Abstract
To maximize the therapeutic efficacy of cardiac muscle constructs produced by stem cells and tissue engineering protocols, suitable scaffolds should be designed to recapitulate all the characteristics of native muscle and mimic the microenvironment encountered by cells in vivo. Moreover, so not to interfere with cardiac contractility, the scaffold should be deformable enough to withstand muscle contraction. Recently, it was suggested that the mechanical properties of scaffolds can interfere with stem/progenitor cell functions, and thus careful consideration is required when choosing polymers for targeted applications. In this study, cross-linked poly-ε-caprolactone membranes having similar chemical composition and controlled stiffness in a supra-physiological range were challenged with two sources of myoblasts to evaluate the suitability of substrates with different stiffness for cell adhesion, proliferation and differentiation. Furthermore, muscle-specific and non-related feeder layers were prepared on stiff surfaces to reveal the contribution of biological and mechanical cues to skeletal muscle progenitor differentiation. We demonstrated that substrate stiffness does affect myogenic differentiation, meaning that softer substrates can promote differentiation and that a muscle-specific feeder layer can improve the degree of maturation in skeletal muscle stem cells.
Keywords: cardiac tissue engineering, mechanobiology, myoblasts, substrate stiffness
1. Introduction
Because of its complexity, cardiac muscle regeneration is considered to be among the most challenging tasks by the tissue engineers. Given the nature of cardiomyocytes as terminally differentiated cells and the low number of resident stem cells in the tissue, self-healing processes occur at a very low efficiency after pathological solicitations [1]. As a result, the infarcted tissue is replaced by non-contractile scar tissue [2], which—together with the severe loss of cardiomyocytes due to the ischemic event—reduces cardiac muscle functionality, eventually leading to heart failure. Given the continuous contractile activity of the heart, the delivery route of cells to the myocardium has a fundamental importance and has been pursued in the literature. However, when stem/progenitor cells were injected into the myocardium or delivered through bloodstream to the heart, few cells were found engrafted within the host tissue few days after the administration [3–7]. For this reason, a number of biomaterials have been proposed as delivery systems to favor stem/progenitor cells engraftment into the cardiac muscle [8–10].
Cardiac-specific scaffolds need to be compliant enough to sustain cardiac contraction [11]. In addition, they should be able to prompt the commitment of stem cell cardiac differentiation, as cell alignment or early differentiation markers expression [12]. Although synthetic polymers are considered biologically inert, they display mechanical properties that could induce specific responses in cells. The sensitivity of living cells to substrate stiffness has been compellingly demonstrated by different research groups showing that natural and synthetic materials can affect cell adhesion, survival, proliferation and differentiation, irrespective from the biological factors supplied [13–15]. In particular, stem and progenitor cells have been shown to sense substrate mechanical properties and activate specific biological responses [16, 17].
While these evidences raise concerns over contractile and undifferentiated cell response to the standard culture protocols used in vitro, they can be taken as an opportunity to define the characteristics of cell- and tissue-specific scaffolds to be used in cardiac tissue engineering applications [18].
A marked sensitivity to the mechanical properties of the substrate is ascribed to contractile cells and their precursors, whose activity and maturation is impaired on stiff substrates. In a recent study, we demonstrated that the substrate stiffness can affect the assembly of the contractile apparatus in cardiac cells. When substrates displaying similar chemical composition but different stiffness values were challenged with neonatal murine cardiomyocytes, the cells showed a differentiated phenotype on softer materials, while failing to complete their maturation on stiffer polymers [19]. This result agrees well with previous studies addressing the effects of matrix compliance on contractile cells [20, 21].
Skeletal myoblasts are adult tissue-resident muscle progenitors that can be easily harvested from skeletal muscle [22–24]. They retain high proliferation and differentiation capacity in vitro and high resistance to ischemia, which makes them interesting candidates for treating cardiac diseases [25]. Recent pre-clinical and clinical trials demonstrated that, although not being able to improve cardiac performance, their implantation in the diseased heart resulted in an anti-remodeling effect [26–29]. Such a beneficial effect has been ascribed to their acknowledged ability to fuse with host cardiomyocytes [30]. Unfortunately, the occurrence of arrhythmogenic events was reported in treated patients, very likely as a result of incomplete electrical coupling of skeletal muscle cells with host cells [31]. Skeletal myoblast differentiation is believed to depend on substrate stiffness, the quality of the contractile apparatus in differentiated cells being directly related to the mechanical properties of substrate surface [32, 33]. Nonetheless, skeletal muscle differentiation has been shown to take place also on very stiff surfaces, like tissue culture polystyrene (TCPS; a surface with virtually infinite stiffness) in response to biological stimuli [34]. These results were obtained when cells were confluent [35, 36], thus suggesting that factors other than stiffness could be involved in the processes.
In the present investigation, poly-ε-caprolactone (PCL) was used to produce cross-linked films displaying different Young moduli in the MPa range and having similar chemical composition. Although several groups have already showed the effect of stiffness on cell differentiation using hydrogels, it is yet unclear how material properties of biodegradable PCL scaffolds, which were approved by the US Food and Drug Administration, affect cell functions. It is difficult to produce a PCL scaffold with wide range of elastic moduli without affecting cell adhesion. We have prepared crosslinked PCLs that offer branch-structure-dependent stiffness without changing the surface wettability. Therefore, the versatility and biologically friendly nature of our PCL samples could be potentially applied in novel and diverse applications, especially biomaterial development and basic cell biology [37]. The Young moduli of the polymers adopted in the study are supra-physiological and supposed to represent a ‘non-permissive environment’ for skeletal progenitor differentiation. It was demonstrated previously that native skeletal muscle tissue has a stiffness ranging from 11.5 to 45.3 kPa, depending on myoblasts differentiation state [48]. Therefore, the films used in this study display supra-physiological stiffness as compared to native tissue.
Thanks to the peculiarities of these polymers, the differentiation ability of skeletal muscle progenitors was studied as an effect of surface stiffness. Moreover, muscle-specific and non-related feeder layers were prepared on stiff surfaces to decouple the contribution of mechanical and biological factors in the skeletal muscle progenitor differentiation. By this means, we confirmed that high substrate stiffness can impair muscle differentiation and demonstrated that the presence of a feeder layer providing a muscle-specific soft substrate can enhance the differentiation rate of skeletal progenitors.
2. Materials and methods
2.1. Cross-linked poly-ε-caprolactone membranes
The PCL films were prepared by cross-linkage of tetra-branched PCL with acrylate end-groups in the presence of linear PCL telechelic diacrylates according to the previously reported protocol [37]. Briefly, two-branched and four-branched PCL were synthesized by ε-caprolactone ring opening polymerization initiated with tetramethylene glycol and pentaerythritol as initiators, respectively. Then, acryloyl chloride was reacted with the ends of the branched chains. PCL membranes were characterized in our previous report [19] (figure 1).
Figure 1.
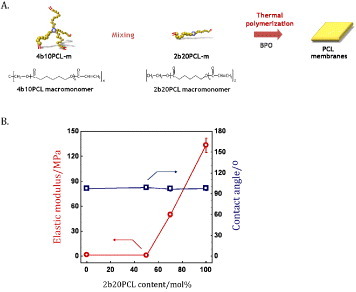
Properties of cross-linked PCL films. Planar PCL layers with various stiffness values were obtained by tuning the concentration of two- and four-branched PCL with acrylate end-groups (A). The percentage of four- and two-branched PCL determines the elastic modulus of the films. Four different preparations were used in the present investigation. (B) Elastic moduli and contact angles of the layers.
2.2. Preparation of PCL films for cell culture
Before cell seeding, PCL films were sterilized in a low-pressure hydrogen peroxide gas plasma system CH-160C (Toho Seisakusho, Tokyo, Japan). Films used for mouse skeletal myoblasts cell line (C2C12; CRL-1772, ATTC, Rockville, MD) were pre-coated for 1 h at 37 °C with a 1 mg mL−1 solution of type-I collagen suspended in 0.1 M acetic acid (Life Laboratories Co., Tokyo, Japan) [38]. Primary myoblasts extracted from rat skeletal muscle were obtained from Primary Cell Co., Ltd (MYB01; F-12, Hokkaido University, Sapporo, Japan). Finally, the films were incubated overnight at 37 °C and in a 5% CO2 atmosphere in the so-called ‘complete medium’, that is, Dulbecco's modified Eagle's medium (DMEM)-High Glucose GLUTAMAX™ (Sigma-Aldrich, St Louis, Missouri, USA) that was supplemented with 10% fetal bovine serum (FBS; Equitech-Bio Inc., Kerrville, Texas, USA) at 5000 U ml−1 and with a 1% penicillin/streptomycin solution at 5000 μg ml−1 (Invitrogen, Carlsbad, California, USA).
2.3. Cell culture
Primary myoblasts extracted from rat skeletal muscle (MYB01; F-12, Primary Cell Co., Ltd, Hokkaido University, Sapporo, Japan) and mouse skeletal myoblasts cell line (C2C12; CRL-1772, ATTC, Rockville, Maryland, USA) were used to study myoblasts function. Undifferentiated MYB01 and C2C12 cells were cultured in the complete medium.
When required, cells were switched to DMEM containing 1% penicillin/streptomycin, 4 mM L-glutamine (Sigma-Aldrich) and 2% horse serum (HS; Life Technologies, Gaithersburg, Maryland, USA) to induce myogenic differentiation as described previously [21]. Normal human dermal fibroblasts (NHDF; ATCC, Manassas, Virginia, USA) and irradiated mouse embryonic fibroblasts (MEF; ATCC) were cultured in DMEM with 10% FBS, 1% penicillin/streptomycin and 4 mM L-glutamine. All cell types were maintained at 37 °C and in 5% CO2. Cells were passaged every third day using a solution containing 0.25% trypsin/1 mM ethylenediaminetetraacetic acid (EDTA; Nacalai Tesque Inc., Kyoto, Japan). Irradiated MEF were purchased from Gibco-BRL (Gaithersburg, Maryland, USA).
2.4. Feeder-layer system
For co-culture experiments, three different direct co-culture systems were prepared. At first, NHDF and MYB01 cells were cultured on PCL polymers and glass slides (BD Biosciences, San Jose, California, USA) at a density of 5.0 × 104 cells cm−2. After 24 h, the medium was removed from culture and 0.1% paraformaldehyde (PFA) in phosphate buffered saline (PBS) was added for 30 min at 37 °C and in 5% CO2. Then, PFA was removed and cells were washed twice with PBS. C2C12 cells were pre-stained with viable red fluorescent dye Vybrant™ DiI (Molecular Probes, Eugene, Oregon, USA), following the manufacturer's specifications, and seeded directly over the layer of inactivated cells at a density of 2.0 × 104 cells cm−2 in differentiation medium. The medium was changed every day throughout the entire experiment and the samples processed after 3 and 5 days for immunofluorescence analysis. Irradiated MEF were seeded on PCL layers at a concentration of 5.0 × 104 cells cm−2. PFA treatment was not necessary for the irradiated MEF.
2.5. AlamarBlue® assay
AlamarBlue® assay is designed to measure quantitatively cell metabolic activity. Thus the assay was used to evaluate cell adhesion and proliferation as an indirect measurement of cell activity. MYB01 and C2C12 were seeded on PCL layers at a concentration of 2.0 × 104 cells cm−2 in 900 μl DMEM supplemented with 10% FBS and 100 μl of AlamarBlue® solution and incubated at 37 °C and in 5% CO2. Floating cells were removed before performing the Alamar Blue assay. ARVO MX1420 multilabel counter (PerkinElmer) was used to calculate the fluorescence at 544 nm excitation wavelength and 590 nm emission wavelength after 3, 8, 24, 48 and 72 h of cell culture.
2.6. Immunofluorescence staining and confocal microscopy
Cells seeded on TCPS dishes or on PCL layers were fixed with 4% PFA in PBS for 15 min at room temperature and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 2 min at room temperature after 1, 3, 5 and 7 day culture. MYB01 and C2C12 cell morphology, cytoskeleton organization and protein expression were detected through immunofluorescence analysis after incubation with tetra-rhodamine-conjugated Phalloidin (1:100; Invitrogen), a drug directed against F-actin, and vinculin (1:50; Sigma-Aldrich) for 1 h at room temperature. The phenotype of undifferentiated and differentiated cells was evaluated by staining with an antibody directed against Myogenesis Differentiation Protein 1 (MyoD; 1:100; Chemicon, Temecula, California, USA) and Alexa Fluor® 488 MF-20 (1:50; eBioscience, San Diego, California, USA). The appropriate secondary antibody (goat-anti-mouse 488, Life Technologies, Gaithersburg, Maryland, USA) was used to detect the specific signals. Nuclei were counterstained with 4–6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Secondary antibody in the absence of a specific primary antibody was used to exclude the occurrence of unspecific signals. The images were taken using Leica SP5 confocal laser scanner microscope (Leica, Wetzlar, Germany).
2.7. Scanning electron microscopy analysis
MYB01 and C2C12 cells were seeded on PCL films in growth and differentiation medium for 3 days, fixed and dried under laminar hood. Once dried, the samples were mounted on aluminum stubs using adhesive conducting carbon tape and then sputtered with gold for 2 min (HITACHI ION Sputter E-1030). The samples were observed in a field-emission scanning electron microscope (SEM; Hitachi S-4800) operated at an acceleration voltage in the range 8–10 kV.
2.8. Statistical analysis
Results are given as mean ± standard deviation of a mean as obtained by counting n fields per group in three independent experiments. The significance of differences in multi-group comparison was derived by unpaired Student's t-test. The values were considered statistically significant when P < 0.05.
3. Results and discussion
3.1. Undifferentiated myoblasts adhere and proliferate on cross-linked PCL membranes with different stiffness values
Substrate stiffness has been shown to affect cell behavior in terms of survival, proliferation and differentiation [16, 17, 19], while the response of cells to matrix elasticity in vitro has been demonstrated to be highly cell specific [13, 39]. To investigate the possibility that matrix compliance could interfere with myoblast function, poly-ε-caprolactone (PCL) layers were produced with overall stiffness values of 0.91 ± 0.08, 1.5 ± 0.2, 50 ± 3 and 133 ± 9 MPa. In previous reports, substrate stiffness has been modulated by varying the crosslinking density or incorporating other components. These approaches, however, showed the difficulty in preserving the surface wettability. As an alternative approach, we have demonstrated control of PCL stiffness by tailoring the semicrystalline structures. Figure 1(B) shows elastic moduli of the films used in this study. AlamarBlue® assay performed on C2C12 myoblast cell line and primary rat skeletal myoblasts (MYB01) revealed no significant differences in myoblast cell adhesion and proliferation on the polymers (figure 2). Interestingly, 72 h after seeding, undifferentiated myoblasts exhibited a typical myoblast-like morphology on all substrates, showing a correct cytoskeleton organization highlighted by F-actin immunostaining, irrespective of the substrate stiffness (figure 3(A), red). Functional adhesion to the substrate was demonstrated by the expression of focal adhesion protein vinculin (figures 3(A) and (B), green) and confirmed by SEM analysis (figure 3(C)). Altogether, these data demonstrate the lack of any specific effect of substrate elasticity on undifferentiated myoblast cells in short-term investigations.
Figure 2.
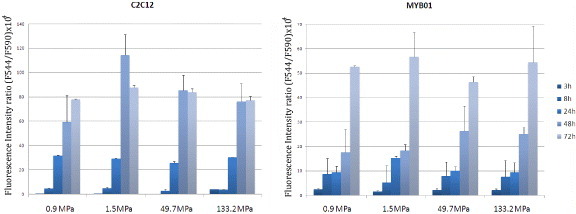
Effect of substrate stiffness on myoblast adhesion and proliferation. Alamar Blue® assay was performed on two sources of skeletal myoblasts (C2C12 cell line and MYB01 ex vivo extracted cells) at 3–8 h to study cell adhesion and at 24, 48 and 72 h to quantify cell proliferation.
Figure 3.
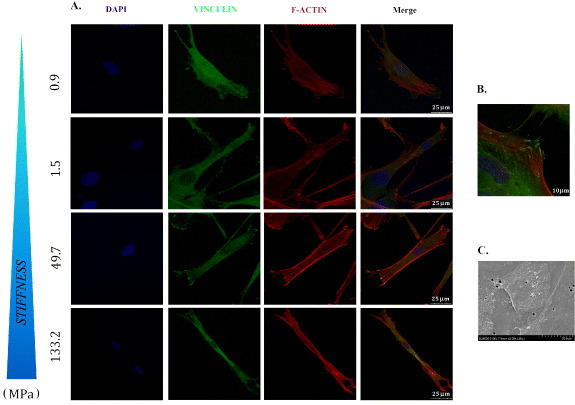
Undifferentiated myoblasts functionally adhere to PCL films. C2C12 and MYB01 cells were cultured on PCL polymers with different stiffness values. Cell morphology and the formation of adhesion processes were detected by immunofluorescence staining after 24 h of culture. To visualize the cytoskeleton organization, F-actin was decorated by Phalloydin-TRITC (A, red). The expression of focal adhesion protein vinculin was detected independently from the layer (A, green). A detail of the focal adhesion processes is shown in (B). SEM analysis confirmed the undifferentiated morphology of adherent cells (C). Scale bars A: 25 μm; B: 10 μm; C: 20 μm.
3.2. Effect of substrate stiffness on myoblast differentiation
To evaluate the occurrence of myogenic differentiation in myoblasts on PCL films with different stiffness values, cells were seeded at 50% confluence to minimize the impact of cell-to-cell contact and switched to differentiation medium for 3, 5 and 7 days.
The first phases of myogenic differentiation are characterized by actin filament remodeling in pre-fusion aligning myoblasts [40]. At day 3, only cells seeded on 0.91 MPa PCL layers appeared aligned and fused into long linear myotubes (figure 4(A)), meaning that cells were already progressing toward mature myogenic differentiation. The differentiation process appeared still in the early phase for myoblasts seeded on the other substrates (figure 4(A)). Instead, after 7 days all the substrates with different stiffness values showed the presence of aligned myoblasts (figure 4(B)). At this time, as shown in figure 4(B), cell density increased accordingly.
Figure 4.
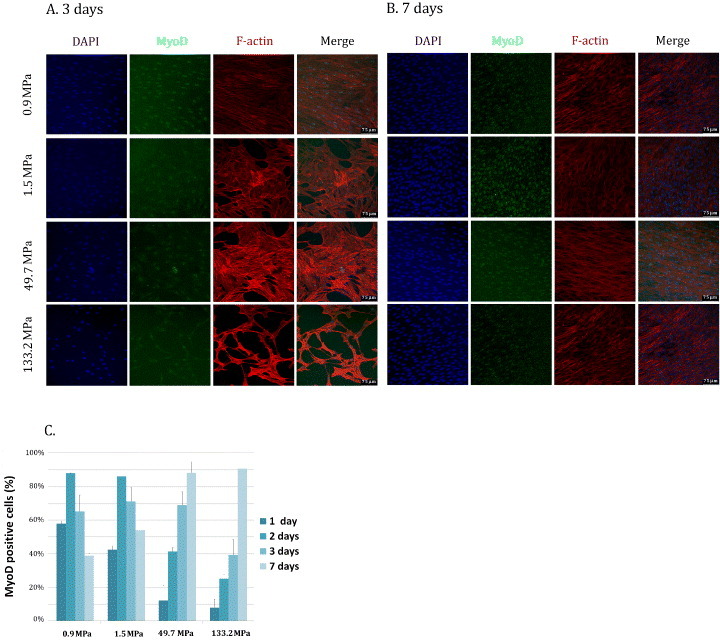
MyoD expression in myoblasts cultured on PCL films. MYB01 were cultured on PCL polymers and showed MyoD expression after 3 (A) and 7 days (B). MyoD-positive cells were quantified by confocal immunofluorescence at the given time-points (C). Nuclei were counterstained in blue (DAPI). Similar results were obtained using C2C12 cell line. Scale bars: 75 μm.
To confirm the hypothesis that the evolution of cell differentiation depends on the substrate stiffness the number of cells expressing the nuclear form of the early myogenic differentiation factor MyoD [41] was assessed after 3 and 7 days of culture.
At day 3, the percentage of MyoD-positive cells on 0.91, 1.5 and 50 MPa films was ranging between 65 and 72% (72 ± 12, 65 ± 12 and 69 ± 8%, respectively), showing no significant differences among these samples. On the contrary, on the stiffest film (133 ± 9 MPa), the number of MyoD-positive cells was significantly smaller (39 ± 10%) than on the other layers (figures 4(A) and (C)). Then, after 7 days of culture, the stiffer films yielded a significantly larger percentage of MyoD-positive cells than softer films (91 ± 5, 88 ± 7, 54 ± 2 and 39 ± 2% for 133, 50, 1.5 and 0.9 MPa, respectively, figures 4(B) and (C)). These results can have two possible explanations. (i) Cells proliferate more on soft substrates and reach confluence to activate cell fusion faster than on the stiff surface. A critical step in skeletal muscle differentiation is cell fusion, a process that requires cells to be confluent enough to contact each other [42]. (ii) MyoD activation occurs earlier on 0.91 and 1.5 MPa than on the stiffer 50 and 133 MPa films. A common understanding of muscle differentiation states that the master gene MyoD is activated in the early phases of the process, when it translocates to the nucleus triggering late muscle gene transcription. Then, MyoD expression is downregulated [43–45]. Our data are consistent with MyoD activation occurring earlier on soft substrates (figure 4(A)) [32]. At day 7, the peak in MyoD nuclear expression on the stiff substrates accounts for a delayed onset of muscle differentiation on the stiff layers. At this time-point, the number of MyoD-positive cells is comparable to that encountered on the soft substrates after 3 days. Consistently, at the last time-point, cells on the stiff substrates start to align, having reached the critical confluence necessary for muscle differentiation (figure 4(B)). The completion of the muscle differentiation process requires cell fusion to form an aligned syncytium and the concomitant assembly of the contractile apparatus [40, 46]. Thus, the expression of contractile myosin in differentiating myoblasts on the polymers was evaluated at day 3 and 5 by MF-20 antibody directed against contractile myosin. On day 3, myosin-positive cells on 0.91, 1.5 and 50 MPa films were already expressed to 99 ± 1, 99 ± 1 and 93 ± 3%, respectively. No significant differences could be detected among these samples. Conversely, only 15 ± 3% of cells were positive for contractile myosin on the stiffest film (figures 5(A) and (B)). In addition, although being multinucleated, cells expressing contractile myosin on the stiffest polymer did not attain the typical elongated shape that could be detected on softer films (figure 5(C)). Also, a striking difference in myosin-positive cell percentage could be found when TCPS was used as a control at day 3 (8 ± 2%). Surprisingly, at day 5, no significant differences could be found in differentiated cell percentage on all the substrates tested, while TCPS yielded 51 ± 5% of myosin-positive cells. Myotube formation on PCL films was confirmed by SEM analysis (figure 5(D)). These data strengthen the hypothesis that myogenic differentiation occurred earlier on softer than stiffer membranes. This event is very likely the result of cells finding softer substrates more suitable for myoblast differentiation, as described previously [47]. Remarkably, these values are still much higher than the elasticity of myoblasts in vivo (11.5 ± 1.3 kPa), a value that can rise up to 45 ± 4 kPa when myotubes are formed [48]. On the stiffest polymer and TCPS, a delayed expression of MyoD (7 days) and MF-20 (5 days) was detected, demonstrating that the differentiation process was hindered when the stiffness was too high.
Figure 5.
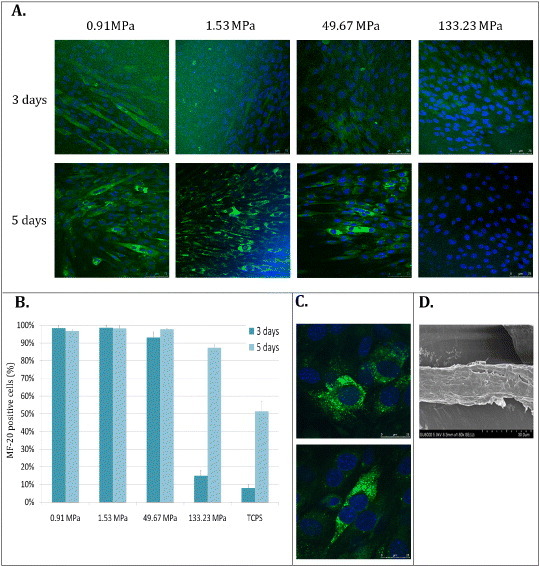
Myogenic differentiation of myoblasts on PCL layers. C2C12 cells were seeded on PCL polymers and switched to differentiation medium for 3 and 5 days. At these time-points myosin heavy chain expression was assessed by immunofluorescence analysis through MF-20 antibody (A, scale bars: 75 μm. Myosin-positive cells were quantified and compared between cells cultured on polymers and on TCPS (B). The morphology of differentiated cells on the stiffest substrate (133 MPa) at day 3 is shown at day 3 (C, upper image) and 7 (C, lower image). Nuclei were counterstained in blue (DAPI). (Scale bars: 25 μm.) Myogenic differentiation was confirmed by SEM analysis (D).
3.3. Cell confluence is essential for myogenic differentiation
Cell confluence is known to cause growth arrest, which is essential for myogenic differentiation [36]. A thorough confocal microscopy analysis of myoblasts differentiated on PCL films revealed that cells were growing in multilayers after reaching complete confluence. Confocal microscopy 3D rendering showed that on the stiffest polymer, only cells located in the top layers could differentiate in myotubes, while those in the bottom layer (in contact with the substrate) could not (figure 6). This result reflects the difficulty of skeletal myoblasts to differentiate when they sense a stiff substrate, as previously indicated [32]. Altogether, these data demonstrate that cell confluence is required to provide a continuous layer (‘feeder layer’) on stiff substrates having an appropriate compliance to allow cell differentiation. This observation is consistent with previous reports showing that substrate elasticity is essential to stem and progenitor cell differentiation [17, 19].
Figure 6.
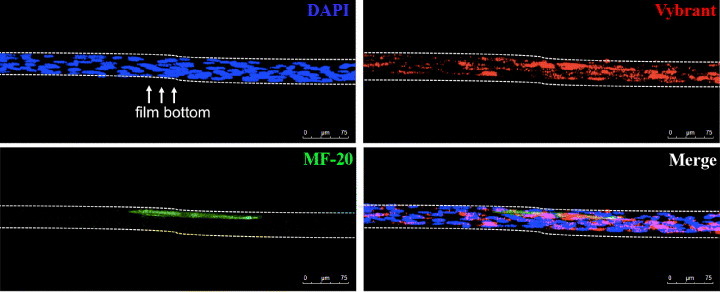
Myotube formation occurred only on upper layer of cells. Myoblasts pre-stained with red Vybrant™ vital die were cultured on stiffest polymer (133 MPa). At day 5, after immunofluorescence staining with MF-20 (green) and nuclear stain DAPI (blue), the sample was observed in a confocal microscope. 3D reconstruction showed that only cells located on top layers could differentiate (green) in myotubes.
3.4. Biochemical signals are critical for myoblast differentiation
To compellingly confirm the abovementioned assumptions, direct co-culture experiments were carried out using non-proliferating undifferentiated skeletal myoblasts (MYB01) and two sources of embryonic (MEF) and adult (NHDF) fibroblasts. Homeostasis of stem and progenitor cells in vivo is tightly controlled by a complex array of mechanical and biological factors [12, 49] concurring to maintain cells plasticity and triggering their differentiation when required. Microenvironment stiffness is thought to be only one of these factors, while the impact of biological cues on stem and progenitor cells is widely recognized [50]. The ‘feeder layer’ experiments were meant to provide differentiating C2C12 cells with appropriate (MYB01) or non-related (MEF, NHDF) feeder layers, so that the biological contribution of cell-to-cell contact could be decoupled from that due to the substrate stiffness. C2C12 cells were pre-stained with red Vybrant™ vital dye and seeded on top of inactivated cells, which have been previously seeded on PCL films, as described in figure 7. Myogenic differentiation was quantified by red Vybrant™/MF-20 co-staining at day 3 and 5. Confocal microscopy showed that, when inactivated MYB01 cells were used as feeder layer (figure 8(A)), C2C12 cell differentiation was more efficient as compared to MEF or NHDF (figures 8(B) and (C)). When MEF and NHDF were used as feeder layers, the percentages of differentiated cells on 133 MPa films at day 3 were 4.4 ± 1.5 and 9 ± 3%, respectively. These percentages were significantly lower than those obtained when MYB01 cells were used as feeder layer (30 ± 8%). As expected, at day 5, the percentage of differentiated C2C12 cells on MYB01 was still significantly higher than that on MEF or NHDF (52 ± 2% versus 30 ± 2% and 14 ± 2%, figure 8(D)). Figure 8 shows results from the films having a stiffness of 133 MPa as a typical example, since similar results were obtained on all the substrates tested (data not shown).
Figure 7.
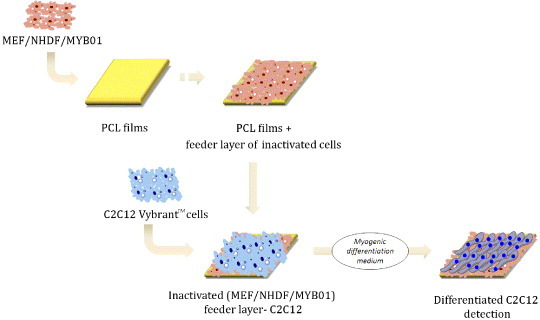
Graphical representation of the feeder-layer setup. First, NHDF or MYB01 or irradiated MEF were seeded onto the PCL films at high confluence to form a feeder layer. After 24 h, feeder layer cells were treated with 0.1% PFA for 20 min and washed extensively in PBS. PFA treatment was not necessary for irradiated MEF. At this point, VybrantTM-labeled C2C12 were seeded onto the feeder layer and switched to myogenic differentiation medium for 3 and 5 days. ChemBioDraw 12.0 software was used for cells animation representation.
Figure 8.
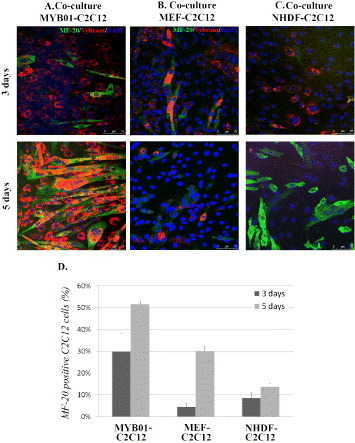
Feeder layer systems. C2C12 cells pre-stained with red Vybrant™ vital dye were cultured directly over a feeder layer of non-proliferating MYB01, MEF and NHDF in 3 distinct co-culture systems. C2C12 differentiation was evaluated by MF-20 immunostaining (A, B, C, green) at day 3 and day 5. The percentage of MF-20 positive cells was calculated for the 3 co-cultures and compared with each other (D). Scale bars: 75 μm.
Altogether, the obtained findings confirmed that the mechanical features of the microenvironment are of paramount importance for addressing muscle differentiation. In this context, the key role of substrate stiffness is demonstrated by the evidence that the first phases of muscle commitment (i.e. MyoD nuclear expression and cell alignment) are delayed on very stiff substrates. More importantly, the late phases of the process (i.e. cell fusion and expression of contractile proteins) are only taking place on stiff surfaces when a critical confluence—providing an appropriate ‘soft’ substrate—is reached on stiff polymers. Nonetheless, there is no preferential effect of substrate stiffness on undifferentiated cell adhesion and proliferation, even though the question whether a supra-physiological compliance of the matrix can affect per se the plasticity of progenitor cells remains to be addressed. Our data seem to corroborate the hypothesis that, beside the mechanical properties of the milieu, the formation of specific cell-to-cell contacts plays a major role in stem and progenitor cell commitment [48]. When the appropriate cell layer (MYB01 cells) is provided to differentiating C2C12 cells, the negative effect of high substrate stiffness on myoblast differentiation is overcome. As expected, when fibroblasts are used as feeder layer, the differentiation rate decreases significantly.
More importantly, our findings argue against the possibility that efficient cell differentiation can be achieved solely by tuning the matrix stiffness [16, 44]. Our evidences point at biological factors and cell-to-cell contact as pivotal determinants of stem cell maturation. These results agree well with what was previously demonstrated by our group in a different cell system [50]. The nature of the biological molecules—be they soluble or exposed on cell membrane—involved in cell commitment to muscle phenotype is still to be clarified and will be the objective of a future investigation. Moreover, the mechanisms responsible for the transduction of the mechanical signals to the nucleus of stem and progenitor cells need further investigation.
4. Conclusions
Our experimental data indicate that, as far as muscle differentiation is concerned, the nature of the substrate on which cells are grown plays a key role. In this context, matrix stiffness appears to be a critical parameter in determining progenitor cell specification. Nevertheless, the completion of muscle cell differentiation requires a contribution of biological factors, which are still unknown and will require further attention.
References
- Bergmann O. Science. 2009;324:98. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer M A. and Braunwald E. Circulation. 1990;81:1161. doi: 10.1161/01.CIR.81.4.1161. [DOI] [PubMed] [Google Scholar]
- Orlic D. Nature. 2001;410:701. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- Zhang M, Methot D, Poppa V, Fujio Y, Walsh K. and Murry C E. J. Mol. Cell. Cardiol. 2001;33:907. doi: 10.1006/jmcc.2001.1367. [DOI] [PubMed] [Google Scholar]
- Muller-Ehmsen J, Whittaker P, Kloner R A, Dow J S, Sakoda T, Long T I, Laird P W. and Kedes L. J. Mol. Cell. Cardiol. 2002;34:107. doi: 10.1006/jmcc.2001.1491. [DOI] [PubMed] [Google Scholar]
- Chong J J H. Heart Lung Circ. 2012;21:532. doi: 10.1016/j.hlc.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Menasché P. J. Cardiovasc. Transl. Res. 2012;5:555. doi: 10.1007/s12265-012-9356-9. [DOI] [PubMed] [Google Scholar]
- Eschenhagen T. and Zimmermann Z. Circ. Res. 2005;97:1220. doi: 10.1161/01.RES.0000196562.73231.7d. [DOI] [PubMed] [Google Scholar]
- Vunjak-Novakovic G, Tandon N, Godier A, Maidhof R, Marsano A, Martens T P. and Radisic M. Tissue Eng. Part B. 2010;16:169. doi: 10.1089/ten.teb.2009.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoashi T. J. Thorac. Cardiovasc. Surg. 2009;138:460. doi: 10.1016/j.jtcvs.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Jawad H, Lyon A R, Harding S E, Ali N N. and Boccaccini A R. Br. Med. Bull. 2008;87:31. doi: 10.1093/bmb/ldn026. [DOI] [PubMed] [Google Scholar]
- Forte G, Pagliari S, Pagliari F, Ebara M, Di Nardo P. and Aoyagi T. Stem Cell Rev. 2011 doi: 10.1007/s12015-011-9325-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Georges P C. and Janmey P A. J. Appl. Physiol. 2005;98:1547. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- Wang H B, Dembo M. and Wang Y L. Am. J. Physiol. Cell Physiol. 2000;279:C1345. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- Peyton S R. and Putnam A J. J. Cell. Physiol. 2005;204:198. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- Engler A J, Sen S, Sweeney H L. and Discher D E. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Forte G. Stem Cells. 2008;26:2093. doi: 10.1634/stemcells.2008-0061. [DOI] [PubMed] [Google Scholar]
- Di Nardo P, Forte G, Ahluwalia A. and Minieri M. J. Cell. Physiol. 2010;224:590. doi: 10.1002/jcp.22165. [DOI] [PubMed] [Google Scholar]
- Forte G. Tissue Eng. Part A. 2012;18:1837. doi: 10.1089/ten.tea.2011.0707. [DOI] [PubMed] [Google Scholar]
- Jacot J G, McCulloch A D. and Omens J H. Biophys. J. 2008;95:3479. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhana B, Iyer R K, Chen W L, Zhao R, Sider K L, Likhitpanichkul M, Simmons C A. and Radisc M. Biotechnol. Bioeng. 2010;105:1148. doi: 10.1002/bit.22647. [DOI] [PubMed] [Google Scholar]
- Rando T A. and Blau H M. J. Cell Biol. 1994;125:1275. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Straeter J, Bran G, Riedel F, Sauter A, Hörmann K. and Goessler U R. Int. J. Mol. Med. 2008;21:49. [PubMed] [Google Scholar]
- Lapan A D. and Gussoni E. Methods Mol. Biol. 2012;798:3. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formigli L. Am. J. Physiol. Cell Physiol. 2005;288:C795. doi: 10.1152/ajpcell.00345.2004. [DOI] [PubMed] [Google Scholar]
- Hata H, Matsumiya G, Miyagawa S, Kondoh H, Kawaguchi N, Matsuura N, Shimizu T, Okano T, Matsuda H. and Sawa Y. J. Thorac. Cardiovasc. Surg. 2006;132:918. doi: 10.1016/j.jtcvs.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Menasché P. J. Mol. Cell. Cardiol. 2008;45:545. doi: 10.1016/j.yjmcc.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Menasché P. Semin Thorac. Cardiovasc. Surg. 2008;20:131. doi: 10.1053/j.semtcvs.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Durranis S, Konoplyannikov M, Ashraf M. and Haider K H. Regen. Med. 2010;5:919. doi: 10.2217/rme.10.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke H, Minami E, Poppa V. and Murry C E. Circ. Res. 2004;94:e56. doi: 10.1161/01.RES.0000125294.04612.81. [DOI] [PubMed] [Google Scholar]
- Menasché P. Circulation. 2008;117:1189. doi: 10.1161/CIRCULATIONAHA.107.734103. [DOI] [PubMed] [Google Scholar]
- Engler A J, Griffin M A, Sen S, Bönnemann C G, Sweeney H L. and Discher D E. J. Cell Biol. 2004;166:877. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Mishali M, Zoldan J. and Levenberg S. Tissue Eng. Part A. 2009;154:935. doi: 10.1089/ten.tea.2008.0111. [DOI] [PubMed] [Google Scholar]
- Linkhart T A, Clegg C H. and Hauschka S D. Dev. Biol. 1981;86:19. doi: 10.1016/0012-1606(81)90311-0. [DOI] [PubMed] [Google Scholar]
- Lawson M A. and Purslow P P. Cells Tissues Organs. 2000;167:130. doi: 10.1159/000016776. [DOI] [PubMed] [Google Scholar]
- Chowdhury S R, Muneyuki Y, Takezawa Y, Kino-Oka M, Saito A, Sawa Y. and Taya M. J. Biosci. Bioeng. 2010;109:310. doi: 10.1016/j.jbiosc.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Ebara M, Uto K, Idota N, Hoffman J M. and Aoyagi T. Adv. Mater. 2011;24:273. doi: 10.1002/adma.201102181. [DOI] [PubMed] [Google Scholar]
- DeQuach J A, Mezzano V, Miglani A, Lange S, Keller G M, Sheikh F. and Christman K L. PLOs ONE. 2010;5:064211. doi: 10.1371/journal.pone.0013039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee I, Yekkala K, Borg T K. and Baudino T A. Ann. N. Y. Acad. Sci. 2006;1080:76. doi: 10.1196/annals.1380.007. [DOI] [PubMed] [Google Scholar]
- Swailes N T, Knight P J. and Peckham M. J. Anat. 2004;205:381. doi: 10.1111/j.0021-8782.2004.00341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J B. J. Cell Biol. 1990;111:1149. doi: 10.1083/jcb.111.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velleman S G. Poult. Sci. 1999;78:778. doi: 10.1093/ps/78.5.778. [DOI] [PubMed] [Google Scholar]
- Eom Y K. Biochem. Biophys. Res. Commun. 2011;408:167. doi: 10.1016/j.bbrc.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Choi Y S, Vincent L G, Lee A R, Dobke M K. and Engler A J. Biomaterials. 2012;33:2482. doi: 10.1016/j.biomaterials.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar S, Panguluri S K. and Kumar A. Methods Mol. Biol. 2012;798:311. doi: 10.1016/j.biomaterials.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H, Ishibashi T, Nagamine K, Kanzaki M. and Nishizawa M. Biomaterials. 2010;31:6981. doi: 10.1016/j.biomaterials.2010.05.071. [DOI] [PubMed] [Google Scholar]
- Guilak F, Cohen D M, Estes B T, Gimble J M, Liedtke W. and Chen C S. Cell Stem Cell. 2009;5:17. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinsworth A M, Zhang S, Kraus W E. and Truskey G A. Am. J. Physiol. Cell Physiol. 2002;283:C1219. doi: 10.1152/ajpcell.00502.2001. [DOI] [PubMed] [Google Scholar]
- Discher D E, Janmey P. and Wang Y. Science. 2005;310:1139. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Pagliari S. Adv. Mater. 2011;23:514. doi: 10.1002/adma.201003479. [DOI] [PubMed] [Google Scholar]


