Abstract
Obtaining a biopsy of solid tumors requires invasive procedures that strongly limit patient compliance. In contrast, a blood extraction is safe, can be performed at many time points during the course disease and encourages appropriate therapy modifications, potentially improving the patient’s clinical outcome and quality of life. Fusion of the tyrosine kinase genes anaplastic lymphoma kinase (ALK), C-ROS oncogen 1 (ROS 1), rearranged during transfection (RET) and neurotrophic tyrosine kinase 1 (NTRK1) occur in 1–5% of lung adenocarcinomas and constitute therapeutic targets for tyrosine kinase inhibitors. In addition, a MET splicing variant of exon 14, has been reported in 2–4% of lung adenocarcinoma and recent studies suggests that targeted therapies inhibiting MET signaling would be beneficial for patients with this alteration. In this review, we will summarize the new techniques recently developed to detect ALK, RET, ROS and NTRK1 fusions and MET exon 14 splicing variant in liquid biopsy using plasma, serum, circulating tumor cells (CTCs), platelets and exosomes as starting material.
Keywords: Lung cancer, splice variant, liquid biopsy, fusion gene
Introduction
Lung cancer is the leading cause of cancer death worldwide and non-small cell lung cancer (NSCLC) accounts for approximately 80–85% of cases (1). The majority of the patients are diagnosed when the disease is locally advanced or metastatic and the overall 5-year survival rate is less than 20% (2). In the last years, driver alterations have been discovered in several oncogenes (3) such as KRAS, EGFR, ALK, MET, BRAF, PIK3CA, ROS1, HER2, RET, PTEN, NRG1, NTRK1, DDR2 or FGFR1. These alterations render the tumor sensitive to targeted agents that, in most cases, have significantly better clinical outcomes compared to standard chemotherapy (4-6).
Among the above mentioned driver alterations, ALK, ROS-1 or RET fusion transcripts occur in 1–5% of stage IIIB-IV lung adenocarcinomas and constitute therapeutic targets for tyrosine kinase inhibitors such as crizotinib or alectinib. The splicing variant of MET exon 14 is present in 2–4% of advanced NSCLC (3,7) and recent studies in small series of patients have demonstrated the efficacy of MET tyrosine kinase inhibitors in this population (8).
Tissue biopsy is the traditional source of material for tumor genotyping. However, tissue specimens are not always available or tumor material may be insufficient for molecular testing, especially when cancer is diagnosed at advanced stages on small biopsy specimens. On other occasions, due to tumor location or small size, tissue sampling can be challenging and potentially risky, particularly in extensively treated patients (9). But, even when available, tissue biopsies have other limitations. A single biopsy at presentation does not necessarily reflect the molecular characteristics of the tumor at the time of disease progression, highlighting the need of additional, more dynamic and less invasive sources for continuous genotyping of cancer patients (10).
Circulating tumor cells (CTCs), exosomes and nucleic acid fragments such as cell-free and platelet-bound RNAs, and circulating cell-free DNA (cfDNA) (11) have emerged as useful biosources that are complementary or even alternative to tissue biopsy when a tissue biopsy is not available. The non-invasive liquid biopsies allow frequent sampling and treatment monitoring (12).
EML4-ALK fusions
The EML4-ALK fusion (13) is the result of a chromosome rearrangement between the N-terminal portion of the EML4 gene and the tyrosine kinase domain of the ALK gene. ALK rearrangements are more prevalent in younger NSCLC patients, females, never or former light smokers (14). EML4-ALK positive patients derive clinical benefit from ALK tyrosine kinase inhibitors such as crizotinib (6), ceritinib (15) or alectinib (16). However, despite initial good responses, all patients eventually relapse due to the emergence of different mechanisms of resistance (17).
EML4-ALK fusion transcripts in CTCs
CTCs are viable cells released into the circulatory system (18), but they constitute a minor fraction of the cell population in the peripheral blood of cancer patients, both in terms of absolute (<10 cells/mL) and relative numbers compared to other blood cells (one CTC per 106–107 leukocytes) (19). CTC counting has been reported to have a prognostic value in metastatic breast (20), colorectal (21) and castration resistant prostate cancer (22). They may also have a role in other solid tumors (20), including NSCLC (23) and SCLC (24).
Several techniques have been developed for the isolation and characterization of CTCs that differ in enrichment, staining and detection methods (25) as well as sensitivity, specificity and reproducibility (26). CTCs enrichment can be based on physical or immunomagnetic approaches. Magnetic beads and ferrofluid-based systems are examples of magnetic affinity cell sorting while filters and density gradients are examples of physical approaches (27). Both of them have been commercialized. The CellSearch System (CSS; Veridex, Warren, USA) is a semi-automated CTCs detection method, approved by the FDA, where CTCs are enriched enrichment using magnetic microbeads covered by antibodies against the epithelial marker EpCAM (epithelial cell adhesion molecule). The Isolation by Size of Epithelial Tumor Cells (ISET) is a kit where CTCs are isolated by a combination of enrichment and filtration. Regarding the identification of CTCs, it can be performed either by direct cytometry techniques like immunocytochemistry, immunofluorescence and flow cytometry or by indirect, nucleic acid-based methods (26,27).
Using the ISET technology for CTCs isolation, Ilie et al. (11) were able to detect ALK-gene rearrangements on CTCs by immunochemistry (IHC) and fluorescence in situ hybridization (FISH). When comparing the results with paired biopsies in 87 lung adenocarcinoma patients, they found a total of five patients with ALK-gene fusions by FISH and strong ALK protein expression by IHC on both CTCs and in the corresponding tumor sample. This seminal study demonstrated that CTCs can be a reliable source for the detection of ALK-gene rearrangement in lung cancer patients (11). Pailler et al. analyzed by filter-adapted FISH the CTCs of 18 ALK-positive patients isolated with the ISET technology, finding four or more ALK-rearranged CTCs per milliliter of blood in all cases (28). In contrast, only one or zero ALK-rearranged CTCs where detected in 14 ALK negative NSCLC patients. In the same study, variations in the levels of ALK-rearranged CTC were detected when monitoring over time five patients treated with crizotinib (28).
Enrichment and characterization techniques other than ISET have also been successfully used for CTC isolation in order to detect EML4-ALK fusions by FISH (29,30). In a recent study, an antibody-independent system was employed for the isolation of CTCs from 31 patients (14 ALK positive, 12 ALK-negative lung cancer and five healthy controls) with paired tissue biopsy (31). Using CTCs of blood samples from healthy donors, a ‘false positive’ cutoff for ALK FISH of ≤2 cells per 1.88 mL of blood was established and validated. Using this cut-off, a ≥90% concordance was found between the detection of ALK rearrangements in CTCs versus tissue biopsy. The same study reported differences in the ALK pattern between CTCs collected at baseline and at the time of disease progression in one patient treated with crizotinib (31). Finally, using the NanoVelcro chip system for CTCs isolation, ≥3 ALK-rearranged CTCs per mL of blood could be detected in ALK-positive patients, versus ≤2 in ALK-negative cases (32).
EML4-ALK fusion transcripts in platelets
Isolation of other blood components like plasma, serum or platelets is simpler than CTCs and can also provide material for biomarker analysis (33). Recent studies have shown that platelets can sequester tumor related RNA by a microvesicle dependent mechanism, resulting in the generation of tumor-educated platelets (TEPs) (34,35). Our group has demonstrated 65% sensitivity and 100% specificity for the detection of EML4-ALK rearrangements in RNA isolated from platelets in patients at presentation (36). During the course of the disease, the presence of EML4-ALK fusion transcripts in platelets correlated with clinical outcome to crizotinib treatment response. Monitoring of EML4-ALK rearrangements in the platelets of one index patient revealed re-appearance of fusion transcripts associated with crizotinib resistance two months prior to radiographic disease progression. Our data showed that platelets are a valuable source for the non-invasive detection of EML4-ALK fusion transcripts in lung cancer patients, useful for predicting and monitoring outcome to ALK targeted therapies (36).
EML4-ALK fusion transcripts in plasma
cfDNA purified from plasma is commonly used for mutational analysis in advanced NSCLC and other malignancies (37). However, RNA circulating in plasma has not been so widely used for gene fusion detection due to a variety of reasons. Unlike cfDNA, cfRNA degrades very quickly and plasma samples need to be processed rapidly after blood extraction. The usual procedure is adding an RNA preservative such as Trizol and immediately freezing the sample at −80 °C. However, this procedure is not easily accessible to many clinical sites.
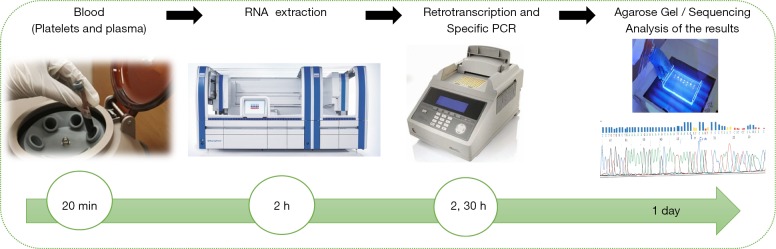
Our research group has 5-year experience in detection of EML4-ALK fusion transcripts by reverse transcription-polymerase chain reaction (RT-PCR) in plasma, and we have demonstrated 22% sensitivity and 100% specificity for the detection of this alteration (36). We are currently working to improve the method for plasma cfRNA extraction and EML4-ALK fusion transcript analysis. Submitting the plasma samples to automated extraction within 24 h after blood collection, plasma cfRNA can be obtained in the shortest possible time avoiding degradation and without the need of an RNA preservative. Using these new protocol, we have analyzed more than 200 plasma samples and we have significantly improved the sensitivity of RT-PCR for the detection of EML4-ALK fusion transcripts (unpublished data) (Figure 1).
Figure 1.
Workflow of the fusion gene analyses in plasma and platelets-derived RNA using automatic extraction followed by RT-PCR. RT-PCR, reverse transcription-polymerase chain reaction.
KIF5B-RET, ROS-1 and NTRK gene fusions
Additional fusions involving ROS1, RET and NTRK oncogenes have been identified in 1–2% of advanced NSCLC patients in the case of RET and ROS1 (3) and 3.3% for NTRK1 (38,39). Despite these low frequencies, these alterations offer the possibility for targeted therapies with better clinical outcomes than chemotherapy.
Similarly to EML4-ALK, ROS1 rearrangements can be detected on the CTCs of NSCLC patients using filter-adapted-fluorescent in situ hybridation (FA-FISH). In a recent study, ROS1 rearrangements were studied in the CTCs of patients with paired tumor tissue (31). The median number of ROS1-rearranged CTCs at baseline was 34.5 per 3 mL blood (range, 24–55) in ROS1 positive patients versus 7.5 per 3 mL blood (range, 7–11) in ROS1-negative patients. New generation platforms, such as Cancer Personalized Profiling by deep Sequencing (CAPP-Seq) (40) or NEOliquid® (41), have also been used to detect gene fusions in the circulating free DNA purified from advanced NSCLC patients. NEOliquid® is a next-generation sequencing assay, based on hybrid-capture, that covers clinically relevant genomic alterations, such as point mutations, small insertions and deletions, selected gene fusions and copy number alterations within a panel of more than 30 genes. Using NEOliquid®, Heukamp et al. identified KIF5B-RET gene fusions in two patients previously tested negative for the rest of mutations (41).
Alternative splicing variants
Alternative splicing in cancer is emerging as a growing and promising field in basic and translational oncology and is an object of active investigation (42,43). Although alternative splicing in several genes have been associated with lung cancer (44), MET 14 is so far the only splicing variant with clinical relevance.
MET splicing variant in exon 14
c-MET alterations, including overexpression and amplification, have been described in a number of solid tumors such as papillary renal, gastric and lung cancer, and hepatocellular carcinoma (45-47). Somatic mutations affecting splice sites of exon 14 of the MET gene (METex14) were first reported in primary lung cancer specimens and in a lung cancer cell line (48,49). These METex14 alterations were shown to promote RNA-splicing-based skipping of MET exon 14, which results in activation of MET kinase activity through a unique mechanism (50).
MET ex14 alterations in NSCLC are not necessarily associated with MET amplification occur in approximately 3% of lung adenocarcinoma cases (51) and have also been observed in neuroblastoma (52) and gastric (53) cancer cell lines. Mutational events of MET leading to exon 14 skipping are frequent and potentially targetable events. In vitro preclinical studies indicated that MET-targeted agents can counteract oncogenesis resulting from MET exon 14 loss and the efficacy of these agents in NSCLC patients with MET 14 positive tumors has recently been demonstrated (50,54).
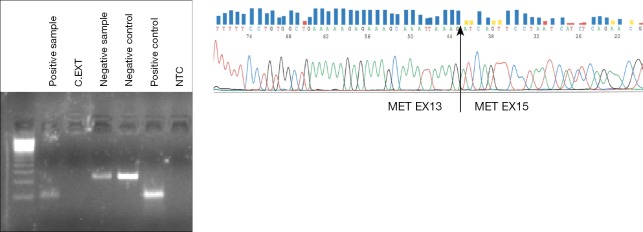
There are no reports so far about detection of exon 14 MET splice variant in liquid biopsies, but great progress is expected in the near future due to the relevance of this alteration for the treatment of lung cancer patients. In our laboratory, we have started to analyze the exon 14 MET splice variant by RT-PCR in RNA isolated from plasma and platelets with the same methodology used for EML4-ALK rearrangements. Recently, we have detected the alteration in the platelet-derived RNA of a NSCLC patient who harbored the MET exon 14 splicing variant in tumor tissue (Figure 2). The patient attained a partial response to crizotinib (unpublished data).
Figure 2.
Agarose gel (left) and sequence (right) of the RT-PCR products products of a patient positive for exon 14 MET splicing variant in platelet-derived RNA. RT-PCR, reverse transcription-polymerase chain reaction.
Conclusions
Gene fusions and splicing variants have been recognized as tumor drivers for more than three decades, providing new targets for personalized therapy. The accurate detection of these alterations in blood will be a significant step for treatment selection at the time of the diagnosis, monitoring treatment outcome and predicting disease progression in NSCLC patients.
Acknowledgements
None.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Sholl LM, Aisner DL, Allen TC, et al. Liquid Biopsy in Lung Cancer: A Perspective From Members of the Pulmonary Pathology Society. Arch Pathol Lab Med 2016;140:825-9. 10.5858/arpa.2016-0163-SA [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Karachaliou N. Large-scale screening for somatic mutations in lung cancer. Lancet 2016;387:1354-6. 10.1016/S0140-6736(15)01125-3 [DOI] [PubMed] [Google Scholar]
- 4.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 6.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. 10.1056/NEJMoa1214886 [DOI] [PubMed] [Google Scholar]
- 7.Seki Y, Mizukami T, Kohno T. Molecular Process Producing Oncogene Fusion in Lung Cancer Cells by Illegitimate Repair of DNA Double-Strand Breaks. Biomolecules 2015;5:2464-76. 10.3390/biom5042464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J Thorac Oncol 2016;11:1493-502. 10.1016/j.jtho.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 9.Vanderlaan PA, Yamaguchi N, Folch E, et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer 2014;84:39-44. 10.1016/j.lungcan.2014.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. 10.1093/annonc/mds137 [DOI] [PubMed] [Google Scholar]
- 12.Marchetti A, Palma JF, Felicioni L, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol 2015;10:1437-43. 10.1097/JTO.0000000000000643 [DOI] [PubMed] [Google Scholar]
- 13.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 14.Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 2011;12:1004-12. 10.1016/S1470-2045(11)70232-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim DW, Mehra R, Tan DS, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol 2016;17:452-63. 10.1016/S1470-2045(15)00614-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ou SH, Ahn JS, De Petris L, et al. Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J Clin Oncol 2016;34:661-8. 10.1200/JCO.2015.63.9443 [DOI] [PubMed] [Google Scholar]
- 17.Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. 10.1126/scitranslmed.3003316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MY, Oskarsson T, Acharyya S, et al. Tumor self-seeding by circulating cancer cells. Cell 2009;139:1315-26. 10.1016/j.cell.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross AA, Cooper BW, Lazarus HM, et al. Detection and viability of tumor cells in peripheral blood stem cell collections from breast cancer patients using immunocytochemical and clonogenic assay techniques. Blood 1993;82:2605-10. [PubMed] [Google Scholar]
- 20.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. 10.1056/NEJMoa040766 [DOI] [PubMed] [Google Scholar]
- 21.Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 22.de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Wang X, He H, et al. Combination of circulating tumor cells with serum carcinoembryonic antigen enhances clinical prediction of non-small cell lung cancer. PLoS One 2015;10:e0126276. 10.1371/journal.pone.0126276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. 10.1200/JCO.2010.33.3716 [DOI] [PubMed] [Google Scholar]
- 25.Riethdorf S, Pantel K. Disseminated tumor cells in bone marrow and circulating tumor cells in blood of breast cancer patients: current state of detection and characterization. Pathobiology 2008;75:140-8. 10.1159/000123852 [DOI] [PubMed] [Google Scholar]
- 26.Andreopoulou E, Yang LY, Rangel KM, et al. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. Int J Cancer 2012;130:1590-7. 10.1002/ijc.26111 [DOI] [PubMed] [Google Scholar]
- 27.Aurilio G, Sciandivasci A, Munzone E, et al. Prognostic value of circulating tumor cells in primary and metastatic breast cancer. Expert Rev Anticancer Ther 2012;12:203-14. 10.1586/era.11.208 [DOI] [PubMed] [Google Scholar]
- 28.Pailler E, Adam J, Barthélémy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. 10.1200/JCO.2012.44.5932 [DOI] [PubMed] [Google Scholar]
- 29.Wu S, Liu Z, Liu S, et al. Enrichment and enumeration of circulating tumor cells by efficient depletion of leukocyte fractions. Clin Chem Lab Med 2015;53:337. 10.1515/cclm-2015-9999 [DOI] [PubMed] [Google Scholar]
- 30.Khoo BL, Warkiani ME, Tan DS, et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PLoS One 2014;9:e99409. 10.1371/journal.pone.0099409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan CL, Lim TH, Lim TKh, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 2016;7:23251-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He W, Xu D, Wang Z, et al. Detecting ALK-rearrangement of CTC enriched by nanovelcro chip in advanced NSCLC patients. Oncotarget 2016. [Epub ahead of print].27029032 [Google Scholar]
- 33.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 34.Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3. 10.1182/blood-2011-03-344408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Best MG, Sol N, Kooi I, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28:666-76. 10.1016/j.ccell.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. 10.1001/jamaoncol.2014.257 [DOI] [PubMed] [Google Scholar]
- 38.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375-7. 10.1038/nm.2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013;19:1469-72. 10.1038/nm.3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heukamp L, Menon R, Müller J, et al. 65P NEOliquid: Detection of KIF5B-RET fusions in liquid biopsy samples. J Thorac Oncol 2016;11:S82-3. 10.1016/S1556-0864(16)30178-227198421 [DOI] [Google Scholar]
- 42.Kalnina Z, Zayakin P, Silina K, et al. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer 2005;42:342-57. 10.1002/gcc.20156 [DOI] [PubMed] [Google Scholar]
- 43.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res 2004;64:7647-54. 10.1158/0008-5472.CAN-04-1910 [DOI] [PubMed] [Google Scholar]
- 44.Pio R, Montuenga LM. Alternative splicing in lung cancer. J Thorac Oncol 2009;4:674-8. 10.1097/JTO.0b013e3181a520dc [DOI] [PubMed] [Google Scholar]
- 45.Wang WL, Healy ME, Sattler M, et al. Growth inhibition and modulation of kinase pathways of small cell lung cancer cell lines by the novel tyrosine kinase inhibitor STI 571. Oncogene 2000;19:3521-8. 10.1038/sj.onc.1203698 [DOI] [PubMed] [Google Scholar]
- 46.Maulik G, Kijima T, Ma PC, et al. Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clin Cancer Res 2002;8:620-7. [PubMed] [Google Scholar]
- 47.Ma PC, Maulik G, Christensen J, et al. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev 2003;22:309-25. 10.1023/A:1023768811842 [DOI] [PubMed] [Google Scholar]
- 48.Ma PC, Kijima T, Maulik G, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res 2003;63:6272-81. [PubMed] [Google Scholar]
- 49.Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res 2006;66:283-9. 10.1158/0008-5472.CAN-05-2749 [DOI] [PubMed] [Google Scholar]
- 50.Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. 10.1158/2159-8290.CD-15-0285 [DOI] [PubMed] [Google Scholar]
- 51.Onozato R, Kosaka T, Kuwano H, et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 2009;4:5-11. 10.1097/JTO.0b013e3181913e0e [DOI] [PubMed] [Google Scholar]
- 52.Yan B, Lim M, Zhou L, et al. Identification of MET genomic amplification, protein expression and alternative splice isoforms in neuroblastomas. J Clin Pathol 2013;66:985-91. 10.1136/jclinpath-2012-201375 [DOI] [PubMed] [Google Scholar]
- 53.Asaoka Y, Tada M, Ikenoue T, et al. Gastric cancer cell line Hs746T harbors a splice site mutation of c-Met causing juxtamembrane domain deletion. Biochem Biophys Res Commun 2010;394:1042-6. 10.1016/j.bbrc.2010.03.120 [DOI] [PubMed] [Google Scholar]
- 54.Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. 10.1158/2159-8290.CD-14-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]