Abstract
Background
Oxytocin has been proposed to mediate amygdala dysfunction associated with altered emotion processing in schizophrenia, but the contribution of oxytocin pathway genes is yet to be investigated.
Aims
To identify potential different contributions of three oxytocin receptor polymorphisms (rs53576, rs237902 and rs2254298) between patients with schizophrenia spectrum disorders (SCZ), affective spectrum disorders (AD) and healthy controls (HC).
Method
In a total of 346 participants (104 with SCZ, 100 with AD, and 142 HC) underwent genotyping and functional magnetic resonance imaging (fMRI) during an emotional faces matching paradigm. Genetic association analyses were performed to test the possible effects on task-induced BOLD amygdala response to fearful/angry faces.
Results
In participants with SCZ, the rs237902 G allele was associated with low amygdala activation (left hemisphere: b=−4.99, Bonferroni corrected P=0.04) and interaction analyses showed that this association was disorder specific (left hemisphere: Bonferroni corrected P=0.003; right hemisphere: Bonferroni corrected P=0.03). There were no associations between oxytocin polymorphisms and amygdala activation in the total sample, among AD patients or HC.
Conclusions
Rs237902 was associated with amygdala activation in response to fearful/angry faces only in patients with SCZ. Our findings indicate that the endogenous oxytocin system could serve as a contributing factor in biological underpinnings of emotion processing and that this contribution is disorder specific.
Declaration of interest
O.A.A. received speaker’s honoraria from GSK, Otsuka, Lundbeck.
Copyright and usage
© The Royal College of Psychiatrists 2016. This is an open access article distributed under the terms of the Creative Commons Non-Commercial, No Derivatives (CC BY-NC-ND) licence.
Social cognitive deficits, including deficits in emotion processing, precede onset of illness, are stable impairments over time and important predictors for functional disabilities in patients with schizophrenia.1,2 Add-on trials with oxytocin administered intranasally in this patient group suggest a treatment effect on these deficits as well as a modulating effect on amygdala response to facial emotion processing.3,4 Trials from healthy subjects show an effect on amygdala activation in a possible sexual dimorphic pattern, but the diverse direction of effects seen are not yet established.5–7 The results from oxytocin trials are inconsistent in general,3,4,8 and this inconsistency has been attributed to individual differences, including genetically based differences in the oxytocin system,9,10 and a poor understanding of drug targets and mechanisms.11 The field is also troubled by small sample sizes with an average sample size in schizophrenia and oxytocin imaging of 16 and 32, respectively.12–15 The endogenous oxytocin system is less explored, and the contribution of oxytocin pathway genes to amygdala activation has yet to be investigated in this patient group. Rs53576, rs2254298 and rs237902 are single nucleotide polymorphisms (SNPs) in the gene coding for the production of the oxytocin receptor (OXTR) and have previously been associated with disease characteristics in patients with schizophrenia, that is, general psychopathology (rs53576 and rs2254298) and negative symptoms (rs237902 and rs53576) as measured by the Positive and Negative Syndrome Scale (PANSS) and empathic concern (rs2254298).16–18 The results have, however, also here been inconsistent.10,18,19 Thus, to get a better understanding of the contribution of oxytocin pathway gene polymorphisms to the amygdala response to fearful/angry faces, we used a functional magnetic resonance imaging (fMRI) paradigm for matching of emotional faces in the largest oxytocin clinical sample size to date,20 consisting of patients with schizophrenia spectrum (SCZ) and affective spectrum disorders (AD) as well as a community representative sample of healthy participants (HC), specifically targeting the SNPs: rs53576, rs2254298 and rs237902. As emotion stimuli processing is particularly altered in patients with schizophrenia, we here hypothesised that the contribution of the three SNPs would be more pronounced in the SCZ group. Because of the reported sex differences in the response to exogenous oxytocin, we additionally investigated interaction effects between SNPs and sex in the case of significant associations between SNPs and amygdala response.
Method
Sample characteristics
Participants, all of Caucasian origin, were included in the Norwegian multicentre Thematically Organized Psychosis (TOP) research study recruiting patients from in- and out-patient clinics in the greater Oslo area in south-eastern Norway. None of the participants had a history of head injury, neurological disorder, autoimmune or infectious disorders or malignancies, or an IQ below 70. Patient assessments included clinical and physical examinations by a physician, neuropsychological testing by a psychologist, collection of blood samples for somatic screening and DNA analyses, and MRI. The Structured Clinical Interview for DSM-IV Axis 1 Disorders (SCID-1) was used for diagnostic purposes. The sample of 104 patients with SCZ included the following diagnoses: schizophrenia (n=62), schizophreniform disorder (n=6), schizoaffective disorder (n=12) and psychotic disorder not otherwise specified (NOS) (n=24). The AD group (n=100) consisted of patients with bipolar disorder I (BD I, n=50), bipolar disorder II (BD II, n=38), bipolar disorder NOS (BD NOS, n=6) and depressive psychosis (n=6) (Table 1). The healthy controls (HC) (n=142) were invited via stratified random selection from statistical records of persons from the same catchment area as the patient groups and excluded in case of the presence of severe mental disorders, including schizophrenia and AD in the participant and in first-degree relatives, or current illegal substance use.
Table 1. Demographic and clinical background of the study sample.
| SCZ n=104 | AD n=100 | HC n=142 | Total N=346 | Test statistic | ||
|---|---|---|---|---|---|---|
| F/χ² | P | |||||
| Gender (m/f) | 63/41 | 43/57 | 84/58 | 190/156 | χ²=8.1 | 0.02 |
| Age, years: mean (s.d.) | 31.5 (8.9) | 33.2 (11.2) | 33.4 (8.4) | 32.8 (9.5) | F=1.44 | 0.24a |
| Left amygdala activation (COPE): mean (s.d.) | 11.6 (13.7) | 12.3 (13.1) | 11.0 (12.9) | 11.6 (12.9) | χ²=0.73 | 0.69b |
| Right amygdala activation (COPE): mean (s.d.) | 12.1 (15.4) | 12.6 (13.6) | 12.3 (11.7) | 12.3 (13.4) | χ²=0.19 | 0.91b |
| Medicatedc N (%) | 91 (87.5) | 80 (80) | – | – | – | – |
| AP/LIT/AE/AD/HYPd (N) | 88/1/21/32/12 | 47/10/39/31/13 | – | – | – | – |
| Rs2254298 (maf, Ae) | 0.11 | 0.19 | 0.10 | 0.10 | – | – |
| Rs53576 (maf, A) | 0.29 | 0.28 | 0.32 | 0.30 | – | – |
| Rs237902 (maf, A) | 0.31 | 0.42 | 0.35 | 0.36 | – | – |
| Behavioural data | ||||||
| Response timef (ms): mean (s.d.) | 1192.8 (314.9) | 1197.6 (339) | 1069.9 (230.9) | 1141 (295.3) | χ²=13.6 | 0.001 |
| Accuracy ratef (%): mean (s.d.) | 98.7 (0.06) | 98.5 (0.09) | 99.2 (0.03) | 98.9 (0.06) | χ²=0.29 | 0.87 |
SCZ, schizophrenia spectrum disorders; AD, affective spectrum disorders; HC, healthy controls; COPE, contrast parameter estimates; maf, minor allele frequency.
Brown–Forsythe test on log-transformed age.
Non-parametric Kruskal–Wallis test.
Regular use of psychopharmacological medication.
Regular use of antipsychotics/lithium/anti epileptics/anti-depressives/hypnotics.
The minor allele frequency of the A allele.
Complete behavioural data (response time and accuracy rate) were available for 89 SCZ, 94 AD and 140 HC. For the remaining individuals (SCZ 15, AD 6, HC 2) an accuracy rate (i.e. combined rate for negative and shapes) was available and was used to confirm that the participants paid attention to the task (accuracy rate 96.2%, 96.3% and 93.5%, respectively).
The study received approval from the Norwegian Scientific-Ethical Committees and the Norwegian Data Protection Agency, and all participants provided written informed consent.
Genotyping
DNA was extracted from blood and rs237902 was genotyped using the Affymetrix Human SNP Array 6.0 (Affymetrix Inc., Santa Clara, CA, USA), as previously reported.21,22 All chips were subjected to the Birdseed-v2 algorithm developed by Affymetrix Inc. and Broad Institute23 and implemented in the software Affymetrix Power Tools (APT v1.10). Samples were excluded through quality control if they had low-yield (call rate below 95%), if they were duplicates of other samples, if they had a sex determined by X chromosome marker homozygosity different from their reported sex, or if they were calculated to have ancestry other than European.21 The markers did not show deviation from Hardy–Weinberg equilibrium (P<0.001), low-yield (<95%) or minor allele frequencies below 0.01. Following the above-mentioned quality control, rs53576 and rs2254298 were imputed with MaCH (http://www.sph.umich.edu/csg/abecasis/MACH/download/1000G-PhaseI-Interim.html) using the European samples in the Phase I release of the 1000 Genomes Project.
Experimental paradigm
A widely used and validated paradigm was employed to elicit amygdala activation.24–27 In this task, participants selected which of two stimuli (displayed at the bottom of the screen) matched a target stimulus (displayed at the top). In the faces-matching task, the images displayed were human faces expressing either anger or fear (‘negative faces’), or happiness (‘positive faces’). In the sensorimotor control task, geometrical shapes were displayed. The negative and positive faces were presented in two separate runs, each with four blocks of faces interleaved by five blocks of geometrical shapes. The order of the runs was counterbalanced among participants. Within blocks of negative faces, fearful and angry faces were intermixed. Each block of faces consisted of six emotion-specific face trios derived from a standard set of facial-affected pictures.28 Each trial (face/shape stimulus) was presented for 5.4 s with no inter-stimulus interval, for a total block length of 32.6 s. Each run lasted 294 s. The E-prime software (version 1.0 Psychology Software Tools, Inc, Pittsburgh, PA, USA) controlled the presentations of the stimuli using VisualSystem (NordicNeuroLab, Bergen, Norway). Measures of reaction times and task accuracy were recorded through MRI-compatible ResponseGrips (NordicNeuroLab, Bergen, Norway).
BOLD fMRI data acquisition
MRI was performed on a 1.5 T Siemens Magnetom Sonata scanner (Siemens Medical Solutions, Erlangen, Germany) supplied with a standard head coil. We collected 152 functional volumes per run using a T2*-weighted echo-planar imaging (EPI) sequence. Each volume consisted of 24 axial slices with a pixel size of 3 mm in the axial plane and a slice thickness of 4 mm with 1 mm gap between slices (TR=2040 ms, TE=50 ms, FA=90°, matrix 64×64, FOV 224 mm). The first seven volumes and the last one were discarded, leaving 144 volumes for analyses. Prior to fMRI, a sagittal T1-weighted 3D Magnetization Prepared Rapid Gradient Echo (MPRAGE) scan was collected (160 slices, TR=2730 ms, TE=3.93 ms, FA=7°, matrix 192×256, FOV 240 mm, voxel size 1.33×0.94×1 mm3), here used for co-registration purposes.
fMRI data analysis
The fMRI data went through an initial quality check procedure, to detect images with poor quality because of excessive head motion, slice dropout, radiofrequency (RF) artefacts or other noise. It was subsequently processed and analysed using FEAT (FMRI Expert Analysis Tool) version 6.0, part of FSL (fMRIB’s Software Library http://www.fmrib.ox.ac.uk/fsl).29 Conventional preprocessing included motion correction,30 non-brain removal,31 spatial smoothing with a 6-mm full-width half-maximum kernel32 and a high-pass filter with a 90-s window. A first-level general linear model (GLM) analysis was performed for each run, where the onset and duration of the on-blocks (positive faces and negative faces, respectively) was modelled with that of the off-blocks (geometrical shapes) as implicit baseline. The design matrix was filtered and convolved with a haemodynamic response function before the model fit. This analysis resulted in individual contrast parameter estimates (COPEs) reflecting the faces versus shapes contrast within runs. In the current study, we focused on participants’ amygdala responses to negative stimuli (the negative faces>shapes contrast), because of the implication of altered threat-related facial affect perception in psychotic disorders33,34 and since this contrast has been reported to reliably engage the amygdala.27,35,36 Amygdala regions of interest (ROIs) were defined in accordance with the probabilistic Harvard–Oxford subcortical atlas provided with FSL, and were thresholded at 25% probability. Mean BOLD signal changes (parameter estimates) across all ROI voxels were extracted from FSL and used in statistical analyses.
Statistics
Group analyses of amygdala activation and behavioural data
Potential differences in amygdala activation, response time and accuracy rate between subgroups (SCZ, AD, and HC) were examined. The distributions were not normal within all subgroups, hence Kruskal–Wallis tests were performed. Sex effects for amygdala activation were tested for in the total group with Mann–Whitney tests because of values without normal distribution across gender. The correlation between right and left amygdala activation was examined using bivariate correlation analyses (Spearman’s rho). These analyses were performed in SPSS version 22 (IBM, Armonk, NY).
Genetic associations analyses
The additive effects of allele dosage on right and left amygdala activation in the total group were investigated for rs237902 (genotyped SNP), as well as rs53576 and rs2254298 (imputed SNPs) with age, sex and diagnosis as covariates in linear regression models. In the follow-up analyses, the genetic association tests for all the three SNPs were repeated in the three subgroups separately, with age and sex as covariates (PLINK Version 1.07). In the case of significant genetic association with amygdala activation, interaction analyses between SNP and diagnosis as well as between SNP and sex in linear regression models were performed (R Version 3.2.2). All analyses were performed with and without outliers. The significance threshold was set to P=0.05/3=0.017 as a result of Bonferroni corrections for three SNPs. Linkage disequilibrium (LD) analyses across the three SNPs were performed (R Version 3.2.2).
Results
Group analyses of behavioural data and amygdala activation
We did not detect differences in amygdala activation (left hemisphere: χ²=0.73, P=0.69; right hemisphere: χ²=0.19, P=0.91) or accuracy rate (χ²=0.29, P=0.87) between diagnostic subgroups, but in response time (χ²=13.6, P=0.001) (Table 1). There were no main effects of sex on amygdala activation in the total sample (left hemisphere: z=−1.72, P=0.09), (right hemisphere: z=−1.17, P=0.24). COPE values from the right and left amygdala showed a strong correlation (rho=0.67, P<0.01).
Genetic associations analyses
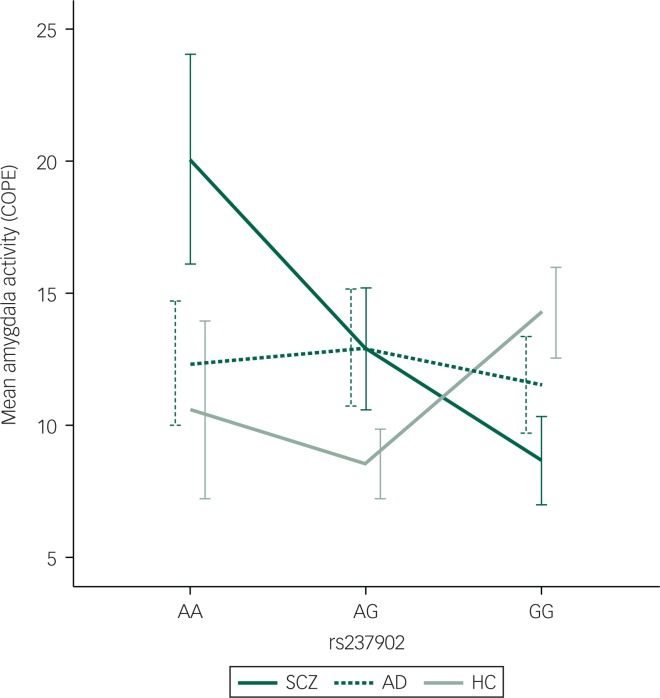
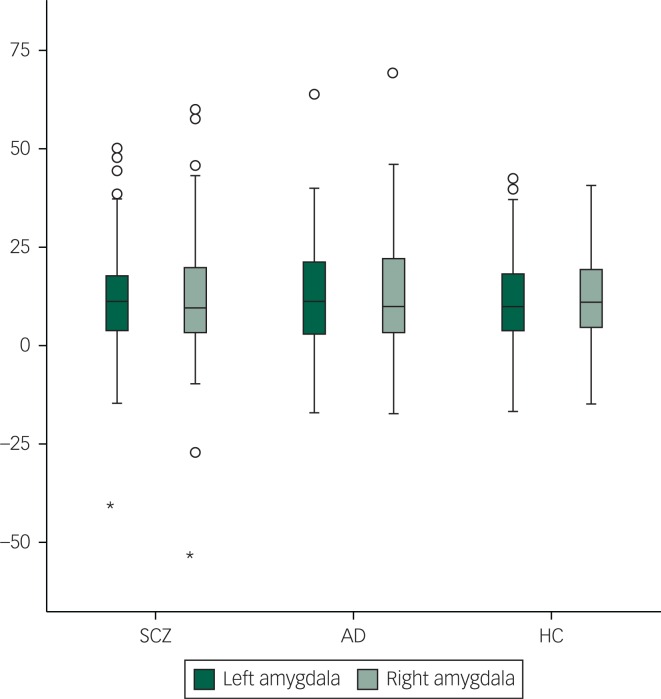
Among patients with SCZ, rs237902 was significantly associated with left amygdala activation, whereas the rs237902G allele displayed increased risk of low amygdala activation (P=0.014, Bonferroni corrected P=0.04) (Table 2, Fig. 1). An interaction analysis of rs237902 and sex was performed in this group, but no effect was detected. No significant associations between oxytocin polymorphisms and amygdala activation were found within patients with AD, HC or the total sample (Table 2). Interaction analyses showed a significant effect of having a diagnosis of SCZ compared with HC on the association between rs237902 and amygdala activation (left hemisphere: b=−8.15, P=0.001, Bonferroni corrected P=0.003; right hemisphere: b=−6.77, P=0.01, Bonferroni corrected P=0.03), while this was not the case for patients with AD (left hemisphere: b=−3.42, P=0.17; right hemisphere: b=−5.58, P=0.03, Bonferroni corrected P=0.09). Twelve individuals with outliers were identified after inspection of boxplots (Fig. 2), and repeating the analyses without them strengthened the results (Table 3). No meaningful LD was found across the three SNPs (r²<0.25).
Table 2. Association analyses between amygdala activity in the Negative Faces > Shapes contrast and oxytocin receptor polymorphisms.
| SNP | Amygdala hemisphere | SCZ n=104 | AD n=100 | HC n=142 | Total N=346 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Effect size | P | Effect size | P | Effect size | P | Effect size | P | ||
| Rs2254298 (G)a | Left | b=0.12 | 0.97 | b=−1.35 | 0.67 | b=1.92 | 0.40 | b=0.72 | 0.66 |
| Right | b=−1.11 | 0.77 | b=4.24 | 0.20 | b=0.45 | 0.84 | b=1.22 | 0.47 | |
| Rs53576 (G) | Left | b=4.93 | 0.026 | b=0.37 | 0.87 | b=−3.36 | 0.06 | b=0.35 | 0.77 |
| Right | b=3.28 | 0.19 | b=0.69 | 0.77 | b=−1.14 | 0.50 | b=0.77 | 0.54 | |
| Rs237902 (G) | Left | b=−4.99 | 0.014b | b=−0.25 | 0.90 | b=2.81 | 0.08 | b=−0.56 | 0.59 |
| Right | b=−3.84 | 0.097 | b=−2.66 | 0.18 | b=2.65 | 0.08 | b=−1.00 | 0.36 | |
SNP, single nucleotide polymorphism; SCZ, schizophrenia spectrum disorders; AD, affective spectrum disorders; HC, healthy controls.
Results for the G allele dosage are displayed.
Remains significant after Bonferroni corrections (3 independent tests).
Fig. 1. Differentiated mean amygdala left hemisphere activation during the Negative Faces > Shapes contrast for patients with SCZ (n=104) with the respective genotypes (n(AA)=11, n(AG)=43, n(GG)=50). Mean hits with one standard error are presented. COPE, contrast parameter estimates; SCZ, schizophrenia spectrum disorders; AD, affective spectrum disorders; HC, healthy controls.
Fig. 2. Differentiated mean amygdala activation (COPE) during the Negative Faces > Shapes contrast for patients with SCZ (n=104), patients with AD (n=100) and healthy subjects (n=142). No statistical significant differences between the groups were detected (left hemisphere: χ²=0.73, P=0.69, right hemisphere: χ²=0.19, P=0.91). COPE, contrast parameter estimates; SCZ, schizophrenia spectrum disorders; AD, affective spectrum disorders; HC, healthy controls.
Table 3. Association analyses performed after excluding outliers between amygdala activation in the Negative Faces > Shapes contrast and oxytocin receptor polymorphisms.
| SNP | Amygdala hemisphere | SCZ n=97 | AD n=99 | HC n=138 | Total N=334 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Effect size | P | Effect size | P | Effect size | P | Effect size | P | ||
| Rs2254298 (G)a | Left | b=0.52 | 0.85 | b=−1.94 | 0.51 | b=1.12 | 0.60 | b=0.37 | 0.80 |
| Right | b=0.66 | 0.81 | b=3.60 | 0.24 | b=−0.13 | 0.95 | b=1.29 | 0.38 | |
| Rs53576 (G)a | Left | b=2.45 | 0.16 | b=−0.46 | 0.83 | b=−2.60 | 0.12 | b=−0.39 | 0.71 |
| Right | b=0.36 | 0.84 | b=−0.24 | 0.91 | b=−0.51 | 0.76 | b=−0.21 | 0.84 | |
| Rs237902 (G)ac | Left | b=−4.56 | 0.004b | b=−0.15 | 0.93 | b=1.93 | 0.19 | b=−0.66 | 0.48 |
| Right | b=−3.69 | 0.03 | b=−2.54 | 0.16 | b=2.09 | 0.16 | b=−1.07 | 0.25 | |
SNP, single nucleotide polymorphism; SCZ, schizophrenia spectrum disorders; AD, affective spectrum disorders; HC, healthy controls.
Results for the G allele dosage are displayed.
Remains significant after Bonferroni corrections (3 independent tests).
Interaction analyses in the total sample showed a significant effect of having a diagnosis of SCZ compared to healthy controls on the association between rs237902 and amygdala activation (left hemisphere; b=−6.95, P=0.002, Bonferroni corrected P=0.006, right hemisphere; b=−6.05 , P=0.009, Bonferroni corrected P=0.03), while this was not the case for patients with AD (left hemisphere; b=−2.46, P=0.26, right hemisphere; b=−4.87, P=0.03, Bonferroni corrected P=0.09).
Discussion
In this study, we show attenuated amygdala activation in response to emotionally negative faces in rs237902G allele carriers among patients with schizophrenia spectrum disorders (SCZ), while not in patients with affective disorders or healthy control participants. Rs237902 is an OXTR synonymous variant located at exon 3 (https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19& lastVirtModeType=default& lastVirtModeExtraState=& virtModeType=default& virtMode=0& nonVirtPosition=& position=chr3%3A8808934%2D8809434& hgsid=480554839_Izi7M39PmzcwlaTbAUfn3La2J6O0), thus with potential influence on protein expression, conformation and function, and relative potential functional and clinical consequences.37 Montag and colleagues found rs237902G to be associated with more negative symptoms in schizophrenia,17 a clinical characteristic found to be correlated with emotion processing and face recognition in a meta-analysis.38 Thus, the main result of the current study is in line with this previous finding. Rs237902 could also serve as a proxy for rs237915 (LD: r²=0.82), the single SNP Loth and colleagues found to be associated with activation in the ventral striatum in response to threat perception in 1445 healthy adolescents, investigating 23 tag SNPs in the OXTR.39 The nucleus accumbens in the ventral striatum plays an important role in the mesocorticolimbic system and has been hypothesised to interplay with the amygdala via dopaminergic–oxytocinergic mechanisms.5 Loth and colleagues did not find any associations between OXTR SNPs and the amygdala, similar to our findings in healthy subjects in the present study. Combined, these findings imply that oxytocin plays an important role in emotion processing and provide additional evidence for alterations in the endogenous oxytocin system in patients with SCZ.40
The influence of rs53576 and rs2254298, two intronic polymorphisms, has been extensively studied across social features, psychiatric symptoms and ethnic groups; however, the results are mixed in spite of large meta-analyses.19,41,42 Tost and colleagues showed associations between rs53576 and rs2254298 and amygdala activation in separate studies in 228 healthy individuals using the same fMRI paradigm as the present study,43,44 findings neither we nor Loth and colleagues were able to replicate. While Loth and colleagues suggest that this may be because of their use of a different emotion processing paradigm and inclusion of adolescents instead of adults, our lack of replication might be because of a smaller sample of healthy individuals and the use of a clinical sample that may not show associations to the same OXTR polymorphisms as healthy individuals.
Another possible explanation for ambiguous genetic associations regarding oxytocin pathway genes is potential interaction effects with sex.45 The expression of central oxytocin is steroid-dependent,46 the central regulation differs between men and women,47 and men and women respond differently to exogenous oxytocin.5 Additionally, several brain imaging studies investigating oxytocin pathway genes in healthy individuals have detected sex-specific effects.44,48,49 However, we were not able to detect any interaction effects between sex and rs237902 in patients with SCZ, and all genetic association analyses included sex as a covariate. Hence, our genetic association results presented are not because of sex differences, and the disorder-specific association with rs237902 did not differ between men and women with SCZ.
As no oxytocin-related polymorphisms have been detected as risk variants for SCZ in large genome-wide association studies (http://www.broadinstitute.org/mpg/ricopili/), we chose a hypothesis-based approach for SNP selection. Rs53576, rs2254298 and rs237902 are the only SNPs previously found associated with disease characteristics in Caucasian schizophrenia samples; 10,16–18 hence, we were interested in these polymorphisms in particular. However, only a few studies have focused on the oxytocin pathway genes in patients with schizophrenia, and the possibility of publication bias must be taken into account. The clinical consequences and functions of these variants are also as yet unknown, and their influence on brain function should be interpreted with caution.
To conclude, we found a disorder-specific contribution of rs237902 to amygdala activation during emotional face processing in patients with SCZ. This is, to our knowledge, the first fMRI study investigating oxytocin pathway polymorphisms in this patient group. Our findings suggest that the endogenous oxytocin system contributes to biological underpinnings of emotion processing, that this contribution differs between patients with SCZ and HC and that exonic regions in OXTR are of interest for future research. Future research should investigate additional oxytocin pathway polymorphisms as well as involving other brain circuits, to better understand the biological mechanisms underlying emotion processing. We encourage replication of our findings in further imaging studies and meta-analyses of oxytocin pathway genes in this patient group.
Acknowledgements
We thank the patients and healthy controls for participating in the study, and the TOP study group members for contributing with data collection. We also thank Eivind Bakken, Monica Aas and Christian Melbø-Jørgensen for administrative and statistical assistance.
Funding
This work was supported by the Kristian Gerhard Jebsen Foundation, the Research Council of Norway (grant numbers 181831, 147787/320, 167153/V50, 204966/F20) and the Regional Health Authority for South-Eastern Norway Health Authority (grant numbers 2014–102, 2014–097, 2010–074, 2006–258). The funders had no role in study design, data collection, analysis and interpretation, writing of the report or the decision to submit the paper for publication.
References
- 1.Corcoran CM, Keilp JG, Kayser J, Klim C, Butler PD, Bruder GE, et al. Emotion recognition deficits as predictors of transition in individuals at clinical high risk for schizophrenia: a neurodevelopmental perspective. Psychol Med 2015; 45: 2959–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green MF, Horan WP, Lee J. Social cognition in schizophrenia. Nat Rev Neurosci 2015; 16: 620–31. [DOI] [PubMed] [Google Scholar]
- 3.Shin NY, Park HY, Jung WH, Park JW, Yun JY, Jang JH, et al. Effects of oxytocin on neural response to facial expressions in patients with schizophrenia. Neuropsychopharmacology 2015; 40: 1919–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feifel D, Shilling PD, MacDonald K. A review of oxytocin’s effects on the positive, negative, and cognitive domains of schizophrenia. Biol Psychiatry 2016; 79: 222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shamay-Tsoory SG, Abu-Akel A. The social salience hypothesis of oxytocin. Biol Psychiatry 2016; 79: 194–202. [DOI] [PubMed] [Google Scholar]
- 6.Rilling JK, DeMarco AC, Hackett PD, Chen X, Gautam P, Stair S, et al. Sex differences in the neural and behavioral response to intranasal oxytocin and vasopressin during human social interaction. Psychoneuroendocrinology 2014; 39: 237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domes G, Lischke A, Berger C, Grossmann A, Hauenstein K, Heinrichs M, et al. Effects of intranasal oxytocin on emotional face processing in women. Psychoneuroendocrinology 2010; 35: 83–93. [DOI] [PubMed] [Google Scholar]
- 8.Heringa SM, Begemann MJ, Goverde AJ, Sommer IE. Sex hormones and oxytocin augmentation strategies in schizophrenia: a quantitative review. Schizophr Res 2015; 168: 603–13. [DOI] [PubMed] [Google Scholar]
- 9.Bartz JA, Zaki J, Bolger N, Ochsner KN. Social effects of oxytocin in humans: context and person matter. Trends Cogn Sci 2011; 15: 301–9. [DOI] [PubMed] [Google Scholar]
- 10.Bartholomeusz CF, Ganella EP, Labuschagne I, Bousman C, Pantelis C. Effects of oxytocin and genetic variants on brain and behaviour: implications for treatment in schizophrenia. Schizophr Res 2015; 168: 614–27. [DOI] [PubMed] [Google Scholar]
- 11.Leng G, Ludwig M. Intranasal oxytocin: myths and delusions. Biol Psychiatry 2016; 79: 243–50. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Chan RC, McAlonan GM, Gong QY. Facial emotion processing in schizophrenia: a meta-analysis of functional neuroimaging data. Schizophr Bull 2010; 36: 1029–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anticevic A, Van Snellenberg JX, Cohen RE, Repovs G, Dowd EC, Barch DM. Amygdala recruitment in schizophrenia in response to aversive emotional material: a meta-analysis of neuroimaging studies. Schizophr Bull 2012; 38: 608–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor SF, Kang J, Brege IS, Tso IF, Hosanagar A, Johnson TD. Meta-analysis of functional neuroimaging studies of emotion perception and experience in schizophrenia. Biol Psychiatry 2012; 71: 136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bethlehem RA, van Honk JF, Auyeung B, Baron-Cohen S. Oxytocin, brain physiology, and functional connectivity: a review of intranasal oxytocin fMRI studies. Psychoneuroendocrinology 2013; 38: 962–74. [DOI] [PubMed] [Google Scholar]
- 16.Montag C, Brockmann EM, Lehmann A, Muller DJ, Rujescu D, Gallinat J. Association between oxytocin receptor gene polymorphisms and self-rated ‘empathic concern’ in schizophrenia. PLoS One 2012; 7: e51882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montag C, Brockmann EM, Bayerl M, Rujescu D, Muller DJ, Gallinat J. Oxytocin and oxytocin receptor gene polymorphisms and risk for schizophrenia: a case-control study. World J Biol Psychiatry 2013; 14: 500–8. [DOI] [PubMed] [Google Scholar]
- 18.Haram M, Tesli M, Bettella F, Djurovic S, Andreassen OA, Melle I. Association between genetic variation in the oxytocin receptor gene and emotional withdrawal, but not between oxytocin pathway genes and diagnosis in psychotic disorders. Front Hum Neurosci 2015; 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakermans-Kranenburg MJ, van Ijzendoorn MH. A sociability gene? Meta-analysis of oxytocin receptor genotype effects in humans. Psychiatr Genet 2014; 24: 45–51. [DOI] [PubMed] [Google Scholar]
- 20.Shilling PD, Feifel D. Potential of oxytocin in the treatment of schizophrenia. CNS Drugs 2016; 30: 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Athanasiu L, Mattingsdal M, Kahler AK, Brown A, Gustafsson O, Agartz I, et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatric Res 2010; 44: 748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djurovic S, Gustafsson O, Mattingsdal M, Athanasiu L, Bjella T, Tesli M, et al. A genome-wide association study of bipolar disorder in Norwegian individuals, followed by replication in Icelandic sample. J Affect Disord 2010; 126: 312–6. [DOI] [PubMed] [Google Scholar]
- 23.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008; 40: 1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carre JM, Fisher PM, Manuck SB, Hariri AR. Interaction between trait anxiety and trait anger predict amygdala reactivity to angry facial expressions in men but not women. Soc Cogn Affect Neurosci 2012; 7: 213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science 2002; 297: 400–3. [DOI] [PubMed] [Google Scholar]
- 26.Ousdal OT, Anand Brown A, Jensen J, Nakstad PH, Melle I, Agartz I, et al. Associations between variants near a monoaminergic pathways gene (PHOX2B) and amygdala reactivity: a genome-wide functional imaging study. Twin Res Hum Genet 2012; 15: 273–85. [DOI] [PubMed] [Google Scholar]
- 27.Tesli M, Kauppi K, Bettella F, Brandt CL, Kaufmann T, Espeseth T, et al. Altered brain activation during emotional face processing in relation to both diagnosis and polygenic risk of bipolar disorder. PLoS One 2015; 10: e0134202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tottenham N, Tanaka JW, Leon AC, McCarry T, Nurse M, Hare TA, et al. The NimStim set of facial expressions: judgments from untrained research participants. Psychiatry Res 2009; 168: 242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004; 23 (Suppl 1): S208–19. [DOI] [PubMed] [Google Scholar]
- 30.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage 2002; 17: 825–41. [DOI] [PubMed] [Google Scholar]
- 31.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002; 17: 143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith SM, Brady JM. SUSAN – A new approach to low level image processing. Int J Comput Vis 23: 45–78. [Google Scholar]
- 33.Kohler CG, Turner TH, Bilker WB, Brensinger CM, Siegel SJ, Kanes SJ, et al. Facial emotion recognition in schizophrenia: intensity effects and error pattern. Am J Psychiatry 2003; 160: 1768–74. [DOI] [PubMed] [Google Scholar]
- 34.Hall J, Whalley HC, McKirdy JW, Romaniuk L, McGonigle D, McIntosh AM, et al. Overactivation of fear systems to neutral faces in schizophrenia. Biol Psychiatry 2008; 64: 70–73. [DOI] [PubMed] [Google Scholar]
- 35.Costafreda SG, Brammer MJ, David AS, Fu CHY. Predictors of amygdala activation during the processing of emotional stimuli: a meta-analysis of 385 PET and fMRI studies. Brain Res Rev 2008; 58: 57–70. [DOI] [PubMed] [Google Scholar]
- 36.Zald DH. The human amygdala and the emotional evaluation of sensory stimuli. Brain Res Rev 2003; 41: 88–123. [DOI] [PubMed] [Google Scholar]
- 37.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 2011; 12: 683–91. [DOI] [PubMed] [Google Scholar]
- 38.Ventura J, Wood RC, Jimenez AM, Hellemann GS. Neurocognition and symptoms identify links between facial recognition and emotion processing in schizophrenia: meta-analytic findings. Schizophr Res 2013; 15: 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loth E, Poline JB, Thyreau B, Jia T, Tao C, Lourdusamy A, et al. Oxytocin receptor genotype modulates ventral striatal activity to social cues and response to stressful life events. Biol Psychiatry 2014; 76: 367–76. [DOI] [PubMed] [Google Scholar]
- 40.Uhrig S, Hirth N, Broccoli L, von Wilmsdorff M, Bauer M, Sommer C, et al. Reduced oxytocin receptor gene expression and binding sites in different brain regions in schizophrenia: a post-mortem study. Schizophr Res 2016. [DOI] [PubMed] [Google Scholar]
- 41.LoParo D, Waldman ID. The oxytocin receptor gene (OXTR) is associated with autism spectrum disorder: a meta-analysis. Mol Psychiatry 2015; 20: 640–6. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Zhao Y, Li R, Broster LS, Zhou C, Yang S. Association of oxytocin receptor gene (OXTR) rs53576 polymorphism with sociality: a meta-analysis. PLoS One 2015; 10: e0131820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tost H, Kolachana B, Hakimi S, Lemaitre H, Verchinski BA, Mattay VS, et al. A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proc Natl Acad Sci USA 2010; 107: 13936–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tost H, Kolachana B, Verchinski BA, Bilek E, Goldman AL, Mattay VS, et al. Neurogenetic effects of OXTR rs2254298 in the extended limbic system of healthy Caucasian adults. Biol Psychiatry 2011; 70: e37–39; author reply e41–32. [DOI] [PubMed] [Google Scholar]
- 45.Feldman R, Monakhov M, Pratt M, Ebstein RP. Oxytocin pathway genes: evolutionary ancient system impacting on human affiliation, sociality, and psychopathology. Biol Psychiatry 2016; 79: 174–84. [DOI] [PubMed] [Google Scholar]
- 46.Insel TR. The challenge of translation in social neuroscience: a review of oxytocin, vasopressin, and affiliative behavior. Neuron 2010; 65: 768–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dumais KM, Veenema AH. Vasopressin and oxytocin receptor systems in the brain: sex differences and sex-specific regulation of social behavior . Front Neuroendocrinol 2016; 40: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Qin W, Liu B, Zhou Y, Wang D, Zhang Y, et al. Neural mechanisms of oxytocin receptor gene mediating anxiety-related temperament. Brain Struct Funct 2014; 219: 1543–54. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Qin W, Liu B, Wang D, Zhang Y, Jiang T, et al. Variant in OXTR gene and functional connectivity of the hypothalamus in normal subjects. NeuroImage 2013; 81: 199–204. [DOI] [PubMed] [Google Scholar]