Abstract
The purpose of this study was to investigate the differential expression and functional roles of long non-coding RNAs (lncRNAs) in neuroblastoma tissue. LncRNA microarrays were used to identify differentially expressed lncRNAs between tumor and para-tumor tissues. In total, in tumor tissues, 3,098 and 1,704 lncRNAs were upregulated and downregulated, respectively. HCN3 and linc01105 exhibited the higher expression (P < 0.05; P < 0.01, respectively) in neuroblastoma tissue, whereas MEG3 displayed the lower expression (P < 0.01). HIF-1α expression was negatively correlated with cell proliferation in the linc01105 KD group. In addition, Noxa and Bid expression was positively correlated with cell apoptosis. Moreover, linc01105 knockdown promoted cell proliferation, whereas MEG3 overexpression inhibited proliferation. Finally, linc01105 knockdown, MEG3 overexpression and HCN3 knockdown all increased apoptosis. The correlation coefficients between those three lncRNAs and the International Neuroblastoma Staging System (INSS) stage were −0.48, −0.58 and −0.55, respectively. In conclusion, we have identified lncRNAs that are differentially expressed in neuroblastoma tissues. The lncRNAs HCN3, linc01105, and MEG3 may be important in biological behaviors of neuroblastoma through mechanisms involving p53 pathway members such as HIF-1α, Noxa, and Bid. The expressions of MEG3, HCN3 and linc01105 are all negatively correlated with the INSS stage.
Neuroblastoma is the most common type of extracranial solid tumor in children; it arises from the neural crest of the sympathetic nervous system and typically occurs in the adrenal medulla1. Neuroblastoma may regress spontaneously or differentiate into a ganglioneuroblastoma or benign ganglioneuroma, particularly in infants. The prognosis is good in children diagnosed with neuroblastoma at 1 year of age; however, undifferentiated neuroblastomas diagnosed after 5 years of age have a poor prognosis2,3.
In addition to protein-coding mRNAs, a large number of non-coding RNAs have been identified with structural, regulatory, and unknown functions. Long non-coding (lnc)RNAs, which are >200 nt in length4,5, have been implicated in gene regulation at the transcriptional and post-transcriptional levels6. The dysregulation of lncRNAs is a primary feature of human cancers, including prostate, breast, colon, and gastric cancers7,8,9,10. LncRNAs are also aberrantly expressed in disorders such as Beckwith-Wiedemann, DiGeorge, and Down’s syndromes11,12,13,14,15,16. However, the role of lncRNAs in neuroblastoma has not been fully investigated. Here, we have investigated the role of lncRNAs in the pathogenesis of neuroblastoma.
Results
LncRNA and mRNA expression profiles
After quantile normalization of the raw data, the expression profiles of 36,178 lncRNAs and 24,992 mRNAs were obtained for tumor and para-tumor tissues (Supplementary Fig. 1). The log2 distributions of the ratios of lncRNAs to mRNAs were similar for the two groups.
lncRNAs and mRNAs are differentially expressed in neuroblastoma tissues and para-tumor tissues
To identify differentially expressed lncRNAs and mRNAs between tumor and para-tumor tissues, we performed fold-change filtering (values ≥ 2). We found that 3,098 lncRNAs and 2,526 mRNAs were upregulated, whereas 1,704 lncRNAs and 2,604 mRNAs were downregulated, in neuroblastoma tissues compared to control samples (Supplementary Fig. 2).
Pathway analysis
The pathway analysis revealed 44 downregulated and 59 upregulated pathways in neuroblastoma tissue compared to control tissue. The p53 signaling pathway, which is associated with tumorigenesis, was among the upregulated pathways (enrichment score = 1.44; P = 0.036).
CNC network analysis
The upregulated p53 signaling pathway genes included BH3-interacting domain death agonist (BID), phorbol-12-myristate-13-acetate-induced protein (PMAIP), mitochondrial ribonuclease P protein 2 precursor 2, sestrin 3, cyclin (CCN) D1, CCNB1, and CCNE2. The CNC analysis revealed that the following lncRNA genes were differentially expressed between tumor and para-tumor tissues: maternally expressed gene (MEG) 3, hyperpolarization-activated cyclic nucleotide (HCN) 3, and long intergenic non-protein-coding RNA (linc) 01105 (P < 0.001). Pearson correlation coefficients were calculated as follows: lncRNA-MEG3 and PMAIP1 (−0.989), lncRNA-HCN3 and BID (0.988) and linc01105 and BID (0.987) (Supplementary Fig. 3).
Validation of differential lncRNA expression by qRT-PCR
Consistent with the microarray results, the qRT-PCR analysis revealed that the expression levels of HCN3 (P < 0.05) and linc01105 (P < 0.01) were higher, whereas MEG3 expression was lower (P < 0.01), in neuroblastoma tissue compared to para-tumor tissue (Supplementary Fig. 4).
Cell proliferation (CCK8 test) results
The change between the optical density (OD) values from 0 to 24 h was 0.064 ± 0.002 in the MEG3 overexpression (OE) group and 0.124 ± 0.01 in the linc01105 knockdown (KD) group. The MEG3 OE group and the linc01105 KD group were significantly different from their blank groups (MEG3 OE, P = 0.002; linc01105 KD P = 0.022) The change between the OD values from 24 to 48 h was 0.173 ± 0.011 in the MEG3 OE group and 0.97 ± 0.076 01 in the linc01105 KD group. Both groups were significantly different from their controls (P < 0.001) There were no differences between the HCN3 KD group and the controls at any time point (0 to 24 h, P = 0.269; 24 to 48 h, P = 0.095) (Fig. 1).
Figure 1. Effect of lncRNA expression on cell proliferation at different time points.
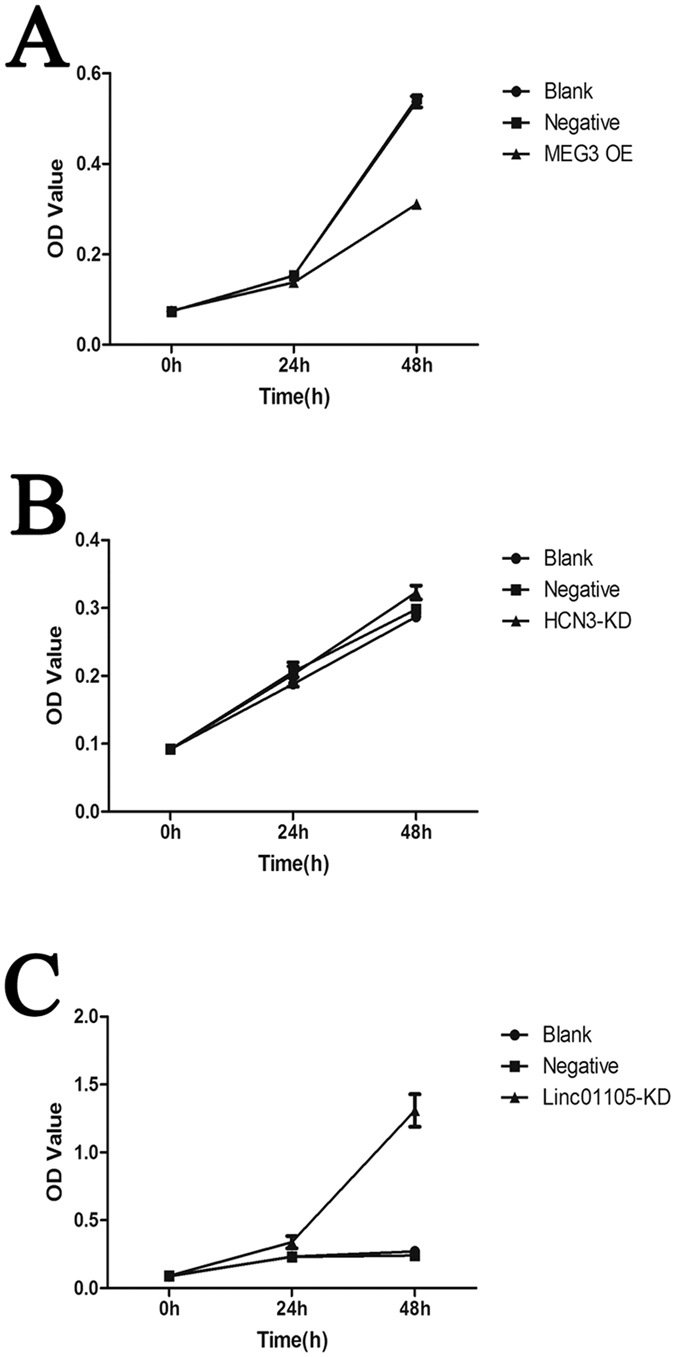
(A) Between 0 and 24 h and from 24 to 48 h, cell proliferation decreased in the MEG3 OE group compared to the blank group (P = 0.002; P < 0.001). (B) Between 0 and 24 h and from 24 to 48 h, cell proliferation increased in the linc01105 KD group compared to the blank group (P = 0.022; P < 0.001). (C) Between 0 and 24 h and from 24 to 48 h, cell proliferation was similar in the HCN3 KD and blank groups (P = 0.269; P = 0.095).
Apoptosis results
The numbers of apoptotic cells were as follows: blank group, 398 (6.7%); MEG3 OE group, 394 (8.7%); HCN3 KD group, 600 (11.4%); and linc01105 KD group, 534 (9.7%). All OE and KD groups had significantly higher levels of apoptosis than the blank group (P < 0.001) (Fig. 2).
Figure 2. Effect of lncRNA expression on apoptosis.
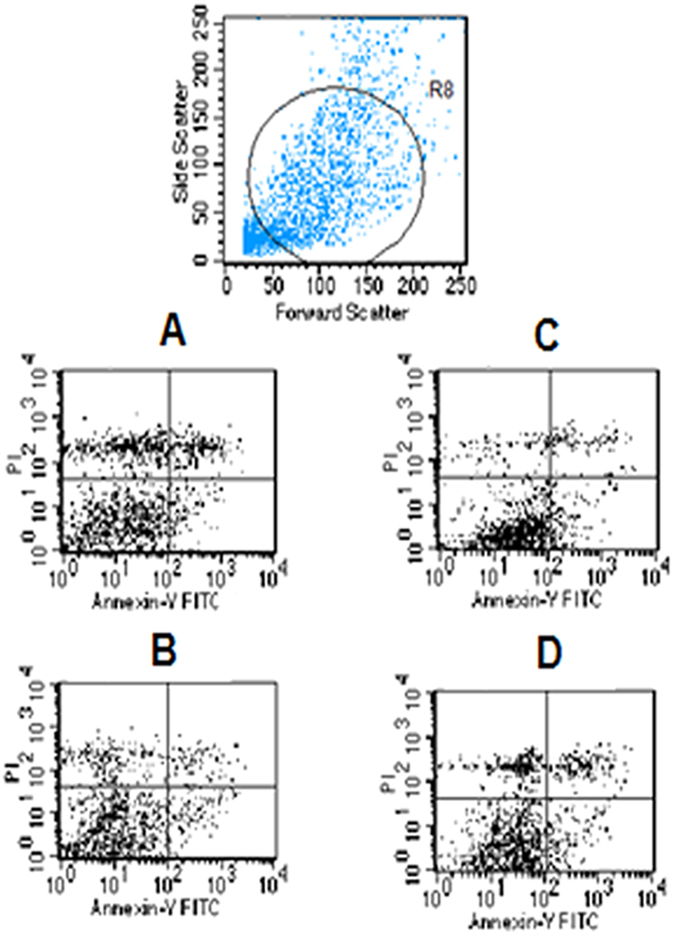
(A) Blank group. (B) MEG3 OE group. (C) HCN3 KD group. (D) linc01105 KD group.
mRNA expression of genes of interest
The relative expression (2−ΔΔCt) of Bid mRNA in the MEG3 OE group was 0.751 ± 0.071, which was lower than the blank group (P = 0.009). The relative expression (2−ΔΔCt) of Noxa mRNA was 1.368 ± 0.118 in the HCN3 KD group and 1.354 ± 0.19 in the linc01105 KD group. The expression in both groups was higher than in the controls (P = 0.009; P = 0.025). In the MEG3 OE group, the relative expression of HIF-1α (P = 0.683) and Noxa mRNA (P = 0.606) was similar to the blank groups. In the HCN3 KD and linc01105 KD groups, the relative expression of HIF-1α and Bid mRNA was similar to the controls (Fig. 3).
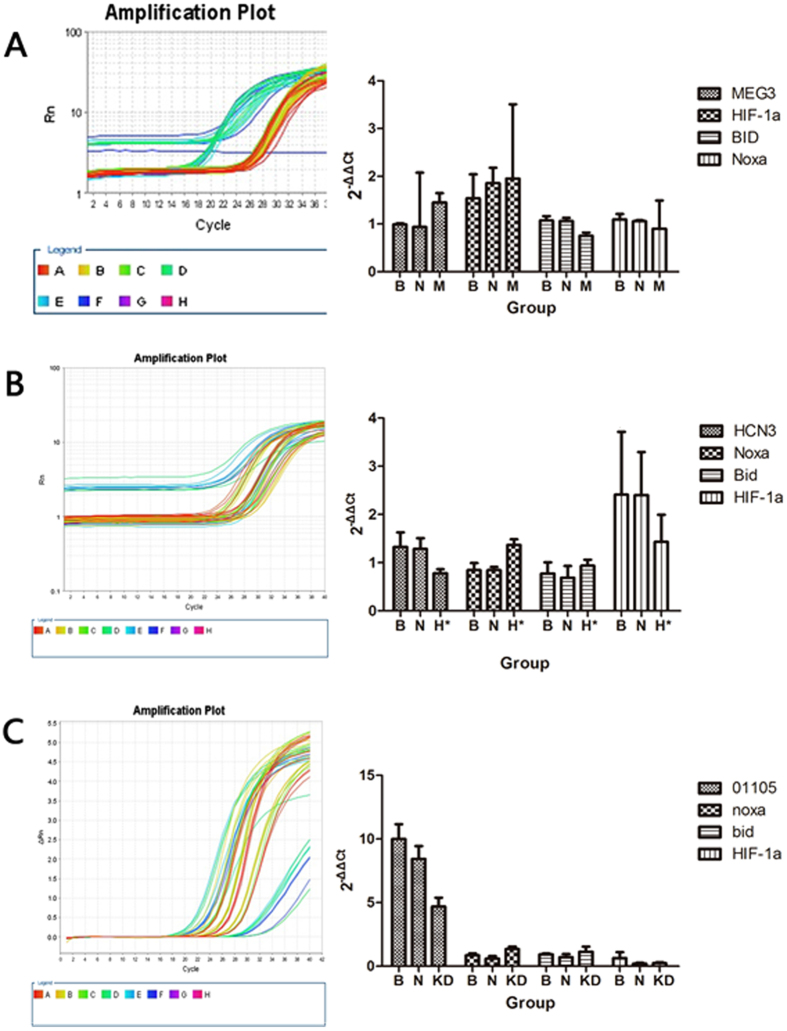
Figure 3. The conditions of target genes and relevant genes in each group.
(A) Amplification plots of lncRNA-MEG3, HIF-1α, Noxa and Bid mRNA expression in the blank, negative control and MEG3 OE groups. (B) Amplification plots of lncRNA-HCN3, HIF-1α, Noxa and Bid mRNA expression in the blank, negative control and HCN3 KD groups. (C) Amplification plots of lncRNA-linc01105, HIF-1α, Noxa and Bid mRNA expression in the blank, negative control and linc01105 KD groups. B, Blank group; N, Negative control group; H, HCN3 KD group; M, MEG3 OE group; KD, 01105 KD group.
Expression of relevant proteins
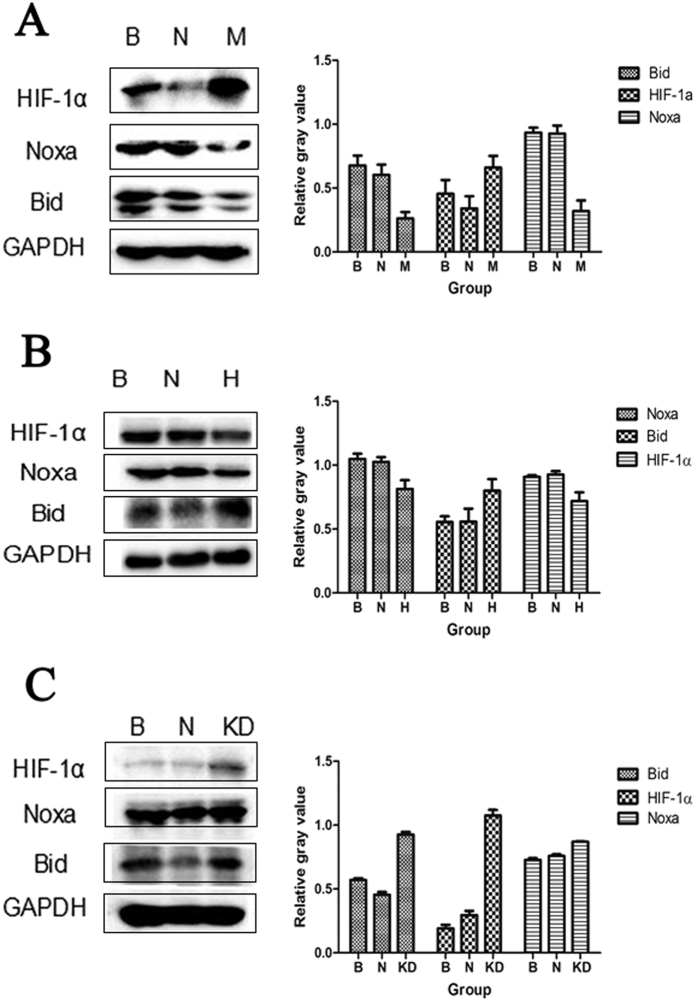
The effects of MEG3, HCN3 and linc01105 OE or KD on the protein expressions of HIF-1α, Noxa and Bid were analyzed. The expression of Noxa protein was 0.319 ± 0.084 in the MEG3 OE group and 0.815 ± 0.068 in the HCN3 KD group, both of which were lower than the blank groups (P < 0.001; P = 0.007). However, the expression of Noxa increased in the linc01105 KD group compared to the blank group (0.871 ± 0.003; P = 0.002). In addition, the expression of Bid protein was 0.8 ± 0.092 in the HCN3 KD group and 0.927 ± 0.021 in the linc01105 KD group, both of which were higher than Bid expression in the blank groups (P = 0.014; P < 0.001). However, Bid expression was reduced in the MEG3 OE group compared to the blank group (0.262 ± 0.05; P = 0.001). The expression of HIF-1α protein decreased in the HCN3 KD group (0.719 ± 0.069, P = 0.009) and increased in the linc01105 KD group (1.075 ± 0.046, P < 0.001). HIF-1α expression was not affected by MEG3 OE (0.661 ± 0.091; P = 0.065) (Fig. 4).
Figure 4. The effect of lncRNAs on HIF-1α, Bid and Noxa protein expression.
(A) Expression of HIF-1α, Noxa and Bid in the blank, negative control and MEG3 OE groups. (B) Expression of HIF-1α, Noxa and Bid in blank, negative control and HCN3 KD groups. (C) Expression of HIF-1α, Noxa and Bid in blank, negative control and linc01105 KD groups. B, Blank group; N, Negative control group; H, HCN3 KD group; M, MEG3 OE group; KD, linc01105 KD group. Full-length blots were shown as supplementary figure 6 in supplementary information.
Immunofluorescence results
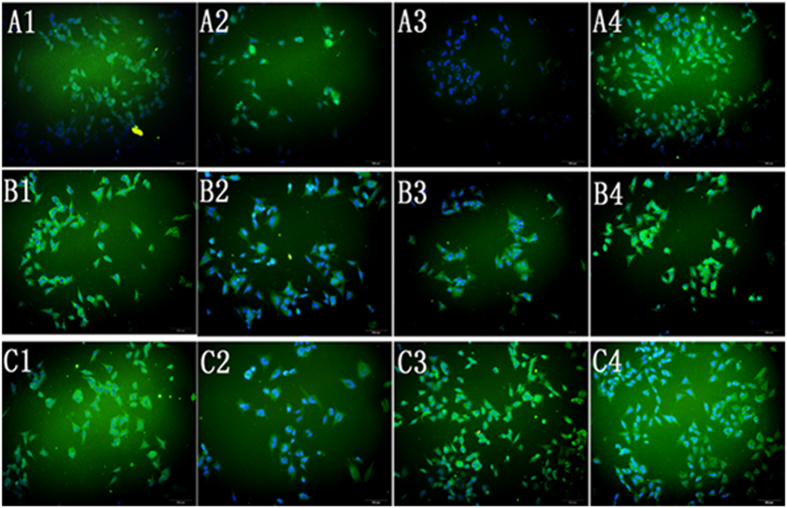
HIF-1α protein was highly expressed in the linc01105 KD group, weakly expressed in the HCN KD group and did not change expression in the MEG3 OE group. Positive HIF-1α staining was observed in the cytoplasm and nucleus of the cells in the blank, MEG3 OE and linc01105 KD groups, but its expression was restricted to the cytoplasm in the HCN3 KD group. Noxa staining was strong in the linc01105 KD group but weak in the MEG3 OE and HCN3 KD groups, and staining was observed in the cytoplasm and nucleus of the cells in all groups. Bid protein was strongly expressed in HCN KD and linc01105 KD cells but was weakly expressed in the cells overexpressing MEG3. Bid staining was observed in the cytoplasm and nucleus of the cells in all groups (Fig. 5).
Figure 5. Immunofluorescence provides information about protein expression and localization.
(A1–A4) Expression of HIF-1α protein in the blank, MEG3 OE, HCN3 KD and linc01105 KD groups. (B1–B4) Expression of Noxa protein in the blank, MEG3 OE, HCN3 KD and linc01105 KD groups. (C1–C4) Expression of Bid protein in the blank, MEG3 OE, HCN3 KD and linc01105 KD groups.
Correlation between lncRNA expression and neuroblastoma International Neuroblastoma Staging System (INSS) stage
The expression of the three lncRNAs (MEG3, HCN3, and linc01105) was negatively correlated with INSS stage to a moderate extent. The Spearman correlation coefficient values were: MEG3, −0.48; HCN3, −0.58; and linc01105, −0.55.
Discussion
Neuroblastoma is the most common type of solid tumor in infants, accounting for 8–10% of all childhood cancers and approximately 15% of all cancer-related deaths in children1,17. The etiology of neuroblastoma is heterogeneous and includes amplification of the MYCN or anaplastic lymphoma kinase oncogenes and the allelic loss of chromosomes 1p, 3p, or 11q. These etiologies are associated with varying clinical outcomes18 and affect chromosomes or protein-coding genes. The dysregulation of lncRNA expression has been linked to various diseases19,20,21,22,23. However, studies of the roles of lncRNAs in neuroblastoma are lacking. In the present study, we identified 4,802 lncRNAs and 5,130 mRNAs that were differentially expressed in neuroblastoma tissues. ENST00000523785, ENST00000393515, uc010fhf.3, ENST00000513011, and uc003wtj.3 were the most significantly upregulated lncRNAs, whereas NR_051975, ENST00000570869, ENST00000555539, uc001lva.4, and ENST00000570945 were the most significantly downregulated.
The p53 signaling pathway has been implicated in the development of various carcinomas, and recent studies suggest that lncRNAs regulate gene expression in some tumors by modulating p53 pathway members. For example, metastasis-associated lung adenocarcinoma transcript 1 has been linked to lung, breast, colon and liver cancer24,25,26,27,28. Here, a CNC network analysis implicated the lncRNAs HCN3, linc01105, and MEG3 in p53 signaling. HCN3 encodes a type of pacemaker channel in spontaneously contractile cells and plays an important role in regulating cell excitability29,30. MEG3 is a maternally expressed imprinted gene that inhibits the growth of ectopic human cancer cells and is considered to be a tumor suppressor31. For example, the downregulation of MEG3 was found to be associated with poor overall survival in osteosarcoma32. linc01105 is an RNA gene of undetermined function. Hypoxia is considered to be an inducer of p53 signaling33, and evidence suggests that increased p53 expression under hypoxic conditions is dependent upon HIF-1α34,35. In this study, we examined whether these three lncRNAs were involved in the pathogenesis of neuroblastoma and whether the underlying mechanisms involved the p53 and HIF-1α pathway.
We identified MEG3, HCN3 and linc01105 as lncRNAs that are relevant to neuroblastoma. MEG3 is expressed in many normal tissues, but its expression is lost in many primary human tumors36. HCN3 represents one of four HCN channel isoforms (HCN1-HCN4)37. Little is known about the role of linc01105. In this study, the overexpression of MEG3 inhibited the proliferation of BE(2)-C neuroblastoma cells but promoted their apoptosis. Knockdown of HCN3 had no effect on cell proliferation but promoted cell apoptosis. Knockdown of linc01105 expression promoted BE(2)-C cell proliferation and apoptosis. Altered lncRNA expression influenced the levels of HIF-1α mRNA and protein in neuroblastoma cells. MEG3 OE had no effect on the levels of HIF-1α mRNA or protein. Knockdown of HCN3 and linc01105 had no effect on the levels of HIF-1α mRNA. HCN3 KD resulted in the downregulation of HIF-1α protein expression, but linc01105 KD led to elevated HIF-1α protein expression. This result suggests that MEG3, HCN3 and linc01105 regulate the expression of HIF-1α by post-transcriptional mechanisms. Evidence suggests that HIF-1α is expressed in several tumor types as a result of hypoxia and the expression of certain oncogenes38,39, and it is involved in cell proliferation40,41. Our study suggests that linc01105 influences proliferation by modulating HIF-1α protein expression. However, it is unknown whether MEG3 and HCN3 control cell proliferation through the regulation of HIF-1α expression.
Noxa is a member of the BH3-only subset of the pro-apoptotic BCL-2 protein family. BCL-2 proteins are important regulators of the intrinsic apoptotic pathway42,43. BH3-interacting death domain agonist (BID) also belongs to the BH3-only subset of BCL-2 proteins, and it plays a pro-apoptotic role in cell stress responses44,45,46,47,48,49. KD of HCN3 increased Bid expression and levels of cell apoptosis. In addition, linc01105 KD increased the protein-level expression of Noxa and Bid and enhanced cell apoptosis. Noxa and Bid are both apoptosis-associated proteins in the p53 pathway, and our findings suggest that HCN3 and linc01105 regulate apoptosis via the Noxa and Bid proteins. MEG3 OE reduced the expression of Noxa and Bid but increased the levels of apoptosis. The abnormal expression of MEG3 was shown to decrease cell growth and promote apoptosis50; therefore, MEG3 may directly promote apoptosis in neuroblastoma cells. Our study also revealed the relationship between Bid and Noxa expression and cell proliferation. Reduced expressions of Bid and Noxa inhibited cell proliferation, whereas increased and decreased Bid and Noxa expressions, respectively, had no effect on cell proliferation. However, given that the increases in Bid and Noxa expressions promoted cell proliferation, it is possible that Bid and Noxa influence cell proliferation cooperatively.
The expressions of MEG3, HCN3 and linc01105 were all correlated negatively with the INSS neuroblastoma stage; low and high expressions of these genes were associated with early and late tumor stages, respectively. These correlations were consistent with the regulatory roles of these lncRNAs in neuroblastoma cells. These findings suggest that MEG3, HCN3 and linc01105 may predict the prognosis of neuroblastoma.
In summary, this study has identified lncRNAs and mRNAs that are differentially expressed in neuroblastoma and that may serve as biomarkers for prognosis or disease progression. HCN3, linc01105, and MEG3 are likely to be important in the biological behaviors and progression (stage) of neuroblastoma and may act via a mechanism involving HIF-1α, Noxa and Bid, and thus the p53 pathway.
Materials and Methods
Clinical samples
Tumor and para-tumor tissue samples (n = 6) were collected from patients who underwent surgery between December 2011 and December 2013; these samples were and stored at −80 °C. Tumor tissue specimens were confirmed as neuroblastomas by a pathologist. Para-tumor tissue samples consisted of adrenal gland tissue, and the absence of tumor cells was confirmed by microscopy. The age of the patients from whom the tissue samples were collected were 2 months, 10 months, 7 years, 3 years, 6 years and 3 months, respectively. We confirm that all of the methods were performed in accordance with relevant guidelines, and all of the experimental protocols were approved by the Ethics Committee of the Children’s Hospital of Fudan University. We also confirm that informed consent was obtained from the parents or legal guardians of the subjects.
RNA isolation
The tissue samples were washed three times with cold phosphate-buffered saline and dissolved in TRIzol reagent (Invitrogen, USA). Total RNA was isolated using the RNeasy Mini kit (Qiagen, Germany) according to the manufacturer’s protocol, which included DNase digestion. The purity and concentration of the RNA samples were determined by measuring absorbance with an ND-10009 NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). Samples with an OD 260/280 nm close to 2.0 and an OD 260/230 nm >1.8 were deemed sufficiently pure for use in the experiments.
RNA labeling and array hybridization
To identify differentially expressed mRNAs, RNA samples were amplified and labeled using the Quick Amp Labeling kit (Agilent Technologies, USA) and hybridized onto whole genome microarrays (Agilent Gene Expression Hybridization kit). The Human LncRNA Microarray v3.0 platform (Agilent Technologies) was used to identify differentially expressed lncRNAs. After hybridization and washing, the arrays were scanned (G2565BA; Agilent Technologies) according to the manufacturer’s recommendations.
Data analysis
Agilent Feature Extraction software was used to analyze the acquired images. Quantile normalization and raw data processing were conducted using GeneSpring GX V12.0 software (Agilent Technologies). LncRNAs and mRNAs with “present” or “marginal” (“all targets value”) flags were selected for further analysis. The differential expression of lncRNAs and mRNAs between tumor and para-tumor samples was determined by fold-change (>2-fold) filtering.
LncRNA classification and pathway analysis
Functional analyses were performed to identify to which Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways the differentially expressed genes belonged. Pathways with a P-value of ≤ 0.05 were considered significant.
Coding/non-coding gene co-expression (CNC) network analysis
A subset of the data was selected according to the coding list. The Pearson correlation coefficients between coding and non-coding RNAs were determined, with an r-value of ≥0.985 indicating a relationship. A CNC network was generated based on the results using Cytoscape (v2.8.1).
Evaluation of lncRNA expression by quantitative real-time (qRT)-PCR
The expression of lncRNAs identified by microarray analysis was confirmed by qRT-PCR. cDNA was synthesized from total RNA using the PrimeScript reverse transcription reagent kit with a genomic DNA Eraser (Takara, Japan). Primers were designed to amplify the three lncRNAs, and amplification was performed using a LightCycler 480 instrument (Roche Applied Science, Switzerland). The 10-μl PCR reactions included 1 μl of cDNA template and 5 μl of SYBR Premix Ex Taq II (Takara). The reaction conditions were as follows: 95 °C for 1 min, followed by 40 cycles of 95 °C for 5 s, 60 °C for 10 s, and 72 °C for 15 s. The reactions were run in triplicate. Threshold cycle (Ct) values were determined using default threshold settings, and the mean Ct was determined from duplicate PCR reactions. LncRNA expression levels were measured and normalized to the expression of human 18s rRNA using the 2−ΔΔCt method.
Functional analyses of target genes
Gene overexpression and knockdown
Lentiviral overexpression and knockdown vectors were constructed for lncRNAs of interest. The BE(2)-C neuroblastoma cell line was purchased from the American Type Culture Collection (ATCC) and cultured in complete Dulbecco’s modified Eagle medium (DMEM) (Gibical, Australia) supplemented with 10% fetal bovine serum (Gibical) in a 37 °C incubator maintained at 5% CO2. BE(2)-C cells were infected with 100 μl of lentiviral vectors in 900 μl of DMEM plus 5 μg/ml polybrene. The cells were incubated with viruses for 12 h at 37 °C and 5% CO2.
Cell proliferation
Three groups were tested using the CCK8 kit (Dojindo, Japan): (1) the overexpression or knockdown group, (2) the negative control group and (3) the blank group. BE(2)-C neuroblastoma cells were seeded in 96-well plates (3,000 cells/well), and the OD values of cells in the three groups were measured at two different time points (24 and 48 h).
Apoptosis detection
The target genes were either overexpressed (OE) or knocked down (KD) in BE(2)-C cells, and apoptosis levels relative to the control cells were quantified using the Annexin V-FITC/PI Apoptosis Detection kit (Dojindo, Japan). Apoptosis was analyzed using a FACScan flow cytometer.
RNA extraction and qRT-PCR
RNA was extracted from BE(2)-C cells using the RNeasy kit (Omega, USA). cDNA was produced using the PrimeScript Reverse Transcriptase reagent kit with genomic DNA Eraser (Perfect Real-Time) (Takara). qRT-PCR was performed using an Applied Biosystems 7900 HT qPCR machine and SYBR Premix Ex Taq (Tli RNaseH Plus) (Takara). The relative abundance of cDNA was calculated using the relative standard curve method.
Western blot analysis
The cells were lysed in radio immunoprecipitation assay (RIPA) buffer for 30 min, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were then blotted with rabbit anti-HIF-1α (Abcam, 1:1000), rabbit anti-noxa (Abcam, 1:1000), rabbit anti-bid (Abcam, 1:500) or mouse monoclonal anti-GAPDH (Abcam, 1:8000) primary antibodies in 5% non-fat dry milk, followed by HRP-conjugated anti-rabbit (Abcam, 1:1000) or anti-mouse secondary antibodies (Abcam, 1:1000).
Immunofluorescence
BE(2)-C cells were seeded in 48-well plates and labeled with the following primary antibodies: anti-HIF-1α (Abcam, 1:500), anti-noxa (Abcam, 1:500) and anti-bid (Abcam, 1:500). Primary antibodies were labeled using fluorescein isothiocyanate (FITC)-conjugated anti-rabbit secondary antibodies (Jackson, 1:1000). The cell nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime, China).
Correlation between lncRNA expression and the INSS stage
Total RNA was extracted from the neuroblastoma tissue of 32 patients, and the expression of target lncRNAs was quantified by qRT-PCR. The INSS stage of each neuroblastoma tissue was confirmed by clinical diagnosis and therapy. Spearman correlation analyses were used to assess correlations between the expressions of lncRNAs of interest and the INSS stage.
Statistics
A t-test was used to analyze the results of the cell proliferation assays and gene and protein expression data. The χ2 test was used analyze the results of the apoptosis assays. Spearman correlation analyses were used to analyze the correlations between lncRNA expression levels and the INSS stage.
Additional Information
How to cite this article: Tang, W. et al. MEG3, HCN3 and linc01105 influence the proliferation and apoptosis of neuroblastoma cells via the HIF-1α and p53 pathways. Sci. Rep. 6, 36268; doi: 10.1038/srep36268 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
I would like to thank the National Natural Science Foundation of China (no. 81572324), National Key Clinical Specialty Construction Programs of China (2014-2016), Shanghai ‘Non key-in-key discipline’ Clinical medical centers (2014–2016), Shanghai Hospital Development Center (SHDC12014106), and The Science Foundation of Shanghai (15ZR1404200).
Footnotes
Author Contributions W.T. participated in the development of the methodology; the acquisition, analysis, and interpretation of data; and writing the manuscript. K.D. participated in study conception and design, the development of methodology, review and revision of the manuscript, and study supervision. S.Z. participated in study supervision. K.L. and R.D. provided materials.
References
- Park J. R., Eggert A. & Caron H. Neuroblastoma: biology, prognosis, and treatment. Hematol Oncol Clin NORTH Am. 24, 65–86 (2010). [DOI] [PubMed] [Google Scholar]
- Hogarty M. D. et al. Analysis of genomic imprinting at 1p35-36 in neurolastoma. Med Pediatr Oncol. Jan. 36(1), 52–55 (2001). [DOI] [PubMed] [Google Scholar]
- Pajtler K. W. et al. Neuroblastoma in dialog with its stroma:NTRK1 is a regulator of cellular cross-talk with Schwann cells. Oncotarget 5(22), 11180–11192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachane A. N. & Harrison P. M. Mining mammalian transcript data for functional Long non-coding RNAs. PloS One 5(4), e10316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E. & Mattick J. S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 10, 155–159 (2009). [DOI] [PubMed] [Google Scholar]
- Guangfu J. et al. Human polymorphisms at long non-coding RNAs(lncRNAs) and association with prostate cancer risk. Carcinogenesis. 32(11), 1655–1659 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M. et al. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Sci Rep. 6, 31038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Long non-coding RNA TUG1 promotes colorectal cancer matastasis via EMT pathway. Oncotarget. 32, 51713–51719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H. et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 10, 18632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Øromua et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 143(1), 46–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi I. A., Mattick J. S. & Mehler M. F. Long non-coding RNAs in nervous system function and disease. Brain Res. 1338, 20–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparago A. et al. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat Genet. 36, 958–960 (2004). [DOI] [PubMed] [Google Scholar]
- Johnson R. et al. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA 15(1), 85–96 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- González W. & Bautista R. E. Seizures and EEG findings in an adult patient with DiGeorge syndrome: a case report and review of the literature. Seizure. 18(9), 648–651 (2009). [DOI] [PubMed] [Google Scholar]
- Willingham A. T. et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309(5740), 1570–1573 (2005). [DOI] [PubMed] [Google Scholar]
- Triche T. J. Neuroblastoma–biology confronts nosology. Arch Pathol Lab Med. 110, 994–996 (1986). [PubMed] [Google Scholar]
- Cheung N. & Dyer M. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 13(6), 397–411 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert L. S. et al. MCT-1 protein interacts with the cap complex and modulates messenger RNA translational profiles. Cancer Res. 66(18), 8994–9001 (2006). [DOI] [PubMed] [Google Scholar]
- Chen L. et al. lncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo. OncoTarget and Therapy 9, 4075–4087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wang D. L. & Pang Q. Long noncoding RNA SPRY4-IT1 is a prognostic factor for poor overall survival and has an oncogenic role in glioma. Eur Rev Med Pharmacol. Sci. 20(14), 3035–3039 (2016). [PubMed] [Google Scholar]
- Gupta R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W. et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 451, 202–206 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P. et al. MALAT-1, a novel noncoding RNA, and thymosin beta 4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 22, 8031–8041 (2003). [DOI] [PubMed] [Google Scholar]
- Yamada K. et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 97, 106–112 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. et al. Alarge noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 26, 851–858 (2007). [DOI] [PubMed] [Google Scholar]
- Guffanti A. et al. A trascriptional sketch of a primary huan breast cancer by 454 deep sequencing. BMC Genomics 10, 163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-327 in liver cancer. Nucleic Acid Res. 38, 5366–5383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFrancesco D. The contribution of the ‘pacemaker’ current (if) to generation of spontaneous activity in rabbit sino-atrial node myocytes. J Physiol. 434, 23–40 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape H. C. Queer current and pacemaker: the hyperpolarizationactivated cation current in neurons. Annu Rev Physiol. 58, 299–327 (1996). [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhang X. & Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 48, R45–R53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z. Z. et al. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of asteosarcoma. Int J Clin Exp Pathol. 8(11), 15138–15142 (2015). [PMC free article] [PubMed] [Google Scholar]
- Hammond E. M. & Giaccia A. J. The role of p53 in hypoxia-induced apoptosis. Biochem Biophys Res Commun. 331, 718–725 (2005). [DOI] [PubMed] [Google Scholar]
- An W. G. et al. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 392, 405–408 (1998). [DOI] [PubMed] [Google Scholar]
- Carmeliet P. et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394, 485–490 (1998). [DOI] [PubMed] [Google Scholar]
- Zhong H. et al. Overexpression of hypoxia-inducible factor 1 alpha in common human cancers and their metastases. Cancer Res. 59, 5830–5835 (1999). [PubMed] [Google Scholar]
- Koong A. C. et al. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 48, 919–922 (2000). [DOI] [PubMed] [Google Scholar]
- Benetatos L., Vartholomatos G., Eleftheria H. & Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer 129(4), 773–779 (2011). [DOI] [PubMed] [Google Scholar]
- Fried H. U., Kaupp U. B. & Muller F. Benjamin Kaupp.Hyperpolarization-activated and cyclic nucleotide-gated channels are differentially expressed in juxtaglomerular cells in the olfactory bulb of mice. Cell Tissue Res. 339, 463–479 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. Expression and correlation of hypoxia-inducible factor-1alpha, vascular endothelial growth factor and microvessel density in experimental rat hepatocarcinogenesis. J Int Med Res. 37, 417–425 (2009). [DOI] [PubMed] [Google Scholar]
- Sasabe E., Tatemoto Y., Yamanoto T. & Osaki T. Mechanism of HIF-1alpha-dependent suppression of hypoxia-induced apoptosis in squamous cell carcinoma cells. Cancer Sci. 96, 394–402 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J. & Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 9, 47–59 (2008). [DOI] [PubMed] [Google Scholar]
- Chen L. et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 7, 393–403 (2005). [DOI] [PubMed] [Google Scholar]
- Bonzon C., Hayes L. B., Pagliari L. D. & Newmeyer D. Caspase-2-induced apoptosis requires bid cleavage: a physiological role for bid in heat shock-induced death. Mol. Biol. Cell. 17, 2150–2157 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhu H., Xu C. J. & Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491–501 (1998). [DOI] [PubMed] [Google Scholar]
- Luo X. et al. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 94, 481–490 (1998). [DOI] [PubMed] [Google Scholar]
- Maas C., Vries E. D., Tait S. W. G. & Borst J. Bid can mediate a pro-apoptotic response to etoposide and ionizing radiation without cleavage in its unstructured loop and in the absence of p53. Oncogene 30, 3636–3647 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesnila N. et al. BID mediates neuronal cell death after oxygen/glucose deprivation and focal cerebral ischemia. Proc. Natl. Acad. Sci. USA 98, 15318–15323 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton J. P. et al. Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol. Cell. Biol. 28, 3943–3951 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. H. et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 13, 461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.