Abstract
Resistance to benzimidazoles (BZs) in trichostrongyloid nematodes is a worldwide problem for livestock production, particularly regarding small ruminants. Sensitive and reliable methods are required to assess anthelmintic resistance status. Currently available methods for BZ resistance detection can be divided into three main groups, in vivo (e.g. faecal egg count reduction test), in vitro (e.g. egg hatch assay) and molecular tests. Three single nucleotide polymorphisms (SNPs) in the isotype-1 β-tubulin gene of various nematode species correlate with BZ resistance. While PCR-based methods have been reported for the three most economically important nematodes of sheep, namely, Trichostrongylus, Haemonchus and Teladorsagia, pyrosequencing assays are so far only available for the latter two. Here, the design and evaluation of pyrosequencing assays for isotype-1 and isotype-2 β-tubulin genes of Trichostrongylus colubriformis are described. PCR fragments carrying the susceptible and corresponding resistant genotype were combined in defined ratios to evaluate assay sensitivity and linearity. The correlation between the given and the measured allele frequencies of the respective SNPs (codons F167Y, E198A and F200Y) was very high. Pyrosequencing assays for Haemonchus, Teladorsagia and Trichostrongylus were subsequently used for a BZ resistance survey, carried out in the three European countries, namely Ireland, Italy and Switzerland. Larval cultures obtained from field survey samples in 2012 and 2013 were used for pyrosequencing. The test was applied when the target species represented at least 10% of the sample. Trichostrongylus and Teladorsagia were detected in all countries' samples whereas Haemonchus was not detected in samples from Ireland. SNPs in isotype-1 associated with resistance were detected for all three species, with frequencies at codon F200Y far exceeding those at codons F167Y and E198A. Elevated SNP frequencies in isotype-2 of Trichostrongylus were only rarely detected. Farms with BZ resistance-associated SNP frequencies above 10% were most often found in Switzerland followed by Ireland and Italy.
Keywords: Trichostrongylus colubriformis, Nematode, Diagnosis, Benzimidazole, Resistance, Single nucleotide polymorphism, Sheep, Europe
Graphical abstract
Highlights
-
•
Successful development of a pyrosequencing assay for Trichostrongylus.
-
•
Resistance survey revealed high number of SNPs present in all three countries.
-
•
SNP at codon 200 most prevalent in European trichostrongyloid populations.
-
•
Low number of SNPs found in Italy corresponds to FECRT data.
-
•
Pyrosequencing successfully employed as a tool for large scale surveys.
1. Introduction
Of the main anthelmintic drug classes, the benzimidazoles (BZs) have been most widely used since their release/launch onto the market in the 1960s for helminth control in livestock. BZs represent a class of broad-spectrum anthelmintic substances including several chemical compounds which all share the same mode of action. They are known to inhibit the polymerisation of α- and β-tubulin dimers to microtubules (Sangster et al., 1985), leading to interference with the formation of the cytoskeleton, the mitotic spindle and intracellular transport. This results in the disruption of the worm's metabolism, and particularly, the inhibition of glucose uptake and intracellular transport leads to death of the worm (Lacey, 1988). Shortly after the introduction of BZs, resistance was detected (Conway, 1964), particularly in parasitic nematodes of small ruminants. The spread of BZ resistance has led to major problems in small ruminant farming industries worldwide (Kaplan, 2004, Howell et al., 2008, Papadopoulos et al., 2012, Torres-Acosta et al., 2012, Falzon et al., 2013). Of the currently available anthelmintics, the mechanism of BZ resistance has been intensively studied in the past decades. First, loss of function mutants (deletions) of the ben-1 gene were identified in an in vitro screening assay using Caenorhabdithis elegans. All mutants successfully developing in the presence of the BZ benomyl were lacking functional ben-1 which encodes a β-tubulin in C. elegans (Driscoll et al., 1989). The first single nucleotide polymorphism (SNP) identified in a parasitic nematode correlating with BZ resistance was found in codon F200Y (TTC to TAC) of isotype-1 β-tubulin of Haemonchus contortus, leading to the expression of tyrosine instead of phenylalanine (Kwa et al., 1994, Kwa et al., 1995). Furthermore, two additional codons, codon F167Y (TTC to TAC) (Prichard, 2001) and codon E198A (GAA to GCA) (Ghisi et al., 2007) have also been reported to be associated with BZ resistance. Until now, these three SNPs in the isotype-1 β-tubulin gene have been described for different parasitic nematodes of ruminants, such as H. contortus, Teladorsagia circumcincta, Ostertagia ostertagi and Cooperia oncophora (von Samson-Himmelstjerna et al., 2007) and Haemonchus placei (Chaudhry et al., 2014). Primarily, codon 200 was thought to play the most important role in BZ resistance, while some studies also reported elevated allele frequencies in codon F167Y (TTC to TAC) and E198A (GAA to GCA) (Silvestre and Cabaret, 2002, von Samson-Himmelstjerna et al., 2007, Chaudhry et al., 2015, Redman et al., 2015). Analyses of field populations are still relatively rare, but data obtained from surveys conducted in Eastern Canada and the USA initially suggested that, particularly in H. contortus but also in H. placei codon F200Y (TAC) is widespread and often highly frequent, however changes in allele frequencies at codon 167 are relatively low (Barrere et al., 2013a, Barrere et al., 2013b, Chaudhry et al., 2014), and codon E198A (GCA) appears to be rarely involved. Moreover, investigations conducted in Brazil revealed a high level in TAC frequencies at codons F167Y and F200Y in H. contortus from field samples (dos Santos et al., 2014). Data from a recent survey in India and Pakistan confirmed the TAC SNP in codon F200Y as the most prevalent one, but in contrast to the results from previous studies in America and Europe, no mutations were found at codon F167Y (TAC) and only a small number of populations displayed the SNP in codon E198A (GCA) in India (Chaudhry et al., 2015). Microsatellite marker analysis of H. contortus populations from Pakistan suggests that frequent BZ drug treatment does not result in a reduction of overall genetic diversity (Chaudhry et al., 2016). There have been very few molecular genotyping studies performed in cattle parasitic nematodes. SNPs at all three codons have been associated with benzimidazole resistance in trichostrongylid parasite species of small ruminants and have been recently reported in three cattle parasites, H. placei, O. ostertagi and C. oncophora (Njue and Prichard, 2003, Winterrowd et al., 2003, Brasil et al., 2012, Demeler et al., 2013, Chaudhry et al., 2014). This diversity complicates the molecular detection and accordingly requires the examination of all three codons when molecular tests are applied to field samples.
To enable effective anthelmintic management programs, regular surveys of drug efficacies on farms are urgently required (Sutherland and Leathwick, 2011, Kaplan and Vidyashankar, 2012). The currently available BZ resistance detection methods can be grouped into three categories: i) in vivo methods, mainly represented by the faecal egg count reduction test (FECRT) or the controlled efficacy test; ii) in vitro methods, particularly the egg hatch assay (EHA) and the larval development assays (LDA) and iii) molecular tools. The FECRT is labour- as well as cost-intensive and can only provide reliable results once the resistant portion of the population has exceeded at least 25% (Martin et al., 1989). However, advanced statistical analysis methods (Torgerson et al., 2014), in combination with the use of more sensitive coproscopical methods such as (Mini-)FLOTAC (Barda et al., 2013), might improve the power of the FECRT. The EHA is relatively simple and inexpensive but requires fresh faecal samples, as the inhibition of hatching is the main determinant. The LDA is very sensitive but more labour-intensive than the EHA and also takes at least a week to provide results. Molecular tests include conventional PCR (Silvestre and Humbert, 2000, Njue and Prichard, 2003, Winterrowd et al., 2003), real-time PCR (Alvarez-Sanchez et al., 2005, Walsh et al., 2007) as well as pyrosequencing (Höglund et al., 2009, von Samson-Himmelstjerna et al., 2009, Demeler et al., 2013) and are all based on the detection of SNPs in the three above-mentioned codons in the isotype-1 β-tubulin gene.
Initial results suggested that selection occurs, at least initially, at the isotype-1 locus (Geary et al., 1992, Kwa et al., 1993), but isolates with the highest levels of BZ resistance have been shown to carry a deletion of isotype-2 (Kwa et al., 1993). However, for these analyses, Southern blots were used to compare the H. contortus β-tubulin genes and direct sequence data of isotype-2 genes of BZ resistant isolates are largely absent. Direct binding studies revealed that recombinant α-tubulin had the highest affinity for mebendazole, followed by β-tubulin isotype-2 and isotype-1 (Oxberry et al., 2001). After the identification of SNPs in isotype-1 correlating with BZ resistance, isotype-2 was largely ignored since no correlation was initially found (von Samson-Himmelstjerna et al., 2009).
The advantage of pyrosequencing is the possibility of quantification of allele frequencies in DNA extracted from field samples, in this case representing multiple pooled individuals. In order to conduct a BZ resistance survey including the three most important trichostrongyloid nematodes of small ruminants, pyrosequencing assays for the quantitative analysis of BZ resistance associated SNPs at the codons F167Y, E198A and F200Y of the isotype-1 and -2 β-tubulin genes of T. colubriformis were developed in the present study. Standardisation of the assays was performed by examining defined mixtures of susceptible and resistant alleles. The BZ resistance survey was conducted using previously developed pyrosequencing assays available for H. contortus, T. circumcincta, as well as the newly developed assay for T. colubriformis for analysis of field samples collected from sheep farms in pilot regions from three European countries (Ireland, Switzerland and Italy), collected as part of the EU-funded GLOWORM project, as recently described in Rinaldi et al. (2015a).
2. Materials and methods
2.1. Collection of field samples
Field samples from sheep were obtained from two cross-sectional copromicroscopical surveys conducted in three pilot areas in Ireland (north-west), Switzerland (north-east) and Italy (south-west), in the years 2012 and 2013 (for details see Rinaldi et al. (2015a)). The sheep farms in the three countries were selected using standardised spatial sampling procedures (Rinaldi et al., 2015a). For each farm, samples of 20 animals (or all animals if fewer than 20) were pooled and faecal cultures prepared and incubated in the dark at 27 °C for 5–7 days, during which time they were checked periodically and moistened if necessary (van Wyk et al., 2004). Third stage larvae (L3) were recovered and identified using the morphological keys proposed by van Wyk et al. (2004). When a coproculture had 100 or fewer L3, all were identified; when more than 100 larvae were present, only the first 100 were identified. It is worth noting that Teladorsagia and Trichostrongylus larvae were difficult to differentiate based on sheath extension length alone. To further refine their differentiation, additional morphological features were required based on the presence of an inflexion at the cranial extremity of Teladorsagia larvae (Roeber et al., 2013).
2.2. Extraction of DNA
DNA was extracted only from those samples in which one of the target species was present at a proportion of at least 10%, according to morphological identification. For extraction of DNA, the NucleoSpin Tissue 8 kit (Macherey-Nagel, Düren, Germany) was used, following the manufacturer's instructions.
2.3. Pyrosequencing assays for determination of β-tubulin allele frequencies in H. contortus and T. circumcincta
Pyrosequencing assays targeting codons F167Y, E198A and F200Y of the isotype-1 β-tubulin gene in H. contortus and T. circumcincta, respectively, were already available (von Samson-Himmelstjerna et al., 2009, Skuce et al., 2010) and were employed in the present study. The respective primer sets are detailed in Table S1.
For the analysis of all three SNPs in T. circumcincta as well as the SNP in codon 167 in H. contortus, PCR reactions were conducted using Novataq Hot Start PCR Mastermix (Merck). Each 50 μl reaction contained 25 μl mastermix buffer, final MgCl2 concentration of 1.5 mM, 0.2 mM biotinylated primer, 0.4 mM of the non-biotinylated primer and 4 μl of template DNA, in this case, genomic DNA extracted from pools of L3 from the study farms in the respective countries. Amplification of template DNA was verified by gel electrophoresis, 5 μl PCR product run on a 2% agarose gel. All pyrosequencing reactions were carried out using 45 μl PCR product and followed the protocols as described previously (von Samson-Himmelstjerna et al., 2009, Skuce et al., 2010).
The primers used for H. contortus pyrosequencing assays for codons 198 and 200 were previously described by von Samson-Himmelstjerna et al. (2009). PCRs and a modified protocol of the pyrosequencing reaction were conducted as recently reported by Ademola et al. (2015). Only the sequencing primer for codon 198 was used to quantify SNP frequencies in both codons E198A and F200Y. Nucleotides were dispensed as provided in Table S2. Pyrosequencing reactions always contained 45 μl of the initial PCR reaction.
2.4. Development of pyrosequencing assays for T. colubriformis
Pyrosequencing assays for codons F167Y, E198A and F200Y of the isotype-1 and -2 β-tubulin genes of T. colubriformis were not available and so were developed specifically for the present study. For isotype-1, three independent assays (based on the same PCR) were developed, while for isotype-2, one assay for codon 167, and a combined assay for codons 198 and 200 were designed based on an alignment of the published T. colubrifomis sequences for isotype-1 (HQ116825) and isotype-2 (L23506) β-tubulin genes. Initially, a BZ susceptible laboratory isolate was used to obtain the respective fragments of the susceptible alleles in the β-tubulin isotype-2 gene. Controls encoding the potentially resistance-associated alleles (isotype-2) or encoding both alleles in isotype-1 were obtained by custom DNA synthesis (ShineGene Molecular Biotech, Shanghai, China).
Plasmid DNAs containing either the susceptibility- or resistance-associated allele in the target codons were mixed in 17 different ratios (100:0, 95:5, 90:10, 85:15, 80:20, 75:25, 70:30, 60:40, 50:50, 40:60, 30:70, 25:75, 20:80, 15:85, 10:90, 5:95, 0:100). PCRs were carried out using Phusion® Hot-Start II High Fidelity DNA polymerase (Thermo Scientific, St. Leon-Rot, Germany) as follows: 50 μl 1 × Phusion HF buffer, 200 μM dNTPs (Thermo Scientific), 250 nM of each primer, 1 M betaine (only for isotype-2), 1 U Phusion polymerase and 5–50 ng genomic DNA or 1 ng plasmid mixture. Primer sequences and corresponding annealing temperatures are listed in the supplemental data (Table S1). Cycling conditions were: initial denaturation at 98 °C for 30 s, 40 cycles with 98 °C for 10 s, a primer pair-specific annealing temperature for 30 s and elongation at 72 °C for 30 s followed by a final elongation at 72 °C for 10 min. Pyrosequencing assays were performed on a PyroMark Q24 instrument as previously described in Demeler et al. (2013) using 22–45 μl of the PCR product. Nucleotides were dispensed as provided in Table S2.
2.5. Statistical analyses
Samples were considered to show an elevated frequency of an allele conferring a susceptible phenotype if this allele represented 10% of the pyrosequencing signal. This threshold was chosen since there is a technical background of 1–5% (rarely up to 9%) in samples that contain only plasmid DNA representing a single allele. In order to obtain a conservative estimate of the prevalence of BZ resistance, this threshold ensured that no false positive samples with low SNP frequency were considered positive. To evaluate the performance of the newly developed T. colubriformis assays, measured allele frequencies were plotted against given frequencies of plasmid mixtures used as templates in GraphPad Prism 5.02. Linear regressions with 95% confidence bands were calculated. For comparison of SNP frequencies at the three codons and between countries, either the Mann Whitney U Test (for comparison between two groups) or the Kruskal-Wallis test, followed by the Dunn's Multiple Comparison test, were performed.
3. Results
3.1. Pyrosequencing assay evaluation for T. colubriformis
The isotype-1 and -2 β-tubulin genes of T. colubriformis were both evaluated. Pyrosequencing assays were successfully developed for all three codons, F167Y, E198A and F200Y, and in both isotypes.
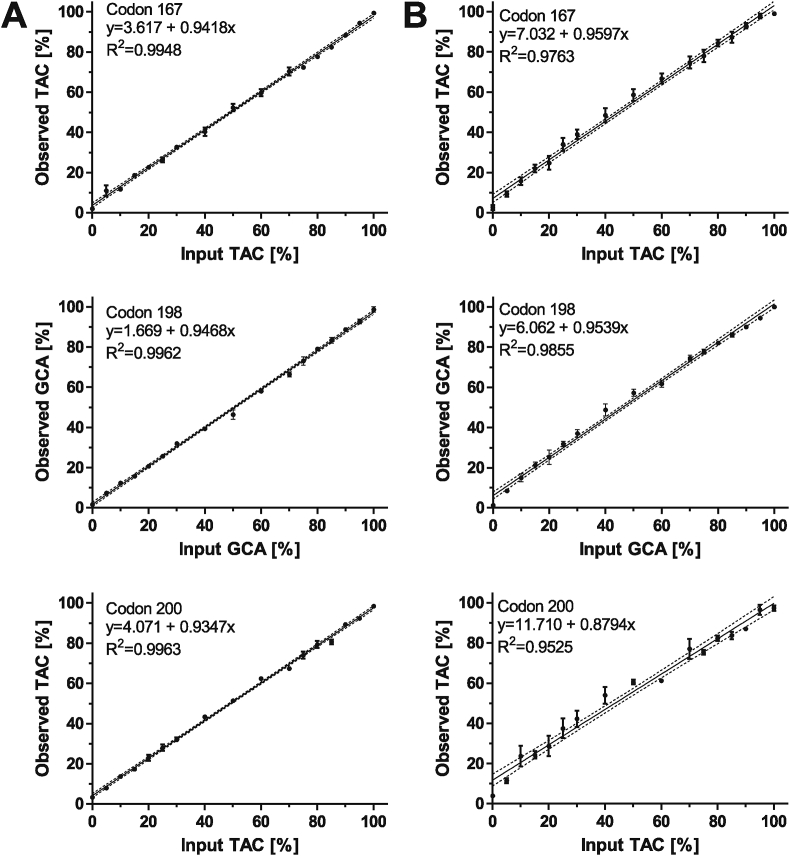
Plasmid DNA containing cDNA inserts encoding the alleles associated with susceptibility and resistance, respectively, were obtained and mixed to obtain defined allele frequencies. Correlations obtained between calculated and observed frequencies for the SNP determination were excellent, with R2 > 0.95 for all 6 SNPs (Fig. 1).
Fig. 1.
Regression analysis of pyrosequencing assays for Trichostrongylus isotype 1 (A) and isotype 2 (B) β-tubulin assays for codons F167Y (TTC to TAC), E198A (GAA to GCA) and F200Y (TTC to TAC). Artificial mixtures of plasmids containing the target sequences as templates were mixed and analysed by pyrosequencing. Observed frequencies (mean of 3–5 replicates ± SEM) were plotted against calculated input frequencies. Regression plots (including linear equation and Pearson correlation coefficient) with 95% confidence bands are shown.
Cross-reactivity was excluded by performance of species-specific assays (Trichostrongylus) with the respective heterologous controls (Haemonchus, Teladorsagia and Cooperia).
3.2. Field samples
In total, field samples from 120 sheep farms were included from the year 2012 and 94 from 2013. The respective number of farms sampled in each country and the final number of samples included in the pyrosequencing analyses are provided in Table 1. T. circumcincta and T. colubriformis were present in all three countries. In some samples, the percentage of the target species was lower than 10% and, accordingly, these were excluded from the respective pyrosequencing survey. This was particularly the case for H. contortus in samples from Ireland, where the total prevalence was very low and the number of L3s was always below 10%. Therefore, no samples from Ireland were analysed for the presence of SNPs in H. contortus. However, for some samples, the amount of DNA was not sufficient to conduct all pyrosequencing assays (Table 1).
Table 1.
Total number of farms sampled per country and year. The number of farms positive for each species as well as the number of pyrosequencing assays performed are provided.
| 2012 |
2013 |
|||||
|---|---|---|---|---|---|---|
| Switzerland | Italy | Ireland | Switzerland | Italy | Ireland | |
| Haemonchusa | 31/63 | 8/39 | 0/18 | 30/56 | 11/26 | 0/15 |
| 167b | 31 | 8 | nd | 29 | 10 | nd |
| 198/200 b |
18 |
7 |
nd |
30 |
11 |
nd |
| Teladorsagiaa | 44/63 | 36/39 | 7/18 | 31/56 | 20/26 | 11/15 |
| 167b | 43 | 36 | 6 | 31 | 19 | 11 |
| 198/200b |
44 |
35 |
6 |
31 |
20 |
11 |
| Trichostrongylusa | 49/63 | 31/38 | 9/18 | 35/56 | 19/26 | 12/15 |
| 167 Iso1b | 27 | 23 | 3 | 22 | 17 | 4 |
| 198 Iso1b | 27 | 23 | 2 | 21 | 14 | 4 |
| 200 Iso1b | 27 | 23 | 3 | 21 | 14 | 4 |
| 167 Iso2b | nd | nd | nd | 33 | 19 | 12 |
| 198 Iso2b | nd | nd | nd | 33 | 19 | 12 |
| 200 Iso2b | nd | nd | nd | 33 | 19 | 12 |
nd, not done.
Number of positive farms per total number of farms.
Number of successful pyrosequencing assays performed.
3.3. Pyrosequencing results
3.3.1. Isotype-1 β-tubulin
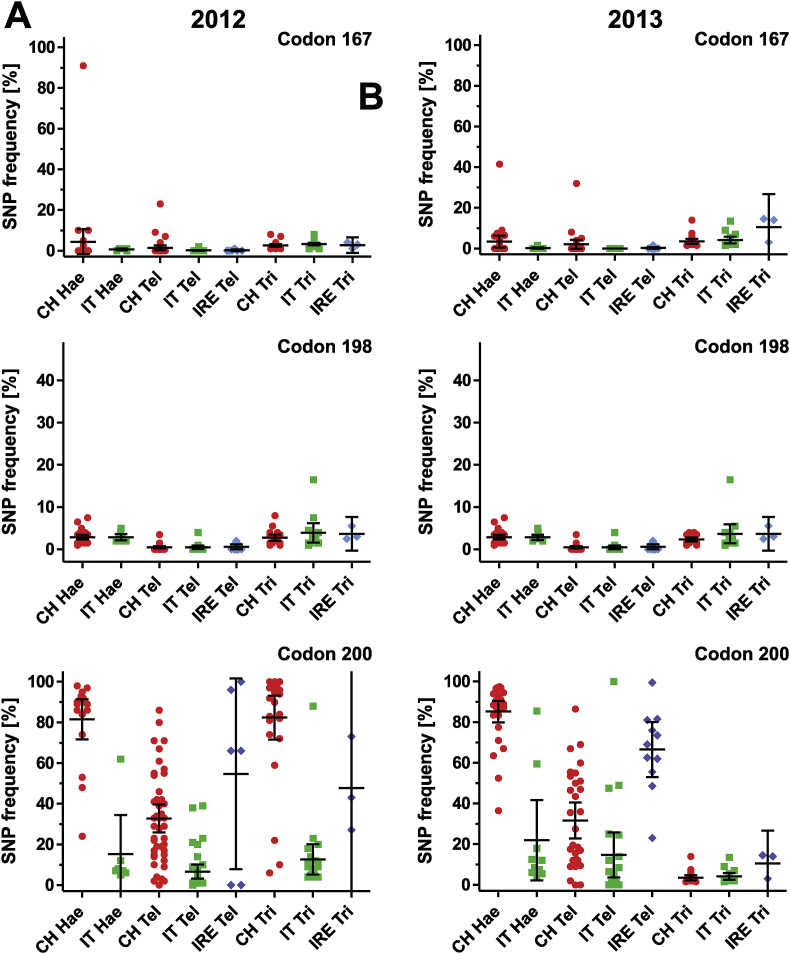
In codons F167Y and E198A, elevated frequencies (≥10%) of the resistance-associated alleles were only rarely observed (Fig. 2). In 2012, codon F167Y TAC frequency was only found to be elevated above background in three samples for Haemonchus (10%, 10% and 91%) and one sample for Teladorsagia (23%), all from Switzerland. In the following year, for both, Haemonchus and Teladorsagia, one sample was found with elevated frequencies (41.5% and 32%, respectively) in Switzerland. Additionally, values above 10% were observed for four samples for Trichostrongylus (13–14%) from all three countries. For codon E198A, only two farms in Switzerland displayed elevated GCA values in assays targeting Haemonchus (11 and 15%) in 2012. In 2013, only one Trichostrongylus sample from Italy had an elevated E198A GCA value (16.5%).
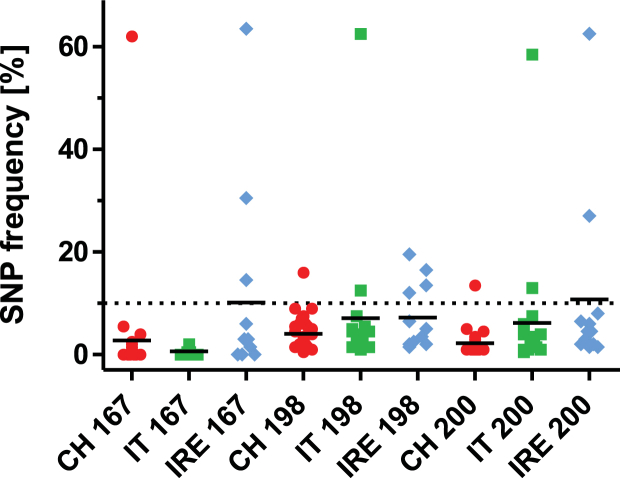
Fig. 2.
Frequency of single nucleotide polymorphisms associated with benzimidazole resistance in isotype 1 β-tubulin in codons F167Y (TAC), E198A (GCA) and F200Y (TAC) in 2012 (A) and 2013 (B). Values represent means of two pyrosequencing assays per farm. The data are shown separately according to country (CH, Switzerland; IT, Italy; IRE, Ireland) and parasite (Hae, Haemonchus spp., Tel, Teladorsagia spp., Tri, Trichostrongylus spp.). The black line represents the overall mean.
In contrast, frequencies of TAC codons associated with BZ resistance at codon F200Y were considerably higher than frequencies in the other two investigated codons in all three countries (Fig. 2) but there were also remarkable differences between the countries. For Haemonchus, TAC frequencies were significantly higher in both years in Switzerland than in Italy (p < 0.0003). For Teladorsagia, elevated TAC frequencies in codon 200 was most frequently observed in Ireland, followed by Switzerland and Italy. All differences were significant (p < 0.05) with the exception being between Ireland and Switzerland in 2012. For Trichostrongylus, frequencies were significantly higher in Switzerland compared to Italy (p < 0.0001). Comparative statistics were not performed for Trichostrongylus samples from Ireland since the total number of samples was very low (2–4 farms).
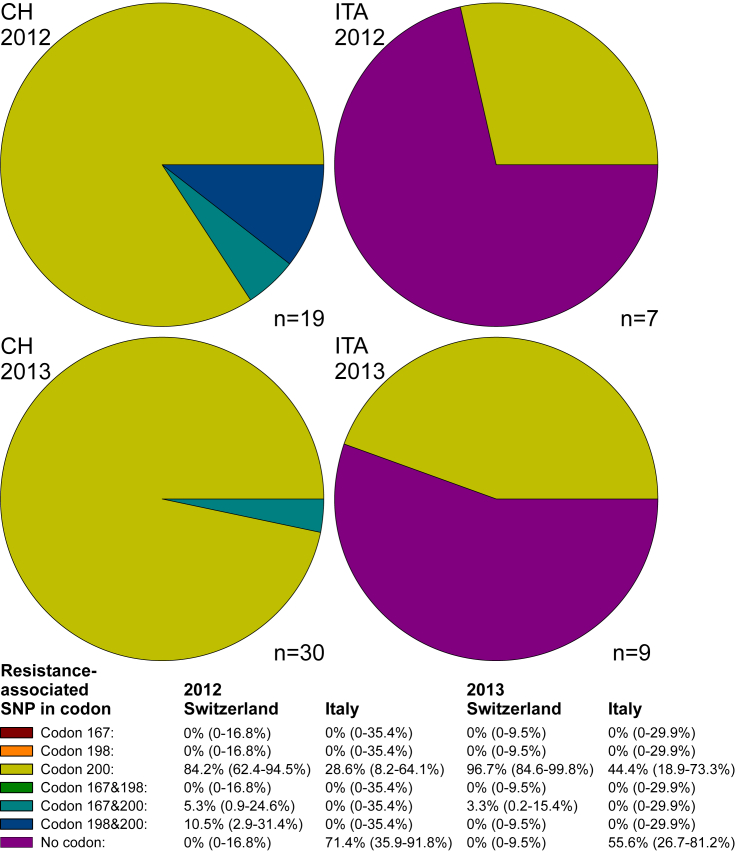
In the following, only those farms for which data for all three codons were available were included. Using a cut-off value of 10% for an elevated SNP frequency, all farms in Switzerland had elevated values for either TAC in codon F200Y alone or TAC in codon F200Y in combination with one of the other resistance-associated codons (Fig. 3) for Haemonchus. In contrast, the majority of farms in Italy (71.4%) did not show any elevated frequencies of resistance-associated codons. The only elevated frequencies were found in codon F200Y.
Fig. 3.
Pie charts showing the prevalence of farms with a frequency ≥10% of single nucleotide polymorphism (SNPs) in isotype 1 β-tubulin of Haemonchus spp, associated with benzimidazole resistance. Only farms with complete datasets for all three SNPs were included in the analysis (numbers of farms are shown at the right bottom of the pie charts). Prevalence of SNPs and combinations of SNPs on the farm level are shown with 95% confidence intervals below the figures. CH, Switzerland; IT, Italy.
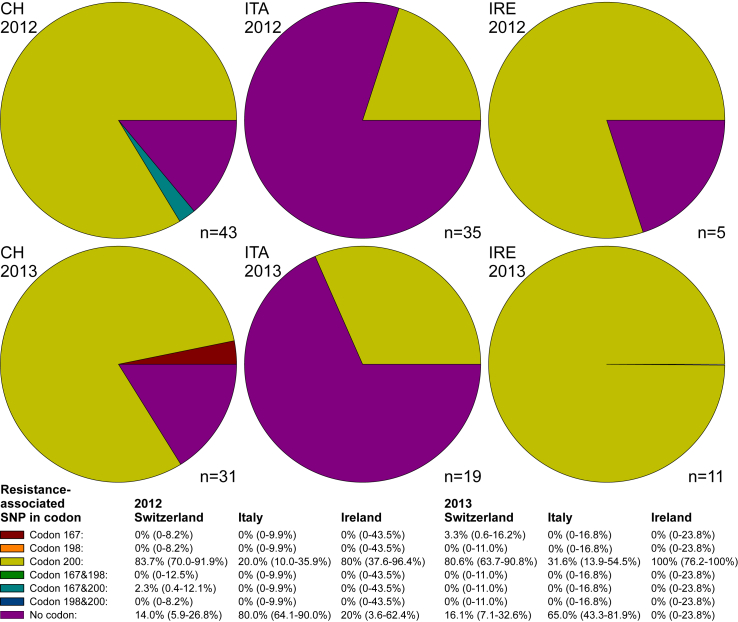
For Teladorsagia, an elevated frequency above background was only observed for TAC in codon F200Y on 80% (2012) and 100% (2013) of the investigated farms in Ireland (Fig. 4). In Switzerland, only approximately 15% of the farms displayed no elevated frequencies, while the majority showed an elevation of TAC in codon 200 and only one farm was found with an elevated frequency of TAC in both, codons F167Y and F200Y. In Italy, again the majority of farms did not show any increase (80% in 2012; 65% in 2013) and those with elevated frequencies were only displayed at codon 200.
Fig. 4.
Pie charts showing the prevalence of farms with a frequency ≥10% of single nucleotide polymorphism (SNPs) in isotype 1 β-tubulin of Teladorsagia spp. associated with benzimidazole resistance. Only farms with complete datasets for all three SNPs were included in the analysis (numbers of farms are shown at the right bottom of the pie charts). Prevalence of SNPs and combinations of SNPs on the farm level are shown with 95% confidence intervals below the figures. CH, Switzerland; IT, Italy; IRE, Ireland.
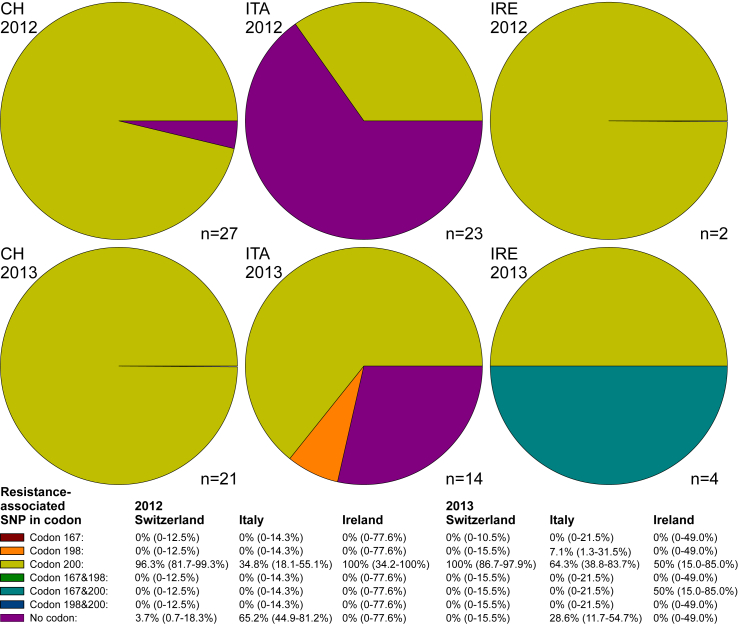
For Trichostrongylus, the situation was slightly different between the two years (Fig. 5). In 2012, one farm in Switzerland showed no elevation of any resistance-associated alleles while, in 2013, on all farms an evelated frequency of TAC in codon F200Y was found. In Ireland, all farms sampled in 2012 displayed the TAC in codon F200Y. In 2013, only half of the farms were found with this SNP while the other half had additionally elevated frequencies of TAC at codon F167Y. However, it needs to be mentioned that the total number of farms in Ireland tested by pyrosequencing was much lower than in the two other countries. In Italy, in 2012, eight farms and in 2013, nine farms showed an elevation of TAC frequency at codon F200Y while no-resistance-associated alleles were detected in codons F167Y or E198A. In 2012, the remaining 15 farms were negative for any resistance-associated alleles. In 2013, one farm had an elevated frequency of GCA in codon E198A and four farms were negative for resistance-associated SNPs.
Fig. 5.
Pie charts showing the prevalence of farms with a frequency ≥10% of single nucleotide polymorphism (SNPs) in isotype 1 β-tubulin of Trichostrongylus spp, associated with benzimidazole resistance. Only farms with complete datasets for all three SNPs were included in the analysis (numbers of farms are shown at the right bottom of the pie charts). Prevalence of SNPs and combinations of SNPs on the farm level are shown with 95% confidence intervals below the figures. CH, Switzerland; IT, Italy; IRE, Ireland.
On 39 farms a total of 48 parasite/farm combinations were investigated in both years (data available for both years for a certain parasite on a particular farmn). In 77.1% of the combinations, the obtained results were qualitatively very similar to almost identical between the years. In 14.6% the respective resistance associated SNP increased in 2013 compared to 2012. On four farms (8.3%) the frequency of the resistance associated allele decreased in 2013 compared to 2012.
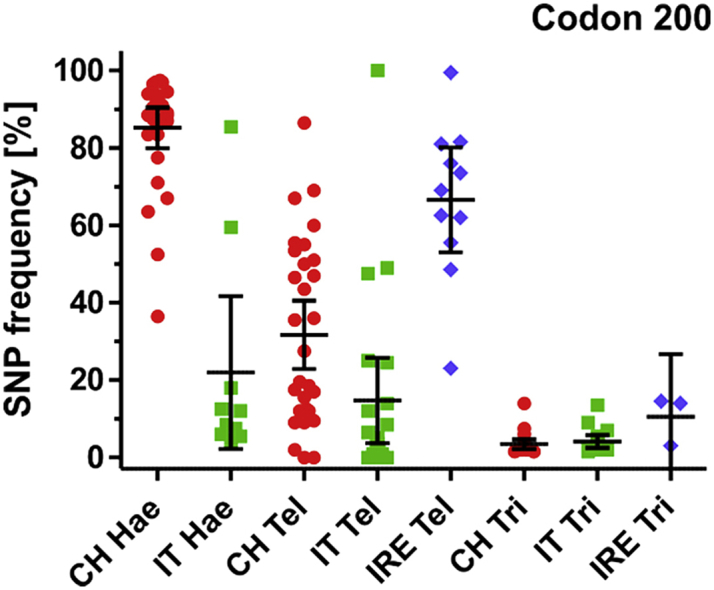
3.3.2. Isotype-2 β-tubulin
SNP frequencies in isotype-2 ß-tubulin were only measured in Trichostrongylus. In comparison to isotype-1, the picture is significantly different. In general, the majority of farms tested showed no or only moderate elevation of F167Y, E198A and F200Y alleles (Fig. 6). However, a few farms showed frequencies of approximately 60%. These five farms were split across the three countries and elevated SNP frequencies were observed at all three codons (TAC in codons F167200 and F200Y and GCA in codon E198A). Additionally, a tendency for higher prevalence of elevated frequencies of these SNPs was observed in Ireland.
Fig. 6.
Frequency of single nucleotide polymorphisms associated with benzimidazole resistance in isotype 2 β-tubulin in codons F167Y (TTC to TAC), E198A (GAA to GCA) and F200Y (TTC to TAC) in 2013. Values represent means of two pyrosequencing assays per farm. The data are shown separately according to country (CH, Switzerland; IT, Italy; IRE, Ireland) and parasite (Hae, Haemonchus spp., Tel, Teladorsagia spp., Tri, Trichostrongylus spp.). The black line represents the overall mean.
4. Discussion
Infection with gastrointestinal nematodes is one of the most important economic constraints on the sheep industry in Europe and possibly worldwide (Sutherland et al., 2010, Miller et al., 2012, Mavrot et al., 2015). One of the aims of the EU-GLOWORM project was to update current knowledge of prevalence of the most important gastrointestinal nematodes, namely, Haemonchus, Teladorsagia and Trichostrongylus, through harmonised sampling and diagnostic procedures in key regions in Europe (Rinaldi et al., 2015c). Overall, it was shown that, in all three pilot countries, Italy, Ireland and Switzerland, prevalences of gastrointestinal nematodes were generally very high, ranging between 91 and 100% (Rinaldi et al., 2015b). However, prevalence of Haemonchus differed considerably between countries, with 77% in Switzerland, 73% in Italy and only 4% in Ireland. These results are in agreement with recent studies in southern Italy, showing high prevalences of H. contortus (based on coprocultures), often associated with other gastrointestinal nematodes and often leading to clinical disease in small ruminants (Musella et al., 2011, Dipineto et al., 2013). Also in Switzerland studies in recent years showed a high prevalence of Haemonchus as well as a high prevalence of anthelmintic resistance in the populations, which was suspected to have spread after the import of animals from South Africa (Artho et al., 2007). For Ireland there is no survey data available other than the study of Rinaldi et al. (2015b) (which is based on the same samples as this study) but it seems that Haemonchus is rarely detected and does no lead to clinical disease. In most local areas, it remains unknown which parasite species are present on a farm. It is particularly unknown, which of these genera have a resistant phenotype and contribute to post-treatment FECs. The current survey used samples derived from this harmonised sampling approach, with the aim to gain an insight into the BZ resistance situation in three countries in Europe, based on a molecular approach. Since the sampling did not involve any pre and post treatment samples and no other methods such as in vitro assays have been used to phenotypically characterise the parasitic nematode populations on these farms, the molecular data reported in the present study cannot be compared to any phenotypic data on resistance level. Since it was not known at the beginning of the study, which of the three codons associated with BZ resistance is mainly involved on European farms, all three codons were investigated. Pyrosequencing assays for Haemonchus and Teladorsagia were already available and, for Trichostrongylus, assays were successfully established.
The results show clearly that, in all populations investigated, codon F200Y in the β-tubulin gene isotype-1 is the most common SNP under selection, while elevated allele frequencies associated with BZ resistance above the 10% threshold were only rarely observed for the other codons, F167Y and E198A. This is in accordance with data published previously. Most studies described in the literature, reporting either results of field studies or the use of laboratory isolates, have been performed with Haemonchus. Comparing laboratory isolates from various geographical origins, von Samson-Himmelstjerna et al. (2009) confirmed the importance of codon 200 relative to codon 167. However, codon 198 was not included at that time. A high prevalence of SNPs at codon 200 has been reported from Pakistan (Chaudhry et al., 2016), where no SNPs were obtained in codon F167Y and F167Y & E198A. Similarly, field studies in Canada (Barrere et al., 2013a, Barrere et al., 2013b) revealed no SNPs in codon 198 but in codon 167 some SNPs were found in Haemonchus. Chaudhry et al. (2015) published data obtained from a survey in India where again F200Y was the most common mutation, E198A was found in one third of the farms but no SNPs were obtained in codon F167Y. In comparison, a similar number of farms in the present study displayed slightly elevated TAC/GCA frequencies in codons F167Y and E198A, respectively. However, TAC in codon 200 was by far the most abundant change observed in Haemonchus. A field study investigating the presence of BZ resistance in Haemonchus in cattle in the US using pyrosequencing, revealed that one out of ten farms was identified as harbouring H. contortus and the remaining nine were all H. placei populations (Chaudhry et al., 2014). While in H. contortus, 91% of analysed individual worms carried BZ resistance associated TAC SNPs at codon F200Y, the frequencies observed in H. placei were much lower (between 1.6 and 9.4%). In the only study from Africa using pyrosequencing, Ademola et al. (2015) identified only BZ susceptible genotypes in H. placei from cattle in Nigeria.
For Teladorsagia, there are even fewer data on BZ-resistant genotypes. PCR-based investigations have almost exclusively been reported for codon F200Y (Elard et al., 1999) and those studies were mainly performed on one or two selected isolates. In one study, codons E198A and F200Y were investigated in field populations in Mexico by PCR, but only TAC SNPs at codon F200Y were found (Liebano-Hernandez et al., 2013). Using pyrosequencing, one initial investigation of a resistant T. circumcincta isolate in Scotland (Skuce et al., 2010), confirmed the importance of the SNP at codon 200. Only one other field study investigating the occurrence of resistance-associated SNPs at all three codons by pyrosequencing has been reported from Spain (Martinez-Valladares et al., 2012), but no resistance-associated SNPs were found. Recently, a study investigating all three resistance-associated codons in H. contortus and T. circumcincta populations from sheep farms in the UK revealed that, for Teladorsagia, again the most abundant SNP was TAC at codon F200Y but, in contrast to Haemonchus, a considerable number of populations also displayed a GCA SNP at codon E198A and only a very low frequency of TCA at codon F167Y was found and only on one farm (Redman et al., 2015).
For Trichostrongylus, data regarding the occurrence of SNPs at all three resistance-associated codons are not available. Only one real time PCR-based investigation, targeting codon F200Y in Trichostrongylus vitrinus, has been reported (Alvarez-Sanchez et al., 2005). To the authors' best knowledge, the current study is the first report using pyrosequencing analysis and comparing frequencies of SNPs at all three codons, F167Y, E198A and F200Y in Trichostrongylus. Even though codon F200Y was found to be the most important codon under selection in Haemonchus and Teladorsagia in the three investigated countries, the different results obtained for Trichostrongylus show that, generally, the other codons should not be neglected.
The data obtained from sheep parasitic nematodes is somewhat different to those obtained for the cattle parasitic nematodes Cooperia and Ostertagia. Recent analyses showed no clear preference for anyof the three SNPs (Demeler et al., 2013, AlGusbi et al., 2014), although it is noteworthy that, in total, a much lower number of populations/isolates has so far been analysed in cattle parasitic nematodes compared to sheep.
The relevance of SNPs in isotype-2 still remains largely unknown. Initially, deletions in the isotype-2 β-tubulin gene, as detected by Southern Blot, have been implicated in a very high BZ resistance phenotype (Kwa et al., 1993). In subsequent studies reporting β-tubulin SNP frequencies, isotype-2 has been largely ignored. von Samson-Himmelstjerna et al. (2009) found only minimal changes in BZ resistance-associated SNPs in H. contortus laboratory isolates. However, due to its high expression level, Saunders et al. (2013) recently suggested to further investigate potential roles of strongylid isotype-2 in BZ resistance. The fact that elevated SNP frequencies in isotype-2 β-tubulin were observed in some field populations in the current study reinforces the view that, even thought it does not appear to play a major role, isotype-2 should not be completely ignored in future anthelmintic resistance research.
In the current study, there were differences in prevalence of BZ resistance alleles between the countries and the genera. For Haemonchus, only Italy and Switzerland were investigated since prevalences and larval counts were too low in Ireland. In Switzerland, every farm showed a considerable elevation in resistance-associated TAC frequency at codon F200Y, while the situation in Italy, with 2 out of 7 farms (28.6%), is still moderate. A similar picture was presented for Teladorsagia in the current study where, in Switzerland and Ireland, elevated SNP frequencies were found on the majority of farms and in Italy on only a few farms. In contrast, a BZ resistance study performed in Spain (Martinez-Valladares et al., 2012) did not reveal any SNPs present at any of the three codons. For Trichostrongylus, all farms in Switzerland were observed to have a significant elevation of TAC frequency in codon 200 above background. Even though a much lower number of farms was investigated in Ireland, the mean frequency of the resistance-associated codons was also high, with no farms classified as genotypically BZ susceptible. As seen for the two other genera, Italy again showed the least problem of the three countries with only a moderate elevation of TAC frequency in codon F200Y in 2012 (34%) but an almost doubling of farms with this allele in 2013 (64%). Although not enough farms were analysed in both years to draw valid conclusions this data suggests that there is currently selection on codon F200Y in Trichostrongylus. The current findings highlight the importance of investigating the BZ resistance situation for this specific parasite.
The prevalence of anthelmintic resistance is much harder to determine than parasite presence, since samples need to be taken more than once, at specific intervals and to be analysed by highly sensitive methods. Accordingly, it remains difficult, if not impossible, to compare the results obtained in the current survey with previous field data from the same regions. Most previously reported studies investigated only a limited number of farms and concentrated mainly on Haemonchus. One reason for this limitation might be that, often, studies have been performed in combination with FECRTs and/or in vitro assays (EHA, LDA), which are laborious procedures. However, BZ resistance in sheep flocks in all of the sampled countries have been previously reported (Hertzberg et al., 2000, Traversa et al., 2007, Good et al., 2012, McMahon et al., 2013, Geurden et al., 2014, Rinaldi et al., 2014, Rose et al., 2015). Particularly in Switzerland it has been assumed that import of ruminants from South Africa carrying resistant nemaoted populations led to the spread of anthelmintice resistance (Artho et al., 2007). The relatively low prevalence of anthelmintice resistance in Italy is in agreement with previously reported studies (Rinaldi et al., 2014, Rose et al., 2015), where low treatment frequencies and particularly the absence of anthelmintic treatment during periods of droughts were determined as the major explanatory factors. Field studies, including FECRT, in vitro assays and molecular tests, revealed an overall good agreement between results obtained from in vivo/in vitro tests and pyrosequencing (Höglund et al., 2009, Martinez-Valladares et al., 2012, Barrere et al., 2013a, Barrere et al., 2013b).
In order to increase the number of farms investigated, the current study aimed to only use pyrosequencing for the assessment of BZ resistance status. It is the first large-scale survey investigating three genera and all three codons associated with BZ resistance on a molecular basis, with samples obtained from a harmonised spatial sampling approach in three different countries. Even though samples were taken in two consecutive years, only limited comparison between years was possible since a) not from every farm larval cultures were available in both years and b) larval cultures revealed different percentages of the respective genera so comparative data were only available for 39 farms and a total of 48 combinations of farm and parasite genus (data available for a particular farm and a particular parasite for both 2012 and 2013). The vast majority of these combinations did not show any difference in frequency of resistance associated alleles between the two years. For a small percentage of comparisons (same parasite and farm) (14.6%), an increase in frequency of any resistance associated allele was observed from 2012 to 2013. The opposite was found in 8.3% (4 comparisons for the same parasite on the same farm), where the frequency of a resistance associated allele obtained in 2013 were lower than in 2012. This can putatively be explained by sampling of different animals, grazing of animals on different pastures in both years or addition of animals purchased from other flocks. However, the results of this survey still show that BZ resistance is a serious issue across all three genera in Switzerland, with BZ resistance-associated SNP frequencies reaching significantly high values. For Ireland, a similar picture was observed. For the two genera investigated, namely, Teladorsagia and Trichostrongylus, high SNP frequencies were detected on the majority of farms. The present study also confirms that BZ resistance is rare in sheep in southern Italy, as shown in FECRT-based field surveys (Rinaldi et al., 2014 ).
5. Conclusion
BZ resistance studies using FECRTs usually have the limitation that mixed infections with different parasite genera are used to calculate one FECR over all trichostrongylid species. At best, larval cultures or PCRs were used to identify those species surviving treatment. In contrast, genus-specific pyrosequencing assays offer the advantage of obtaining detailed information for each investigated genus. In addition, it appears that detection of BZ resistance alleles using pyrosequencing is more sensitive than FECRTs or even EHAs, even for mono-specific infections. The use of pooled pyrosequencing assays in the current study enabled the inclusion of a high number of farms. One of the most important constraints in regard to frequent use of pyrosequencing for routine anthelmintic resistance diagnostics is the fact that it is still limited to the few respectively equipped laboratories (due to the high cost of the pyrosequencer). However, once the technology is more widely available, the cost per sample is considerably less expensive than a FECRT, since it does not require (repeated) farm visits and analyses of numerous individual faecal samples and is additionally independent of previously applied treatments. In comparison to in vitro assays, genotyping/pyrosequencing does not require the availability of fresh faeces containing undeveloped eggs, DNA can be extracted directly from the faecal sample and/or stored for later analysis.
Overall, the results here confirm previous findings of BZ resistance in the three investigated countries. Additionally, it also highlights the importance of inclusion of Teladorsagia spp and Trichostrongylus spp.
Acknowledgements
The authors greatly acknowledge the financial supported by the EU-funded FP7 project GLOWORM (KBBE 2011.1.3–04, No 288975), the intensive support by Maike Boje in molecular work and all farmers and colleagues who helped in collecting and preparing samples.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ijpddr.2016.10.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ademola I.O., Krücken J., Ramünke S., Demeler J., von Samson-Himmelstjerna G. Absence of detectable benzimidazole-resistance associated alleles in Haemonchus placei in cattle in Nigeria revealed by pyrosequencing of beta-tubulin isotype 1. Parasitol. Res. 2015;114:1997–2001. doi: 10.1007/s00436-015-4406-2. [DOI] [PubMed] [Google Scholar]
- AlGusbi S., Krücken J., Ramünke S., von Samson-Himmelstjerna G., Demeler J. Analysis of putative inhibitors of anthelmintic resistance mechanisms in cattle gastrointestinal nematodes. Int. J. Parasitol. 2014;44:647–658. doi: 10.1016/j.ijpara.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Alvarez-Sanchez M.A., Perez-Garcia J., Cruz-Rojo M.A., Rojo-Vazquez F.A. Real time PCR for the diagnosis of benzimidazole resistance in trichostrongylids of sheep. Vet. Parasitol. 2005;129:291–298. doi: 10.1016/j.vetpar.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Artho R., Schnyder M., Kohler L., Torgerson P.R., Hertzberg H. Avermectin-resistance in gastrointestinal nematodes of Boer goats and Dorper sheep in Switzerland. Vet. Parasitol. 2007;144:68–73. doi: 10.1016/j.vetpar.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Barda B.D., Rinaldi L., Ianniello D., Zepherine H., Salvo F., Sadutshang T., Cringoli G., Clementi M., Albonico M. Mini-FLOTAC, an innovative direct diagnostic technique for intestinal parasitic infections: experience from the field. PLoS Negl. Trop. Dis. 2013;7:e2344. doi: 10.1371/journal.pntd.0002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrere V., Falzon L.C., Shakya K.P., Menzies P.I., Peregrine A.S., Prichard R.K. Assessment of benzimidazole resistance in Haemonchus contortus in sheep flocks in Ontario, Canada: comparison of detection methods for drug resistance. Vet. Parasitol. 2013;198:159–165. doi: 10.1016/j.vetpar.2013.07.040. [DOI] [PubMed] [Google Scholar]
- Barrere V., Keller K., von Samson-Himmelstjerna G., Prichard R.K. Efficiency of a genetic test to detect benzimidazole resistant Haemonchus contortus nematodes in sheep farms in Quebec. Can. Parasitol. Int. 2013;62:464–470. doi: 10.1016/j.parint.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Brasil B.S., Nunes R.L., Bastianetto E., Drummond M.G., Carvalho D.C., Leite R.C., Molento M.B., Oliveira D.A. Genetic diversity patterns of Haemonchus placei and Haemonchus contortus populations isolated from domestic ruminants in Brazil. Int. J. Parasitol. 2012;42:469–479. doi: 10.1016/j.ijpara.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Miller M., Yazwinski T., Kaplan R., Gilleard J. The presence of benzimidazole resistance mutations in Haemonchus placei from US cattle. Vet. Parasitol. 2014;204:411–415. doi: 10.1016/j.vetpar.2014.05.025. [DOI] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Ashraf K., Shabbir M.Z., Rashid M.I., Ashraf S., Gilleard J.S. Microsatellite marker analysis of Haemonchus contortus populations from Pakistan suggests that frequent benzimidazole drug treatment does not result in a reduction of overall genetic diversity. Parasit. Vectors. 2016;9:349. doi: 10.1186/s13071-016-1624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry U., Redman E.M., Raman M., Gilleard J.S. Genetic evidence for the spread of a benzimidazole resistance mutation across southern India from a single origin in the parasitic nematode Haemonchus contortus. Int. J. Parasitol. 2015;45:721–728. doi: 10.1016/j.ijpara.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Conway D.P. Variance in the effectiveness of thiabendazole against Haemonchus contortus in sheep. Am.J. Vet. Res. 1964;25:844–846. [PubMed] [Google Scholar]
- Demeler J., Krüger N., Krücken J., von der Heyden V.C., Ramünke S., Küttler U., Miltsch S., Lopez Cepeda M., Knox M., Vercruysse J., Geldhof P., Harder A., von Samson-Himmelstjerna G. Phylogenetic characterization of beta-tubulins and development of pyrosequencing assays for benzimidazole resistance in cattle nematodes. PloS One. 2013;8:e70212. doi: 10.1371/journal.pone.0070212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipineto L., Rinaldi L., Bosco A., Russo T.P., Fioretti A., Cringoli G. Co-infection by Escherichia coli O157 and gastrointestinal strongyles in sheep. Vet. J. 2013;197:884–885. doi: 10.1016/j.tvjl.2013.03.025. [DOI] [PubMed] [Google Scholar]
- dos Santos J.M., Monteiro J.P., Ribeiro W.L., Macedo I.T., Camurca-Vasconcelos A.L., Vieira Lda S., Bevilaqua C.M. Identification and quantification of benzimidazole resistance polymorphisms in Haemonchus contortus isolated in Northeastern Brazil. Vet. Parasitol. 2014;199:160–164. doi: 10.1016/j.vetpar.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Driscoll M., Dean E., Reilly E., Bergholz E., Chalfie M. Genetic and molecular analysis of a Caenorhabditis elegans β-tubulin that conveys benzimidazole sensitivity. J. Cell Biol. 1989;109:2993–3003. doi: 10.1083/jcb.109.6.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elard L., Cabaret J., Humbert J.F. PCR diagnosis of benzimidazole-susceptibility or -resistance in natural populations of the small ruminant parasite, Teladorsagia circumcincta. Vet. Parasitol. 1999;80:231–237. doi: 10.1016/s0304-4017(98)00214-3. [DOI] [PubMed] [Google Scholar]
- Falzon L.C., Menzies P.I., Shakya K.P., Jones-Bitton A., Vanleeuwen J., Avula J., Stewart H., Jansen J.T., Taylor M.A., Learmount J., Peregrine A.S. Anthelmintic resistance in sheep flocks in Ontario, Canada. Vet. Parasitol. 2013;193:150–162. doi: 10.1016/j.vetpar.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Geary T.G., Nulf S.C., Favreau M.A., Tang L., Prichard R.K., Hatzenbuhler N.T., Shea M.H., Alexander S.J., Klein R.D. Three beta-tubulin cDNAs from the parasitic nematode Haemonchus contortus. Mol. Biochem. Parasitol. 1992;50:295–306. doi: 10.1016/0166-6851(92)90227-b. [DOI] [PubMed] [Google Scholar]
- Geurden T., Hoste H., Jacquiet P., Traversa D., Sotiraki S., Frangipane di Regalbono A., Tzanidakis N., Kostopoulou D., Gaillac C., Privat S., Giangaspero A., Zanardello C., Noe L., Vanimisetti B., Bartram D. Anthelmintic resistance and multidrug resistance in sheep gastro-intestinal nematodes in France, Greece and Italy. Vet. Parasitol. 2014;201:59–66. doi: 10.1016/j.vetpar.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144:313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Good B., Hanrahan J.P., de Waal D.T., Patten T., Kinsella A., Lynch C.O. Anthelmintic-resistant nematodes in Irish commercial sheep flocks- the state of play. Ir. Vet. J. 2012;65:21. doi: 10.1186/2046-0481-65-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg H., Rossmann J., Kohler L., Willi U. Benzimidazole-resistance in gastrointestinal nematodes of sheep and goats in Switzerland. Wien. Tierarztl. Mon. 2000;87:3–9. [Google Scholar]
- Höglund J., Gustafsson K., Ljungstrom B.L., Engstrom A., Donnan A., Skuce P. Anthelmintic resistance in Swedish sheep flocks based on a comparison of the results from the faecal egg count reduction test and resistant allele frequencies of the β-tubulin gene. Vet. Parasitol. 2009;161:60–68. doi: 10.1016/j.vetpar.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Howell S.B., Burke J.M., Miller J.E., Terrill T.H., Valencia E., Williams M.J., Williamson L.H., Zajac A.M., Kaplan R.M. Prevalence of anthelmintic resistance on sheep and goat farms in the southeastern United States. J. Am. Vet. Med. Assoc. 2008;233:1913–1919. doi: 10.2460/javma.233.12.1913. [DOI] [PubMed] [Google Scholar]
- Kaplan R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kaplan R.M., Vidyashankar A.N. An inconvenient truth: global worming and anthelmintic resistance. Vet. Parasitol. 2012;186:70–78. doi: 10.1016/j.vetpar.2011.11.048. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Kooyman F.N., Boersema J.H., Roos M.H. Effect of selection for benzimidazole resistance in Haemonchus contortus on β-tubulin isotype 1 and isotype 2 genes. Biochem. Biophys. Res. Commun. 1993;191:413–419. doi: 10.1006/bbrc.1993.1233. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in β-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Van Dijk M., Roos M.H. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J. Mol. Biol. 1995;246:500–510. doi: 10.1006/jmbi.1994.0102. [DOI] [PubMed] [Google Scholar]
- Lacey E. The role of the cytoskeletal protein, tubulin, in the mode of action and mechanism of drug resistance to benzimidazoles. Int. J. Parasitol. 1988;18:885–936. doi: 10.1016/0020-7519(88)90175-0. [DOI] [PubMed] [Google Scholar]
- Liebano-Hernandez E., Gonzalez-Olvera M., Vazquez-Pelaez C., Mendoza-de-Gives P., Ramirez-Vargas G., Peralta-Lailson M., Reyes-Garcia M.E., Osorio J., Sanchez-Pineda H., Lopez-Arellano M.E. Benzimidazole-resistant gastrointestinal nematodes in indigenous Chiapas and Pelibuey sheep breeds from Chiapas, Mexico. J. Helminthol. 2013:1–6. doi: 10.1017/S0022149X13000618. [DOI] [PubMed] [Google Scholar]
- Martin P.J., Anderson N., Jarrett R.G. Detecting benzimidazole resistance with faecal egg count reduction tests and in vitro assays. Aust. Vet. J. 1989;66:236–240. doi: 10.1111/j.1751-0813.1989.tb13578.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Valladares M., Donnan A., Geldhof P., Jackson F., Rojo-Vazquez F.A., Skuce P. Pyrosequencing analysis of the beta-tubulin gene in Spanish Teladorsagia circumcincta field isolates. Vet. Parasitol. 2012;184:371–376. doi: 10.1016/j.vetpar.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Mavrot F., Hertzberg H., Torgerson P. Effect of gastro-intestinal nematode infection on sheep performance: a systematic review and meta-analysis. Parasit. Vectors. 2015;8:557. doi: 10.1186/s13071-015-1164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon C., Bartley D.J., Edgar H.W., Ellison S.E., Barley J.P., Malone F.E., Hanna R.E., Brennan G.P., Fairweather I. Anthelmintic resistance in Northern Ireland (I): prevalence of resistance in ovine gastrointestinal nematodes, as determined through faecal egg count reduction testing. Vet. Parasitol. 2013;195:122–130. doi: 10.1016/j.vetpar.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Miller C.M., Waghorn T.S., Leathwick D.M., Candy P.M., Oliver A.M., Watson T.G. The production cost of anthelmintic resistance in lambs. Vet. Parasitol. 2012;186:376–381. doi: 10.1016/j.vetpar.2011.11.063. [DOI] [PubMed] [Google Scholar]
- Musella V., Catelan D., Rinaldi L., Lagazio C., Cringoli G., Biggeri A. Covariate selection in multivariate spatial analysis of ovine parasitic infection. Prev. Vet. Med. 2011;99:69–77. doi: 10.1016/j.prevetmed.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Njue A.I., Prichard R.K. Cloning two full-length beta-tubulin isotype cDNAs from Cooperia oncophora, and screening for benzimidazole resistance-associated mutations in two isolates. Parasitology. 2003;127:579–588. doi: 10.1017/s0031182003004086. [DOI] [PubMed] [Google Scholar]
- Oxberry M.E., Gear T.G., Prichard R.K. Assessment of benzimidazole binding to individual recombinant tubulin isotypes from Haemonchus contortus. Parasitology. 2001;122:683–687. doi: 10.1017/s0031182001007788. [DOI] [PubMed] [Google Scholar]
- Papadopoulos E., Gallidis E., Ptochos S. Anthelmintic resistance in sheep in Europe: a selected review. Vet. Parasitol. 2012;189:85–88. doi: 10.1016/j.vetpar.2012.03.036. [DOI] [PubMed] [Google Scholar]
- Prichard R.K. Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends Parasitol. 2001;17:445–453. doi: 10.1016/s1471-4922(01)01983-3. [DOI] [PubMed] [Google Scholar]
- Redman E., Whitelaw F., Tait A., Burgess C., Bartley Y., Skuce P.J., Jackson F., Gilleard J.S. The emergence of resistance to the benzimidazole anthlemintics in parasitic nematodes of livestock is characterised by multiple independent hard and soft selective sweeps. PLoS Negl. Trop. Dis. 2015;9:e0003494. doi: 10.1371/journal.pntd.0003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L., Biggeri A., Musella V., De Waal T., Hertzberg H., Mavrot F., Torgerson P.R., Selemetas N., Coll T., Bosco A., Grisotto L., Cringoli G., Catelan D. Sheep and Fasciola hepatica in Europe: the GLOWORM experience. Geospat. Health. 2015;9:309–317. doi: 10.4081/gh.2015.353. [DOI] [PubMed] [Google Scholar]
- Rinaldi L., Catelan D., Musella V., Cecconi L., Hertzberg H., Torgerson P.R., Mavrot F., De Waal T., Selemetas N., Coll T., Bosco A., Biggeri A., Cringoli G. Haemonchus contortus: spatial risk distribution for infection in sheep in Europe. Geospat. Health. 2015;9:325–331. doi: 10.4081/gh.2015.355. [DOI] [PubMed] [Google Scholar]
- Rinaldi L., Hendrickx G., Cringoli G., Biggeri A., Ducheyne E., Catelan D., Morgan E., Williams D., Charlier J., Von Samson-Himmelstjerna G., Vercruysse J. Mapping and modelling helminth infections in ruminants in Europe: experience from GLOWORM. Geospat. Health. 2015;9:257–259. doi: 10.4081/gh.2015.347. [DOI] [PubMed] [Google Scholar]
- Rinaldi L., Morgan E.R., Bosco A., Coles G.C., Cringoli G. The maintenance of anthelmintic efficacy in sheep in a Mediterranean climate. Vet. Parasitol. 2014;203:139–143. doi: 10.1016/j.vetpar.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Roeber F., Jex A.R., Gasser R.B. Advances in the diagnosis of key gastrointestinal nematode infections of livestock, with an emphasis on small ruminants. Biotechnol. Adv. 2013;31:1135–1152. doi: 10.1016/j.biotechadv.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose H., Rinaldi L., Bosco A., Mavrot F., de Waal T., Skuce P., Charlier J., Torgerson P.R., Hertzberg H., Hendrickx G., Vercruysse J., Morgan E.R. Widespread anthelmintic resistance in European farmed ruminants: a systematic review. Vet. Rec. 2015;176:546. doi: 10.1136/vr.102982. [DOI] [PubMed] [Google Scholar]
- Sangster N.C., Prichard R.K., Lacey E. Tubulin and benzimidazole-resistance in Trichostrongylus colubriformis (nematoda) J. Parasitol. 1985;71:645–651. [PubMed] [Google Scholar]
- Saunders G.I., Wasmuth J.D., Beech R., Laing R., Hunt M., Naghra H., Cotton J.A., Berriman M., Britton C., Gilleard J.S. Characterization and comparative analysis of the complete Haemonchus contortus beta-tubulin gene family and implications for benzimidazole resistance in strongylid nematodes. Int. J. Parasitol. 2013;43:465–475. doi: 10.1016/j.ijpara.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Silvestre A., Cabaret J. Mutation in position 167 of isotype 1 beta-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Mol. Biochem. Parasitol. 2002;120:297–300. doi: 10.1016/s0166-6851(01)00455-8. [DOI] [PubMed] [Google Scholar]
- Silvestre A., Humbert J.F. A molecular tool for species identification and benzimidazole resistance diagnosis in larval communities of small ruminant parasites. Exp. Parasitol. 2000;95:271–276. doi: 10.1006/expr.2000.4542. [DOI] [PubMed] [Google Scholar]
- Skuce P., Stenhouse L., Jackson F., Hypsa V., Gilleard J. Benzimidazole resistance allele haplotype diversity in United Kingdom isolates of Teladorsagia circumcincta supports a hypothesis of multiple origins of resistance by recurrent mutation. Int. J. Parasitol. 2010;40:1247–1255. doi: 10.1016/j.ijpara.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Sutherland I.A., Bailey J., Shaw R.J. The production costs of anthelmintic resistance in sheep managed within a monthly preventive drench program. Vet. Parasitol. 2010;171:300–304. doi: 10.1016/j.vetpar.2010.03.035. [DOI] [PubMed] [Google Scholar]
- Sutherland I.A., Leathwick D.M. Anthelmintic resistance in nematode parasites of cattle: a global issue? Trends Parasitol. 2011;27:176–181. doi: 10.1016/j.pt.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Torgerson P.R., Paul M., Furrer R. Evaluating faecal egg count reduction using a specifically designed package “eggCounts” in R and a user friendly web interface. Int. J. Parasitol. 2014;44:299–303. doi: 10.1016/j.ijpara.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Torres-Acosta J.F., Mendoza-de-Gives P., Aguilar-Caballero A.J., Cuellar-Ordaz J.A. Anthelmintic resistance in sheep farms: update of the situation in the American continent. Vet. Parasitol. 2012;189:89–96. doi: 10.1016/j.vetpar.2012.03.037. [DOI] [PubMed] [Google Scholar]
- Traversa D., Paoletti B., Otranto D., Miller J. First report of multiple drug resistance in trichostrongyles affecting sheep under field conditions in Italy. Parasitol. Res. 2007;101:1713–1716. doi: 10.1007/s00436-007-0707-4. [DOI] [PubMed] [Google Scholar]
- van Wyk J.A., Cabaret J., Michael L.M. Morphological identification of nematode larvae of small ruminants and cattle simplified. Vet. Parasitol. 2004;119:277–306. doi: 10.1016/j.vetpar.2003.11.012. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Blackhall W.J., McCarthy J.S., Skuce P.J. Single nucleotide polymorphism (SNP) markers for benzimidazole resistance in veterinary nematodes. Parasitology. 2007;134:1077–1086. doi: 10.1017/S0031182007000054. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Walsh T.K., Donnan A.A., Carriere S., Jackson F., Skuce P.J., Rohn K., Wolstenholme A.J. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009;136:349–358. doi: 10.1017/S003118200800543X. [DOI] [PubMed] [Google Scholar]
- Walsh T.K., Donnan A.A., Jackson F., Skuce P., Wolstenholme A.J. Detection and measurement of benzimidazole resistance alleles in Haemonchus contortus using real-time PCR with locked nucleic acid Taqman probes. Vet. Parasitol. 2007;144:304–312. doi: 10.1016/j.vetpar.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Winterrowd C.A., Pomroy W.E., Sangster N.C., Johnson S.S., Geary T.G. Benzimidazole-resistant β-tubulin alleles in a population of parasitic nematodes (Cooperia oncophora) of cattle. Vet. Parasitol. 2003;117:161–172. doi: 10.1016/j.vetpar.2003.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.