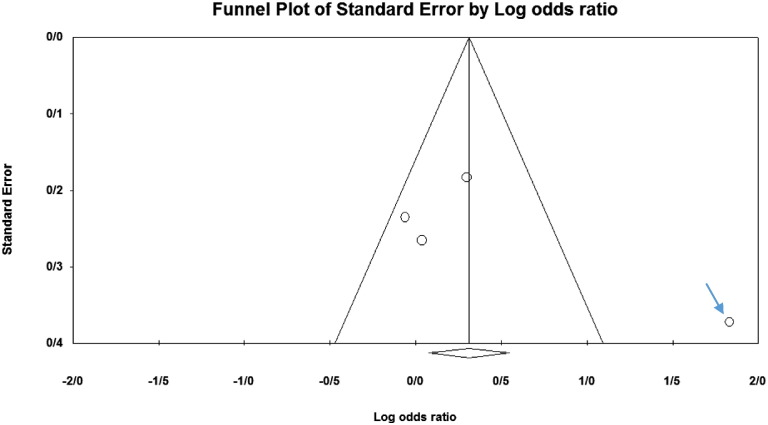
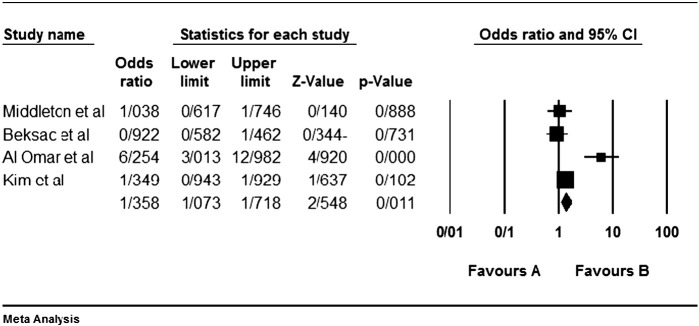
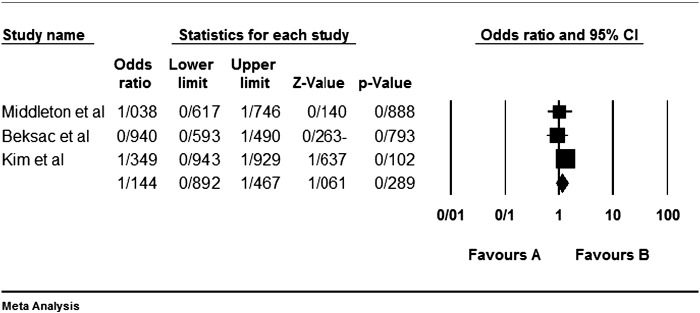
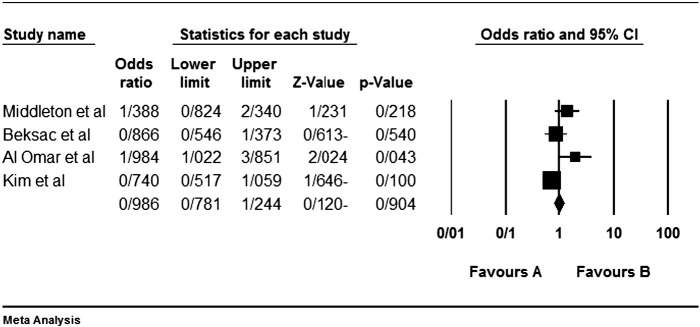
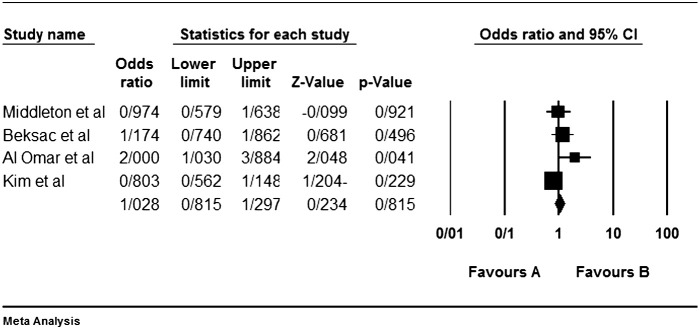
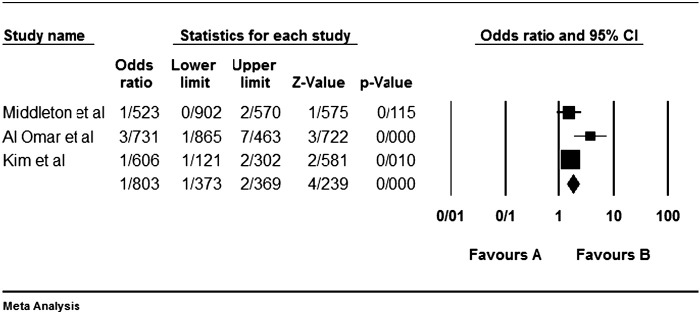
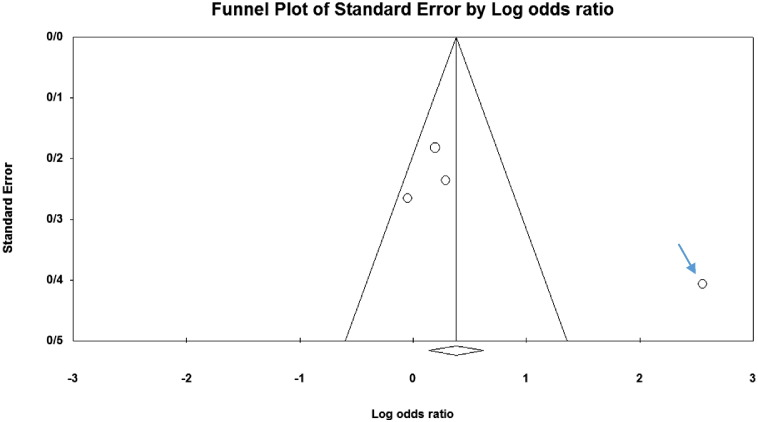
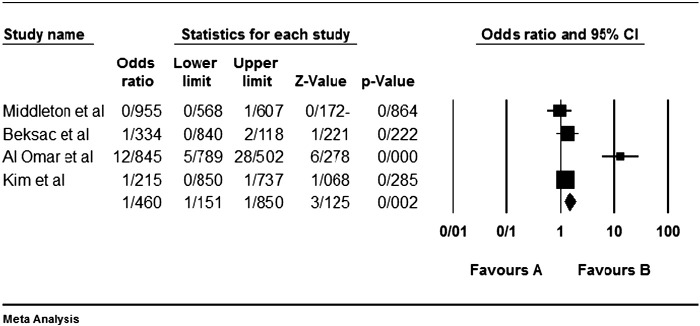
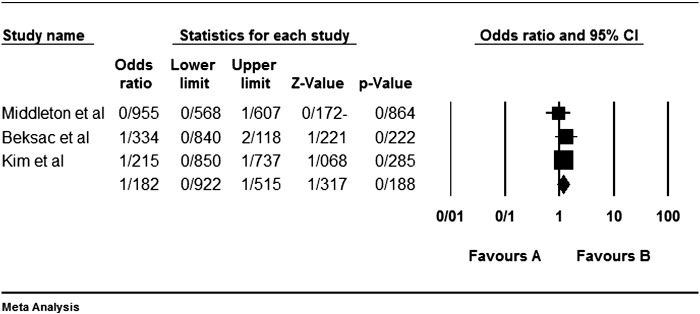
Among the four studies analyzed by us (Table 2), the three genes 2DL4, 3DL2 and 3DL3, and both pseudogenes were present in approximately all participants of the studies, so these genes were excluded from our meta-analyses. Among the other genes, at first, the meta-analyses showed a significant association for the genes 2DS1, 3DS1 and 2DS5 (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11, Fig. 12, Fig. 13, Fig. 14, Fig. 15); but because of high odds ratio of study of Al Omar et al. in Saudi Arabia [61] for the genes 2DS1 and 3DS1 (Fig. 6, Fig. 13), we were supposed to exclude it from the analysis of these two genes. After the exclusion, only association of KIR2DS5 remained significant (Fig. 7, Fig. 8, Fig. 12, Fig. 14, Fig. 15).
Table 2.
Data and meta-analysis. The ED stands for effective direction and the nagative and possitive each one respectively means protective effect and risk factor.
| Gene | Middleton et al. [58] |
Beksac et al. [60] |
Al Omar et al. [61] |
Kim et al. [57] |
Meta-analysis |
||||
|---|---|---|---|---|---|---|---|---|---|
| Cancer N = 90 | Control N = 100 | Cancer N = 87 | Control N = 154 | Cancer N = 52 | Control N = 70 | Cancer N = 241 | Control N = 159 | P value (ED) | |
| 2DL1 | 86 | 94 | 77 | 147 | 51 | 70 | 241 | 159 | 0.32 (−) |
| 2DL2 | 61 | 57 | 68 | 103 | 47 | 55 | 27 | 20 | 0.07 (+) |
| 2DL3 | 69 | 85 | 48 | 56 | 240 | 159 | 0.99 (+) | ||
| 2DL4 | 52 | 70 | 241 | 159 | Excluded | ||||
| 2DL5 | 50 | 52 | 45 | 53 | 107 | 59 | 0.09 (+) | ||
| 3DL1 | 83 | 91 | 51 | 66 | 229 | 156 | 0.45 (−) | ||
| 3DL2 | 52 | 70 | 241 | 159 | Excluded | ||||
| 3DL3 | 50 | 70 | 241 | 159 | Excluded | ||||
| 2DS1 | 36 | 39 | 40 | 75 | 43 | 25 | 106 | 56 | 0.28 |
| 2DS2 | 61 | 58 | 54 | 103 | 46 | 50 | 31 | 31 | 0.09 (−) |
| 2DS3 | 29 | 34 | 34 | 52 | 40 | 40 | 32 | 29 | 0.81 (+) |
| 2DS4 | 82 | 91 | 51 | 70 | 229 | 156 | 0.41 (−) | ||
| 2DS5 | 34 | 26 | 35 | 22 | 80 | 33 | 0.0001(+)**** | ||
| 3DS1 | 34 | 39 | 35 | 76 | 43 | 16 | 99 | 56 | 0.18 (+) |
| 2DP1 | 52 | 70 | 241 | 159 | Excluded | ||||
| 3DP1 | 52 | 70 | 241 | 159 | Excluded | ||||
Significance at P < 0.05 level.
Fig. 2.
Colorectal cancer is not statistically affected or protected by the KIR2DL2.
Fig. 3.
Colorectal cancer is not statistically affected or protected by the KIR2DL3.
Fig. 4.
Colorectal cancer is not statistically affected or protected by the KIR2DL5.
Fig. 5.
Colorectal cancer is not statistically affected or protected by the KIR3DL1.
Fig. 6.
KIR2DS1. The arrowed population in for the study of Al Omar et al.
Fig. 7.
KIR2DS1 meta-graph, before exclusion of Al Omar's et al. study.
Fig. 8.
KIR2DS1 meta-graph, after exclusion of Al Omar's et al. study.
Fig. 9.
Colorectal cancer is not statistically affected or protected by the KIR2DS2.
Fig. 10.
Colorectal cancer is not statistically affected or protected by the KIR2DS3.
Fig. 11.
Colorectal cancer is not statistically affected or protected by the KIR2DS4.
Fig. 12.
KIR2DS5 meta-graph. Even with exclusion of the study of Al Omar's et al., the P value would be as 0.003. For this gene, the mentioned study did not have an odds ratio bias.
Fig. 13.
KIR3DS1 and that study of Al Omar's et al.
Fig. 14.
KIR3DS1 meta-graph, before exclusion of Al Omar's et al study.
Fig. 15.
KIR3DS1 meta-graph, after exclusion of Al Omar's et al study.
Another justification for this paradox is different basis of different cancers. As we published before, breast cancer is affected by KIR2DL2 which is an inhibitory gene [62]. In verse, colorectal cancer is affected by an activating KIR as we found in the present meta-analysis. Although at the first glance it seems that inhibitory interactions of KIR-HLA may be associated with the more susceptibility to colorectal cancer because of this fact that NKs play a key role against the cancerous cells escaped from toxic activity of T cells because of their loss of HLA expression, but this rule is not true for such inflammation-based cancers.
Other than the justification above, another justification is that the ligands of activating KIRs are not necessarily HLAs; rather, they are still unknown [36], [53]. This, can be pointed out as the limitation of the previous studies.
4. Conclusion
Finally we concluded that colorectal cancer is affected by KIR2DS5 and also there were no protecting gene. This result shows the inflammatory basis of this cancer. In other words, in contrast to leukemia and blood cancers, colorectal cancers seem to be affected by hyper activity of natural killer-cells (NKs). Whys and therefore of this paradox, is suggested to be investigated further.
Conflict of interest
We declare that there is no conflict of interest.
References
- 1.Coates A. Cancer Forum is Produced by Cancer Council Australia for Health Professionals Working in Cancer Control. It is the Official Journal of the Clinical Oncological Society of Australia. 2013. Most common cancers; p. 54. [Google Scholar]
- 2.Atashkhoei S., Fakhari S. Management of breast cancer related lymphoedema. Crescent J. Med. Biol. Sci. 2016;3:111–112. [Google Scholar]
- 3.Farshbaf-Khalili A., Shahnazi M., Vahed L., Javadi L. Status of breast self-examination performance among women referring to health centers of Tabriz, Iran. Crescent J. Med. Biol. Sci. 2014;1:90–96. [Google Scholar]
- 4.Asghari E., Nahamin M., Khoshtarash M., Ghanbari A., Parizad N., Mahdavi N., Asgarlo Z. The relationship between health belief and breast self-examination among Iranian university students. Int. J. Wom. Health Reprod. Sci. 2016;4:110–113. [Google Scholar]
- 5.Shafaie F.S., Nagizadeh S., Valizadeh S. Breast cancer screening tests in Tabriz Behbood hospital. Int. J. Wom. Health Reprod. Sci. 2016;4:134–140. [Google Scholar]
- 6.Jafari-Shobeiri M., Jangi M., Tabrizi A.D., Sayyah-Melli M., Mostafa-Gharabaghi P., Ouladsahebmadarek E., Neginfar E., Pouraliakbar Y. Diagnostic value of novel biomarker human epididymis protein 4 (HE4) in detecting endometrial cancer. Int. J. Wom. Health Reprod. Sci. 2016;4:29–33. [Google Scholar]
- 7.Sadeghi L., Dastranj A., Gharabaghi P.M., Sheikhzadeh F., Zamanvand S., Jafari-Shobeiri M. The impact of parity on the number of ovarian cortical inclusion cysts. Int. J. Womens Health Reprod. Sci. 2016;4:73–76. [Google Scholar]
- 8.Ghanadi K., Valizadeh J., Hasanvand A. Vol. 5. Springer Plus; 2016. Epidemiological and Clinical Aspects of Ulcerative Colitis in West of Iran: A Cross Sectional Study; p. 1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grothey A., Van Cutsem E., Sobrero A., Siena S., Falcone A., Ychou M., Humblet Y., Bouché O., Mineur L., Barone C. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:303–312. doi: 10.1016/S0140-6736(12)61900-X. [DOI] [PubMed] [Google Scholar]
- 10.Ghanadi K., Anbari K., Obeidavi Z., Pournia Y. Characteristics of colorectal cancer in Khorramabad, Iran during 2013. Middle East J. Dig. Dis. 2014;6:81. [PMC free article] [PubMed] [Google Scholar]
- 11.Baena R., Salinas P. Diet and colorectal cancer. Maturitas. 2015;80:258–264. doi: 10.1016/j.maturitas.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Coppedè F., Lopomo A., Spisni R., Migliore L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J. Gastroenterol. 2014;20:943–956. doi: 10.3748/wjg.v20.i4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegel R., DeSantis C., Jemal A. Colorectal cancer statistics, 2014. CA Cancer J. Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 14.Tabung F.K., Steck S.E., Ma Y., Liese A.D., Zhang J., Caan B., Hou L., Johnson K.C., Mossavar-Rahmani Y., Shivappa N. The association between dietary inflammatory index and risk of colorectal cancer among postmenopausal women: results from the Women's Health Initiative. Cancer Causes Control. 2015;26:399–408. doi: 10.1007/s10552-014-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carr P.R., Walter V., Brenner H., Hoffmeister M. Meat subtypes and their association with colorectal cancer: systematic review and meta-analysis. Int. J. Cancer. 2016;138:293–302. doi: 10.1002/ijc.29423. [DOI] [PubMed] [Google Scholar]
- 16.Fearon E.R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol.: Mech. Dis. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 17.Franken M.D., van Rooijen E.M., Uyl-de Groot C.A., van Oijen M.G., Koopman M. Cost-effectiveness in colorectal cancer: challenges on quality and comparability. Colorectal Cancer. 2016;5:21–31. [Google Scholar]
- 18.Lugli A., Langer R. Vol. 39. 2015. Toward a Molecular Classification of Colorectal Cancer: the Role of BRAF, Towards a Molecular Classification of Colorectal Cancer. [Google Scholar]
- 19.Tomasetti C., Vogelstein B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rennoll S.A., Eshelman M.A., Raup-Konsavage W.M., Kawasawa Y.I., Yochum G.S. The MYC 3′ Wnt-responsive element drives oncogenic MYC expression in human colorectal cancer cells. Cancer. 2016;8:52. doi: 10.3390/cancers8050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maughan T.S., Adams R.A., Smith C.G., Meade A.M., Seymour M.T., Wilson R.H., Idziaszczyk S., Harris R., Fisher D., Kenny S.L. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–2114. doi: 10.1016/S0140-6736(11)60613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veronese N., Nottegar A., Pea A., Solmi M., Stubbs B., Capelli P., Sergi G., Manzato E., Fassan M., Wood L.D. Prognostic impact and implications of extracapsular lymph node involvement in colorectal cancer: a systematic review with meta-analysis. Ann. Oncol. 2016;27:42–48. doi: 10.1093/annonc/mdv494. [DOI] [PubMed] [Google Scholar]
- 23.Bordonaro M., Shirasawa S., Lazarova D.L. In hyperthermia increased ERK and WNT signaling suppress colorectal cancer cell growth. Cancer. 2016;8:49. doi: 10.3390/cancers8050049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lui C., Mok M.T., Henderson B.R. Characterization of adenomatous polyposis coli protein dynamics and localization at the centrosome. Cancer. 2016;8:47. doi: 10.3390/cancers8050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Sousa e Melo F., Vermeulen L. Wnt Signaling in Cancer Stem Cell Biology. Cancer. 2016;8:60. doi: 10.3390/cancers8070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravindranath A.J., Cadigan K.M. The role of the C-clamp in Wnt-related colorectal cancers. Cancer. 2016;8:74. doi: 10.3390/cancers8080074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vatandoost N., Ghanbari J., Mojaver M., Avan A., Ghayour-Mobarhan M., Nedaeinia R., Salehi R. Early detection of colorectal cancer: from conventional methods to novel biomarkers. J. Cancer Res. Clin. Oncol. 2016;142:341–351. doi: 10.1007/s00432-015-1928-z. [DOI] [PubMed] [Google Scholar]
- 28.Zauber A.G., Winawer S.J., O'Brien M.J., Lansdorp-Vogelaar I., van Ballegooijen M., Hankey B.F., Shi W., Bond J.H., Schapiro M., Panish J.F. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N. Engl. J. Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soleimani A., Ghanadi K., Noormohammadi Z., Irani S. The correlation between miR-146a C/G polymorphism and UHRF1 gene expression level in gastric tumor. J. Dig. Dis. 2016;17:169–174. doi: 10.1111/1751-2980.12329. [DOI] [PubMed] [Google Scholar]
- 30.Calon A., Lonardo E., Berenguer-Llergo A., Espinet E., Hernando-Momblona X., Iglesias M., Sevillano M., Palomo-Ponce S., Tauriello D.V., Byrom D. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015;47:320–329. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 31.Giardiello F.M., Allen J.I., Axilbund J.E., Boland C.R., Burke C.A., Burt R.W., Church J.M., Dominitz J.A., Johnson D.A., Kaltenbach T. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US multi-society task force on colorectal cancer. Gastroenterology. 2014;147:502–526. doi: 10.1053/j.gastro.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Verberne C.J., Wiggers T., Grossmann I., Bock G., Vermeulen K. Cost-effectiveness of a carcinoembryonic antigen (CEA) based follow-up programme for colorectal cancer (the CEA watch trial) Color. Dis. 2016;18:O91–O96. doi: 10.1111/codi.13273. [DOI] [PubMed] [Google Scholar]
- 33.Palos G., Gilmore K.R., Chapman P., Lewis-Patterson P., Bi W., Rodriguez M.A. Low concordance with CEA tumor marker monitoring in colorectal cancer survivors. Cancer Res. 2016;76:3464. [Google Scholar]
- 34.Menon A.G., Janssen-van Rhijn C.M., Morreau H., Putter H., Tollenaar R.A., van de Velde C.J., Fleuren G.J., Kuppen P.J. Immune system and prognosis in colorectal cancer: a detailed immunohistochemical analysis. Lab. Investig. 2004;84:493–501. doi: 10.1038/labinvest.3700055. [DOI] [PubMed] [Google Scholar]
- 35.Mousavi T., Shahsavar F., Farnia P., Tajik N., Soofi M. Study of KIR expression and HLA ligands in CD56 + lymphocytes of drug resistant tuberculosis patients. Iran. J. Allergy Asthma Immunol. 2011;10:189–194. [PubMed] [Google Scholar]
- 36.Shahsavar F., Mapar S., Ahmadi S.A.Y. Multiple sclerosis is accompanied by lack of KIR2DS1 gene: a meta-analysis. Genomics Data. 2016;10:75–78. doi: 10.1016/j.gdata.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajik N., Shahsavar F., Mousavi T., Radjabzadeh M.F. Distribution of KIR genes in the Iranian population. Tissue Antigens. 2009;74:22–31. doi: 10.1111/j.1399-0039.2009.01263.x. [DOI] [PubMed] [Google Scholar]
- 38.Varzi A.M., Shahsavar F., Tarrahi M.J. Distribution of HLA-DRB1 and HLA-DQB1 alleles in Lak population of Iran. Hum Immunol. 2016;77:580–583. doi: 10.1016/j.humimm.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 39.Mousavi T., Poormoghim H., Moradi M., Tajik N., Shahsavar F., Asadifar B. Inhibitory killer cell immunoglobulin-like receptor KIR3DL1 in combination with HLA-B Bw4iso protect against Ankylosing spondylitis. Iran. J. Immunol. 2010;7:88–95. [PubMed] [Google Scholar]
- 40.Shahsavar F., Tajik N., Entezami K.Z., Radjabzadeh M.F., Asadifar B., Alimoghaddam K., Dahaghi M.O., Jalali A., Ghashghaie A., Ghavamzadeh A. KIR2DS3 is associated with protection against acute myeloid leukemia. Iran. J. Immunol. 2010;7:8–17. [PubMed] [Google Scholar]
- 41.Augusto D.G., Hollenbach J.A., Petzl-Erler M.L. A deep look at KIR–HLA in Amerindians: comprehensive meta-analysis reveals limited diversity of KIR haplotypes. Hum. Immunol. 2015;76:272–280. doi: 10.1016/j.humimm.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 42.Tajik N., Shahsavar F., Poormoghim H., Radjabzadeh M.F., Mousavi T., Jalali A. KIR3DL1 + HLA-B Bw4 Ile80 and KIR2DS1 + HLA-C2 combinations are both associated with ankylosing spondylitis in the Iranian population. Int. J. Immunogenet. 2011;38:403–409. doi: 10.1111/j.1744-313X.2011.01024.x. [DOI] [PubMed] [Google Scholar]
- 43.Tajik N., Shahsavar F., Nasiri M., Radjabzadeh M.F. Compound KIR-HLA genotype analyses in the Iranian population by a ovel PCR-SSP assay. Int. J. Immunogenet. 2010;37:159–168. doi: 10.1111/j.1744-313X.2010.00906.x. [DOI] [PubMed] [Google Scholar]
- 44.Ahmadi S.A.Y., Shahsavar F., Akbari S. A review on controversies about the role of immune and inflammatory systems in implantation process and durability of pregnancy. Int. J. Wom. Health Reprod. Sci. 2016;4:96–102. [Google Scholar]
- 45.Beigi Boroujeni N., Beigi Boroujeni M., Rafiei Alavi E., Shafiei A. The effect of ethanolic extract of Salvia officinalis on the uterine natural killer cells population at day 7 of pregnancy. J. Med. Plant. 2015;14:15–24. [Google Scholar]
- 46.Shahsavar F., Mousavi T., Azargoon A., Entezami K. Association of KIR3DS1 + HLA-B BW4lle8 combination with susceptibility to tuberculosis in lur population of Iran. Iran. J. Immunol. 2012;9:39–47. [PubMed] [Google Scholar]
- 47.Shahsavar F., Asadifar B., Jafarzadeh M., Forutani S. Distribution of KIR genes in the Lur population of Iran. Life Sci. J. 2013;10:11–16. [Google Scholar]
- 48.Rajaei S., Zarnani A.H., Jeddi-Tehrani M., Tavakoli M., Mohammadzadeh A., Dabbagh A., Mirahmadian M. Cytokine profile in the endometrium of normal fertile and women with repeated implantation failure. Iran. J. Immunol. 2011;8:201. [PubMed] [Google Scholar]
- 49.Chen C.-P., Piao L., Chen X., Yu J., Masch R., Schatz F., Lockwood C.J., Huang S.J. Expression of interferon γ by decidual cells and natural killer cells at the human implantation site implications for preeclampsia, spontaneous abortion, and intrauterine growth restriction. Reprod. Sci. 2015;22:1461–1467. doi: 10.1177/1933719115585148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shahsavar F., Mousavi T., Entezami K. Association of KIR-HLA interactions with diseases. Yafteh. 2011;13:82–96. [Google Scholar]
- 51.Shahsavar F., Azargoon A., Jafarzadeh M., Forutani S., Asadifar B. Distribution of KIR genes in the Lur population of Iran. Life Sci. J. 2013;10:11–16. [Google Scholar]
- 52.Ghafourian M., Band N.A., Pour A.F., Kooti W., Rad M.F., Badiee M. The role of CD16 +, CD56 +, NK (CD16 +/CD56 +) and B CD20 + cells in the outcome of pregnancy in women with recurrent spontaneous abortion. Int. J. Wom. Health Reprod. Sci. 2015;3:61–66. [Google Scholar]
- 53.De Re V., Caggiari L., De Zorzi M., Talamini R., Racanelli V., D'Andrea M., Buonadonna A., Zagonel V., Cecchin E., Innocenti F. Genetic diversity of the KIR/HLA system and outcome of patients with metastatic colorectal cancer treated with chemotherapy. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lauterbach N., Wieten L., Popeijus H.E., Voorter C.E.M., Tilanus M.G.J. HLA-E regulates NKG2C + natural killer cell function through presentation of a restricted peptide repertoire. Hum. Immunol. 2015;76:578–586. doi: 10.1016/j.humimm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Castelli E.C., Mendes-Junior C.T., Sabbagh A., Porto I.O.P., Garcia A., Ramalho J., Lima T.H.A., Massaro J.D., Dias F.C., Collares C.V.A., Jamonneau V., Bucheton B., Camara M., Donadi E.A. HLA-E coding and 3′ untranslated region variability determined by next-generation sequencing in two West-African population samples. Hum. Immunol. 2015;76:945–953. doi: 10.1016/j.humimm.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 56.Gonçalves A.S., Oliveira J.P., Oliveira C.F.P., Silva T.A., Mendonça E.F., Wastowski I.J., Batista A.C. Relevance of HLA-G, HLA-E and IL-10 expression in lip carcinogenesis. Hum. Immunol. 2016;77:785–790. doi: 10.1016/j.humimm.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Kim H.-J., Choi H.-B., Jang J.-P., Baek I.-C., Choi E.-J., Park M., Kim T.-G., Oh S.-T. HLA-Cw polypmorphism and killer cell immunoglobulin-like receptor (KIR) gene analysis in Korean colorectal cancer patients. Int. J. Surg. 2014;12:815–820. doi: 10.1016/j.ijsu.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 58.Middleton D., Vilchez J., Cabrera T., Meenagh A., Williams F., Halfpenny I., Maleno I., Ruiz-Cabello F., Lopez-Nevot M., Garrido F. Analysis of KIR gene frequencies in HLA class I characterised bladder, colorectal and laryngeal tumours. Tissue Antigens. 2007;69:220–226. doi: 10.1111/j.1399-0039.2006.00792.x. [DOI] [PubMed] [Google Scholar]
- 59.Sanchez C., Baier C., Colle J.G., Chelbi R., Rihet P., Le Treut T., Imbert J., Sébahoun G., Venton G., Costello R.T. Natural killer cells in patients with polycythemia vera. Hum. Immunol. 2015;76:644–650. doi: 10.1016/j.humimm.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 60.Beksac K., Beksac M., Dalva K., Karaagaoglu E., Tirnaksiz M.B. Impact of “Killer immunoglobulin-like receptor/ligand” genotypes on outcome following surgery among patients with colorectal cancer: activating KIRs are associated with long-term disease free survival. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al Omar S.Y., Mansour L., Dar J.A., Alwasel S., Alkhuriji A., Arafah M., Al Obeed O., Christmas S. The relationship between killer cell immunoglobulin-like receptors and HLA-C polymorphisms in colorectal cancer in a Saudi population. Genet. Test. Mol. Biomarkers. 2015;19:617–622. doi: 10.1089/gtmb.2015.0105. [DOI] [PubMed] [Google Scholar]
- 62.Shayanrad B., Ahmadi S.A.Y., Shahsavar F. Breast cancer is protected by the KIR gene 2DL1 and affected by 2DL2: a systematic review. Der Pharmacia Lettre. 2016;8:22–25. [Google Scholar]