A ratiometric, quantitative, and genetically encoded strigolactone sensor, StrigoQuant, opens new avenues in SL signaling studies.
Keywords: Strigolactone signaling, strigolactone quantitative biosensor, plant synthetic biology, genetically encoded strigolactone biosensor, strigolactone perception
Abstract
Strigolactones are key regulators of plant development and interaction with symbiotic fungi; however, quantitative tools for strigolactone signaling analysis are lacking. We introduce a genetically encoded hormone biosensor used to analyze strigolactone-mediated processes, including the study of the components involved in the hormone perception/signaling complex and the structural specificity and sensitivity of natural and synthetic strigolactones in Arabidopsis, providing quantitative insights into the stereoselectivity of strigolactone perception. Given the high specificity, sensitivity, dynamic range of activity, modular construction, ease of implementation, and wide applicability, the biosensor StrigoQuant will be useful in unraveling multiple levels of strigolactone metabolic and signaling networks.
INTRODUCTION
Strigolactones (SLs) are a novel class of phytohormones that regulate different aspects of plant development, such as shoot branching (1, 2), and mediate plant adaptations to nutrient availability (3, 4). Moreover, SLs are released to the rhizosphere to promote symbiosis with arbuscular mycorrhizal fungi, recruited to provide the plant with minerals (5). However, SLs also mediate the recognition of host roots by parasitic weeds of the genera Striga and Orobanche, which cause severe yield losses in cereals and other crops in the Mediterranean, Africa, and Asia (6). Because of these versatile and important biological functions, elucidating SL biosynthesis and understanding their signaling mechanisms have become major research areas in plant sciences (7–12). SLs are a diverse class of carotenoid-derived molecules containing a butenolide ring (D-ring) linked to a less conserved second moiety that corresponds to the tricyclic lactone ring (ABC-ring) in canonical SLs (Fig. 1A). SL biosynthesis is initiated by the all-trans/9-cis-carotene isomerase DWARF27 forming 9-cis-β-carotene that is converted by the carotenoid cleavage dioxygenases 7 and 8 into carlactone (13–16). Carlactone is oxidized by cytochrome P450 enzymes (clade711; MAX1 in Arabidopsis) to carlactonoic acid (17), to 4-deoxyorobanchol (4DO), and further to orobanchol (18). After methylation, carlactonoic acid is converted in Arabidopsis by an oxoglutarate-dependent dioxygenase (lateral branching oxidoreductase) into a yet unidentified product needed to inhibit shoot branching (19).
Fig. 1. StrigoQuant design, characterization, and analysis of receptor complex requirements.
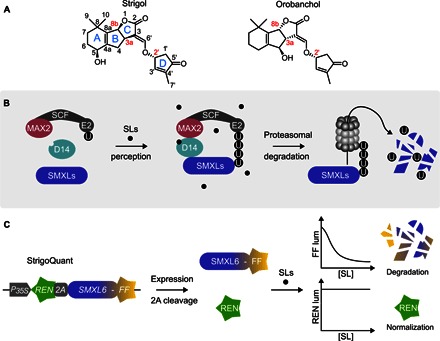
(A) General structure and configuration of the canonical SLs, strigol and orobanchol. Note the stereocenters at the BC-junction (carbons 3a and 8b) and in the D-ring (carbon 2′). (B) Scheme of the SL perception machinery in Arabidopsis. In the presence of SLs, receptor complex formation takes place, with SLs binding to the receptor protein AtD14, with recruitment of SMXLs to the MAX2 and SKP1/CUL1/F-box E2 ubiquitin ligase complex (SCFMAX2). SMXLs are ubiquitinated (U) and consequently degraded by the 26S proteasome. (C) StrigoQuant construct expressing a renilla luciferase (REN; green) connected via a 2A peptide to the sensor module (SM), AtSMXL6 (SMXL6), fused to a firefly luciferase (FF; yellow), under the control of a constitutive 35S promoter. The 2A peptide in the synthetic construct leads to stoichiometric coexpression of REN (normalization element) and SM (SMXL6-FF). Upon the addition of SLs, SMXL6-FF becomes ubiquitinated and degraded, whereas REN expression remains constant, leading to a decrease in the FF/REN ratio.
On the basis of the stereochemistry of their BC-ring junction (carbon atoms 3a and 8b), the approximately 20 known canonical SLs are divided into the strigol- and orobanchol-like subfamilies (Fig. 1A), with the C-ring in the β [(+), up] or α [(−), down] orientation, respectively (12). Modifications of the ABC-ring, such as hydroxylation, provide for the diversity within each group of SLs (12). In addition, there are noncanonical SLs, such as carlactonoic acid, that are characterized by the absence of the B- and C-rings. However, because of the lack of molecular tools for the targeted monitoring of signaling processes mediated by individual SLs, our understanding of the functional diversity that may explain why plants produce various SLs is limited.
SL perception and early signaling are mediated by a degradation-based mechanism similar to other phytohormones, such as auxins, gibberellins, and jasmonates (20). Binding of SLs to the α/β-fold hydrolase D14/AtD14 (21, 22) in rice and Arabidopsis leads to the formation of a co-receptor complex also comprising the F-box protein D3/MAX2 and the target regulator proteins of the D53/SMXL family (23–26). D3/MAX2 engages in a SKP1/CUL1/F-box E2 ubiquitin ligase protein complex (SCFD3/MAX2), leading to the proteolytic degradation of the D53 target proteins, thus triggering SL-regulated responses (Fig. 1B). Notably, D3/MAX2 is also required for the perception of smoke-derived karrikins, compounds present in burned or charred plant material that may mimic an as of yet unidentified growth regulator (10). Karrikins have a butenolide ring similar to the D-ring of SLs and are perceived by the D14 paralog KAI2, with downstream signaling requiring the SMXL family member SMAX1 (10). In light of the structural SL diversity, the overlapping signaling mechanisms with karrikins, and the apparent redundancy of signaling components, current biochemical and genetic approaches to quantitatively analyze SL signaling networks remain challenging. Hence, novel molecular tools for investigating SL perception and signaling mechanisms are needed and would provide an experimental quantum leap for the understanding of plant development, growth, and environmental interactions.
To monitor the SL dynamics in plant cells, we have developed the first genetically encoded quantitative biological sensor for this class of hormones. Using this molecular tool in plant protoplasts, we demonstrate the structural specificity and sensitivity of the SL perception complex toward various natural and synthetic SLs. By combining the quantitative sensor with SL perception mutants, we further analyze the activity of components of the co-receptor complex. Moreover, we reveal quantitative insights into the stereoselective nature of the signaling process mediated by natural and synthetic SL enantiomers.
RESULTS
Design and engineering of the genetically encoded quantitative SL biosensor StrigoQuant
We have engineered the degradation-based luminescent ratiometric SL sensor StrigoQuant using the intrinsic perception machinery for SLs in Arabidopsis. As a sensor module (SM), the full-length complementary DNA (cDNA) of SMXL6, a close homolog to rice D53 (23, 24) that has been shown to mediate SL signaling in Arabidopsis (25, 26), was implemented. This SM was fused to a firefly luciferase (FF) to monitor its degradation (Fig. 1C and table S1). Furthermore, StrigoQuant also comprises a renilla luciferase (REN), incorporated as a normalization element. The synthetic construct encodes REN and the SM-FF fusion protein separated by a self-processing 2A peptide, allowing for cotranslational cleavage and resulting in stoichiometric expression of the sensor elements from a single transcript (27, 28). By monitoring the activity of both luciferases of StrigoQuant, it becomes possible to observe the initial steps of SL signaling (namely, perception) over the SMXL6 SM and to translate substrate specificity, activity, and concentration to SM-FF degradation, with the unaffected REN activity for normalization (Fig. 1C and table S1). The ratiometric design of StrigoQuant makes it a useful and robust tool, unaffected by differences in expression levels between plant genotypes or single experiments. As a control, an additional sensor, CtrlQuant, was designed. For this, the SM was replaced by a small repeated GA sequence (GAGAGAGAGAGAGA amino acid sequence) that is not expected to show hormone-dependent SM-FF degradation (28).
Characterization and application of StrigoQuant to study SL perception complex and signaling processes
To verify the functionality and sensitivity of the sensor, we transiently expressed StrigoQuant and CtrlQuant constructs in wild-type (WT) Arabidopsis protoplasts and applied increasing concentrations of the synthetic strigol-like SL analog racemic GR24 (rac-GR24) to the transformed protoplast suspensions. Following incubation, luciferase activity was determined by simultaneously measuring each of the luciferases, FF and REN, in separate replicate samples (see “Treatment with SLs and luminescence analysis” for further details), and the ratio of the FF and REN luminescence was analyzed. Experiments using WT protoplasts transformed with StrigoQuant showed a rac-GR24–dependent decrease in the FF-to-REN ratio (FF/REN), with a significant reduction at concentrations as low as 10 pM (Fig. 2A), whereas cells expressing CtrlQuant did not show any reduction (fig. S1). The degradation of the SM-FF was dependent on the 26S proteasome, as suggested by treatment with the proteasomal inhibitor MG132 (fig. S2). To study the minimal components necessary for the perception of GR24 by StrigoQuant (Fig. 1B) (25, 26), we also introduced the sensor in protoplasts isolated from the mutants Atd14 or max2. In both genotypes, StrigoQuant failed to show a rac-GR24–dependent FF/REN signal reduction, confirming the AtD14 and MAX2 dependency of SMXL6 degradation (Fig. 2, A and B). We additionally tested the sensor in dlk2 and kai2 protoplasts affected in the AtD14 homolog DLK2 and in the karrikin responsive paralog KAI2, respectively; however, we observed similar degradation profiles as in WT protoplasts (Fig. 2B). These results suggest the dependency of rac-GR24 perception and signaling over SMXL6 being mediated primarily through AtD14. Expression of rice D14 (OsD14) in protoplasts isolated from Atd14 (col) plants could rescue SMXL6 degradation of StrigoQuant upon induction with rac-GR24 (Fig. 2C). This depicts the utility of the sensor to study the activity and specificity of heterologous components of the SL sensing machinery upon transient expression in Arabidopsis protoplasts.
Fig. 2. StrigoQuant biosensor for the study of the SL perception machinery.
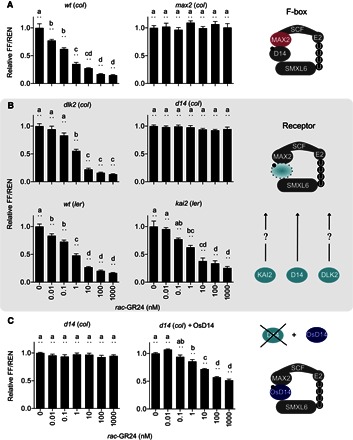
(A) StrigoQuant biosensor for the study of the SL perception machinery Characterization of the StrigoQuant sensor construct in protoplasts isolated from WT [Columbia (col-0), ecotype] and max2 (col) mutant Arabidopsis backgrounds upon addition of increasing concentrations of rac-GR24 (left). Protoplasts were isolated from the seedlings of each genotype and transformed with StrigoQuant. Twenty-four hours after transformation, protoplasts were supplemented with increasing concentrations of a rac-GR24 serial dilution for 2 hours before luciferase activity determination. On the right, the schematic principle of the experiment is shown, where the functionality of the F-box protein MAX2 for SL-mediated SMXL6 degradation was analyzed. (B) Characterization of the StrigoQuant sensor construct in protoplasts isolated from WT and mutant Arabidopsis backgrounds for potential SL receptors upon addition of increasing concentrations of rac-GR24 (left). Protoplasts were isolated from Atd14 (col), dlk2 (col), WT [Landsberg erecta (ler), ecotype], and kai2 (ler) seedlings and transformed with StrigoQuant. Twenty-four hours after transformation, protoplasts were supplemented with increasing concentrations of a rac-GR24 serial dilution for 2 hours before luciferase activity determination. On the right, the schematic principle of the experiment is shown, where the functionality of AtD14, DLK2, and KAI2 in mediating SMXL6 degradation was tested. (C) Characterization of the effect of overexpression of OsD14 in Atd14 (col) protoplasts. Protoplasts isolated from Atd14 (col) seedlings were transformed either only with StrigoQuant and a stuffer plasmid (pGEN16; with equal amounts of 15 μg each) or with StrigoQuant and a plasmid harboring OsD14 (with equal amounts of 15 μg each) and induced with rac-GR24 for 2 hours before luminescence determination. On the right, a schematic principle of the experiment is shown, where OsD14 was overexpressed in protoplasts lacking AtD14 to rescue functional SMXL6 degradation. Results for each panel are averaged FF/REN ratios, normalized to the sample without addition of any inducer substrate for each genotype. The data shown correspond to one representative experiment of four replicated experiments for (A) and (B) and three experiments for (C). Error bars represent SEM from the individual experimental data shown. n = 6. Statistical significance between the tested concentrations within a genotype is indicated with lowercase letters above each bar, where “a” significantly differs from “b,” “b” from “c,” and so on. One-way analysis of variance (ANOVA), P < 0.01.
Analysis of the specificity and sensitivity to various strigol- and orobanchol-like SLs
With the functionality of StrigoQuant being established for rac-GR24 and its dependency on MAX2 and AtD14 being verified, we further analyzed the specificity and sensitivity of the sensor upon induction with SL species that are commonly used in SL signaling studies (1, 2, 4, 5). To this aim, we treated WT protoplasts transformed with StrigoQuant and CtrlQuant with different concentrations of racemic mixtures of the (i) strigol-like SLs [strigol and 5-deoxystrigol (5DS)] and (ii) orobanchol-like SLs (orobanchol and 4DO). Results showed varying degrees of degradation of StrigoQuant among the chemical substrates, with large differences in sensitivity (Fig. 3). Both strigol-like and orobanchol-like substrates lead to SM-FF degradation, but among the compounds tested, rac-5DS showed the largest FF/REN decrease at low substrate concentrations. Additional experiments were conducted with rac-5DS, revealing that StrigoQuant responded to 5DS concentrations as low as 100 fM (relative FF/REN, 0.92 ± 0.01; P < 0.01; fig. S3). Our results show a first approximation to the physiological concentrations required for each of these substrates tested to induce degradation of SMXL6, triggering SL signaling in Arabidopsis cells. With the aid of this sensor, quantitative information can be gained on the processes of perception, making it a powerful tool to screen natural and synthetic SL substrates for their signaling capacity. In conjunction with phenotypic analyses using these substrates, data obtained using StrigoQuant will deliver information while using lower substrate concentrations, delivering quick results, with unprecedented molecular and quantitative resolution.
Fig. 3. Specificity and sensitivity of the sensor to strigol- and orobanchol-like SLs.
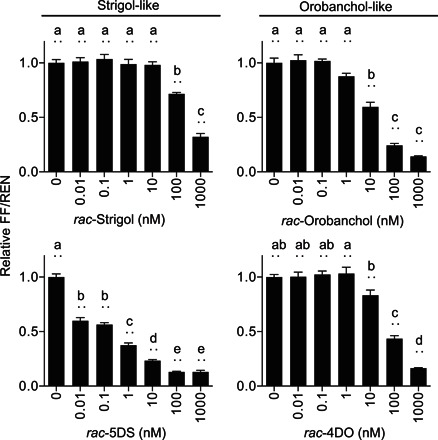
WT Arabidopsis protoplasts transformed with StrigoQuant were induced with racemic mixtures of strigol-like (rac-strigol and rac-5DS) and orobanchol-like (rac-orobanchol and rac-4DO) SLs for 2 hours before luminescence activity determination. Results are averaged FF/REN ratios, normalized to the sample without addition of inducer for each substrate tested. The data shown correspond to one representative experiment of three replicated experiments. Error bars represent SEM from the individual experimental data shown. n = 6. Statistical significance between the tested concentrations for each substrate is indicated with lowercase letters above each bar, where “a” significantly differs from “b,” “b” from “c,” and so on. One-way ANOVA, P < 0.01.
Quantitative determination of the stereoselectivity of SL perception
One important question in SL signaling pertains to the stereochemistry of the butenolide D-ring. Recent works in Arabidopsis based on phenotypic and downstream signaling analyses point to 2′R stereoisomers of SLs, like in natural SLs, as being the species mainly detected by canonical SL receptor complexes, with experimental indications that synthetic 2′S SLs also have signaling effects (29–32). The StrigoQuant sensor constitutes the suitable means to quantitatively analyze this at the level of SL perception and signal induction, uncoupled from downstream events, by separately screening enantiomers of GR24, 5DS, and 4DO. Both 5DS and 4DO are naturally occurring SLs (2′R configuration), whereas ent-5DS and ent-4DO, exhibiting 2′S stereochemistry in the D-ring, have not been shown to occur naturally (30). GR24 and ent-GR24 exhibit the same configurations as 5DS and ent-5DS, both at the BC-ring junction and in the D-ring, respectively (Fig. 4A). Results from these experiments indicated that all 2′R enantiomers of 5DS, GR24, and 4DO induced a much greater reduction in FF/REN of StrigoQuant at lower concentrations when compared to their respective 2′S enantiomers (Fig. 4B and fig. S4). However, higher concentrations of the latter, with nonnatural D-ring conformations, ultimately affected the sensor stability in this assay, inducing SMXL6 degradation. Phenotypic data and downstream transcript level analyses suggest that 2′R enantiomers are perceived by AtD14 as the SL signaling receptor, whereas 2′S enantiomers are primarily perceived by the KAI2 receptor (29, 30). Assuming that SL-induced SMXL6 degradation is primarily mediated over AtD14 (Fig. 2A), these results may suggest some flexibility for the receptor toward both 2′R and 2′S SL substrates. Recent reports in literature presenting in vitro hydrolysis rates for SL substrates by AtD14 have indicated that both 2′R and 2′S SLs can be bound and hydrolyzed by the receptor, albeit at different rates dependent on the SL, stereochemistry, and the study (22, 32, 33). We show here that the naturally occurring orientation of the D-ring, 2′R, indeed confers a higher SL signaling capacity, ultimately extending to the possibility of analyzing the relevance of SL stereochemistry for the level of receptor complex formation and SL signaling initiation over SMXL6 at high quantitative resolution.
Fig. 4. Stereoselectivity analysis of SL species.
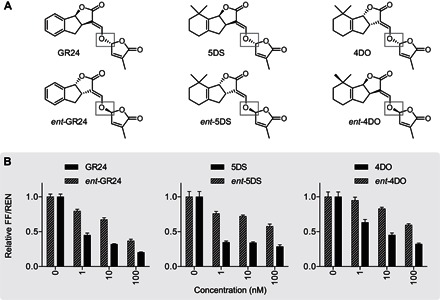
(A) Chemical structures of enantiomers of selected SLs. The differential stereochemistry at the carbon 2′ in the D-ring is highlighted (gray boxes). (B) WT protoplasts were transformed with StrigoQuant and induced with the individual enantiomers GR24 and ent-GR24, 5DS and ent-5DS, and 4DO and ent-4DO for 2 hours before luminescence activity determination. Dose-response curves for substrates with the 2′R (+) D-ring configuration are shown with dashed lines, whereas those for 2′S (−)–configured substrates are in solid black. The data shown correspond to one representative experiment of three replicated experiments. Results are averaged FF/REN ratios, normalized to the sample without addition of inducer for each substrate tested. Error bars represent SEM from the individual experimental data shown. n = 6.
DISCUSSION
The study of SL regulatory roles has currently gained much momentum. However, most of the available knowledge on the structure-function relationship of SLs and on the signaling processes triggered by these hormones is based on physiological and genetic studies in planta (25, 26, 30). Here, we have developed the first SL sensor, translating SL concentrations into a direct readout with high quantitative resolution. This allows dissecting the initial events of hormone perception and the resulting outcomes. In selected proof-of-principle applications, we have implemented the genetically encoded quantitative sensor StrigoQuant to study the components of the sensing complex involved in SL perception in Arabidopsis, including MAX2, D14, KAI2, and DLK2 (Fig. 2A). Moreover, we show the applicability of StrigoQuant in the Arabidopsis protoplast system for the straightforward study of the activity of heterologous components of SL signaling, as demonstrated in this work for rice D14 (OsD14) (Fig. 2B). We further analyzed the specificity of recognition for different synthetic and natural SLs, achieving the closest approximation to physiological conditions ever performed in terms of sensitivity: StrigoQuant can detect up to 100 fM changes in vivo upon exogenous addition of SLs (fig. S3). It shows a broad range of recognition capacity both in specificity (both families of SLs, strigol- and orobanchol-like) and in sensitivity (high dynamic range, spanning several orders of magnitude in concentration). Finally, this sensor confirmed the importance of stereochemistry at the butenolide D-ring in a direct, quantitative manner, demonstrating that 2′R-configured SLs affect the initial steps of SL signaling to a much higher degree than their nonnatural counterparts (Fig. 4).
The quantitative nature of the here-introduced SL sensor, its modular construction that can be used to incorporate other SL signaling (for example, different SMXLs) or readout proteins (fluorescent), and the potential to apply the simple DNA constructs used here in multiple plant species render this principle ideally suited to experimentally address multiple levels of SL signaling. In principle, this tool can be used as a molecular proxy in the protoplast system to complete a full quantitative, comparative characterization of (i) SL substrate signaling capacity [identifying the bioactive SL(s) in Arabidopsis and screening of SLs from other species or endogenous or synthetic compounds], (ii) receptor-substrate binding specificity (screening of D14s, endogenous and heterologous, in combination with different SLs), and (iii) specific substrate, receptor, and signaling capacity/function of D53/SMXL family members (by incorporation of other SL-responsive D53/SMXLs into the sensor platform). Combination of the sensor with genetic tools, such as mutants in metabolic and signaling components, will open up novel avenues for the study of SL biosynthesis as well as complex SL regulatory and defense networks.
Furthermore, StrigoQuant has been developed in this study and optimized for use in protoplasts with luminescent readout proteins, allowing for quick and sensitive detection. However, through replacing the luciferases with fluorescent proteins, this tool can be expanded for use in plant tissues and whole plants, both transiently and stably transformed. This can provide complementary information in terms of spatial resolution, as has been illustrated for auxin and jasmonic acid semiquantitative sensors (34, 35).
The sensor allows the rapid and highly sensitive analysis of SL signaling processes without the need for time-consuming, intrinsically qualitative phenotypic analyses. In summary, the timely development of this genetically encoded quantitative SL sensor with unprecedented versatility and sensitivity will allow breakthrough insights in the highly active field of SL research.
MATERIALS AND METHODS
Plasmid construction
The StrigoQuant sensor plasmid pHB1105 was constructed as follows: renilla and firefly-myc tag were PCR (polymerase chain reaction)–amplified from the L2min17-Luc auxin sensor (table S1) (28) using the oligonucleotides oHB556/oHB562 and oHB565/oHB557, respectively (table S2). cDNA for AtD53Like1/SMXL6 (At1g07200) was synthesized and PCR-amplified using the oligonucleotides oHB563FP and oHB564RP. Products were assembled via Gibson cloning into pGEN16 (received from M. Rodriguez-Franco, University of Freiburg, Germany) digested with Hind III/Age I. The CtrlQuant control sensor plasmid pSW209 was constructed by Gibson cloning of PCR-amplified renilla-2A-(GA)7-firefly-myc tag, using the oligonucleotides oSW237 and oSW238 and the plasmid Ctrl-Luc as template (28) and pGEN16 digested with Not I/Age I. The construct harboring OsD14 (pHB1111) was constructed as follows: the cDNA of OsD14 (Q10QA5) was synthesized and PCR-amplified using the oligonucleotides oHB567/oHB577 and assembled into pGEN16 digested with Hind III/Age I via AQUA cloning (36).
Plant material
Arabidopsis thaliana seeds were surface-sterilized with 5% (w/v) calcium hypochlorite and 0.02% (v/v) Triton X-100 in 80% (v/v) ethanol solution and seeded either (i) on sterile filter paper strips with 200 to 300 seeds per strip on 12-cm2 plates (Greiner Bio-One), two strips per plate, or (ii) in Magenta vessels (Sigma-Aldrich) with 4 to 6 seeds evenly dispersed and with both Magenta vessels and plates containing 50 ml of SCA (seedling culture Arabidopsis) growth medium [0.32% (w/v) Gamborg B5 basal salt powder with vitamins (bioWORLD), 4 mM MgSO4·7H2O, 43.8 mM sucrose, 0.1% (v/v) Gamborg B5 Vitamin Mix (bioWORLD), and 0.8% (w/v) phytoagar in H2O (pH 5.8)]. Seedlings were grown in either a Sanyo/Panasonic MLR-352-PE or a Binder APT.line KBWF 240 or KBWF 720 (E5.3) growth chamber with a 16-hour light regime at 21°C. Those grown on plates were harvested for protoplast isolation 1.5 to 2.5 weeks after seeding. Magenta vessel–grown seedlings were harvested 3 to 4 weeks after seeding. WT A. thaliana (Col-0 and Ler) seeds and Atd14, max2, kai2, and dlk2 mutants were used for this study.
Protoplast isolation and transformation
Protoplast isolation and transformation were completed as described previously (37). Briefly, plant material was dissected with a sterile scalpel, cut into small pieces, and incubated overnight in 10 ml of MMC [MES, mannitol, and calcium; 10 mM MES, 40 mM CaCl2·H2O, 550 mosmol with mannitol (pH 5.8), sterile-filtered] with 1 ml of a 5% cellulose and macerozyme solution (1 g of cellulase Onozuka R10 and 1 g of macerozyme R10 from SERVA Electrophoresis GmbH in 20 ml of MMC). Isolation of protoplasts was performed via flotation in MSC solution [MES, sucrose, and calcium; 10 mM MES, 0.4 M sucrose, 20 mM MgCl2·6H2O, 550 mosmol with mannitol (pH 5.8)] overlaid with MMM solution [MES, mannitol, and magnesium; 15 mM MgCl2, 5 mM MES, 550 mosmol with mannitol (pH 5.8)]. Protoplasts were collected and counted in W5 solution [2 mM MES, 154 mM NaCl, 125 mM CaCl2·2H2O, 5 mM KCl, 5 mM glucose (pH 5.8)] using a Rosenthal counting chamber. Transformations of 500,000 protoplasts in 100 μl of MMM were completed by mixing the protoplasts with 20 μg of plasmid DNA (in the case of double-plasmid transformations for OsD14 overexpression, 15 μg of each plasmid was transformed for a total of 30 μg of DNA per transformation) in a volume of 20 μl (volume was adjusted with MMM solution), then adding a polyethylene glycol (PEG) solution (2.5 ml of 0.8 M mannitol, 1 ml of 1 M CaCl2, 4 g of PEG4000 from Sigma-Aldrich, and 3 ml of H2O, made fresh for each experiment) in a dropwise manner, and incubating for 8 min. After this time, 120 μl of MMM was added to the protoplasts and, immediately thereafter, overlaid with PCA (protoplast culture Arabidopsis) to a final volume of 1.8 ml per reaction [0.32% (w/v) Gamborg B5 basal salt powder with vitamins from bioWORLD, 2 mM MgSO4·7H2O, 3.4 mM CaCl2·2H2O, 5 mM MES, 0.342 mM l-glutamine, 58.4 mM sucrose, 550 mosmol with glucose, 8.4 μM Ca-pantothenate, 2% (v/v) biotin from a biotin solution of 0.02% (w/v) in H2O, 0.1% (v/v) Gamborg B5 Vitamin Mix (pH 5.8), and 1:2000 ampicillin]. Multiple transformations of the same DNA were completed in this manner, and the protoplasts were pooled 20 to 24 hours after transformation, before induction with substrates.
Treatment with SLs and luminescence analysis
The SL substrates (±)-GR24, (±)-5DS, (±)-2′-epi-5DS, (±)-strigol, (±)-orobanchol, (+)-5DS, (−)-5DS, (+)-2′-epi-5DS, and (−)-2′-epi-5DS were obtained from OlChemIm Ltd. For (+)-GR24 and (−)-GR24, (±)-GR24 was purchased from OlChemIm Ltd. and separated on an isocratic (97% methanol/water) high-performance liquid chromatography (HPLC) using a chiral column (4.6 × 150 mm, 3.0 μm; CHIRALPAK AD-3R, DAICEL Corp.) at a flow rate of 0.5 ml/min, and the column temperature was set to 30°C. To determine the stereochemistry structure of the two purified enantiomers, they were separated on an Astec Cellulose DMP Chiral HPLC column (4.6 × 150 mm, 3.0 μm), using a 14:3:3 ratio of hexane/methanol/methyl tert-butyl ether as the mobile phase, and then compared with previously reported HPLC data (30). Stock solutions of either 5 or 10 mM in methanol were prepared. For treatment with the proteasomal inhibitor MG132, stock solutions with a concentration of 40 mM were prepared in dimethyl sulfoxide and added directly to the protoplasts 2 hours before induction with SLs.
After pooling replicate transformations, 960-μl aliquots of the protoplast suspension were pipetted into the wells of a 2-ml deep-well storage plate (Corning) for each concentration of SL inducer substrate tested. Serial dilutions of inducer substrates were prepared in PCA with an 11-fold concentration of the desired final experimental concentration, and a volume of 96 μl was added into a well of 960 μl of protoplasts, carefully mixed via gentle pipetting up and down. After 2 hours of incubation with the corresponding SL substrate, luminescence analysis was performed. For this, two samples of induced protoplasts with a volume of 80 μl (approximately 20,000 cells) were pipetted per replicate into two separate white 96-well assay plates to simultaneously determine the luminescence of both luciferases. REN and FF activities were determined by adding 20 μl of either coelenterazine (472 mM coelenterazine stock solution in methanol, diluted directly before use, 1:15 in phosphate-buffered saline) or firefly substrate [0.47 mM d-luciferin (Biosynth AG), 20 mM tricine, 2.67 mM MgSO4·7H2O, 0.1 mM EDTA·2H2O, 33.3 mM dithiothreitol, 0.52 mM adenosine 5′-triphosphate, 0.27 mM acetyl–coenzyme A, 5 mM NaOH, 264 μM MgCO3·5H2O, in H2O], respectively. Because of the large number of samples per experiment (six replicates per sample, including both FF and REN samples for each replicate, totaling 12 measurements per condition), induction and measurement of all samples within a single experiment were conducted in a time-staggered manner. All concentrations of a single SL inducer substrate were measured at once, and the substrates or plant genotypes tested in a given experiment were induced and measured consecutively. REN luminescence was determined with a BioTek Synergy 4 or a Berthold TriStar2 S LB 942 multimode plate reader. FF luminescence was measured with a Tecan Infinite M200 PRO multimode reader or a BERTHOLD Centro XS3 LB 960 microplate luminometer.
Statistical analysis
Ordinary one-way ANOVAs and multiple comparisons for statistical significance were performed with GraphPad Prism 6 for Mac OS X version 6.0h. Corresponding P values/levels of significance are indicated in the figure captions.
Supplementary Material
Acknowledgments
We thank S. Knall, E. Wehinger, and R. Wurm for valuable experimental assistance; M. Kollmann, R. Ochoa-Fernandez, and J. Braguy for the helpful comments on the manuscript; R. Engesser for statistical analysis support; M. Rodriguez-Franco for providing the plasmid pGEN16; O. Leyser for providing the max2 seeds; and S. Smith for the d14, dlk, and kai2 Arabidopsis seeds. Funding: This research was supported by funding from the excellence initiatives of the German federal and state governments (DFG EXC-1028-CEPLAS, EXC 294-BIOSS, and GSC 4-SGBM) and the King Abdullah University of Science and Technology (KAUST). Author contributions: S.L.S. designed, performed, and analyzed experiments and made figures. H.M.B. designed and analyzed experiments and made figures. X.G., K.-P.J., and L.B. prepared experimental material. M.A. designed and prepared constructs and performed experiments. O.E. analyzed experiments. P.B. and W.W. designed and analyzed experiments and supervised the project. S.A.-B. and M.D.Z. conceived and supervised the project and designed and analyzed experiments. S.L.S., S.A.-B., and M.D.Z. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/11/e1601266/DC1
fig. S1. CtrlQuant activity upon incubation with racemic SLs in WT and mutant Arabidopsis protoplasts.
fig. S2. SL-dependent degradation of the SMXL6-FF sensor component is mediated by the 26S proteasome.
fig. S3. Specificity and sensitivity of StrigoQuant to 5DS.
fig. S4. Stereoselectivity analysis of SL species with CtrlQuant.
table S1. Amino acid sequences of the components of the StrigoQuant and CtrlQuant sensors.
table S2. Oligonucleotides used for the cloning of the sensor constructs.
REFERENCES AND NOTES
- 1.Gomez-Roldan V., Fermas S., Brewer P. B., Puech-Pagès V., Dun E. A., Pillot J.-P., Letisse F., Matusova R., Danoun S., Portais J.-C., Bouwmeester H., Bécard G., Beveridge C. A., Rameau C., Rochange S. F., Strigolactone inhibition of shoot branching. Nature 455, 189–194 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S., Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Mayzlish-Gati E., De-Cuyper C., Goormachtig S., Beeckman T., Vuylsteke M., Brewer P. B., Beveridge C. A., Yermiyahu U., Kaplan Y., Enzer Y., Wininger S., Resnick N., Cohen M., Kapulnik Y., Koltai H., Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol. 160, 1329–1341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun H., Tao J., Liu S., Huang S., Chen S., Xie X., Yoneyama K., Zhang Y., Xu G., Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 65, 6735–6746 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akiyama K., Matsuzaki K.-i., Hayashi H., Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Parker C., Observations on the current status of Orobanche and Striga problems worldwide. Pest Manage. Sci. 65, 453–459 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Ruyter-Spira C., Al-Babili S., van der Krol S., Bouwmeester H., The biology of strigolactones. Trends Plant Sci. 18, 72–83 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Brewer P. B., Koltai H., Beveridge C. A., Diverse roles of strigolactones in plant development. Mol. Plant 6, 18–28 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Seto Y., Yamaguchi S., Strigolactone biosynthesis and perception. Curr. Opin. Plant Biol. 21, 1–6 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Smith S. M., Li J., Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 21, 23–29 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Obando M., Ligerot Y., Bonhomme S., Boyer F.-D., Rameau C., Strigolactone biosynthesis and signaling in plant development. Development 142, 3615–3619 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Al-Babili S., Bouwmeester H. J., Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S., The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Bruno M., Hoffman M., Vermathen M., Alder A., Beyer P., Al-Babili S., On the substrate- and stereospecificity of the plant carotenoid cleavage dioxygenase 7. FEBS Lett. 588, 1802–1807 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Bruno M., Al-Babili S., On the substrate specificity of the rice strigolactone biosynthesis enzyme DWARF27. Planta 243, 1429–1440 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Seto Y., Sado A., Asami K., Hanada A., Umehara M., Akiyama K., Yamaguchi S., Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. U.S.A. 111, 1640–1645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe S., Sado A., Tanaka K., Kisugi T., Asami K., Ota S., Kim H. I., Yoneyama K., Xie X., Ohnishi T., Seto Y., Yamaguchi S., Akiyama K., Yoneyama K., Nomura T., Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. U.S.A. 111, 18084–18089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., van Dijk A. D. J., Scaffidi A., Flematti G. R., Hofmann M., Charnikhova T., Verstappen F., Hepworth J., van der Krol S., Leyser O., Smith S. M., Zwanenburg B., Al-Babili S., Ruyter-Spira C., Bouwmeester H. J., Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 10, 1028–1033 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Brewer P. B., Yoneyama K., Filardo F., Meyers E., Scaffidi A., Frickey T., Akiyama K., Seto Y., Dun E. A., Cremer J. E., Kerr S. C., Waters M. T., Flematti G. R., Mason M. G., Weiller G., Yamaguchi S., Nomura T., Smith S. M., Yoneyama K., Beveridge C. A., LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 6301–6306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett T., Leyser O., Strigolactone signalling: Standing on the shoulders of DWARFs. Curr. Opin. Plant Biol. 22, 7–13 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Hamiaux C., Drummond R. S. M., Janssen B. J., Ledger S. E., Cooney J. M., Newcomb R. D., Snowden K. C., DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Nakamura H., Xue Y.-L., Miyakawa T., Hou F., Qin H.-M., Fukui K., Shi X., Ito E., Ito S., Park S.-H., Miyauchi Y., Asano A., Totsuka N., Ueda T., Tanokura M., Asami T., Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4, 2613 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., Meng X., Liu G., Yu H., Yuan Y., Yi W., Zhao L., Ma H., He Y., Wu Z., Melcher K., Qian Q., Xu H. E., Wang Y., Li J., DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., Wu F., Mao H., Dong W., Gan L., Ma W., Gao H., Chen J., Yang C., Wang D., Tan J., Zhang X., Guo X., Wang J., Jiang L., Liu X., Chen W., Chu J., Yan C., Ueno K., Ito S., Asami T., Cheng Z., Wang J., Lei C., Zhai H., Wu C., Wang H., Zheng N., Wan J., D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J. P., Abbas A., Leyser O., Nelson D. C., SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., Meng X., Wang Y., Smith S. M., Li J., Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan M. D., Drew J., Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 13, 928–933 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wend S., Bosco C. D., Kämpf M. M., Ren F., Palme K., Weber W., Dovzhenko A., Zurbriggen M. D., A quantitative ratiometric sensor for time-resolved analysis of auxin dynamics. Sci. Rep. 3, 1–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson D. C., Scaffidi A., Dun E. A., Waters M. T., Flematti G. R., Dixon K. W., Beveridge C. A., Ghisalberti E. L., Smitha S. M., F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 8897–8902 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scaffidi A., Waters M. T., Sun Y. K., Skelton B. W., Dixon K. W., Ghisalberti E. L., Flematti G. R., Smith S. M., Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165, 1221–1232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artuso E., Ghibaudi E., Lace B., Marabello D., Vinciguerra D., Lombardi C., Koltai H., Kapulnik Y., Novero M., Occhiato E. G., Scarpi D., Parisotto S., Deagostino A., Venturello P., Mayzlish-Gati E., Bier A., Prandi C., Stereochemical assignment of strigolactone analogues confirms their selective biological activity. J. Nat. Prod. 78, 2624–2633 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Fle matti G. R., Scaffidi A., Waters M. T., Smith S. M., Stereospecificity in strigolactone biosynthesis and perception. Planta 243, 1361–1373 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Waters M. T., Scaffidi A., Moulin S. L. Y., Sun Y. K., Flematti G. R., Smith S. M., A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27, 1925–1944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunoud G., Wells D. M., Oliva M., Larrieu A., Mirabet V., Burrow A. H., Beeckman T., Kepinski S., Traas J., Bennett M. J., Vernoux T., A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Larrieu A., Champion A., Legrand J., Lavenus J., Mast D., Brunoud G., Oh J., Guyomarc’h S., Pizot M., Farmer E. E., Turnbull C., Vernoux T., Bennett M. J., Laplaze L., A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat. Commun. 6, 6043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beyer H. M., Gonschorek P., Samodelov S. L., Meier M., Weber W., Zurbriggen M. D., AQUA cloning: A versatile and simple enzyme-free cloning approach. PLOS ONE 10, e0137652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochoa-Fernandez R., Samodelov S. L., Brandl S. M., Wehinger E., Müller K., Weber W., Zurbriggen M. D., Optogenetics in plants: Red/far-red light control of gene expression. Methods Mol. Biol. 1408, 125–139 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/11/e1601266/DC1
fig. S1. CtrlQuant activity upon incubation with racemic SLs in WT and mutant Arabidopsis protoplasts.
fig. S2. SL-dependent degradation of the SMXL6-FF sensor component is mediated by the 26S proteasome.
fig. S3. Specificity and sensitivity of StrigoQuant to 5DS.
fig. S4. Stereoselectivity analysis of SL species with CtrlQuant.
table S1. Amino acid sequences of the components of the StrigoQuant and CtrlQuant sensors.
table S2. Oligonucleotides used for the cloning of the sensor constructs.


