Abstract
MicroRNAs (miRNAs) play crucial regulatory roles in various human diseases including cancer, making them promising biomarkers. However, given the low levels of miRNAs present in blood, their use as cancer biomarkers requires the development of simple and effective analytical methods. Herein, we report the development of a highly sensitive dual mode electrochemical platform for the detection of microRNAs. The platform was developed using peptide nucleic acids as probes on gold electrode surfaces to capture target miRNAs. A simple amplification strategy using gold nanoparticles has been employed exploiting the inherent charges of the nucleic acids. Electrochemical impedance spectroscopy was used to monitor the changes in capacitance upon any binding event, without the need for any redox markers. By using thiolated ferrocene, a complementary detection mode on the same sensor was developed where the increasing peaks of ferrocene were recorded using square wave voltammetry with increasing miRNA concentration. This dual-mode approach allows detection of miRNA with a limit of detection of 0.37 fM and a wide dynamic range from 1 fM to 100 nM along with clear distinction from mismatched target miRNA sequences. The electrochemical platform developed can be easily expanded to other miRNA/DNA detection along with the development of microarray platforms.
MicroRNAs (miRNAs) are a class of noncoding genes that are transcribed as RNA sequences of around 22 nucleotides long1. More than twenty years after the first article about miRNA, there have been many reports on the role of miRNAs in gene expression and regulation2,3. Many reports have shown their importance in various biological processes in the human body including regulation of target gene4, cellular proliferation and differentiation5, cell death6, translational and transcriptional regulation of their expression7. It was only in the last decade that miRNAs have been identified for their importance in human diseases such as cancer8,9,10. For instance, miR-155 and miR-21 are up-regulated in breast cancer, but miR-91 has been found to be down-regulated in breast cancer. Other miRNAs can be related with more than one type of cancer. Many recent studies show that miR-145 is a tumour-suppressive miRNA, that is downregulated in several cancer types, including bladder cancer11, colon cancer12, breast cancer13, ovarian cancer14 and prostate cancer (PCa)5.
For PCa, miR-145 is a well-characterised tumour-suppressor with an important regulatory role since it can protect from cancer cell invasion and metastasis15. miRNAs have been reported to act as fingerprints of a disease, including PCa, making them a promising tool as a biomarker. Conventionally, technologies used for miRNA study include northern blotting16, in situ hybridization17, quantitative polymerase-chain reaction18 and miRNA microarrays. All these techniques are powerful but laborious and hence restricted to central laboratories. Consequently, there is a pressing need to develop simple and sensitive techniques to quantify levels of miRNAs in a portable and inexpensive way9,19.
To take up such a challenge, electrochemical biosensors find their right application, thanks to their varied advantages such as specificity, portability and low cost20. There is a handful of reports on detection of miRNAs using electrochemical approaches. Electrochemical impedance spectroscopy (EIS) using redox markers has been previously used to quantify target miRNA hybridisation down to femtomolar levels with complex amplification techniques19,21. Other electrochemical techniques such as differential pulse voltammetry (DPV) have also been used to detect miRNAs with a limit of detection (LOD) over picomolar levels22.
However, the developed biosensors reported lack simplicity in their fabrication process. To address this issue, we report the development of a simple and sensitive dual mode electrochemical detection platform for miRNA using peptide nucleic acids (PNA) as probes. PNA was used since it presents many advantages such as neutral charge and higher stability than its biological counterparts. Also, the PNA/miRNA duplex with mismatches is less stable than a DNA/miRNA duplex with the same mismatches23,24. Furthermore, a simple and powerful amplification strategy using positively charged gold nanoparticles (AuNPs) has been employed. EIS was used without redox markers to monitor the changes in the dielectric properties of the bilayer through capacitance changes. Such an approach further simplifies the detection strategy making it suitable for easy integration into the system. Our group previously reported the development of a capacitive and potentiometric sensor for DNA detection based on the same strategy25. In this work, the detection strategy was applied to detect miR-145 sequence using EIS. In order to reduce false positives, a complementary detection technique was developed on the same sensors using square wave voltammetry for validation of the signals. Such a step was performed by exploiting the availability of AuNPs on the surface with PNA/miRNA duplex to attach thiolated ferrocene in order to provide a voltammetric detection using square wave voltammetry (SWV). With the developed dual detection system on the same sensor, a LOD of 0.37 fM was achieved with a wide dynamic range from 1 fM to 100 nM.
Results and Discussion
Characterisation of PNA-based sensor
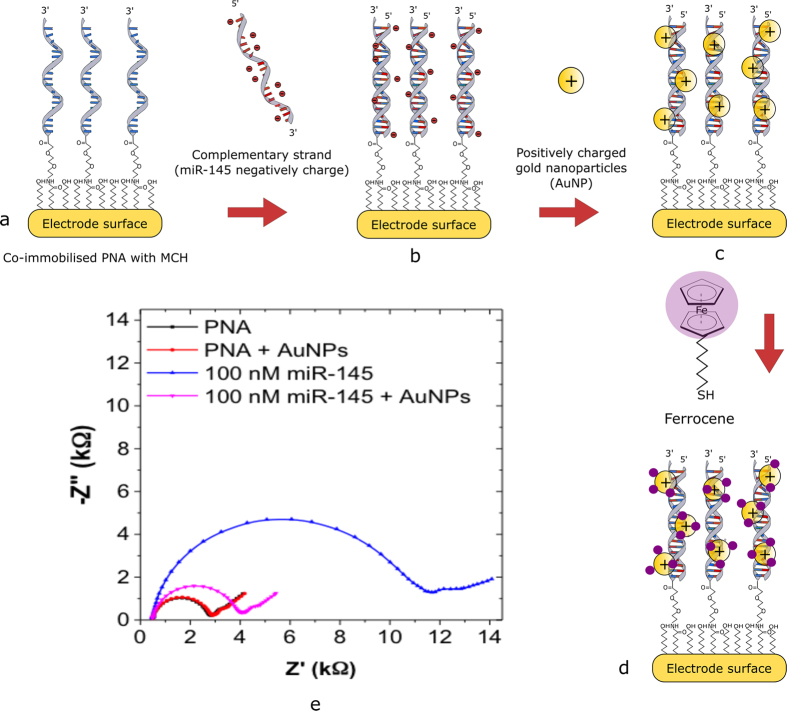
Figure 1 provides a schematic of the detection strategy (a–d) and a Nyquist plot illustrating changes in the electrical properties of the gold electrode during the modification process (e). EIS in a Faradaic mode was used to characterise any molecular binding event. A relatively low Rct of 2.9 kΩ was recorded due to physical barrier imposed by the presence of the self-assembled monolayer (SAM) comprised of co-immobilised uncharged PNA and 6-mercapto 1-hexanol (MCH) on the surface. Initially, the PNA immobilised biosensor was tested with positively charged AuNPs to study non-specific interactions. As seen in Fig. 1 a negligible change in the Rct (ca. 2.5%) of the system was observed, indicating that no AuNPs bind to the system. The PNA-based biosensor was then incubated with 100 nM miR-145 for 30 minutes, where upon hybridisation a significant increase in Rct from 2.9 kΩ to 11.9 kΩ was recorded. An increase of Rct is attributed to the formation of a PNA/miR145 duplex, making the electrode surface highly negatively charged and creating an electrostatic barrier to the negatively charged redox marker in the measurement solution.
Figure 1.
(a) Typical Cole-Cole plot obtained with PNA-based sensor before and after hybridisation with 1 fM miR-145 (black and blue respectively) and AuNP attachment (pink). A zoom-in version of Cole-Cole plot shows the changes in capacitance upon a molecular binding event. (b) Calibration curve and data points represent average changes in capacitance from a minimum of three independent samples upon binding with AuNPs at different concentration of miR-145.
Finally, AuNPs modified with poly(ethylenimine) (PEI) were added, which electrostatically bind to the PNA/miR145 duplex because of the positive charges of the amine groups on the modified AuNPs, resulting in a decrease in the Rct of the system from 11.9 kΩ to 4 kΩ. Such a decrease could be attributed to the screening of negative charges of the PNA/DNA duplex with positively charged AuNPs. It is worth mentioning that the Rct of the system does not return to its initial value. Such an observation could be attributed to the fact that although AuNPs are screening the charges, the mass loading effect still causes an increased physical barrier to the redox couple.
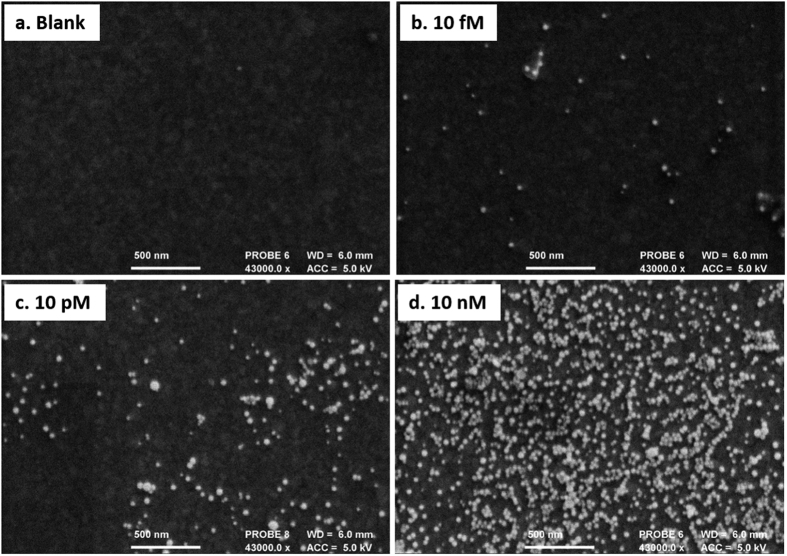
The molecular interaction after incubation with AuNPs was also examined using scanning electron microscopy (SEM) – see Fig. 2. For the blank sample (i.e. no miRNA), no AuNPs are observed showing that there is almost no non-specific interactions between the AuNPs and the PNA SAM. Upon increasing concentrations of target miR-145, more nanoparticles are visible in the samples. It is worth mentioning that aggregation of AuNPs is significantly observed at high concentrations (Fig. 2d: 10 nM).
Figure 2.
SEM micrograph showing the distribution of AuNPs after hybridisation with (a). Blank, (b): 10 fM, (c): 10 pM and (d): 10 nM of mir-145.
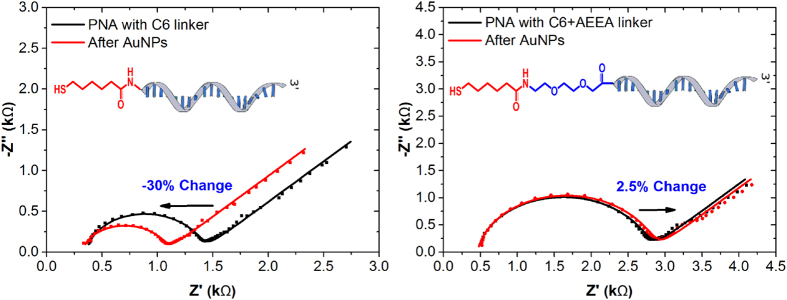
In our previous work, we reported small signal changes in capacitance upon hybridisation with complementary DNA due to the length of the polyethylene glycol (PEG)-like linker with PNA probe25. Therefore, in order to increase the signal change with hybridisation, we studied the effect of the size of the PNA linker. A C6 thiol linker can be used in place of the C6-AEEA (where AEEA is a 8-amino-3,6-dioxaoctanoic acid glycol linker) linker as a PNA modification that can be co-immobilised on the gold surface (see Fig. 3). By reducing the linker length from C6-AEEA to C6, a significant variation in the capacitance of over 5% was observed upon hybridisation with the complementary miRNA strand. However, what was more important in this particular electrochemical platform development, was to test its selectivity efficiency. For the same, the biosensor was constructed using both modifications of PNA (C6 and C6-AEEA) with MCH on the gold surface. Electrodes were subsequently tested with just AuNPs without any hybridisation. EIS in Faradaic mode was used to monitor the changes in Rct. Figure 3 illustrates impedance plots recorded from the EIS experiment. A significant non-specific interaction of around 30% was observed as a result of the reduction in Rct after incubation of PNA-based biosensor with C-6 linker with AuNPs. A decrease in Rct is a result of attachment of positively charged AuNPs to the electrode surface which consequently results in a decrease in the resistance to negatively changed redox couple in the solution.
Figure 3. Faradaic EIS plots of PNA based biosensor.
(Left) Represents C6-Linker modified PNA as a probe for biosensor fabrication and the change in Rct after incubating with AuNPs. (Right) Represents C6-AEEA Linker modified PNA as a probe for biosensor fabrication and the change in Rct after incubating with AuNPs.
Such an observation indicates that the SAM formed on the gold surface is not efficient enough to prevent non-specific interactions. As compared to the C6 linker, when a C6-AEEA linker was employed, a significant reduction in the non-specific interaction to less than 3% was observed showing an enhanced anti-fouling efficiency, thanks to the PEG-like linker. Such an enhanced antifouling effect could be predominantly due to the creation of a hydration layer by the presence of the AEEA. PEG coatings have been previously reported to prevent non-specific binding as each PEG ether group can H-bound with two water molecules creating a strong hydration shell26. Such a hydration layer prevents proteins and other charged molecules to penetrate through the water layer and adsorb on the surface27,28,29. As a result, only C6-AEEA modification of PNA probes was used in our sensors from here on.
Capacitive Sensing
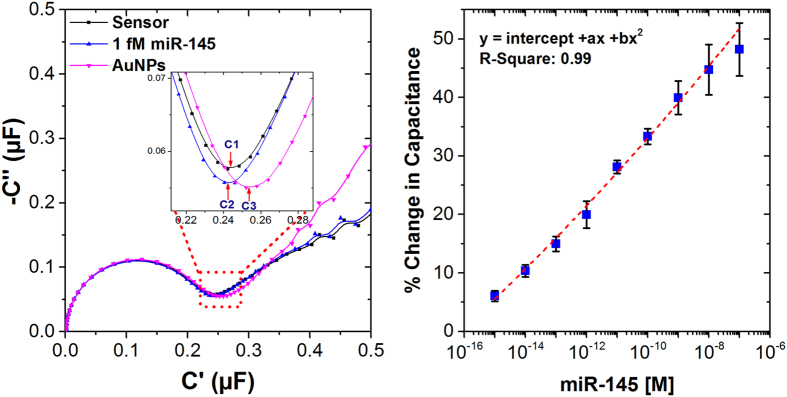
The detection of different concentrations of miR-145 was successfully monitored using EIS in non-Faradaic mode by estimating the changes in the capacitance upon a molecular binding event25,30. The main advantage of using a non-Faradaic mode EIS measurement technique is the simple and easy execution of the measurements compared to any labelled biosensors. In order to evaluate the capacitance of the system, a complex capacitance was defined (see supplementary information, S-1). From the capacitance data, Cole-Cole capacitance plots can be obtained (see supplementary information, Figure S-1). With these plots, the diameter of the obtained semicircle corresponds the capacitance of the system. Figure 4 (left) depicts an example of Cole-Cole plots obtained from non-Faradaic EIS measurements of the electrode modified with a PNA sequence which was used to detect 1 fM of miR-145. An initial capacitance of 0.243 μF was observed with PNA-based sensor (C1) which upon hybridisation with 1 fM of target miR-141, decreased to 0.241 μF (C2), leading to a change of −0.82%. As reported earlier, only a slight variation in capacitance is observed upon miR-145 hybridisation25. The probable reason for this small signal is that the PNA has a linker which is a derivate of PEG (AEEA, C21H23NO6) with a length of 1.4 nM. As a result, the binding event is happening far away from the surface and well above the Debye length of the system, leading to screening of the DNA charges by the electrolyte. However, when incubated with positively charged AuNPs, a significant increase in capacitance was observed due to disruption of the electrochemical double layer. From Fig. 4 (left), incubating with AuNPs leads to a capacitance increase to 0.254 μF (C3), leading to more than 6 folds increase due to AuNPs amplification with a percentage change of +5.4%. Such an amplified response could be attributed to the molecular interactions with AuNPs near the electrode surface where redistribution of the solvent molecules and of ions is initiated, which causes changes in the electrochemical double layer.
Figure 4.
(a) Typical Cole-Cole plot obtained with PNA-based sensor before and after hybridisation with 1 fM miR-145 (black and blue respectively) and AuNP attachment (pink). A zoom-in version of Cole-Cole plot shows the changes in capacitance upon a molecular binding event. (b) Calibration curve and data points represent average changes in capacitance from a minimum of three independent samples upon binding with AuNPs at different concentration of miR-145.
The fabricated PNA-based biosensor was tested with a wide range of miR-145 concentrations from 1 fM to 100 nM in 10 mM PB pH 7.4, where an increase in the change in the capacitance upon AuNP incubation was observed with increasing miRNA concentrations. It is worth mentioning that the electrode was used only once with each concentration. After 30 minutes incubation at each concentration, the variation in capacitance was proportional to the amount of captured miR-145. Based on the variation of capacitance corresponding to attachment of AuNPs, a calibration curve was obtained as a function of miR-145 concentration. The biosensor demonstrated an excellent response (Fig. 4, right). The sensor could potentially detect miR-145 down to 1 fM with a nearly 6% change. The obtained dose-response for the specific capture of miR-145 presented in Fig. 3 follows indeed a typical second order polynomial fit (quadrant equation) with a root mean square value (R-Square) of 0.99 and an intercept of 5.60 + 0.58%. A very low limit of detection (LOD) of 0.37 fM was calculated using 4 independent samples as described by Armbruster et al.31. The demonstrated LOD with the current detection technique is better or comparable to the state-of-art technologies reported for electrochemical detection of miRNAs. Furthermore, the electrochemical sensor presents a simple and inexpensive detection strategy as compared to using complex fabrication steps or expensive enzymes (see Table 1).
Table 1. Comparison with the reported state-of-art electrochemical platforms for miRNA detection.
| Detection Method | Electrode Surface | MicroRNA | LOD | Reference |
|---|---|---|---|---|
| CV & EIS | Gold electrode | miR-21 | 3.96 pM | Zhou et al.39 |
| DPV & EIS | Gold electrode modified with AuNP | miR-21 | 6 fM | Meng et al.40 |
| CV & DPV | Gold electrode | miR-21 | 45 fM | Xia et al.41 |
| EIS | Gold electrode and duplex specific nuclease as an amplification tool | let-7a, let-7b, let-7c | 1 fM | Ren et al.42 |
| DPV & EIS | Gold electrode modified with AuNP | miR-159a | 3.5 fM | Wang et al.43 |
| CV | Gold electrode | miR-21 | 3 fM | Liu et al.44 |
| DPV & EIS | Gold electrode | let-7a, let-7b, let-7c | 99.2 fM | Peng et al.45 |
| CV, DPV & EIS | Gold electrode modified with AuNPs | miR-21 | 0.17 fM | Wang et al.46 |
| DPV | Gold electrode modified with AuNP | miR-139a | 1.7 fM | Zhou et al.47 |
| DPV | Gold electrode modified with AuNP | miR-21 | 0.36 fM | Li et al.48 |
| EIS & SWV | Gold Electrode | mir145 | 0.37 fM | This study |
CV: Cyclic voltammetry; DPV: Differential Pulse Voltammetry; EIS: Electrochemical Impedance Spectroscopy; SWV: Square Wave Voltammetry.
Voltammetric Biosensor
With successful application of a simple amplification strategy to detect miR-145 using capacitance measurements, the setup was explored for additional applications. One such application explored the availability of AuNPs on the surface of the electrode32. Formisano et al. demonstrated the use of AuNPs on the electrode surface as a possible platform to immobilise thiolated ferrocene molecules33; such a modified AuNP with ferrocene was used as a redox indicator by monitoring the oxidation peaks of ferrocene.
Since detecting ultra-low levels of miRNAs can introduce false positives in the signal, a complementary voltammetry technique was developed in the present study by using thiolated ferrocene molecules (6-(ferrocenyl)hexanethiol), which will serve as a validation technique to confirm the binding event. After specific interaction of AuNPs with PNA/miR-145 duplex, 50 μM thiolated ferrocene was flowed through the system, allowing the thiolated ferrocene to covalently bind to the free available spaces of AuNPs. The PNA-based biosensor was tested with a wide range of miR-145 concentrations from 1 fM to 100 nM in 10 mM PB pH 7.4, where an increasing oxidation peak of ferrocene was observed (Fig. 5) after the final step of thiolated ferrocene flow using square wave voltammetry (SWV).
Figure 5. Dose response.
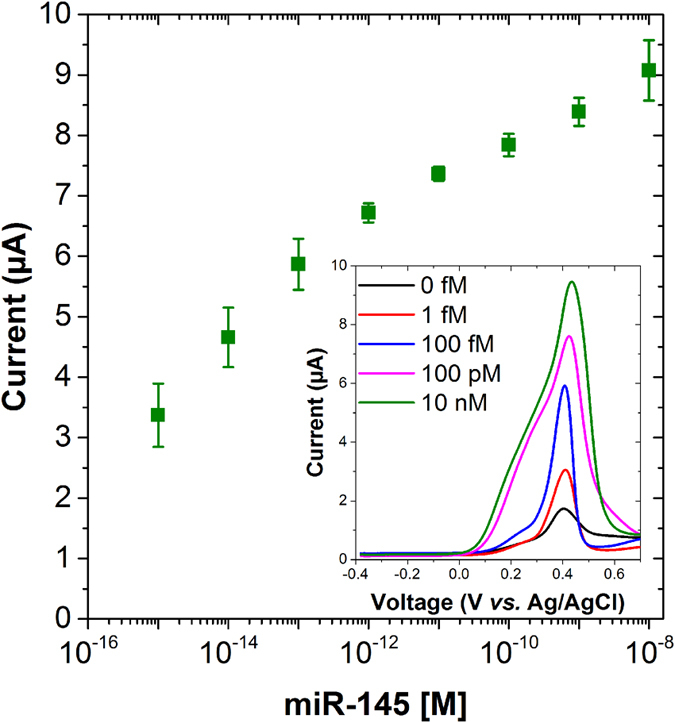
Data points represent average ferrocene peaks recorded using square wave voltammetry (SWV) from three independent samples on the binding of thiolated ferrocene with AuNPs at different concentration of mir145 (left). Inset shows the SWV curves recorded at different miR-145 concentrations.
From Fig. 5, it can be seen that the peak current increases for increasing miR-145 from 1 fM to 1 pM, after which the peak current starts saturating, resulting in similar peak currents. Such saturation could be due to aggregation of AuNPs at a higher miR-145 concentration resulting in decreased charge transfer. A similar effect could be seen in the SWV recording (inset Fig. 5) where as we move towards higher miR-145 concentration, a wider ferrocene peak was observed which could be attributed to the non-homogeneous assembly of ferrocene near the electrode surface. As a result, two processes could be seen due to diffusion effects of electrons; one is the fast oxidation response due to presence of some amount of ferrocene closer to the electrode surface and later a relatively slower response due to presence of ferrocene further away from the gold electrode surface34. The ferrocene peaks are also affected by the environment where, for example, the presence of amine groups could shift the peaks towards more positive potential35. It is worth mentioning that a high background current of nearly 1 μA was recorded. Such a current could be due to two probable reasons. Firstly, due to the non-specific interaction of ferrocene to the electrode surface and secondly, due to the presence of even a small amount of AuNPs due to non-specific interaction which could be used as a platform where ferrocene can attach. Nevertheless, when the blank measurement was compared with the lowest miR-145 concentration used, a six time higher ferrocene peak current was recorded (Fig. 5).
With the application of ferrocene, unlike any other reported biosensor for miRNA detection, a simple and powerful dual detection technique could be performed where the signals from ferrocene can be used as an alternative detection technique to validate the results obtained from the capacitive measurements. As a result, more reliable data could be generated for any molecular event and thereby, reducing overall false positives.
Selectivity Study
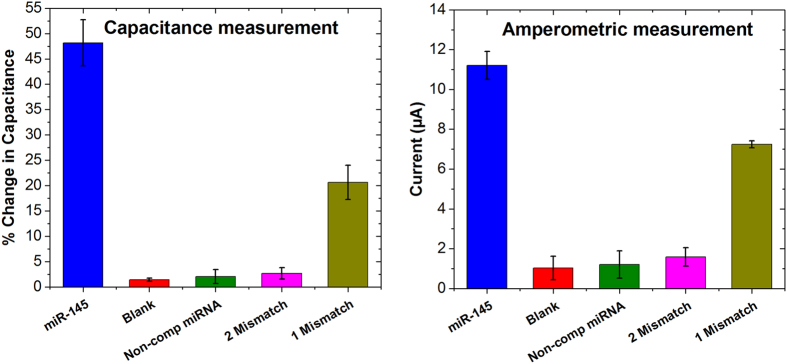
The developed PNA-based biosensor was challenged with stringent controls in order to investigate the selectivity of the sensor. For the study, 100 nM concentration of miR-145 was compared with 100 nM of RNA sequence with 1 mismatch, 2 mismatches, and a non-complementary RNA. The results from both capacitance measurement and SWV are presented in Fig. 6.
Figure 6. Selectivity study with mismatched miRNA sequences.
All the RNAs concentration were 100 nM.
In both types of measurements, the signal change observed with 100 nM of RNA sequence with 2 mismatches and a non-complementary RNA showed negligible signal change when compared with 100 nM miR-145. However, with 1 mismatch sequence, a noteworthy change in the signal was observed with both detection techniques. Such an interaction could be due to the choice of the mismatch. As shown in Table 2, the mismatch is in the middle of the sequence, which is considered the most stable position in PNA/DNA or PNA/miRNA hybridisation36. Nevertheless, the signal change observed was still 50% lower than what was observed with miR-145. The non-specific interaction observed with mismatched sequences could be reduced by adding another parameter of temperature. By using a temperature close to the melting point of the PNA/miR-145 duplex, the non-specific binding of mismatches can be greatly reduced.
Table 2. miRNA sequences and PNA probes used in this work.
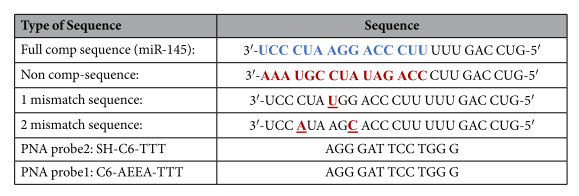
The full complementary sequence in the first row corresponds to the mir-145 sequence. AEEA is a glycol linker of nine atoms (8-amino-3,6-dioxaoctanoic acid).
Conclusions
In the current work, a highly sensitive dual-mode electrochemical biosensor with a simple AuNP amplification for the detection of miR-145 is presented. It has been shown how AuNPs can disrupt the dielectric biolayer of the sensor, leading to significant changes in the capacitance of the system. Also, a complementing detection technique was demonstrated by exploiting the availability of AuNPs on the surface with PNA/miRNA duplex; thiolated ferrocene was used to provide a voltammetric detection using SWV. A dual-mode approach can be used as a validation detection technique to compare the sensor’s performance and reduce inconsistencies in output signals. With the current PNA-based system, a wide dynamic range from 1 fM to 100 nM with a LOD of 0.37 fM was achieved. Of significant importance is that the detection technique reported can easily be expanded into arrays for the parallel screening of panels of DNAs or miRNAs.
Methods
Electrochemical setup
The experiments were carried out with a CompactStat potentiostat (Ivium Technologies, The Netherlands) and a three-electrode cell system: Ag/AgCl (KCl) reference electrode (BASi, USA) connected via a salt bridge filled with 10 mM phosphate buffer (PB, pH 7.4), Pt counter electrode (ALS, Japan) and gold working electrode (2.0 mm diameter, CH Instruments, USA). The non-Faradaic impedance spectrum was conducted in 10 mM PB (pH 7.4) measurement buffer over a frequency range from 100 kHz to 100 mHz, with a 10 mV a.c. voltage superimposed on a bias d.c. voltage of 0 V with respect to the open circuit potential25. The Faradaic impedance spectrum was conducted in 10 mM PB (pH 7.4) measurement buffer containing 10 mM of the ferro/ferricyanide [Fe(CN)6]3−/4− redox couple (potassium hexacyanoferrate II/III, Sigma, UK) over a frequency range from 100 kHz to 100 mHz, with a 10 mV a.c. voltage superimposed on a bias d.c. voltage of 0.2 V vs. Ag/AgCl (which corresponds to the formal potential of the redox couple). The square wave voltammetry technique (SWV) was used to monitor the oxidation peaks of the ferrocene attached to the AuNPs. SWV was performed in 10 mM PB (pH 7.4) in the potential range from −0.4 V to 0.7 V vs. Ag/AgCl with conditioning time of 120 s, modulation amplitude of 20 mV and frequency of 50 Hz.
For characterisation of AuNPs, stock AuNPs were diluted ten times in MilliQ water, and optical absorption spectra were measured using a UV–vis spectrophotometer (UV-1800/TMSPC-8, Shimadzu, UK) using a 8-cell quartz cuvette with 150 μL sample size. The wavelength for absorbance analysis was scanned from 800 nM to 400 nM.
Oligonucleotides
HPLC purified synthetic oligonucleotides were purchased from Sigma-Aldrich, UK in lyophilised form, while PNA probe sequences were purchased from Cambridge Research Biochemical, UK. The sequences are shown in Table 2.
Biosensor fabrication
Gold working electrodes were cleaned using a protocol previously described in Jolly et al.25 electrodes were first mechanically polished for 2 minutes with 1 μM diamond solution (Buehler, USA) and thereafter for 5 minutes with 50 nM alumina slurry (Buehler, USA) on polishing pads (Buehler, USA). In between each step, 10 minutes sonication and rinsing in Milli-Q water were performed to remove any remaining particle residues. Additionally, electrodes were exposed to chemical cleaning using piranha solution (3 parts of concentrated H2S04 with 1 part of H202) for 10 minutes. Electrodes were then rinsed with ultra-pure water and thereafter electrochemically cleaned in 0.5 M H2S04 (Sigma-Aldrich, UK) by scanning the potential between the oxidation and reduction of gold, 0 V and +1.5 V vs. an Ag/AgCl reference electrode, for 50 cycles until no further changes in the voltammogram were noticed. Finally, the electrodes were rinsed with Milli-Q water.
Clean gold electrodes were then co-immobilized with a thiolated ssPNA probe sequence and 6-mercapto-1-hexanol (MCH, Sigma-Aldrich, UK) in 50% dimethyl sulfoxide (DMSO, Sigma-Aldrich, UK), 50% ultra-pure water (v/v) immobilisation solution for 16 h in a humidity chamber. An optimized ratio of 1:5 was adopted from the literature25. After immobilisation, electrodes were rinsed with surplus MilliQ water to remove any unattached thiols. In order to ensure complete thiol coverage of the gold surface, the electrodes were backfilled with 1 mM MCH for 1 hour. Electrodes were then rinsed with MilliQ water and placed in the measurement buffer for 1 hour to stabilise the SAM. The functionalised electrodes were then used to detect target miR-145.
Preparation of positively charged AuNPs was adopted as reported by Kim et al.36. Briefly, an optimised ratio of hydrogen tetrachloroaurate (HAuCl4, Sigma-Aldrich, UK) and branched poly-(ethylenimine) (PEI, MW ~ 25 kDa, Sigma-Aldrich, UK) was mixed overnight using vigorous stirring to produce AuNPs of average size of 20 nM. Before the experiments, the AuNPs were washed three times using centrifugation and re-dispersion in Milli-Q water. Optical characterisation of PEI coated AuNPs was performed using a spectrophotometer where a characteristic absorbance peak at 523 nM (see supplementary information S-2) was observed, which is similar to the previously reported results for AuNPs with 20 nM size37,38.
For the binding studies, different concentrations of target miR-145, non-complementary miRNAs and 1- or 2-mismatched miRNA sequences were prepared in 10 mM PB, pH 7.4.
Surface Characterisation
50 nM gold coated SPR gold chips, supplied from Reichert Technologies were used for studying the reaction. SPR chips were modified with PNA probes as described in the biosensor fabrication section. Binding study with 0 fM, 10 fM, 10 pM and 10 nM miR-145 was performed on the chip followed by incubation with AuNPs. Thereafter, the chips were dried with N2 flow and were left in vacuum overnight. Surface characterization of SPR chips were performed using a scanning electron microscopy (JSM-6480Jeol, Japan) at 43000X magnification with acceleration voltage of 5 kV to acquire SEM images.
Additional Information
How to cite this article: Jolly, P. et al. Highly sensitive dual mode electrochemical platform for microRNA detection. Sci. Rep. 6, 36719; doi: 10.1038/srep36719 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was funded by the European Commission FP7 Programme through the Marie Curie Initial Training Network PROSENSE (grant no. 317420, 2012–2016) and by a Cancer Research at Bath (CR@B) seedcorn grant. MRB is funded by FAPESP (process no. 2015/14403-5). MM and PE acknowledge support from FAPESP and the University of Bath through the SPRINT program.
Footnotes
Author Contributions P.J. conceived, designed and performed the experiments and analysed data. M.R.B., A.M. and P.Z. helped with capacitive and voltammetric measurements. The paper was written by P.J. and M.R.B. with contributions by A.M. and P.Z. and revised by P.E. The work was supervised by M.M., M.A.L. and P.E.
References
- Calin G. A. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. P Natl Acad Sci USA 99(24), 15524–15529 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Horvitz H. R. & Sulston J. E. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 24(1), 59–69 (1981). [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L. & Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75(5), 843–854 (1993). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116(2), 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A. & Slack F. J. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 6(4), 259–269 (2006). [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A. & Enright A. J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieglschmid V., Hollmann C. & Bocher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Cl Lab Sci. 42(2), 155–196 (2005). [DOI] [PubMed] [Google Scholar]
- He L. et al. A microRNA polycistron as a potential human oncogene. Nature. 435(7043), 828–833 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. MicroRNA expression profiles classify human cancers. Nature. 435(7043), 834–838 (2005). [DOI] [PubMed] [Google Scholar]
- Ramaswamy S. et al. Multiclass cancer diagnosis using tumor gene expression signatures. P Natl Acad Sci USA 98(26), 15149–15154 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimi T. et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 125(2), 345–352 (2009). [DOI] [PubMed] [Google Scholar]
- Akao Y., Nakagawa Y. & Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 26(5), 311–320 (2007). [DOI] [PubMed] [Google Scholar]
- Spizzo R. et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ 17(2), 246–254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam E. J. et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 14(9), 2690–2695 (2008). [DOI] [PubMed] [Google Scholar]
- Wang C. et al. Tumor-suppressive microRNA-145 induces growth arrest by targeting SENP1 in human prostate cancer cells. Cancer Sci. 106(4), 375–382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoczi A. et al. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 32(22), e175 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catuogno S. et al. Recent Advance in Biosensors for microRNAs Detection in Cancer. Cancers. 3(2), 1877–1898 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33(20), e179 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Deng H., Shen W. & Gao Z. A highly sensitive and selective electrochemical biosensor for direct detection of microRNAs in serum. Anal Chem. 85(9), 4784–4789 (2013). [DOI] [PubMed] [Google Scholar]
- Wang J. Electrochemical biosensors: towards point-of-care cancer diagnostics. Biosens Bioelectron. 21(10), 1887–1892 (2006). [DOI] [PubMed] [Google Scholar]
- Peng Y. & Gao Z. Amplified detection of microRNA based on ruthenium oxide nanoparticle-initiated deposition of an insulating film. Anal Chem. 83(3), 820–827 (2011). [DOI] [PubMed] [Google Scholar]
- Kilic T. et al. Electrochemical based detection of microRNA, mir21 in breast cancer cells. Biosens Bioelectron. 38(1), 195–201 (2012). [DOI] [PubMed] [Google Scholar]
- Demidov V. V. et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol. 48(6), 1310–1313 (1994). [DOI] [PubMed] [Google Scholar]
- Ray A. & Norden B. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J. 14(9), 1041–1060 (2000). [DOI] [PubMed] [Google Scholar]
- Jolly P., Wong L., Miodek A., Lindsay M. A. & Estrela P. A simple and highly sensitive electrochemical platform for detection of MicroRNAs. P IEEE Sensors. 1–4 (2015). [Google Scholar]
- Wang R. L. C., Kreuzer H. J. & Grunze M. Molecular conformation and solvation of oligo (ethylene glycol)-terminated self-assembled monolayers and their resistance to protein adsorption. J Phys Chem B. 101(47), 9767–9773 (1997). [Google Scholar]
- Balamurugan S., Obubuafo A., Soper S. A., McCarley R. L. & Spivak D. A. Designing highly specific biosensing surfaces using aptamer monolayers on gold. Langmuir. 22(14), 6446–6453 (2006). [DOI] [PubMed] [Google Scholar]
- Tegoulia V. A., Rao W., Kalambur A. T., Rabolt J. F. & Cooper S. L. Surface properties, fibrinogen adsorption, and cellular interactions of a novel phosphorylcholine-containing self-assembled monolayer on gold. Langmuir. 17(14), 4396–4404 (2001). [Google Scholar]
- White A. & Jiang S. Local and bulk hydration of zwitterionic glycine and its analogues through molecular simulations. J Phys Chem B. 115(4), 660–667 (2010). [DOI] [PubMed] [Google Scholar]
- Jolly P. et al. Label-free impedimetric aptasensor with antifouling surface chemistry: A prostate specific antigen case study. Sensors Actuat B-Chem. 209, 306–312 (2015). [Google Scholar]
- Armbruster D. A., Tillman M. D. & Hubbs L. M. Limit of blank, limit of detection and limit of quantitation. Clin. Chem. 40(7), 1233–1238 (2008). [PubMed] [Google Scholar]
- Kwon D., Jeong H. & Chung B. H. Label-free electrochemical detection of human α-thrombin in blood serum using ferrocene-coated gold nanoparticles. Biosens Bioelectron. 28(1), 454–458 (2011). [DOI] [PubMed] [Google Scholar]
- Formisano N. et al. Multimodal electrochemical and nanoplasmonic biosensors using ferrocene-crowned nanoparticles for kinase drug discovery applications. Electrochem Commun. 57, 70–73 (2015). [Google Scholar]
- Song P. et al. A mechanistic study of the EC′ mechanism–the split wave in cyclic voltammetry and square wave voltammetry. RSC Adv. 6(74), 70237–70242 (2016). [Google Scholar]
- Ho M. Y., Li P., Estrela P., Goodchild S. & Migliorato P. Detection of molecular interactions with modified ferrocene self-assembled monolayers. J Phys Chem B. 114(32), 10661–10665 (2010). [DOI] [PubMed] [Google Scholar]
- Igloi G. L. Variability in the stability of DNA–peptide nucleic acid (PNA) single-base mismatched duplexes: Real-time hybridization during affinity electrophoresis in PNA-containing gels. P Natl Acad Sci USA 95(15), 8562–8567 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Lee H. B., Lee J. W., Park H. K. & Shin K. S. Self-assembly of poly (ethylenimine)-capped Au nanoparticles at a toluene– water interface for efficient surface-enhanced Raman scattering. Langmuir. 24(14), 7178–7183 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang Xiao-Dong et al. Size-dependent radiosensitization of PEG-coated gold nanoparticles for cancer radiation therapy. Biomaterials. 33(27), 6408–6419 (2012). [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wang M., Meng X., Yin H. & Ai S. Amplified electrochemical microRNA biosensor using a hemin-G-quadruplex complex as the sensing element. RSC Adv. 2(18), 7140–7145 (2012). [Google Scholar]
- Meng X. et al. Electrochemical determination of microRNA-21 based on bio bar code and hemin/G-quadruplet DNAenzyme. Analyst. 138(12), 3409–3415 (2013). [DOI] [PubMed] [Google Scholar]
- Xia N., Zhang L., Wang G., Feng Q. & Liu L. Label-free and sensitive strategy for microRNAs detection based on the formation of boronate ester bonds and the dual-amplification of gold nanoparticles. Biosens Bioelectron. 47, 461–466 (2013). [DOI] [PubMed] [Google Scholar]
- Ren Y., Deng H., Shen W. & Gao Z. A highly sensitive and selective electrochemical biosensor for direct detection of microRNAs in serum. Anal Chem. 85(9), 4784–4789 (2013). [DOI] [PubMed] [Google Scholar]
- Wang M. et al. A label-free electrochemical biosensor for microRNA detection based on apoferritin-encapsulated Cu nanoparticles. J Solid State Electrochem. 18(10), 2829–2835 (2014). [Google Scholar]
- Liu L. et al. Highly sensitive and label-free electrochemical detection of microRNAs based on triple signal amplification of multifunctional gold nanoparticles, enzymes and redox-cycling reaction. Biosens Bioelectron. 53, 399–405 (2014). [DOI] [PubMed] [Google Scholar]
- Peng Y., Jiang J. & Yu R. A sensitive electrochemical biosensor for microRNA detection based on streptavidin–gold nanoparticles and enzymatic amplification. Anal Method Instrum. 6(9), 2889–2893 (2014). [Google Scholar]
- Wang M. et al. One-step, ultrasensitive, and electrochemical assay of microRNAs based on T7 exonuclease assisted cyclic enzymatic amplification. Anal Chem. 86(12), 5606–5610 (2014). [DOI] [PubMed] [Google Scholar]
- Zhou Y. et al. Electrochemical biosensor for microRNA detection based on poly (U) polymerase mediated isothermal signal amplification. Biosens Bioelectron. 79, 79–85 (2016). [DOI] [PubMed] [Google Scholar]
- Li B. et al. Two-stage cyclic enzymatic amplification method for ultrasensitive electrochemical assay of microRNA-21 in the blood serum of gastric cancer patients. Biosens Bioelectron. 79, 307–312 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.