Abstract
Genomic analysis of circulating tumor DNA (ctDNA) released from cancer cells into the bloodstream has been proposed as a useful method to capture dynamic changes during the course of the disease. In particular, the ability to monitor epidermal growth factor receptor (EGFR) mutation status in cell-free circulating DNA (cfDNA) isolated from advanced non-small cell lung cancer (NSCLC) patients EGFR can help to the correct management of the disease and overcome the challenges associated with tumor heterogeneity and insufficient biopsied material to perform key molecular diagnosis. Here, we report a case of long term monitorization of EGFR mutation status in cfDNA from peripheral blood in an NSCLC patient in, with excellent correlation with clinical evolution.
Keywords: circulating free DNA (cfDNA), epidermal growth factor receptor (EGFR), non-small cell lung cancer (NSCLC)
Introduction
Activating mutations in the epidermal growth factor receptor (EGFR) gene are the first validated biomarker to predict significant progression-free survival (PFS) benefit with treatment with EGFR tyrosine kinase inhibitors (TKIs) in non-small cell lung cancer (NSCLC) (1-5).
EGFR mutations are found in 10–15% of lung cancer patients in European populations, particularly non-smokers with adenocarcinoma histology. They occur in exons 18 to 21, which encode the EGFR kinase domain; and approximately 90% are exon 19 deletions or exon 21 L858R point mutations. The alteration of these exons causes constitutive activation of the receptor and a pathological activation of downstream molecular pathways, leading to proliferation and survival of cancer cells. The first TKIs developed were gefinitib and erlotinib, which are highly active in EGFR mutant lung tumors, but other drugs such as afatinib have been also approved after showing comparable activity. In spite of the high response rates to these drugs in EGFR mutant lung cancer patients, resistance invariably occurs after 12 to 24 months on treatment (6). Acquisition of the T790M ‘gatekeeper’ mutation is one of the most frequent mechanisms of resistance to the TKIs gefitinib and erlotinib. With the recent development the so-called third-generation EGFR TKIs, such as osimertinib, that effectively target T790M, the need for effective methods to identify this mutation has become ever more pressing (7-9).
EGFR mutation analysis at diagnosis is not always possible in all patients, often due to suboptimal quantity or quality of biopsied material. In addition, assessing dynamic changes in the tumor during treatment would require serial biopsies, but this is an invasive, expensive and time-consuming process, that it is often not feasible.
Peripheral blood samples, taken in a simple, non-invasive blood draw, provide a source of cancer-derived material such as circulating tumor DNA (ctDNA) which could provide insights into the status of the primary tumor and metastases in real time. These non-invasive ‘liquid biopsies’ are easier to obtain than traditional tissue biopsies and repeated samples can readily be taken at different time points in order to monitor disease progression and treatment response, overcoming the problems of tumor heterogeneity and scarceness of material associated with biopsy sampling (10,11).
We developed an in-house, highly sensitive and specific method for analysis of EGFR and other mutations in cell-free circulating DNA (cfDNA) based on a modified real-time PCR analysis using a peptide-nucleic acid (PNA) polymer in order to increase sensitivity (12,13). Here we present a case of long-term monitorization of a NSCLC patient through analysis of EGFR mutations in ctDNA.
Case presentation
The patient, a 57-year-old woman, former light smoker, was diagnosed in January 2012 with L858R EGFR mutation-positive metastatic lung adenocarcinoma by direct sequencing analysis on tumor tissue. Radiologic studies showed a lung mass in the right upper lobe, malignant right pleural effusion, mediastinal lymph nodes, lung carcinomatous lymphangitis and two subcentimetric subpleural lung nodules. A single right cerebellar lesion of 2.8 cm × 2.2 cm was detected on brain MRI.
The patient was initially treated by surgical resection of the cerebellar metastasis. Pathologic examination reported a poorly differentiated lung adenocarcinoma (immunohistochemistry positive for cytokeratins AE1/AE3, CK8/18and TTF-1).
Treatment was initiated on March 18th, 2012, with erlotinib 100 mg per day plus bevacizumab 500 mg. The patient had a complete response on CT scan of May 17th, 2012.
After sixteen months on treatment, in July 2013, the patient underwent the first blood extraction for the analysis of EGFR mutations. Unexpectedly, we detected in ctDNA the presence of a L858R mutations in exon 21 of EGFR and also the resistance mutation T790M. A confirmatory radiologic evaluation by PET/CT scan was performed on August 30th showing disease progression with hypermetabolic thickening of the pleura combined with reappearance of right pleural effusion, as well as an increase in the size of the lesion on the right lung. A pleural biopsy by thoracoscopy was performed in August 2013. Histological and molecular analyses of the pleural biopsy confirmed a poorly differentiated lung adenocarcinoma, harboring both the L858R and the T790M resistance mutation (Figures 1,2). The patient started second line treatment on October 3rd 2013 with cisplatin (75 mg/m2) and pemetrexed (500 mg/m2). In total, she received four cycles of this treatment, with partial response and rapid disappearance of circulating EGFR mutations (L858R and T790M) in peripheral blood. After that, she continued maintenance therapy with erlotinib + avastin + pemetrexed.
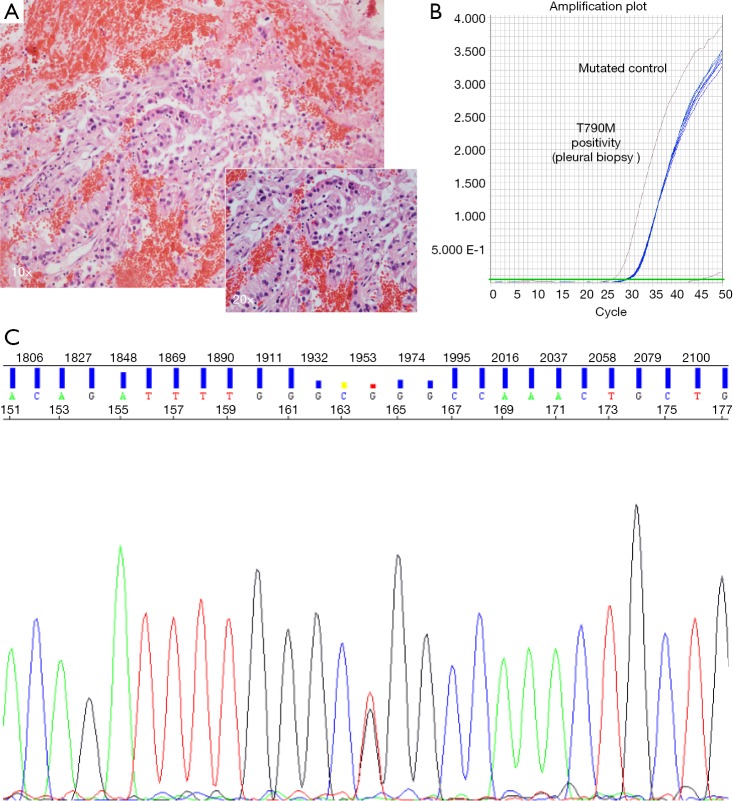
Figure 1.
Histological and molecular analysis of pleural biopsy at progression disease. (A) H&E stain. Below and to the right (magnification, ×20); (B) T790M positivity: amplification curve using a PNA-mediated 5’ nuclease real time Taqman PCR, blue amplification line corresponds to the patient; (C) L858R mutation confirmed by Sanger sequencing. PNA, peptide-nucleic acid.
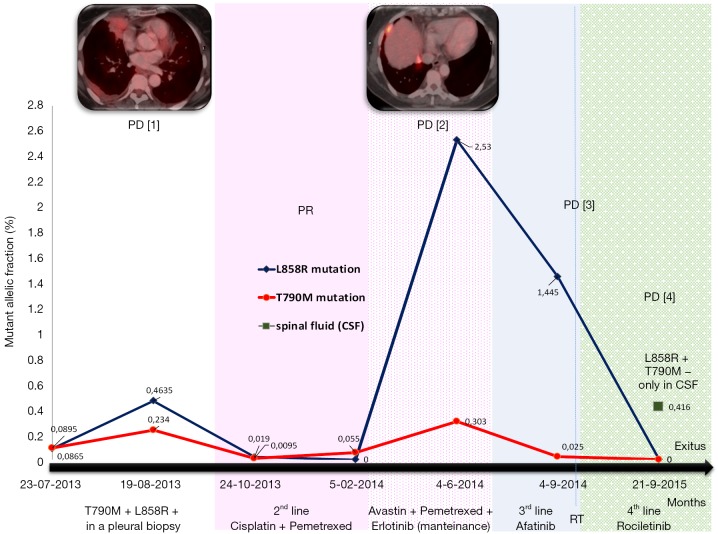
Figure 2.
Summary of evolution: correlation of clinical data and EGFR mutations status [L858R mutation, in blue; T790M mutation, in red, spinal fluid (CSF) in green] in a serial ctDNA analysis. PD [1]: pleural progression, PD [2]: pleural and lung progression, PD [3]: lung and brain progression, PD [4]: brain progression. Different lines of treatments were indicated with specific colors. PD, progression disease; PR, partial response; CSF, cerebrospinal fluid; EGFR, epidermal growth factor receptor; ctDNA, circulating tumor DNA.
In June 2014, a new cfDNA analysis showed a significant increase in L858R and T790M mutations in blood. A PET/CT scan confirmed a new progression of pleural and lung disease. Consequently, a third line therapy was started with afatinib in July 2014 with a rapid progression of the disease that included an increased size of lung tumor and the appearance of two new brain lesions. On Sept 2014, a new bronchoscopy was performed that confirmed the presence of adenocarcinoma upon pathological examination. After local treatment of the brain metastases with Gamma knife, the patient was enrolled in the CO-1831 trial on January 23rd 2015 and treated with rociletinib. Within three weeks she improved dramatically, and the CT on February 26th showed a partial response. In spite of this initial partial response, the brain MR performed 2 months later showed new multiple brain metastases, while lung disease response was maintained. In consequence, the patient received treatment with palliative whole brain irradiation from June 22nd and July 14th.
At this point, no EGFR mutations were detected in cfDNA purified from blood, but the analysis of ctDNA obtained from cerebrospinal fluid (CSF) determined the presence of the L858R mutation. The T790M mutation was absent. Interestingly, the cytological analysis was negative for adenocarcinoma infiltration on CSF.
The patient developed symptoms of meningeal carcinomatosis, leading to exit 2 months later, December 2015, under supportive care at hospice.
Discussion
The acquisition of new molecular alterations has been reported as a key mechanism in the development of acquired resistance to specific TKIs, such as EGFRi. EGFR mutant patients treated with first or second generation TKIs frequently develop resistance through T790M mutation in the exon 20 of EGFR (14). Therefore, serial tumor biopsies would be necessary for the identification of underlying drug resistance mechanisms in order to select specifically directed therapeutic options following disease progression. However, detection of these alterations in tumor tissue samples from NSCLC patients has significant limitations, including tumor heterogeneity and the difficulties associated with obtaining samples by invasive procedures. Since cfDNA is a BioSource that enables noninvasive identification of molecular changes, genomic analysis of ctDNA released from cancer cells into the blood has been proposed as a useful and practical method to detect dynamic molecular changes in the disease.
Data from different studies recently published suggest that clinical monitoring using cfDNA is useful for lung cancer patients, not only in terms of diagnosis, but also for evaluation of drug response and acquired resistance (15-17). In all of these studies, the use of highly sensitive and specific techniques is essential for the reliable detection in cfDNA of somatic mutations that can be potential targets for therapy. In the clinical case presented here, we used an in-house, highly sensitive (75.8% vs. paired tissue) and specific (100%) methodology for the analysis of EGFR mutations in cfDNA (PNA-mediated 5’ nuclease real time Taqman PCR) (12). This methodology also allows quantifying the mutant allelic fractions.
In the present case report, a good correlation was observed between the clinical evolution of the patient and the monitorization of EGFR mutations on ctDNA isolated from serum/plasma or CSF, as summarized in Figure 2. Using a similar highly sensitive methodology (Droplet digital PCR), another group of investigators recently demonstrated the feasibility of serial monitoring of EGFR mutations in cfDNA, and correlated it with the clinical evolution of the disease (15-17). However, there are some areas of concern still exist and this case report casts light on some of them.
First, the possibility of detecting an early progression or relapse of the disease in asymptomatic patients using ctDNA for analysis of EGFR mutations. In the present case, progression to the first generation TKI erlotinib was established by detecting L858R and T790M mutations in ctDNA, which was later confirmed by image techniques. Also, after second line treatment response, progression was manifested as a reappearance of L858R and T790M mutations in ctDNA isolated from blood.
Second, early identification of response, with rapid disappearance of EGFR mutations in cfDNA was found, matching response periods to chemotherapy and to rocelitinib.
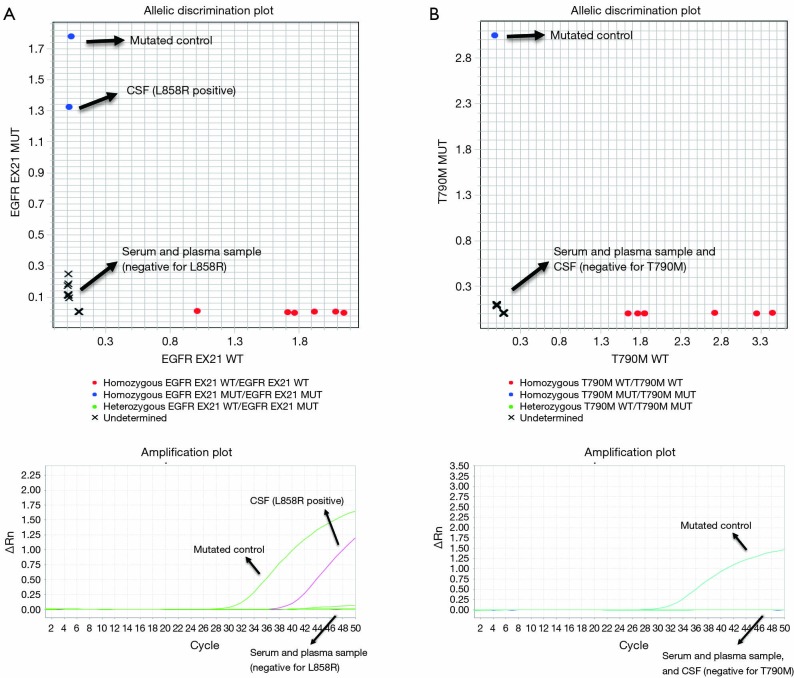
Third, the identification of progression exclusively at CNS, while the response was maintained in extracranial metastases, is a challenging situation. Analysis of ctDNA from CSF could be helpful, especially as it has been demonstrated that this technique has higher sensitivity for capturing molecular alterations than analysis in ctDNA obtained from blood. In the case reported here, we were able to detect the presence of L858R mutation exclusively in ctDNA obtained from CSF analysis. T790M was not found, as it has been described in patients under treatment with third-generation TKIs (Figure 3) (7,9,18).
Figure 3.
Analysis of ctDNA from CSF. (A) Allelic discrimination plots and amplification curves of L858R mutation in ctDNA from serum and plasma samples versus CSF, showing a positive amplification only in CSF; (B) allelic discrimination plots and amplification curves of T790M mutation in ctDNA from serum and plasma samples versus CSF, without positive amplification in none of them. CSF, cerebrospinal fluid; ctDNA, circulating tumor DNA.
Conclusions
Analyses of circulating EGFR mutations have important clinical implications and can be a useful surrogate biomarker of response to therapy and early detection of mechanisms of resistance to TKIs, in advance of clinically detection. Specifically, the status of EGFR T790M can be identified non-invasively (by sensitive methodologies), and potentially used to guide subsequent treatment decisions. In addition, CSF can be employed as an alternative ‘liquid biopsy source’ with applications for the management of brain tumors and metastases.
Acknowledgements
The authors thanks Kate Williams for the English corrections support, and Ana Perez-Rosado, for processing blood samples.
Informed Consent: Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
Footnotes
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- 1.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. 10.1056/NEJMoa0904554 [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Gervais R, Vergnenegre A, et al. Erlotinib vs chemotherapy (CT) in advanced non-small-cell lung cancer (NSCLC) patients (p) with epidermal growth factor receptor (EGFR) activating mutations: Interim results of the European Tarceva® vs Chemotherapy (EURTAC) phase III randomized trial. J Clin Oncol 2011;29:abstr 7503.
- 3.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 6.Lee CK, Brown C, Gralla RJ, et al. Impact of EGFR inhibitor in non-small cell lung cancer on progression-free and overall survival: a meta-analysis. J Natl Cancer Inst 2013;105:595-605. 10.1093/jnci/djt072 [DOI] [PubMed] [Google Scholar]
- 7.Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med 2015;372:1700-9. 10.1056/NEJMoa1413654 [DOI] [PubMed] [Google Scholar]
- 8.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. 10.1158/2159-8290.CD-15-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. 10.1038/nature12065 [DOI] [PubMed] [Google Scholar]
- 11.Spindler KL, Pallisgaard N, Andersen RF, et al. Changes in mutational status during third-line treatment for metastatic colorectal cancer--results of consecutive measurement of cell free DNA, KRAS and BRAF in the plasma. Int J Cancer 2014;135:2215-22. 10.1002/ijc.28863 [DOI] [PubMed] [Google Scholar]
- 12.Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. 10.1001/jamaoncol.2014.257 [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Cao M, Mayo-de-Las-Casas C, Molina-Vila MA, et al. BRAF mutation analysis in circulating free tumor DNA of melanoma patients treated with BRAF inhibitors. Melanoma Res 2015;25:486-95. 10.1097/CMR.0000000000000187 [DOI] [PubMed] [Google Scholar]
- 14.Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med 2013;368:842-51. 10.1056/NEJMra1204892 [DOI] [PubMed] [Google Scholar]
- 15.Wei Z, Shah N, Deng C, et al. Circulating DNA addresses cancer monitoring in non small cell lung cancer patients for detection and capturing the dynamic changes of the disease. Springerplus 2016;5:531. 10.1186/s40064-016-2141-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JY, Qing X, Xiumin W, et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02). Oncotarget 2016;7:6984-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. 10.1158/1078-0432.CCR-13-2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Mattos-Arruda L, Mayor R, Ng CK, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 2015;6:8839. 10.1038/ncomms9839 [DOI] [PMC free article] [PubMed] [Google Scholar]