Abstract
The acknowledged ability of synthetic materials to induce cell-specific responses regardless of biological supplies provides tissue engineers with the opportunity to find the appropriate materials and conditions to prepare tissue-targeted scaffolds. Stem and mature cells have been shown to acquire distinct morphologies in vitro and to modify their phenotype when grown on synthetic materials with tunable mechanical properties. The stiffness of the substrate used for cell culture is likely to provide cells with mechanical cues mimicking given physiological or pathological conditions, thus affecting the biological properties of cells. The sensitivity of cells to substrate composition and mechanical properties resides in multiprotein complexes called focal adhesions, whose dynamic modification leads to cytoskeleton remodeling and changes in gene expression. In this study, the remodeling of focal adhesions in human mesenchymal stem cells in response to substrate stiffness was followed in the first phases of cell–matrix interaction, using poly-ε-caprolactone planar films with similar chemical composition and different elasticity. As compared to mature dermal fibroblasts, mesenchymal stem cells showed a specific response to substrate stiffness, in terms of adhesion, as a result of differential focal adhesion assembly, while their multipotency as a bulk was not significantly affected by matrix compliance. Given the sensitivity of stem cells to matrix mechanics, the mechanobiology of such cells requires further investigations before preparing tissue-specific scaffolds.
Keywords: substrate stiffness, stem cell-matrix interaction, mesenchymal stem cells
1. Introduction
The translation of tissue engineering protocols from the laboratory bench to the bedside calls for full understanding of the interaction between living cells and the synthetic materials to be used as cell delivery systems. Indeed, cellular processes, including cell survival, adhesion, migration, proliferation and differentiation depend on the tissue or matrix microenvironment [1]. For stem and differentiated cells, matrix stiffness and nanopatterning are known to influence the formation of focal adhesions and cytoskeletal structures [2–6], i.e. matrix structural parameters directly affect cell functions. As a result, cell response to mechanical forces is crucial in embryo development and pathogenesis in the adult [7–9].
Cells sense matrix elasticity and nanostructure by pulling against their extracellular matrix (ECM), after which outside-in mechanotransduction starts, inducing the cells to generate the counteracting force (traction force), and remodeling cell morphology and gene expression [8]. Substrate stiffness has been demonstrated to be a key parameter in the control of cell spreading and cytoskeleton assembly [10], as well as directional motility [2]. Remarkably, changes in cell sensitivity to matrix resistance are considered prognostic of tumor transformation, a metastatic phenotype being required for tumor cells to migrate [11]. When cells adhere to 2D solid surfaces in vitro, force transduction is achieved by the formation of focal adhesions, bridging between ECM and actin cytoskeleton [12] (figure 1). On the contrary, differences in focal adhesions formation and turnover have been demonstrated in 3D cultures, such arrangement supposed to be more similar to that experienced by the cells in vivo [13].
Figure 1.
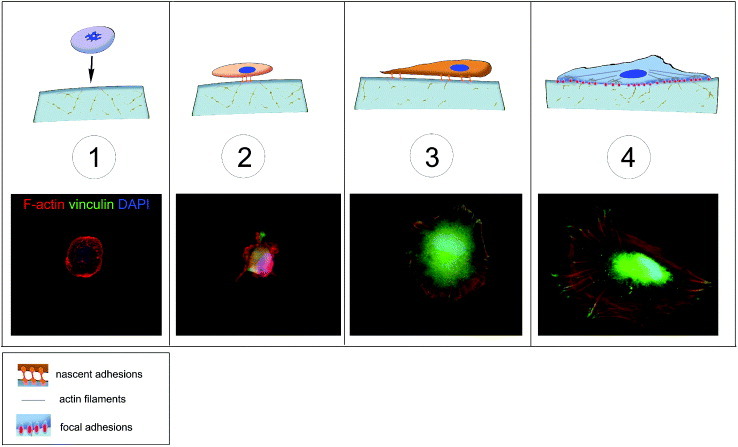
Schematic of cell adhesion. Cell adhesion is a complex process that can be divided into different phases. A floating cell (1) can physically contact with the substrate. In the first phase of cell–matrix interaction (2) the cell adheres passively and starts sensing the substrate. This phase is guided by gravity and followed by cell reshaping and the concomitant adhesion processes (3), resulting in an immature form of the focal adhesions (4). These structures are macromolecular complexes connecting the extracellular matrix to the cytoskeleton, the scaffold of the cell. A number of docking and shuttling proteins are involved in such a process.
Focal adhesions act as an integrated center for cell signaling, mechanosensing and force transduction to mediate cell attachment, spreading and motility in response to ECM composition [14]. They orchestrate adhesion maturation via their multiprotein complexes (e.g. talin, vinculin and paxillin), transducing the mechanical response to dynamic remodeling of actin cytoskeleton. Variations in composition or physical parameters of ECM, like elasticity and topography, influence cell shape and functions, because cell behavior depends on the mechanical feedback from the ECM stiffness [15]. Focal adhesions exhibit continuous remodeling and complex mechanosensitivity such that they form or enlarge when mechanical force increases, and shrink or disassemble when it decreases [14]. Hence, focal adhesions serve as indicators of cell–matrix interaction, and cell response to matrix composition can be studied dynamically by following the recruitment of proteins like vinculin, talin, paxillin to the basal surface of adherent cells.
The interaction between living cells and the biomaterials to be used in tissue engineering applications represents a crucial step in the definition of suitable scaffolds for targeted organ repair. The materials so far proposed as delivery systems for tissue engineering applications are of natural origin (e.g. collagen, laminin, fibrin and alginate) or synthesized on purpose. Among the synthetic materials, poly-ε-caprolactone (PCL) [16], poly(glycerol) sebacate (PGS) [17], polyethylene glycol (PEG) [18], polylactic acid (PLA), polyglycolic acid (PGA), and their copolymers such as poly-lactic-co-glycolic acid (PLGA) exhibit high processability, which allows an easy tuning of their mechanical and structural properties. PLGA [19], PLA [20] and PCL, [12, 21] have been widely used for cell growth [22, 23]. As a bulk, polyesters display mechanical properties that are very different from those encountered in vivo, thus they could be perceived as foreign materials by living cells.
PCL is an elastomeric polyester, which has been approved for biomedical applications by the US Food and Drug Administration (FDA) and proven to be suitable for cell culture. We have designed and synthesized biocompatible PCL planar films that exhibit similar chemical composition and surface stiffness in a supra-physiological range from 0.9 to 133 MPa at 37 °C [24, 25]. These films were used in this study to investigate cellular responses to matrix stiffness in terms of focal adhesion maturation. Therefore, the expression of proteins involved in the formation and stabilization of focal adhesions was followed in human mesenchymal stem cells (hMSCs) in the first phases of cell–matrix interaction to exclude the contributions of cell-to-cell contact and cell proliferation. As a control, the maturation of focal adhesions in differentiated normal human dermal fibroblasts (NHDFs) was also monitored as a function of substrate stiffness.
2. Materials and methods
2.1. Preparation of PCL layers with different stiffnesses
PCL planar layers were produced as previously described [24]. The films were composed of 2 mixed macromonomers (2-branched and 4-branched PCL) in different concentrations, as to obtain 4 distinct formulations displaying Young moduli of 0.9 ± 0.1, 1.5 ± 0.2, 50 ± 3 and 133 ± 9 MPa and a constant contact angle of 97 ± 3°.
2.2. Cell culture
NHDFs (PromoCell GmbH, Heidelberg, Germany) were maintained in growth medium consisting of Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, California, US) supplemented with 10% fetal bovine serum (FBS, Invitrogen), 1% Antibiotic-Antimycotic (Gibco Invitrogen). hMSCs (Lonza, Walkersville , Maryland, US) were grown in fully supplemented Mesenchymal Stem Cell Basal Medium (Lonza) according to the manufacturer's instructions. Early passages of both cell types were used in experiments (passage 3–7). Cell culture medium was changed every third day. To obtain cells expressing talin fused with a green-fluorescent protein (GFP), hMSCs or NHDFs were transduced with CellLight® talin-GFP (Molecular Probes Invitrogen) according to manufacturer's instructions.
2.3. Matrigel assays
hMSCs were seeded at a concentration of 3.0 × 104 cells cm−2 in growth-factor-reduced MATRIGEL™ (BD Biosciences, Franklin Lakes, New Jersey, US) and allowed to branch and migrate for 3, 8 and 24 h. At these time-points the samples were fixed and prepared for immunofluorescence analysis as described in section 2.4. To assess the multipotency of hMSCs, cells previously cultured for 7 days on 0.9 or 133 MPa PCL were detached and seeded on growth-factor-reduced MATRIGEL™ for 3 h. The process of tubulogenesis was monitored with a light microscope. The number of tubules formed and the average tubule length were calculated by manual identification of single tubules using ImageJ software.
2.4. Cell adhesion analyses
AlamarBlue® assay was used to study the adhesion rate of cells to PCL substrates with different stiffness. Fluorescence intensity of AlamarBlue® (Invitrogen) was directly proportional to the number of cells attached to the substrate, while floating cells did not contribute significantly. NHDFs and hMSCs were seeded at low density (2 × 103 cells cm−2) in growth medium with 10% AlamarBlue® onto tissue culture polystyrene (TCPS) or PCL substrates. Cells detached from the substrates were measured by AlamarBlue® after 3 and 8 h of culture and used as internal control. Fluorescence intensity was measured by an ARVO MX1420 multilabel counter (Perkin Elmer, Waltham, Massachusetts, US) at F544/F590. Intensity readings were normalized by background intensity and calculated as percentage of the intensity obtained for TCPS at 3 h.
2.5. Differentiation assay
hMSCs were seeded on the PCL films at 2 × 104 cells cm−2. After 24 h the medium was replaced with differentiation medium (osteogenic, adipogenic, or chondrogenic, Lonza) and cultured for 14 days. Adipogenic differentiation was assessed by staining for the fatty acid binding protein 4 (FABP-4; R&D Systems, Minneapolis), while osteogenic differentiation was confirmed by Alizarin Red S staining (Sigma-Aldrich, Missouri, US). The occurrence of chondrogenic differentiation was assessed by Masson's trichrome staining (Sigma-Aldrich).
2.6. Immunofluorescence staining and microscopy
hMSCs and NHDFs on PCL were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for 5 min at room temperature. Following blocking for 30 min in 5% bovine serum albumin (BSA), the substrates were incubated with primary antibodies (vinculin produced in mouse, paxillin produced in rabbit; Sigma-Aldrich) for 1 h at room temperature, and stained with goat anti-mouse Alexa Fluor 488 (Invitrogen) for green, goat anti-mouse Alexa Fluor 546, or goat anti-rabbit Alexa Fluor 546 (Invitrogen) for red where appropriate for 1 h at room temperature (1:300). To visualize F-actin and nucleus, we used rhodamine-conjugated phalloidin (Invitrogen) and 4,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich), respectively. Samples were mounted on glass slides with 50% glycerol. Images were captured using a confocal laser scanner microscope (Leica SF5), after excitation at 405, 488 and 543 nm wavelengths for blue, green and red channels, respectively.
2.7. Statistical analysis
Results are presented as mean ± standard error, and are averages of the means of duplicated assays in at least three independent experiments. GraphPad Prism version 5.0 (GraphPad Software, Inc., San Diego, California, US) was used to plot the graphs of AlamarBlue® assay and to perform statistical analyses using one-way ANOVA followed by Dunnett's test. Differences were considered significant for P < 0.05.
3. Results and discussion
3.1. Soft biomaterials enhance adhesion of mesenchymal stem cells and fibroblast
Matrix elasticity has been shown to affect cell adhesion in a cell-specific manner [10]. As illustrated in figure 1, cell adhesion is a multistep process: in the first phase, the cell adheres passively and starts to sense the substrate. This phase is followed by a remodeling in cell shape and the concomitant formation of the adhesion complexes, which are immature forms of the focal adhesions. These structures are macromolecular complexes connecting the extracellular matrix to the cytoskeleton and have been proposed to be responsible for sensing the substrate composition and mechanical properties (figure 2).
Figure 2.
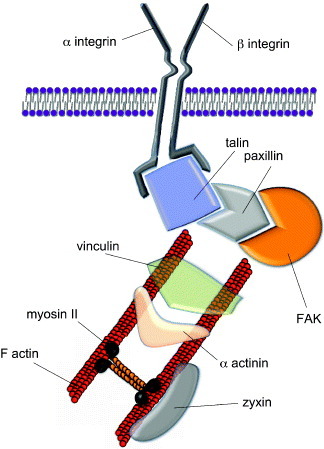
Structure of mature focal adhesions. Mature focal adhesions are macromolecular complexes composed of a number of proteins bridging between the extracellular matrix (ECM) and the cytoskeleton. The mechanical signals arising from the ECM are transduced via the integrins and a number of docking proteins (vinculin, talin, paxillin, zyxin etc) to the actin filaments. Myosin II is involved in cell tension sensing. Some of the proteins found in the focal adhesions can shuttle to the nucleus to act as co-activators of gene transcription. A prominent role in integrin signaling is played by focal adhesion kinase (FAK), a protein tyrosine kinase recruited at an early stage of focal adhesion formation and directly involved in the recruitment of proteins containing SH-2 and SH-3 domains.
To study the response of undifferentiated and mature cells to the substrate stiffness, hMSCs and NHDFs were cultured on PCL, and cell adhesion rates were measured using AlamarBlue® after 3 and 8 h of cell seeding (figure 3). The experimental setup was chosen on the basis of previous experiments showing that the first phase of cell–matrix interaction occurred at these time-points (data not shown). Our results showed that, when compared to the number of cells adhering to TCPS after 3 h of cell seeding, cell adhesion was most enhanced on 0.9 MPa PCL for both hMSCs and NHDFs. When the cell adhesion rates were compared between hMSCs and NHDFs, the number of cells attached to the substrates was approximately 4 times higher than on TCPS at 3 h for hMSCs, but only a 2.5-fold increase was found in NHDFs. The overall enhanced cell attachment indicated a higher sensitivity towards substrate stiffness for hMSCs than NHDFs. This dissimilar response of hMSCs and NHDFs could be attributed to differences in cell plasticity: hMSCs are undifferentiated cells that exhibit multipotency, whereas NHDFs are differentiated cells that have lost plasticity.
Figure 3.
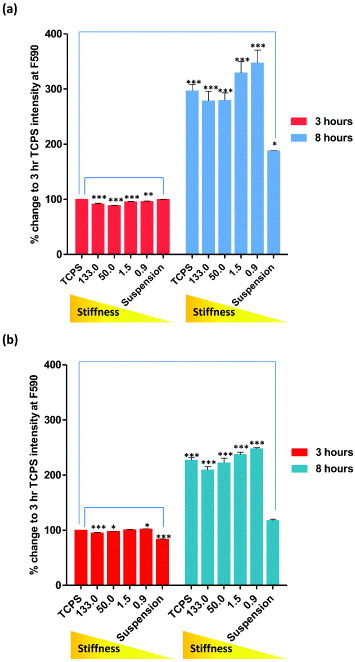
Cell adhesion rates of (a) hMSCs and (b) NHDFs assessed by AlamarBlue® assay. Cells were seeded onto PCL of increasing stiffness (0.9, 1.5, 50 and 133 MPa) and TCPS. Cells grown on low-adhesion plates were used as control (suspension). Fluorescent intensity of each sample was measured after 3 and 8 h of culture at F544/F590. Readings were normalized to the background intensity and presented as percentage change of TCPS after 3 h of culture.
3.2. Differential regulation of focal adhesion maturation in hMSCs and dermal fibroblasts on substrates with different stiffness
To further understand the differential adhesion rates observed in hMSCs and NHDFs, focal adhesion proteins were visualized using fluorescent immunostaining and confocal microscopy. The docking protein vinculin is preferentially found in the focal adhesions in cells grown on stiff substrates, while its cytoplasmic form prevails in cells embedded in 3D soft substrates [13]. Indeed, when hMSCs were cultured on growth-factor-reduced MATRIGEL™ for 3, 8 and 24 h, the cells branched and migrated inside the 3D hydrogel. In migrating cells embedded in this very soft gel (Young modulus in the Pa range), a clear rearrangement of cell cytoskeleton occurred, while vinculin was not detected in focal adhesions and in the cytoplasm (figure 4). A similar pattern was detected for paxillin and talin expression (data not shown). Consistent with previous reports, vinculin recruitment to focal adhesions occurs on substrates with a Young modulus higher that 200 kPa in epithelial cells [26].
Figure 4.
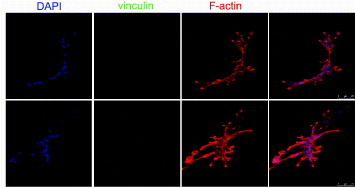
Human mesenchymal stem cells do not express vinculin protein when grown on a very compliant substrate. The cells were grown on growth-factor-reduced MATRIGEL™ for 8 h (upper line) and 24 h (central line) and stained for F-actin (red) and vinculin (green). Nuclei are counterstained with 4',6-diamidino-2-phenylindole (DAPI). The formation of tubules within the 3D microenvironment is accompanied by clear cytoskeletal rearrangement and morphology change, while vinculin expression could not be detected, as shown by higher-magnification images (bottom row).
On stiff substrates, vinculin recruitment to cell edges is associated with the stabilization of the adhesion processes, while migrating cells mainly express the cytoplasmic form of vinculin [27]. When stiff PCL polymers were challenged with human cells, we found that both hMSCs and NHDFs could spread fully within 8 h after seeding, with the formation of focal adhesions, and assembled cortical F-actin cytoskeleton on all PCL substrates tested (figure 5). Among the focal adhesion proteins examined, vinculin (figures 5(a), (b) and (d), paxillin and talin (figure 5(d)) formed streak-like aggregates at cell–matrix focal contacts in hMSCs on all the PCL substrates tested. This expression pattern showing paxillin, vinculin and talin co-localizing at cell edge and stabilizing cell–matrix interaction [13] was not paralleled by zyxin, a focal adhesion protein that can shuttle to the nucleus in response to mechanical stimuli [28, 29]. Our data showed that zyxin was localized exclusively in the nucleus of all PCL cultured–hMSCs (figure 5(d)). However, the formation of protein clusters and the degree of maturation at the cell–matrix contacts showed stiffness and time dependence.
Figure 5.
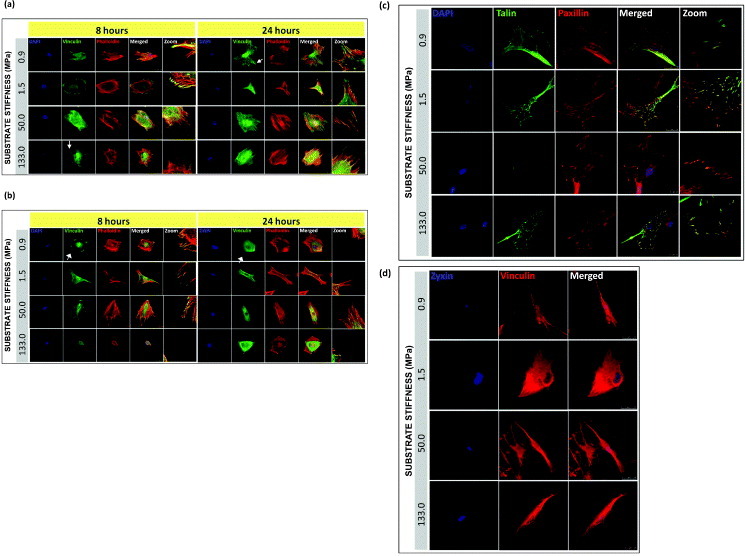
Localization and maturation of focal adhesions, actin cytoskeleton in hMSCs and NHDFs cultured on PCL. Confocal fluorescence micrographs of F-actin (phalloidin staining), vinculin, and nuclei (DAPI) for (a) hMSC and (b) NHDF, 8 and 24 h after seeding. White arrows indicate the formation of vinculin-rich spikes. Representative immunofluorescence images of single hMSC plated on PCL for 24 h, showing (c) co-localization of talin-GFP with paxillin and counterstained with DAPI, and (d) zyxin with vinculin. Scale bars: 25 μm.
In figure 5(a), when comparing vinculin distribution across the stiffness range at 8 and 24 h, we found that after the initial 8 h culturing, on the stiffest PCL (133 MPa), vinculin-containing dot-like adhesions were most abundant along the cell edge organized in typical spikes (indicated by the white arrows), whereas such pattern was found on the softest PCL (0.9 MPa) after 24 h of culturing. As a marker for focal complexes and focal adhesions [30], after the formation of initial adhesions [31, 32], vinculin was recruited to the focal complexes which were later matured to form focal adhesions at the cell–matrix contacts. Since the size of vinculin aggregates correlates with the amount of force generated at stable focal adhesions in stationary cells [33], cells cultured on stiffer substrates would experience a greater force exerted on them by the ECM, resulting in a higher rate of vinculin accumulation at the cellular periphery as compared to the softer substrates. However, after 24 h of culturing, the delayed vinculin accumulation and maturation on softer PCL films appeared gradually over time, which strengthened the connection between the cytoskeleton and the ECM at focal complexes [30, 34].
Since our data on cell adhesion rate suggested that a differential ability to interact with the substrate could be related to cell plasticity, mature NHDFs were cultured in parallel with the hMSCs to monitor a potentially different response towards changing the matrix elasticity. No clear dependence of the formation and maturation of focal adhesions on the substrate stiffness was observed in NHDFs although a stiffness-dependent increase in the cell adhesion rates of NHDFs could be demonstrated (figure 5(b)). In figure 5(b), regardless of the culturing time and the stiffness of the matrix, vinculin-rich patches were detected at sites of cell–substratum as well as the cytoplasm in NHDFs. When we examined the cytoskeleton organization of hMSCs and NHDFs at 8 and 24 h, actin filament organization had already completed after 8 h of seeding, and the fibers appeared evenly distributed throughout the cell in addition to the cell periphery, and were not significantly affected by the substrate stiffness. For hMSCs, it has been proposed that F-actin reorganization in response to change of matrix elasticity is a structural-remodeling process that shifts the sensitivity peak towards the new value of matrix elasticity; this proposal agrees with the regulatory role of scaffold stiffness for cell differentiation [1, 35, 36]. Cells of all types tend to move in a slower pace and exert higher traction forces on stiffer 2D substrates as well as increase their cortical stiffness by changing the amount and pattern of polymerized cytoskeletal actin [7, 37–39]. Stiffness of the ECM generates a force or builds up a tension like pulling, which occurs at the cell edge on a thin lamella, consisting mainly of the actin cortex coupled to the plasma membrane, where the force is transmitted to the focal complexes and developed into focal contacts by stretching a cortical sheet [12]. Thus it is likely that F-actin and focal adhesion proteins work in a synchronized fashion in accommodating the force imposed on the cells by the ECM.
To further investigate the focal adhesion processes in hMSCs, talin and paxillin expression was followed 24 h after cell seeding (figure 5). Co-localization of talin and paxillin was found at the protrusions of hMSCs on all PCL substrates. Hence, apart from vinculin, talin and paxillin are likely to contribute actively to the cell–matrix interaction and facilitate cell attachment to the PCL substrates. Zyxin does not seem to be involved in stem cell interaction with PCL substrates under these experimental conditions (figure 5(d)).
3.3. High substrate stiffness does not affect human mesenchymal stem cell multipotency
Since hMSCs have been reported to differentiate in response to altered substrate stiffness [8], we addressed the hMSC multipotency on PCL films with different stiffness by inducing specific lineage commitment. In particular, Engler and colleagues showed that physiologically relevant values of substrate stiffness (kPa range) were sufficient to induce at least hMSC commitment in the absence of biological stimuli [8].
Under the experimental conditions of this investigation, no spontaneous differentiation was noticed in hMSCs cultured on the polymers. This result can be ascribed to the elasticity of the substrates, being far stiffer than the ones used by Engler and colleagues [8]. Surprisingly, when the hMSCs that were initially cultured on the PCL films with supraphysiological Young moduli were challenged with specific differentiation stimuli, they were able to complete adipogenic, osteogenic and chondrogenic differentiation processes (figure 6). These data suggested that cells as a bulk preserved their multipotency independently of the substrate used, hence matrix stiffness is not the only determinant of cell fate in progenitors. One plausible explanation would be that, within hMSC population, different subsets of cells with distinct sensitivity to matrix stiffness exist. As a result, the impact of microenvironment composition would be more striking on a particular subset, while the other cells could still retain their multipotency and differentiate upon specific stimulation. Therefore, independently of microenvironment changes, stem cell homeostasis preserves a reservoir of undifferentiated progenitors, and the composition of stem cell compartment is tightly regulated [40].
Figure 6.
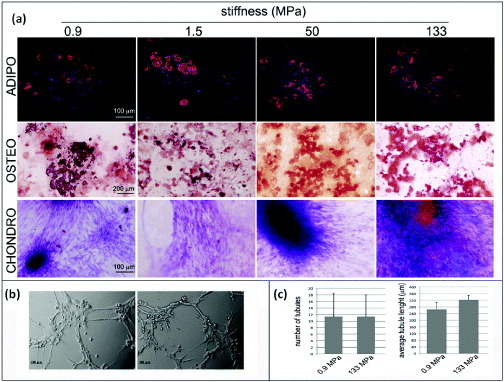
Human mesenchymal stem cell multipotency on PCL films with different stiffnesses. hMSCs were cultured on PCL films and switched to specific differentiation media for 14 days. The formation of lipid vacuoles (FABP4 staining, red), calcium deposits (ALIZARIN RED S staining, red) is shown in cells cultured in adipogenic (ADIPO) or osteogenic media (OSTEO). The occurrence of chondrogenic (CHONDRO) differentiation after 14 days in differentiation medium was confirmed by Masson's trichrome staining. (a) hMSCs grown for 1 week on the softest (0.9 MPa, left) or the stiffest (133 MPa, right) substrates were detached and seeded onto growth-factor-reduced MATRIGEL™ for 3 h. (b) Formation of tubular structures (Scale bars: 25 μm). (c) Number of tubules and the average tubule length measured by ImageJ software.
To address this hypothesis, cells cultured for 7 days on substrates with different Young moduli (0.9 or 133 MPa) were switched to growth-factor-reduced MATRIGEL™. As previously stated, such substrates display Young moduli in the Pa range and allow cells to migrate inside the 3D microenvironment. Interestingly, cells that were previously cultured on the stiffest PCL substrate displayed the ability to complete tubulogenesis (figures 6(b) and (c)), which is an established indicator of undifferentiated cells, demonstrating that despite being exposed to an extremely stiff milieu, hMSCs could preserve their plasticity.
4. Conclusions
We have demonstrated that undifferentiated cells, represented by hMSCs, have a marked sensitivity to substrate stiffness, and their adhesion is responsive to the matrix composition. Cell adhesion, mediated by the formation and maturation of focal adhesions, relies on the mechanical feedback of the microenvironment. The ability of stem cells that were previously exposed to stiff ECM to differentiate supported the current model: stem cell compartment is composed of different cells subsets in which the core is preserved in the undifferentiated state. While the different cell subsets within the compartment possess a distinct sensitivity to different substrate stiffness, the differentiation potential of the bulk population is maintained dynamically by the core in a homeostatic fashion.
Acknowledgments
The present work was supported by the Japan Society for the Promotion of Science (JSPS) through the “Funding for World-Leading Innovative R&D on Science and Technology (FIRST Program)”, and from the World Premiere International (WPI) Research Center Initiative of MEXT, Japan.
References
- Trichet L, Le Digabel J, Hawkins R J, Vedula S R, Gupta M, Ribrault C, Hersen P, Voituriez R. and Ladoux B. Proc. Natl Acad. Sci. USA. 2012;109:6933. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R J., Jr and Wang Y. Proc. Natl Acad. Sci. USA. 1997;94:13661. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C M, Wang H B, Dembo M. and Wang Y L. Biophys. J. 2000;79:144. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens D R. and Yamada K M. Science. 2001;294:1708. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Bershadsky A D, Balaban N Q. and Geiger B. Annu. Rev. Cell Dev. Biol. 2003;19:677. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- Engler A J, Griffin M A, Sen S, Bonnemann C G, Sweeney H L. and Discher D E. J. Cell Biol. 2004;166:877. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D E, Janmey P. and Wang Y L. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Engler A J, Sen S, Sweeney H L. and Discher D E. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Hoffman B D, Grashoff C. and Schwartz M A. Nature. 2011;475:316. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges P C. and Janmey P A. J. Appl. Physiol. 2005;98:1547. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- Welch D R. Cancer Res. 2000;60:1552. [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban N Q, Schwarz U S, Ishizaki T, Narumiya S, Kam Z, Geiger B. and Bershadsky A D. J. Cell Biol. 2001;153:1175. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley S I, Feng Y, Krishnamurthy R, Kim D H, Celedon A, Longmore G D. and Wirtz D. Nature Cell Biol. 2010;12:598. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C. Nature. 2010;466:263. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trappmann B. Nature Mater. 2012;11:642. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- Shin M, Ishii O, Sueda T. and Vacanti J P. Biomaterials. 2004;25:3717. doi: 10.1016/j.biomaterials.2003.10.055. [DOI] [PubMed] [Google Scholar]
- Engelmayr G C, Jr, Cheng M, Bettinger C J, Borenstein J T, Langer R. and Freed L E. Nature Mater. 2008;7:1003. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D H, Lipke E A, Kim P, Cheong R, Thompson S, Delannoy M, Suh K Y, Tung L. and Levchenko A. Proc. Natl Acad. Sci. USA. 2010;107:565. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B K. and West J L. Mater. Res. 2002;60:86. doi: 10.1002/jbm.10042. [DOI] [PubMed] [Google Scholar]
- Hutmacher D W, Goh J C. and Teoh S H. Ann. Acad. Med. Singap. 2001;30:183. [PubMed] [Google Scholar]
- Ng K W, Hutmacher D W, Schantz J T, Ng C S, Too H P, Lim T C, Phan T T. and Teoh S H. Tissue Eng. 2001;7:441. doi: 10.1089/10763270152436490. [DOI] [PubMed] [Google Scholar]
- Forte G. J. Biomed. Biotechnol. 2009;2009:064205. doi: 10.1155/2009/910610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte G. Stem Cells. 2008;26:2093. doi: 10.1634/stemcells.2008-0061. [DOI] [PubMed] [Google Scholar]
- Ebara M, Uto K, Idota N, Hoffman J M. and Aoyagi T. Adv. Mater. 2012;24:273. doi: 10.1002/adma.201102181. [DOI] [PubMed] [Google Scholar]
- Forte G. Tissue Eng. Part A. 2012 doi: 10.1089/ten.TEA.2011.0707. [DOI] [PubMed] [Google Scholar]
- Kocqozlu L, Lavalle P, Koenig G, Senger B, Haikel Y, Schaaf P, Voegel J C, Tenenbaum H. and Vautier D. J. Cell Sci. 2010;123:29. doi: 10.1242/jcs.053520. [DOI] [PubMed] [Google Scholar]
- Mierke C T. Cell Biochem. Biophys. 2009;53:115. doi: 10.1007/s12013-009-9047-6. [DOI] [PubMed] [Google Scholar]
- Nix D A. and Beckerle M C. J. Cell Biol. 1997;138:1139. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervy M, Hoffman L. and Beckerle M C. Curr. Opin. Cell Biol. 2006;18:524. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Galbraith C G, Yamada K M. and Sheetz M P. J. Cell Biol. 2002;159:695. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePasquale J A. and Izzard C S. J. Cell Biol. 1987;105:2803. doi: 10.1083/jcb.105.6.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzard C S. Cell Motil Cytoskeleton. 1988;10:137. doi: 10.1002/cm.970100118. [DOI] [PubMed] [Google Scholar]
- Balaban N Q. Nature Cell Biol. 2001;3:466. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- Choquet D, Felsenfeld D P. and Sheetz M P. Cell. 1997;88:39. doi: 10.1016/S0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- Zemel A, Rehfeldt F, Brown A E, Discher D E. and Safran S A. Nature Phys. 2010;6:468. doi: 10.1038/nphys1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santis G, Lennon A B, Boschetti F, Verhegghe B, Verdonck P. and Prendergast P J. Eur. Cell Mater. 2011;22:202. doi: 10.22203/ecm.v022a16. [DOI] [PubMed] [Google Scholar]
- Ingber D E. Int. J. Dev. Biol. 2006;50:255. doi: 10.1387/ijdb.052044di. [DOI] [PubMed] [Google Scholar]
- von Dassow M. and Davidson L A. Birth Defects Res. C, Embryo Today. 2007;81:253. doi: 10.1002/bdrc.20108. [DOI] [PubMed] [Google Scholar]
- Zoldan J, Karagiannis E D, Lee C Y, Anderson D G, Langer R. and Levenberg S. Biomaterials. 2011;32:9612. doi: 10.1016/j.biomaterials.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte G. Stem Cells. 2011;29:2051. doi: 10.1002/stem.763. [DOI] [PubMed] [Google Scholar]


