Abstract
Introduction of clonal reproduction through seeds (apomixis) in crops has the potential to revolutionize agriculture by allowing self-propagation of any elite variety, in particular F1 hybrids. In the sexual model plant Arabidopsis thaliana synthetic clonal reproduction through seeds can be artificially implemented by (i) combining three mutations to turn meiosis into mitosis (MiMe) and (ii) crossing the obtained clonal gametes with a line expressing modified CENH3 and whose genome is eliminated in the zygote. Here we show that additional combinations of mutations can turn Arabidopsis meiosis into mitosis and that a combination of three mutations in rice (Oryza sativa) efficiently turns meiosis into mitosis, leading to the production of male and female clonal diploid gametes in this major crop. Successful implementation of the MiMe technology in the phylogenetically distant eudicot Arabidopsis and monocot rice opens doors for its application to any flowering plant and paves the way for introducing apomixis in crop species.
Keywords: meiosis, mitosis, apomixis, rice, MiMe
Introduction
Asian rice (Oryza sativa L.), the first cereal of human consumption, is the staple food for more than half of the mankind. Current rice production (738 million tons of paddy rice over 160 million hectares) has to rise by more than 20% in the next 20 years1 on the same arable land surface, to meet the demand of an increasing world population mainly residing in rice-eating countries. Enhancing yield potential, limiting inputs and enabling adaptation to unstable biotic and abiotic environments are current breeding targets. Due to the autogamous mode of reproduction, rice varieties are generally pure lines. However, occurrence of heterosis in rice hybrids that translates into a 15%-20% increase in yield potential2,3 prompted the development of F1 hybrids. Hybrid rice has been successfully deployed in India, Indonesia, Vietnam, the Philippines, and in China it is grown in more than 50% of the rice-grown areas4. However, the seed cost and the necessity to buy new seeds every year prevent its large adoption by subsistence farmers5. Creating hybrids that can breed true through a clonal mode of reproduction would result in major agricultural benefits for many crops, including rice, allowing the full harnessing of hybrid vigor6.
Apomixis is an asexual reproductive process through seeds that bypasses meiosis and fertilization, to produce offspring genetically identical to the mother6. Apomixis occurs in over 400 angiosperm species and is notably common in the Poacae family, but is absent from the most important crops7. Genetic analyses of the inheritance of apomixis in several species including the cereal and grass relatives of the genera Tripsacum, Pennisetum, Panicum, Bracchiaria and Paspalum have identified a single-chromosome segment that can induce apomixis (reviewed in8). However, attempts to transfer this chromosomal segment to their respective cultivated relatives have been unsuccessful9,10. The existence of apomictic relatives in cereals and grasses has prompted the screening of wild rice relatives but no natural source of apomixis was discovered11,12. One possible alternative to create apomixis in crop plants, and notably in rice, is to de novo engineer clonal reproduction by targeted modification of the sexual reproduction mode, which has been shown to be possible in Arabidopsis13 (Supplementary information, Figure S1).
The first component of apomixis is apomeiosis, which is the conversion of the meiotic division into a mitotic-like division, leading to the formation of functional, diploid clonal gametes. Several meiotic mutants have been identified in Arabidopsis, maize or rice that produce apomeiosis-derived gametes14. However, most of these mutations lead to almost complete male and female sterility, and the production of unreduced gametes occurs at a low frequency (e.g.,15). One exception is the Arabidopsis MiMe (Mitosis instead of Meiosis) genotype, in which meiosis is turned into a mitotic-like division, associated with both high fertility and production of clonal diploid gametes at a very high frequency (virtually 100%)16,17. MiMe is the combination of mutations in three genes (SPO11-1, REC8 and OSD1), each mutation impairing one of the three main processes that distinguish meiosis from mitosis. First, spo11-1 abolishes meiotic recombination18. Second, the mutation of REC8 causes the separation of sister chromatids at first meiotic division, instead of the distribution of homologous chromosomes19. Last, osd1 causes the skipping of the second meiotic division16. Therefore, meiosis in MiMe occurs without recombination and distributes sister chromatids in a single round of division, mimicking a mitotic division16. This results in the production of clonal male and female gametes, but leads to doubling of ploidy at each generation when self-fertilized. Crossing a MiMe plant as male or female with a line whose genome is eliminated following fertilization (lines expressing modified CENH3) leads to the production of clonal offspring13,20.
The original MiMe genotype is a triple mutant Atspo11-1 Atrec8 osd116. To facilitate the transfer of the MiMe technology to crops, it would be useful to have more potential target genes, notably when the mutant resource is limited or when the genes are represented by several paralogs. For AtREC8, there is currently no alternative as the single other gene known to be required for monopolar orientation of kinetochores is SCC3, which is also expressed mitotically and thus the corresponding mutation would affect regular development19. Two alternatives to osd1 can be used to create MiMe, tam and TDMΔD17,21. Finally, besides AtSPO11-1, several genes have been shown to be essential for initiation of meiotic recombination through DNA double-strand break formation (AtSPO11-2, PRD1, PRD2, PRD3/PAIR1, DFO and MTOPVIB)22,23,24,25,26, but are not yet tested to create the MiMe phenotype.
Given the potential interest in developing apomictic plants and establishing the MiMe genotype in crops, the objective of this study was three-fold: (i) extend the catalog of genes that can be mutated to create the MiMe phenotype; (ii) identify the ortholog of the key OSD1 gene in rice; (iii) determine whether one could establish the MiMe phenotype in rice.
Results
Several mutant combinations give rise to the MiMe phenotype in Arabidopsis
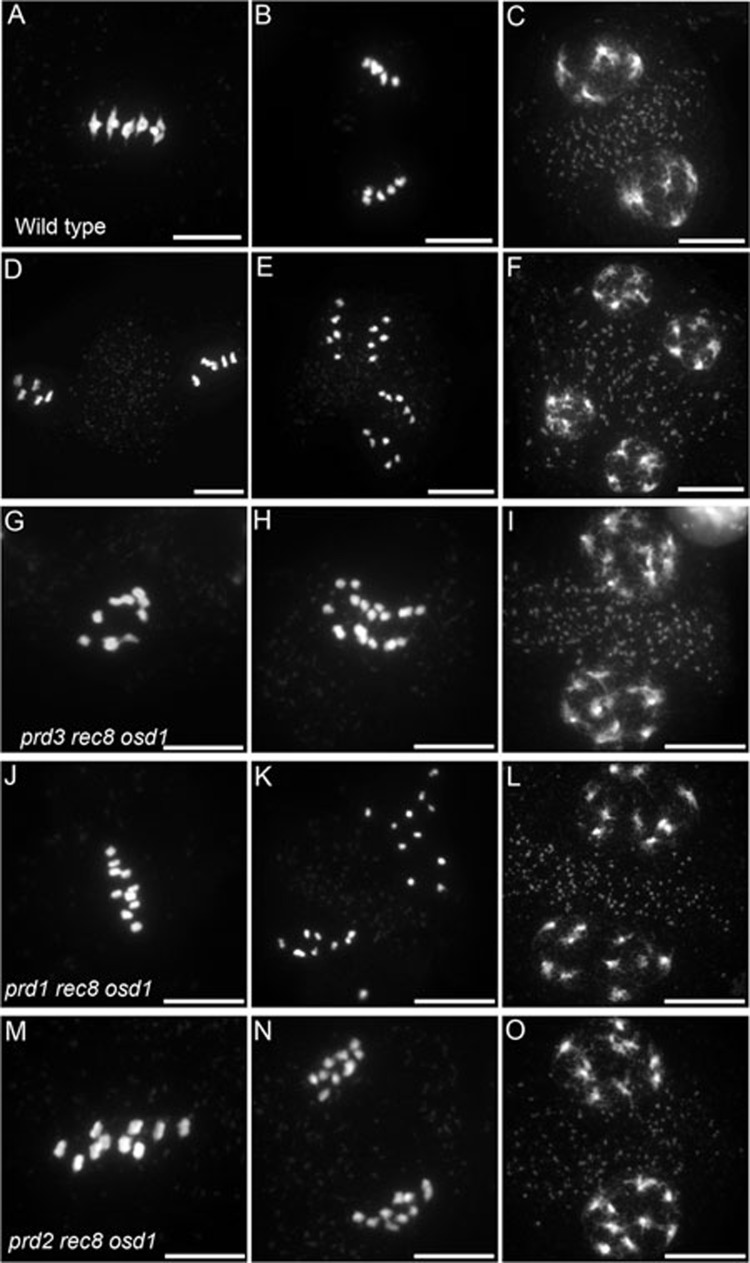
To test the ability of other mutations than spo11-1 to generate MiMe, we constructed triple mutants prd1 Atrec8 osd1, prd2 Atrec8 osd1 and prd3 Atrec8 osd1 and analyzed their meiotic behavior (Figure 1). In wild type, five bivalents align at metaphase I (Figure 1A), and segregate into two groups of five chromosomes at anaphase I (Figure 1B). After a short interkinesis (Figure 1C), the five pairs of chromatids align on the two metaphase II plates (Figure 1D). The sister chromatids segregate at anaphase II (Figure 1E), giving rise to four spores each containing five chromatids (Figure 1F). In each of the triple mutants, meiosis was modified toward a mitotic-like division: ten univalents aligned on the metaphase I plate (Figure 1G, 1J and 1M) and 10 pairs of sister chromatids segregated evenly at anaphase I (Figure 1H, 1K and 1N). No second division occurred, resulting in the production of two spores each containing 10 chromatids (Figure 1I, 1L and 1O). Thus in this series of triple mutants, meiosis recapitulated the MiMe phenotype. It is likely that mutations in any of the genes required for double-strand break formation, in combination with osd1 and Atrec8, can cause the MiMe phenotype, enlarging the list of genes than can be targeted.
Figure 1.
Chromosome spreads of male meiosis in wild-type Arabidopsis and Arabidopsis MiMe genotypes. (A-F) Wild type. (A) Metaphase I with five aligned bivalents. (B) Anaphase I. (C) Telophase I. (D) Metaphase II. (E) Anaphase II. (F) Telophase II. (G-I) prd3 rec8 osd1 triple mutant (n = 17). (J-L) prd1 rec8 osd1 triple mutant (n = 21). (M-O) prd2 rec8 osd1 triple mutant (n = 27). (G, J, M) Metaphase I with 10 aligned univalents. (H, K, N) Anaphase I with segregation of 10 pairs of chromatids. (I, L, O) Telophase. Scale bar = 10 μm.
Identification of OsOSD1
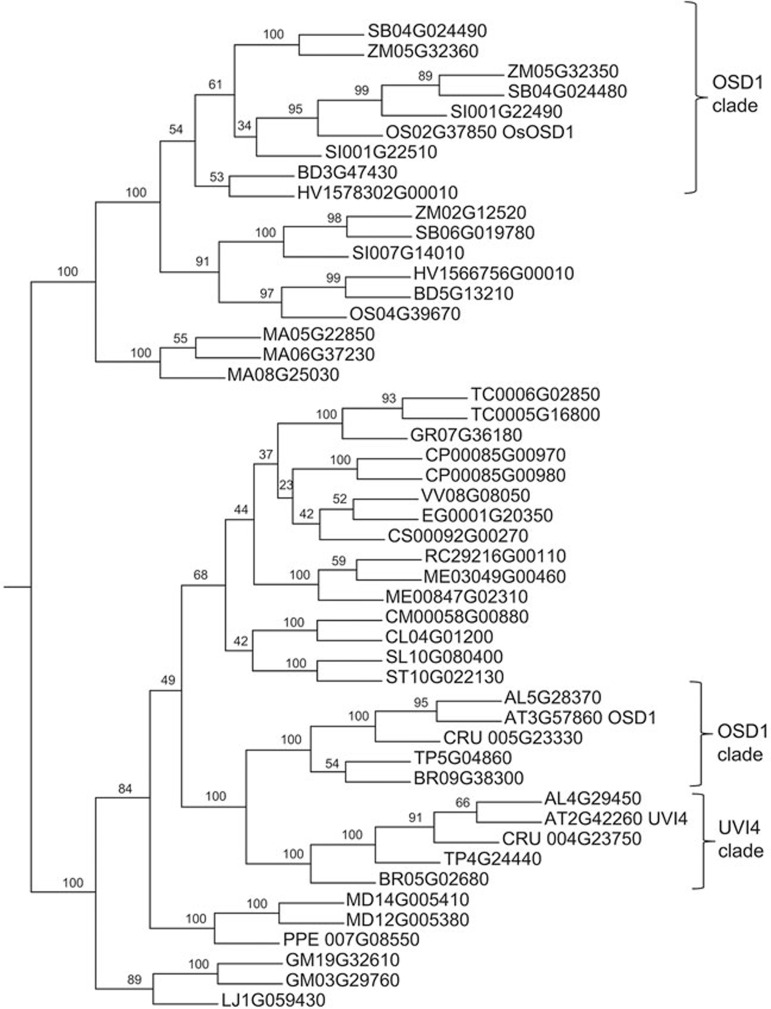
There is one OSD1 homologue in the Arabidopsis genome, UVI4. Mutation of UVI4 affects the somatic cell cycle but not meiosis27. These two genes originated from a whole-genome duplication that is shared by all Brassicaceae28. Accordingly, two genes representative of this family are typically found in Brassicaceae species and sequence similarity analysis clearly distinguishes the OSD1 proteins from the UVI4 proteins (Figure 2). In other eudicots, many species have only one representative of the gene family, and when two representatives are present they result from a recent whole-genome duplication28 and are not shared with the closest diploid species analyzed (see, for example, GM Glycine max or ME Manihot esculenta in Figure 2). In Poaceae, an independent whole-genome duplication28 led to two subgroups of genes, with each species having at least one representative of both (e.g., OS Oryza sativa, HV Hordeum vulgare, ZM Zea mays in Figure 2). This raised the possibility that these two Poaceae gene families could represent OSD1 and UVI4 functional homologues, even though they originated from distinct duplications from their Brassicaceae counterparts. With the aim of exploring the function of these genes in Poaceae, we searched for mutants in the international rice collections. No insertion was identified in Os04g39670, hindering the study of this gene in the present work. However, three insertions were identified in Os02g37850. These three insertions, one T-DNA (AMBA12) and two Tos17 inserts (AMQF10 and ALJA10), are in the second intron (Supplementary information, Figure S2).
Figure 2.
Phylogram of OSD1 and UVI4 homologues from flowering plants. Consensus tree of OSD1 and UVI4 protein family inferred from a Muscle alignment. Analyses were performed using the MPI Bioinformatic Toolkit50, Muscle and Phylip-neighbor with default parameters, enabling boostrapping with 100 replicates. The numbers of forks indicate the number of times the group consisting of the species that are to the right of that fork occurred among the trees, out of 100 trees. AT, Arabidopsis thaliana; AL, Arabidopsis lyrata; BD, Brachypodium distachyon; BR, Brassica rapa (turnip mustard); CL, Citrullus lanatus (water melon); CM, Cucumis melo (muskmelon); CP, Carica papaya (papaya); CRU, Capsella rubella; CS, Citrus sinensis (orange); EG, Eucalyptus grandis (eucalyptus); GM, Glycine max (soybean); GR, Gossypium raimondii (cotton); HV, Hordeum vulgare (barley); LJ, Lotus japonicus; MA, Musa acuminata (banana); MD, Malus domestica (apple); ME, Manihot esculenta (cassava); OS, Oryza sativa (rice); PPE Prunus persica (peach); RC Ricinus communis (castor bean plant); SB, Sorghum bicolor (sorghum); SI, Setaria italica (foxtail millet); SL Solanum lycopersicum (tomato); ST, Solanum tuberosum (potato); TC, Theobroma cacao (cacao); VV, Vitis vinifera (grape vine); ZM, Zea mays (maize).
Because of omission of the second meiotic division, Arabidopsis osd1 produces diploid male and female gametes and its self-fertilization generates tetraploid progeny. To address the function of Os02g37850, we used flow cytometry to determine the ploidy of the progeny that are homozygous for the AMBA12, AMQF10 and ALJA10 mutations. This revealed the presence of 100% (n = 82), 37% (n = 27) and 5% (n = 20) of tetraploid plants in the respective progeny, the rest being diploid. This suggests that mutations of Os02g37850 can provoke the production of male and female diploid gametes. AMBA12 appears to be a null mutant, while AMQF10 and ALJA10 appear partially affected. Consistently, RT-PCR experiments showed that the OSD1 spliced mRNA is barely detectable in AMBA12 homozygotes, and is only reduced in abundance in AMQF10 (Supplementary information, Figure S3). We then compared meiosis in wild type and AMBA12. First, observation of male meiotic products revealed the presence of 100% dyads (n = 500) in AMBA12 homozygotes, instead of tetrads in wild type (Figure 3), showing that a single meiotic division occurs in AMBA12. In AMQF10, a mixture of dyads (29%) and tetrads were observed (n = 130), in accordance with the mixed ploidy in offspring of this mutant. Chromosome spreads showed that the first meiotic division in AMBA12 was indistinguishable from wild type (Figure 4), suggesting that crossover formation and homologous chromosome segregation were unaffected, but no second division took place. Thus, a single first meiotic division occurred in AMBA12. In summary, disruption of Os02g37850 recapitulated the Arabidopsis osd1 meiotic defect: the omission of the second meiotic division which leads to the production of functional diploid gametes. We therefore named Os02g37850 OsOSD1 and AMBA12 Ososd1-1.
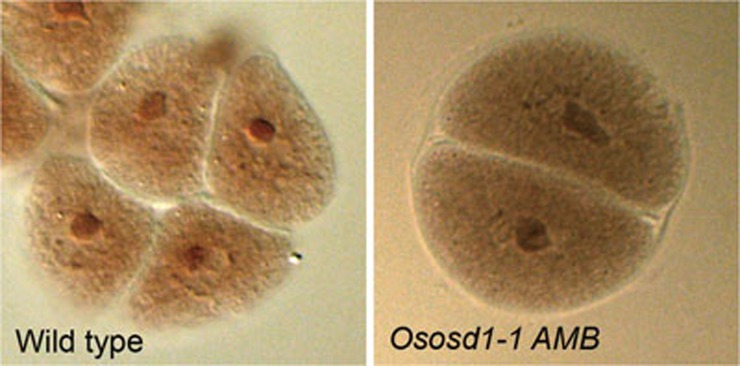
Figure 3.
Male meiotic products in wild-type rice and Ososd1. Fresh anthers squashed in acetocarmine. (A) Tetrads of spores in wild type. (B) Dyad of spores in Ososd1-1 (n = 500).
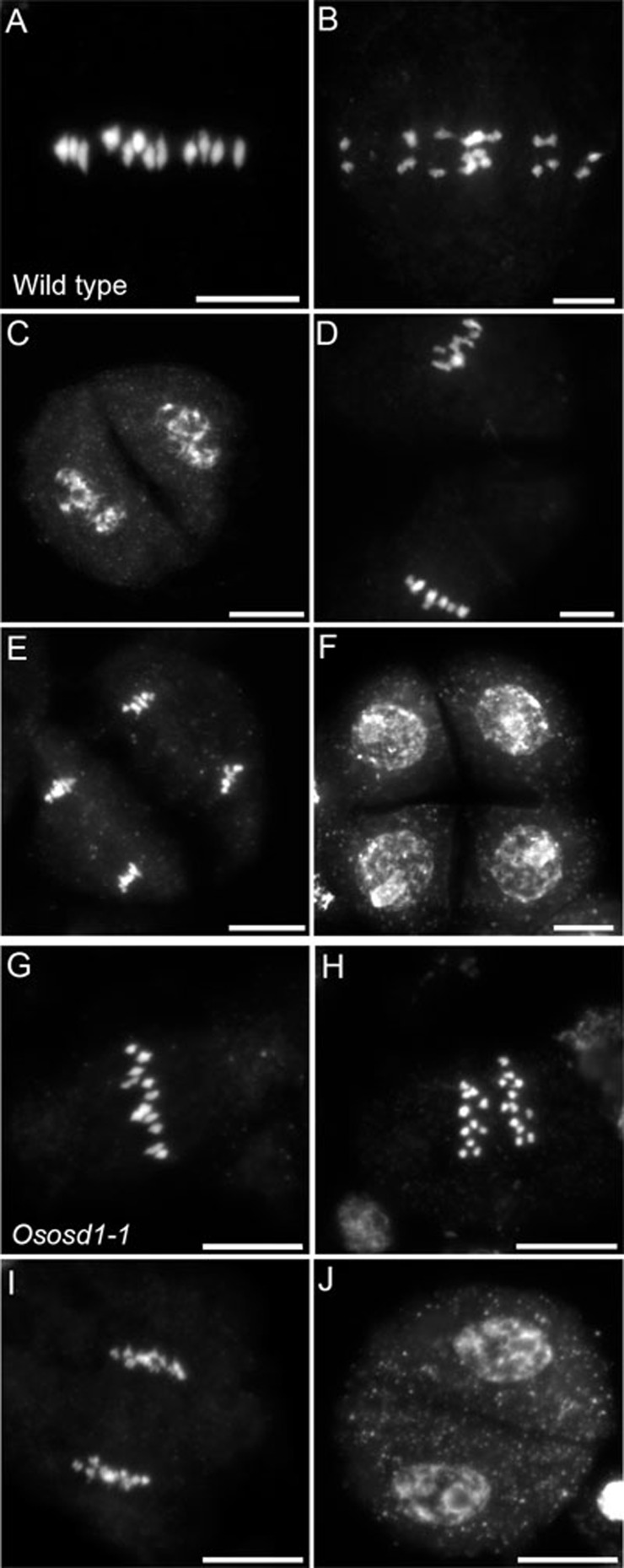
Figure 4.
Chromosome spreads of male meiosis in wild-type rice and Ososd1. (A-F) Wild type. (A) Metaphase I with 12 aligned bivalents. (B) Anaphase I. (C) Telophase I. (D) Metaphase II. (E) Anaphase II. (F) Telophase II. (G-H) Ososd1-1 (n = 62). (G) Metaphase I with 12 aligned bivalents. (H) Anaphase I. (I) Late anaphase I. (J) Telophase I. No second division was observed. Scale bar = 10 μm.
Frequencies of diploid gametes in Ososd1-1 mutant
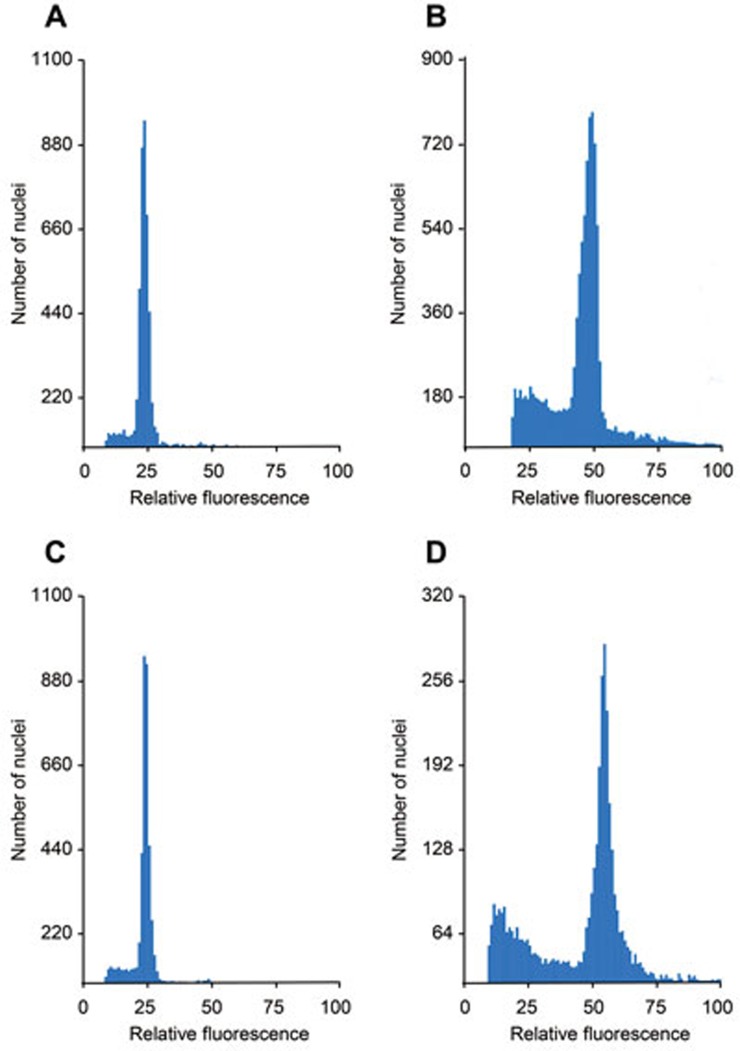
To directly determine the ploidy of male gametes in the Ososd1-1 mutant, we examined the DNA content of nuclei isolated from mature pollen grains with flow cytometry (Figure 5). Pollen nuclei isolated from OsOSD1+/+ (n = 5) and OsOSD1+/− plants (n = 6) exhibited the expected haploid DNA content (Figure 5A and 5C), which was half of the DNA content in leaf nuclei (Figure 5B). In contrast, pollen nuclei isolated from Ososd1−/− plants (n = 3) formed a unique peak corresponding to a diploid DNA content. This indicates that all pollen grains produced by Ososd1 mutants are diploid. We then examined the female meiosis product of Ososd1−/− plants by fertilizing them with wild-type pollen. Among 112 seeds obtained, 104 were shrunken and unable to germinate, whereas 8 developed normally and germinated. The obtained eight offspring were diploid and heterozygous for the Ososd1-1 mutation, indicating they were the result of the fertilization of haploid female gametes produced by Ososd1-1. To recover more offspring, we repeated the experiment, by rescuing the embryo at 5 days post fertilization and transferring the resulting plants in soil. Among 47 successfully hybridized flowers, 40 exhibited early signs of abnormal development, including absence of albumen, and developed into triploid plants after embryo rescue. The seven remaining plants, derived from normally developed seeds, were diploid and Ososd1+/−. Altogether, these results show that ∼91% of female gametes produced by Ososd1-1 were diploid and ∼9% (15/159) were haploid. This is reminiscent of the effect of the osd1 mutation in Arabidopsis, in which ∼100% of male gametes and ∼85% of female gametes are diploid16. However, in contrast to the situation in Arabidopsis, seeds resulting from the fertilization of a haploid female gamete by a diploid male gamete in rice are not viable and require embryo rescue, due to abnormal endosperm development29. The identification of the OSD1 ortholog in rice and the high frequency of diploid gametes produced by the corresponding mutant open the way to creating MiMe in this species.
Figure 5.
Ploidy of Ososd1-1 pollen grains. The ploidy was determined using staining of isolated nuclei with propidium iodide followed by flow cytometry. (A) Wild-type Nipponbare pollen grains. A single peak is observed, corresponding to haploid nuclei. (B) Wild-type Nipponbare leaf. A single peak, corresponding to diploid nuclei. (C) Ososd1+/+ pollen grains. A single peak is observed, corresponding to haploid nuclei. (D) Ososd1−/− pollen grains. A single peak is observed, corresponding to diploid nuclei.
Generating MiMe in rice
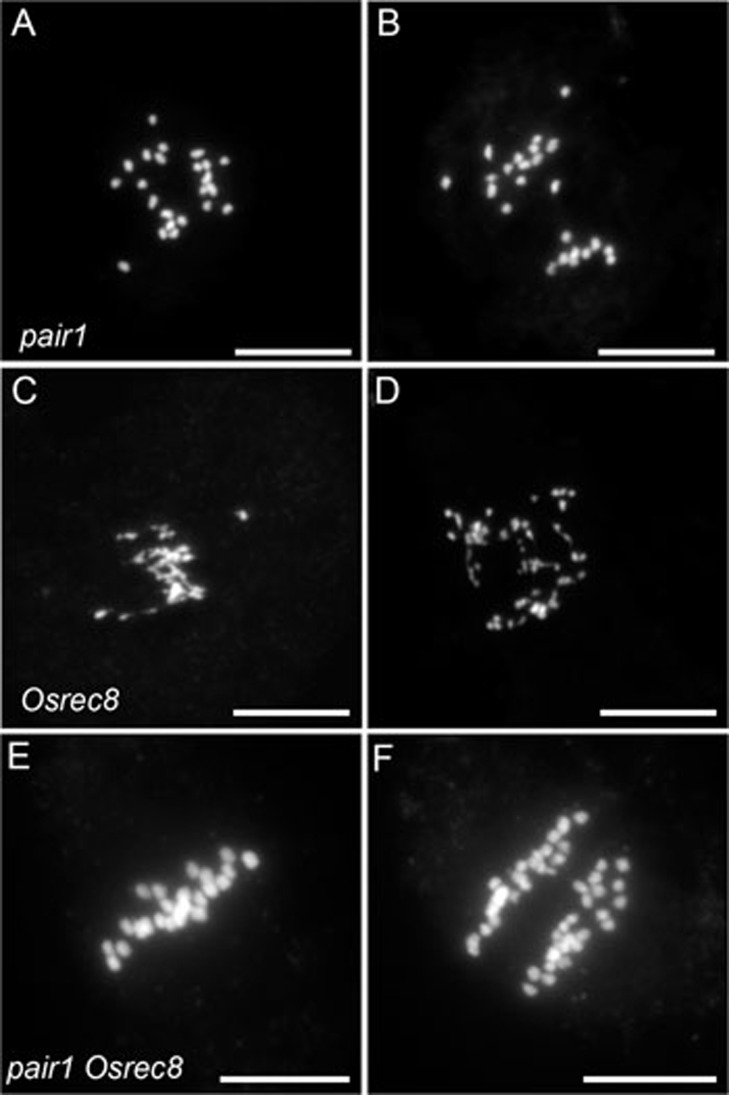
To generate the MiMe genotype, Ososd1 mutation must be combined with a mutation that abolishes recombination, and Osrec8 that modifies chromatid segregation. The PAIR1 gene has been previously shown to be required for meiotic recombination initiation in rice30. One allele (Supplementary information, Figure S2) was selected in a Nipponbare background (AQUG12/pair1-4). As previously described for pair1 alleles, plants homozygous for this insertion were fully sterile. In pair1-4 meiocytes, 24 univalents were observed at metaphase I instead of 12 bivalents in wild type, followed by unbalanced distribution of univalents at anaphase I, suggesting that recombination was completely abolished by this allele (Figure 6A and 6B). OsREC8 has also been functionally characterized31. Osrec8 mutation leads to sister chromatid cohesion defect and chromosome fragmentation, similar to Atrec819,32,33. We identified a novel T-DNA allele in the Hwayoung genetic background (Postech mutant line B01997/Osrec8-3) which caused full sterility (Supplementary information, Figure S2). Meiotic chromosome spreads from Osrec8-3 meiocytes displayed entangled mass of DNA at metaphase I, followed by chromosome fragmentation and missegregation (Figure 6C and 6D), similar to effects of other Osrec8 alleles. In summary, three single mutants pair1-4, Osrec8-3 and Ososd1-1 exhibited the same phenotypes as the three mutants used to build the MiMe genotype in Arabidopsis.
Figure 6.
Male meiosis I in rice pair1, Osrec8 and pair1 Osrec8 mutants. (A-B) pair1 (n = 154). (A) Metaphase I with 24 unaligned univalents. (B) Anaphase I with unbalanced segregation of univalents. (C, D) Osrec8 (n = 222). (C) Metaphase I with abnormal chromosomes. (D) Anaphase I with chromosome fragmentation. (E, F) pair1 Osrec8 (n = 154). (E) Metaphase I with 24 aligned univalents. (F) Anaphase I with segregation of 24 pairs of chromatids. Scale bar = 10 μm.
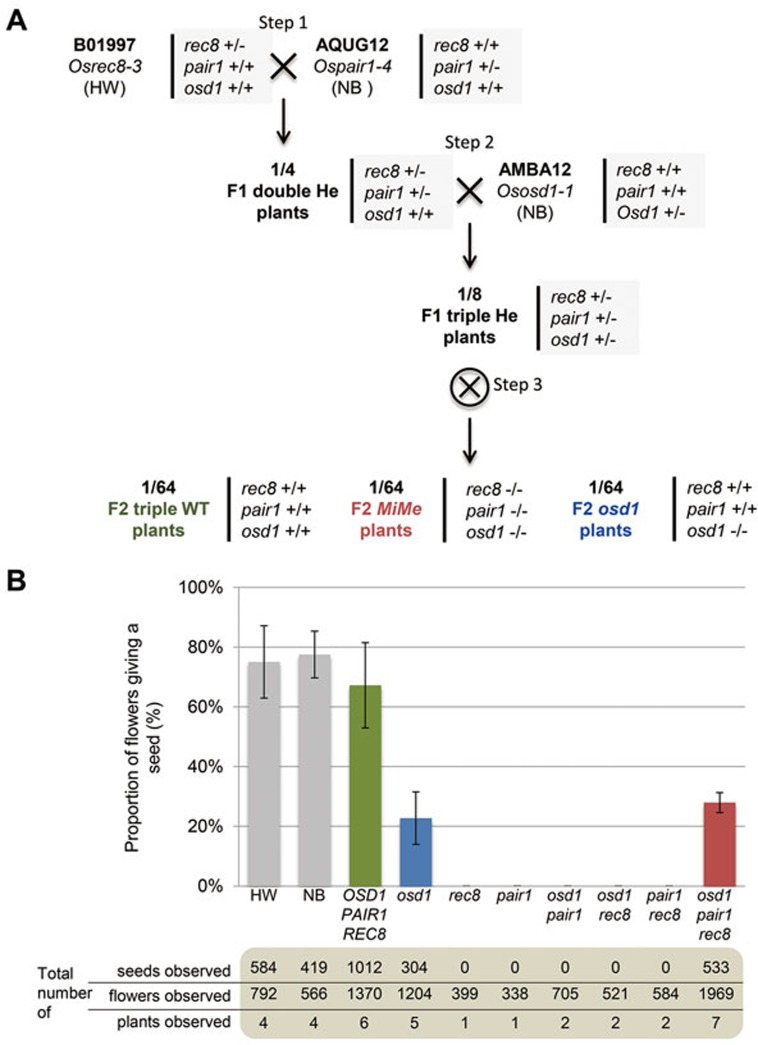
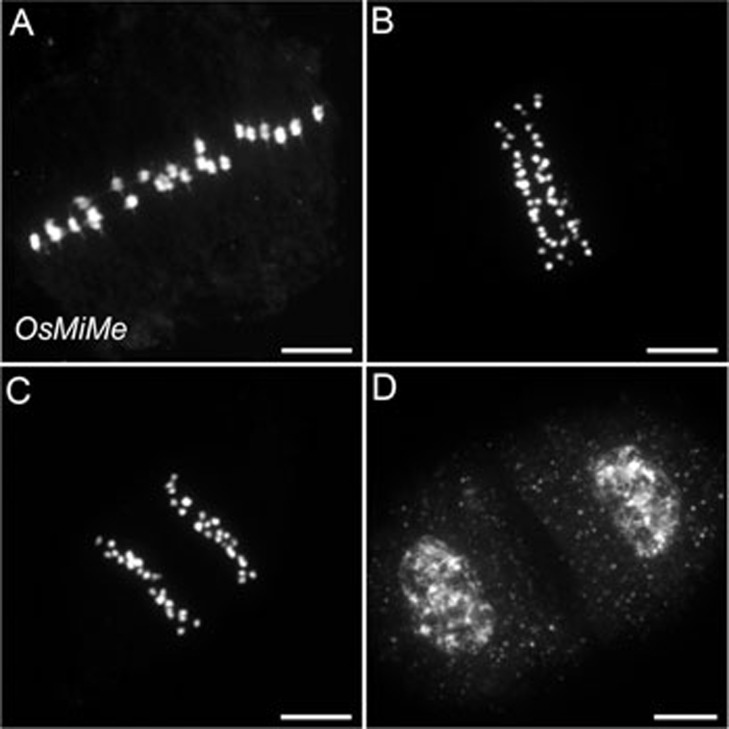
Combination of pair1, Osrec8 and Ososd1 mutations in rice was performed through sequential crossing of heterozygous plants (Figure 7A). In each generation, the segregation of mutant alleles followed the expected Mendelian segregation pattern. The pair1 Osrec8 double mutant was sterile. In this double mutant, 24 univalents were observed, which were aligned on the metaphase plate and were segregated into two groups of 24 chromatids (Figure 6E and 6F). This shows that the pair1 mutation abolishes chromosome fragmentation of Osrec8, indicating that OsREC8 is required for efficient DNA double-strand break repair. It also shows mutating OsREC8 in pair1 modifies the mode of segregation of chromosomes: in pair1 the two sister chromatids segregate as a single unit (monopolar orientation), leading to unbalanced segregation of univalents at anaphase I, while in pair1 Osrec8 the pairs of chromatids segregate to opposite poles (bipolar orientation), leading to a balanced mitotic-like distribution of sister chromatids at anaphase I. Hence, like in Arabidopsis19 and fission yeast34, OsREC8 is required for monopolar orientation of sister kinetochores at meiosis I in rice. In pair1 Osrec8, the free chromatids resulting from the mitotic-like segregation at meiosis I did not segregate properly at second division, producing unbalanced spores. The single mutants pair1, Osrec8 and the double mutants pair1 Osrec8 and pair1 Ososd1 plants were fully sterile. In contrast, the pair1 Osrec8 Ososd1 triple mutant showed higher fertility, similar to Ososd1 single mutant (Figure 7B). Examination of the male meiotic chromosome behavior in pair1 Osrec8 Ososd1 (Figure 8) revealed the presence of 24 univalents that aligned at metaphase I, and segregated into two groups of 24 chromatids to produce dyads of spores. This behavior resembles a mitotic division, suggesting that, like in Arabidopsis, the combination of pair1, rec8 and osd1 mutations turns meiosis into mitosis. We therefore term this genotype OsMiMe.
Figure 7.
Creating OsMiMe. (A) Crossing scheme. OsMiMe mutant was created first by crossing B01997 plants heterozygous for the Osrec8 mutation with AQUG12 plants heterozygous for the pair1 mutation (step 1). 1/4 of F1 were double heterozygous plants and were in turn crossed with AMBA12 plants heterozygous for the Ososd1 mutation to produce 1/8 of F1 triple heterozygous plants (Osrec8+/−; pair1+/−; Ososd1+/−) (step 2). These plants were selfed (step 3) and the progeny were genotyped to identify 1/64 triple homozygous OsMiMe plants and relevant control segregants. HW, Hwayoung; NB, Nipponbare; He, heterozygous. (B) Fertility in MiMe and related mutants. The histogram represents the percentage of flowers giving a seed by self-fertilization. Numbers of observed flowers and seeds are indicated. Errors bars correspond to the SD for 4-7 plants.
Figure 8.
Male meiosis I in OsMiMe. (A) Metaphase I with 24 aligned univalents. (B, C) Anaphase I with segregation of 24 pairs of chromatids. (D) Telophase I. n = 43. Scale bar = 10 μm.
To gather further support that OsMiMe turns meiosis into mitosis, we asked if these plants produce diploid clonal gametes. Ploidy of seedling progeny from self-pollination was estimated using flow cytometry, for both Ososd1 and OsMiMe plants. All the progeny (from two and three mother plants, n = 50 each) were tetraploid, indicating that both Ososd1 and OsMiMe diploid plants produce diploid male and female gametes. As the chromosome spreads suggested that gametes are derived from a first meiotic division in Ososd1, and from a mitotic-like division in OsMiMe, their genetic make-up should be different (Supplementary information, Figure S1). OsOsd1 should produce recombined gametes, while pair1 Osrec8 Ososd1 should produce clonal gametes. More specifically, segregation of a heterozygous marker (Aa) following meiosis I depends on its position along the chromosome35. A marker at the centromere would be homozygous in each diploid gamete (AA or aa), because of co-segregation of sister chromatids. This would render the tetraploid progeny after selfing having AAAA (¼ of plants), aaaa (¼ of plants), and AAaa (½ of plants). Markers distant from the centromere would segregate with different frequencies because of recombination35. The situation following a mitotic-like division is simpler: all heterozygosity would be maintained in each diploid gamete (Aa) because of separation of sister chromatids, and absence of recombination.
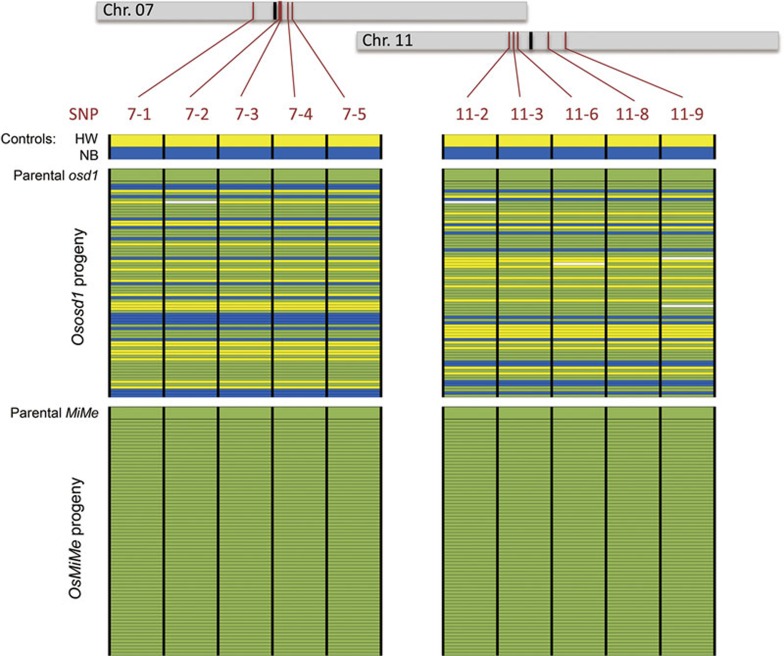
To analyze the segregation pattern in Ososd1 and pair1 Osrec8 Ososd1, we took advantage of the fact that the genetic background of the segregating population is derived from a cross between two polymorphic strains Hwayoung (B01997/Osrec8-3) and Nipponbare (AQUG12/pair1-4), which was backcrossed to Nipponbare (AMBA12/Ososd1-1) (Figure 7A). We selected markers that were heterozygous in the Ososd1 and pair1 Osrec8 Ososd1 diploid plants. The genotypes of tetraploid progeny siblings of Ososd1 and pair1 Osrec8 Ososd1 diploid parents at diagnostic markers are shown in Figure 9. In Ososd1 progeny (n = 77), the expected segregation was observed with the loss of heterozygosity at centromeric markers in ∼50% of the plants. In sharp contrast, all the diagnostic markers heterozygous in the OsMiMe diploid parents were also found heterozygous in all the tetraploid progeny siblings (n = 73). This indicates that all OsMiMe gametes fully retained the parental plant genotype.
Figure 9.
Genotypes of OsOsd1 and OsMiMe diploid parentals and their respective tetraploid progeny siblings. Tetraploid offpring of OsOsd1 and OsMiMe plants (refer to Figure 7 for scheme of creation of plant materials) were genotyped using 10 SNP markers residing on rice chromosomes 7 and 11. Positions of markers (brown) and centromeres (black) are indicated along the chromosomes. Each line represents a plant. For each marker, plants carrying only the Hwayoung allele are in yellow, plants carrying only the Nipponbare allele are in blue, while plants with both the Hwayoung and Nipponbare alleles appear in green. Osd1 and OsMiMe parental plants (top line of upper and lower panels, respectively) are heterozygous at the 10 markers. While the offspring of OsOsd1 exhibit segregation of parental alleles at the 10 markers, all the offspring of OsMiMe are heterozygous at the same 10 markers and exhibit the same genotype like the parental plants.
Discussion
Engineering apomixis in sexual crops would allow the self-propagation of F1 hybrids, which would have immense interest in agriculture by giving access to hybrid vigor in more species and to resource-poor farmers. Apomixis can be separated into two developmental components, the absence of meiosis or its modification into a mitosis-like division and the development of the embryo and its nourishing tissue from the female gametophyte without contribution of a male genome. The most efficient strategy to date to modify meiosis into mitosis has been obtained by combining mutations in three genes SPO11-1, REC8 and OSD1 (called the MiMe genotype) in the model species Arabidopsis thaliana. These three mutations affect, respectively, the three pillars of meiosis, homologous recombination, monopolar orientation of sister chromatids and the occurrence of a second division. Here we show that the mutation of SPO11-1 can be efficiently replaced by mutation in other recombination initiation factors PRD1, PRD2 or PRD3/PAIR1 to create the MiMe phenotype. This suggests that mutation in any gene essential for double-strand break formation, such as SPO11-2, DFO or MTOPVIB, may be used instead of SPO11-122,23,24,25,26. This observation may allow us to build MiMe in other species, in which mutant collection is limited or in which some genes could be present in more than one copy.
Although MiMe technology has been proven to be very efficient in the model species Arabidopsis, a major question has been about its transferability to crop species, especially the distant cereal species. SPO11-1 and REC8 are widely conserved among eukaryotes, both in sequence and in function, making the identification of their homologs in other plant species easily feasible. Other genes known to be essential for recombination initiation are also well conserved among plants and all these meiosis genes are typically represented by a single copy28. Indeed, mutating REC8 or PAIR1/PRD3 in rice recapitulates the phenotypes observed in Arabidopsis. In contrast, OSD1 appears to be a plant-specific gene and exhibits a more complex phylogeny (Figure 2). There is one OSD1 paralog in the Arabidopsis genome, UVI4, which has a distinct function in regulating somatic cell cycle27. osd1 uvi4 double mutant confers very strong gametophyte and developmental defect, suggesting that these two genes have redundant functions in mitosis36,37. Sequence analysis clearly distinguishes OSD1 from the UVI4 protein families in Brassicaceae species, but not in more distant flowering plants (Figure 2). In cereals, there are clearly two subgroups of genes, with one or two representatives of both groups in each species. The Oryza sativa genome contains two genes, one in each group, and we showed that the single mutation of Os02g37850 is sufficient to give rise to the meiotic defects observed in the Arabidopsis osd1 mutant. We consequently called this gene OsOSD1. Meiosis in Ososd1 skips the second division, producing diploid spores and gametes. These gametes are functional, leading to increased ploidy in the offspring by selfing. The existence and identification of the functional homolog of OSD1 in rice suggests that it could be identified in other cereals. Synteny and protein sequence similarity indicate ortholog of OSD1 as a single gene in Hordeum vulgare, Brachypodium distachyon and as a tandem duplication in Zea mays, Sorghum bicolor and Setaria italica28. This suggests that diploid gamete production through osd1 mutations could be obtained in cereals, but may require the use of RNAi or genome editing technologies in the case of tandem duplications.
We identified three meiotic mutations that respectively abolish recombination (pair1), allow the separation of sister chromatids (Osrec8) and provoke the omission of second division (Ososd1). Next, we combined them to construct the MiMe genotype. In this triple mutant, meiosis is converted into a mitotic-like division with balanced segregation of sister chromatids in a single division event. OsMiMe plants produce diploid gametes that are genetically identical to their parents. Our study thus establishes that the MiMe technology is transferable from the model plant Arabidopsis to the distant crop Oryza sativa, and suggests that it may be adopted in other important crops. The creation of the rice OsMiMe triple mutant was time-consuming and the approach remains so far restricted to genotypes/species with large associated mutant resources. However, recent breakthrough in genome editing by CRISPR/Cas9 may remove many obstacles38, making apomeiotic conversion a feasible reality in many crops.
Rice OsMiMe plants produce functional clonal embryos. However, fertilization still occurs normally, leading to the doubling of ploidy in the next generation. The next challenge is to trigger the development of a MiMe gamete into an embryo and then a plant without the contribution of another gamete. One approach is to use genome elimination, where the chromosome set from one parent is removed after fertilization in the zygote. MiMe combined with genome elimination indeed leads to clonal offspring, as previously demonstrated in Arabidopsis13. However, the CENH3 manipulation used in Arabidopsis to provoke genome elimination has not been transferred in other species to date20,39,40,41, except in maize but with a lower frequency42. The frequency of genome elimination can be genetically controlled in cereals, notably in maize43,44,45. The gynogenesis inducer 1 (ggi1) locus has been shown to induce genome elimination and is widely used in maize breeding to produce haploids and the identification of the underlying gene(s) will open exciting new avenues to clonal reproduction46. Alternatively, parthenogenic development of embryo could be induced, and recent progress has been made in the understanding of its genetic control47,48.
Materials and Methods
Phylogeny
Protein sequences from the OSD1/UVI4 family were collected using PLAZZA DICOTS3.0 and PLAZZA Monocots 3.0 (http://bioinformatics.psb.ugent.be/plaza/)49 and correspond to family HOM03M004665, HOM03M067707 and HOM03D003883. Some species were removed from the analysis for simplicity. BD5G13210 sequence was replaced by Bradi5g13212.1 (Phytozome 10.3.). Three proteins (SI005G06210, ZM10G18380 and GM11G21435) were excluded because, though they share sequence similarity with the OSD1/UVI4 proteins, they lack most of the conserved domains. Multi-protein alignment (Muscle v3.8) followed by phylogeny inference (PHYLIP Neighbor-Joining/UPGMA method version 3.696) was performed using the MPI toolkit with default parameters50.
Genetic material
The Arabidopsis mutants used were prd1-2, prd2-1, prd3-322,23, osd1-1 and atrec8-316. Rice (Oryza sativa L.) mutants used in this study were the following: AMBA12 T-DNA insertion line and two Tos17 insertion mutant AMQF10 and ALJA10 lines for OsOSD1 (Os02g37850); AQUG12 Tos17 insertion line for PAIR1 (Os03g01590); B01997 T-DNA insertion line for OsREC8 (Os05g50410). All insertion lines were generated in the Nipponbare background and are from the Oryza Tag Line insertion line library51,52 except B01997, which has a Hwayoung background and is from the POSTECH Rice Insertion Database53. Position of the inserts in the respective genes is shown in Supplementary information, Figure S2. The exact junction of the T-DNA insert with the rice genome in the mutants was ascertained by Sanger sequencing.
Genotyping of T-DNA and Tos17 inserts
Genotyping primers are listed in Supplementary information, Table S1. Primers were designed using the “Genotyping Primer Designer” tool of OryGenesDB (http://www.orygenesdb.cirad.fr)54,55. In these standard conditions, the expected PCR product is ∼800 bp for a mutant allele and ∼1 000 bp for a wild-type allele. PCR was performed following the protocol described in56.
SNP genotyping
Single-nucleotide polymorphism genotyping was performed using Kompetitive Allele Specific PCR (KASP) following the LGC group recommendations for the use of KASP technology on Roche LC480.
Cytological analyses
Arabidopsis chromosome spreads and observation were carried out using the technique described in18.
Conditions of growth and crossing of the plants
Rice plants were grown under containment greenhouse condition (temperature of 28 °C day and 24 °C night, 60% hygrometry) under natural light that could be assisted by artificial sodium light (light intensity of 700 μmol/m2/s). Crossing was accomplished through manual castration of florets and pollination according to the standard procedures, followed by bagging to avoid pollen contamination.
Embryo rescue
Five days after pollination, fertilized flowers were harvested, their lemma and palea removed under a stereo microscope and soaked in 70% ethanol for 5 min. The caryopses were then washed thrice in sterile distilled water. Embryos were isolated following two distinct procedures depending on the presence or absence of endosperm42,43. Immature embryos were plated on MS basal medium44 supplemented with 50 g/l sucrose and 6 g/l phytagel. Following a 10-day growth period in the dark at 26 °C, the seedlings were transferred to a fresh MS solid medium (2.6 g/l of phytagel; 30 g/l of sucrose) and in a growth chamber (photoperiod 16:8), 28 °C and 60% humidity for 3 weeks.
Flow cytometry determination of DNA content in pollen and leaf cell nuclei
Ploidy was determined by estimating nuclear DNA content by flow cytometry. Pollen grains from flowers at anthesis were collected by introducing the panicle into a 50 ml tube and gently shaking allowing the shedding of pollen from anthers. One milliliter of LB01 buffer was added to humidify the pollen followed by a mild centrifugation (10 min at 150× g) allowing collection of the pollen pellet. The supernatant was discarded and the pellet was resuspended with 1 ml of a fresh LB01. The nucleus extraction from pollen grains was performed according to Kron et al.57 bursting method. Due to the size of rice pollen grains and nuclei, we used Partec CellTrics at 100 μm for the first filtration and 30 μm for the second one. For leaf nucleus extraction, ∼0.5 cm2 of leaf blade tissue was chopped in 250 μl of LB01 extraction buffer. The suspension was filtered (30 μm Partec CellTrics filters) to eliminate cell debris. After filtration, 700 μl of LB01 buffer was added. After DNA nucleus extraction from pollen or leaf tissue, DNA was stained by addition of propidium iodide (40 μl of PI at 0.2 mg/ml). The samples were analyzed with UV excitation in a Partec flow cytometer (PA-I; Partec, Munster, Germany).
Author Contributions
EG and RM conceived the project. DM, EG and RM designed experiments. DM, SJ, MR, LC, AV, PM, LP, GD and BD performed experiments and analysed the data. DM, EG and RM wrote the paper with the input from other authors.
Competing Financial Interests
The authors declare no competing financial interests
Acknowledgments
This work has been funded by the ANR Emergence DiplOSD (ANR-09-EBIO-011) to RM and EG. Part of this work has been supported by the RicE FUnctional GEnomics (REFUGE) platform funded by Agropolis Fondation. We thank Eve Lorenzini, Christian Chaine and Rémy Michel for providing technical assistance, notably in greenhouse. We thank the guidance of Dr Frédéric Bakry, Cirad, for flow cytometric analyses. We thank Marta Cifuentes, Eric Jenczewski, Wayne Crismani, Rajeev Kumar and Anne Diévart for critical reading of the manuscript. The IJPB benefits from the support of the Labex Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Shematic representation of synthetic apomeiosis and apomixis.
Positions of T-DNA or Tos17 insertions in A: OsOSD1, B: PAIR1 and C: OsREC8.
Accumulation of OsOSD1 transcripts in Ososd1-1 (AMBA12) and Ososd1-2 (AMQF10) plants (−/−) and their wild type siblings (+/+).
Primer sequences used for genotyping and SNP position in the rice genome (MSU v7.0)
References
- Rejesus RM, Mohanty S, Balagtas JV. Forecasting global rice consumption. 2012. http://www.agecon.purdue.edu/staff/balagtas/rice_timeseries_v6.pdf. Accessed 01 March 2014
- Virk PS, Khush GS, Virmani SS. Breeding strategies to enhance heterosis in rice. 2003.
- Virmani SS, Mao CX HBE. In: Virmani SS, Mao CX, Hardy B, eds. Hybrid Rice For Food Security, Poverty Alleviation, and Environmental Protection. International Rice Research Institute, Manila, Philippines, 2003.
- Ma GH, Yuan LP. Hybrid rice achievements and development in China. In: Virmani SS, Mao CX, Hardy B, ed. Hybrid Rice For Food Security, Poverty Alleviation, and Environmental Protection. International Rice Research Institute, Manila, Philippines, 2003.
- Virmani SS. Heterosis and Hybrid Rice Breeding. Berlin, Heidelberg: Springer, 1994.
- Spillane C, Curtis MD, Grossniklaus U. Apomixis technology development-virgin births in farmers' fields? Nat Biotechnol 2004; 22:687–691. [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Grossniklaus U. Apomixis: a developmental perspective. Annu Rev Plant Biol 2003; 54:547–574. [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P, van Dijk PJ. Mendelian genetics of apomixis in plants. Annu Rev Genet 2007; 41:509–537. [DOI] [PubMed] [Google Scholar]
- Barcaccia G, Albertini E. Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reprod 2013; 26:159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidan Y. Transfer of apomixis through wide crosses. In: Savidan Y, Carman JG, Dresselhaus T, eds. The Flowering of Apomixis: From Mechanisms to Genetic Engineering. Mexico: CIMMYT, IR; 2001:153–167.
- Rutger JN. Searching for apomixis in rice. In: Proceedings of Apomixis Workshop. Beltsville, MD, USA: US Department of Agriculture; 1992:36–39.
- Khush GS. Apomixis: Exploiting Hybrid Vigor in Rice. International Rice Research Institute, Manila, Philippines, 1994.
- Marimuthu MP, Jolivet S, Ravi M, et al. Synthetic clonal reproduction through seeds. Science 2011; 331:876. [DOI] [PubMed] [Google Scholar]
- Ronceret A, Vielle-Calzada JP. Meiosis, unreduced gametes, and parthenogenesis: implications for engineering clonal seed formation in crops. Plant Reprod 2015; 28:91–102. [DOI] [PubMed] [Google Scholar]
- Ravi M, Marimuthu MPA, Siddiqi I. Gamete formation without meiosis in Arabidopsis. Nature 2008; 451:1121–1124. [DOI] [PubMed] [Google Scholar]
- D'Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R. Turning meiosis into mitosis. PLoS Biol 2009; 7:e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Erfurth I, Cromer L, Jolivet S, et al. The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet 2010; 6:e1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J 2001; 20:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 2005; 118:4621–4632. [DOI] [PubMed] [Google Scholar]
- Ravi M, Chan SWL. Haploid plants produced by centromere-mediated genome elimination. Nature 2010; 464:615–618. [DOI] [PubMed] [Google Scholar]
- Cifuentes M, Jolivet S, Cromer L, et al. TDM1 regulation determines the number of meiotic divisions. PLoS Genet 2016; 12:e1005856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Vezon D, Gendrot G, Gallois JL, Stevens R, Grelon M. AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J 2007; 26:4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pereira L, Vezon D, et al. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet 2009; 5:e1000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey NJ, Kuromori T, Azumi Y, et al. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J 2006; 48:206–216. [DOI] [PubMed] [Google Scholar]
- Zhang C, Song Y, Cheng Z, et al. The Arabidopsis thaliana DSB formation (AtDFO) gene is required for meiotic double-strand break formation. Plant J 2012; 72:271–281. [DOI] [PubMed] [Google Scholar]
- Vrielynck N, Chambon A, Vezon D, et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 2016; 351:939–943. [DOI] [PubMed] [Google Scholar]
- Hase Y, Trung KH, Matsunaga T, Tanaka A. A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J 2006; 46:317–326. [DOI] [PubMed] [Google Scholar]
- Lloyd AH, Ranoux M, Vautrin S, et al. Meiotic gene evolution: can you teach a new dog new tricks? Mol Biol Evol 2014; 31:1724–1727. [DOI] [PubMed] [Google Scholar]
- Sekine D, Ohnishi T, Furuumi H, et al. Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J 2013; 76:792–799. [DOI] [PubMed] [Google Scholar]
- Nonomura K, Nakano M, Fukuda T, et al. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 2004; 16:1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T, Tang D, Wang K, et al. OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 2011; 156:1386–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Peirson BN, Dong F, Xue C, Makaroff CA. Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 1999; 11:417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, et al. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 1999; 19:463–472. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol 2003; 23:3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W, Girard C, Mercier R. Tinkering with meiosis. J Exp Bot 2013; 64:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer L, Heyman J, Touati S, et al. OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genet 2012; 8:e1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata E, Ikeda S, Matsunaga S, et al. GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 2011; 23:4382–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicotplants. Mol Plant 2015; 8:1274–1284. [DOI] [PubMed] [Google Scholar]
- Karimi-Ashtiyani R, Ishii T, Niessen M, et al. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc Natl Acad Sci USA 2015; 112:1504333112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Tan EH, West A, Franklin FCH, Comai L, Chan SWL. Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet 2015; 11:e1004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppu S, Tan EH, Nguyen H, et al. Point mutations in centromeric histone induce post-zygotic incompatibility and uniparental inheritance. PLoS Genet 2015; 11:e1005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Wang W, et al. Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize. Front Plant Sci 2016; 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle J. Androgenesis conditioned by a mutation in maize. Science 1969; 217:446–448. [DOI] [PubMed] [Google Scholar]
- Eder J, Chalyk S. In vivo haploid induction in maize. Theor Appl Genet 2002; 104:703–708. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Qiu F, Liu Y, Ma K, Li Z, Xu S. Chromosome elimination and in vivo haploid production induced by Stock 6-derived inducer line in maize (Zea mays L.). Plant Cell Rep 2008; 27:1851–1860. [DOI] [PubMed] [Google Scholar]
- Barret P, Brinkmann M, Beckert M. A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor Appl Genet 2008; 117:581–594. [DOI] [PubMed] [Google Scholar]
- Conner JA, Mookkan M, Huo H, Chae K, Ozias-Akins P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc Natl Acad Sci USA 2015; 112:11205–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst NA, Katz A, Pereman I, Decker EL, Ohad N, Reski R. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat Plants 2016; 2:15209. [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Vaneechoutte D, et al. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 2015; 43:D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegert A, Mayer C, Remmert M, Söding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res 2006; 34:W335–W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larmande P, Gay C, Lorieux M, et al. Oryza Tag Line, a phenotypic mutant database for the Génoplante rice insertion line library. Nucleic Acids Res 2008; 36:1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Gay C, Larmande P, et al. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J 2004; 39:450–464. [DOI] [PubMed] [Google Scholar]
- An S, Park S, Jeong DH, et al. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol 2003; 133:2040–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droc G, Ruiz M, Larmande P, et al. OryGenesDB: a database for rice reverse genetics. Nucleic Acids Res 2006; 34:D736–D740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droc G, Périn C, Fromentin S, Larmande P. OryGenesDB 2008 update: database interoperability for functional genomics of rice. Nucleic Acids Res 2009; 37:D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieulet D, Diévart A, Droc G, Lanau N, Guiderdoni E. Reverse genetics in rice using Tos17. Methods Mol Biol 2013; 1057:205–221. [DOI] [PubMed] [Google Scholar]
- Kron P, Husband BC. Using flow cytometry to estimate pollen DNA content: improved methodology and applications. Ann Bot 2012; 110:1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shematic representation of synthetic apomeiosis and apomixis.
Positions of T-DNA or Tos17 insertions in A: OsOSD1, B: PAIR1 and C: OsREC8.
Accumulation of OsOSD1 transcripts in Ososd1-1 (AMBA12) and Ososd1-2 (AMQF10) plants (−/−) and their wild type siblings (+/+).
Primer sequences used for genotyping and SNP position in the rice genome (MSU v7.0)