Abstract
The rabbit has great commercial importance as a source of meat and fur, as well as its uses as a laboratory animal for the production of antibodies, used to detect the presence or absence of disease and for research in infectious diseases and immunology. One of the most critical problems in immunology is to understand how the immune system detects the presence of infectious agents and disposes the invader without destroying the self-tissues. Genetic characterization of Toll-like receptors has established that innate immunity is a skillful system that detects invasion of microbial pathogens. Our work aimed to identify, clone and express the Oryctolagus cuniculus (rabbit) TLR-1 mRNA and its encoding protein. We cloned the complete mRNA sequence of Oryctolagus cuniculus TLR-1 and deposit it in the GenBank under accession number (KC349941), which has 2388 base pair and it encodes encode an open reading frame (ORF) translated into 796 amino acids mRNA and consist of 20 types of amino acids. The analysis of amino acid sequence revealed that the rabbit TLR-1 has a typical protein components belonging to the TLR family. Rabbit TLR-1 was expressed in a wide variety of rabbit tissues, which indicate an important role in immune system in different organs.
Keywords: rabbit, Toll-like receptor, cloning, molecular characterization
Introduction
Knowledge of the rabbit’s immune system is of vital importance where rabbit is used as a meat source, fur source and as a model animal for infectious diseases and other inflammatory responses [1]. The rabbit has been a model for immunology studies since 1960s [2, 3]. In the last years, several studies have been performed in the European rabbit and other leporid’s innate immune system [4–9]. However, only one was about TLRs [6]. Toll-like receptors in fishes, birds and mammals have been evolving under positive selection [10–18]. However, relatively little immunological knowledge exists for the rabbit, compared to that available for the human or mouse. Innate immunity is believed to be essential for the removal of invading microbes as well as the control of their systemic spreading. Mammalian toll-like receptors are members of the pattern recognition receptor (PRR) family and play a major role in the initiation of innate immune responses and the subsequent adaptive immune responses against the invading pathogens [19, 20]. Members of the TLR family are necessary for the immune function through the sensing of pathogenic agents and initiation of a proper immune response. More specifically, tissue and cell specific TLR expression patterns have been correlated with the ability to respond to different pathogenic challenges [16, 21–23]. Detection of pathogens by the acquired immune system requires their prior recognition and presentation by cells of the innate immune system, so it plays a major role during outbreaks of new infectious threats mainly when pathogens are able to avoid the immune detection [24]. TLRs help the body to distinct foreign from self-tissue, so their function begins at the earliest stages of immune development [16, 25–30]. In addition, TLRs are able to differentiate between pathogenic agents and commensals. Factors such as the site and amount of each TLR are mainly involved in this process [16, 21, 31]. Recognition of foreign molecules is based on the detection of molecular patterns. These pathogen-associated molecular patterns include molecules such as flagellin, lipopolysaccharide (LPS), lipoproteins, bacterial DNA and unmethylated dinucleotide. The ability of TLRs to recognize a broad spectrum of microbial molecules enables the host to detect the presence of pathogens rapidly, before a more widespread infection occurs [32–38]. Several studies had proved an association between the TLR gene polymorphisms and the occurrence of the disease [15–17, 39–41]. In addition to their role in both bacterial and viral infections, TLRs also recognize and are activated by endogenous molecules associated with damaged cells and tissues [42–44]. TLR1 is a member of the Toll-like receptor family of pattern recognition receptors of the innate immune system. TLR1 recognizes the pathogen-associated molecular pattern with a specificity for gram-positive bacteria. TLR1 has been designated as CD281 [20]. The TLR2/TLR1 form a heterodimer that recognizes a variety of lipoproteins, including those from mycobacteria and meningococci, TLR1-deficient macrophages show normal responses to triacyl lipopeptides but not to diacyl lipopeptides [45, 46].
Our work was aimed to identify, characterize, clone and detect tissue expression of the rabbit (Oryctolagus cuniculus) TLR-1.
Material and methods
Samples
Spleen, duodenum, lung, ileum, colon, kidney, liver, skin, cecum, heart and brain samples were collected from live mature rabbits from common farms in Egypt and kept under inspection for 5 days to be sure that it is free from any clinical infection. The samples were collected and stored at –80°C. RNA was extracted from organs with Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions; both the quantity and quality of total RNA were assessed at OD260 and OD280 using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, USA). Preparation of cDNA library from RNA was performed by AMV reverse transcriptase (Bioer, China) according to the manufacturer’s instructions. The PCR was performed to amplify the target gene using specific primers using Ex Taq polymerase (Takara Bio, Japan) according to the manufacturer’s instructions.
Primers
To identify the TLR-1 from rabbit tissue; degenerative primers were designed based on the alignment of the protein sequence of the ortholog TLR-1 gene; of Pongo pygmaeus (Bornean orangutan, accession no. AB445621), Macaca mulatta (rhesus monkey, accession no. AC204076), Homo sapiens (human, accession no. DQ012261). We used iCODEHOP v1.1 (interactive program for creating Consensus Degenerate Hybrid Oligonucleotide Primers) web-based software, at University of Pittsburgh website, to detect the conservative sites between the different species and then to design primers based on it. The degenerative primers (d-rTLR-1 forward and d-rTLR-1 reverse) were designed based on the conserved sites to the clone short sequence. RACE primers designed toward 3’ end (r-rTLR-1 sense) and 5’ end (r-rTLR-1 antisense) were followed by nested PCR primers (nr-TLR-1 sense, nr-TLR-1 antisense) to use in the RACE system to get the full sequence. Primers were to detect the expression level by qRT-PCR(q-rTLR-1 Forward, q-rTLR-1 Reverse); all the primers were designed by primer premier 5.1 software (PREMIER Biosoft, Palo Alto, CA, USA) (Table 1).
Table 1.
Primers used for the rabbit (Oryctolagus cuniculus) TLR-1 cloning and expression analyses
| Primer | Accession no. | Sequence (5′ to 3′) | Primer type or use |
|---|---|---|---|
| GAPDH Sense primer | L23961 | GAATCCACTGGCGTCTTCAC | Housekeeping gene primers |
| GAPDH Anti-sense primer | CGTTGCTGACAATCTTGAGAGA | ||
| d-rTLR-1 Forward | – | ATGCTGTAGTAGGCTCAG | Degenerative primers |
| d-rTLR-1 Reverse | – | ATTCATCTGCGTGGTCAT | |
| r-rTLR-1 Forward | KC349941 | TATAAACCAATTTTGCAGTAGTACGCT | RACE primers |
| r-rTLR-1 Reverse | TGGGAGGTAAACATCTGAAAACAGAGTC | ||
| nr-rTLR-1 Forward | AGTCTTTCCCAAGGTAGCATCTACG | ||
| nr-rTLR-1 Reverse | CGTGTCCGTCAAGAGGTTGTTGG | ||
| q-rTLR-1 Forward | AATGCGTTTGATGCTCTGCC | Tissue expression primers | |
| q-rTLR-1 Reverse | CCTCCGTGCCATGAGGGTTT | ||
| rTLR-1 Forward | TGACCATCACTATTTCTAGCAGCTT | Full length primers | |
| rTLR-1 Reverse | CCATGTTTAGTCTTCTCCTTCGGC |
Cloning of rabbit TLR-1(rTLR-1) cDNA sequence
The PCR product was purified and ligated into pMD18-T easy vector, transformed into the competent Escherichia coli DH5a cell and plated on the LB-agar Petri dish. Positive clones containing the expected-size inserts were screened by colony PCR. Plasmid DNA was extracted using an Axyprep plasmid miniprep kit (Axygen Biosciences, USA) according to the manufacturer’s instructions. Three representative plasmid DNAs were sequenced. Rapid amplification of cDNA ends (RACE) system, was done by SMARTer™ RACE cDNA Amplification Kit (Clontech, USA) to get the full length of the required sequences according to the manufacturer’s instructions.
Sequence analyses and alignment
The BLAST tool (http://www.ncbi.nlm.nih.gov/blast) was used to detect sequence homology. The translated amino acid sequences were analyzed with the Expert Protein Analysis System (http://www.expasy.org/) and the protein domain features in the translated amino acids were predicted by Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/). Phylogenetic and molecular evolution analysis was conducted by MEGA 5 software [47] and optimized manually.
Rabbit TLR-1 expression
The real-time quantitative polymerase chain reaction was used to quantify the rTLR-1 gene expressions using an ABI Prism 7000 Sequence Detection Systems and TaqMan 2× PCR Master Mix Reagents Kit following the manufacturer’s instructions (Applied Biosystems; Life Technologies, USA). The housekeeping gene (GAPDH) primers [48] were used as an internal control for cDNA normalization where the unit number showing relative mRNA levels in each sample was determined as a value of mRNA normalized against GAPDH. The expression data obtained from three independent biological replicate, RT-PCR data were analyzed by using the 2−ΔΔCT method as described [49].
Nucleotide sequence deposition
The BankIt tool was used to deposit the analyzed sequence (rTLR-1) in the GenBank (http://www.ncbi.nlm.nih.gov/genbank/submit/).
Results and discussion
Identification and characterization of rTLR-1
The complete mRNA sequence of Oryctolagus cuniculus TLR-1 was deposited in the NCBI GeneBank database under accession no. KC349941. While it consists of 2388 nucleotides and the consensus cDNA sequence showed identity with the TLR-1 mRNA sequences Chinese hamster TLR-1 (Cricetulus griseus), of 69%; nine-banded armadillo TLR-1 (Dasypus novemcinctus) – 47% identity, cattle TLR-1 (Bos taurus) – 39% identity; mice (Mus musculus) – 65% identity.
The encoded polypeptide consists of 796 amino acids; we detect the site of variation in the encoded polypeptide sequence for the cloned rabbit TLR-1 (UniProt ID: M9T155) and the predicted Rabbit TLR-1 (UniProt ID: G1TIU2) (Table 2). The identity degree in the encoded polypeptide amino acids sequences around 97% between the cloned rabbit TLR-1 and the predicted rabbit TLR-1, where the variation occurs in 24 amino acids (Table 3). While the cloned rabbit TLR-1 showed a degree of identity with the rodents; where it shows 73% identity with the Mole rat, 71% identity with the guinea pigs , 68% identity with the house mouse and 65% identity with the rat.
Table 2.
Illustrate transmembrane structure of Oryctolagus cuniculus TLR-1 showing confidently predicted domains, repeats and motifs done by SMART analysis web based application
| Name | Start | End | E-value |
|---|---|---|---|
| Signal peptide | 1 | 28 | N/A |
| LRR | 72 | 95 | 7.36 |
| LRR | 375 | 397 | 33.6 |
| Low complexity | 428 | 437 | N/A |
| LRR | 471 | 494 | 52.7 |
| LRRCT | 528 | 582 | 0.00000654 |
| Transmembrane region | 586 | 608 | N/A |
| TIR | 640 | 783 | 1.45e-42 |
Table 3.
The site of variation in the encoded polypeptide sequence for the cloned rabbit TLR-1 (UniProt ID: M9T155) and the predicted rabbit TLR-1 (UniProt ID: G1TIU2)
| Amino acid sites | 3 | 10 | 20 | 22 | 44 | 84 | 180 | 181 | 204 | 231 | 262 | 302 | 312 | 325 | 339 | 368 | 377 | 459 | 567 | 568 | 740 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cloned rabbit TLR-1 M9T155 | I | Q | L | S | G | T | R | V | N | E | S | C | C | E | R | T | A | S | N | R | H |
| Predicted rabbit TLR-1 G1TIU2 | N | H | V | R | P | R | G | E | K | D | W | S | S | K | G | I | G | N | K | G | Q |
Analysis of the rTLR-1 sequence
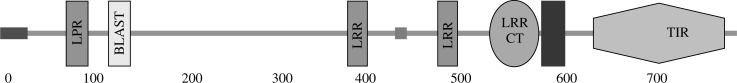
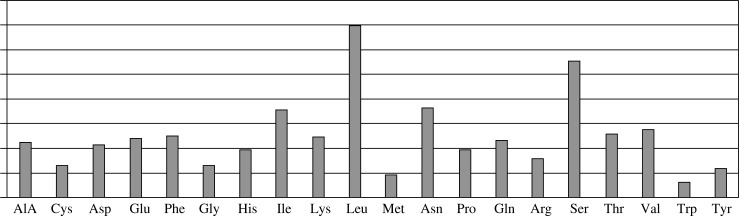
The predicted protein encoded by Oryctolagus cuniculus TLR-1 mRNA sequence is composed of 796 amino acids and shows a typical toll-like receptor structure. The Oryctolagus cuniculus TLR-1 domain structure has been estimated using the SMART web tool. The rTLR-1 started by signal peptide (28 amino acids from 1-28) followed by three leucine rich repeat (LRR) domains (at position 72-95, 375-395, 471-491) and one C-terminal LRR domain around 54 amino acids (LRR-CT, residues 528-582) in the extracellular region, then transmembrane region around 22 amino acids (at position 586-608) and a TIR domain around 143 amino acids (residues 640-783) in the cytoplasmic region as shown in Fig. 1, Table 3 and Fig. 2. The encoded amino acids vary among different available amino acids where it has 20 different amino acids; the highest amino acid encoded is leucine while the lowest is tryptophan as shown in Fig. 2.
Fig. 1.
The transmembrane structure of rabbit TLR-1; started by signal peptide (28 amino acids from 1-28) followed by three lucien rich repeat (LRR) domains (at position 72-95, 375-395, 471-491) and one C-terminal LRR domain around 54 amino acids (LRR-CT, residues 528-582) in the extracellular region then transmembrane region around 22 amino acids (at position 586-608) and a TIR domain around 143 amino acids (residues 640-783) in the cytoplasmic region
Fig. 2.
Oryctolagus cuniculus TLR-1 amino acid composition and percentage; where it has 20 different amino acids; the highest amino acid encoded is leucine while the lowest is tryptophan
We detect the site of variation in the encoded polypeptide sequence for the cloned rabbit TLR-1 (UniProt ID: M9T155) and the predicted Rabbit TLR-1 (UniProt ID: G1TIU2). The identity degree in the encoded polypeptide amino acids sequences around 97% between the cloned rabbit TLR-1 and the predicted rabbit TLR-1, where the variation occurs in 24 amino acids (Table 3) while showed 73% identity with the Mole rat, 71% identity with the guinea pigs, 68% identity with the house mouse and 65% identity with the rat.
Phylogenetic analysis with other TLRs
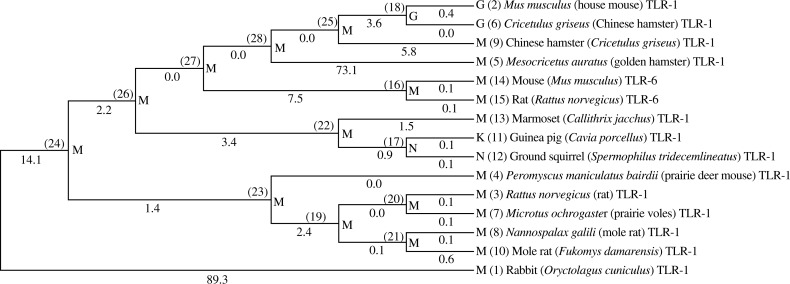
Two methods were used to construct a phylogenetic tree (neighbour-joining and maximum parsimony) based on the amino acid of TLR-1, which was downloaded from the GenBank. The phylogenetic analysis performed using the translated Oryctolagus cuniculus amino acid sequence with almost all the known amino acid sequences found in the GenBank, both phylogenetic methods provided almost the same results, where the phylogenetic analysis showed that Oryctolagus cuniculus TLR-1 is closely related to mole rat, guinea pig and house mouse TLR-1. The structure of Oryctolagus cuniculus TLR-1 amino acids is in general similar to other identified TLR1 sequences in different mammalians and birds as shown in Fig. 3.
Fig. 3.
Phylogenetic tree of rabbit TLR-1 against the available rodents TLR-1 sequence in the Genebank
Expression pattern of Oryctolagus cuniculus TLR-1
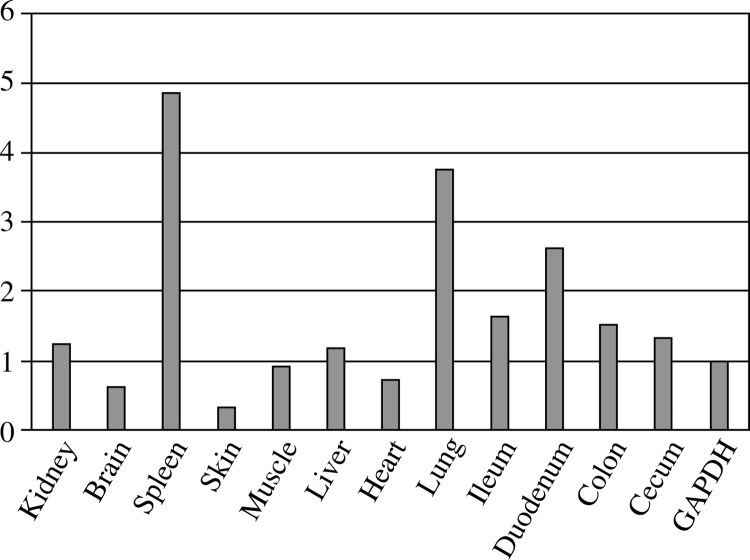
The rabbit TLR-1 mRNA was constitutively expressed in all selected tissues, with higher levels detected in spleen, duodenum, and lung; moderate levels in ileum, kidney, liver, cecum, colon; and low levels in heart, skin and brain; as shown in Figure 4 and the level of expression measured by RT-PCR as shown in Table 4 which similar to the distinct distribution of specific tissue expression in mice, mouse, human, chicken and fish TLR-1 models.
Fig. 4.
rTLR-1 expression throughout the tissue cells (spleen, skin, duodenum, lung, ileum, colon, kidney, liver, cecum, heart and brain). TLR expression counts were normalized to GAPDH using the formula 2− (TLR−GAPDH). All values represent mean ± standard error of the mean of triplicate from quantitative real-time PCR assays
Table 4.
rTLR-1 expression throughout the tissue cells (spleen, skin, duodenum, lung, ileum, colon, kidney, liver, cecum, heart and brain). TLR expression counts were normalized to GAPDH using the formula 2−(TLR−GAPDH). All values represent mean ± standard error of the mean of triplicate from quantitative real-time PCR assays
| Tissue | TLR-1 relative expression level (mean ± SD) |
|---|---|
| Kidney | 1.23 ±0.33 |
| Brain | 0.64 ±0.03 |
| Spleen | 4.87 ±0.78 |
| Skin | 0.32 ±0.01 |
| Muscle | 0.92 ±0.15 |
| Liver | 1.20 ±0.25 |
| Heart | 0.71 ±0.04 |
| Lung | 3.76 ±0.34 |
| Ileum | 1.64 ±0.17 |
| Duodenum | 2.62 ±0.31 |
| Colon | 1.52 ±0.29 |
| Cecum | 1.32 ±0.27 |
| GAPDH | 1.00 ±0.08 |
Conclusions
This is the first study to make actual cloning and characterization to the rabbit toll-like receptor type 1. The rTLR-1 encodes an open reading frame (ORF) translated into 796 amino acids mRNA and consists of 20 types of amino acids. The structure of Oryctolagus cuniculus TLR-1 amino acids is in general similar to other identified TLR1 sequences in different mammalians and birds. Rabbit TLR-1 was expressed in a wide variety of rabbit tissues, which indicates an important role in the immune system in different organs.
The authors declare no conflict of interest.
References
- 1.Abdel-Daim MM, Ghazy EW. Effects of Nigella sativa oil and ascorbic acid against oxytetracycline-induced hepato-renal toxicity in rabbits. Iran J Basic Med Sci. 2015;18:221–227. [PMC free article] [PubMed] [Google Scholar]
- 2.Gertz EM, Schäffer AA, Agarwala R, et al. Accuracy and coverage assessment of Oryctolagus cuniculus (Rabbit) Genes Encoding Immunoglobulins in the Whole Genome Sequence Assembly (OryCun2.0) and Localization of the IGH Locus to Chromosome 20. Immunogenetics. 2013;65:749–762. doi: 10.1007/s00251-013-0722-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinheiro A, Lanning D, Alves PC, et al. Molecular bases of genetic diversity and evolution of the immunoglobulin heavy chain variable region (IGHV) gene locus in leporids. Immunogenetics. 2011;63:397–408. doi: 10.1007/s00251-011-0533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neves F, Abrantes J, Lissovsky AA, Esteves PJ. Pseudogenization of CCL14 in the Ochotonidae (pika) family. Innate Immunity. 2015;21:647–654. doi: 10.1177/1753425915577455. [DOI] [PubMed] [Google Scholar]
- 5.Neves F, Abrantes J, Pinheiro A, et al. Convergent evolution of IL-6 in two leporids (Oryctolagus and Pentalagus) originated an extended protein. Immunogenetics. 2014;66:589–595. doi: 10.1007/s00251-014-0787-0. [DOI] [PubMed] [Google Scholar]
- 6.Abrantes J, Areal H, Esteves PJ. Insights into the European rabbit (Oryctolagus cuniculus) innate immune system: genetic diversity of the toll-like receptor 3 (TLR3) in wild populations and domestic breeds. BMC Genet. 2013;14:73. doi: 10.1186/1471-2156-14-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Matos AL, van der Loo W, Areal H, et al. Study of Sylvilagus rabbit TRIM5alpha species-specific domain: how ancient endoviruses could have shaped the antiviral repertoire in Lagomorpha. BMC Evol Biol. 2011;11:294. doi: 10.1186/1471-2148-11-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siewe BT, Kalis SL, Esteves PJ, et al. A novel functional rabbit IL-7 isoform. Dev Comp Immunol. 2010;34:828–836. doi: 10.1016/j.dci.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmo CR, Esteves PJ, Ferrand N, van der Loo W. Genetic variation at chemokine receptor CCR5 in leporids: alteration at the 2nd extracellular domain by gene conversion with CCR2 in Oryctolagus, but not in Sylvilagus and Lepus species. Immunogenetics. 2006;58:494–501. doi: 10.1007/s00251-006-0095-4. [DOI] [PubMed] [Google Scholar]
- 10.Heffelfinger C, Pakstis AJ, Speed WC, et al. Haplotype structure and positive selection at TLR1. Eur J Hum Genet. 2014;22:551–557. doi: 10.1038/ejhg.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Z, Sun Y, Wang R, Xu T. Evolutionary analysis of TLR9 genes reveals the positive selection of extant teleosts in Perciformes. Fish Shellfish Immunol. 2013;35:448–457. doi: 10.1016/j.fsi.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 12.Grueber CE, Wallis GP, Jamieson IG. Episodic positive selection in the evolution of avian toll-like receptor innate immunity genes. PLoS One. 2014;9:e89632. doi: 10.1371/journal.pone.0089632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Temperley ND, Ren L, et al. Molecular evolution of the vertebrate TLR1 gene family – a complex history of gene duplication, gene conversion, positive selection and co-evolution. BMC Evol Biol. 2011;11:149. doi: 10.1186/1471-2148-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wlasiuk G, Nachman MW. Adaptation and constraint at Toll-like receptors in primates. Mol Biol Evol. 2010;27:2172–2186. doi: 10.1093/molbev/msq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elfeil W, Soliman E, Sobeih M. Epidemiological Studies on Environmental Pollution in Poultry Farms. Munich: GRIN Publishing GmbH; 2011. p. 233. [Google Scholar]
- 16.Elfeil WK, Abouelmaatti RR, Sun CJ, et al. Identification, Cloning, Expression of a Novel Functional Anas platyrhynchos mRNA TLR4. J Anim Vet Adv. 2012;11:1727–1733. [Google Scholar]
- 17.Elfeil WKM. Newcastle-Avian flu recombinant vaccine in embryonated eggs and chicks. Germany: LAP LAMBERT Academic Publishing; 2012. p. 80. [Google Scholar]
- 18.Abouelmaatti RR, Algammal AM, Li X, et al. Cloning and analysis of Nile tilapia Toll-like receptors type-3 mRNA. Centr Eur J Immunol. 2013;38:277–282. [Google Scholar]
- 19.Chaudhary PM, Ferguson C, Nguyen V, et al. Cloning and characterization of two Toll/Interleukin-1 receptor-like genes TIL3 and TIL4: evidence for a multi-gene receptor family in humans. Blood. 1998;91:4020–4027. [PubMed] [Google Scholar]
- 20.Rock FL, Hardiman G, Timans JC, et al. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci U S A. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abouelmaatti RR, Elfeil W, Wang Y, Liu S. Pattern recognition receptors mini review. GASJ. 2013;1(1) [Google Scholar]
- 22.O’Bryan MK, Sebire KL, Gerdprasert O, et al. Cloning and regulation of the rat activin betaE subunit. J Mol Endocrinol. 2000;24:409–418. doi: 10.1677/jme.0.0240409. [DOI] [PubMed] [Google Scholar]
- 23.Prince LR, Whyte MK, Sabroe I, Parker LC. The role of TLRs in neutrophil activation. Curr Opin Pharmacol. 2011;11:397–403. doi: 10.1016/j.coph.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sebastiani G, Leveque G, Larivière L, et al. Cloning and characterization of the murine toll-like receptor 5 (Tlr5) gene: sequence and mRNA expression studies in Salmonella-susceptible MOLF/Ei mice. Genomics. 2000;64:230–240. doi: 10.1006/geno.2000.6115. [DOI] [PubMed] [Google Scholar]
- 26.Zarember KA, Godowski PJ. Tissue expression of human toll-like receptors and differential regulation of toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J Immunol. 2002;168:554–561. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 27.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 28.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 29.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 30.Jault C, Pichon L, Chluba J. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol Immunol. 2004;40:759–771. doi: 10.1016/j.molimm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Abreu MT. Immunologic regulation of toll-like receptors in gut epithelium. Curr Opin Gastroenterol. 2003;19:559–564. doi: 10.1097/00001574-200311000-00008. [DOI] [PubMed] [Google Scholar]
- 32.Roach JC, Glusman G, Rowen L, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci U S A. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moynagh PN. Toll-like receptor signalling pathways as key targets for mediating the anti-inflammatory and immunosuppressive effects of glucocorticoids. J Endocrinol. 2003;179:139–144. doi: 10.1677/joe.0.1790139. [DOI] [PubMed] [Google Scholar]
- 34.Kaisho T, Akira S. Regulation of dendritic cell function through Toll-like receptors. Curr Mol Med. 2003;3:373–385. doi: 10.2174/1566524033479726. [DOI] [PubMed] [Google Scholar]
- 35.Kaisho T, Akira S. Critical roles of toll-like receptors in host defense. Crit Rev Immunol. 2000;20:393–405. [PubMed] [Google Scholar]
- 36.Kaisho T, Akira S. Toll-like receptor function and signaling. J Allergy Clin Immunol. 2006;117:979–87. doi: 10.1016/j.jaci.2006.02.023. quiz 88. [DOI] [PubMed] [Google Scholar]
- 37.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Uematsu S, Sato S, Yamamoto M, et al. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-{alpha} induction. J Exp Med. 2005;2005:915–923. doi: 10.1084/jem.20042372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hawn TR, Misch EA, Dunstan SJ, et al. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur J Immunol. 2007;37:2280–2289. doi: 10.1002/eji.200737034. [DOI] [PubMed] [Google Scholar]
- 40.Bochud PY, Bochud M, Telenti A, Calandra T. Innate immunogenetics: a tool for exploring new frontiers of host defence. Lancet Infect Dis. 2007;7:531–542. doi: 10.1016/S1473-3099(07)70185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bochud PY, Hersberger M, Taffe P, et al. Polymorphisms in Toll-like receptor 9 influence the clinical course of HIV-1 infection. AIDS. 2007;21:441–446. doi: 10.1097/QAD.0b013e328012b8ac. [DOI] [PubMed] [Google Scholar]
- 42.Kariko K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 43.Karikó K, Weissman D, Welsh FA. Inhibition of toll-like receptor and cytokine signaling – a unifying theme in ischemic tolerance. J Cereb Blood Flow Metab. 2004;24:1288–1304. doi: 10.1097/01.WCB.0000145666.68576.71. [DOI] [PubMed] [Google Scholar]
- 44.Areal H, Abrantes J, Esteves PJ. Signatures of positive selection in Toll-like receptor (TLR) genes in mammals. BMC Evol Biol. 2011;11:368. doi: 10.1186/1471-2148-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi O, Akira S. Genetic approaches to the study of Toll-like receptor function. Microbes Infect. 2002;4:887–895. doi: 10.1016/s1286-4579(02)01615-5. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espino AM, Rivera F. Quantitation of cytokine mRNA by real-time RT-PCR during a vaccination trial in a rabbit model of fascioliasis. Vet Parasitol. 2010;169:82–92. doi: 10.1016/j.vetpar.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CTmethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]