Abstract
Neurons have a central role in the systemic coordination of mitochondrial unfolded protein response (UPRmt) and the cell non-autonomous modulation of longevity. However, the mechanism by which the nervous system senses mitochondrial stress and communicates to the distal tissues to induce UPRmt remains unclear. Here we employ the tissue-specific CRISPR-Cas9 approach to disrupt mitochondrial function only in the nervous system of Caenorhabditis elegans, and reveal a cell non-autonomous induction of UPRmt in peripheral cells. We further show that a neural sub-circuit composed of three types of sensory neurons, and one interneuron is required for sensing and transducing neuronal mitochondrial stress. In addition, neuropeptide FLP-2 functions in this neural sub-circuit to signal the non-autonomous UPRmt. Taken together, our results suggest a neuropeptide coordination of mitochondrial stress response in the nervous system.
Keywords: mitochondria, unfolded protein response, cell non-autonomous
Introduction
To cope with a challenging and unpredictable environment, living systems have evolved several organelle-specific stress responses, e.g., the cytosolic heat shock response (HSR), and the endoplasmic reticulum or mitochondrial unfolded protein response (UPRER or UPRmt). Failure to appropriately respond to different stresses and maintain cellular homeostasis could result in multiple diseases such as neurodegenerative and metabolic diseases1,2,3. UPRmt monitors mitochondrial function in general: it is responsible for buffering the mitochondrial protein folding environment4,5, and controlling mitochondria-nuclear balance of the electron transport chain (ETC) components6. One major component of UPRmt is ATFS-1, a transcription factor that induces the expression of mitochondrial-specific chaperones HSP-6 and HSP-60 during mitochondrial stress7,8. ATFS-1 contains an N-terminal mitochondrial targeting sequence as well as a C-terminal nuclear localization signal. Under normal conditions, ATFS-1 is imported into mitochondria and degraded by the mitochondrial protease Lon. However, when mitochondria are damaged, the import efficiency of mitochondria is decreased, leading to a nuclear accumulation of ATFS-1 and hsp-6 upregulation8. Once activated, UPRmt not only upregulates mitochondrial protective proteins such as chaperones and proteases, but also resets metabolic state through the regulation of genes involved in glycolysis and oxidative phosphorylation7,8. In Caenorhabditis elegans, activation of UPRmt also alters the organismal investment strategy to delay reproduction and development, and to extend lifespan9. These physiological changes must involve the coordination of multiple tissues and organs.
The decision to shift the resource allocation strategy in C. elegans seems to be made and coordinated by the nervous system9. RNAi knockdown of ETC components only in neurons induced the expression of mitochondrial chaperones in distal tissues and caused a lifespan extension9, suggesting a cell non-autonomous regulation of mitochondrial stress response. Intriguingly, the nervous system also exerts tight control over the coordination of HSR and UPRER activation. For instance, the AFD thermosensory neuron and the AIY interneuron are responsible for behavioral changes of C. elegans upon temperature shift10,11. Moreover, neuronal expression of a constitutive active form of XBP-1 is sufficient to induce UPRER in distal tissues12. Although the molecular mediators of cell non-autonomous regulation of stress response have not yet been determined, certain evidence suggested a role of neuropeptide-like molecules11.
To understand how mitochondrial stress is sensed in the nervous system, and transmitted throughout the organism to induce UPRmt in the peripheral cells, we disrupted mitochondrial function only in neurons with the expression of reactive oxygen species (ROS)-generating fluorescent protein KillerRed, uncoupling protein UCP-4, or with endonuclease Cas9 to knock out mitochondrial gene spg-7 or cco-1. We found that the expression of KillerRed or knocking out spg-7 or cco-1 in neurons is sufficient to elicit a cell non-autonomous UPRmt. However, expression of UCP-4 only induces cell autonomous UPRmt to activate mitochondrial chaperone gene expression in the nervous system, suggesting that different types of mitochondrial perturbation elicit different organismal responses. Induction of the UPRmt in distal tissues requires functional UPRmt pathway components in both neurons and the receiving cells. Constitutive activation of UPRmt in the nervous system is sufficient to drive cell non-autonomous UPRmt. Furthermore, we found that neuropeptide FLP-2 has a role in neurons to mediate cell non-autonomous UPRmt. Loss of FLP-2 significantly blocks cell non-autonomous UPRmt signaling, whereas restricted expression of FLP-2 only in neurons is sufficient to induce UPRmt in peripheral tissues.
Results
Different forms of mitochondrial perturbation in the nervous system elicit different UPRmt responses
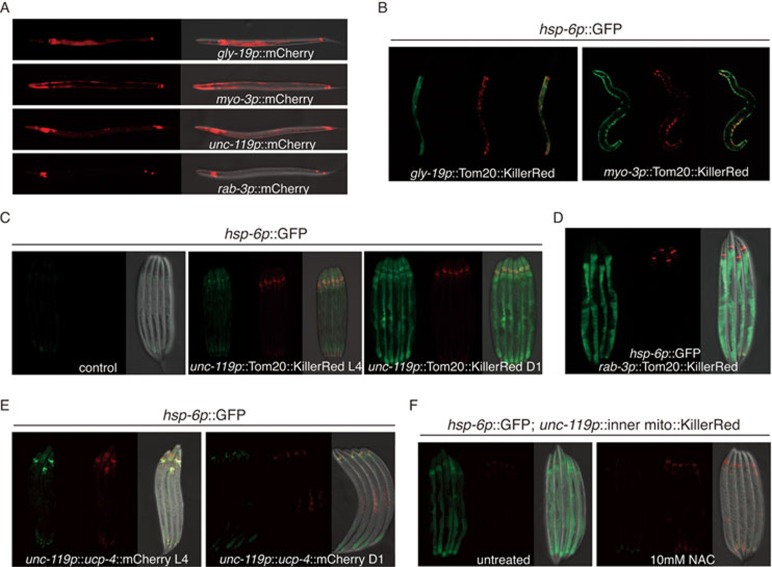
In an effort to reveal the molecular elements involved in cell non-autonomous regulation of mitochondrial UPRmt, we expressed two types of mitochondrial disrupting proteins in various tissues driven by C. elegans tissue-specific promoters, i.e., intestine-specific promoter gly-19, muscle-specific promoter myo-3 and neural-specific promoters unc-119 and rab-3 (Figure 1A). KillerRed is a dimeric red fluorescent protein that can produce large amount of ROS13. Expression of a mitochondrial outer membrane targeted Tom20::KillerRed using the gly-19 intestine-specific promoter induced UPRmt and activated the expression of hsp-6 chaperone gene, a hallmark of the UPRmt4,14, in the intestine (Figure 1B). Consistently, muscle-specific expression of Tom20::KillerRed driven by a myo-3 promoter solely activated UPRmt in muscle cells (Figure 1B). However, when the mitochondrial localized KillerRed was driven by a pan-neural promoter unc-119, it not only activated UPRmt in neurons but also in distal tissues (Figure 1C). It should be noted that the activation of hsp-6 expression was not discernible until worms reached day 1 adult stage (Figure 1C). Moreover, the endogenous levels of known UPRmt response genes15 such as irg-1 (pathogen response gene) and cyp-14A1 (detoxification gene) were also elevated in neuronal KillerRed overexpression animals (Supplementary information, Figure S1A). Consistent with this result, expression of mitochondrial localized KillerRed driven by the rab-3 neural-specific promoter also induced UPRmt in the periphery (Figure 1D). Antioxidants NAC or ascorbic acid treatment suppressed KillerRed-induced mitochondrial damage, and, therefore, abolished non-autonomous UPRmt (Supplementary information, Figure S1B). Collectively, these data suggested that disrupting mitochondrial function in neurons with the expression of KillerRed is sufficient to activate a cell non-autonomous UPRmt.
Figure 1.
Different forms of mitochondrial perturbation in the nervous system elicit different UPRmt responses. (A) Representative fluorescent images testing the expression pattern of tissue-specific promoters (gly-19p: intestine; myo-3p: muscle; unc-119p or rab-3p: neuron). (B) Expression of a mitochondrial outer membrane-targeted KillerRed in the intestine or muscle cells induces UPRmt reporter gene hsp-6 cell autonomously. (C, D) Neural expression of mitochondrial outer membrane-targeted KillerRed induces UPRmt in distal tissues when worms reach day 1 adult stage. (E) Neural expression of UCP-4 fails to activate cell non-autonomous UPRmt. (F) NAC treatment suppresses the activation of UPRmt in distal tissues induced by neural expression of mitochondrial inner membrane-targeted KillerRed. Representative images of adult animals are shown. Mitochondrial inner membrane-targeting sequence contains the first exon and part of second exon of sdhb-1.
Uncoupling proteins (UCPs) are a family of proton carriers localized in the inner mitochondrial membrane where they uncouple electron transport from ATP production16. UCP-4, which is one of the UCPs, is highly expressed in the brain17. We expressed UCP-4 with the use of pan-neural promoter unc-119 to disrupt mitochondrial function only in neurons. To our surprise, unlike KillerRed, expression of UCP-4 failed to induce cell non-autonomous UPRmt in C. elegans (Figure 1E). However, the mitochondria were most likely defective because activation of cell autonomous UPRmt could be observed in the nervous system. To exclude the possibility that the different outcomes between KillerRed and UCP-4 expression is due to their different localizations within mitochondria, we targeted KillerRed to the inner membrane of mitochondria, but again observed cell non-autonomous UPRmt (Figure 1F). Collectively, these results point to two principal findings: (1) C. elegans nervous system is the central tissue to sense and transduce mitochondrial stress signal to coordinate whole-organism physiology; (2) Not all types of mitochondrial perturbation in neurons could induce cell non-autonomous UPRmt.
Neural-specific knockout of mitochondrial genes induces cell non-autonomous UPRmt
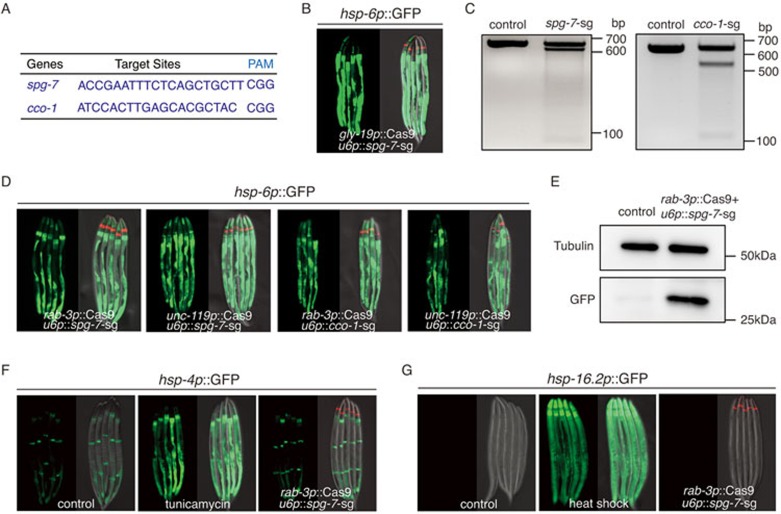
Spatially controlled CRISPR-Cas9 system18,19 was also employed to access non-autonomous UPRmt (Supplementary information, Figure S2A). We expressed Cas9 endonuclease tissue specifically to knock out two nuclear-encoded mitochondrial genes, the mt-AAA protease spg-7 and the complex IV subunit cco-1 (Figure 2A and Supplementary information, Figure S2C). RNAi knockdown of these two genes have been reported to activate C. elegans UPRmt9,20. Consistent with previous results, intestine-specific knockout of spg-7 using gly-19 promoter robustly induced the expression of hsp-6 reporter cell autonomously (Figure 2B). In contrast, neural-specific knockout of spg-7 or cco-1 (Figure 2C and Supplementary information, Figure S2B) induced cell non-autonomous UPRmt in the periphery to activate the expression of UPRmt reporter hsp-6p::GFP (Figure 2D and 2E). To exclude the possibility of off-targeting effect, we used a second sgRNA sequence to knock out spg-7 or cco-1 and observed the same result (Supplementary information, Figure S2D). Moreover, the endogenous levels of UPRmt response genes irg-1 and cyp-14A1 were upregulated in spg-7 knockout strain (Supplementary information, Figure S2E). Neuronal knockout of spg-7 or cco-1 had a specific effect on mitochondrial stress response, as stress reporters for UPRER (hsp-4p::GFP) and heat shock (hsp-16.2p::GFP) were not induced (Figure 2F and 2G). Therefore, the Cas9 system provided us a robust and specific model to dissect the molecular elements of cell non-autonomous UPRmt.
Figure 2.
Neural-specific knockout of mitochondrial genes induces cell non-autonomous UPRmt. (A) Targeting sequences for CRISPR-Cas9 knockout of spg-7 or cco-1, together with their PAM sequences. (B) Knockout of spg-7 in intestine activates hsp-6p::GFP reporter cell autonomously. odr-1p::dsRed is used as the co-injection marker. (C) Deletions of spg-7 or cco-1 by CRISPR/Cas9 are detected by T7E1 assay. Representative DNA gels of T7E1 assay show spg-7 or cco-1 PCR products amplified from genomic DNA of hsp-6 p::GFP worms (control), or unc-119p::Cas9+u6p::spg-7-sg(left), rab-3p::Cas9+u6p::cco-1-sg(right) worms. (D) Neural-specific knockout of spg-7 or cco-1 activates hsp-6p::GFP in distal tissues. (E) Western blotting shows the increasing level of hsp-6p::GFP in rab-3p::Cas9+u6p::spg-7-sg strain. The control lysate is from hsp-6p::GFP strain. Cell lysates are probed with GFP antibody. Tubulin is used as a loading control. (F, G) Neural knockout of spg-7 fails to induce UPRER or HSR in distal tissues. As positive controls, tunicamycin treatment robustly induces hsp-4p::GFP reporter, heat shock 1 h at 34 °C robustly induces hsp-16.2p::GFP reporter.
Different types of mitochondrial perturbation affect the fitness of animals differently
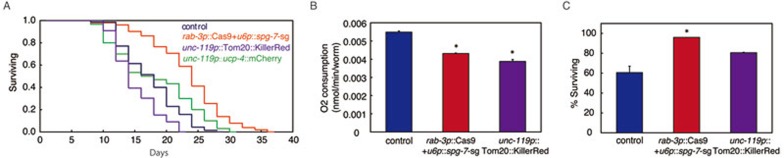
Intrigued by the different UPRmt outputs caused by KillerRed or UCP-4 expression, and CRISPR-Cas9 knockout of spg-7, we asked if the three types of mitochondrial disruption affect animal fitness similarly. We first conducted lifespan analysis: spg-7 knockout in the nervous system alone significantly extended lifespan, consistent with previous findings that tissue-specific RNAi of nuclear encoded mitochondrial genes only in neurons resulted in longevity9; neuronal expression of UCP-4 had no obvious effect on mean lifespan; whereas neuronal expression of KillerRed reduced lifespan (Figure 3A). Both spg-7 knockout and KillerRed expression in the nervous system clearly impaired mitochondrial function and resulted in lowered respiration rates (Figure 3B). When treated with mitochondrial inhibitor antimycin A, neural spg-7 knockout exhibited an advantage in fitness (Figure 3C). These results suggested that three types of mitochondrial perturbation affected the fitness of animals differently.
Figure 3.
Different types of mitochondrial perturbation affect the fitness of animals differently. (A) Lifespan analysis of three independent transgenic lines rab-3p::Cas9+u6p::spg-7-sg, unc-119p::Tom20::KillerRed or unc-119p::ucp-4::mCherry. (B) Oxygen consumption is detected in transgenic strain containing rab-3p::Cas9+u6p::spg-7-sg or unc-119p::Tom20::KillerRed. n ≥ 1 200. (C) Survival analyses of transgenic worms exposed to 25 μg/ml antimycin A is performed as described in the Materials and Methods. n = 30. Error bars indicate mean ± SE. A Student's t-test is used to assess significance: *P < 0.05.
A neural sub-circuit is required for sensing and transducing mitochondrial stress
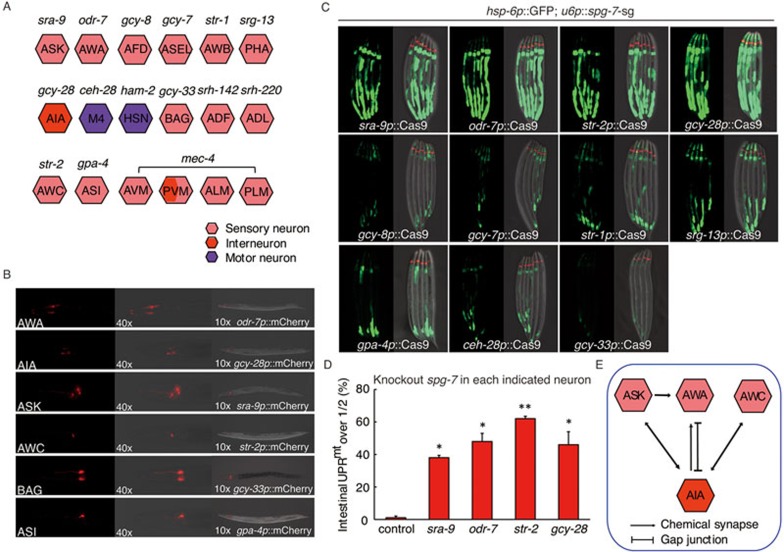
To understand the molecular basis of cell non-autonomous UPRmt induced by neural mitochondrial stress, we decided to first determine which neurons are essential for sensing and transducing the stress signal. We picked a panel of neuron-specific promoters (including sensory neurons, interneurons and motor neurons) (Figure 4A) and verified their expression pattern with the use of fluorescent protein mCherry (Figure 4B). The expression of Cas9 endonuclease was then driven by each neural-specific promoter to knock out mitochondrial protease spg-7 in each indicated neuron and tested for their abilities to induce hsp-6p::GFP reporter in peripheral cells. In summary, spg-7 deletion in three types of sensory neurons ASK, AWA and AWC, and one interneuron AIA was sufficient to induce UPRmt in the intestine, whereas spg-7 mutation in other neurons failed to induce cell non-autonomous UPRmt (Figure 4C). The robustness of cell non-autonomous UPRmt induction was quantified by counting the percentage of day 1 adult worms that carry GFP signal over the half-length of their intestine (Figure 4D and Supplementary information, Figure S3A).
Figure 4.
A neural sub-circuit is required for sensing and transducing mitochondrial stress. (A) A schematic graph depicting each tested neuron and its promoter. (B) Representative fluorescent images showing the expression pattern of each indicated neuronal-specific promoter. (C-D) Representative fluorescent images (C) or quantification (D) of hsp-6p::GFP reporter expression in animals containing Cas9-spg-7-sg in each indicated neuron. Analysis is performed as described in Materials and Methods. n ≥ 30, error bars indicate mean ± SE. A Student's t-test is used to assess significance: *P < 0.05, **P < 0.01. (E) A schematic graph of chemical synapses or gap junctions between ASK, AWA, AWC sensory neurons and AIA interneuron.
On the basis of the neuronal wiring diagram of C. elegans21, all three types of sensory neurons form chemical synapses or gap junction with AIA interneuron (Figure 4E), suggesting a central role of AIA interneuron in the regulation of cell non-autonomous UPRmt. The combination of spg-7 deletion in two types of sensory neurons ASK, AWA and AWC did not further enhance cell non-autonomous UPRmt (Supplementary information, Figure S3B). Moreover, knocking out spg-7 in ASK, AWA, AWC or AIA neuron alone did not result in lifespan extension (Supplementary information, Figure S3C).
A functional UPRmt pathway in neurons is required for peripheral induction of UPRmt
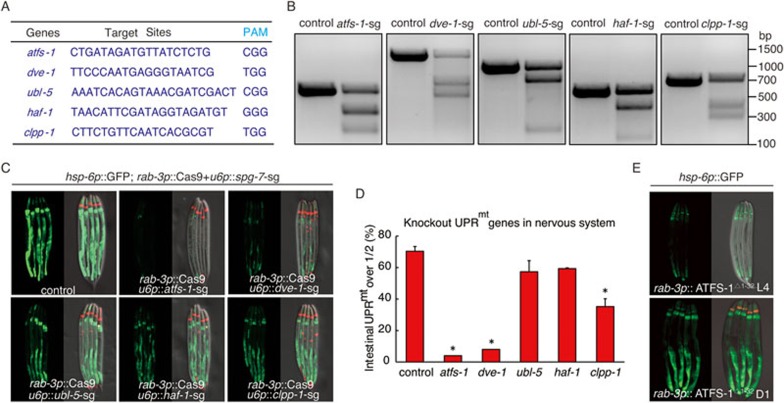
The UPRmt consists of a signaling cascade that results in mitochondria-to-nucleus communication, and the induction of nuclear-encoded genes to alleviate the stress. Previous works have reported several proteins required for the UPRmt4,5,8,14,15,20, such as transcription factor ATFS-1, homeodomain-containing transcription factor DVE-1 and its co-activator UBL-5. In addition, mitochondrial protease CLPP-1 has been identified as another component of UPRmt14, possibly by generating the mitochondrial derived peptides, which might be transported by the matrix peptide transporter HAF-1 to signal UPRmt20.
We predicted that the induction of cell non-autonomous UPRmt in peripheral cells most likely require one or more UPRmt components. To determine which of the UPRmt factors are critical in the receiving cells to induce the expression of mitochondrial chaperones, we treated neuronal spg-7 deletion animals with RNAi directed toward the known pathway components of the UPRmt(Supplementary information, Figure S4A). RNAi against atfs-1 robustly blocked the neuronal spg-7 deletion, and neuronal KillerRed expression induced hsp-6 induction, whereas RNAi of dve-1 partially suppressed the induction of hsp-6p::GFP (Supplementary information, Figure S4B and S4D). In contrast, ubl-5, haf-1 and clpp-1 knockdowns had no obvious effects on the induction of UPRmt in distal tissues (Supplementary information, Figure S4B and S4C). Because RNAi efficiency is low in C. elegans neurons, the phenotypes that we saw were most likely due to RNAi of UPRmt components in the intestine. Therefore, these results suggest that atfs-1 and dve-1 in the receiving cells are required for the induction of non-autonomous UPRmt.
To test the requirement of the UPRmt pathway components in neurons for signaling cell non-autonomous UPRmt, we utilized again tissue-specific CRISPR-Cas9 approach to express Cas9 endonuclease specifically in neurons using a neural-specific promoter rab-3. Small-guide (sg) RNA target sites were carefully designed to achieve efficient UPRmt gene deletions (Figure 5A and 5B). Deletion of atfs-1 or dve-1 only in the nervous system significantly suppressed the induction of cell non-autonomous UPRmt, whereas clpp-1 deletion in neurons partially blocked the induction of hsp-6p::GFP (Figure 5C and 5D). Consistently, RNAi or knockout of atfs-1 also abolished KillerRed overexpression-induced cell non-autonomous UPRmt (Supplementary information, Figure S4D). Moreover, deletion of atfs-1 in ASK, AWA and AWC sensory neurons, and AIA interneuron suppressed the induction of non-autonomous UPRmt caused by spg-7 deletion in the same neuron (Supplementary information, Figure S4E). In contrast, ubl-5 and haf-1 deletion in neurons did not affect cell non-autonomous UPRmt (Figure 5C and 5D). Taken together, these data indicate that a functional UPRmt pathway in the nervous system is required for the cell non-autonomous induction of UPRmt in distal tissues.
Figure 5.
A functional UPRmt pathway in neurons is required for peripheral induction of UPRmt. (A) Targeting sequences of UPRmt pathway genes for CRISPR-Cas9 knockout, together with their PAM sequences. (B) Deletions of UPRmt pathway genes produced by CRISPR/Cas9 are detected by T7E1 assay. Representative DNA gels of T7E1 assay reveals the PCR products amplified from genomic DNA of control worms, or worms with UPRmt pathway gene deletion in the nervous system. (C, D) Neural knockout of atfs-1 or dve-1 suppresses the UPRmt in distal tissues in rab-3p::Cas9+u6p::spg-7-sg worms. Representative fluorescent images (C) and quantification of hsp-6p::GFP reporter expression (D) in animals containing rab-3p::Cas9+u6p::spg-7-sg (control) or rab-3p::Cas9+u6p::spg-7-sg with neural-specific knockout of UPRmt genes are shown. mec-7p::RFP is used as co-injection marker. n ≥ 28, error bars indicate mean ±SE. A Student's t-test is used to assess significance: *P < 0.05. (E) Neural expression of ATFS-1Δ1-32 induces UPRmt in distal tissues when worms reach day 1 adult stage. odr-1 p::dsRed is used as co-injection marker.
We next determined if the activation of UPRmt in the nervous system is sufficient to induce hsp-6p::GFP reporter in the periphery. We expressed an N-terminal 1-32 amino acids deletion of ATFS-1 (ATFS-1Δ1-32) driven by the neural-specific promoter rab-3. Deletion of the N-terminal 1-32 amino acids abolishes the mitochondrial targeting sequence and leads to constitutive nuclear accumulation of ATFS-18,15. Neural expression of ATFS-1Δ1-32 was sufficient to induce hsp-6p::GFP reporter in distal tissues (Figure 5E), suggesting that the cell non-autonomous UPRmt is regulated by the UPRmt pathway in neurons.
Neuropeptide regulates cell non-autonomous UPRmt signaling
We have found that neuronal knockout of spg-7 or cco-1 disrupts mitochondrial function in neurons and induces a cell non-autonomous activation of the UPRmt in the intestine. Because the nervous system does not physically interact and innervate the intestine, it is likely a neuroendocrine signal is secreted to signal the cell non-autonomous UPRmt in distal tissues. Previous studies reported a role of dense core vesicle release on the effect of the AFD thermosensory neurons on HSR, suggesting a neuropeptide-like molecule may function in this process11. We next took a hypothesis-driven approach to test if neuropeptide is required to induce UPRmt in the periphery.
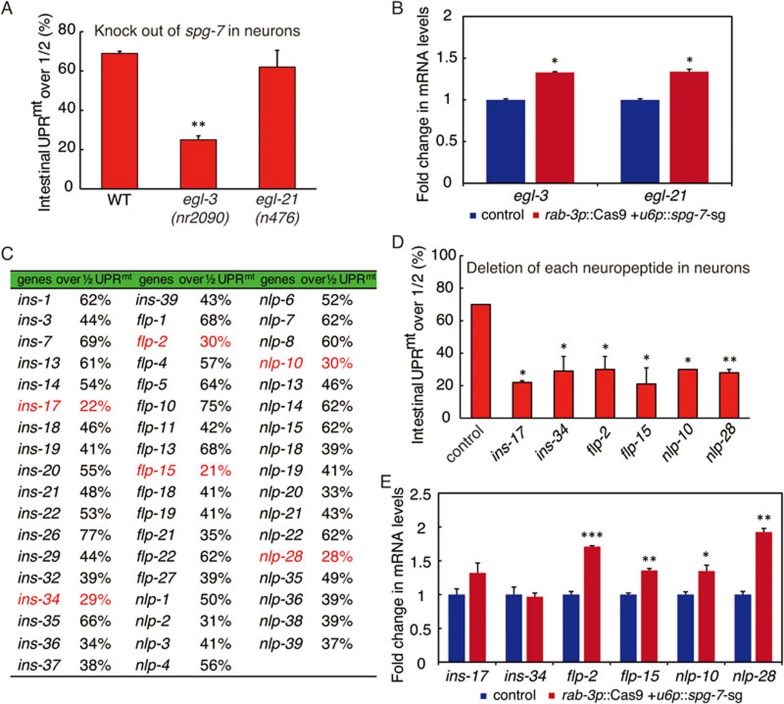
Neuropeptides are short peptides that modulate synaptic activity22,23. Their precursors are typically posttranslationally cleaved by a series of enzymes to yield the mature neuropeptides24,25,26,27. EGL-3 is one of the major enzymes responsible for the initial endoproteolytic cleavage of pro-neuropeptides28,29. Once the pro-neuropeptides are cleaved, EGL-21, a neural-specific carboxypeptidase then cleaves the C-terminal basic residues from the peptide sequences30. To examine the functional connection between neuropeptides and cell non-autonomous UPRmt, we tested the ability of worms to induce peripheral hsp-6p::GFP in mutant strains carrying egl-3 or egl-21 deletion (Supplementary information, Figure S5A). We found that egl-3 mutation was able to suppress cell non-autonomous UPRmt induction (Figure 6A). However, egl-21 mutation had no such effect (Figure 6A). In addition, both egl-3 and egl-21 transcripts were induced when spg-7 was deleted in the nervous system (Figure 6B). The lack of peripheral induction of UPRmt in egl-3 mutant indicates that neuropeptide might be required to mediate cell non-autonomous UPRmt.
Figure 6.
Neuropeptide regulates cell non-autonomous UPRmt signaling. (A) Cell non-autonomous UPRmt is suppressed in egl-3 mutants. Peripheral expression of hsp-6p::GFP induced by neuronal knockout of spg-7 in each strain is quantified. n ≥ 30. (B) qRT-PCR measures transcript levels of egl-3 and egl-21 in rab-3p::Cas9+u6p::spg-7-sg; hsp-6p::GFP worms. Fold changes are compared with the control hsp-6p::GFP worms. (C) hsp-6p::GFP induction in rab-3p::Cas9+u6p::spg-7-sg animals with the neural deletion of each indicated neuropeptide genes is quantified. The numbers of percentage under 30% are marked in red. (D) Individual deletion of six indicated neuropeptide genes in the nervous system suppresses the peripheral induction of hsp-6p::GFP. n ≥ 20. (E) Transcript levels of six indicated neuropeptide genes in rab-3p::Cas9+u6p::spg-7-sg; hsp-6p::GFP worms are measured by qRT-PCR. Fold changes are compared with the control hsp-6p::GFP worms. Error bars indicate mean ± SE. A Student's t-test is used to assess significance: *P < 0.05, **P < 0.01,***P < 0.001.
Over a hundred neuropeptide genes encoding more than 250 neuropeptides have been reported in C. elegans22,23. To identify which neuropeptide plays a key role in the cell non-autonomous regulation of mitochondrial stress signaling, we employed the CRISPR-Cas9 system and designed sgRNA sequences to specifically knock out all 103 reported neuropeptide genes in C. elegans (Supplementary information, Table S1). For the initial experiments, we randomly grouped three Cas9 plasmids against three neuropeptide genes, and tested if deletion of three neuropeptide genes together would block cell non-autonomous UPRmt (Supplementary information, Figure S5B). After the first round of test, we picked the groups that showed significant suppression of peripheral UPRmt induction, and knocked out those neuropeptide genes individually to test their efficiencies for blocking cell non-autonomous UPRmt (Figure 6C). From the 53 neuropeptide genes that we deleted individually, 6 of them showed promising results: deletion of individual neuropeptide genes including ins-17, ins-34, flp-2, flp-15, nlp-10 or nlp-28 strongly inhibited the peripheral induction of UPRmt (Figure 6C and 6D, Supplementary information, Figure S5C and S5D). The transcript levels of flp-2, flp-15, nlp-10 and nlp-28 were significantly increased when neuronal spg-7 was deleted (Figure 6E). Interestingly, the upregulation of nlp-28, but not the other three, depended on the transcription factor ATFS-1 (Supplementary information, Figure S5E). Collectively, these data indicate that neuropeptide indeed has an important role in signaling cell non-autonomous UPRmt.
Neuropeptide FLP-2 mediates the cell non-autonomous UPRmt
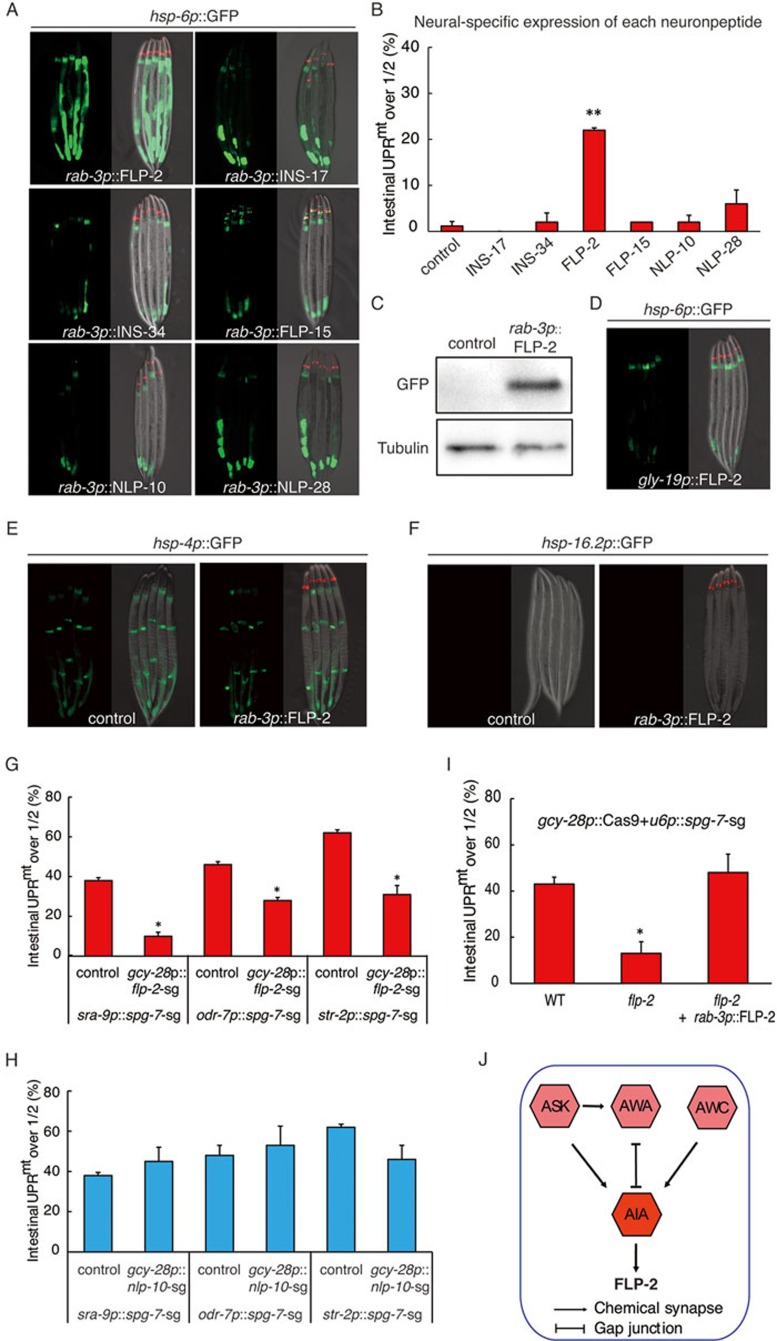
To determine which of the six neuropeptides we identified directly signals cell non-autonomous UPRmt, we overexpressed each neuropeptide in the nervous system and tested whether expression of any neuropeptide could directly induce peripheral hsp-6p::GFP reporter expression. Intriguingly, we found that overexpression of FLP-2 but not other neuropeptides in neurons directly induced peripheral hsp-6p::GFP expression (Figure 7A and 7C), and the endogenous levels of UPRmt response genes irg-1 and cyp-14A1 (Supplementary information, Figure S6A). Consistent with a role of FLP-2 in mediating the peripheral induction of mitochondrial stress response, deletion of spg-7 in FLP-2-expressing neurons driven by an flp-2 promoter activated the cell non-autonomous expression of hsp-6p::GFP (Supplementary information, Figure S6B and S6C). FLP-2 only functions in the nervous system to signal cell non-autonomous UPRmt, as intestine-specific expression of FLP-2 driven by a gly-19 promoter did not induce UPRmt (Figure 7D). Moreover, the other five neuropeptides that we identified from the screen were not required for FLP-2 overexpression induced cell non-autonomous UPRmt (Supplementary information, Figure S6D).
Figure 7.
Neuropeptide FLP-2 functions in AIA interneuron to mediate cell non-autonomous UPRmt. (A, B) Representative fluorescent images (A) and quantification (B) of hsp-6p::GFP reporter induction in animals expressing each indicated neuropeptide under a neural-specific promoter. odr-1 p::dsRed is used as co-injection marker. n ≥ 35, error bars indicate mean ± SE. A Student's t-test is used to assess significance: **P < 0.01. (C) Induction of hsp-6p::GFP in rab-3p::FLP-2 strain is assayed by western blotting. (D) Overexpression of FLP-2 in intestine fails to activate UPRmt. (E, F) Overexpression of FLP-2 in the nervous system fails to activate UPRER or HSR. (G, H) Knockout of flp-2(G), but not control neuropeptide gene nlp-10 (H), in AIA interneuron suppresses the peripheral induction of hsp-6p::GFP in animals with spg-7 knockout in each indicated sensory neurons. (I) Peripheral induction of hsp-6p::GFP by spg-7 knockout in AIA interneuron is significantly alleviated in flp-2 null allele, and rescued by neural expression of FLP-2. (J) A schematic graph showing a neural network sub-circuit and the secretion of neuropeptide FLP-2 for signaling cell non-autonomous UPRmt. n ≥ 30, error bars indicate mean ± SE. A Student's t-test is used to assess significance: *P < 0.05.
FLP-2 has a specific role in UPRmt, because stress reporters for UPRER (hsp-4p::GFP) or heat shock (hsp-16.2p::GFP) were not activated when FLP-2 was overexpressed (Figure 7E and 7F). Interestingly, flp-2 was not required for KillerRed-induced cell non-autonomous UPRmt (Supplementary information, Figure S6E), further strengthening the idea that different types of mitochondrial perturbation affect animal fitness and the corresponding signaling cascades differently. Although overexpression of FLP-2 in the nervous system activates cell non-autonomous UPRmt, expression of FLP-2 in neurons did not extend worm lifespan (Supplementary information, Figure S6F). Accumulating evidence suggested that the relationship between the UPRmt and longevity is complicated. Activation of UPRmt is not necessarily associated with an increased lifespan31. Thus, while FLP-2 is required for cell non-autonomous activation of UPRmt, additional signaling might be required for the lifespan extension in neuronal spg-7 deletion model.
FLP-2 functions in AIA interneuron to mediate cell non-autonomous UPRmt
We reported a neural sub-circuit (ASK, AWA, AWC and AIA) regulating cell non-autonomous induction of UPRmt (Figure 4E), and discovered a role of neuropeptide FLP-2 in signaling peripheral UPRmt. Interestingly, AIA interneuron is one of the neurons expressing and releasing neuropeptide FLP-232. flp-2 knockout in AIA neuron driven by a gcy-28 promoter significantly suppressed spg-7 deletion in AIA neuron induced cell non-autonomous UPRmt(Supplementary information, Figure S7A). As a control, nlp-10 knockout in AIA neuron did not suppress spg-7 deletion in AIA neuron-induced cell non-autonomous UPRmt (Supplementary information, Figure S7B), suggesting that the secretion of FLP-2 from AIA interneuron, indeed, functions to signal UPRmt.
Finally, because ASK, AWA and AWC sensory neurons form chemical synapses or gap junction with AIA interneuron, we thought to understand if the secretion of neuropeptide FLP-2 from AIA neuron has a role upstream or downstream of sensory neuron activation. We blocked FLP-2 secretion from AIA interneuron by deletion of flp-2 using gcy-28 promoter and asked if it would suppress the cell non-autonomous UPRmt induced by mitochondrial perturbation in ASK/AWA/AWC neurons. In all three cases, deletion of flp-2 but not nlp-10 in AIA interneuron significantly suppressed the abilities of ASK/AWA/AWC sensory neuron to induce UPRmt in distal tissues (Figure 7G and 7H). In addition, flp-2 mutant allele suppressed spg-7 deletion in AIA interneuron-induced cell non-autonomous UPRmt, which could be rescued with the expression of FLP-2 in the nervous system (Figure 7I). Collectively, these results suggest a model that once ASK/AWA/AWC neurons sense mitochondrial dysfunction, they communicate with AIA interneuron. AIA interneuron then releases neuropeptide FLP-2 to signal downstream neurons and eventually signals peripheral tissues to activate cell non-autonomous UPRmt (Figure 7J).
Discussion
Organisms constantly face a dynamic and complex environment. To cope with the ever-changing surroundings, living systems evolve several organelle-specific stress responses to maintain cellular homeostasis. Intriguingly, all these stress responses seem to be controlled by only a subset of cells, particularly neurons, suggesting a cell non-autonomous coordination of multiple tissues to meet environmental challenges.
Consistent with previous reports, we find that expression of ROS-generating protein KillerRed, or endonuclease Cas9 to knock out mitochondrial gene spg-7 or cco-1, only in the nervous system is sufficient to activate cell non-autonomous UPRmt. However, expression of uncoupling protein UCP-4 only induces cell autonomous UPRmt in neurons. It is not clear why certain mitochondrial perturbation in neurons activates cell non-autonomous UPRmt, but others do not. It is possible that the UPRmt pathway could distinguish different forms of mitochondrial dysfunction and initiate different organismal responses. It will be of special interest in the future to understand which types of mitochondrial perturbation (e.g., changes in mitochondrial membrane potential, metabolic state, mitochondrial protein folding capacity or stoichiometric of large multimeric complexes) would invoke the induction of cell non-autonomous UPRmt.
Mild inhibition of mitochondrial function has been associated with longevity. For example, RNAi knockdown of nuclear-encoded mitochondrial genes in C. elegans is sufficient to extend the animals' lifespan9,33,34. Consistently, studies in Drosophila and rodents also confirm the role of mitochondria in regulating lifespan6,35,36,37,38. One model for the robust induction of longevity by mitochondrial gene inactivation is that these perturbations mimic the action of natural xenobiotics that C. elegans encounters in the soil, and thus are subjected to evolutionary selection for regulation of lifespan15,39,40,41,42. If xenobiotics are the triggers of a long-lived state, it is likely that mitochondrial inactivation takes place initially in the cells with direct environment exposure, such as sensory neurons and intestinal cells. The deficiency in mitochondria sensed in those cells would induce an endocrine signal to the rest of the animal to suspend further development toward reproduction and to extend lifespan. Consistent with this idea, inhibition of mitochondrial ETC function only in the nervous system of C. elegans is able to induce lifespan extension to the level comparable to the whole organismal mitochondrial inhibition9. In addition, our results also strengthen the idea that neurons have a central role in sensing and transmitting a stress cue to other tissues. In particular, we uncover a neuronal sub-circuit including ASK, AWA and AWC sensory neurons, and AIA interneuron for sensing and signaling mitochondrial stress. These results also suggest that organelle-specific stress responses employ different neural circuit to sense and transduce the stress signal, as HSR in the cytosol depends primarily on the AFD sensory neuron and AIY interneuron10,11. It will be interesting in the future to explore the broader neural circuit for UPRmt and determine how each neuron communicates with others.
A functional UPRmt pathway is required in both neurons and the receiving cells for a cell non-autonomous induction of UPRmt. Two mechanisms have been proposed for the activation of UPRmt upon mitochondrial dysfunction. First, mitochondrial import efficiency, which is affected by mitochondrial inhibition, determines the localization of transcription factor ATFS-1 and the induction of UPRmt8. This process also requires the transcription factors DVE-1 and its co-factor UBL-514. In the second model, the UPRmt might be induced by peptides generated by the mitochondrial matrix protease CLPP-114. The peptide transporter HAF-1 is also required for the activation of UPRmt, possibly by transporting peptides to the cytosol where they signal downstream events20. Of the current identified UPRmt pathway members, ATFS-1 is essential for the cell non-autonomous induction of UPRmt, whereas DVE-1 is partially required. However, CLPP-1 or HAF-1 does not significantly affect peripheral induction of UPRmt in our system. It is also interesting to note that expression of ATFS-1Δ1-32, which constitutively activates the UPRmt pathway, only in the nervous system is sufficient to induce hsp-6p::GFP in distal tissues. It indicates that the neuronal UPRmt pathway directly regulates the neuroendocrine mediators of non-autonomous UPRmt.
We find a dependency of non-autonomous signaling of UPRmt on neuropeptide and perform a large-scale CRISPR-Cas9 screen to identify neuropeptide FLP-2 as a potential mediator of cell non-autonomous UPRmt. Loss of flp-2 suppresses the peripheral induction of hsp-6p::GFP. More importantly, restricted expression of FLP-2 in neurons directly induced hsp-6p::GFP expression in distal tissues. Because neuropeptides are highly conserved among C. elegans and vertebrates43, it is possible that the neuropeptides that we discovered in C. elegans may have a similar role to signal cell non-autonomous UPRmt in mammals. We do not yet understand the detailed mechanism by which FLP-2 is perceived at the current stage. More efforts are warranted to identify FLP-2 receptor and its receiving cells in the future. Furthermore, intestinal expression of FLP-2 does not induce hsp-6p::GFP, indicating that FLP-2 acts locally in the nervous system upon release. Therefore, identification of secondary long-range acting molecules to signal the peripheral induction of UPRmt will be of particular interest. Such molecules may resemble the characteristics of endocrine signal or metabolic regulator, like the recent identified cell non-autonomous stress signal molecule serotonin or FGF2144,45,46.
Mitochondrial dysfunction has been associated with multiple neurodegenerative disorders. Intriguingly, cell non-autonomous effects such as metabolic decline have been observed in these diseases47. Therefore, understanding the cell non-autonomous aspects of mitochondrial stress response may allow future control of this protective response, which will have therapeutic potential in the treatment of the symptoms of mitochondrial disorders and neurodegenerative diseases.
Materials and Methods
Strains
egl-3(nr2090) and egl-21(n476) were a generous gift from Dr Joshua Kaplan. atfs-1(tm4525) and ATFS-1Δ1-32 containing vector were a gift from Dr Cole Haynes. hsp-6p::GFP(zcIs13), N2 and flp-2(ok3351) strains were obtained from the Caenorhabditis Genetic Center.
For tissue-specific mCherry expressing strains, each promoter was PCR amplified from worm genomic DNA. Transgenic lines were generated by microinjecting plasmid DNA containing 50-100 ng/μl plasmid DNA with 35 ng/μl co-injection marker pRF4(rol-6).
For generation of Tom20::KillerRed strain, the Tom20:: KillerRed sequence was PCR amplified from a plasmid provided by Dr Marc Hammarlund and inserted into vectors containing myo-3, gly-19, unc-119 or rab-3 promoter and unc-54 3′-UTR sequence. For generation of UCP-4::mCherry strains, the ucp-4 sequence was PCR amplified from worm genomic DNA and inserted into the corresponding vector containing unc-119 promoter and the mCherry sequence. Transgenic lines were generated by microinjecting plasmid DNA containing 35 ng/μl KillerRed or UCP-4 construct into hsp-6p::GFP strain.
The CRISPR-Cas9 vector was obtained from Addgene (#47549)18. The eft-3 promoter in the original vector was replaced with each tissue-specific promoter to express Cas9 endonuclease tissue specifically. The sg RNA target sequences following GN19-25NGG(N = A, C, G or T) pattern were designed via http://tools.flycrispr.molbio.wisc.edu/targetFinder/. The uniqueness of each target site was confirmed by BLAST search targeting sequence against the C. elegans genome (http://www.wormbase.org/tools/blast_blat). Conditional knockout strains were generated by microinjecting 50 ng/μl Cas9-sgRNA plasmids with 35 ng/μl co-injection marker odr-1p::dsRed or mec-7p::RFP. rab-3p::Cas9+u6p::spg-7-sg; hsp-6p::GFP; odr-1p::dsRed transgenic line was integrated by gamma irradiation, and backcrossed three times with wild-type N2 strain.
Neuropeptides DNA sequence were inserted downstream of the neural-specific promoter rab-3 and upstream of unc-54 3′-UTR into pPD49.26 vector. Transgenic animals expressing each neuropeptides were obtained by microinjecting of 50 ng/μl neuropeptide expression plasmid with 50 ng/μl hsp-6p::GFP and 35 ng/μl co-injection marker odr-1p::dsRed.
Unless otherwise stated, all strains were raised on plates with the Escherichia coli strain OP50 at 20 °C.
Antioxidants treatment
NAC or ascorbic acid was added into RNAi plates' medium with a final concentration of 10 mM before pouring of the plates. Synchronized L4 worms with desired genotype were transferred to fresh plates seeded with HT115, and UPRmt phenotype was scored after 24 h. Representative images showed in the figure were repeated at least three times. It should be noted that KillerRed does not need to be light activated in our experiment. KillerRed expression on the inner or outer mitochondrial membrane alone is sufficient to induce non-autonomous UPRmt.
Lifespan measurement
Synchronized L1 worms were raised on fresh plates with the E. coli strain OP50 at 20 °C. After growing into day 1 adult stage (with a well-formed vulva but without eggs in the uterus), worms with desired genotype were transferred to fresh plates every 2 days and monitored for survival. A worm did not response to three gentle touches on head and displayed no pharyngeal pumping was considered dead. Those that died due to internal hatching or ruptured vulvae, or crawling off the agar were censored.
Oxygen consumption
Synchronized L4 worms were collected, washed with M9 buffer for three times, resuspended to a final volume of 2.1 ml, and then loaded into the chamber of Oroboros Oxygraph-2K (Oroboros Instruments). Measurements were performed for 15 min at 20 °C. Oxygen concentration was analyzed using DatLab software. Consumption rates were normalized to the number of worms.
Antimycin survival assay
Antimycin A was evenly spread on the fresh worm plates seeded with OP50 at a final concentration of 25 μg/ml. L4 worms were picked onto plates and survival rates were assayed after 24 h raising at 20 °C.
Genotyping
Worms were placed in 10 μl of worm lysis buffer (50 mM KCl, 10 mM Tris-HCl pH 8.0, 2.5 mM MgCl2, 0.45% IGEPAL CA-630, 0.45% Tween-20, 0.2 μg/μl Proteinase K), incubated at 60 °C for 1 h and 95 °C for 15 min. 1 μl supernatant was taken and performed for PCR analysis with the following primers.
egl-3: forward 5′-CGAGAGATGGGAAGTCACGG-3′, reverse 5′-CCCCAATGTTCTGCTGCTCT-3′
egl-21: forward 5′-ACGGGGCATGATCGTTCTTT-3′, reverse 5′-CGTTTCTTTCACGTCCGCTC-3′
To detect indels produced by CRISPR/Cas9 knockout, T7 endonuclease I (T7E1) assay was performed19,48. DNA fragments containing target sites were PCR amplified and digested with T7E1 (NEB, #M0302) and analyzed via agarose gel electrophoresis. Non-perfectly matched DNA would be cleaved by T7E1. To sequence the indels, PCR fragments were cloned into pEASY blunt Cloning Vectors (Transgen, #CB111), and the colonies were sequenced individually.
spg-7: forward 5′-TGTTTGCGCAGTGCATGATT-3′, reverse 5′-AAAATACGGCCCGGGAAACC-3′
cco-1: forward 5′-GCTCAACTTGCTAAGACGGC-3′, reverse 5′-TGTCCGGAATCTTGCTCACC-3′
atfs-1: forward 5′-TTTCAGGACCCAAAAACCCAC-3′, reverse 5′-TCCGAAGCTGGTCCTGTAAG-3′
dve-1: forward 5′-ACTTTCGCTCCATTCGTCGT-3′, reverse 5′-TGAGTGGGTAGGGTCGTCTT-3′
ubl-5: forward 5′-CATTGCAAGTCTGCATTTGCG-3′, reverse 5′-CATCACCAAGAATGGCGGTC-3′
haf-1: forward 5′-TCACTTGAGAGCACGGTGAG-3′, reverse 5′-GCAGCATTACGTCTGTCAACTC-3′
clpp-1: forward 5′-CCCCCTGTTCTGCTCTGTGA-3′, reverse 5′-GGTTCGGCCCATTCTTTTGTT-3′
flp-2: forward 5′-ATGCAAGTTTCTGGAATCCTATCTG-3′, reverse 5′-TTATTGGAAGTCGTAATCTGGCAGC-3′.
Imaging and analysis
Worms were anesthetized with 50 mM sodium azide in M9 (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4, H2O to 1 L), mounted on 2% agarose pads, and imaged by a Zeiss Axio Imager M2 microscope. Comparable GFP reporter images were captured with the same exposure time and magnification.
For quantifying periphery induction of UPRmt, percentage of the transgenic lines with intestinal hsp-6p::GFP induction was measured. Day 1 adult worms with intestinal UPRmt over the half of full-length intestine were scored. Over 30 transgenic worms were tested for each experiment. Each experiment had been done with at least two biological replicates.
Western blotting
Day 1 adult worms were washed off from plates by M9 and resuspended with SDS loading buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue), and boiled at 95 °C for 10 min. Samples containing the same amount of total protein were loaded onto SDS-PAGE and transferred onto PVDF membrane (Bio-Rad). After blocked with 5% milk-TBST, membrane was probed with the designated primary and secondary antibodies (anti-GFP antibody: sungene, #KM8009; anti-tubulin antibody: abcam, #ab6161), developed with the enhanced chemiluminescence method (Thermo) and visualized by Tanon 5200 chemical luminescence imaging system.
RNA interference
RNAi clones were grown in LB containing 50 μg/ml carbenicillin at 37 °C overnight and seeded on to worm plates with IPTG. Dried plates were kept at room temperature overnight to allow IPTG induction of dsRNA expression.
RNA isolation and real-time PCR
Synchronized young adult worms were washed with M9 and resuspended with trizol reagent, frozen and homogenized three times by liquid nitrogen. Total RNA was isolated by chloroform extraction, followed by isopropanol precipitation. cDNA was synthesized by reverse transcription kit (Transgen, AT311). Quantitative PCR was carried out using SYBR Green PCR Master Mix (Bio-Rad). Quantification of transcripts was normalized to rpl-32.
Primer sequences for real-time PCR:
rpl-32: forward 5′-AGGGAATTGATAACCGTGTCCGCA-3′, reverse 5′-TGTAGGACTGCATGAGGAGCATGT-3′
egl-3: forward 5′-CAACACACACATGGGGAGAG-3′, reverse 5′-ACGGTGTCGAGCTTCTTGTT-3′
egl-21: forward 5′-TATTCCAACGAAACCGGAAG-3′, reverse 5′-GGTTCATGGATGGAAGGATG-3′
ins-17: forward 5′-TGGCTGCTGTAGCCGCATT-3′, reverse 5′-CCACCGCCGTCGTTGTAGT-3′
ins-34: forward 5′-TCGGGAATCGACAGTCTTCCA-3′, reverse 5′-GCACGGCTGTCTACAGACTC-3′
flp-2: forward 5′-AGCTTCAGCAATGGCAAAAC-3′, reverse 5′-CAGCGGATTGAATCGTTTTC-3′
flp-15: forward 5′-GCAGTTCTCCACGCTTATTCGAG-3′, reverse 5′-GTCCAGATGGTCCACGTCGT-3′
nlp-10: forward 5′-CAACGGCGGTATGTACGGCA-3′, reverse 5′-GAAAACGGCATTGCTCCTCGT-3′
nlp-28: forward 5′-CTGATTCTCGTCTTCCTTCTCGC-3′, reverse 5′-ACTTTCCCCATCCTCCGTACA-3′
atfs-1: forward 5′-CGTGACGCTGGAAGCATG-3′, reverse 5′-GGTGGCATTCCGTACTCATC-3′
dve-1: forward 5′-CTCGACCTCGGAGCATTCA-3′, reverse 5′-CGCGCATTGAGTTTGCCATATC-3′
ubl-5: forward 5′-CGAATCAAGTGCAATCCATC-3′, reverse 5′-TCATTGGTAGTAGAGCTCG-3′
haf-1: forward 5′-GTTAATCAGACTGCGATCGAGCG-3′, reverse 5′-GCTCTCCATATGTGATATGTCCCG-3′
clpp-1: forward 5′-AACAGTCCAGGCGGCAGTGT-3′, reverse 5′-ACCACCACTCGGCTGGTGAA-3′
irg-1: forward 5′-TAAGGCGTTGCTAGTTGGGG-3′, reverse 5′-CCAAACATGCGCGGTACATT-3′
cyp-14A1: forward 5′-ACGATGGGGAGAGTGAACCT-3′, reverse 5′-TGATGGCAAACGTGCTGTTC-3′.
Author Contributions
LS and YL designed the experiments; LS and RN carried out the experiments. LS and YL wrote the paper. YL supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Dr Cole Haynes and the Caenorhabditis Genetics Center for providing strains. We thank Drs Guangshuo Ou and Dong Liu for technical support. The work is supported by grants from the National Natural Science Foundation of China (grants 31422033 and 31471381) and the Ministry of Science and Technology of China (973 grants 2013CB910104) awarded to YL.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Overexpression of KillerRed in neurons induces cell-non-autonomous UPRmt.
Neuronal deletion of spg-7 or cco-1 by CRISPR-Cas9 induces UPRmt in distal tissues.
A neural sub-circuit is required for sensing and transducing mitochondrial stress.
atfs-1 in neurons is required for the peripheral induction of UPRmt.
Cell non-autonomous UPRmt signaling requires neuropeptides.
FLP-2 participates in cell-non-autonomous UPRmt signaling.
FLP-2 participates in cell-non-autonomous UPRmt signaling.
References
- Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol 2014; 217:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Ozcan U. Unfolded protein response signaling and metabolic diseases. J Biol Chem 2014; 289:1203–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol 2012; 197:857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 2006; 174:229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci 2004; 117:4055–4066. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 2013; 497:451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol Cell 2015; 58:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 2012; 337:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 2011; 144:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 2008; 320:811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Morimoto RI. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc Natl Acad Sci USA 2011; 108:14204–14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 2013; 153:1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DC, Bejjani RE, Ramirez PM, et al. Rapid and permanent neuronal inactivation in vivo via subcellular generation of reactive oxygen with the use of KillerRed. Cell Rep 2013; 5:553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell 2007; 13:467–480. [DOI] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 2014; 508:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Liu D. Mitochondrial potassium channels and uncoupling proteins in synaptic plasticity and neuronal cell death. Biochem Biophys Res Commun 2003; 304:539–549. [DOI] [PubMed] [Google Scholar]
- Ji C, Guo W, Zhang M, Lu X, Ni Y, Guo X. Caenorhabditis elegans ucp-4 regulates fat metabolism: suppression of ucp-4 expression induced obese phenotype and caused impairment of insulin like pathway. Gene 2012; 491:158–164. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 2013; 10:1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Zhang X, Chai Y, et al. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Dev Cell 2014; 30:625–636. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell 2010; 37:529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 1986; 314:1–340. [DOI] [PubMed] [Google Scholar]
- Li C, Kim K. Neuropeptide gene families in Caenorhabditis elegans. Adv Exp Med Biol 2010; 692:98–137. [DOI] [PubMed] [Google Scholar]
- Li C, Nelson LS, Kim K, Nathoo A, Hart AC. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann NY Acad Sci 1999; 897:239–252. [DOI] [PubMed] [Google Scholar]
- Husson SJ, Clynen E, Baggerman G, De Loof A, Schoofs L. Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem Biophys Res Commun 2005; 335:76–86. [DOI] [PubMed] [Google Scholar]
- Marks NJ, Maule AG, Geary TG, et al. APEASPFIRFamide, a novel FMRFamide-related decapeptide from Caenorhabditis elegans: structure and myoactivity. Biochem Biophys Res Commun 1997; 231:591–595. [DOI] [PubMed] [Google Scholar]
- Marks NJ, Shaw C, Halton DW, et al. Isolation and preliminary biological assessment of AADGAPLIRFamide and SVPGVLRFamide from Caenorhabditis elegans. Biochem Biophys Res Commun 2001; 286:1170–1176. [DOI] [PubMed] [Google Scholar]
- Rosoff ML, Doble KE, Price DA, Li C. The flp-1 propeptide is processed into multiple, highly similar FMRFamide-like peptides in Caenorhabditis elegans. Peptides 1993; 14:331–338. [DOI] [PubMed] [Google Scholar]
- Kass J, Jacob TC, Kim P, Kaplan JM. The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J Neurosci 2001; 21:9265–9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker C, Rose AM. A look at the Caenorhabditis elegans Kex2/Subtilisin-like proprotein convertase family. Bioessays 2000; 22:545–553. [DOI] [PubMed] [Google Scholar]
- Jacob TC, Kaplan JM. The EGL-21 carboxypeptidase E facilitates acetylcholine release at Caenorhabditis elegans neuromuscular junctions. J Neurosci 2003; 23:2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Kaeberlein M. The mitochondrial unfolded protein response and increased longevity: cause, consequence, or correlation? Exp Gerontol 2014; 56:142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol 2004; 475:540–550. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, et al. Rates of behavior and aging specified by mitochondrial function during development. Science 2002; 298:2398–2401. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet 2003; 33:40–48. [DOI] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T Jr, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol 2009; 19:1591–1598. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev 2005; 19:2424–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 2013; 154:430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell 2013; 155:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 2012; 149:452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 2014; 516:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz AM, Haynes CM. UPR(mt)-mediated cytoprotection and organismal aging. Biochim Biophys Acta 2015; 1847:1448–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore DE, Carr CE, Ruvkun G. Induction of cytoprotective pathways is central to the extension of lifespan conferred by multiple longevity pathways. PLoS Genet 2012; 8:e1002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SS. Neuroscience: as the worm turns. Nature 2013; 494:296–299. [DOI] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med 2013; 19:83–92. [DOI] [PubMed] [Google Scholar]
- Suomalainen A1, Elo JM, Pietiläinen KH, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 2011; 10:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen KM, Durieux J, Shao LW. Neuroendocrine coordination of mitochondrial stress signaling and proteostasis. Cell 2016; 166:1553–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Schuck PF, Wenk GL, Ferreira GC. Metabolic disturbances in diseases with neurological involvement. Aging Dis 2014; 5:238–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashal RD, Koontz J, Sklar J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat Genet 1995; 9:177–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overexpression of KillerRed in neurons induces cell-non-autonomous UPRmt.
Neuronal deletion of spg-7 or cco-1 by CRISPR-Cas9 induces UPRmt in distal tissues.
A neural sub-circuit is required for sensing and transducing mitochondrial stress.
atfs-1 in neurons is required for the peripheral induction of UPRmt.
Cell non-autonomous UPRmt signaling requires neuropeptides.
FLP-2 participates in cell-non-autonomous UPRmt signaling.
FLP-2 participates in cell-non-autonomous UPRmt signaling.